Abstract
Deregulation of miR-153 has recently been observed in several common human cancer, while miR-153 serves an oncogene or tumor suppressive role in different cancer types. Previously, miR-153 has been identified to be overexpressed in prostate cancer. miR-153 played an important role in promoting proliferation of human prostate cancer cells and presented a novel mechanism of microRNA-mediated direct suppression of phosphatase and tensin homolog (PTEN) expression in prostate cancer cells. Until now, little is known about the clinical significance of miR-153 expression in prostate cancer.
The miR-153 expression in 143 pairs of prostate cancer and adjacent non-cancerous prostate tissues was measured by quantitative reverse transcription polymerase chain reaction (qRT-PCR) analysis. Student t test was conducted for intergroup comparison. Pearson correlation test was used for correlation analysis. Survival curves were carried out by the Kaplan–Meier method and evaluated using the log-rank test. Multivariable Cox proportional hazard risk regression model was performed to screen the independent factor affected the prognosis of prostate cancer patients.
qRT-PCR analysis showed that the expression of miR-153 was significantly increased in the prostate cancer tissues in comparison with the adjacent noncancerous prostate tissues (P < .001). The high expression of miR-153 in prostate cancer tissues is closely correlated with aggressive clinical pathological parameters such as lymph node metastasis (P = .001); bone metastasis (P < .001); Gleason score (P < .001); and tumor-node-metastasis (TNM) stage (P < .001). Prostate cancer patients with a high expression of miR-153 had an evidently lower 5-year overall survival as compared with those with a low expression of miR-153 (P = .019). Notably, the multivariate Cox regression analysis indicated that miR-153 expression was an independent factor for predicting the 5-year overall survival of prostate cancer patients (hazard ratio [HR] = 2.481, 95% confidence interval [CI]: 1.582–10.727; P = .018).
Our study demonstrated that high miR-153 expression was significantly associated with a poor overall survival independently of other factors in prostate cancer. Therefore, miR-153 may be an available biomarker for prostate cancer prognosis.
Keywords: biomarker, microRNA-153, prognostic value, prostate cancer
1. Introduction
Prostate cancer ranks the most commonly diagnosed cancer among male population, and ranks the second leading cause of cancer deaths rate in the United States.[1,2] At present, open, laparoscopic, or robotic assistance radical prostatectomy are the standard treatment for localized prostate cancer.[3] Prostate-specific antigen (PSA) is the main factor to predict the recurrence after operation. However, PSA is not sensitive enough.[4] Therefore, it is necessary to find new factors to predict the recurrence after operation and guide the treatment after operation.
MicroRNAs (miRNAs) are single-stranded, short (about 22 nucleotides in length) RNAs that interact with downstream messenger RNAs (mRNAs), resulting in their translational repression or degradation.[5,6] Accelerating evidence links miRNAs to the initiation, development, promotion, and progression of malignancies.[7,8] Dysregulation of miRNA expression has been identified in various cancers, and accumulating data suggest that miRNAs function as classical oncogenes or tumor suppressor genes.[9]
Deregulation of miR-153 has recently been observed in several common human cancer, while miR-153 serves an oncogene[10,11] or tumor suppressive[12–17] role in different cancer types. Previously, miR-153 has been identified to be overexpressed in prostate cancer. miR-153 played an important role in promoting proliferation of human prostate cancer cells and presented a novel mechanism of microRNA-mediated direct suppression of phosphatase and tensin homolog (PTEN) expression in prostate cancer cells.[18] Until now, little is known about the clinical significance of miR-153 expression in prostate cancer. In the present study, we aimed to investigate the expression level of miR-153 in human prostate cancer tissue, and the associations between miR-153 expression and clinicopathological factors and prognosis of patients with prostate cancer.
2. Materials and methods
2.1. Patients
In the present study, a total of 143 prostate cancer tissues and paired adjacent normal tissues were obtained from patients who underwent radical prostatectomy between April 2014 and March 2018 at Yunnan Cancer Hospital (Third Affiliated Hospital of Kunming Medical University). None of the patients had received androgen deprivation treatment, chemotherapy, or radiation therapy prior to the surgery. The prostate cancer specimens were subjected to histological examination by 2 expert pathologists for confrmation of the Gleason grading, WHO classifcation of the tumor, and staging according to the tumor-node-metastasis (TNM) system. Subject information is shown in Table 1. The approval of the present study protocol was obtained from the Ethics Committee of Yunnan Cancer Hospital (Third Affiliated Hospital of Kunming Medical University), and the written informed consent was provided from all patients.
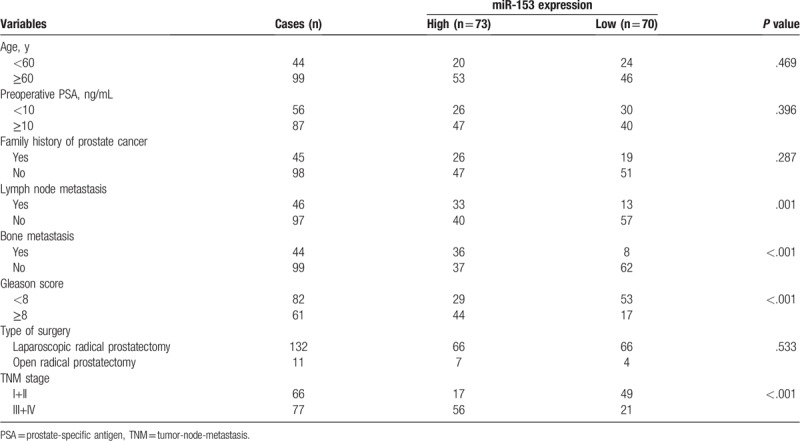
Table 1.
Relationship between miR-153 expression and clinicopathological characteristics of patients with prostate cancer.
2.2. Quantitative real-time RT-PCR
Total RNA was extracted using Trizol Reagent according to the manufacturer's protocol. cDNA synthesis was performed using TaqMan MicroRNA Array kit (Applied Biosystems, CA). Real-time quantitative PCR was performed using primers at 95 °C for 3 minutes, then 40 cycles of 95 °C for 12 seconds, and 60 °C for 40 seconds. U6 was used as an endogenous control to normalize expression. Each experiment was performed in triplicate, and all the relative expression levels were measured by the 2–ΔΔCT method. The primers were listed as follows: miR-153, 5′-ACACTCCAGCTGGGTTGCATAGTCACAAA-3′ (forward) and 5′-CAGTGCGTGTCGTGGAGT-3′ (reverse); U6, 5′-CCCTTCGGGGACATCCGATA-3′(forward) and 5′-TTTGTGCGTGTCATCCTTGC-3′ (reverse).
2.3. Statistical analyses
Student t test was conducted for intergroup comparison. Pearson correlation test was used for correlation analysis. Survival curves were carried out by the Kaplan–Meier method and evaluated using the log-rank test. Multivariable Cox proportional hazard risk regression model was performed to screen the independent factor affected the prognosis of prostate cancer patients. All the statistical analysis was performed on SPSS 18.0 software (SPSS Inc., Chicago, IL). All tests were 2-sided, and P < .05 was considered as statistically significant.
3. Results
3.1. The tissue expression of miR-153 was increased in prostate cancer
The miR-153 expression in 143 pairs of prostate cancer and adjacent non-cancerous prostate tissues was measured by qRT-PCR analysis. The results showed that the expression of miR-153 was significantly increased in the prostate cancer tissues in comparison with the adjacent noncancerous prostate tissues (P < .001, shown in Fig. 1). The expression of miR-153 was considered as either high (n = 73) or low (n = 70) according to the cut-off value, which was defined as the median of the cohort.
Figure 1.
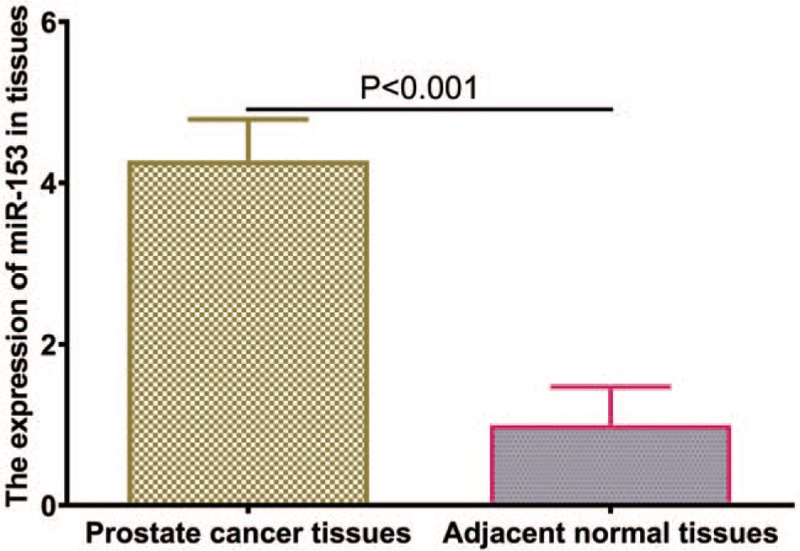
The expression levels of miR-153 in prostate cancer tissues and matched nontumor tissues.
3.2. Relationship between miR-153 expression level and clinical features of prostate cancer patients
To investigate the clinical significance of miR-153, we assessed the association between miR-153 expression and clinicalpathological features. As shown in Table 1, the high expression of miR-153 in prostate cancer tissues is closely correlated with aggressive clinical pathological parameters such as lymph node metastasis (P = .001); bone metastasis (P < .001); Gleason score (P < .001); and TNM stage (P < .001). No association was found between the expression level of miR-153 and other clinical factors including age, preoperative PSA, family history of prostate cancer, and type of surgery (all P > .05).
3.3. miR-153 expression level and prostate cancer prognosis
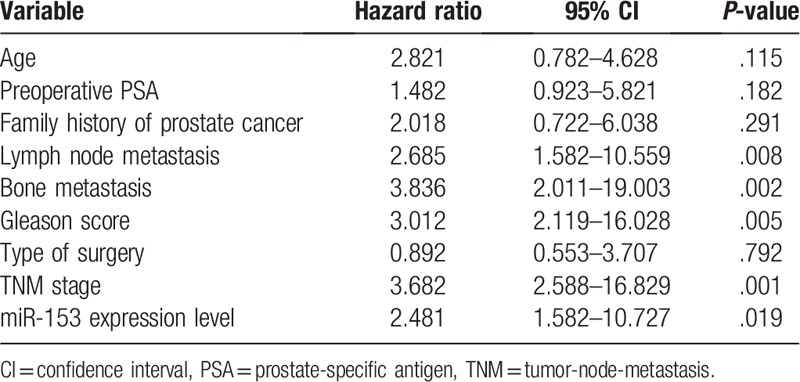
The Kaplan–Meier survival analysis was performed to determine the prognostic value of miR-153 in prostate cancer patients. Prostate cancer patients with a high expression of miR-153 had an evidently lower 5-year overall survival as compared with those with a low expression of miR-153 (P = .019, shown in Fig. 2). Notably, the multivariate Cox regression analysis indicated that miR-153 expression was an independent factor for predicting the 5-year overall survival of prostate cancer patients (HR = 2.481, 95% CI: 1.582–10.727; P = .018, shown in Table 2). These data indicated that miR-153 might be a potent biomarker for predicting the prognosis of prostate cancer patients.
Figure 2.
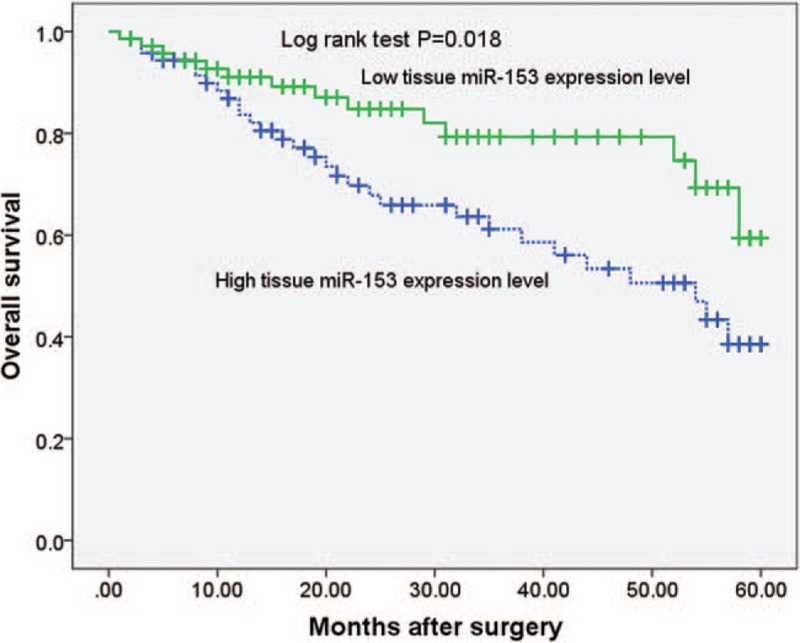
According to the level of miR-153 expression, Kaplan–Meier 5-year overall survival of prostate cancer patients showed that high expression of miR-153 was significantly correlated with poor prognosis (P = .018).
Table 2.
Multivariate analyses for overall survival by Cox regression model.
4. Discussion
The prostate-specific antigen (PSA), digital rectal examination (DRE), and ultrasonography of prostate are now employed for early detection of prostate cancer. Prostate biopsy is considered as a gold standard in patients with raised suspicion from digital rectal examination or increased PSA levels.[4] At present, open, laparoscopic or robotic assistance radical prostatectomy are the standard treatment for localized prostate cancer. However, about 25% to 50% postsurgery patients on long-term follow-up have biochemical recurrence. This is generally the earliest indicator of the recurrence. Therefore, the early risk detection of biochemical recurrence with sensitive methods and specific tumor markers may effectively improve the prognosis of patients with prostate cancer.[19,20]
miRNAs are transcribed genes processed to single-stranded regulatory RNA of approximately 22 nucleotides and regulate target mRNAs.[5] miRNAs effect gene silencing via both translational inhibition and mRNA degradation. An individual miRNA is capable of regulating several distinct mRNAs, and together >1000 human miRNAs are believed to modulate more than one-third of the mRNA species encoded in the genome. Accelerating evidence links miRNAs to the initiation, development, promotion, and progression of malignancies.[7,8] Dysregulation of miRNA expression has been identified in various cancers, and accumulating data suggest that miRNAs function as classical oncogenes or tumor suppressor genes.[9]
Deregulation of miR-153 has recently been observed in several common human cancer, while miR-153 serves an oncogene or tumor suppressive role in different cancer types. In some cancer types, the expression of miR-153 is up-regulated and plays an oncogene role. For example, in the study by Zhang et al,[10] miRNA profiling revealed that miR-153 was highly expressed in a cellular model of advanced stage colorectal cancer. Its upregulation was also noted in primary human colorectal cancer compared with normal colonic epithelium and in more advanced colorectal cancer stages compared with early stage disease. Functional studies revealed that miR-153 supported colorectal cancer progression via pleiotropic effects that enhance invasion and chemotherapeutic resistance. Hua et al[11] found that miR-153 was up-regulated in hepatocellular carcinoma (HCC) tissues and correlated with poor survival of HCC patients. Overexpression of miR-153 was able to promote β-catenin transcriptional activity, leading to cell-cycle progression, proliferation, and colony formation of HCC cells. However, in some other cancer types, the expression of miR-153 is decreased, playing a role of tumor suppressor. For example, Zhao et al[12] found that miR-153 was down-regulated in glioma tissues and cells versus paracancerous tissues and normal immortalized gliocyte HEB cells. They verified that SNAI1 was a target of miR-153 and had a negative association with miR-153 detected by luciferase reporter assay. Additionally, miR-153 suppressed cell invasive ability by regulating SNAI1 expression, whose partial function was reversed by SNAI1. Guo et al[13] found that the expression of miR-153 in patients with nasopharyngeal cancer was markedly decreased compared with that in paracarcinoma tissue. miR-153 affected the progression of nasopharyngeal cancer by targeting the TGF-β2/Smad2 signaling pathway. Wang et al[14] found that the expression of miR-153 in breast cancer tissue specimens and MDA-MB-231 cells was significantly lower than that in nonmalignant counterparts. Furthermore, miR-153 inhibited migration, invasion and epithelial-mesenchymal transition of breast cancer by regulating transforming growth factor beta (TGF-β) signaling pathway. Zhang et al[15] found that miR-153 suppressed human laryngeal squamous cell carcinoma migration and invasion by targeting the Snail family transcriptional repressor 1 (SNAI1) gene.
Previously, miR-153 has been identified to be overexpressed in prostate cancer. miR-153 played an important role in promoting proliferation of human prostate cancer cells and presented a novel mechanism of microRNA-mediated direct suppression of PTEN expression in prostate cancer cells.[18] Until now, little is known about the clinical significance of miR-153 expression in prostate cancer. In the present study, we aimed to investigate the expression level of miR-153 in human prostate cancer tissue, and the associations between miR-153 expression and clinicopathological factors and prognosis of patients with prostate cancer. The miR-153 expression in 143 pairs of prostate cancer and adjacent non-cancerous prostate tissues was measured by qRT-PCR analysis. The results showed that the expression of miR-153 was significantly increased in the prostate cancer tissues in comparison with the adjacent noncancerous prostate tissues. The expression of miR-153 was considered as either high or low according to the cut-off value, which was defined as the median of the cohort. To investigate the clinical significance of miR-153, we assessed the association between miR-153 expression and clinicalpathological features. We found that the high expression of miR-153 in prostate cancer tissues was closely correlated with aggressive clinical pathological parameters such as lymph node metastasis, bone metastasis, Gleason score, and TNM stage. The Kaplan–Meier survival analysis was performed to determine the prognostic value of miR-153 in prostate cancer patients. Prostate cancer patients with a high expression of miR-153 had an evidently lower 5-year overall survival as compared with those with a low expression of miR-153. Notably, the multivariate Cox regression analysis indicated that miR-153 expression was an independent factor for predicting the 5-year overall survival of prostate cancer patients. These data indicated that miR-153 might be a potent biomarker for predicting the prognosis of prostate cancer patients. The limitation of the present study is that we have not done depth analysis on the gene ontology, browser tracks, and we have not investigated the targets of onco-miR-153 such as PTEN, FOXOM1, or Akt in prostate cancer cells. More in-depth study is needed in the future to clarify the role of miR-153 in prostate cancer.
In conclusion, our study demonstrated that high miR-153 expression was significantly associated with a poor overall survival independently of other factors in prostate cancer. Therefore, miR-153 may be an available biomarker for prostate cancer prognosis.
Author contributions
Conceptualization: Cheng-wei Bi, Guo-ying Zhang, Hong Yang.
Data curation: Cheng-wei Bi, Guo-ying Zhang, Yu Bai, Bin Zhao, Hong Yang.
Investigation: Cheng-wei Bi, Guo-ying Zhang, Yu Bai, Bin Zhao.
Methodology: Cheng-wei Bi, Yu Bai, Bin Zhao.
Project administration: Cheng-wei Bi.
Writing – original draft: Cheng-wei Bi, Guo-ying Zhang, Bin Zhao, Hong Yang.
Writing – review & editing: Cheng-wei Bi, Hong Yang.
Footnotes
Abbreviations: DRE = digital rectal examination, HCC = hepatocellular carcinoma, miRNAs = MicroRNAs, PSA = prostate-specific antigen, SNAI1 = Snail family transcriptional repressor 1, TGF-β = transforming growth factor beta.
How to cite this article: Bi Cw, Zhang Gy, Bai Y, Zhao B, Yang H. Increased expression of miR-153 predicts poor prognosis for patients with prostate cancer. Medicine. 2019;98:36(e16705).
C-wB and G-yZ have contributed equally.
The authors have no conflicts of interest to disclose.
References
- [1].Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394–424. [DOI] [PubMed] [Google Scholar]
- [2].Hsing AW, Chokkalingam AP. Prostate cancer epidemiology. Front Biosci 2006;11:1388–413. [DOI] [PubMed] [Google Scholar]
- [3].Wilt TJ, Ahmed HU. Prostate cancer screening and the management of clinically localized disease. BMJ 2013;346:f325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Mottet N, Bellmunt J, Bolla M, et al. EAU-ESTRO-SIOG guidelines on prostate cancer. Part 1: Screening, diagnosis, and local treatment with curative Intent. Eur Urol 2017;71:618–29. [DOI] [PubMed] [Google Scholar]
- [5].Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell 2009;136:215–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Ambros V. The functions of animal microRNAs. Nature 2004;431:350–5. [DOI] [PubMed] [Google Scholar]
- [7].Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer 2006;6:857–66. [DOI] [PubMed] [Google Scholar]
- [8].McManus MT. MicroRNAs and cancer. Semin Cancer Biol 2003;13:253–8. [DOI] [PubMed] [Google Scholar]
- [9].Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer 2006;6:259–69. [DOI] [PubMed] [Google Scholar]
- [10].Zhang L, Pickard K, Jenei V, et al. miR-153 supports colorectal cancer progression via pleiotropic effects that enhance invasion and chemotherapeutic resistance. Cancer Res 2013;73:6435–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Hua HW, Jiang F, Huang Q, et al. MicroRNA-153 promotes Wnt/beta-catenin activation in hepatocellular carcinoma through suppression of WWOX. Oncotarget 2015;6:3840–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Zhao W, Yin CY, Jiang J, et al. MicroRNA-153 suppresses cell invasion by targeting SNAI1 and predicts patient prognosis in glioma. Oncol Lett 2019;17:1189–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Guo G, Zhang Y, Hu L, et al. MicroRNA-153 affects nasopharyngeal cancer cell viability by targeting TGF-beta2. Oncol Lett 2019;17:646–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Wang J, Liang S, Duan X. Molecular mechanism of miR-153 inhibiting migration, invasion and epithelial-mesenchymal transition of breast cancer by regulating transforming growth factor beta (TGF-beta) signaling pathway. J Cell Biochem 2019;120:9539–46. [DOI] [PubMed] [Google Scholar]
- [15].Zhang B, Fu T, Zhang L. MicroRNA-153 suppresses human laryngeal squamous cell carcinoma migration and invasion by targeting the SNAI1 gene. Oncol Lett 2018;16:5075–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Ouyang Y, Yuan W, Qiu S. MicroRNA-153 functions as a tumor suppressor in gastric cancer via targeting Kruppel-like factor 5. Exp Therap Med 2018;16:473–82. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [17].Wu X, Li L, Li Y, et al. MiR-153 promotes breast cancer cell apoptosis by targeting HECTD3. Am J Cancer Res 2016;6:1563–71. [PMC free article] [PubMed] [Google Scholar]
- [18].Wu Z, He B, He J, et al. Upregulation of miR-153 promotes cell proliferation via downregulation of the PTEN tumor suppressor gene in human prostate cancer. Prostate 2013;73:596–604. [DOI] [PubMed] [Google Scholar]
- [19].Witte JS. Prostate cancer genomics: towards a new understanding. Nat Rev Genet 2009;10:77–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Lee JT, Lehmann BD, Terrian DM, et al. Targeting prostate cancer based on signal transduction and cell cycle pathways. Cell Cycle 2008;7:1745–62. [DOI] [PMC free article] [PubMed] [Google Scholar]


