Supplemental Digital Content is available in the text.
Background.
Delayed graft function (DGF) after kidney transplantation is negatively associated with long-term graft function and survival. Kidney function after transplantation depends on multiple factors, both donor- and recipient-associated. Prediction of posttransplantation graft function would allow timely intervention to optimize patient care and survival. Currently, graft-based predictions can be made based on histological and molecular analyses of 0-hour biopsy samples. However, such analyses are currently not implemented, as biopsy samples represent only a very small portion of the entire graft and are not routinely analyzed in all transplantation centers. Alternatives are thus required.
Methods.
We analyzed whether donor organ preservation fluid contain small extracellular vesicles (sEV) and whether the RNA content of these vesicles could be used as a source for potential biomarkers for posttransplantation kidney function.
Results.
We provide proof of principle that sEVs are present in preservation fluid, which contain RNAs associated with donor origin. Furthermore, sEV micro RNA profiles could be associated with graft function during the first 7 days posttransplantation, but no significant correlation with DGF could be established based on the current dataset.
Conclusions.
Overall, the predictive potential of sEV RNA biomarkers together with relatively easy and noninvasive sample collection and analysis methods could pave the way towards universal screening of donor kidney-associated risk for DGF, optimized patient treatment, and subsequently improved short- and long-term graft function and survival.
With the increased life expectancy and concomitant incidence of vascular diseases in the Western world, chronic kidney disease has an enormous social and economic impact. With an estimated 30 million adult patients, healthcare costs are estimated to be close to 100 billion dollars annually.1 Kidney transplantation remains the preferred treatment for patients with end stage kidney disease, and the shortage of donor organs will further increase in the upcoming years. To optimize donor potential, it is important to be able to predict posttransplantation function. Prior knowledge on organ function allows for patient-tailored care, such as adaptation of the immunosuppressive regimen and planning of dialysis to bridge the period to full graft function. In addition, assays that provide information on graft function could be used to screen kidneys that are currently rejected for transplantation to identify additional organs that are suitable for transplantation, thereby increasing the availability of donor kidneys.
Currently, allograft quality is judged by donor age, donor plasma creatinine and urea values (when available), visual inspection, and warm and cold ischemia times. This procedure includes little information on the individual kidney and in many cases information may be incomplete, especially in donation after circulatory death donors. Graft function and survival can be monitored by analyzing biomarkers in urine and blood (after transplantation)2-4 or by taking biopsy samples, which can be done at the time of transplantation (0-h biopsies)5-8 or after transplantation.9,10 In these samples, morphological and genetic markers that represent kidney function and survival can be identified. A great disadvantage of above-mentioned approaches is that they can either only take place after transplantation (blood/urine sampling) or that the transplanted kidney has to be intentionally damaged (biopsies). Furthermore, it is under debate whether the small piece of tissue retrieved in a renal biopsy is representative of the entire organ. Altogether, despite the identification of markers associated with transplant outcome, no routine testing on predictive biomarkers is currently performed on donor kidneys, illustrating the limited value of these markers in clinical practice and the urge for the identification of markers in representative samples.
Communication between cells, tissues, and organs is crucially important to maintain homeostasis of an organism. It allows for proper responses to endogenous and environmental cues and occurs through different pathways, such as electrical, chemical, and hormonal signaling. Especially in cases of stress (ie, ischemia-reperfusion injury), cytokine signaling plays an important role, and recently we demonstrated that cytokines secreted into donor kidney preservation fluid contain donor organ-derived information allowing to predict delayed graft function (DGF).11 Besides cytokines and growth factors, extracellular vesicles (EVs) gained interest for their unique potential in intercellular signaling, since they can contain complex sets of signals that can be transferred to target cells in a cell-specific manner.12-16 Small extracellular vesicles (sEVs), also termed exosomes, are 50- to 150-nm membrane vesicles that are secreted into the environment by cells upon fusion of the limiting membrane of multivesicular bodies with the plasma membrane.17 They arise from inward budding of endosomal membranes and, as they are formed at the late endosomes, capture a small portion of the cytoplasm, including proteins and RNA molecules.18,19
We hypothesize that sEVs secreted into transplant kidney preservation fluid by the renal endothelium reflect the stress, and thereby the condition, of the donor organ and may therefore serve as biomarkers to assess donor kidney quality and predict posttransplantation function. To investigate this, we collected perfusion fluid after static cold storage from living, donation after brain (DBD), and donation after circulatory death (DCD) donor kidneys and investigated whether sEVs were secreted into this fluid and whether their content reflects donor organ condition. We show for the first time that sEVs are secreted into the preservation fluid. We identified an optimal procedure for the isolation of sEVs from preservation fluid and determined that they contain small RNAs that reflect donor type (living, DBD, or DCD) and associate with graft function.
MATERIALS AND METHODS
Sample Collection
Donor Kidney Fluid
In agreement with the declaration of Helsinki and with approval of the University Medical Center Utrecht Ethical Committee, intravascular perfusion fluid was retrieved as described.11 Briefly, after static cold storage donor kidneys were perfused with 40–60 mL of physiological salt solution, supplemented with 20 U/mL heparin into the renal artery. Intravascular perfusion fluid, flowing out from the renal vein, was collected in a sterile manner. Blood cells in the collected fluid were counted using a CELL DYN 1800 Hematology Analyzer (Abbott, Abbott Park, IL). Cells were removed by centrifugation at 2000g for 30 minutes, after which the supernatant was used for exosome isolation. Samples for analysis of sEV presence were collected based on availability (n = 8), and at later stages, samples from specific subgroups (n = 19), based on donor type and posttransplantation function (n = 16), were collected sequentially for retrospective analyses.
Transplantations were performed in the University Medical Center Utrecht (The Netherlands) between January 2013 and July 2015. All recipients provided informed consent to retrieve data from medical records for the Netherlands Organ Transplantation Registry.
Cell Culture Supernatant
HMEC-120 cells (CDC; Atlanta, GA) were used for control experiments. They were maintained up to passage 27 at 37°C, 5% CO2 in MCDB131 medium (Life Technologies, Grand Island, NY) supplemented with 10% fetal calf serum (FCS), 100 U/mL penicillin, 100 µg/mL streptomycin, 50 nmol/L hydrocortisone, 10 ng/mL human recombinant epidermal growth factor, and 10 mmol/L l-glutamine. Exosome-free medium was prepared using FCS centrifuged for at least 1 hour at 200 000g and filter-sterilized using a 0.20-µm filter.
Serum
Venous blood was drawn from healthy volunteers using BD Vacutainer CAT blood collection tubes. After gentle shaking and incubation at room temperature (RT) for 1.5 hours, samples were centrifuged at 1500g for 15 minutes. The supernatant consisting of the serum was used for further experiments.
sEV Isolation
Ultracentrifugation
sEVs were collected by differential centrifugation.21 Samples—either cell culture supernatant, serum, or donor kidney perfusion fluid—were subsequently centrifuged for 30 minutes at 2000g, 30 minutes at 10 000g, and 60 minutes at 100 000g. Pelleted EVs were resuspended in phosphate buffered saline (PBS), pelleted again, and finally resuspended in 60 µL PBS (for Nanoparticle tracking, sucrose density centrifugation, or electron microscopy analysis) or 100 µL lysis buffer (for immunoblotting).
Precipitation
Samples—cell culture supernatant, serum, or donor kidney perfusion fluid—were centrifuged subsequently for 30 minutes at 2000g, after which the supernatant was mixed with Total Exosome Isolation (TexIs) reagent (Invitrogen, Carlsbad, CA) following manufacturer instructions. Pelleted sEVs were resuspended in 60 µL PBS (for Nanoparticle tracking, sucrose density centrifugation, electron microscopy analysis, and RNA qPCR analysis) or 100 µL lysis buffer (for immunoblotting of RNA sequencing analysis).
Sucrose Gradient Analysis
Pellets obtained after centrifugation at 100 000g were resuspended in 250 µL 2.5M sucrose, 20 mmol/L Tris HCl pH 7.4 and floated in a SW60 tube for 16 hours at 190 000g using a linear sucrose gradient (2.0–0.25M sucrose, 20 mmol/L Tris HCl, pH 7.4). Gradient fractions (250 μL) were collected from the top and used for subsequent immunoblot and electron microscopy analyses.
Electron Microscopy
Transmission electron microscopy was performed as described.22 Briefly, carbon-coated Formvar filmed grids were placed on exosome suspension for 20 minutes and washed with 0.15% glycin in PBS 3 times followed by a 0.1% bovine serum albumin (BSA) in PBS wash. Vesicles were fixed in 1% glutaraldehyde in PBS for 5 minutes and washed twice with PBS. For CD63 labeling, grids were placed on 5 μL 1% BSA in PBS containing 5 μg/mL anti-human CD63 (CLB, Amsterdam, The Netherlands) for 20 minutes, washed 4 times with 0.1% BSA in PBS, and incubated on a drop of 1% BSA in PBS containing rabbit anti-mouse polyclonal antibody (Dako, Glostrup, Denmark) for 20 minutes. Secondary antibodies were labeled by incubation on PBS containing 1% BSA and 10 nm gold particles coupled to Protein A in for 10 minutes. Specimen were fixed in 1% glutaraldehyde in PBS for 5 minutes and washed twice with PBS. After 4 washes on distilled water, grids were placed on ice-cold 1.8% methylcellulose (25 Ctp)/0.4% Uranyl acetate (MC-AU) for 5 minutes, and, after drying, vesicles were visualized using a FEI Tecnai 12 (FEI, Hillsboro, OR) transmission electron microscope.
Immunoblotting
sEVs were lysed in lysis buffer (0.1% sodium dodecyl sulphate [SDS], 1% Triton X-100 with complete protease inhibitor cocktail [Sigma] in PBS) and concentrations were determined using a bicinchoninic acid protein assay (Pierce). Equal sample amounts were subjected to 12% SDS-PAGE electrophoresis and transferred to polyvinylidene difluoride (PVDF) membranes (Thermo Scientific, Waltham, MA). PVDF membranes were blocked in 5% low-fat dry milk powder (Campina, Amersfoort, The Netherlands) in tris buffered saline (TBS) with 0.1% Tween-20 (blocking buffer). PVDF membranes were incubated with primary antibodies in blocking buffer, washed in TBS with 0.1% Tween-20, followed by incubation with horse radish peroxidase (HRP)-conjugated secondary antibodies in blocking buffer. Proteins were detected with Chemiluminescent Peroxidase Substrate (Sigma) and imaged on the Molecular Image ChemiDoc XRS system (Biorad, Hercules, CA). The primary antibodies used were mouse-anti-β-actin (A5441, 1:15 000, Sigma) and rabbit-anti-Flotillin-1 (sc-25506, 1:500, Santa Cruz Biotechnologies [Santa Cruz, CA]). Secondary antibodies were HRP-conjugated Swine anti-Rabbit (P0399, 1:2000, Dako) and Rabbit anti-Mouse (P0260, 1:2000, Dako).
Nanoparticle Tracking Analysis
Size distribution of vesicles in 100 000g pellets were analyzed using a Nanosight LM14C (NanoSight, Amesbury, UK) with a 532 nm laser-equipped sample flow-chamber. Shutter and gain were manually adjusted for optimal detection and were kept at this setting for all samples. A 1-minute audio video interleave file was recorded and analyzed using nanoparticle tracking analysis (version 2.3, build 0025, Nanosight) software to calculate size distributions and vesicle concentrations using the following settings: blur: auto; detection threshold: 7, minimum track length: auto, viscosity: 0.929 cP. Each observation represents the average of 5 measurements.
Small RNA Sequencing Analysis
After exosome precipitation using the TExIs from serum reagent (Invitrogen), total RNA was isolated using the Total exosome RNA and protein isolation kit (Invitrogen). Subsequently, small RNA libraries were prepared, sequenced, and bioinformatically processed as extensively described by Schageman et al.23
RNA Isolation
Total RNA was isolated using the Qiagen miRNeasy Mini Kit (QIAgen, Hliden, Germany). After lysis in 700 μL Qiazol lysis reagent, the lysate was transferred to a new tube with 140 μL chloroform, mixed, incubated for 2 minutes at room temperature and centrifuged at 12 000g for 15 minutes at 4°C. The aqueous phase was transferred to a new tube and 1.5 volume of 100% ethanol was added. After mixing, the sample was transferred to a Qiagen RNeasy Mini spin column in a collection tube followed by centrifugation at 8000g for 15 seconds at RT. Following a rinse with 700 μL Qiagen RWT buffer and centrifuged at 8000g for 15 seconds at RT, filters were rinsed with 500 μL Qiagen RPE buffer and centrifuged as above. A rinse step (500 μL Qiagen RPE buffer) was repeated twice. The Qiagen RNeasy Mini spin column was transferred to a new collection tube and centrifuged at 15000g for 2 minutes at RT. The Qiagen RNeasy Mini spin column was transferred to a new microcentrifuge tube with the lid open for 1 minute to allow the column to dry. Total RNA was eluted by adding 50 μL of RNase-free water to the spin column and centrifugation at 15000g for 1 minute after incubating at RT for 1 minute.
microRNA Real-time qPCR Arrays
RNA was isolated from sEVs precipitated from 2.0 mL perfusion fluid (where possible, 1.5 mL for one sample) and reverse transcribed in 50 μL reactions using the miRCURY LNA Universal RT microRNA polymerase chain reaction (PCR), Polyadenylation and cDNA synthesis kit (Exiqon, Vedbaek, Denmark). cDNA was diluted 50× and assayed in 10 μL PCR reactions according to the protocol for miRCURY LNA Universal RT microRNA PCR; each microRNA was assayed once by qPCR on the microRNA Ready-to-Use PCR, Human Panel I using ExiLENT SYBR Green master mix. Amplification was performed in a LightCycler 480 Real-Time PCR System (Roche, Basel, Switzerland) in 384 well plates. Amplification curves were analyzed using the Roche LC software, both for determination of Cq (by the second derivative method) and melting curve analysis. Amplification efficiency was calculated using algorithms similar to the LinReg software. All assays were inspected for distinct melting curves and the Tm was checked to be within known specifications for the assay. Furthermore assays must be detected with 5 Cqs less than the negative control and Cq<37 to be included in the data analysis. Cq was calculated as the second derivative. Using NormFinder the best normalizer was found to be the average of assays detected in all samples. All data were normalized to the average of assays detected in all samples (average–assay Cq)
Statistics
Differences between groups were assessed using an unpaired samples Student t-test with Benjamini-Hochberg post hoc testing. Missing, undetected values were set at 9 for calculation purposes.
Unsupervised clustering methods were used to identify patterns in the microRNA expression patterns across samples in an unbiased fashion and results from different methods were set side-by-side to check for internal consistency. For hierarchical cluster analysis, we used Euclidean distance as distance metric and the unweighted pair group method with arithmetic mean as linkage criterion. Results were verified by visual inspection of the plot of first two principal components and cluster number was assessed by partitioning around medoids with estimation of number of clusters. R version 3.5.0 was used for all analyses, with additional use of the Flexible Procedures for Clustering package.
RESULTS
Donor Kidney Preservation Fluid Contains sEVs
To investigate whether donor kidney preservation fluid contains sEVs, freshly collected fluid was subjected to the standard differential centrifugation protocol for the isolation of such vesicles21 and analyzed for the presence of sEVs using sucrose density centrifugation, nanoparticle tracking analysis, and electron microscopy. The density of the collected particles was determined at 1.127 ± 0.025 g/mL (n = 3), with a representative immunoblot of density gradient fractions displayed in Figure 1A. Transmission electron microscope of the peak fraction revealed the presence of 50–200 nm particles of different electron densities, among which cup-shaped vesicles, the typical appearance of sEVs in such analyses (Figure 1B). Nanoparticle tracking analysis confirmed the electron microscopic observations, showing a modal size of 83.7±10 nm and mean size of 128.3±10.5 nm (n = 3). Combined, these observations demonstrate that donor kidney preservation fluid accumulates renal sEVs.
FIGURE 1.
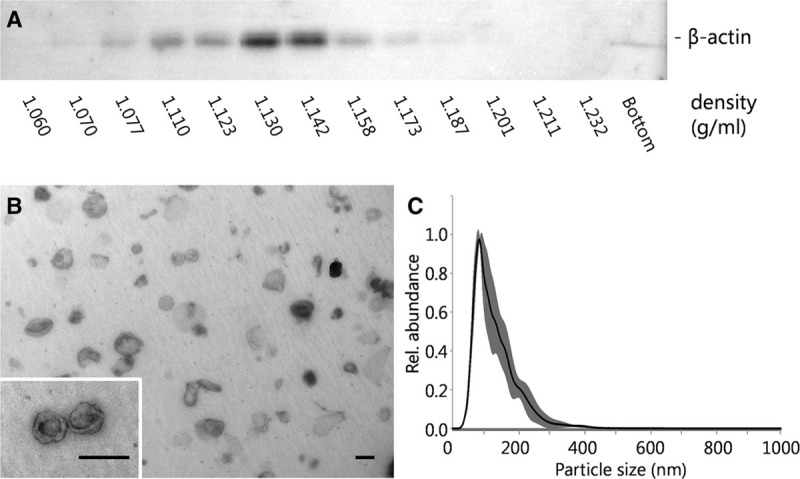
sEVs from donor kidney perfusate. A, A representative immunoblot of sucrose density gradient fractions shows a peak signal at a density of 1.130 g/mL. B, Transmission electron microscope analysis of the peak fraction (scale bar = 100 nm) and (C) NTA analysis demonstrate the presence of 50–150 nm vesicles (SD in grey). NTA, nanoparticle tracking analysis; SD, standard deviation; sEVs, small extracellular vesicles.
A Precipitation Method for Renal Perfusate sEVs
For future potential use in a diagnostic setting, isolation of sEVs by differential (ultra-) centrifugation cannot be performed because this approach is time-consuming, and taking into account that the centrifuge rotor can hold only a few samples at a time—incompatible with high throughput sample processing in a clinical laboratory. We therefore aimed to precipitate a representative exosome fraction from the preservation fluid, an approach for which currently several approaches are available.24-27 We compared the TExIs reagents for serum and for cell culture supernatant28 for their efficiency in isolating sEVs from the corresponding biofluids and compared their performance with that of sequential ultra-centrifugation. Using sucrose density gradient and nanoparticle tracking analyses, we could demonstrate that centrifugation and precipitation of endothelial cell culture medium yields particles with identical characteristics, displaying a density and size corresponding to endothelial cell-derived sEVs22,29 (Figure 2A, C). Using serum as a source, the TExIs reagent (for serum) precipitated particles with a slightly smaller size than that observed for particles isolated by centrifugation, whereas the density appeared higher, given that the peak intensity shifted by 1 fraction (Figure 2B, D).
FIGURE 2.
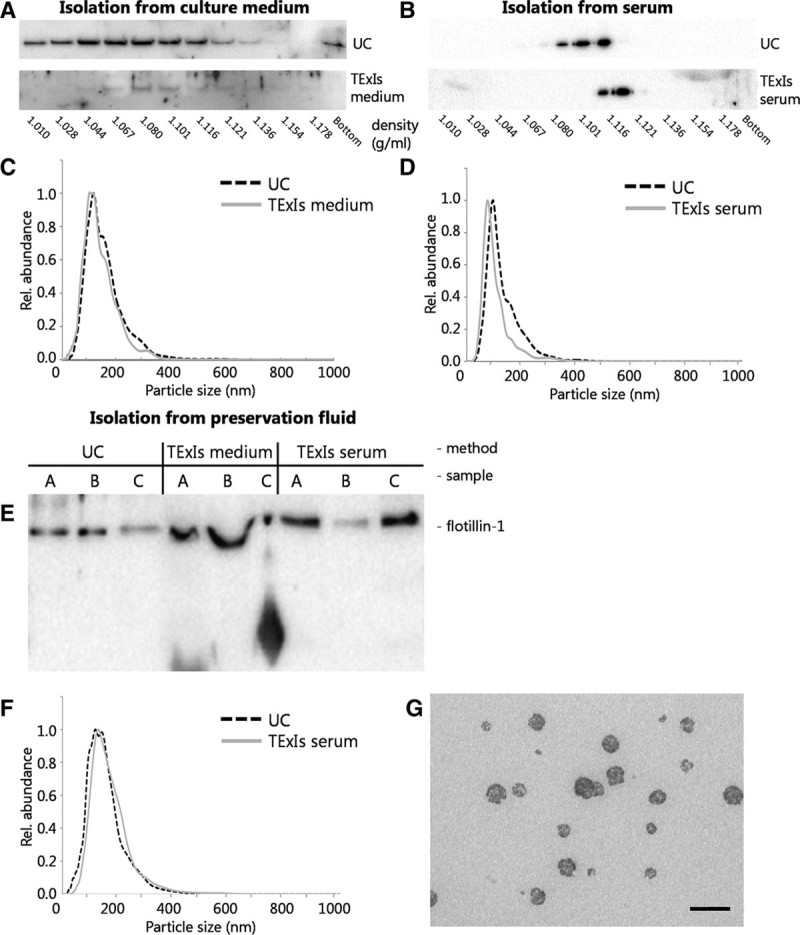
A precipitation method for perfusion fluid sEVs. sEVs were isolated from human microvascular endothelial cells-1-conditioned culture medium or human serum using sequential UC or precipitation (using the TExIs reagent for culture medium or serum, respectively) and subsequently analyzed by sucrose density gradient ultracentrifugation and immunoblotting for β-actin. sEVs isolated from culture medium by precipitation (A, TExIs medium) appeared slightly more dense than those isolated by ultracentrifugation (A, UC). Also, sEVs isolated from serum using the TExIs reagent for serum (B, TExIs serum) appeared slightly more dense than those isolated by ultracentrifugation. Size distribution of the sEVs from either source and by either method did not significantly differ (C,D). Immunoblotting of sEVs from 3 different donor fluids isolated by ultracentrifugation (UC) or TExIs reagents for culture media (TExIs medium) or serum (TExIs serum) show a disturbed running pattern for those isolated using the reagent for serum (E). sEVs were isolated from preservation fluid by UC or using the TExIs reagent for serum (TExIs serum). F, Subsequent NTA analysis shows that both sEV preparations contain equally sized particles, consistent with the size of small EVs. G, TEM analysis of the precipitated sEVs confirms the presence of sEVs, with signs of co-isolated reagent (scale bar = 200 nm). EVs, extracellular vesicles; NTA, nanoparticle tracking analysis; sEVs, small extracellular vesicles; TExIs, Total Exosome Isolation.
The use of reagents thus appears a promising approach for rapid, benchtop isolation of sEVs from donor kidney preservation fluid. As the preservation fluid resembles neither cell culture medium nor serum, we first investigated which of the 2 reagents could be used to isolate sEVs from preservation fluid and directly compared this with ultracentrifugation (UC)-isolated sEVs. Isolated sEVs from 3 donor kidneys were immunoblotted for the exosome marker protein flotillin-1, and based on the obtained band pattern the TExIs reagent for serum was chosen for subsequent exosome isolations from preservation fluid (Figure 2E). Nanoparticle tracking analysis comparing sEVs from preservation fluid isolated by UC or reagent for serum (TExIs serum) shows that particles of equal size are isolated by each method (Figure 2F). Electron microscopic analysis of these sEV isolated using the TExIs serum reagent confirms their size (Figure 2G), though their appearance differs from the UC-isolated sEVs from preservation fluid (Figure 1B).
Exosomal Small RNAs Reflect Donor Type
As we previously described that exosomal RNAs reflect the condition of the exosome-producing cells,22 we hypothesized that the RNA content of sEVs from preservation fluids from DCD, DBD, and living donors will reflect the donor type and may provide prognostic information about the donor kidney quality and posttransplantation kidney function. To investigate this, RNA was isolated from 19 precipitated perfusion fluid exosome samples representing three donor types (6 × DCD, 6 × DBD, and 7 × living donor) and subjected to small RNA sequencing. Due to low RNA yields (which is typical for EV preparations), sequence analysis was successful for 11 of these 19 samples, including 1 living donor sample, 4 DCD samples, and 6 DBD samples. In total, 2111 small RNAs were identified among all samples, including 1526 microRNAs (miRNAs). Identified miRNAs were ranked based on their observed relative abundances across samples, and subsequent unsupervised clustering based on the top 400 most abundant miRNAs, which represent 99.21% of all miRNA reads and 68.61% of the total number of reads across samples, clearly distinguished donor types, with the profile of the living donor clustering with the DCD group (Figure 3).30,31 As the perfusion fluid from which the sEVs were isolated is in direct contact with the renal vascular endothelium, we compared the identified miRNA profile with the miRNA profile of sEVs from cultured endothelial cells.32 Out of the 155 miRNAs detected with an average >10 reads in cultured endothelial cell sEVs, 140 were present among the 500 most abundant miRNAs identified in perfusion fluid. Furthermore, there was a significant association between the rank-order of expression in the 2 data sets (Spearman’s rho = 0.31, P = 0.0002), suggesting that at least part of the isolated sEVs are of endothelial origin (Table S1, SDC, http://links.lww.com/TXD/A219).
FIGURE 3.
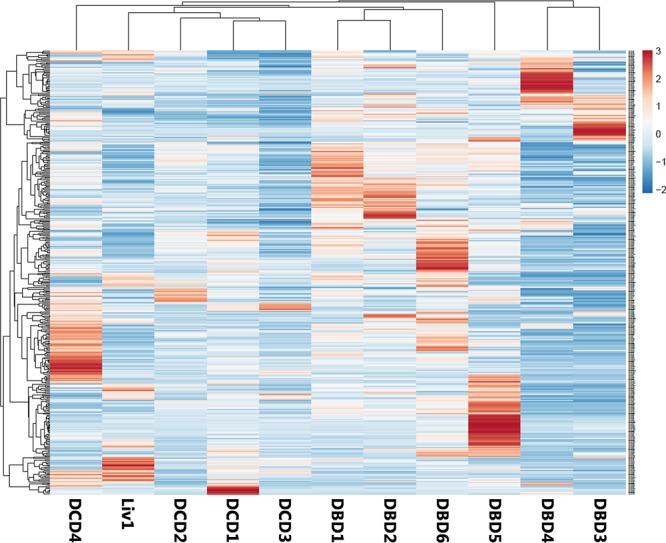
Unsupervised clustering differentiates donor types. Unsupervised hierarchical clustering of samples based on the 400 most abundant miRNA groups all DBD samples together. The second group consists of all DCD-derived samples combined with the sample from the living donors. DBD, donation after brain death; DCD, deceased donation after circulatory death; miRNA, micro RNA.
Exosome RNA Profiles Are Associated With Posttransplantation Kidney Function
We further explored whether sEV-contained RNAs could serve as a source for biomarkers predicting posttransplantation kidney function. As RNA profiles differ between donor type, we collected 16 additional samples, only from DCD donors, with known short-term outcome (8 × DGF, 8 × immediate function [IF]) Donor, recipient, and organ characteristics are listed in Table 1, sample characteristics are listed in Table 2. RNA was isolated and analyzed using the Exiqon miRCURY LNATM Universal RT microRNA PCR Human Panel. In this miRNA PCR array analysis, 223 out of 332 miRNAs were detected in 12 or more samples. Direct comparison of miRNA abundances, normalized for on-chip control small RNA SNORD49A in sEVs from kidneys that showed DGF versus IF revealed 10 miRNAs that significantly differentiate between the 2 groups; however, none of these remained significant after multiple testing correction (Table S2, SDC, http://links.lww.com/TXD/A220). To further investigate whether miRNA profiles are associated any posttransplantation kidney function parameter, a principal component analysis was performed. Clearly, 2 distinct clusters can be observed: the first cluster consisting of 5 samples and a second cluster containing 10 samples (Figure 4A). Each cluster consists of samples from kidneys that showed DGF and IF in recipients, and although no correlation to this outcome parameter could be confirmed, average plasma creatinine levels in cluster 1 remained significantly higher from day 2 until day 7 after transplantation (Figure 4B).
TABLE 1.
Donor, recipient, and organ characteristics discovery panel
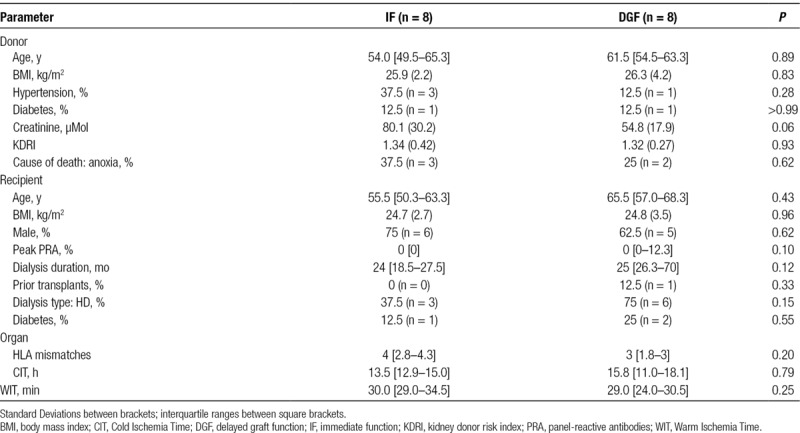
TABLE 2.
Sample characteristics IF and DGF cohort
FIGURE 4.
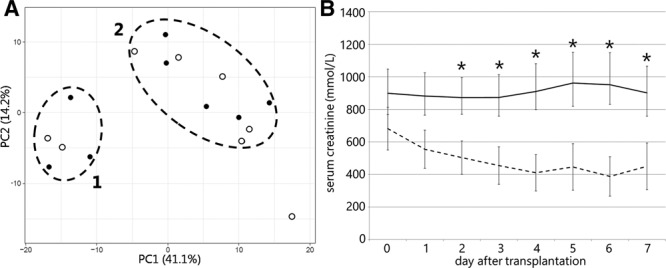
Analysis of sEV-miRNA abundance from preservation fluid of 16 DCD kidneys. A, Principal Component analysis of miRNA PCR array data clearly distinguishes 2 groups, both containing samples from kidney that show IF (open circles) or DGF (closed circles) in recipients. B, Average short-term graft function in the 2 identified clusters, as assessed by serum creatinine levels. DCD, deceased donation after circulatory death; DGF, delayed graft function; IF, immediate function; miRNA, microRNA; PCR, polymerase chain reaction; sEVs, small extracellular vesicles.
DISCUSSION
Here, we demonstrate that donor kidney preservation fluid contains sEVs and these sEVs harbor a small RNA repertoire that is associated with immediate posttransplantation graft function, thereby providing a novel source for potential biomarkers to predict graft function after transplantation. Our analysis of sEV-contained small RNAs provides a strong indication that sEVs in preservation fluid are derived from the endothelium, with which this fluid is in direct contact. Most miRNA identified in sEVs from cultured endothelial cell were represented in the top 500 miRNA identified in our analysis.
Prediction of posttransplantation kidney function is currently based on the assessment of renal (0-h) biopsies and predictive algorithms.5,6,33-35 Putative biomarkers have been identified in blood, urine, tissue biopsies, and preservation fluid,2-4,8,36,37 but clinical application of any biomarker is still hampered. sEVs are vastly explored for diagnostic information in various diseases38-41 and harbor early biomarkers for acute kidney injury.42-46 Lozano-Ramos et al47 examined the protein and RNA content of EVs obtained from urine of (deceased) kidney donors. Their approach allowed for an elegant molecular insight into the donor kidney without the need for a biopsy, although the population of kidneys investigated was insufficient to identify biomarkers associated with either graft function or donor type. Based on our earlier analysis of proteins in donor kidney preservation fluid, we explored whether sEVs from this fluid would be a more potent biomarker source.
Indeed, we could demonstrate the sEVs are present in preservation fluid and they can be isolated using a method compatible with a clinical diagnostics setting. Analysis of small RNAs in these sEVs allowed to discriminate between DBD and DCD donors, clearly underlining the diagnostic potential of sEV-contained RNAs. Interestingly, samples from living donors contained insufficient amounts of RNA for this analysis, which may imply that accumulation of sEV in the preservation fluid is a continuous process, even during cold storage.
We have previously used donor kidney preservation fluid as a source for biomarkers to assess posttransplantation kidney function, identifying Leptin and granulocyte-macrophage colony-stimulating factor as predictive biomarkers for DGF.11 In our analysis of sEV-contained miRNAs, we could observe miRNAs that significantly differed in abundance between DGF and IF; however, unsupervised clustering analysis could not distinguish these groups. Instead, we observed a significant relation of a cluster of 5 samples with kidney function assessed by serum creatinine levels during day 2–7 after transplantation, which limits clinical applicability at this moment. The significance of the 2 identified clusters is at present unclear, although the same clustering was consistently identified using different clustering algorithms and parameter settings. We did not identify a relationship with DGF, which was defined as the start of dialysis within 7 days of transplantation. This definition is increasingly criticized, as the decision to initiate dialysis in the early phase after transplantation is dependent on many factors, many of which are related to the recipient and not necessarily to graft quality.48 We did observe a significant association between posttransplantation creatinine levels and the 2 clusters, suggesting at least some relationship to graft function.
Recently, cell-free circulating miR-505-3p in organ preservation fluid was identified as a predictor of DGF.49 As a lysis step was performed before RNA analysis, it is likely that EV-contained RNAs were among the pool of detected RNAs. In our analysis however, miR-505-3p did not discriminate between DGF and IF (P value 0.45), which suggests that this miRNA mainly resides in a cell- and EV-free fraction of the preservation fluid. Our approach, with the identified optimal procedure for rapid precipitation of sEV and subsequent RNA analysis is likely to render a different, more specific pool of RNAs.
There are several limitations to our study that obscure the full potential of EVs as a source for biomarkers to assess posttransplantation kidney function. Importantly, the limited group size used in this study should be vastly expanded, regarding both number and donor type. Such an extension of this study provides more power to the generated data and will allow further analyses on the relation of EV-harbored RNAs with short- and long-term kidney function in recipients. Additionally, we may have missed other RNA classes that distinguish between DGF and IF which were not included in the PCR array. With current advances in RNA sequencing technology, a broader and unbiased range of RNAs should be interrogated in an extended set of samples.
Overall, we have demonstrated here that RNA-containing sEVs can be isolated from donor kidney preservation fluid. Our RNA sequencing data demonstrate that these RNAs, to a certain extent reflect the donor organ and differentiate between kidneys from DCD and DBD donors. An extended study is required to demonstrate whether specific RNAs could be used as prognostic biomarkers in kidney transplantation.
ACKNOWLEDGMENTS
The authors would like to thank all transplant surgeons at the University Medical Center Utrecht, Utrecht, the Netherlands involved in the collection of preservation fluids.
Supplementary Material
Footnotes
Published online 12 August, 2019.
M.C.V. and B.W.M.v.B. contributed equally.
H.G., M.C.V., and B.W.M.v.B. participated in research design and the writing of the article. H.G., O.G.d.J., R.J.T., L.M., A.D.v.Z., AV.V., and B.W.M.v.B. performed experiments and contributed to samples and data collection and analyses.
The authors declare no conflicts of interest.
This work was supported by an unrestricted research grant from Shire (AR 2014–118).
Supplemental digital content (SDC) is available for this article. Direct URL citations appear in the printed text, and links to the digital files are provided in the HTML text of this article on the journal’s Web site (www.transplantationdirect.com).
REFERENCES
- 1.Saran R, Robinson B, Abbott KC, et al. US Renal Data System 2017 Annual Data Report: epidemiology of kidney disease in the United States Am J Kidney Dis. 2018;713S1A7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anglicheau D, Suthanthiran M. Noninvasive prediction of organ graft rejection and outcome using gene expression patterns. Transplantation. 2008;86:192–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hartono C, Muthukumar T, Suthanthiran M. Noninvasive diagnosis of acute rejection of renal allografts. Curr Opin Organ Transplant. 2010;15:35–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mannon RB. Immune monitoring and biomarkers to predict chronic allograft dysfunction Kidney Int Suppl. 2010119:S59–S65. [DOI] [PubMed] [Google Scholar]
- 5.Kotsch K, Kunert K, Merk V, et al. Novel markers in zero-hour kidney biopsies indicate graft quality and clinical outcome. Transplantation. 2010;90:958–965. [DOI] [PubMed] [Google Scholar]
- 6.Bodonyi-Kovacs G, Putheti P, Marino M, et al. Gene expression profiling of the donor kidney at the time of transplantation predicts clinical outcomes 2 years after transplantation. Hum Immunol. 2010;71:451–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kainz A, Perco P, Mayer B, et al. Gene-expression profiles and age of donor kidney biopsies obtained before transplantation distinguish medium term graft function. Transplantation. 2007;83:1048–1054. [DOI] [PubMed] [Google Scholar]
- 8.Korbély R, Wilflingseder J, Perco P, et al. Molecular biomarker candidates of acute kidney injury in zero-hour renal transplant needle biopsies. Transpl Int. 2011;24:143–149. [DOI] [PubMed] [Google Scholar]
- 9.Anglicheau D, Sharma VK, Ding R, et al. MicroRNA expression profiles predictive of human renal allograft status. Proc Natl Acad Sci U S A. 2009;106:5330–5335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hauser P, Schwarz C, Mitterbauer C, et al. Genome-wide gene-expression patterns of donor kidney biopsies distinguish primary allograft function. Lab Invest. 2004;84:353–361. [DOI] [PubMed] [Google Scholar]
- 11.van Balkom BWM, Gremmels H, Ooms LSS, et al. Proteins in preservation fluid as predictors of delayed graft function in kidneys from donors after circulatory death. Clin J Am Soc Nephrol. 2017;12:817–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Denzer K, Kleijmeer MJ, Heijnen HF, et al. Exosome: from internal vesicle of the multivesicular body to intercellular signaling device. J Cell Sci. 2000;113pt 193365–3374. [DOI] [PubMed] [Google Scholar]
- 13.Smalheiser NR. Exosomal transfer of proteins and RNAs at synapses in the nervous system. Biol Direct. 2007;2:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Février B, Vilette D, Laude H, et al. Exosomes: a bubble ride for prions? Traffic. 2005;6:10–17. [DOI] [PubMed] [Google Scholar]
- 15.Valadi H, Ekström K, Bossios A, et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–659. [DOI] [PubMed] [Google Scholar]
- 16.Yu S, Liu C, Su K, et al. Tumor exosomes inhibit differentiation of bone marrow dendritic cells. J Immunol. 2007;178:6867–6875. [DOI] [PubMed] [Google Scholar]
- 17.Stoorvogel W, Kleijmeer MJ, Geuze HJ, et al. The biogenesis and functions of exosomes. Traffic. 2002;3:321–330. [DOI] [PubMed] [Google Scholar]
- 18.Wubbolts R, Leckie RS, Veenhuizen PT, et al. Proteomic and biochemical analyses of human B cell-derived exosomes. Potential implications for their function and multivesicular body formation. J Biol Chem. 2003;278:10963–10972. [DOI] [PubMed] [Google Scholar]
- 19.Taylor DD, Gercel-Taylor C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol Oncol. 2008;110:13–21. [DOI] [PubMed] [Google Scholar]
- 20.Ades EW, Candal FJ, Swerlick RA, et al. HMEC-1: establishment of an immortalized human microvascular endothelial cell line. J Invest Dermatol. 1992;99:683–690. [DOI] [PubMed] [Google Scholar]
- 21.Théry C, Amigorena S, Raposo G, et al. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr Protoc Cell Biol. 2006;Chapter 3:Unit 3.22. [DOI] [PubMed] [Google Scholar]
- 22.de Jong OG, Verhaar MC, Chen Y, et al. Cellular stress conditions are reflected in the protein and RNA content of endothelial cell-derived exosomes. J Extracell Vesicles. 2012;1:18396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schageman J, Zeringer E, Li M, et al. The complete exosome workflow solution: from isolation to characterization of RNA cargo. Biomed Res Int. 2013;2013:253957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alvarez ML, Khosroheidari M, Kanchi Ravi R, et al. Comparison of protein, microRNA, and mRNA yields using different methods of urinary exosome isolation for the discovery of kidney disease biomarkers. Kidney Int. 2012;82:1024–1032. [DOI] [PubMed] [Google Scholar]
- 25.Momen-Heravi F, Balaj L, Alian S, et al. Current methods for the isolation of extracellular vesicles. Biol Chem. 2013;394:1253–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haqqani AS, Delaney CE, Tremblay TL, et al. Method for isolation and molecular characterization of extracellular microvesicles released from brain endothelial cells. Fluids Barriers CNS. 2013;10:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Musante L, Saraswat M, Ravidà A, et al. Recovery of urinary nanovesicles from ultracentrifugation supernatants. Nephrol Dial Transplant. 2013;28:1425–1433. [DOI] [PubMed] [Google Scholar]
- 28.Zeringer E, Li M, Barta T, et al. Methods for the extraction and RNA profiling of exosomes. World J Methodol. 2013;3:11–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van Balkom BW, de Jong OG, Smits M, et al. Endothelial cells require miR-214 to secrete exosomes that suppress senescence and induce angiogenesis in human and mouse endothelial cells. Blood. 2013;121:3997S1. [DOI] [PubMed] [Google Scholar]
- 30.Babicki S, Arndt D, Marcu A, et al. Heatmapper: web-enabled heat mapping for all. Nucleic Acids Res. 2016;44W1W147–W153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Metsalu T, Vilo J. Clustvis: a web tool for visualizing clustering of multivariate data using principal component analysis and heatmap. Nucleic Acids Res. 2015;43W1W566–W570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van Balkom BW, Eisele AS, Pegtel DM, et al. Quantitative and qualitative analysis of small RNAs in human endothelial cells and exosomes provides insights into localized RNA processing, degradation and sorting. J Extracell Vesicles. 2015;4:26760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Irish WD, Ilsley JN, Schnitzler MA, et al. A risk prediction model for delayed graft function in the current era of deceased donor renal transplantation. Am J Transplant. 2010;10:2279–2286. [DOI] [PubMed] [Google Scholar]
- 34.Rao PS, Schaubel DE, Guidinger MK, et al. A comprehensive risk quantification score for deceased donor kidneys: the kidney donor risk index. Transplantation. 2009;88:231–236. [DOI] [PubMed] [Google Scholar]
- 35.Balaz P, Rokosny S, Wohlfahrtova M, et al. Identification of expanded-criteria donor kidney grafts at lower risk of delayed graft function. Transplantation. 2013;96:633–638. [DOI] [PubMed] [Google Scholar]
- 36.Field M, Dronavalli V, Mistry P, et al. Urinary biomarkers of acute kidney injury in deceased organ donors–kidney injury molecule-1 as an adjunct to predicting outcome. Clin Transplant. 2014;28:808–815. [DOI] [PubMed] [Google Scholar]
- 37.Cantaluppi V, Dellepiane S, Tamagnone M, et al. Neutrophil gelatinase associated lipocalin is an early and accurate biomarker of graft function and tissue regeneration in kidney transplantation from extended criteria donors. PLOS One. 2015;10:e0129279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wermuth PJ, Piera-Velazquez S, Rosenbloom J, et al. Existing and novel biomarkers for precision medicine in systemic sclerosis. Nat Rev Rheumatol. 2018;14:421–432. [DOI] [PubMed] [Google Scholar]
- 39.Xu R, Rai A, Chen M, et al. Extracellular vesicles in cancer - implications for future improvements in cancer care. Nat Rev Clin Oncol. 2018;15:617–638. [DOI] [PubMed] [Google Scholar]
- 40.Mann J, Reeves HL, Feldstein AE. Liquid biopsy for liver diseases. Gut. 2018;67:2204–2212. [DOI] [PubMed] [Google Scholar]
- 41.Gámez-Valero A, Beyer K, Borràs FE. Extracellular vesicles, new actors in the search for biomarkers of dementias. Neurobiol Aging. 2019;74:15–20. [DOI] [PubMed] [Google Scholar]
- 42.du Cheyron D, Daubin C, Poggioli J, et al. Urinary measurement of Na+/H+ exchanger isoform 3 (NHE3) protein as new marker of tubule injury in critically ill patients with ARF. Am J Kidney Dis. 2003;42:497–506. [DOI] [PubMed] [Google Scholar]
- 43.Zhou H, Pisitkun T, Aponte A, et al. Exosomal Fetuin-A identified by proteomics: a novel urinary biomarker for detecting acute kidney injury. Kidney Int. 2006;70:1847–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sonoda H, Yokota-Ikeda N, Oshikawa S, et al. Decreased abundance of urinary exosomal aquaporin-1 in renal ischemia-reperfusion injury. Am J Physiol Renal Physiol. 2009;297:F1006–F1016. [DOI] [PubMed] [Google Scholar]
- 45.Zhou H, Cheruvanky A, Hu X, et al. Urinary exosomal transcription factors, a new class of biomarkers for renal disease. Kidney Int. 2008;74:613–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen HH, Lai PF, Lan YF, et al. Exosomal ATF3 RNA attenuates pro-inflammatory gene MCP-1 transcription in renal ischemia-reperfusion. J Cell Physiol. 2014;229:1202–1211. [DOI] [PubMed] [Google Scholar]
- 47.Lozano-Ramos SI, Bancu I, Carreras-Planella L, et al. Molecular profile of urine extracellular vesicles from normo-functional kidneys reveal minimal differences between living and deceased donors. BMC Nephrol. 2018;19:189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Siedlecki A, Irish W, Brennan DC. Delayed graft function in the kidney transplant. Am J Transplant. 2011;11:2279–2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Roest HP, Ooms LSS, Gillis AJM, et al. Cell-free microRNA miR-505-3p in graft preservation fluid is an independent predictor of delayed graft function after kidney transplantation. Transplantation. 2019;103:329–335. [DOI] [PubMed] [Google Scholar]


