ABSTRACT
Mitochondria contain their own genome (mtDNA), encoding 13 proteins of the enzyme complexes of the oxidative phosphorylation. Synthesis of these 13 mitochondrial proteins requires a specific translation machinery, the mitoribosomes whose RNA components are encoded by the mtDNA, whereas more than 80 proteins are encoded by nuclear genes. It has been well established that mitochondrial topoisomerase I (TOP1MT) is important for mtDNA integrity and mitochondrial transcription as it prevents excessive mtDNA negative supercoiling and releases topological stress during mtDNA replication and transcription. We recently showed that TOP1MT also supports mitochondrial protein synthesis, and thus is critical for promoting tumor growth. Impaired mitochondrial protein synthesis leads to activation of the mitonuclear stress response through the transcription factor ATF4, and induces cytoprotective genes in order to prevent mitochondrial and cellular dysfunction. In this perspective, we highlight the novel role of TOP1MT in mitochondrial protein synthesis and as potential target for chemotherapy.
KEYWORDS: Mitochondrial topoisomerase I, mitochondria, mitochondrial translation, mitoribosome, mitochondrial targeted therapy, cancer
Graphical Abstract
Mitochondrial topoisomerases
Mitochondria are central hubs for numerous metabolic pathways, providing cellular energy, supplying macromolecules for cellular proliferation, and controlling redox homeostasis [1]. Likely derived from a bacterial ancestor, mitochondria possess their own circular double-stranded genome of about 16.6 kb. Human mitochondrial DNA (mtDNA) encodes 2 mitochondrial ribosomal RNAs, 22 mitochondrial transfer RNAs and 13 mitochondrial messenger RNAs encoding a small but essential fraction of the subunits of the electron transport chain [2]. MtDNA lacks introns, and contains only one noncoding region (NCR), which harbors the promoters for transcription of both strands and the replication origin for the heavy (H) strand (OH) [3,4]. Replication of mtDNA from this origin is usually strand-synchronous, with the displaced lagging strand template being covered with RNA, protein or a combination of the two [5] until second-strand DNA synthesis begins, typically from a site located 11 kbp downstream of OH, known as OL [6].
Mitochondrial transcription proceeds from the H-strand promoters (HSP1 and HSP2) and L-strand promoter (LSP) for the heavy and light strand, respectively. The resulting polycistronic transcripts are processed according to the “tRNA punctuation model” whereby 22 interspersed tRNAs are excised to concomitantly release rRNAs and mRNAs [7,8]. Translation of the mitochondrial encoded genes requires further RNA processing by polyadenylation, methylation, or pseudouridinylation, and in case of tRNAs, CCA addition and aminoacylation [9].
Due to its circular structure and attachment to the mitochondrial inner membrane [10,11], mtDNA is readily supercoiled as transcription and replication generate torsional stress by unwinding the DNA. Critical factors in releasing topological stress are topoisomerases, which transiently cleave the DNA-backbone, allowing DNA untwisting and passage of a DNA segment through another [12]. Among the six human topoisomerases, three possess dual localizations in the nucleus and the mitochondria: topoisomerase IIα (TOP2A) [13], topoisomerase IIβ (TOP2B) [14] and topoisomerase IIIα (TOP3A) [15]. Mitochondrial topoisomerase I (TOP1MT) is the only topoisomerase exclusively devoted to mitochondria [16]. TOP1MT is critical for limiting mtDNA negative supercoiling and for the maintenance of mtDNA integrity after doxorubicin treatment [13,17]. In Top1mt knockout cells mtDNA is in an underwound state, which might cause the observed increase in steady state levels of mitochondrial RNA [18,19].
In contrast to TOP2A, TOP2B and TOP3A, TOP1MT is dispensable in mice with no obvious phenotype of Top1mt knockout mice under basal conditions, suggesting that its function can be compensated by other topoisomerases [13,17,20]. Yet, loss of TOP1MT attenuates cell proliferation by limiting mtDNA expansion in cardiomyocytes after doxorubicin treatment and under high-energy demand during liver regeneration [17,21].
Role of TOP1MT in mitochondrial protein synthesis
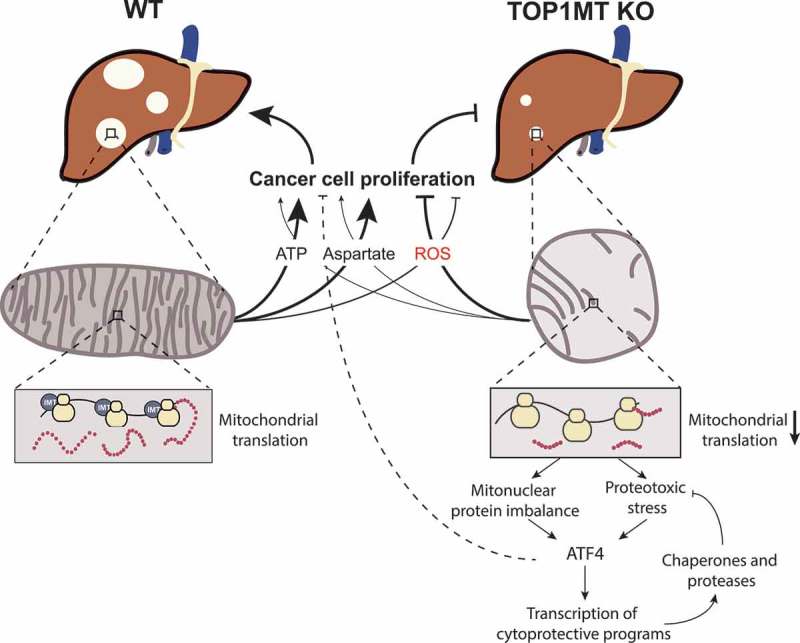
We recently reported that, in mouse models of colon and liver cancer, TOP1MT promotes tumor growth and frequency by sustaining cancer cell proliferation in a metabolically compromised environment [22]. This conclusion tempers the Warburg effect hypothesis by demonstrating the importance of mitochondria for cancer cell proliferation. Consistent with this possibility, analyses of The Cancer Genome Atlas (TCGA) databases show elevated expression of TOP1MT in hepatocellular carcinomas [22]. Besides its role in enabling mtDNA replication and expansion, we demonstrated that TOP1MT possesses a non-canonical function in facilitating mitochondrial translation [22].
Mammalian mitoribosomes consist of a large 39S subunit (mtLSU) and a small 28S (mtSSU) subunit, containing the 16S and 12S mitochondrial rRNA, and mt-tRNA (Val), respectively [23,24]. Approximately 80 nuclear-encoded proteins are imported to mitochondria and are assembled in the mitoribosome with the RNA components as scaffolds [25]. Based on steady-state rRNA levels, it has been estimated that there are 100 mitoribosomes per mitochondrion in rat hepatocytes [3]. Similar to cytosolic ribosomes, mitochondrial ribosomes carry out translation, which encompasses initiation, elongation, termination and recycling. These processes depend on the expression, synthesis and import of nuclear-encoded proteins [26,27]. Thus, protein synthesis in the cytosol and mitochondria warrants temporal and spatial coordination. While the molecular details of mitoribosome biogenesis are currently incomplete [28], the initial steps of mitochondrial assembly have been suggested to occur at mitochondrial nucleoids [25,29,30]. Recent advances indicate that initial and partly co-transcriptional RNA processing is organized in distinct foci, called mitochondrial RNA granules (MRGs) that form in close proximity to mitochondrial nucleoids [31,32]. MRGs contain proteins involved in RNA processing, mitochondrial ribosomal proteins and translation-associated factors, indicating that the early steps of the mitoribosome subunit assembly might take place in these foci. It has been proposed that newly transcribed rRNAs and/or early mitoribosome assembly intermediates are transferred from nucleoids to the MRGs where mitoribosome assembly is completed [28]. However, the proteome of MRGs varies within different subpopulations suggesting that these are dynamic structures [28,33]. TOP1MT has been found to localize to MRGs [34,35] and pulldown experiments have shown that TOP1MT associates with mitochondrial ribosomal proteins [22]. Consistent with these results, we observed that mitochondrial protein synthesis is significantly decreased in Top1mt KO murine embryonic fibroblasts (MEF) and in tumor models lacking TOP1MT [22].
Recent experiments further elaborate on the role of TOP1MT in mitochondrial translation.
Figure 1 shows that transient depletion of mtDNA with ethidium bromide significantly diminishes the steady state protein levels of the electron transport chain and that the rate of mtDNA copy number expansion in the recovery phase is decreased in cells lacking TOP1MT (Figure 1a), which is in line with our previous studies [36]. Despite elevated mitochondrial transcript levels (Figure 1b), the recovery rate of the electron transport chain proteins was markedly delayed in cells lacking TOP1MT as compared to WT controls. While the electron transport chain subunits fully recovered in the WT cells 6 days after ethidium bromide release, protein levels only started to reappear in the TOP1MT KO cells after 3 additional days (Figure 1c). Several possibilities may explain how TOP1MT impacts mitochondrial translation. First, defects in mitochondrial RNA processing could account for the downstream defects in mitochondrial protein synthesis. However, maturation and polyadenylation of the mitochondrial transcriptome occurred normal in TOP1MT KO MEFs [19]. Second, TOP1MT might play a role in the temporal and spatial coordination of the mitoribosomal assembly through its interaction with both the transcription machinery and mitoribosomal proteins. Another possibility is that mitochondrial translation is regulated by a long noncoding RNA spanning the D-loop, which was strongly upregulated in the absence of TOP1MT [19]. Notably, the steady state levels of the same RNA were found strongly increased in cells where mitochondrial protein synthesis was inhibited [37]. Further investigations are warranted to elucidate these possibilities.
Figure 1.
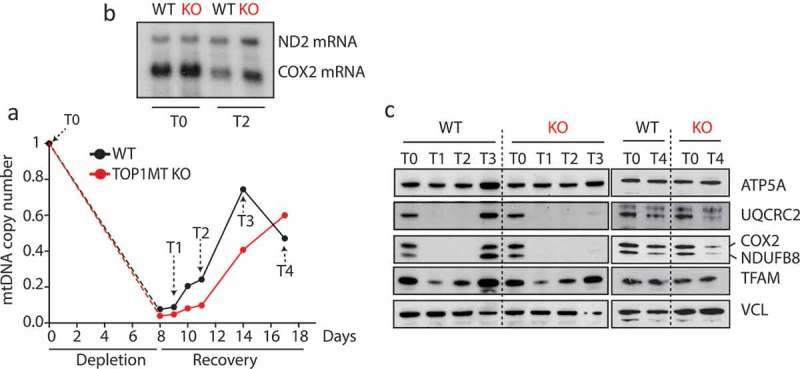
Role of TOP1MT in mitochondrial translation. (a) Transient mtDNA depletion followed by recovery in HCT-116 WT and TOP1MT KO cells: mtDNA was depleted by treatment with 50 ng/mL ethidium bromide (EB) for 8 days and its recovery was followed for 10 days after EB wash-out. Mitochondrial copy number was quantified at each timepoint as described in [69]. (b) Steady state levels of mtRNAs at the indicated timepoints: total RNA was isolated by standard procedures and levels of ND2 and COX2 mRNAs was assessed by Northern Blotting using P32 DNA labeled probes spanning regions of about 500 bp of the respective transcripts. (c) Delayed recovery of electron transport chain subunits in TOP1MT KO cells: Whole cell lysates from different timepoints were subject to SDS-PAGE and Western blotting as described in [69]. Antibodies were purchased from abcam (anti-NDUFB8, Ab110242; anti-MTCO2, Ab110258; anti-UQCRC2, Ab14745; anti-ATP5A, Ab14748; anti-TFAM, ab131607; anti-VCL, Ab238075).
Mitochondrial translation and coupling with cellular proliferation
Mitochondria fulfill essential cellular functions ensuring bioenergetic supply and metabolic processes that are essential for cell growth and survival as well as cell death (cytochrome C release for apoptosis). Consequently, mitochondria are in constant crosstalk with the nucleus and the cytosol to regulate cellular homeostasis and adaption to mitochondrial stress [38]. Proteostatic stress in the mitochondria can initiate feedback responses to block cell proliferation, such as the mitochondrial unfolded protein response, the proteolytic stress response and the heat shock response [39]. Stalling of mitochondrial translation has been demonstrated to trigger antiproliferative signals by activating mitochondrial release factors with concomitant signaling to halt cell proliferation, rather than loss of respiratory function, which has been suggested to be a downstream effect [40,41]. Failure of this initial rescue attempt triggers the mitochondrial ribosomal and RNA decay pathway, ensuring degradation of the majority of mitoribosomes and mitochondrial rRNA and mRNA [40].
A multiomics approach identified activating transcription factor 4 (ATF4) as the main regulator of the mitonuclear stress response [42]. ATF4 activatio reduces cytosolic translation and induces cytoprotective genes, with a concomitant decrease of mitochondrial ribosomal protein levels induced by mitochondrial stress. Thus, ATF4 and mitochondrial ribosomal proteins are the main effectors in mitonuclear stress pathways [42]. Notably, we identified ATF4 as upstream regulator in the transcriptome analysis of chemically induced WT and Top1mt KO liver tumors [22] suggesting that impaired mitochondrial translation activates ATF4, which in turn diminishes total protein synthesis and therefore halts cell proliferation in Top1mt deficient tumors.
Figure 2 summarizes our model linking TOP1MT with cellular proliferation for tissue regeneration and tumor growth [21,22]. Coupling of TOP1MT with cellular proliferation is exemplified by our prior finding that MYC upregulates the transcription of TOP1MT [43].
Figure 2.
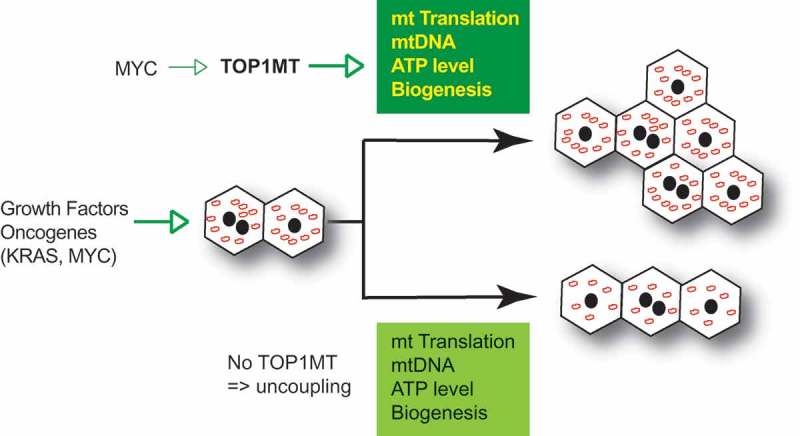
Role of TOP1MT in tissue regeneration and tumorigenesis. TOP1MT couples mitochondrial functions (translation, mtDNA expansion, ATP generation and biogenesis) with cellular proliferation in response to growth factors and oncogenetic stimuli.
Mitochondrial translation as therapeutic target
Mitochondria are essential for cancer cell growth, to meet their anabolic and bioenergetic needs in challenging tumor microenvironments (Figure 2) [44–47]. Hence, mitochondria represent potential targets for the development of novel anticancer agents [48,49]. Recent studies indicate that inhibition of mitochondrial protein synthesis is a potential therapeutic strategy [50], particularly against quiescent cancer stem cells that strictly depend on mitochondrial biogenesis [51]. The FDA-approved antibiotic tigecycline has been demonstrated to sensitize non-small cell lung cancer [52], hepatocellular carcinoma [53], high-grade B-cell lymphoma [54] and renal cell carcinoma to chemotherapy [55]. In line with these findings, tigecycline treatment in combination with daunorubicin and cytarabine displayed additive or syntergistic effects in primary human acute myeloid leukemia (AML) cells [56]. Similarly, the FDA-approved therapeutic doxycycline has been shown to inhibit mitochondrial protein translation and biogenesis in cancer stem cells [51,57]. Oral doxycycline treatment decreased the expression of cancer stem cell markers in a clinical pilot study in early breast cancer patients [58]. In line with these findings, a mitochondrial gene signature has been proposed to stratify patients with high mitochondrial markers to receive mitochondrial-targeted therapies in addition to conventional therapy [57].
A genome-wide CRISPR and shRNA screen revealed that K-Ras mutant tumors are sensitive to mitochondrial translation inhibitors, and combinational therapy with BRAF and MEK inhibitors together with tigecycline might be a sustainable strategy against K-Ras tumors [59]. Thus, the survival of a subpopulation of dormant cells, responsible for relapse of K-Ras mutant pancreatic ductal adenocarcinoma, depends on mitochondrial activity [60] (Figure 2). MYC has also been shown to directly activate TOP1MT [43]. Mouse models of MYC-driven lymphoma showed that MYC activation sensitizes cancer cells to inhibition of mitochondrial translation with tigecycline without affecting normal tissue [61]. Notably, treatment with mitochondria-targeted drugs might enhance mitophagy and TOP1MT-deficient MEFs have been found to activate autophagy [20]. Similar to authophagy, the role of mitophagy in carcinogenesis depends on the cellular context and stage of tumorigenesis [62,63]. Thus, autophagy has been shown to possess both pro- and anti-tumorigenic effects [62]. In a KRAS mutant colorectal cancer cell model, mitochondrial targeted treatment induces autophagy, thereby suppressing cancer cell proliferation [64]. However, autophagy facilitates survival of Ras-driven lung tumor cells by preventing nucleotide depletion and energy crisis in a nutrient deprived environment [65]. Thus, the contextual role of mitophagy might be an important determinant for the success of mitochondrial-targeted therapies.
Based on its role in mitochondrial protein synthesis and due to its upregulation in a wide range of tumors, TOP1MT might represent a druggable target for cancer therapy [66]. TOP1MT inhibition might present an effective strategy in the eradication of cancer stem cells, as these cells are highly dependent on mitochondrial biogenesis [57]. In particular, tumor cells in a hypoglycemic or hypoxic microenvironment might be susceptible to TOP1MT inhibition, since cancer cell growth of TOP1MT deficient cells was significantly reduced in nutrient deprived environment compared to WT cells [22]. However, TOP1MT deficiency did not alter cell growth under standard cell culture conditions, suggesting that it becomes a limiting factor for cell proliferation under nutrient starvation in vivo [22]. Due to mitochondrial impairment, tumor cells might enhance glycolysis to sustain their viability. Notably, the glycolytic rate was unaltered in TOP1MT-deficient cancer cells despite the upregulation of key enzymes involved in the glycolytic pathway, suggesting that glycolysis operated at its maximum [22]. Alternatively, clearance of defective mitochondria by increased autophagy might allow for cancer cell survival [62]. The mediating role of autophagy in the observed antitumorigenic effects in TOP1MT-deficient tumor cells needs to be determined in future studies.
Given the current lack of a selective TOP1MT inhibitor [66], we propose that engineering clinically used topoisomerase I inhibitors with a mitochondrial targeting peptide [67] might be a promising approach for the development of TOP1MT targeting drugs. This strategy has been successfully employed for the targeted delivery of doxorubicin to the mitochondria [68]. Whether TOP1MT inhibition can augment additional targeted treatments to achieve a sustainable response and prevent tumor relapse needs to be explored in the future [22].
Conclusion
Similar to other topoisomerases, TOP1MT releases mtDNA topological stress generated during mitochondrial replication and transcription. Although TOP1MT is a non-essential gene and TOP1MT-deficient mice are viable [22], it becomes important under stress conditions. TOP1MT protects doxorubicin-induced cardiotoxicity [17], enables liver regeneration [36] and promotes cancer cell proliferation in metabolically challenging microenvironments [22]. We propose that TOP1MT exerts a pleiotropic function, enabling mtDNA replication and expansion, but also possesses a noncanonical role in facilitating mitochondrial translation [22]. The decrease in steady-state levels of the oxidative phosphorylation proteins in turn leads to a drop in cellular energy, metabolic intermediates, enhanced oxidative stress and the activation of ATF4 to induce the mitonuclear stress response, ultimately delaying cellular proliferation. Inhibition of mitochondrial protein synthesis has been found to be a promising treatment strategy for a variety of cancers [51–56]. Based on its role in mitochondrial protein synthesis and given its upregulation in cancer, we propose that TOP1MT could be a potential target for intervention in combinational therapies.
Funding Statement
Our studies are supported by the Center for Cancer Research, the Intramural Program of the National Cancer Institute, NIH [BC Z01 006161]. AS is supported by the UK Medical Research Council with a Senior Non-Clinical Fellowship [MC_PC_13029].
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- [1].Chandel Navdeep S. Evolution of mitochondria as signaling organelles. Cell Metab. 2015;22(2):204–206. [DOI] [PubMed] [Google Scholar]
- [2].Anderson S, Bankier AT, Barrell BG, et al. Sequence and organization of the human mitochondrial genome. Nature. 1981;290(5806):457–465. [DOI] [PubMed] [Google Scholar]
- [3].Taanman J-W. The mitochondrial genome: structure, transcription, translation and replication. Biochim Biophys Acta Bioenerg. 1999;1410(2):103–123. [DOI] [PubMed] [Google Scholar]
- [4].Hallberg BM, Larsson NG. Making proteins in the powerhouse. Cell Metab. 2014;20(2):226–240. [DOI] [PubMed] [Google Scholar]
- [5].Holt IJ. The mitochondrial R-loop. Nucleic Acids Res. 2019;47(11):5480–5489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Falkenberg M. Mitochondrial DNA replication in mammalian cells: overview of the pathway. Essays Biochem. 2018;62(3):287–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Mercer Tim R, Neph S, Dinger Marcel E, et al. The human mitochondrial transcriptome. Cell. 2011;146(4):645–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Ojala D, Montoya J, Attardi G. tRNA punctuation model of RNA processing in human mitochondria. Nature. 1981;290(5806):470–474. [DOI] [PubMed] [Google Scholar]
- [9].Pearce SF, Rebelo-Guiomar P, D’Souza AR, et al. Regulation of mammalian mitochondrial gene expression: recent advances. Trends Biochem Sci. 2017;42(8):625–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Bogenhagen DF, Rousseau D, Burke S. The layered structure of human mitochondrial DNA nucleoids. J Biol Chem. 2008;283(6):3665–3675. [DOI] [PubMed] [Google Scholar]
- [11].He J, Mao CC, Reyes A, et al. The AAA+ protein ATAD3 has displacement loop binding properties and is involved in mitochondrial nucleoid organization. J Cell Biol. 2007;176(2):141–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Pommier Y, Sun Y, Huang SN, et al. Roles of eukaryotic topoisomerases in transcription, replication and genomic stability. Nat Rev Mol Cell Biol. 2016;17(11):703–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Zhang H, Zhang Y-W, Yasukawa T, et al. Increased negative supercoiling of mtDNA in TOP1mt knockout mice and presence of topoisomerases IIα and IIβ in vertebrate mitochondria. Nucleic Acids Res. 2014;42(11):7259–7267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Low RL, Orton S, Friedman DB. A truncated form of DNA topoisomerase IIβ associates with the mtDNA genome in mammalian mitochondria. Eur J Biochem. 2003;270(20):4173–4186. [DOI] [PubMed] [Google Scholar]
- [15].Nicholls TJ, Nadalutti CA, Motori E, et al. Topoisomerase 3alpha Is required for decatenation and segregation of human mtDNA. Mol Cell. 2018;69(1):9–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Zhang H, Barceló JM, Lee B, et al. Human mitochondrial topoisomerase I. Proc Nat Acad Sci. 2001;98(19):10608–10613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Khiati S, Dalla Rosa I, Sourbier C, et al. Mitochondrial topoisomerase I (top1mt) is a novel limiting factor of Doxorubicin cardiotoxicity. Clin Cancer Res off J Am Assoc Cancer Res. 2014;20(18):4873–4881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Dalla Rosa I, Goffart S, Wurm M, et al. Adaptation of topoisomerase I paralogs to nuclear and mitochondrial DNA. Nucleic Acids Res. 2009;37(19):6414–6428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Dalla Rosa I, Zhang H, Khiati S, et al. Transcription profiling suggests that mitochondrial topoisomerase IB acts as a topological barrier and regulator of mitochondrial DNA transcription. J Biol Chem. 2017;292(49):20162–20172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Douarre C, Sourbier C, Dalla Rosa I, et al. Mitochondrial topoisomerase I is critical for mitochondrial integrity and cellular energy metabolism. PloS One. 2012;7(7):e41094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Khiati S, Baechler SA, Factor VM, et al. Lack of mitochondrial topoisomerase I (TOP1mt) impairs liver regeneration. Proc Nat Acad Sci. 2015;112(36):11282–11287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Baechler SA, Factor VM, Dalla Rosa I, et al. The mitochondrial type IB topoisomerase drives mitochondrial translation and carcinogenesis. Nat Commun. 2019;10(1):83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].D’Souza AR, Minczuk M. Mitochondrial transcription and translation: overview. Essays Biochem. 2018;62(3):309–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Amunts A, Brown A, Toots J, et al. Ribosome. The structure of the human mitochondrial ribosome. Science. 2015;348(6230):95–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Bogenhagen DF, Martin DW, Koller A. Initial steps in RNA processing and ribosome assembly occur at mitochondrial DNA nucleoids. Cell Metab. 2014;19(4):618–629. [DOI] [PubMed] [Google Scholar]
- [26].Mai N, Chrzanowska-Lightowlers ZM, Lightowlers RN. The process of mammalian mitochondrial protein synthesis. Cell Tissue Res. 2017;367(1):5–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Christian BE, Spremulli LL. Mechanism of protein biosynthesis in mammalian mitochondria. Biochim Biophys Acta. 2012;1819(9–10):1035–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].De Silva D, Tu YT, Amunts A, et al. Mitochondrial ribosome assembly in health and disease. Cell Cycle (georgetown, Tex). 2015;14(14):2226–2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Dalla Rosa I, Durigon R, Pearce SF, et al. MPV17L2 is required for ribosome assembly in mitochondria. Nucleic Acids Res. 2014;42(13):8500–8515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].He J, Cooper HM, Reyes A, et al. Human C4orf14 interacts with the mitochondrial nucleoid and is involved in the biogenesis of the small mitochondrial ribosomal subunit. Nucleic Acids Res. 2012;40(13):6097–6108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Jourdain AA, Koppen M, Wydro M, et al. GRSF1 regulates RNA processing in mitochondrial RNA granules. Cell Metab. 2013;17(3):399–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Iborra FJ, Kimura H, Cook PR. The functional organization of mitochondrial genomes in human cells. BMC Biol. 2004;2(1):9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Jourdain AA, Boehm E, Maundrell K, et al. Mitochondrial RNA granules: compartmentalizing mitochondrial gene expression. J Cell Biol. 2016;212(6):611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Sobek S, Dalla Rosa I, Pommier Y, et al. Negative regulation of mitochondrial transcription by mitochondrial topoisomerase I. Nucleic Acids Res. 2013;41(21):9848–9857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Antonicka H, Shoubridge EA. Mitochondrial RNA granules are centers for posttranscriptional RNA processing and ribosome biogenesis. Cell Rep. 2015;10(6):920–932. [DOI] [PubMed] [Google Scholar]
- [36].Khiati S, Baechler SA, Factor VM, et al. Lack of mitochondrial topoisomerase I (TOP1mt) impairs liver regeneration. Proc Natl Acad Sci U S A. 2015;112(36):11282–11287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Selwood SP, McGregor A, Lightowlers RN, et al. Inhibition of mitochondrial protein synthesis promotes autonomous regulation of mtDNA expression and generation of a new mitochondrial RNA species. FEBS Lett. 2001;494(3):186–191. [DOI] [PubMed] [Google Scholar]
- [38].D’Amico D, Sorrentino V, Auwerx J. Cytosolic proteostasis networks of the mitochondrial stress response. Trends Biochem Sci. 2017;42(9):712–725. [DOI] [PubMed] [Google Scholar]
- [39].Quirós PM, Mottis A, Auwerx J. Mitonuclear communication in homeostasis and stress [Review Article]. Nat Rev Mol Cell Biol. 2016;17:213. [DOI] [PubMed] [Google Scholar]
- [40].Richter U, Lahtinen T, Marttinen P, et al. A mitochondrial ribosomal and RNA decay pathway blocks cell proliferation. Curr Biol. 2013;23(6):535–541. [DOI] [PubMed] [Google Scholar]
- [41].Richter U, Lahtinen T, Marttinen P, et al. Quality control of mitochondrial protein synthesis is required for membrane integrity and cell fitness. J Cell Biol. 2015;211(2):373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Quirós PM, Prado MA, Zamboni N, et al. Multi-omics analysis identifies ATF4 as a key regulator of the mitochondrial stress response in mammals. J Cell Biol. 2017;216(7):2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Zoppoli G, Douarre C, Dalla Rosa I, et al. Coordinated regulation of mitochondrial topoisomerase IB with mitochondrial nuclear encoded genes and MYC. Nucleic Acids Res. 2011;39(15):6620–6632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Porporato PE, Filigheddu N, Pedro JMB-S, et al. Mitochondrial metabolism and cancer [Review]. Cell Res. 2017;28:265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Moreno-Sanchez R, Rodriguez-Enriquez S, Marin-Hernandez A, et al. Energy metabolism in tumor cells. Febs J. 2007;274(6):1393–1418. [DOI] [PubMed] [Google Scholar]
- [46].Wallace DC. Mitochondria and cancer. Nat Rev Cancer. 2012;12(10):685–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Vyas S, Zaganjor E, Haigis MC. Mitochondria and Cancer. Cell. 2016;166(3):555–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Zong WX, Rabinowitz JD, White E. Mitochondria and Cancer. Mol Cell. 2016;61(5):667–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Greer YE, Porat-Shliom N, Nagashima K, et al. ONC201 kills breast cancer cells in vitro by targeting mitochondria. Oncotarget. 2018;9(26):18454–18479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Heerma van Voss MR, Vesuna F, Bol GM, et al. Targeting mitochondrial translation by inhibiting DDX3: a novel radiosensitization strategy for cancer treatment. Oncogene. 2018;37(1):63–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Lamb R, Ozsvari B, Lisanti CL, et al. Antibiotics that target mitochondria effectively eradicate cancer stem cells, across multiple tumor types: treating cancer like an infectious disease. Oncotarget. 2015;6(7):4569–4584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Jia X, Gu Z, Chen W, et al. Tigecycline targets nonsmall cell lung cancer through inhibition of mitochondrial function. Fundam Clin Pharmacol. 2016;30(4):297–306. [DOI] [PubMed] [Google Scholar]
- [53].Tan J, Song M, Zhou M, et al. Antibiotic tigecycline enhances cisplatin activity against human hepatocellular carcinoma through inducing mitochondrial dysfunction and oxidative damage. Biochem Biophys Res Commun. 2017;483(1):17–23. [DOI] [PubMed] [Google Scholar]
- [54].Rava M, D’Andrea A, Nicoli P, et al. Therapeutic synergy between tigecycline and venetoclax in a preclinical model of MYC/BCL2 double-hit B cell lymphoma. Sci Transl Med. 2018;10:426. [DOI] [PubMed] [Google Scholar]
- [55].Wang B, Ao J, Yu D, et al. Inhibition of mitochondrial translation effectively sensitizes renal cell carcinoma to chemotherapy. Biochem Biophys Res Commun. 2017;490(3):767–773. [DOI] [PubMed] [Google Scholar]
- [56].Skrtic M, Sriskanthadevan S, Jhas B, et al. Inhibition of mitochondrial translation as a therapeutic strategy for human acute myeloid leukemia. Cancer Cell. 2011;20(5):674–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Sotgia F, Ozsvari B, Fiorillo M, et al. A mitochondrial based oncology platform for targeting cancer stem cells (CSCs): MITO-ONC-RX. Cell Cycle (georgetown, Tex). 2018;17(17):2091–2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Scatena C, Roncella M, Di Paolo A, et al. Doxycycline, an inhibitor of mitochondrial biogenesis, effectively reduces cancer stem cells (CSCs) in early breast cancer patients: a clinical pilot study. Front Oncol. 2018;8:452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Martin TD, Cook DR, Choi MY, et al. A role for mitochondrial translation in promotion of viability in K-ras mutant cells. Cell Rep. 2017;20(2):427–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Viale A, Pettazzoni P, Lyssiotis CA, et al. Oncogene ablation-resistant pancreatic cancer cells depend on mitochondrial function. Nature. 2014;514(7524):628–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].D’Andrea A, Gritti I, Nicoli P, et al. The mitochondrial translation machinery as a therapeutic target in Myc-driven lymphomas. Oncotarget. 2016;7(45):72415–72430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].White E. Deconvoluting the context-dependent role for autophagy in cancer. Nat Rev Cancer. 2012;12(6):401–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Mancias JD, Kimmelman AC. Mechanisms of selective autophagy in normal physiology and cancer. J Mol Biol. 2016;428(9, Part A):1659–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Boyle KA, Van Wickle J, Hill RB, et al. Mitochondria-targeted drugs stimulate mitophagy and abrogate colon cancer cell proliferation. J Biol Chem. 2018;293(38):14891–14904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Guo JY, Teng X, Laddha SV, et al. Autophagy provides metabolic substrates to maintain energy charge and nucleotide pools in Ras-driven lung cancer cells. Genes Dev. 2016;30(15):1704–1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Khiati S, Seol Y, Agama K, et al. Poisoning of mitochondrial topoisomerase I by lamellarin D. Mol Pharmacol. 2014;86(2):193–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Chamberlain GR, Tulumello DV, Kelley SO. Targeted delivery of doxorubicin to mitochondria. ACS Chem Biol. 2013;8:1389–1395. [DOI] [PubMed] [Google Scholar]
- [68].Jean SR, Tulumello DV, Riganti C, et al. Mitochondrial targeting of doxorubicin eliminates nuclear effects associated with cardiotoxicity. ACS Chem Biol. 2015;10(9):2007–2015. [DOI] [PubMed] [Google Scholar]
- [69].Dalla Rosa I, Camara Y, Durigon R, et al. MPV17 loss causes deoxynucleotide insufficiency and slow DNA replication in mitochondria. PLoS Genet. 2016;12(1):e1005779. [DOI] [PMC free article] [PubMed] [Google Scholar]


