Abstract
BACKGROUND.
How prostate epithelial cells differentiate and how dysregulation of this process contributes to prostate tumorigenesis remain unclear. We recently identified a Myc target and chromatin reader protein, ING4, as a necessary component of human prostate luminal epithelial cell differentiation, which is often lost in primary prostate tumors. Furthermore, loss of ING4 in the context of oncogenic mutations is required for prostate tumorigenesis. Identifying the gene targets of ING4 can provide insight into how its loss disrupts differentiation and leads to prostate cancer.
METHODS.
Using a combination of RNA-Seq, a best candidate approach, and chromatin immunoprecipitation (ChIP), we identified Miz1 as a new ING4 target. ING4 or Miz1 overexpression, shRNA knock-down, and a Myc-binding mutant were used in a human in vitro differentiation assay to assess the role of Miz1 in luminal cell differentiation.
RESULTS.
ING4 directly binds the Miz1 promoter and is required to induce Miz1 mRNA and protein expression during luminal cell differentiation. Miz1 mRNA was not induced in shING4 expressing cells or tumorigenic cells in which ING4 is not expressed. Miz1 dependency on ING4 was unique to differentiating luminal cells; Miz1 mRNA expression was not induced in basal cells. Although Miz1 is a direct target of ING4, and its overexpression can drive luminal cell differentiation, Miz1 was not required for differentiation.
CONCLUSIONS.
Miz1 is a newly identified ING4-induced target gene which can drive prostate luminal epithelial cell differentiation although it is not absolutely required.
Keywords: chromatin, integrins, RNA-Seq, Myc, human
INTRODUCTION
The manner in which prostate epithelial cells differentiate, that is, how cells in the prostate epithelium transition from basal to sejcretory luminal cells, still remains to be fully elucidated. The process is one that demands attention since dysregulated differentiation is implicated in prostate oncogenesis [1]. Several different models have been used to investigate prostate epithelial differentiation, the most common being in vivo mouse models [2–4]. We developed an in vitro differentiation model using human basal prostate epithelial cells to better assess both differentiation and oncogenesis in a human model [5]. Stimulation of human basal cells with KGF and DHT for 14–18 days results in a bilayer culture with fully differentiated AR-positive secretory luminal cells sitting atop basal cells that mimics human prostate histology. Utilizing this model, we identified the chromatin binding protein and Myc target, ING4 [6–8], as a major luminal cell determinant [1]. ING4 is induced down-stream of Myc and required for luminal differentiation, and its induction is coincident with integrin loss within the luminal cell population.
Introduction of oncogenes, that is, overexpression of Erg and Myc and knock-down of Pten, into differentiating basal cells generated AR-positive luminal-like tumorigenic cells that retained some basal markers including integrin α6β1 analogous to what is seen in human tumors [1,9]. These same tumorigenic cells lost their ability to fully differentiate and this was shown to be due to loss of ING4. To better understand how ING4 drives integrin loss during normal differentiation, we sought to identify the gene targets of ING4.
The loss of integrin expression during epithelial cell differentiation has been studied in other contexts including mammary and skin [10,11]. In addition to Notch being a strong suppressor of both integrins and matrix [11], a Myc/Miz1 (ZBTB17) repressive complex, which binds integrin α6 and β1 promoters, was shown to be necessary for Myc-induced differentiation of keratinocytes [10]. Miz1 is also necessary in the mammary gland for proper transitioning from late stage pregnancy to early lactation [12]. ING4 expression is also lost in some breast cancers [13] where it may suppress NF-κB signaling [14], and elevated expression of an ING4 E3-ligase, SCF(JFK), promotes breast cancer metastasis [15]. Interestingly, expression of an ING4 mutant unable to bind chromatin induced integrin expression in a mouse breast cancer model [8]. Thus, we hypothesized that Miz1 might be the link between ING4 and α6ß1 integrin that could explain its loss during normal differentiation and its retention in tumor cells.
MATERIALS AND METHODS
Cell Lines
Immortalized human basal prostate epithelial cells (iPrEC) were generated from primary clinical prostectomies as previously described [1,5]. Cultures were validated to be Mycoplasma-free and express only basal epithelial cell markers [5]. Tumorigenic iPrEC-EMP (Erg/Myc/shPten overexpression) and ING4 or shING4 overexpressing (iPrEC-ING4; iPrEC-shING4) cells were generated as previously described [1,8]. All lines were maintained and passaged in keratinocyte serum-free media (Invitrogen) [1,5].
Differentiation Protocol
Differentiation and layer separation protocols were detailed previously [5]. Briefly, iPrECs at confluency were treated in complete growth medium with 2 ng/ml keratinocyte growth factor (KGF) (Cell Sciences) and 5nM R1881 (PerkinElmer) every other day for up to 18 days. For biochemical analysis, the differentiated luminal layer was separated from the basal layer using disassociation buffer (Invitrogen) as previously described [5].
Constructs
The pLKO vector containing Pten shRNA was generated by subcloning the oligo 5’- CCGGTGGGCT TTAACTGTAGTATTTGTACTAGTCAAATACTACA GTTAAAGCCCTTTTTG-3’, complementary to the 3’- UTR of Pten, into a lentiviral vector to generate pLKO.1-shPten. The shPten in the iPrEC-EMP cells reported here contain the above targeting sequence, which is more stable and generated subsequent to the initial report on iPrEC-EMP [1]. The pLKO vector containing ING4 shRNA was purchased from Sigma-Aldrich (Clone ID:NM_016162.3–522s21c1) and used to generate the iPrEC-shING4 cells. ING4 shRNA targeting sequence: 5’-CCGGTTAAAG CTCGTGCGCACAAGTCTCGAGACTTGTGCGCAC- GAGCTTTAATTTTTTG-3’. The pLKO-TetON-shMiz1 constructs were generated by subcloning each of two oligos into the pLKO-TetON vector purchased from Addgene [16]. The Tet-pLKO-Puro vector was first modified, EZ-Tet-pLKO-Puro, to contain a shortened stuffer region by inserting an EcoRI site at base 222 of the stuffer (primer 5’-GCTACTCCACCACTT-GAATTCCTAAGCGGTCAGC-3’). The vector was then digested with EcoRI and re-ligated. Mutagenesis was then used to mutate the AgeI site to NheI (primer 5 ‘-TATCAGTGATAGAGACGCTAGCGTGTTGTAAA TGAGCA-3’). shMiz1 oligo sequences were as follows: 5’-CTAGTGTCCAAGCACATCATCATT- CAACTAGTGAGAATGATGATGTGCTTGGACATTT TT-3’ (5730), 5’-CTAGGTTCACTTTAAGGCTCA- TAAAAACTAGTGATTTATGAGCCTTAAAGTGAAC TTTTT-3’ (5729). Wild-type Miz1 (pLenti-Myc-DDK- ZBTB17)(PS100064) was purchased from OriGene Technologies (Rockville, MD). Wild-type c-Myc (pMSCV-c-Myc-GFP) and Myc-Miz1 binding mutant (pMSCV-c-Myc-V394D-RFP) were generous gifts from Dr. Martine Roussel [10,17].
Virus Generation and Infection
Lentiviruses expressing shRNAs or Miz1 cDNA were generated by co-transfecting the 293FT packaging cell line with 6 μg each of the lentiviral packaging plasmids, pVSVG, pLP1, and pLP2 with Lipofectamine 2000 (ThermoFisher) following manufacturers recommended protocol. Virus was harvested 3 days later and immediately used to infect iPrECs. Pooled cells were selected and maintained in 0.75 μg/ml puromycin. Retroviruses expressing Myc or MycV394D were generated by transfecting Phoenix cells (National Gene Vector Biorepository) using Lipofectamine 2000 following manufacturers recommended protocol, harvesting 2 days later and immediately infecting iPrECs. Pools of Myc expressing cells were selected and maintained in 0.75 μg/ml puromycin.
Antibodies
Immunofluorescence: AR (C-19) and Miz1 (H-190) were purchased from Santa Cruz. ITGα6 (GoH3) antibody was purchased from BD Pharmingen. Immunoblotting: Myc (N-term) and ING4 (EP3804) antibodies were purchased from Abcam. Polyclonal Miz1 antibody was purchased from GeneTex. Tubulin antibody (DM1A) was purchased from Sigma and GAPDH (6CS) from Millipore.
Immunostaining and Microscopy
Differentiated cultures were fixed in 4% paraformaldehyde and permeabilized with 0.2% Triton-X 100 for 5 min. After washing with PBS, cells were blocked with 4% normal goat serum for 2 hr. Primary antibodies, diluted in 1% BSA/PBS, were applied to samples overnight at 4°C. After washing, secondary conjugated antibodies diluted in 1% BSA/PBS were incubated for 1–2 hr. Nuclei were stained with Hoechst 33258 (Sigma) for 10 min at room temperature. Coverslips were mounted using Fluoromount-G (SouthernBiotech). Epifluorescent images were acquired on a Nikon Eclipse TE300 fluorescence microscope using OpenLab v5.5.0 image analysis software (Improvision).
Immunoblotting
Total cell lysates were prepared for immunoblotting as previously described [18]. Briefly, cells were lysed in RIPA buffer and 30–50 μg total protein was separated on SDS polyacrylamide gels and transferred to PVDF membranes. Membranes were blocked in 5% BSA in TBST overnight at 4°C then probed with primary antibody, and HRP-conjugated secondary antibodies (Bio-Rad) in TBST + 5% BSA. Signals were visualized by chemiluminescence reagent with a CCD camera in a Bio-Rad Chemi-Doc Imaging System using Quantity One software v4.5.2 (Bio-Rad).
qRT-PCR
Total RNA was isolated and purified using RNase- free DNase and Life Technology’s RNeasy PureLink Kits. For qRT-PCR, 0.5 μg RNA was reversed transcribed using a reverse transcription system (Promega). Synthesized cDNA was amplified for qRT-PCR using SYBR green master mix (Roche) with gene-specific primers and an ABI 7500 RT-PCR system (Applied Biosystems). Gene expression was normalized to 18 sec rRNA by the 2-ΔΔCt method (Livak, 2001). qRT-PCR primers for Miz1 were as follows: Miz1 Fwd: 5’-CTACTCTTTTCTGACAGTTTGCC-3’, Miz1 Rev: 5’-CCTTTGTCTGCTCTGGAGT-3’.
Chromatin Immunoprecipitations
Cells (3.0 × 106) were fixed in 1% formaldehyde (Thermo Scientific) for 1–5 min and washed 3× with ice cold calcium-magnesium free PBS (CMF-PBS) supplemented with protease inhibitors: pepstatin, aprotinin, leupeptin, and phenylmethylsulfonyl (PMSF). Cells were scraped and pelleted at 2,000 rpm for 8 min at 4°C. Pellet was resuspended in swelling buffer (5 mM PIPES pH 8.0, 85 mM KCl, 0.5% IGEPAL, and incubated on ice for 30 min). Nuclei were dounce homogenized and then pelleted at 4,000 rpm for 10 min, 4°C. Nuclei were resuspended in sonication buffer (0.1% SDS, 10mM EDTA, 50mM Tris-HCl pH 8.1) and incubated on ice for 10 min prior to sonication. Chromatin was sheared at 4°C using the Covaris E220 Ultra Sonicator following manufacturer’s suggested settings of 2% Duty Cycle, 105 Watt Peak Intensity, 200 Cycles/Burst. Chromatin was sonicated for 10 min to achieve 300–500 bp fragments.
Chromatin immunoprecipitations (ChIPs) were performed with 1 million cells/IP using magnetic beads (NEB). The following antibodies were used: ING4 (EP3804) and anti-HBO1 (ab70183) from Abcam. Chromatin was incubated with 6 μg of appropriate antibody overnight at 4°C with rotation. Following incubation, magnetic beads blocked with 1% BSA supplemented with 10 μg/ml salmon sperm, were added to samples and incubated at 4°C with rotation for 6 hr. Following immunoprecipitations, beads were washed in the following buffers at 4°C for 10 min with rotation: Triton Wash Buffer (50 mM Tris-HCl pH 7.4, 150 mM NaCl, 1% Triton X-100), followed by Lysis Buffer 500 (0.1% NaDOC, 1mM EDTA, 50mM HEPES pH 7.5, 500mM NaCl, 1% Triton X-100), LiCl Detergent buffer (0.5% NaDOC, 1 mM EDTA, 250 mM LiCl, 0.5% IGEPAL, 10mM Tris-HCl pH 8.1), and Tris-EDTA pH 8.1. Chromatin was eluted from beads in Elution Buffer (10 mM EDTA, 1% SDS, Tris-HCl pH 8.0) for 30 min at 65°C. Samples were then treated with 20 μg proteinase K, and 10 μg RNase A, and NaCl (200 mM) was added and incubated at 65°C overnight to reverse cross-links. DNA was purified using phenol/chloroform extraction followed by ethanol precipitation.
ChIP Primer Sequences
Primers were designed referencing the UCSC Genome Browser to determine transcriptional start sites of promoters, and in the case of ING4-ChIP, regions within the promoter region with high H3K4me3 were used to design targeting primers. Primer sequences are as follows: Miz1: Fwd: 5’-AACAGTCTCCCC ACTGCATA-30, Rev: 5’-GTAGCTCTAGGCCACTG ACT-30; Histone 3: Fwd: 5’-TTTTGTTΓTCCA AAGCGCCC-3’, Rev: 5’-TCAGATTGTTCCCTTTC CGC-30; SAT2: Fwd: 50-ATCGAATGGAAATGAAAG- GAGTCA-30, Rev: 5’-GACCATTGGATGATTGCAG TCA-30.
RNA-Sequencing
The iPrEC and EMP lines were grown and differentiated as described above and harvested at days 0 (basal), 4, 8, 11, 14, and 17. At day 14 and 17, iPrEC differentiated cultures were treated with CFM-PBS supplemented with 1 mM EDTA and dissociation buffer for 40–45 min to isolate the luminal cells. Total RNA was isolated and purified using Life Technologies RNeasy and Purelink RNA mini kits. TruSeq mRNA libraries were prepared for sequencing using standard Illumina protocols from PolyA-enriched RNA. Illumina RNAseq—single read, 50 bp, approximately 30 million reads per sample. Sequenced reads were mapped to the hg19 whole genome using the Subread aligner (v1.4.3). Reads were assigned to genes using featureCounts. Raw read counts were voom transformed and differential expression performed using limma.
NCBI GEO Database Access to RNA-Sequencing
http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=utahcoigpdubpcj&acc=GSE77460
RESULTS
Miz1 Expression Is Increased During Prostate Luminal Cell Differentiation
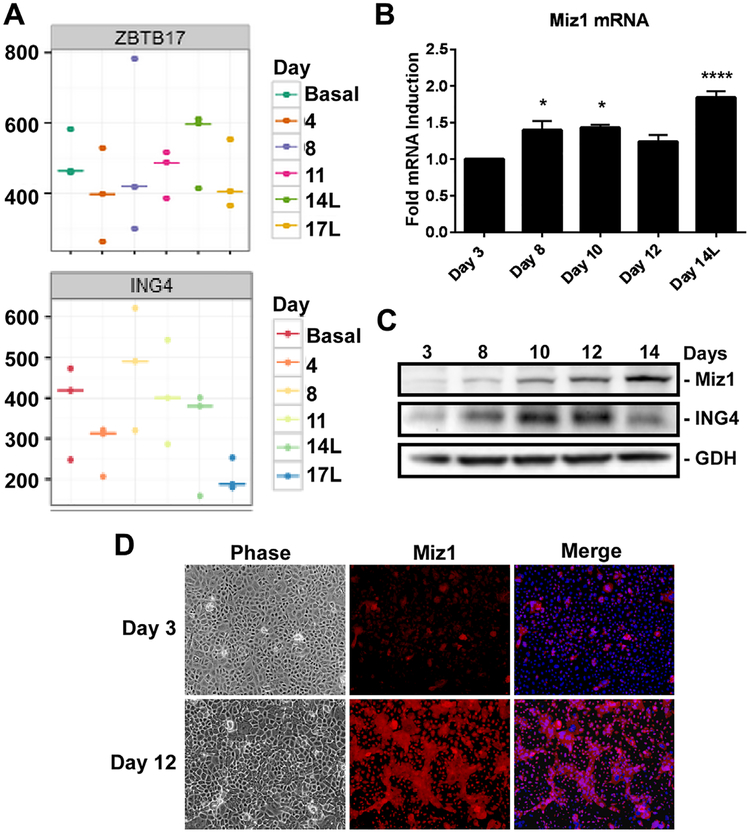
When grown to confluency and treated with KGF plus androgen, basal prostate epithelial cells (iPrEC) undergo differentiation such that a second suprabasal luminal layer forms on top of the basal layer in 14–18 days [1,5]. To identify genes associated with luminal cell differentiation, iPrECs differentiated for 0–17 days were subjected to RNA sequencing. Miz1 (ZBTB17) transcript levels first dropped and then increased over the course of differentiation, peaking at day 14 in the luminal cells (Fig. 1A top panel). ING4 expression (Fig. 1A bottom panel) followed a similar trend, except that it peaked earlier at day 11. This trend of increased Miz1 mRNA expression over time was further validated by qRT-PCR (Fig. 1B), with a 1.8-fold peak in expression occurring after 14 days of differentiation in the luminal cells; paralleling the RNA-Seq data. There was a similar steady and significant increase in Miz1 protein expression over time, subsequent to the induction of ING4 expression (Fig. 1C). Highest expression of Miz1 was at day 14, mirroring the mRNA. Immunofluorescence imaging at day 3 versus day 12 revealed Miz1 was dramatically induced in the luminal cells (Fig. 1D). It should be noted that Miz1 expression is not restricted to the luminal cells; Miz1 staining is also seen in the basal cells of differentiated cultures, where its expression also increased but less dramatically.
Fig. 1.
L Mizl expression increasing during prostate luminal cell differentiation. Confluent immortalized prostate basal epithelial cells (iPrECs) were induced to differentiate with 2ng/ml KGF and 5 nM R1881 for 0–17 days. “L” denotes isolated luminal-specific cells.(A) RNA sequencing was performed on samples isolated at the specified time points during iPrEC differentiation. Raw transcript counts are shown for Miz1 (ZBTBI7) and ING4. (B) qRT-PCR was used to validate Miz1 mRNA expression. Data is normalized to 18sec rRNA and expressed as fold induction relative to day 3 of differentiation. Error bars denote S.D. One-way ANOVA multiple comparisons t-test was used to calculate significance relative to day 3; *P< 0.05; ****P < 0.0001. (C) Miz1 and ING4 protein levels were measured by immunoblotting. GAPDH (GDH) served as a loading control. (D) iPrECs differentiated for 3 or 12 days were immunostained for Miz1 (red), nuclei stained with Dapi (blue), and imaged by phase and epifluorescence microscopy.
ING4 Induces MIz1 Expression in Prostate Luminal Cells and Binds Directly to its Promoter
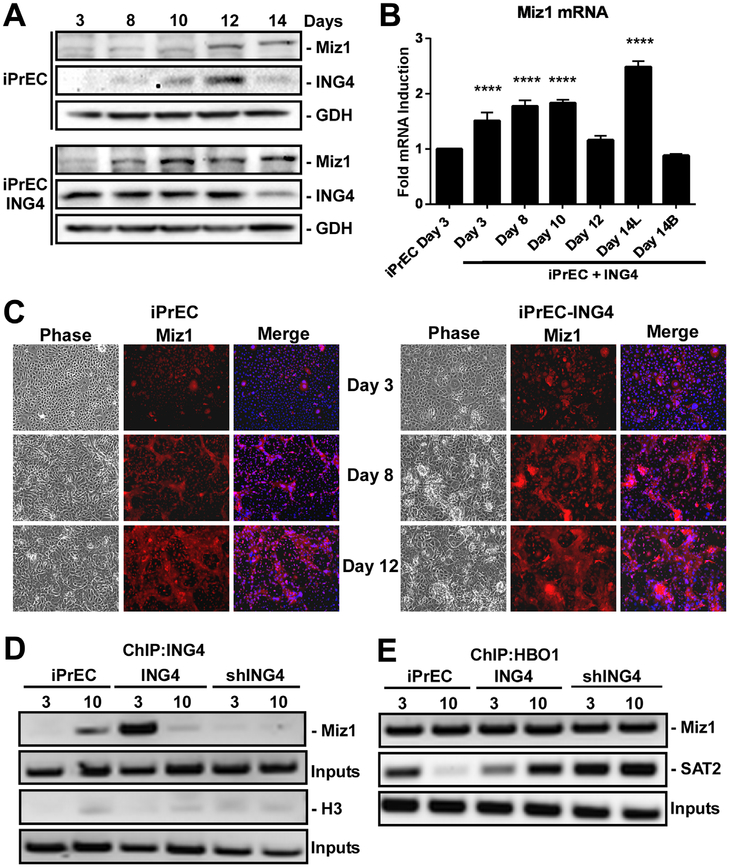
Because Miz1 was induced subsequent to ING4, we tested the effect of constitutive ING4 expression on Miz1. ING4 overexpression (iPrEC-ING4), resulted in an earlier and more robust induction of Miz1 expression around day 8 of differentiation, compared to a modest increase at day 12 in normal iPrECs (Fig. 2A). Constitutive ING4 expression also resulted in a sustained ~ 1.8-fold induction in Miz1 mRNA over the course of differentiation (Fig. 2B), which peaked at 2.5-fold in the luminal cells. Despite ING4 overexpression in the basal cells, Miz1 mRNA was not constitutively expressed in basal cells. Thus, the effect of ING4 on Miz1 is limited to luminal cells. We previously demonstrated that ING4 overexpression accelerates luminal cell differentiation [1] and as seen before there is significantly more luminal cells at day 8 in the ING4 overexpressing cells compared to normal iPrECs (Fig. 2C). There is also a concomitant increase in Miz1 expression in this luminal population as seen by immunostaining (Fig. 2C). These data indicate that ING4 overexpression in the luminal population enhances induced Miz1 expression.
Fig. 2.
ING4 is sufficient to induce Mizl expression in prostate luminal cells and binds directly to its promoter. Confluent iPrEC, iPrEC-ING4, and shING4-iPrEC cell lines were induced to differentiate with 2ng/ml KGF and 5 nM R1881 for 3–14 days. (A) Miz1 and ING4 protein levels were measured by immunoblotting. GAPDH (GDH) served as a loading control. (B) qRT-PCR analysis of Miz1 mRNA isolated from iPrECs + ING4 over the indicated time course of differentiation. Data normalized to 18sec rRNA and expressed as fold induction relative to day 3 of iPrEC differentiation. (L and B) denotes isolated luminal and basal cell populations. Error bars denote S.D. One-way ANOVA multiple comparisons t-test was used to calculate significance relative to day 3 of iPrEC differentiation; ****P < 0.0001. (C) Cells differentiated for indicated times were immunostained for Miz1 (red), nuclei stained with Dapi (blue), and imaged by phase and epifluorescence microscopy. (D) Chromatin immunoprecipitation (ChIP) of ING4 on the Miz1 promoter at day 3 or 10 of differentiation. Histone 3 served as a negative control and shING4 controlled for antibody specificity. (E) ChIP of HBO1 on the Miz1 and SAT2 promoters at day 3 or 10 of differentiation.
Since constitutive ING4 expression was able to enhance the induction of Miz1 expression in luminal cells, we tested whether Miz1 is a direct target of ING4 using chromatin immunoprecipitation (ChIP). For these experiments, we compared iPrEC, iPrEC-ING4, and iPrEC-shING4 cells. Cell lines were differentiated for 3 days (low ING4) or 10 days (high ING4). ChIP of ING4 in normal iPrECs revealed that after 10 days of differentiation, ING4 was inducibly bound to the Miz1 promoter (Fig. 2D). ING4 over-expression resulted in its constitutive association at the Miz1 promoter at day 3. Cells lacking ING4 ablated its ChIP at the Mizl promoter (Fig. 2D) and ING4 did not bind the Histone 3 promoter. ING4 is thought to recruit the histone acetyltransferase HBOl to chromatin [7]; however, we found HBO1 to be constitutively bound to the Miz1 promoter independent of ING4 (Fig. 2E). On the other hand, a gene known to be repressed during differentiation, SAT2 [19], lost HBO1 association at day 10 of normal differentiation and at day 3 in ING4 overexpressing cells (Fig. 2E).
ING4 is Necessary for Mizl mRNA Induction in Luminal Cells
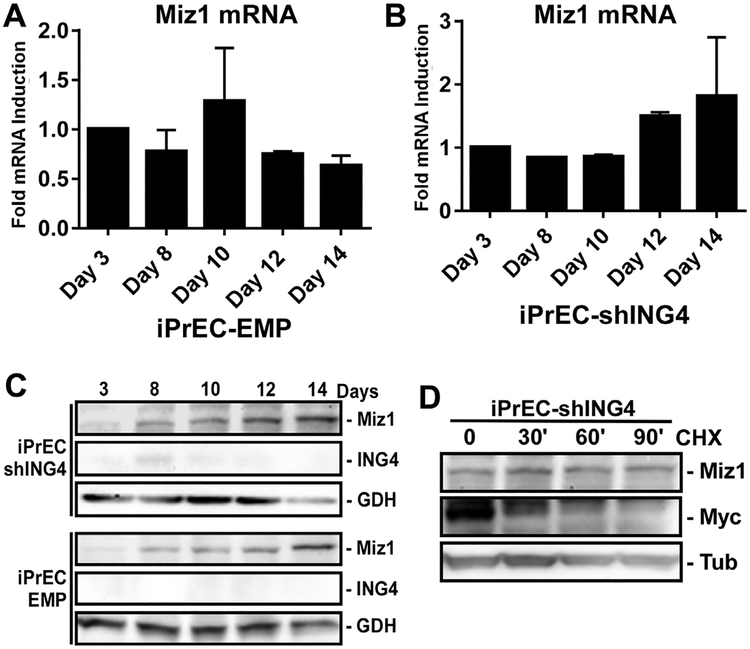
To determine if ING4 is necessary for Miz1 induction, we utilized two cell lines that do not express ING4 and do not differentiate; iPrEC-shING4 and tumorigenic iPrEC-EMP [1]. RNA-Seq data from iPrEC-EMP cells indicated Miz1 mRNA was not induced during the differentiation time course (not shown), and this was similarly observed by qRT-PCR (Fig. 3A). Miz1 mRNA also was not significantly induced in the iPrEC-shING4 cells (Fig. 3B). Thus, ING4 expression is necessary for Miz1 mRNA induction during luminal cell differentiation.
Fig. 3·.
ING4 is necessary for Mizl induction in luminal cells, while Mizl protein is stabilized in basal cells. iPrECs, iPrEC-shING4, and EMP-iPrECs were induced to differentiate with 2 ng/ml KGF and 5 nM R1881 for 3–17 days. (A and B) qRT-PCR analysis of Miz1 mRNA isolated from iPrEC-shING4 and EMP cells. Error bars denote S.D. No statistical difference between time points. (C) Miz1 and ING4 protein levels were measured by immunoblotting. GAPDH (GDH) served as a loading control. (D) iPrEC-shING4 cells were differentiated for 12 days and then treated with 50 μg/ml cycloheximide (CHX) for 0–90 min and levels of Miz1 and Myc protein measured by immunoblotting. Tubulin (Tub) served as a loading control.
Mizl Protein is Stabilized in Basal Cells
Despite the lack of significant Miz1 mRNA induction, Miz1 protein was still induced by the differentiation conditions in the undifferentiated “basal-like” EMP and shING4 cells in absence of ING4 (Fig. 3C). We attempted to measure Miz1 protein stability in iPrEC-shING4 cells by treating with cycloheximide for different times. Regardless of the length of time of CHX treatment (up to 6 hr), we were unable to detect a change in Miz1 protein expression (Fig. 3E), suggesting Miz1 protein in basal cells is very stable. Under the same conditions, Myc protein was rapidly lost. Thus, ING4 expression is necessary for Miz1 mRNA induction in luminal cells, but Miz1 protein stability may be enhanced independent of ING4 in basal cells.
Constitutive Mizl Expression Is Sufficient to Drive Luminal Cell Differentiation
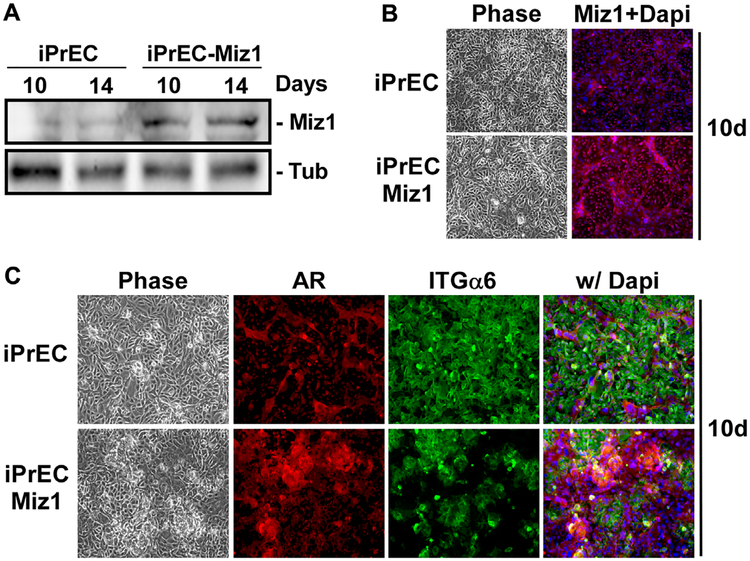
If Miz1 is a primary target of ING4 required for differentiation, then overexpression of Miz1 could be sufficient to drive iPrEC differentiation. To address Mizl sufficiency, we generated iPrECs that constitutively overexpress Miz1 (iPrEC-Miz1) as assessed by immunoblotting and immunostaining (Fig. 4A and B). Compared to normal iPrECs differentiated for 10 days, when a few AR-positive, integrin α6-negative luminal cells appear, there was considerably more of these luminal cells in the iPrEC-Miz1 cultures (Fig. 4C). This is the same phenotype that is observed in ING4 overexpressing cells (see Fig. 2C). Thus, overexpression of Miz1 is sufficient to induce luminal cell differentiation equivalent to that observed when ING4 is overexpressed.
Fig. 4·.
Constitutive Mizl expression is sufficient to drive luminal cell differentiation. iPrECs overexpressing Mizl (iPrEC-MizI) were induced to differentiate with 2 ng/ml KGF and 5 nM R1881 for 10–14 days. (A) Level of Miz1 expression was measured by immunoblotting. Tubulin (Tub) served as a loading control. (B) Cells differentiated for 10 days were immunostained for Miz1 (red) and nuclei were stained with Dapi (blue). (C) Differentiation was measured after 10 days by immunostaining of basal cells for integrin α6 (ITGα6; green) and luminal cells for AR (red), and nuclei were stained with Dapi (blue). All cells were visualized by phase and epifluorescence microscopy.
Miz1 Is Not Required for Luminal Cell Differentiation
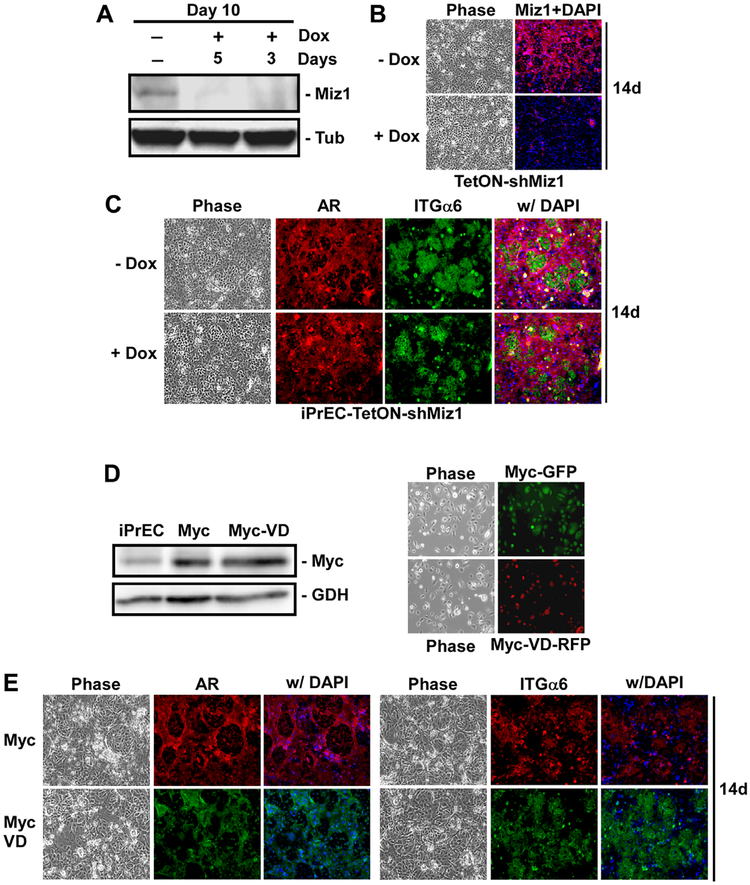
To determine if Miz1 is necessary for luminal cell differentiation, we generated cells that express a Tet-inducible shRNA targeting Miz1. This allowed us to selectively inhibit Miz1 expression late in differentiation, when it is maximally induced in the luminal cells. To test Miz1 knock-down, iPrEC-TetON-shMiz1 cells were differentiated for 10 days, and Miz1 expression suppressed by Dox induction of Miz1 shRNA during the last 5 or 3 days of differentiation. Miz1 expression was significantly reduced in as little as 3 days (Fig. 5A). To test the dependency on Miz1, iPrEC-TetON-shMiz1 cells were differentiated for 8 days, then treated with doxycycline for an additional 6 days of differentiation (14 days total). Miz1 expression was effectively inhibited under these conditions (Fig. 5B). The same amount of AR-positive, integrin α6-negative luminal cells were induced in the presence or absence of Miz1 (Fig. 5B), indicating that the cells were fully able to differentiate without Miz1. This was observed with two different shRNAs (not shown).
Fig. 5.
Mizl is not required for luminal cell differentiation. iPrECs overexpressing Mizl Tet-inducible shRNA (iPrEC-TetON-shMizI), Myc (Myc-GFP), or MycV394D mutant (Myc-V394D-RFP) were induced to differentiate with 2 ng/ml KGF and 5 nM R1881 (A) Cells were differentiated for 10 days and treated with 100 ng/ml doxycycline (Dox) during the last 5 or 3 days of differentiation. Miz1 and tubulin (Tub) expression were measured by immunoblotting. (B and C) Cells were differentiated for l4 days and treated with Dox during the last 6 days of differentiation. (B) Control cultures were immunostained for Miz1 (red) and nuclei stained with Dapi (blue).(C) Differentiation was measured by immunostaining of basal cells for integrin α6 (ITGα6; green) and luminal cells for AR (red), and nuclei were stained with Dapi (blue). All cells were imaged by phase and epifluorescence microscopy. (D) Myc-GFP or Myc-VD-RFP expression was assessed in undifferentiated cells by western blot (left panel), with GAPDH (GDH) as a loading control, and epifluorescence microscopy (right panel). (E) Differentiation was measured after l4 days by immunostaining of luminal cells for AR (red-Myc; green-MycVD) and basal cells for integrin α6 (ITGα6; red-Myc; green-MycVD), and nuclei were stained with Dapi (blue). All cells were visualized by phase and epifluorescence microscopy.
This lack of necessity was surprising given previous findings indicating Miz1 is required for differentiation in other models [10,12,20]. Furthermore, one of these studies indicated Miz1 interaction with Myc was required for differentiation and a Myc mutant which cannot bind Miz1, MycV394D, blocked differentiation [10]. We previously showed that Myc is required for ING4 induction and luminal cell differentiation [1]. Therefore, we generated iPrECs that over-express Myc-GFP or Myc-V394D-RFP (Fig. 5D). Expression of MycV394D had no impact on the ability of these cells to differentiate; inducing comparable levels of AR-positive and integrin α6-negative luminal cells (Fig. 5E).
DISCUSSION
We set out to determine how ING4 induction during prostate luminal cell differentiation leads to the suppression of integrin expression by defining ING4 target genes that might control integrin expression. Our studies successfully identified Miz1 as a direct downstream target of ING4 in luminal cells, and demonstrated that Miz1 overexpression is sufficient to mimic the differentiation phenotype induced by ING4 including loss of integrin α6 expression. However, Miz1 is not necessary for luminal cell differentiation as determined by shRNA knock-down or expression of a dominant Myc-Miz1 binding mutant and therefore is not absolutely necessary for integrin loss during luminal cell differentiation.
ING4 is a chromatin binding protein that specifically recognizes and binds the H3K4me3 chromatin mark [21]. It has been shown to recruit HBO1, an acetyltransferase that can acetylate histone H4 or H3 to promote transcription of target genes [7,22]. However, the exact targets that ING4 actually binds have largely not been identified, and are limited to Smc4, Egln1, Ext1 in HT1080 cells [22], and a few NF-κB targets such as Cox2 and MMP9 [23]. Using RNA-Seq, a best candidate approach, and ChIP, we identified the Miz1 promoter as a binding target of ING4 during prostate luminal epithelial differentiation. We further demonstrate that genetically increasing or decreasing ING4 expression results in a concomitant increase or decrease in Miz1 mRNA expression, respectively. However, this coordinated expression is not present in basal cells, even when ING4 is constitutively overexpressed, being restricted to the luminal cells. Thus, there are likely to be other “competency” factors required, that is, a signal that defines a pre-luminal state induced by the differentiation conditions of KGF and androgen. Similarly, we demonstrate that overexpression of Miz1 is sufficient to robustly accelerate luminal cell differentiation to the same degree seen with ING4 overexpression. However, this effect is still dependent on the differentiation factors; constitutive Miz1 overexpression in the absence of KGF and androgen is not sufficient to induce differentiation on its own.
Given the reported ability of ING4 to specifically recruit HBO1 to H3K4me3-marked promoters [7,21,22], we were surprised to find that HBO1 was constitutively bound to the Miz1 promoter. This was not true for all genes, as we saw HBO1 loss at SAT2, a gene that is down regulated upon luminal cell differentiation in our model. We noted that Miz1 is also expressed in basal cells, where it is not subject to ING4 regulation. These data are consistent with Miz1 being an already active gene in prostate epithelial cells. Thus, ING4 may be acting to enhance transcription via recruitment of other factors specifically during luminal cell differentiation. ChIP-Seq experiments are underway to define other ING4 targets and associated chromatin modifiers.
We previously showed that oncogenic conversion of human iPrECs by Erg, Myc, and shPten over-expression (EMP cells), generates tumorigenic cells that are arrested in differentiation and fail to induce ING4 expression [1]. We also demonstrated Erg/Myc overexpressing cells are not tumorigenic, but loss of ING4 is sufficient to transform them and that ING4 is lost in over 60% of human primary prostate tumors [1]. Consistent with Miz1 dependency on ING4 expression, the EMP cells also did not induce Miz1 expression in response to the differentiation conditions. Correspondingly, these cells do not properly differentiate as defined by a lack of a distinct AR-positive and integrin α6-negative population.
Loss of Miz1 expression per se is not likely to be a good distinguishing marker for prostate cancer as basal cells and EMP cells still express Miz1; it is just not induced or regulated by ING4. Nonetheless, this allowed us to identify at least two mechanisms by which Miz1 expression is regulated. However, the most striking finding was the apparent lack of dependency on Miz1 for luminal cell differentiation. Despite the presence of distinct mechanisms for regulating Miz1 mRNA and protein expression, cells were still capable of differentiating without Miz1. This indicates there are other, potentially compensatory, factors that control differentiation, and integrin expression in particular. It also indicates there are other ING4 targets required for luminal cell differentiation. One potential target could be Notch signaling [24]. Studies are currently underway to determine the relationship between ING4, Notch, and luminal cell differentiation.
CONCLUSIONS
The Myc repressor, Miz1, is a direct target of the chromatin binding protein ING4, whose induction during luminal cell differentiation is dependent on ING4. Miz1 is capable of accelerating luminal cell differentiation when overexpressed, but is not absolutely required for differentiation.
ACKNOWLEDGMENTS
We thank the Michigan State University Research Technology Support Core for the RNA sequencing. Dr. Sander Frank generated the modified pLKO-TetON-Puro shRNA vector, which was originally purchased from Addgene and Dr. Martine Roussel provided the wild-type Myc and MycV394D mutant constructs. This project was supported by funding from the Department of Defense Prostate Cancer Research Program W81XWH-14-1-0479 (PLB), NIH/NCI CA154835 (CKM; PLB), and the Van Andel Research Institute (CKM, MEW).
Grant sponsor: Department of Defense Prostate Cancer Research Program; Grant number: W81XWH-14-1-0479; Grant sponsor: NIH/NCI; Grant number: CA154835; Grant sponsor: Van Andel Research Institute.
Footnotes
Conflicts of interest: The authors have no conflicts to disclose.
REFERENCES
- 1.Berger PL, Frank SB, Schulz VV, Nollet EA, Edick MJ, Holly B, Chang TT, Hostetter G, Kim S, Miranti CK. Transient induction of ING4 by Myc drives prostate epithelial cell differentiation and its disruption drives prostate tumorigenesis. Cancer Res 2014;74(12):3357–3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li X, Wang Y, Sharif-Afshar AR, Uwamariya C, Yi A, Ishii K, Hayward SW, Matusik RJ, Bhowmick NA. Urothelial transdifferentiation to prostate epithelia is mediated by paracrine TGF-beta signaling. Differentiation 2009;77(1): 95–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu CT, Altuwaijri S, Ricke WA, Huang SP, Yeh S, Zhang C, Niu Y, Tsai MY, Chang C. Increased prostate cell proliferation and loss of cell differentiation in mice lacking prostate epithelial androgen receptor. Proc Natl Acad Sci USA 2007;104(31): 12679–12684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Luo W, Rodriguez M, Valdez JM, Zhu X, Tan K, Li D, Siwko S, Xin L, Liu M. Lgr4 is a key regulator of prostate development and prostate stem cell differentiation. Stem cells 2013;31(11): 2492–2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lamb LE, Knudsen BS, Miranti CK. E-cadherin-mediated survival of androgen-receptor-expressing secretory prostate epithelial cells derived from a stratified in vitro differentiation model. J Cell Sci 2010;123(2):266–276. [DOI] [PubMed] [Google Scholar]
- 6.Coles AH, Jones SN. The ING gene family in the regulation of cell growth and tumorigenesis. J Cell Physiol 2009;218(1):45–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Doyon Y, Cayrou C, Ullah M, Landry A-J, Côté V, Selleck W, Lane WS, Tan S, Yang X-J, Côte J. ING tumor suppressor proteins are critical regulators of chromatin acetylation required for genome expression and perpetuation. Mol Cell 2006;21(1): 51–64. [DOI] [PubMed] [Google Scholar]
- 8.Kim S, Welm AL, Bishop JM. A dominant mutant allele of the ING4 tumor suppressor found in human cancer cells exacer-bates MYC-initiated mouse mammary tumorigenesis. Cancer Res 2010;70(12):5155–5162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cress AE, Rabinovitz I, Zhu W, Nagle RB. The α6b1 and α6b4 integrins in human prostate cancer progression. Cancer Metastasis Rev 1995;14(3):219–228. [DOI] [PubMed] [Google Scholar]
- 10.Gebhardt A, Frye M, Herold S, Benitah SA, Braun K, Samans B, Watt FM, Elsässer H-P, Eilers M. Myc regulates keratinocyte adhesion and differentiation via complex formation with Miz1. J Cell Biol 2006;172(1):139–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mazzone M, Selfors LM, Albeck J, Overholtzer M, Sale S, Carroll DL, Pandya D, Lu Y, Mills GB, Aster JC, Artavanis-Tsakonas S, Brugge JS. Dose-dependent induction of distinct phenotypic responses to Notch pathway activation in mammary epithelial cells. Proc Natl Acad Sci USA 2010;107(11): 5012–5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sanz-Moreno A, Fuhrmann D, Wolf E, von Eyss B, Eilers M, Elsäasser H-P. Miz1 deficiency in the mammary gland causes a lactation defect by attenuated stat5 expression and phosphorylation. PloS ONE 2014;9(2):e89187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tapia C, Zlobec I, Schneider S, Kilic E, Guth U, Bubendorf L, Kim S. Deletion of the inhibitor of growth 4 (ING4) tumor suppressor gene is prevalent in human epidermal growth factor 2 (HER2)-positive breast cancer. Hum Pathol 2011;42(7): 983–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Byron SA, Min E, Thal TS, Hostetter G, Watanabe AT, Azorsa DO, Little TH, Tapia C, Kim S. Negative regulation of NF- kappaB by the ING4 tumor suppressor in breast cancer. PloS ONE 2012;7(10):46823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yan R, He L, Li Z, Han X, Liang J, Si W, Chen Z, Li L, Xie G, Li W, Wang P, Lei L, Zhang H, Pei F, Cao D, Sun L, Shang Y. SCF(JFK) is a bona fide E3 ligase for ING4 and a potent promoter of the angiogenesis and metastasis of breast cancer. Genes Dev 2015;29(6):672–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wiederschain D, Susan W, Chen L, Loo A, Yang G, Huang A, Chen Y, Caponigro G, Yao Y-m, Lengauer C, Sellers WR, Benson JD. Single-vector inducible lentiviral RNAi system for oncology target validation. Cell Cycle 2009;8(3):498–504. [DOI] [PubMed] [Google Scholar]
- 17.Herold S, Wanzel M, Beuger V, Frohme C, Beul D, Hillukkala T, Syvaoja J, Saluz HP, Haenel F, Eilers M. Negative regulation of the mammalian UV response by Myc through association with Miz-1. Mol Cell 2002;10(3):509–521. [DOI] [PubMed] [Google Scholar]
- 18.Edick MJ, Tesfay L, Lamb LE, Knudsen BS, Miranti CK. Inhibition of integrin-mediated crosstalk with epidermal growth factor Receptor/Erk or src signaling pathways in autophagic prostate epithelial cells induces caspase-independent death. Mol Biol Cell 2007;18(7):2481–2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen Y, Vujcic S, Liang P, Diegelman P, Kramer DL, Porter CW. Genomic identification and biochemical characterization of a second spermidine/spermine N1-acetyltransferase. Biochem J 2003;373(Pt 3):661–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kerosuo L, Bronner ME. Biphasic influence of Miz1 on neural crest development by regulating cell survival and apical adhesion complex formation in the developing neural tube. Mol Biol Cell 2014;25(3):347–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Culurgioni S, Munoz IG, Moreno A, Palacios A, Villate M, Palmero I, Montoya G, Blanco FJ. Crystal structure of inhibitor of growth 4 (ING4) dimerization domain reveals functional organization of ING family of chromatin-binding proteins. J Biol Chem 2012;287(14):10876–10884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hung T, Binda O, Champagne KS, Kuo AJ, Johnson K, Chang HY, Simon MD, Kutateladze TG, Gozani O. ING4 mediates crosstalk between histone H3 K4 trimethylation and H3 acetylation to attenuate cellular transformation. Mol Cell 2009;33(2): 248–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nozell S, Laver T, Moseley D, Nowoslawski L, De Vos M, Atkinson GP, Harrison K, Nabors LB, Benveniste EN. The ING4 tumor suppressor attenuates NF-kappaB activity at the promoters of target genes. Mol Cell Biol 2008;28(21):6632–6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chakrabarti R, Wei Y, Romano RA, DeCoste C, Kang Y, Sinha S. Elf5 regulates mammary gland stem/progenitor cell fate by influencing notch signaling. Stem Cells 2012;30(7): 1496–1508. [DOI] [PMC free article] [PubMed] [Google Scholar]