Abstract
The human cytochrome P450 CYP3A7, once thought to be an enzyme exclusive to fetal livers, has more recently been identified in neonates and developing infants as old as 24 months post-gestational age. CYP3A7 has been demonstrated to metabolize two endogenous compounds that are known to be important in the growth and development of the fetus and neonate, namely dehydroepiandrosterone sulfate (DHEA-S) and all-trans retinoic acid (atRA). In addition, it is also known to metabolize a variety of drugs and xenobiotics, albeit generally to a lesser extent relative to CYP3A4/5. CYP3A7 is an important component in the development and protection of the fetal liver and additionally plays a role in certain disease states, such as cancer and adrenal hyperplasia. Ultimately, a full understanding of the expression, regulation, and metabolic properties of CYP3A7 is needed to provide neonates with appropriate individualized pharmacotherapy. This article summarizes the current state of knowledge of CYP3A7, including its discovery, distribution, alleles, RNA splicing, expression and regulation, metabolic properties, substrates, and inhibitors.
1. Introduction
The human cytochrome P450s (CYP450s) consist of a superfamily of heme-thiolate monooxygenases that metabolize a diverse array of exogenous and endogenous substrates, including hormones, drugs, toxins, and certain fatty acids [1-3]. The CYP3A subfamily is a major subfamily of human CYP P450, representing approximately 30% of all hepatic cytochrome P450 content, and it is involved in the metabolism of most drugs used clinically [1,4-6]. The human CYP3A subfamily is constituted of four functional isoforms: CYP3A4 [7,8], CYP3A5 [9,10], CYP3A7 [11,12], and CYP3A43 [13,14], as well as three CYP3A pseudogenes [15,16].
In adults, CYP3A4 is the predominant CYP3A isoform, accounting for 10–50% of total hepatic CYP, and metabolism of nearly 50% of currently administered drugs [5,17]. While CYP3A5 and CYP3A43 are primarily extrahepatic enzymes, they also exist in the liver, but at lower levels (10–30% of total CYP content) [10,18] and only 0.2–5% [16] that of CYP3A4, respectively.
CYP3A7 is found predominantly in human fetal livers and presents in embryonic hepatic tissues as early as 50–60 days of gestation [11,19-22], accounting for up to 50% of the total fetal hepatic CYP content and up to 87–100% of total fetal hepatic CYP3A content [23-26]. However, its expression has also been reported in other fetal tissue types, including the adrenal gland, kidney and lung [27]. When originally described by Kitada and Kanakubo [11], CYP3A7 was thought to be an exclusively fetal enzyme. Yet, more recent studies with neonatal and pediatric livers have identified significant levels of CYP3A7 up to 24 months post-gestational age [23,26,28,29]. Unique for any known human cytochrome P450, CYP3A7 begins to gradually decrease during the third trimester of pregnancy but remains active in many individuals during the first year of life, concomitant with gradual increases in CYP3A4 beginning after birth [23,26,28,30,31]. That is, CYP3A7 and CYP3A4 appear to exhibit a mirror image pattern of expression, with CYP3A7 levels declining after birth even as CYP3A4 levels increase.
This isoform switching phenomenon has profound implications for drug and xenobiotic metabolism in the neonate and developing infant. This is due to the fact that, while CYP3A4 and CYP3A7 share 87% amino acid sequence identity [12,32], and a wide overlapping substrate range [33], CYP3A7 is known to be substantially less catalytically active and can generate an altered profile of regiospecificity compared with CYP3A4/5 [29,33,34]. These differences in metabolism can lead to profound differences in drug efficacy and toxicity in the neonate and developing infant [29,35-37].
Interestingly, CYP3A7 expression has also been reported in a minority of adult human livers, human placenta, endometrium [38,39], and hepatocarcinoma [40], indicating that its presence is not exclusive to either the fetus or the developing infant. Furthermore, an allelic variant with a mutation in the promoter region, known as CYP3A7*1C, occurs with 5% frequency in the population and can lead to high levels of expression of the enzyme well into adulthood [31,37,40]. While it is unclear what role CYP3A7 plays in adult extrahepatic tissue, given its established biological importance in the infant, this may be an active area of future study.
Because CYP3A7 mediates the biotransformation of a wide variety of exogenous and endogenous compounds and is involved in the safety and efficacy of a number of drugs, as well as certain disease states in the adult, there has been increased interest in its biological role(s) in humans. In order to provide insight on our current level of understanding of this important CYP enzyme, this review summarizes the available knowledge on the biological role, substrates, inhibitors, activators, expression and regulation patterns, and general relevance to disease, of human CYP3A7.
2. Discovery
At least since the early 1970's it has been known that human fetal liver had the capacity to metabolize many xenobiotic compounds, even at early stages of gestation [41]. Additionally, Cresteil et al. observed that fetal livers exhibited higher dehydroepiandrosterone (DHEA) 16α-hydroxylase activity than adult livers [42]. In the fetus and developing infant, DHEA is a hormone that functions as a precursor for androgens and estrogens that are important for growth and development.
Subsequent to these initial observations, a CYP isoform, originally referred to as P450 HFLalpha, was isolated from fetal livers [11,27], and found to be the major enzyme responsible for the 16α-hydroxylation of DHEA sulfate (DHEA-S), the storage form of DHEA [43]. This enzyme was later designated CYP3A7 [44]. In 1989, the entire CYP3A7 cDNA was cloned from a fetal library [12]. CYP3A7 was found to share 95% nucleotide identity and 87% amino acid identity with CYP3A4, suggesting that the two isozymes might share an overlapping substrate range [45].
3. Gene location
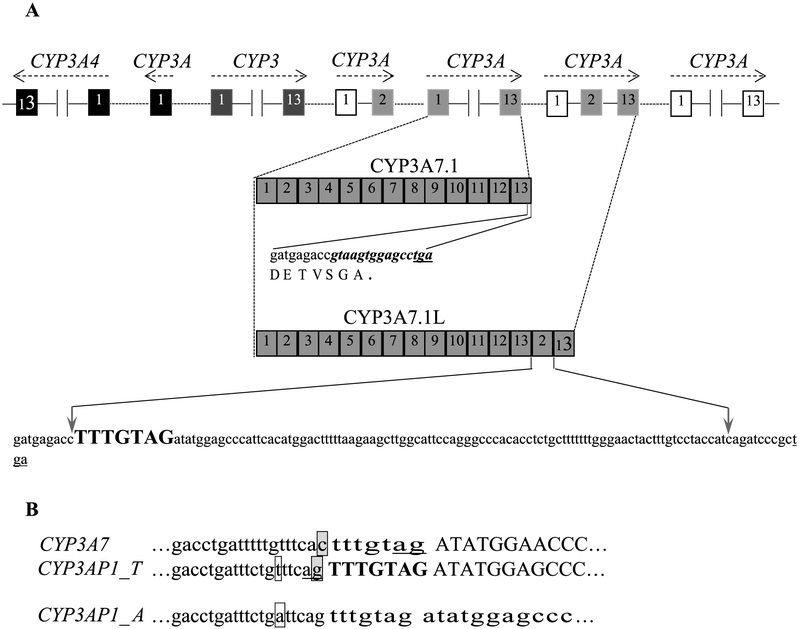
CYP3A7 and the other CYP3A genes form a gene cluster which is in the order of 3A43, 3A4, 3A7 and 3A5 on chromosome 7q21-q22.1. In the intergenic regions, several additional cytochrome CYP3A exons form three pseudogenes (CYP3APs) (Fig. 1A) [15,16,46].
Fig. 1.
Location of CYP3A7 and its alternatively spliced variants. A: organization of CYP3A locus, sequences of the 3′ ends of CYP3A7 and its variant CYP3A7-3AP1. The canonical CYP3A7 last 15 bp (bold and italic) are replaced by the exons 2 and 13 of pseudogene CYP3AP1, generating a longer mRNA species CYP3A7-3AP1 with extra shift-mutated sequence. B: Intron 1-exon 2 boundaries in CYP3A7 and CYP3AP1. Introns and exons are shown in lowercase and in uppercase, respectively. The AG splice site of canonical CYP3A7 exon 2 is located in the 3′-end of the heptanucleotide spacer (tttgtag)(underlined). The 5′-flanking nucleotide of the heptanucleotide spacer (bold and uppercase) is a G (boxed and shaded) in CYP3AP1 rather than a C (boxed and shaded) in CYP3A7. This nucleotide variation shifts the AG acceptor splice site to an alternative site in CYP3AP1. The polymorphism (T > A, boxed) in CYP3AP1 prevents the pseudogene splicing.
4. Distribution
CYP3A7 protein is mainly expressed in fetal livers with much lower levels in extrahepatic tissues including kidney, lung, stomach, intestine, adrenal, thymus, placenta, Jejunum and prostate [39,47-54]. However, the data on CYP3A7 expression in kidney, endometrium, and lung are somewhat conflicting. Haehner et al. (1996) found CYP3A7 mRNA in only one out of 25 kidneys [18], and Koch et al. (2002) described only a marginal expression of CYP3A7 in the kidney [50]. Conversely, Murray and colleagues detected CYP3A7 transcripts in all kidneys they investigated [52]. Also, while Schuetz et al. [39] and Sarkar et al. [55] found that CYP3A7 was expressed in the endometrium; Hukkanen et al. [56] and Williams et al. [57] did not detect it there. Finally, Anttila et al. [58] could not detect CYP3A7 mRNA in lung samples, but Kivisto et al. could [59]. Many of these studies illustrate the difficultly of trying to immunochemically detect CYP3A7 with a less than specific CYP3A antibody [31,60]. This has been an issue that has stymied quantification of CYP3A7 in fetal tissues for some time. However, newer proteomic based methods have recently shown promise [29] and some of the previous studies may be revisited with these newer methods in the future. In any case, it seems likely that the fetal liver remains the main location of CYP3A7 expression in the fetus and neonate.
5. Substrates of CYP3A7
5.1. Typical substrates
As mentioned in the introduction, CYP3A7 metabolizes numerous exogenous and endogenous compounds (Table 1).
Table 1.
Known substrates of the CYP3A7 enzyme.
| Substrate | System | Reaction/product | Reference |
|---|---|---|---|
| Midazolam | cDNA | 1- and 4-OH | [30,33] |
| Pentoxyresorufin | cDNA | Dealkylation | [30] |
| Tacrolimus | cDNA | 13-O-demethylation | [61] |
| Retinoic Acid | cDNA/HFLM | 4-OH | [62,63] |
| Estrone | cDNA | 2-, 4-, 16α- and 6β-OH | [64] |
| Estradiol | cDNA | 2-, 4-, 16α-, y- and 6β-OH | [33,64] |
| Alprazolam | cDNA | 1-and 4-OH | [33] |
| Triazolam | cDNA | 1-and 4-hydroxylation | [33] |
| Diltiazem | cDNA | N-Desmethyl | [33] |
| Clarithromycin | cDNA | N-Desmethyl, 14-OH | [33] |
| Nifedipine | cDNA | Oxidized | [33] |
| Trifluoromethylcoumarin | cDNA | 7-OH | [33] |
| Tamoxifen | cDNA | N-Desmethyl | [33] |
| Sildenafil | cDNA | N-Demethylation | [65] |
| Tadalafil | cDNA | Demethylenation | [65] |
| Cisapride | fHLM/cDNA | Norcisapride, 3-fluoro-4-hydroxycisapride, 4-fluoro-2-hydroxycisapride | [66] |
| Luciferin 6′ benzylether | HepG2 | O-debenzylation | [67] |
| Carbamazepine | cDNA | 10,11-epoxidation | [68] |
| Zonisamide | cDNA | Sulfamoylacetylphenol | [68] |
| Glyburide | fHLMs/cDNA | Ethylene-hydroxylation | [29] |
| Reduced haloperidol pyridinium | Human placenta microsomes | Oxidation | [69] |
| Dimethyl benzoylphenylurea | cDNA | Monomethyl Dimethyl benzoylphenylurea | [70] |
| 17α-hydroxyprogesterone caproate | cDNA/HFL | Uncharacterized metabolites M1, M2 and M3 | [71] |
| Alfentanil | cDNA | Noralfentanil, N-phenylpropionamide | [72] |
| Triamcinolone acetonide | cDNA | 6β-OH, Δ6-dehydrogenation | [73] |
| Budesonide | cDNA | 6β-OH, Δ6-dehydrogenation | [73] |
| Fluticasone propionate | cDNA | 17β-carboxy fluticasone propionate | [73] |
| Oleic acid | cDNA | cis-9,10-epoxyoctadecanoic acid | [74] |
| Carbamazepine | cDNA | Protein-reactive metabolite | [75] |
| 3-hydroxycarbamazepine | cDNA | 2,3-dihydroxycarbamazepine | [75] |
| Diltiazem | cDNA | N-demethylation | [76] |
| Cyclosporine | cDNA | ND | [77] |
| N-methylaniline | HFL/cDNA | N-demethylation | [11,78-80] |
| Ethylmorphine | N-demethylation | ||
| N,N-dimethylnitrosamine | N-demethylation | ||
| Benzphetamine | N-demethylation | ||
| Meperidine | N-demethylation | ||
| Aminopyrine | N-demethylation | ||
| N,N-dimethylaniline | N-demethylation N-oxidation |
||
| Chlorpromazine | N-demethylation Hydroxylation N-oxidation |
||
| Diazepam | N-demethylation Hydroxylation |
||
| Medazepam | N-demethylation Hydroxylation |
||
| Prazepam | N-demethylation Hydroxylation |
||
| Aniline | Hydroxylation | ||
| Desmethylimipramine | Hydroxylation | ||
| Hexobarbital | Hydroxylation | ||
| Caffeine | Hydroxylation | ||
| Diphenylhydantoin | Hydroxylation | ||
| Biphenyl | 4-hydroxylation 2-hydroxylation |
||
| 2,5-diphenyloxazole | Hydroxylation | ||
| 2-acetylaminofluorene | Hydroxylation N-hydroxylation |
||
| Benzo(a)pyrene | Hydroxylation Epoxidation Quinone formation |
||
| Aldrin | Epoxidation | ||
| Carbamazepine | Epoxidation | ||
| p-nitrobenzoic acid | Nitro reduction | ||
| p-nitroanisole | O-deethylation | ||
| 7-ethoxycoumarin | O-deethylation, N-demethylation | ||
| 6-aminochrysene | Metabolic activation | ||
| Aflatoxin B1 | Metabolic activation | ||
| Sterigmatocystin | Metabolic activation | ||
| 2-amino-3-methylimidazo-(4,5-f)quinoline | Metabolic activation | ||
| 2-amino-6-methyldipyrido-(1,2-a:3′2′-d)imidazole | Metabolic activation | ||
| Progesterone | HLM/cDNA | 6β-OH, 16α-OH | [81] |
| Bile acid | cDNA | 1β–OH–deoxycholic acid | [82] |
| Sildenafil | cDNA | N-demethylation | [65] |
| Tadalafil | cDNA | Demethylenation | [65] |
cDNA, protein produced by recombinant complementary DNA system; HFL, human fetal liver; fHLMs, human fetal liver microsomes; Hep G2, hepatocellular carcinoma cell line G2; HFL, human fetal liver; ND, not determined.
In many cases, CYP3A7 has similar characteristics to CYP3A4 and CYP3A5 regarding substrate specificity, regiospecificity, and Km; likely due to the high similarity in their overall structures (there are only 56 individual amino acid differences between CYP3A4 and CYP3A7). However, as described by Williams et al. (2002), the catalytic efficiency of the enzyme is generally greatly diminished for the turnover of most substrates when compared with CYP3A4/5 [33]. It is worth noting that CYP3A7 exhibits a striking catalytic preference for at least two substrates: DHEA-S and all-trans retinoic acid (atRA), with higher catalytic efficiency for both over CYP3A4/5, suggesting an important evolutionary role for this enzyme in the oxidation of these two hormones that are important in growth and development [28,68,83]. Additionally, other studies have demonstrated that CYP3A7 shows a preference in oxidation of 16α-hydroxylation activity of estrone over 2-hydroxylation, but no oxidation product preference for 17β-estradiol [64]. However, for the remaining substrates held in common, CYP3A7 generally exerts much less catalytic efficacy than CYP3A4/5 (Table 1).
5.1.1. DHEA
CYP3A7 converts DHEA-S and DHEA to the major metabolite 16α-OH DHEA-(S) [28,68] and minor metabolite 7β-OH DHEA-(S) [28], respectively. In contrast, 7β-hydroxylation of DHEA-(S) is the preferred reaction for CYP3A4 and 7β-OH DHEA-(S) is the major metabolite [28,30,43]. Based on the catalytic differences in DHEA-S oxidation by both enzymes, DHEA has been applied to differentiate the developmental expression of human hepatic CYP3A isoforms. For exclusively fetal liver samples, the CYP3A7 protein level correlated directly with DHEA 16α-hydroxylase activity due to the negligible presence of CYP3A4 enzymatic activity [28]. For differentiation of CYP3A4 and CYP3A7 in postnatal samples, although the amount of 16α-OH DHEA formed by CYP3A4 is relatively low, it is sufficient to impact CYP3A7 enzymatic activity assays used to determine the amount of CYP3A7 content [28,84]. Stevens et al. developed a DHEA 16α-/7β-metabolite profile-based multivariate regression model to simultaneously calculate CYP3A4 and CYP3A7 protein present in an individual liver [28]. The validity of this novel indirect approach was assayed by using both fetal and postnatal liver samples and indicated some promise in estimating CYP3A7 activity [28,31].
5.1.2. All-trans-retinoic acid
CYP3A7 metabolizes all-trans-retinoic acid (atRA) to 4-OH-, 18-OH-, 4-oxo- and 5,6-epoxy-RA, with 4-OH-RA being the major metabolite. Conversely, the contribution of CYP3A4 to the formation of 4-OH-, 18-OH-, and 4-oxo-RA is regarded to be minimal [62,83]. In particular, atRA metabolism by hepatic CYPs demonstrated that CYP3A4 could not catalyze the conversion of atRA to 18-OH- or 4-oxo-RA when the atRA concentration was below 50 μM, whereas the formation of these two metabolites was preferentially catalyzed by CYP3A7 at these concentrations [83], implying that atRA 4-OH, 18-OH, 4-oxo may have potential as in vivo CYP3A7 biomarkers. More recently, Topletz and colleagues (2019) have shown that while CYP3A7 is the predominant CYP responsible for clearance of atRA in the fetal liver, it does not contribute substantially to clearance of maternal atRA during pregnancy [85]. Given this evidence, CYP3A7 is not likely to provide a sufficient maternal-fetal barrier for atRA exposure during fetal development, suggesting that regulation of total atRA exposure of the fetus is controlled by other mechanisms.
5.1.3. Testosterone
In the adult, CYP3A4/5 are the CYPs responsible for the formation of 6β-OH testosterone from testosterone [30,68]. In contrast, in the fetus, neonate, and developing infant, 2α-OH testosterone is the major testosterone metabolite produced by CYP3A7, the most abundant CYP3A isoform in these individuals [33,34,60], while CYP3A4/5 contribute only minimally to the formation of 2α-OH testosterone in the adult. As noted by originally by Leeder et al. (2005) and more recently by Kandel et al. (2017), there exists a strong divergence in the ratio of 2α-/6β hydroxylation activities between the three CYP3A isoforms (CYP3A7, 1.19; CYP3A5, 0.079; CYP3A4, 0.007), that is, the ratio of 2α-/6β-OH activities of CYP3A7 was one order of magnitude greater than CYP3A5, and two orders of magnitude greater than CYP3A4 [34,60]. The variation of this ratio between CYP3A7 and other CYP3A isoforms originally suggested that it may be a unique endogenous biomarker for discriminating CYP3A isoform activity or identifying CYP3A7 [60]. However, more recently, Kandel et al. (2017) observed that the 2α-/6β-OH ratio varied with testosterone concentration, suggesting that the correlation between the 2α-/6β-OH ratio and CYP3A7 activity may be weak in individuals with higher levels of testosterone [34].
5.1.4. Imipramine
Imipramine is a tricyclic antidepressant and its N-demethylation product has been employed as probe for CYP3A7 activity. However, a complicating factor is that in addition to CYP3A7 other CYP isoforms, particularly CYP1A2 and CYP3A4, also contribute significantly to the metabolic N-demethylation reaction [86]. In an attempt to resolve this, selective inhibitors have been used to exclude imipramine N-demethylation enzymatic activity due to CYP isoforms other than CYP3A7 [19]. In fetal liver, CYP3A7 is the dominant CYP3A isoform and CYP3A4/5 levels are negligible to non-existent [28,31], and therefore a limited number of isoform specific inhibitors is required. Using imipramine as the probe substrate, Chen et al. confirmed that CYP3A7 is the major CYP isoform in human prenatal hepatic tissues [19]. However, it seems that imipramine may not be an ideal probe in analyzing postnatal liver samples due to the confounding effects from other CYP isoforms capable of carrying out the N-demethylation reaction.
5.2. Absolute CYP3A7 quantification and the search for (potential) biomarker substrates
Commercial polyclonal antibodies raised against a CYP3A5 peptide or a CYP3A4 peptide (recognizing both CYP3A4 and CYP3A7) are available. However, while CYP3A5 protein expression can be distinguished from CYP3A7 by immunoquantitation based on Western blot, no effective immunological approaches have been developed for discriminating between CYP3A4 and CYP3A7 due to the lack of availability of isoform-specific antibodies, as noted above. Additionally, exclusive marker enzyme activities or specific inhibitors still do not exist for CYP3A7 as of our writing [28]. In this light, the routine assessment of CYP3A7 concentration in fetal human liver microsomes (fHLMs) and post-natal human liver microsomes (HLMs) using proteomics methods holds great promise [29]. This, combined with approaches to measure the specific enzymatic activity of CYP3A7 in HLMs could help improve the predictivity of neonatal and pediatric pharmacokinetic models of drug disposition. However, for this to be the case, endogenous biomarkers of CYP3A7 activity need to be established and validated. In furtherance of this goal, approximative approaches based on the combination of substrate regioselectivity of the isoforms with other associated strategies have been reported and may yet prove of some value [28]. In conjunction with multivariate regression model or inhibitors, DHEA-S 16α-hydroxylation, testosterone 6β-hydroxylation, and testosterone 2α-hydroxylation have all been used as marker reactions in an attempt to identify the catalytic activity of CYP3A7 (Table 2), as discussed above.
Table 2.
(Potential) biomarker compounds of CYP3A7.
6. Inhibitors/activators of enzymatic activity of CYP3A7
Discovery of an isoform-specific inhibitor is vital for identifying CYP3A7 activity in vivo and elucidating its catalytic mechanism. An additional aspect that makes the study of CYP3A7 inhibitors attractive is that both endogenous and xenobiotic inhibitors could be involved in the regulation of CYP3A7 metabolism in vivo. To date, although a variety of CYP3A7 inhibitors (many of which are drugs) have been reported (Table 3), important parameters such as the type of inhibition and specificity of isoform interaction need further investigation to find a CYP3A7-specific inhibitor. In a recent publication, Godamudunage et al. examined the differential inhibition and binding affinity between CYP3A7 and CYP3A4 for thirteen different azole antifungal compounds, several of which are commonly prescribed in the neonatal intensive care unit [87]. The authors observed that all the imidazole-containing azoles bound to both enzymes via a typical type II coordination to the heme iron. Additionally, they were found to inhibit both enzymes with IC50 values ranging from 180 nM for clotrimazole, the most potent inhibitor, up to the millimolar range for imidazole. For most of the azoles tested, CYP3A4 was inhibited more strongly than CYP3A7, with clotrimazole being the least selective inhibitor examined (less than 1.5-fold), and econazole the most selective (more than 12-fold). Of the 1,2,4-triazole-containing azoles examined in this study, only posaconazole inhibited CYP3A7, again less potently than CYP3A4. In general, the same azole compounds were observed to inhibit both enzymes, albeit more weakly with CYP3A7 than with CYP3A4 [87]. Given the results of this study, and the other information available in the literature, it is clear that the inhibition profile of CYP3A7 is highly similar to that of CYP3A4/5, certainly making finding an isoform specific inhibitor a difficult task for the foreseeable future.
Table 3.
Inhibitors of human CYP3A7 activity.
| Inhibitor | Substrate reaction | Reference |
|---|---|---|
| Triacetyloleandomycin | N-demethylation | [19] |
| α-naphthoflavone | Imipramine N-demethylation | [19] |
| Troleandomycin | all-trans-retinoic acid 4-hydroxylation | [62] |
| Estradiol | Carbamazepine 10,11-epoxidation | [32] |
| Triazolam | Testosterone 6β-hydroxylation | [68] |
| Amprenavir | Testosterone 6β-hydroxylation | [36] |
| Indinavir | Testosterone 6β-hydroxylation | [36] |
| Nelfinavir | Testosterone 6β-hydroxylation | [36] |
| Ritonavir | Testosterone 6β-hydroxylation | [36] |
| Saquinavir | Testosterone 6β-hydroxylation | [36] |
| Azamulin | 7-benzyloxy-4-trifluoromethylcoumarin | [88] |
| Ketoconazole | 7-benzyloxy-4-trifluoromethylcoumarin | [87,88] |
| Δ9-tetrahydrocannabinol | Diltiazem N-demethylation | [76] |
| Cannabidiol | Diltiazem N-demethylation | [76] |
| Cannabinol | Diltiazem N-demethylation | [76] |
| Olivetol | Diltiazem N-demethylation | [76] |
| Hop-Containing products | Dibenzylfluorescein, 7-ethoxy-3-cyanocoumarin, 7-methoxy-4-trifluoromethylcoumarin | [89] |
| West African medicinal and food plants | Dibenzylfluorescein | [90] |
| Melatonin products | NA | [91] |
| Astemizole Cisapride Clotrimazole Cyclosporine A Erythromycin Ketoconazole Mibefradil Midazolam Nicardipine Nifedipine Nimodipine Terfenadine Troleandomycin Verapamil |
7-benzyloxy-4-trifluoromethylcoumarin O-dealkylation | [92] |
NA, not available.
In contrast, several compounds enhancing CYP3A7 enzyme activities have also been reported. This is perhaps not surprising, given the well-known and long described nature of allosteric enzyme activation in the CYP3A family [93-97].
While not tested against an exhaustive set of substrates, these compounds were found to activate at least two reactions, that is: 10,11-epoxidation of carbamazepine and O-dealkylation of 7-benzyloxy-4-trifluoromethylcoumarin (Table 4).
Table 4.
Activators of human CYP3A7.
It should be recognized that, in at least a few cases, a molecule can act either as an inhibitor or activator, depending on the substrates and reactions considered. For instance, α-naphthoflavone inhibited CYP3A7-dependent imipramine demethylation [19] while activating CYP3A7-mediated 7-benzyloxy-4-trifluoromethylcoumarin metabolism [92]. This curious type of behavior is likely due to the nature of the multiple ligand binding sites within the CYP3A7 active site cavity, as has been demonstrated for CYP3A4 [98-100].
7. Inducers/suppressors of the expression of CYP3A7
Expression of CYP3A7 can be induced or inhibited in response to the presence of various compounds, thereby affecting the catalytic activities of CYP3A7 in the liver at any particular point in time. The reported inducers/suppressors of CYP3A7 are summarized in Table 5. Interestingly, the majority of compounds described in the literature tend to have the effect of increasing the expression of the CYP3A7 gene. Some natural products can also exhibit either induction or suppression of the CYP3A7 gene. For instance, apple polyphenols upregulated, whereas St. John's wort downregulated, the expression of CYP3A7 [101,102] (Table 5).
Table 5.
Inducers and suppressors of the expression of CYP3A7.
| Chemical | Host | Up/Down regulation of expression | Reference |
|---|---|---|---|
| Dexamethasone | HepG2/fetal hepatocytes | Up | [101,103] |
| Betamethasone | Fetal hepatocytes | Up | [103] |
| Prednisolone | Fetal hepatocytes/HepG2 | Up | [103,104] |
| Methylprednisolone | Fetal hepatocytes | Up | [103] |
| Fludrocortisone | Fetal hepatocytes | Up | [103] |
| Rifampicin | HepG2 | Up | [104] |
| Phenytoin | HepG2 | Up | [104] |
| Clotrimazole | HepG2 | Up | [104] |
| Cyclosporine | HepG2 | Up | [104] |
| Carbamazepine | HepG2 | Up | [104] |
| Phenobarbital | HepG2 | Up | [104] |
| 4-monochloropheno | HepG2 | Up | [105] |
| Beclomethasone dipropionate | DPX2 | Up | [106] |
| Endosulfan | HepG2 | Up | [107] |
| Chemotherapeutic agents (cisplatin, etoposide or doxorubicin) | HepG2 | Up | [108] |
| Pentachlorophenol | HepG2 | Up | [109] |
| U0126 | HepG2 | Up | [110] |
| Maternal smoking | Fetal livers | Up | [111] |
| Apple polyphenols extract | Colon adenoma LT97 cell | Up | [102] |
| Neonicotinoids (thiacloprid, thiamethoxam, imidacloprid) | H295R cells | Up | [112] |
| Pargyline | Human primary fetal liver cells, HepG2 | Up | [113] |
| Chronic cadmium exposure | HepG2 | Up | [114] |
| Rifampicin | HepG2, hepatocytes | Up | [115,116] |
| Troleandomycin | HepG2 | Up | [116] |
| Erythromycin | HepG2 | Up | [116] |
| Phenobarbital | HepG2 | Up | [116] |
| Phenobarbital-like inducers | HepG2 | Up | [116] |
| Lovastatin | HepG2 | Up | [116] |
| 5-aza-2′-deoxycytidine | HepG2 | Up | [117] |
| St. John's wort | HepG2 | Down | [101] |
| Troglitazone | HepG2 | Down | [118] |
| Acute cadmium exposure | HepG2 | Down | [114] |
Up: upregulated; down: downregulated.
8. Physiological roles
8.1. Relevance to development and fetal protection
CYP3A7 catalyzes numerous reactions involving both xenobiotic and endogenous substrates and hence has potential roles in both protecting the fetus from exposure to drugs and maintaining the balance of various steroid hormones [62,119].
In regards to its role in regulating endogenous steroids, it has been known for some time that excessive DHEA-S is involved in intrauterine growth retardation, which often leads to premature birth [120,121]. Since CYP3A7 has a high catalytic activity for the 16α-hydroxylation of DHEA-S [43,64,84], it can protect the fetus from the buildup of toxic levels of this steroid and prolong the fetal maturation process. CYP3A7 also participates in the synthesis of estrogen, a primary female hormone during pregnancy, that requires oxidized DHEA-S as a precursor [122]. Through adjustment of CYP3A7 enzymatic activity, the fetus can regulate the placental production of estradiol from DHEA-S in order to protect it from exposure to excessive estradiol during development [123]. Consistent with this, DHEA-S and other sulfate conjugated steroids have been reported to activate the catalytic activity of CYP3A7, but not CYP3A4 [32]. Therefore, in general, elevating CYP3A7 activity provides a protection against hyperestrogenization by 16α-hydroxylation of estrogen precursors and estrogens themselves [124]. As a corollary, it follows that changes in the expression and activity of CYP3A7 in utero may have a profound influence on steroid biosynthesis, thereby affecting normal fetal development and the maintenance of pregnancy.
Similarly, inhibition of CYP3A7's role in atRA metabolism can have drastic effects on the fetus since atRA is essential for normal human growth and development. Both excessive and insufficient atRA can lead to birth defects. Indeed, excessive atRA in particular has be demonstrated to exhibit potent embryotoxicity and teratogenicity [125,126]. CYP3A7 catalyzes atRA to form several much less toxic hydroxylated metabolites, including the major metabolite 4-OH-tRA. In this regard, CYP3A7 has important implications in protecting the fetus against atRA-induced embryotoxicity and teratogenicity and, therefore, interference in this function can have profoundly negative consequences on the fetus, up to and including premature termination of the pregnancy [62].
8.2. CYP3A7 interaction with environmental xenobiotics
Given its important role in the metabolism of endogenous hormones that are essential for fetal growth and development, and the promiscuous nature of the CYP3A family, it should come as no surprise that many environmental xenobiotics can have deleterious effects on the metabolism of these same CYP3A7 substrates. In a seminal study on the effects of insecticides on fetal development, Caron-Beaudoin et al. used a unique co-culture model of fetoplacental steroidogenesis and found that metabolism of neonicotinoids (thiacloprid, thiamethoxam and imidacloprid) by CYP3A7 impeded DHEA-S 16α-hydroxylation, which is normally transformed into estriol by placental aromatase, thereby presenting risks for normal fetal development in pregnant mothers that might have been exposed to excessive amounts of these pesticides [112].
Likewise, other work has demonstrated that certain environmental pollutants, such as procarcinogens, can also be metabolically activated by CYP3A7. Many carcinogens exhibit their corresponding toxic roles via bioactivation conducted by biotransformation enzymes, particularly CYPs. For example, aflatoxin B1 (AFB1) exerted its carcinogenic toxicity after being metabolized to the reactive AFB1-8,9-exoepoxide [127,128]. In the fetus, CYP3A7 was found to be predominantly responsible for the bioactivation of some carcinogens such as AFB1, sterigmatocystin, and AFG [129-133]. Additionally, the overexpression of human CYP3A7 has been observed to increase aflatoxin-induced mutations in murine models [134]. When taking these data into account, Wells et al. postulated that the high expression of CYP3A7 in fetal livers, coupled with a low capacity in detoxification of AFB1, could result in a high risk for a fetus via transplacental exposure [135].
8.3. Relevance to disease
CYP3A7 has also been reported to be associated with several disease states, including congenital adrenal hyperplasia [136], chronic lymphocytic leukemia, both breast and lung cancer [137], ovarian endometriosis [138], polycystic ovary syndrome [139], respiratory distress syndrome [140], congenital anomalies [60], endometrial cancer [141], and end-stage liver diseases [142], primarily ascribed to the phenotypic modulation of DHEA-S levels by CYP3A7 variant alleles (especially CYP3A7*1C).
In contrast, although the CYP3A7*1C allele was associated with decreased bone mass at the lumbar spine in postmenopausal women, no direct association was found between CYP3A7*1C and serum DHEA-S levels, implying this genetic variation might influence bone mass via other CYP3A7 hormonal substrates known to protect bone [143]. Gender may play a role in the relationship between the CYP3A7*1C polymorphism and serum DHEAS level. In line with Bacsi et al., Smit et al. found that in a separate analysis for females no statistically significant difference was reached, although CYP3A7*1C was associated with lower serum DHEA-S levels in the whole cohort overall [143,144]. In addition, females showed significantly lower DHEA-S levels than males regardless of wild type or CYP3A7*1C variant [144]. The gender differences in the association between CYP3A7 expression and DHEA-S levels suggests that the impact of CYP3A7 mutations on serum DHEA-S levels is stronger in males than in females, for as yet unknown reasons [143].
Genetic variation in CYP3A7 has also been described to be involved in daily drug dose requirements as well as having a profound effect on levels of endogenous hormones [77,136,145,146]. CYP3A7*1C carriers required a 1.4- to 1.6-fold higher cyclosporine daily dose during the first year after transplantation in a group of Caucasian renal or lung transplant recipients, due to increased cyclosporine metabolism [77]. In another study, the CYP3A7*1C carriers required a significantly decreased glucocorticoid dose ascribable to CYP3A7*1C lowering androgen levels [136].
In adults harboring the CYP3A7*1C allele, CYP3A7 could be responsible for up to 80% of the total biotransformation of retinoic acid [47] and, therefore, in these individuals hepatic CYP3A7 expression could be a determinant for the outcome of retinoic acid therapy. In premenopausal women, CYP3A7*1C is associated with lower urinary estrone levels, and thus may be related to increased breast cancer risk [146,147]. Finally, CYP3A7*1C expression in a cohort of adult women has also recently been ascribed to increase the risk of birth control hormone failure due to increased hormone metabolism [148].
9. Genetic regulation of CYP3A7 expression
The enzymatic activity of CYP3A7 is developmentally differential with high variability among individuals, and most of this variability has been attributed to genetic factors [46,53,144]. The regulation of CYP3A7 has turned out to be quite complex and involves a number of genetic control elements. Indeed, Sp1, Sp3, HNF-3β, USF1, XREM, and NFκB have all been implicated in regulation of the CYP3A7 gene [149,150].
9.1. Variant alleles
Several variant alleles of the CYP3A7 gene have been identified, including 2 coding region and 4 noncoding region variants (Table 6).
Table 6.
Alleles of CYP3A7.
| Allele | Protein | SNPa | Position | Effect | Ethnic and frequency % |
Reference |
|---|---|---|---|---|---|---|
| CYP3A7*1A | CYP3A7.1, CYP3A7.1L | None | [12,46] | |||
| CYP3A7*1B | CYP3A7.1 | −211 C > T | Promoter | Increased expression | Brazilian, 0.4 Caucasians, 1.0 African Americans, 0 |
[151] [47,152] [152] |
| CYP3A7*1C | CYP3A7.1 | −186G > T; −179T > A; −177T > C; −176A > T; −165T > G; −157T > A; −127 A > C | Promoter | Increased expression | Brazilian, 1.2–3.3 Caucasians, 2.7–3.9 African Americans, 6.0 Korean, 0 Jordanian, 1.7 Samples Brussels, 4 Liver samples from UK, 6 |
[136,151] [47,77,139,143,144,152] [152] [153] [154] [155] [156] |
| CYP3A7*1D | CYP3A7.1 | +13 G > A | +13 | Brazilian, 0.4 Caucasian, 1 African Americans, 0 |
[151] [47,152] [152] |
|
| CYP3A7*1E | CYP3A7.1 | +55 G > A | +55 | Brazilian,2.2 Fetal livers, 3.7 African Americans, 8 Caucasians, 0 Caucasian-Americans, 2.6 |
[151] [60] [153] [152,153] [60] |
|
| CYP3A7*2 | CYP3A7.2 | 26041C > G | Exon 11 | T409R | Koreans, 26.0 Caucasians, 8.0–10.5 Tanzanians, 62.0 Chinese, 28 Saudi Arabian, 17 Brazilian, 25 Fetal livers, 37.8 |
[153] [53,157] [53] [53] [53] [151] [60] |
| CYP3A7*3 | CYP3A7.3 | 4011insT | Exon 2 | Frameshift | Korean, 0.21 | [153] |
The numbers are relative to the transcription start site in the CYP3A7 gene, which is defined as +1.
Most of these variants appear as heterozygous with the wild-type allele and some of them have been associated with altered drug clearance and response and disease susceptibility [47,77,136,142]. The most frequent allele, CYP3A7*2, is caused by C > G transversion in exon 11 which results in the replacement of Thr409 with Arg, with highly variable interethnic occurrence frequencies varying from approximately 8% in Caucasians to 62% in Tanzanians. The frequency of CYP3A7*2 in Asians was found to be 26–28% [53,153]; statistically higher than in Caucasians, but lower than that in Tanzanians. From the data acquired to date, it appears that the CYP3A7*2 allele is the only allele that occurs more frequently than the wild-type, but this has only been found to be the case in the Tanzanian population [53]. In HEK293 cells, there were no significant discrepancies in expression levels between CYP3A7.1 and CYP3A7.2. Fetal livers homozygous for CYP3A7*2 had similar or higher CYP3A7 protein contents than CYP3A7*1 carriers. CYP3A7.2 was found to be more active in the metabolism of DHEA, luciferin BE, and alprazolam [53]. In contrast, Leeder et al. did not see significant differences in DHEA metabolism between CYP3A7*1 and CYP3A7*2 using fetal liver microsomes [60]. Differences in ethnicity, sample size and analytical methods may have led to this discrepancy between the two studies. Because CYP3A7*2 possibly leads to higher enzymatic activity, ethnic differences in CYP3A7-mediated fetal drug metabolism or detoxification merit further study.
The CYP3A7*3 allele is derived from a thymidine insertion (4011insT) in exon 2 of CYP3A7, leading to a truncated CYP3A7 protein at the 55th residue, thus possibly producing a null phenotype. The CYP3A7*3 allele seems to only occur in Korean subjects, and with a very low frequency (0.21%). No reports have linked CYP3A7*3 with alterations in CYP3A7 expression or catalytic activity [153]. The rarity in loss-of-function mutations demonstrates the functional conservation and crucial role that CYP3A7 plays in the developing fetus.
None of the variant alleles with SNPs in the coding regions of CYP3A7 described thus far have accounted for the extent of observed variability in expression [158]. These coding variants may contribute to but are not likely to be a major driver of the inter-individual differences observed in CYP3A7 expression and/or catalytic activity.
The CYP3A7*1B allele has frequencies of 1% in Caucasians and < 1% in other ethnic groups (Table 6) [47,151,152]. Although less frequent, CYP3A7*1B was associated with increased CYP3A7 expression in livers, but the mechanism of action is at present unknown [47].
The CYP3A7*1C allele occurs in the proximal promoter of CYP3A7 through replacing an approximately 60-bp stretch (−127/−186) of the CYP3A7 promoter with the corresponding region in CYP3A4 [152]. The CYP3A7*1C allele thus possesses the transcription binding site CYP3A4-ER6 which has a higher affinity for PXR and CAR, resulting in a significant expression of CYP3A7 well into adulthood [47]. In general, CYP3A7*1C allele frequencies range from approximately 1.7%–6% depending on the ethnic group being considered (Table 6). Interestingly, CYP3A7*1C was not found to occur in Koreans [153].
CYP3A7 has been shown to be expressed in more than 10% of adult livers and intestines [47,50,115,152,156,159]. However, not all of these individuals expressing CYP3A7 carry the CYP3A7*1C allele [47,152,156]. Meanwhile, some CYP3A7*1C carriers exhibited no CYP3A7 expression at all [152,156]. These data imply that CYP3A7*1C carriage is neither necessary nor sufficient for the expression of CYP3A7 in adult livers.
CYP3A7*1D is a rare variant with allele frequencies similar to those of CYP3A7*1B (1%≤). Unlike CYP3A7*1C, CYP3A7*1D was not associated with high CYP3A7 expression in adults [47]. No further information on CYP3A7*1D is available at present.
CYP3A7*1E showed the highest allele frequency in African Americans (8%). Although Leeder et al. found one Caucasian sample was heterozygous for the CYP3A7*1E allele, Kuehl et al. and Lee et al. did not detect the CYP3A7*1E allele in Caucasians [60,152,153]. No apparent relationship existed between CYP3A7*1E and DHEA16α-OH activity [60]. However, neonatal CYP3A7*1E resulted in higher rate of respiratory distress syndrome [140].
9.2. RNA splicing
A pseudogene, CYP3AP1, is located between CYP3A7 and CYP3A5 (Fig. 1A). Exons 2 and 13 of CYP3AP1 splice to the 3’ end of canonical CYP3A7 at 12 bp upstream from the translational stop codon, generating a novel CYP3A7 variant, CYP3A7-3AP1. The CYP3A7-3AP1 allele contains 15 exons encoding a longer CYP3A7 protein (CYP3A7.1L). In the putative protein, the last 4 canonical amino acids are replaced by a completely different stretch of 36 amino acids. Therefore, CYP3A7.1L is 32 amino acids longer than the product (CYP3A7.1) of canonical CYP3A7. As shown in Fig. 1B, intron 1-exon 2 region of 3AP1 was believed to be derived from the corresponding region of CYP3A7. However, the C flanking the heptanucleotide spacer (TTTGTAG) of CYP3A7 intron 1 is a G in CYP3AP1. This nucleotide change creates an alternative splicing acceptor site of 3AP1 exon 2; thus, it converts the heptanucleotide spacer into a part of the 3AP1 exon 2, leading to a frameshift mutation of the two captured exons and thus coding a unique 36 amino acid sequence rather than the canonical CYP3A exons 2 and 13. Heterologous expression in yeast demonstrated that CYP3A7.1L has a specific activity similar to that of CYP3A7.1; whereas in the yeast expressing yeast P450 reductase, CYP3A7.1L efficiently catalyzes DHEA 7α-, 16α-, and 7β-hydroxylations, while CYP3A7.1 preferentially catalyzes DHEA 16α-hydroxylation, possibly due to the difference in the C-terminals of two enzymes [15,46].
The CYP3A7-3AP1 transcript occurs in various adult and fetal tissues and the splicing is regulated in a developmental- and tissue-specific manner. Furthermore, CYP3A7-3AP1 mRNA levels tend to be higher in fetal than in adult livers. In most cases, CYP3A7-3AP1 expression is lower than that of CYP3A7, but the relative levels can vary between tissues [15,46]. The CYP3A7-3AP1 allele is polymorphic because the splicing is abrogated by one base mutation (T > A) at position −6 upstream from the heptanucleotide spacer (TTTGTAG) (Fig. 1B). The frequency of the allele exhibits large interethnic differences: Caucasians (8%), Chinese (28%), and African Americans (59%) [53]. Although CYP3A7.1L is assumed to have a vital role in developmental, physiological and toxicological processes [53], there is little empirical evidence to support this idea. Currently, the real functions of CYP3A7.1L and its contribution to drug metabolism remain elusive.
In addition to the CYP3A7-3AP1 allele, in fetal brain another CYP3A7 variant was also discovered, which contains sequences similar to the 3′ end of CYP3A4 intron 12 located upstream of the novel exon 13. However, no further information on this CYP3A7 variant has been reported since its initial discovery [15]. Based on the data obtained from the two variants, it is reasonable to infer that there may be additional human CYP3A7 variants awaiting discovery. The identification of potential CYP3A7 splice variants and investigation of their role in drug metabolism and fetal protection will no doubt be an intriguing topic for future research efforts.
9.3. Epigenetics
Epigenetics is an important factor in modulating the expression of many genes involved in essential liver processes [117]. By analyzing epigenetic modifications in liver tissues, He et al. illustrated that the dynamic variations of dimethylation of histone H3 at lysine 4 (H3K4me2) and H3K27me3 play a crucial role in the developmental transition of CYP3A7 to CYP3A4 [160]. Occupancy of H3K4me2 on the human CYP3A7 promoter (−163/+103) and enhancer region (−4054/−3421 and −6265/−6247) overlapped with the glucocorticoid receptor (GR) binding site. The enriched H3K4me2 in these regions was induced by pargyline with an HNF4α or GR binding site in the CYP3A4/3A7 gene to transactivate the corresponding genes [113].
Furthermore, cytosines in the proximal CYP3A7 promoter were found to be hypomethylated in neonates relative to adolescents. In contrast, a −383 cytosine of CYP3A4 was hypermethylated in liver samples from neonates compared with adolescents. The methylation status of cytosine in the CYP3A4 and CYP3A7 proximal promoters was observed to correlate with changes in developmental expression of mRNA for the two enzymes [161].
9.4. Cis- and trans-elements
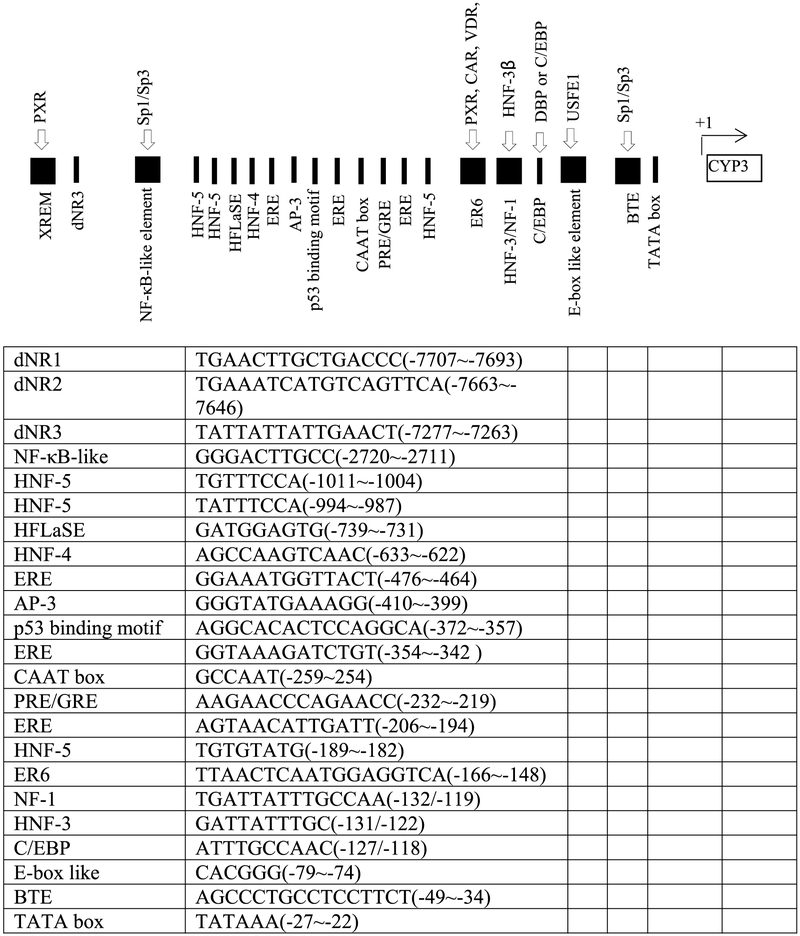
The developmental expression of transcription factors (for instance PXR, CAR, Sp1/Sp3, USF1, NF1, HNF-3β, GR and C/EBP) as well as the interaction between these factors with the CYP3A7 promoter may contribute to the developmental expression pattern observed with the CYP3A7 gene [150,162-166]. The putative responsive motifs of transcription factors in the promoter region of CYP3A7 are shown in Fig. 2.
Fig. 2.
Putative or confirmed CYP3A7 transcriptional factors and their responsive elements. The data are collected from Refs. [45,149,150,164,167,168], respectively.
9.4.1. Ontogeny of CYP3A7 and CYP3A4
In HepG2 cells, a nuclear factor κB (NF-κB)-like element (GGGACTTGCC) (−2326/−2297) imparted transactivation of the CYP3A7 promoter via responsiveness to specificity proteins Sp1 and Sp3. Conversely, a CYP3A4 NF-κB-like element (GGGACTTGAC) did not exhibit transactivation due to lacking the ability to bind Sp1/Sp3, as ascribed to a one base mutation (underlined) relative to that of CYP3A7. Sp1/Sp3 acted as activators of the CYP3A7 gene via the NF-κB-like element [150]. Saffer et al. described the expression of CYP3A7 under the control of Sp1 as regulated in a cell type- and differentiation-specific manner, with a high level in fetal livers compared with adult livers [166], implying that Sp1/Sp3 and the CYP3A7 NF-κB-like motif may contribute to the fetal-specific expression of CYP3A7. It should be noted that full activation of CYP3A7 expression also required participation of the proximal promoter. The mechanism by which Sp1/Sp3 transactivate the CYP3A7 promoter from circa −2.3 kb remains elusive. The CYP3A7 NFB-like element might also be recognized by other yet to be identified factors, such as complex D, which may modulate the binding activity of Sp1 for the NFB-like element [150]. Therefore, other control elements interacting with the NFB-like motif may also be essential for distal activation.
In all cell lines tested (HepG2, WRL68 and Caco-2TC7), CYP3A4 but not CYP3A7 exhibited responsiveness to the D-element-binding protein (DBP) [164], in combination with the exclusive expression of DBP in adult liver [169], this may account for the occurrence of CYP3A7 and CYP3A4 in fetal and postnatal liver, respectively [164].
He et al. discovered that the expression level of HNF4α was most correlated with CYP3A4 expression in the adult liver [160]; however, the expression level of GR was highly associated with CYP3A7 expression in fetal liver. Pang et al. suggested that CYP3A7 is developmentally regulated in mouse liver primarily by glucocorticoids through the GR [170]. In fetal hepatocytes, CYP3A7 was found to be induced by dexamethasone but not by rifampicin, suggesting that induction was mediated by GR rather than pregnane X receptor (PXR) [103,171]. These data demonstrate that HNF4α, GR, and epigenetic changes of H3K4me2 and H3K27me3 are all associated with the ontogenic expressions of CYP3A4/3A7, and they simultaneously demonstrate that the regulation of CYP3A7 expression is very complex. Further work regarding the expression and regulation of CYP3A7 should shed some light on the mechanisms responsible for the ontological switch between CYP3A7 and CYP3A4.
As a classical example of sequential regulation of liver-enriched transcriptional factors, NF1, C/EBPα, as well as liver activator protein (LAP) and DBP, respectively, were active in early fetal, late fetal and postnatal liver, where these factors sequentially activated the human class I alcohol dehydrogenase (ADH) gene family members ADH1, ADH2 and ADH3 at the corresponding liver development stage. LAP and DBP maintained high-level ADH gene family transcription in postnatal liver [172]. Similar mechanisms may also control CYP3A7 expression at various stages of hepatic growth and development.
Taking all of these data into account, we can speculate that Sp1/Sp3, GR and their respective responsive motifs may be responsible for the ontogeny of CYP3A7; DBP, HNF4α and their respective responsive elements may be involved in the ontogeny of CYP3A4; and DBP and its responsive DNA may turn on and maintain the expression of CYP3A4 in postnatal livers. The epigenetic changes of H3K4me2 and H3K27me3 may also have some influence on the ontogeny of CYP3A4/CYP3A7.
9.4.2. Interindividual variation in expression of CYP3A7
The PXR and constitutive androstane receptor (CAR) are hitherto the most intensively studied transcription factors in modulating CYP3A7. Both were expressed at highly variable levels during early fetal life, which correlates with the highest level of CYP3A7 expression [158,162]. PXR binds as a heterodimer with the retinoic acid receptor (RXR) to the distal xenobiotic responsive enhancer module (XREM) and proximal responsive motif (ER6), and thus activates the transcription of CYP3A7 [149]. Similar to CYP3A4, the distal CYP3A7 XREM encompasses two conserved PXR binding sites (dNR1 and dNR2), followed by a third (dNR3), located 368 base pairs downstream of XREM. The three distal PXR binding-sites are significantly conserved between the CYP3A4 and CYP3A7 promoters with only two base pair differences [149,173]. In C3A cells, the 5′ truncated promoters (containing ER6 alone, or dNR3 plus ER6) conferred only minimal PXR-mediated induction. However, the inducibility of the promoter containing dNR1, dNR2 plus ER6 was equivalent to that of the contact promoter with all four elements (dNR1, dNR2, dNR3 and ER6), implying the importance of the XREM for PXR dependent response to xenobiotics. The dNR3 element seemed to be negligible for xenobiotic responsiveness [149]. Interestingly, in a human colon carcinoma cell line, Burk et al. found that the CYP3A7 proximal promoter alone was all that was required to induce CYP3A7 gene expression [47]. This discrepancy may be ascribable to the difference in the cell systems examined, where each have different expression patterns of transcriptional factors. The induction functionality of CYP3A7 XREM alone has not yet been examined, whereas CYP3A4 XREM alone showed only partial activation [173]. It should be noted that the 5′-flanking regions up to approximately −8.8 kb are highly conserved between CYP3A4 and CYP3A7 [149]. We infer from this that there is cooperativity between the XREM and the ER6 of CYP3A7, which mediate the transcriptional response to xenobiotics through PXR, similarly to CYP3A4 [173].
Two kinds of ER6 elements [ER6-JMP (TTAACTcaatggAGGTCA) [174] and ER6-Itoh (TTAACTcaatggAGG-CA) [174,175] have been reported, which differ in only one base at the 3’ half site. The PXR:RXR complex can bind ER6-JMP but not ER6-Itoh [174,175], and the binding has been regarded as the mechanism underlying CYP3A7 expression [47]. This suggests a possible polymorphism of CYP3A7-ER6 responsiveness to activators.
The CYP3A7*1C allele is characterized by replacing 60 bp of the CYP3A7 proximal promoter with the corresponding region of the CYP3A4 promoter. This replacement produces variation in seven bases within the proximal CYP3A7 promoter [152], two (−165T > G; −157T > A) of which fall into ER6 motif [47], one (−127A > C) lies within the NF1/HNF3 binding site [176], and the remaining four occur in the vicinity of ER6. Therefore, the CYP3A7*1C promoter contains both the ER6 and NF1/HNF3 binding sites of the CYP3A4 gene. Postnatal CYP3A7 expression associated with the CYP3A7*1C allele may be ascribable to the higher affinity of binding of PXR for CYP3A4-ER6 compared with CYP3A7-ER6 [47,176]. The variation occurring in the CYP3A7*1C promoter results in increased activation by PXR and CAR in vitro and aberrant postnatal expression of CYP3A7 in vivo, suggesting that this region may be important for developmentally differential regulation [47]. CYP3A7*1C ER6 was indistinguishable, in terms of PXR regulation, from the CYP3A7*1C promoter with all seven mutations. The presence of CYP3A7*1C ER6 (CYP3A4-ER6) was necessary, and sufficient, for PXR-dependent activation of CYP3A7*1C. Song et al. suggested that PXR binding is dependent on the genomic context and PXR activators may modulate such binding [177].
In contrast to PXR, CYP3A4-ER6 alone is necessary, but not sufficient, for CAR-mediated activation of CYP3A7*1C. The full CAR-dependent activation of CYP3A7 promoters requires the extra participation of mutated bases in the vicinity of ER6, suggesting the implication of flanking bases in the transcriptional role of CAR and additional transcriptional activators are important for CYP3A4 or CYP3A7 expression that interact specifically with CAR [47]. Contentious with the above findings that ER6 induced the transcription of CYP3A7 [47,174]. Saito et al. confirmed that PXR-responsive ER6 acted as a negative regulatory motif in HepG2 cells [150]. The authors suggested that a transcriptional repressor, COUP-TF, may also bind the CYP3A7-ER6 motif, which possibly gave rise to the negative effect observed with ER6. These data suggest that additional protein-protein or protein-DNA interactions that occur through other elements may be required for full activation.
With respect to other activators, 1,25-dihydroxyvitamin D3 [1,25(OH)2D3] only weakly induced the expression of CYP3A7, but markedly induced CYP3A4. The reason for this difference was due to the fact that the binding of VDR to the proximal CYP3A7 ER6 region was significantly less than that to the proximal CYP3A4-ER6 [178], similar to PXR and CAR.
The developmental NFI are also capable of interacting with the CYP3A4 and CYP3A7 proximal promoters [150,176]. However, the effect is dependent on which isoform gene is examined. NF1 can repress the transcription of CYP3A7 via the CYP3A7 promoter, but can also transactivate CYP3A4 gene [150]. In addition, the expression of NF1 was found to be up-regulated postnatally [163,165]. NF1 may also play a role for the differential expression of CYP3A7 and CYP3A4 [176]. The YY1 element can bind to the CYP3A4 proximal promoter and interact with NF1, but does not bind the CYP3A7 proximal promoter [150]. Therefore, YY1 seems to be involved in the differential regulation of CYP3A4 and CYP3A7 through either activating or repressing transcription in a cell- and promoter-specific manner [176].
In the HepG2 cell line, HNF3 interacted with CYP3A7 but not CYP3A4 [150,176], which is in accordance with the fact that over-expression of HNF3 showed no effect on the CYP3A4 proximal promoter in transfections of HepG2 or HeLa cells [168]. HNF3 was detected in all prenatal liver nuclear extracts and seemed to decrease with increasing gestational age [176]. Furthermore, it was not detected in any postnatal liver samples. This suggests that HNF3 may play a prominent part in prenatal expression of CYP3A7. In another study, the authors demonstrated that, in both hepatic HepG2 and non-hepatic HeLa cells, although HNF-3γ was unable to enhance the expression of CYP3A7, it strengthened the enhancement effect of C/EBPα in the adenoviral infected cells, which was observed to be hepato-specific. The synergism between C/EBPα and HNF-3γ in the CYP3A4 gene has been reported to be possibly attributable to chromatin remodeling by HNF-3γ. The proximal C/EBPα site of CYP3A7 has only one nucleotide variation relative to CYP3A4, and the distal C/EBPα and HNF-3γ binding sites are identical, implying that CYP3A7 may share a similar mechanism with CYP3A4 [168].
In HepG2 cells, USF1 was found to be the major protein bound to the E-box of CYP3A7 or CYP3A4, and functioned as an activator of the CYP3A7 proximal promoter [150]. C/EBP enhanced the binding activity of USF1 to E-box [179], and the level of C/EBP was high in mature hepatocytes relative to hepatoma cells [180]. These data may account for the differential expression of CYP3A4 and CYP3A7 in HepG2 cells.
When the available data is considered en masse, the regulation of CYP3A7 is understood to be very complex, involving a number of transcriptional factors and responsive motifs (as summarized in Table 7). The unknown interactions among these nuclear factors and DNA elements further contribute to the regulation complexity. RNA splicing, allelic genotypes, and developmental changes in the interactions of transcription factors with the CYP3A7 promoter may also be involved in the variable CYP3A7 activity observed.
Table 7.
Genetic regulatory mechanisms: CYP3A4 vs. CYP3A7.
| Nuclear Receptor/Transcription Factor NRNC Abbreviation: | Nuclear Receptor/Transcription Factor Full Name: | CYP3A4 | CYP3A7 |
|---|---|---|---|
| PXR | Pregnane X receptor | Yes | Yes |
| CAR | Constitutive Androstane receptor | Yes | No |
| FXR | Farnesoid X receptor | Yes | No |
| Sp1 | Specificity protein 1 | ? | Yes |
| Sp3 | Specificity protein 3 | ? | Yes |
| USF1 | Upstream stimulatory factor 1 | ? | Yes |
| NF1 | Nuclear factor 1 | Yes | Yes |
| HNF-3β | Hepatocyte nuclear factor 3β | Yes | Yes |
| HNF-4α | Hepatocyte nuclear factor 4α | Yes | Yes |
| GR | Glucocorticoid receptor | No | Yes |
| C/EBP | CCAAT enhancer binding protein | Yes | Yes |
| NFkB | Nuclear factor kappa light chain enhancer of activated B cells | Yes | Yes |
| XREM | Xenobiotic response enhancer module | Yes | Yes |
| DBP | D-element binding protein | Yes | No |
10. Outlook
In the ensuing decades since its original discovery, we have learned that CYP3A7 is a unique member of the CYP3A cytochrome P450 subfamily that predominates in fetal and neonatal livers, as well as some adults expressing the CYP3A7*1C allele. It is involved in the metabolism of endogenous hormones and numerous drugs and xenobiotics, exhibits great interindividual variability, and gradually decreases after birth as the developing infant liver switches to express CYP3A4. The mechanisms underlying the above-mentioned phenomena, however, remain poorly understood. In addition, some results pertaining to the developmentally differential expression of CYP3A7 have yet to be confirmed. Despite this, there is an increasing awareness of the role epigenetics plays in the expression of CYP3A7 and how environmental factors may impact that [181]. Given the strong relationship between premature birth and environmental pollution, future studies may focus on links between CYP3A7 inhibition by pollutants and pre-term and/or low birthweight infants.
Due to the limited availability of fetal and neonatal liver samples, CYP3A7 has not been investigated in depth and has largely lagged behind the study of CYP3A4/5. Clearly, suitable replacements are sorely needed in order to study CYP3A7 and its role in growth and development as well as metabolism. In this regard, the development of new iPS hepatic stem cell line may yet prove valuable. While it has been difficult to produce hepatic iPS cell lines that accurately recapitulate adult hepatic function (primarily due to the presence of a number of fetal markers), they may more accurately represent the function of the developing neonate liver. A hepatic iPS cell line that bore the functions of the developing infant liver could prove invaluable for evaluating new or off-label use of pharmaceuticals in the neonatal population.
Regardless of the model system used, the development of a specific method for discriminating between CYP3A4/5 and CYP3A7 activities and protein content is necessary for the further study of CYP3A7 in the context of fetal liver hepatocytes or microsomes. Newer proteomics methods hold some promise in this regard. Additionally, all-trans-retinoic acid and testosterone have shown some potential application as possible non-invasive endogenous biomarkers of CYP3A7 activity.
Due to the significance of CYP3A7 in estriol biosynthesis during pregnancy and its potential to protect against exogenous toxins, a deep understanding of alleles, alternative RNA splicing, and cis- and trans-regulatory factors potentially involved in modulating CYP3A7 expression is warranted. However, there is little concrete knowledge concerning the molecular mechanisms for regulation of CYP3A7 gene expression. The current available data are inconclusive but imply a hitherto undefined complexity in the molecular regulation of CYP3A7 expression that was unexpected. However, given the clear importance of the glucocorticoid receptor, future work may focus on the possibilities of drug-drug or drug-endogenous metabolite interactions that may inhibit or induce CYP3A7 expression. This is an area of CYP3A7 research that remains underexplored and potentially may reveal some underlying mechanism of toxicity and/or developmental regulation. Additional work is also required to clarify the hierarchy of the signaling cascade regulating developmental changes and variability in the expression of nuclear factors regulating CYP3A7.
The modulation of CYP3A7 expression cannot be fully elucidated unless many aspects like epigenetics, RNA splicing, developmental and differential expression of nuclear factors, the interactions between these factors and the responsive elements are taken into account. Unfortunately, until now, there has been no effective in vitro model for examining the regulation mechanism. Although HepG2 cells have been extensively used to investigate the mechanism of CYP3A7 expression, HepG2 cells are subject to the limitations of the delineating roles of the transcriptional factors and response elements during development. The current available data we have obtained from in vitro cell models likely does not reflect conditions that actually exist in fetal livers and, hence, has only limited utility in understanding CYP3A7 expression in the native context. A rational approach might be to initially understand the developmental expression pattern of the transcription factors in postnatal liver samples derived from different development stages, then employ various cell lines to clarify the interactions between the factors and their respective responsive elements. This strategy may facilitate elucidation of the ontogeny and differential expression of CYP3A7. In any case, given both its importance in human fetal development and drug metabolism, as well as a variety of disease states, the study of this important member of the CYP3A gene family will only increase in the future, and will likely return to us many satisfying intellectual and practical rewards.
Funding Acknowledgements
The research reported in this publication was supported by the National Institutes of Health National Institute of General Medical Sciences (NIH-NIGMS) Award Number R01 GM128508 (JNL) and the National Institute of Allergy and Infectious Diseases Award Number R01 AI150494 (JNL). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Abbreviations:
- CYP3A7
cytochrome P450 3A7
- DHEA-S
dehydroepiandrosterone sulfate
- atRA
and all-trans retinoic acid
- cDNA
protein produced by recombinant complementary DNA system
- HFL
human fetal liver
- fHLMs
human fetal liver microsomes
- Hep G2
hepatocellular carcinoma cell line G2
- Sp1
specificity protein 1
- Sp3
specificity protein 3
- HNF-3β
hepatocyte nuclear factor 3β
- USF1
upstream stimulatory factor 1
- XREM
xenobiotic response enhancer module
- NF-κB
nuclear factor kappa-light-chain-enhancer of activated B cells
- HEK293 cells
human embryonic kidney cell line 293
- SNP
single nucleotide polymorphism
- ER6
nuclear hormone receptor binding motif 6
- PXR
pregnane X receptor
- CAR
constitutive androstane receptor
- GR
glucocorticoid receptor
- HNF-4α
hepatocyte nuclear factor 4α
- NF1
nuclear factor 1
- C/EBP
CCAAT-enhancer-binding protein
- DBP
D-element binding protein
- LAP
liver activator protein
- COUP-TF
chicken ovalbumin upstream promoter-transcription factor (orphan nuclear receptor)
- YY1
yin and yang 1 protein (transcription factor)
Footnotes
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.abb.2019.108078.
References
- [1].Guengerich FP, Cytochrome P450s and other enzymes in drug metabolism and toxicity, AAPS J 8 (1) (2006) E101–E111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Denisov IG, Makris TM, Sligar SG, Schlichting I, Structure and chemistry of cytochrome P450, Chem. Rev 105 (6) (2005) 2253–2277. [DOI] [PubMed] [Google Scholar]
- [3].Ingelman-Sundberg M, Sim SC, Gomez A, Rodriguez-Antona C, Influence of cytochrome P450 polymorphisms on drug therapies: pharmacogenetic, pharmacoepigenetic and clinical aspects, Pharmacol. Ther 116 (3) (2007) 496–526. [DOI] [PubMed] [Google Scholar]
- [4].Ortiz de Montellano PR, Cytochrome P450: Structure, Mechanism, and Biochemistry, 3rd ed., Kluwer Academic/Plenum Publishers, New York, 2005. [Google Scholar]
- [5].Wilkinson GR, Drug therapy - drug metabolism and variability among patients in drug response, N. Engl. J. Med 352 (21) (2005) 2211–2221. [DOI] [PubMed] [Google Scholar]
- [6].Zanger UM, Schwab M, Cytochrome P450 enzymes in drug metabolism: regulation of gene expression, enzyme activities, and impact of genetic variation, Pharmacol. Ther 138 (1) (2013) 103–141. [DOI] [PubMed] [Google Scholar]
- [7].Guengerich FP, Martin MV, Beaune PH, Kremers P, Wolff T, Waxman DJ, Characterization of rat and human liver microsomal cytochrome P-450 forms involved in nifedipine oxidation, a prototype for genetic polymorphism in oxidative drug metabolism, J. Biol. Chem 261 (11) (1986) 5051–5060. [PubMed] [Google Scholar]
- [8].Molowa DT, Schuetz EG, Wrighton SA, Watkins PB, Kremers P, Mendez-Picon G, Parker GA, Guzelian PS, Complete cDNA sequence of a cytochrome P-450 inducible by glucocorticoids in human liver, Proc. Natl. Acad. Sci. U.S.A 83 (14) (1986) 5311–5315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Aoyama T, Yamano S, Waxman DJ, Lapenson DP, Meyer UA, Fischer V, Tyndale R, Inaba T, Kalow W, Gelboin HV, et al. , Cytochrome P-450 hPCN3, a novel cytochrome P-450 IIIA gene product that is differentially expressed in adult human liver. cDNA and deduced amino acid sequence and distinct specificities of cDNA-expressed hPCN1 and hPCN3 for the metabolism of steroid hormones and cyclosporine, J. Biol. Chem 264 (18) (1989) 10388–10395. [PubMed] [Google Scholar]
- [10].Wrighton SA, Ring BJ, Watkins PB, VandenBranden M, Identification of a polymorphically expressed member of the human cytochrome P-450III family, Mol. Pharmacol 36 (1) (1989) 97–105. [PubMed] [Google Scholar]
- [11].Kitada M, Kamataki T, Itahashi K, Rikihisa T, Kato R, Kanakubo Y, Purification and properties of cytochrome P-450 from homogenates of human fetal livers, Arch. Biochem. Biophys 241 (1) (1985) 275–280. [DOI] [PubMed] [Google Scholar]
- [12].Komori M, Nishio K, Ohi H, Kitada M, Kamataki T, Molecular cloning and sequence analysis of cDNA containing the entire coding region for human fetal liver cytochrome P-450, J. Biochem 105 (2) (1989) 161–163. [DOI] [PubMed] [Google Scholar]
- [13].Domanski TL, Finta C, Halpert JR, Zaphiropoulos PG, cDNA cloning and initial characterization of CYP3A43, a novel human cytochrome P450, Mol. Pharmacol 59 (2) (2001) 386–392. [DOI] [PubMed] [Google Scholar]
- [14].Westlind A, Malmebo S, Johansson I, Otter C, Andersson TB, Ingelman-Sundberg M, Oscarson M, Cloning and tissue distribution of a novel human cytochrome p450 of the CYP3A subfamily, CYP3A43, Biochem. Biophys. Res. Commun 281 (5) (2001) 1349–1355. [DOI] [PubMed] [Google Scholar]
- [15].Finta C, Zaphiropoulos PG, The human cytochrome P450 3A locus. Gene evolution by capture of downstream exons, Gene 260 (1–2) (2000) 13–23. [DOI] [PubMed] [Google Scholar]
- [16].Gellner K, Eiselt R, Hustert E, Arnold H, Koch I, Haberl M, Deglmann CJ, Burk O, Buntefuss D, Escher S, Bishop C, Koebe HG, Brinkmann U, Klenk HP, Kleine K, Meyer UA, Wojnowski L, Genomic organization of the human CYP3A locus: identification of a new, inducible CYP3A gene, Pharmacogenetics 11 (2) (2001) 111–121. [DOI] [PubMed] [Google Scholar]
- [17].Thummel KE, Wilkinson GR, In vitro and in vivo drug interactions involving human CYP3A, Annu. Rev. Pharmacol. Toxicol 38 (1998) 389–430. [DOI] [PubMed] [Google Scholar]
- [18].Haehner BD, Gorski JC, Vandenbranden M, Wrighton SA, Janardan SK, Watkins PB, Hall SD, Bimodal distribution of renal cytochrome P450 3A activity in humans, Mol. Pharmacol 50 (1) (1996) 52–59. [PubMed] [Google Scholar]
- [19].Chen H, Brzezinski MR, Fantel AG, Juchau MR, Catalysis of drug oxidation during embryogenesis in human hepatic tissues using imipramine as a model substrate, Drug Metab. Dispos 27 (11) (1999) 1306–1308. [PubMed] [Google Scholar]
- [20].Hakkola J, Raunio H, Purkunen R, Saarikoski S, Vahakangas K, Pelkonen O, Edwards RJ, Boobis AR, Pasanen M, Cytochrome P450 3A expression in the human fetal liver: evidence that CYP3A5 is expressed in only a limited number of fetal livers, Biol. Neonate 80 (3) (2001) 193–201. [DOI] [PubMed] [Google Scholar]
- [21].Komori M, Nishio K, Kitada M, Shiramatsu K, Muroya K, Soma M, Nagashima K, Kamataki T, Fetus-specific expression of a form of cytochrome P-450 in human livers, Biochemistry 29 (18) (1990) 4430–4433. [DOI] [PubMed] [Google Scholar]
- [22].Yang HY, Lee QP, Rettie AE, Juchau MR, Functional cytochrome P4503A isoforms in human embryonic tissues: expression during organogenesis, Mol. Pharmacol 46 (5) (1994) 922–928. [PubMed] [Google Scholar]
- [23].Hines RN, Ontogeny of human hepatic cytochromes P450, J. Biochem. Mol. Toxicol 21 (4) (2007) 169–175. [DOI] [PubMed] [Google Scholar]
- [24].Strougo A, Yassen A, Monnereau C, Danhof M, Freijer J, Predicting the "First dose in children" of CYP3A-metabolized drugs: evaluation of scaling approaches and insights into the CYP3A7-CYP3A4 switch at young ages, J. Clin. Pharmacol 54 (9) (2014) 1006–1015. [DOI] [PubMed] [Google Scholar]
- [25].Wrighton SA, Vandenbranden M, Isolation and characterization of human fetal liver cytochrome P450HLp2: a third member of the P450III gene family, Arch. Biochem. Biophys 268 (1) (1989) 144–151. [DOI] [PubMed] [Google Scholar]
- [26].Zane NR, Chen Y, Wang MZ, Thakker DR, Cytochrome P450 and flavin-containing monooxygenase families: age-dependent differences in expression and functional activity, Pediatr. Res 83 (2) (2018) 527–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Kitada M, Kamataki T, Itahashi K, Rikihisa T, Kato R, Kanakubo Y, Immunochemical examinations of cytochrome P-450 in various tissues of human fetuses using antibodies to human fetal cytochrome P-450, P-450 HFLa, Biochem. Biophys. Res. Commun 131 (3) (1985) 1154–1159. [DOI] [PubMed] [Google Scholar]
- [28].Stevens JC, Hines RN, Gu C, Koukouritaki SB, Manro JR, Tandler PJ, Zaya MJ, Developmental expression of the major human hepatic CYP3A enzymes, J. Pharmacol. Exp. Ther 307 (2) (2003) 573–582. [DOI] [PubMed] [Google Scholar]
- [29].Shuster DL, Risler LJ, Prasad B, Calamia JC, Voellinger JL, Kelly EJ, Unadkat JD, Hebert MF, Shen DD, Thummel KE, Mao Q, Identification of CYP3A7 for glyburide metabolism in human fetal livers, Biochem. Pharmacol 92 (4) (2014) 690–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Lacroix D, Sonnier M, Moncion A, Cheron G, Cresteil T, Expression of CYP3A in the human liver - evidence that the shift between CYP3A7 and CYP3A4 occurs immediately after birth, Eur. J. Biochem 247 (2) (1997) 625–634. [DOI] [PubMed] [Google Scholar]
- [31].Stevens JC, New perspectives on the impact of cytochrome P450 3A expression for pediatric pharmacology, Drug Discov. Today 11 (9–10) (2006) 440–445. [DOI] [PubMed] [Google Scholar]
- [32].Nakamura H, Torimoto N, Ishii I, Ariyoshi N, Nakasa H, Ohmori S, Kitada M, CYP3A4 and CYP3A7-mediated carbamazepine 10,11-epoxidation are activated by differential endogenous steroids, Drug Metab. Dispos 31 (4) (2003) 432–438. [DOI] [PubMed] [Google Scholar]
- [33].Williams JA, Ring BJ, Cantrell VE, Jones DR, Eckstein J, Ruterbories K, Hamman MA, Hall SD, Wrighton SA, Comparative metabolic capabilities of CYP3A4, CYP3A5, and CYP3A7, Drug Metab. Dispos 30 (8) (2002) 883–891. [DOI] [PubMed] [Google Scholar]
- [34].Kandel SE, Han LW, Mao Q, Lampe JN, Digging deeper into CYP3A testosterone metabolism: kinetic, regioselectivity, and stereoselectivity differences between CYP3a4/5 and CYP3A7, Drug Metab. Dispos.: the biological fate of chemicals 45 (12) (2017) 1266–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Kearns GL, Robinson PK, Wilson JT, Wilson-Costello D, Knight GR, Ward RM, van den Anker JN, Cisapride disposition in neonates and infants: in vivo reflection of cytochrome P450 3A4 ontogeny, Clin. Pharmacol. Ther 74 (4) (2003) 312–325. [DOI] [PubMed] [Google Scholar]
- [36].Granfors MT, Wang JS, Kajosaari LI, Laitila J, Neuvonen PJ, Backman JT, Differential inhibition of cytochrome P450 3A4, 3A5 and 3A7 by five human immunodeficiency virus (HIV) protease inhibitors in vitro, Basic Clin. Pharmacol. Toxicol 98 (1) (2006) 79–85. [DOI] [PubMed] [Google Scholar]
- [37].Buratti FM, Leoni C, Testai E, Foetal and adult human CYP3A isoforms in the bioactivation of organophosphorothionate insecticides, Toxicol. Lett 167 (3) (2006) 245–255. [DOI] [PubMed] [Google Scholar]
- [38].Okajima Y, Inaba N, Fukazawa I, Ota Y, Hirai Y, Sato N, Yamamoto G, Itahashi K, Kitada M, Kamataki T, Immunohistochemical and immunoelectron microscopic study of cytochrome P-450 of human fetal livers (P-450HFLa): implications for an onco-feto-placental enzyme, Asia-Oceania J. Obstet. Gynaecol 19 (3) (1993) 329–341. [DOI] [PubMed] [Google Scholar]
- [39].Schuetz JD, Kauma S, Guzelian PS, Identification of the fetal liver cytochrome CYP3A7 in human endometrium and placenta, J. Clin. Investig 92 (2) (1993) 1018–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Kondoh N, Wakatsuki T, Ryo A, Hada A, Aihara T, Horiuchi S, Goseki N, Matsubara O, Takenaka K, Shichita M, Tanaka K, Shuda M, Yamamoto M, Identification and characterization of genes associated with human hepatocellular carcinogenesis, Cancer Res 59 (19) (1999) 4990–4996. [PubMed] [Google Scholar]
- [41].Yaffe SJ, Rane A, Sjoqvist F, Boreus LO, Orrenius S, The presence of a monooxygenase system in human fetal liver microsomes, Life sciences. Pt. 2, Biochemistry, general and molecular biology 9 (20) (1970) 1189–1200. [DOI] [PubMed] [Google Scholar]
- [42].Cresteil T, Beaune P, Kremers P, Flinois JP, Leroux JP, Drug-metabolizing enzymes in human foetal liver: partial resolution of multiple cytochromes P 450, Pediatr. Pharmacol 2 (3) (1982) 199–207. [PubMed] [Google Scholar]
- [43].Kitada M, Kamataki T, Itahashi K, Rikihisa T, Kanakubo Y, P-450 HFLa, a form of cytochrome P-450 purified from human fetal livers, is the 16 alpha-hydroxylase of dehydroepiandrosterone 3-sulfate, J. Biol. Chem 262 (28) (1987) 13534–13537. [PubMed] [Google Scholar]
- [44].Nelson DR, Cytochrome P450 Homepage, (2007) http://drnelson.utmem.edu/CytochromeP450.html (Accessed 07/17/07 2007).
- [45].Hashimoto H, Toide K, Kitamura R, Fujita M, Tagawa S, Itoh S, Kamataki T, Gene structure of CYP3A4, an adult-specific form of cytochrome P450 in human livers, and its transcriptional control, Eur. J. Biochem 218 (2) (1993) 585–595. [DOI] [PubMed] [Google Scholar]
- [46].Rodriguez-Antona C, Axelson M, Otter C, Rane A, Ingelman-Sundberg M, A novel polymorphic cytochrome P450 formed by splicing of CYP3A7 and the pseudogene CYP3AP1, J. Biol. Chem 280 (31) (2005) 28324–28331. [DOI] [PubMed] [Google Scholar]
- [47].Burk O, Tegude H, Koch I, Hustert E, Wolbold R, Glaeser H, Klein K, Fromm MF, Nuessler AK, Neuhaus P, Zanger UM, Eichelbaum M, Wojnowski L, Molecular mechanisms of polymorphic CYP3A7 expression in adult human liver and intestine, J. Biol. Chem 277 (27) (2002) 24280–24288. [DOI] [PubMed] [Google Scholar]
- [48].Canaparo R, Finnstrom N, Serpe L, Nordmark A, Muntoni E, Eandi M, Rane A, Zara GP, Expression of CYP3A isoforms and P-glycoprotein in human stomach, jejunum and ileum, Clin. Exp. Pharmacol. Physiol 34 (11) (2007) 1138–1144. [DOI] [PubMed] [Google Scholar]
- [49].Fakhoury M, Litalien C, Medard Y, Cave H, Ezzahir N, Peuchmaur M, Jacqz-Aigrain E, Localization and mRNA expression of CYP3A and P-glycoprotein in human duodenum as a function of age, Drug Metab. Dispos 33 (11) (2005) 1603–1607. [DOI] [PubMed] [Google Scholar]
- [50].Koch I, Weil R, Wolbold R, Brockmoller J, Hustert E, Burk O, Nuessler A, Neuhaus P, Eichelbaum M, Zanger U, Wojnowski L, Interindividual variability and tissue-specificity in the expression of cytochrome P450 3A mRNA, Drug Metab. Dispos 30 (10) (2002) 1108–1114. [DOI] [PubMed] [Google Scholar]
- [51].Miyauchi E, Tachikawa M, Decleves X, Uchida Y, Bouillot JL, Poitou C, Oppert JM, Mouly S, Bergmann JF, Terasaki T, Scherrmann JM, Lloret-Linares C, Quantitative atlas of cytochrome P450, UDP-glucuronosyltransferase, and transporter proteins in jejunum of morbidly obese subjects, Mol. Pharm 13 (8) (2016) 2631–2640. [DOI] [PubMed] [Google Scholar]
- [52].Murray GI, McFadyen MC, Mitchell RT, Cheung YL, Kerr AC, Melvin WT, Cytochrome P450 CYP3A in human renal cell cancer, Br. J. Canc 79 (11–12) (1999) 1836–1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Rodriguez-Antona C, Jande M, Rane A, Ingelman-Sundberg M, Identification and phenotype characterization of two CYP3A haplotypes causing different enzymatic capacity in fetal livers, Clin. Pharmacol. Ther 77 (4) (2005) 259–270. [DOI] [PubMed] [Google Scholar]
- [54].Wang H, Ping J, Peng RX, Yue J, Xia XY, Li QX, Kong R, Hong JY, Changes of multiple bio transformation phase I and phase II enzyme activities in human fetal adrenals during fetal development, Acta Pharmacol. Sin 29 (2) (2008) 231–238. [DOI] [PubMed] [Google Scholar]
- [55].Sarkar MA, Vadlamuri V, Ghosh S, Glover DD, Expression and cyclic variability of CYP3A4 and CYP3A7 isoforms in human endometrium and cervix during the menstrual cycle, Drug Metab. Dispos 31 (1) (2003) 1–6. [DOI] [PubMed] [Google Scholar]
- [56].Hukkanen J, Mantyla M, Kangas L, Wirta P, Hakkola J, Paakki P, Evisalmi S, Pelkonen O, Raunio H, Expression of cytochrome P450 genes encoding enzymes active in the metabolism of tamoxifen in human uterine endometrium, Pharmacol. Toxicol 82 (2) (1998) 93–97. [DOI] [PubMed] [Google Scholar]
- [57].Williams ET, Leyk M, Wrighton SA, Davies PJ, Loose DS, Shipley GL, Strobel HW, Estrogen regulation of the cytochrome P450 3A subfamily in humans, J. Pharmacol. Exp. Ther 311 (2) (2004) 728–735. [DOI] [PubMed] [Google Scholar]
- [58].Anttila S, Hukkanen J, Hakkola J, Stjernvall T, Beaune P, Edwards RJ, Boobis AR, Pelkonen O, Raunio H, Expression and localization of CYP3A4 and CYP3A5 in human lung, Am. J. Respir. Cell Mol. Biol 16 (3) (1997) 242–249. [DOI] [PubMed] [Google Scholar]
- [59].Kivisto KT, Griese EU, Fritz P, Linder A, Hakkola J, Raunio H, Beaune P, Kroemer HK, Expression of cytochrome P 450 3A enzymes in human lung: a combined RT-PCR and immunohistochemical analysis of normal tissue and lung tumours, Naunyn Schmiedebergs Arch Pharmacol 353 (2) (1996) 207–212. [DOI] [PubMed] [Google Scholar]
- [60].Leeder JS, Gaedigk R, Marcucci KA, Gaedigk A, Vyhlidal CA, Schindel BP, Pearce RE, Variability of CYP3A7 expression in human fetal liver, J. Pharmacol. Exp. Ther 314 (2) (2005) 626–635. [DOI] [PubMed] [Google Scholar]
- [61].Kamdem LK, Streit F, Zanger UM, Brockmoller J, Oellerich M, Armstrong VW, Wojnowski L, Contribution of CYP3A5 to the in vitro hepatic clearance of tacrolimus, Clin. Chem 51 (8) (2005) 1374–1381. [DOI] [PubMed] [Google Scholar]
- [62].Chen H, Fantel AG, Juchau MR, Catalysis of the 4-hydroxylation of retinoic acids by cyp3a7 in human fetal hepatic tissues, Drug Metab. Dispos 28 (9) (2000) 1051–1057. [PubMed] [Google Scholar]
- [63].Shimshoni JA, Roberts AG, Scian M, Topletz AR, Blankert SA, Halpert JR, Nelson WL, Isoherranen N, Stereoselective formation and metabolism of 4-hydroxy-retinoic Acid enantiomers by cytochrome p450 enzymes, J. Biol. Chem 287 (50) (2012) 42223–42232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Lee AJ, Conney AH, Zhu BT, Human cytochrome P450 3A7 has a distinct high catalytic activity for the 16 alpha-hydroxylation of estrone but not 17 beta-estradiol, Cancer Res 63 (19) (2003) 6532–6536. [PubMed] [Google Scholar]
- [65].Takahiro R, Nakamura S, Kohno H, Yoshimura N, Nakamura T, Ozawa S, Hirono K, Ichida F, Taguchi M, Contribution of CYP3A isoforms to dealkylation of PDE5 inhibitors: a comparison between sildenafil N-demethylation and tadalafil demethylenation, Biological & pharmaceutical bulletin 38 (1) (2015) 58–65. [DOI] [PubMed] [Google Scholar]
- [66].Pearce RE, Gotschall RR, Kearns GL, Leeder JS, Cytochrome P450 Involvement in the biotransformation of cisapride and racemic norcisapride in vitro: differential activity of individual human CYP3A isoforms, Drug Metab. Dispos 29 (12) (2001) 1548–1554. [PubMed] [Google Scholar]
- [67].Miranda SR, Meyer SA, Cytotoxicity of chloroacetanilide herbicide alachlor in HepG2 cells independent of CYP3A4 and CYP3A7, Food Chem. Toxicol 45 (5) (2007) 871–877. [DOI] [PubMed] [Google Scholar]
- [68].Ohmori S, Nakasa H, Asanome K, Kurose Y, Ishii I, Hosokawa M, Kitada M, Differential catalytic properties in metabolism of endogenous and exogenous substrates among CYP3A enzymes expressed in COS-7 cells, Biochim. Biophys. Acta 1380 (3) (1998) 297–304. [DOI] [PubMed] [Google Scholar]
- [69].Fang J, Song J, In vitro characterization of the oxidation of a pyridinium metabolite of haloperidol by human placenta: the effect of smoking, J. Pharm. Pharm. Sci 15 (4) (2012) 538–547. [DOI] [PubMed] [Google Scholar]
- [70].Rudek MA, Zhao M, Smith NF, Robey RW, He P, Hallur G, Khan S, Hidalgo M, Jimeno A, Colevas AD, Messersmith WA, Wolff AC, Baker SD, In vitro and in vivo clinical pharmacology of dimethyl benzoylphenylurea, a novel oral tubulin-interactive agent, Clin. Cancer Res 11 (23) (2005) 8503–8511. [DOI] [PubMed] [Google Scholar]
- [71].Sharma S, Ellis EC, Dorko K, Zhang S, Mattison DR, Caritis SN, Venkataramanan R, Strom SC, Metabolism of 17alpha-hydroxyprogesterone caproate, an agent for preventing preterm birth, by fetal hepatocytes, Drug Metab. Dispos 38 (5) (2010) 723–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Klees TM, Sheffels P, Dale O, Kharasch ED, Metabolism of alfentanil by cytochrome p4503a (cyp3a) enzymes, Drug Metab. Dispos 33 (3) (2005) 303–311. [DOI] [PubMed] [Google Scholar]
- [73].Moore CD, Roberts JK, Orton CR, Murai T, Fidler TP, Reilly CA, Ward RM, Yost GS, Metabolic pathways of inhaled glucocorticoids by the CYP3A enzymes, Drug Metab. Dispos 41 (2) (2013) 379–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Thum T, Batkai S, Malinski PG, Becker T, Mevius I, Klempnauer J, Meyer HH, Frolich JC, Borlak J, Tsikas D, Measurement and diagnostic use of hepatic cytochrome P450 metabolism of oleic acid in liver disease, Liver Int. 30 (8) (2010) 1181–1188. [DOI] [PubMed] [Google Scholar]
- [75].Pearce RE, Lu W, Wang Y, Uetrecht JP, Correia MA, Leeder JS, Pathways of carbamazepine bioactivation in vitro. III. The role of human cytochrome P450 enzymes in the formation of 2,3-dihydroxycarbamazepine, Drug Metab. Dispos 36 (8) (2008) 1637–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Yamaori S, Ebisawa J, Okushima Y, Yamamoto I, Watanabe K, Potent inhibition of human cytochrome P450 3A isoforms by cannabidiol: role of phenolic hydroxyl groups in the resorcinol moiety, Life Sci 88 (15–16) (2011) 730–736. [DOI] [PubMed] [Google Scholar]
- [77].Crettol S, Venetz JP, Fontana M, Aubert JD, Pascual M, Eap CB, CYP3A7, CYP3A5, CYP3A4, and ABCB1 genetic polymorphisms, cyclosporine concentration, and dose requirement in transplant recipients, Ther. Drug Monit 30 (6) (2008) 689–699. [DOI] [PubMed] [Google Scholar]
- [78].Kitada M, Taneda M, Itahashi K, Kamataki T, Four forms of cytochrome P-450 in human fetal liver: purification and their capacity to activate promutagens, Jpn. J. Cancer Res. : Gann 82 (4) (1991) 426–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Pelkonen O, Biotransformation of xenobiotics in the fetus, Pharmacol. Ther 10 (2) (1980) 261–281. [DOI] [PubMed] [Google Scholar]
- [80].Shimada T, Yamazaki H, Mimura M, Wakamiya N, Ueng YF, Guengerich FP, Inui Y, Characterization of microsomal cytochrome P450 enzymes involved in the oxidation of xenobiotic chemicals in human fetal liver and adult lungs, Drug Metab. Dispos 24 (5) (1996) 515–522. [PubMed] [Google Scholar]
- [81].Quinney SK, Benjamin T, Zheng X, Patil AS, Characterization of maternal and fetal CYP3a-mediated progesterone metabolism, Fetal Pediatr. Pathol 36 (5) (2017) 400–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Hayes MA, Li XQ, Gronberg G, Diczfalusy U, Andersson TB, CYP3A specifically catalyzes 1beta-hydroxylation of deoxycholic acid: characterization and enzymatic synthesis of a potential novel urinary biomarker for CYP3A activity, Drug Metab. Dispos 44 (9) (2016) 1480–1489. [DOI] [PubMed] [Google Scholar]
- [83].Marill J, Cresteil T, Lanotte M, Chabot GG, Identification of human cytochrome P450s involved in the formation of all-trans-retinoic acid principal metabolites, Mol. Pharmacol 58 (6) (2000) 1341–1348. [DOI] [PubMed] [Google Scholar]
- [84].Miller KK, Cai J, Ripp SL, Pierce WM Jr., Rushmore TH, Prough RA, Stereo- and regioselectivity account for the diversity of dehydroepiandrosterone (DHEA) metabolites produced by liver microsomal cytochromes P450, Drug Metab. Dispos 32 (3) (2004) 305–313. [DOI] [PubMed] [Google Scholar]
- [85].Topletz AR, Zhong G, Isoherranen N, Scaling in vitro activity of CYP3A7 suggests human fetal livers do not clear retinoic acid entering from maternal circulation, Sci. Rep 9 (1) (2019) 4620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Lemoine A, Gautier JC, Azoulay D, Kiffel L, Belloc C, Guengerich FP, Maurel P, Beaune P, Leroux JP, Major pathway of imipramine metabolism is catalyzed by cytochromes P-450 1a2 and P-450 3a4 in human liver, Mol. Pharmacol 43 (5) (1993) 827–832. [PubMed] [Google Scholar]
- [87].Godamudunage MP, Grech AM, Scott EE, Comparison of antifungal azole interactions with adult cytochrome P450 3A4 versus neonatal cytochrome P450 3A7, Drug Metab. Dispos 46 (9) (2018) 1329–1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Stresser DM, Broudy MI, Ho T, Cargill CE, Blanchard AP, Sharma R, Dandeneau AA, Goodwin JJ, Turner SD, Erve JCL, Patten CJ, Dehal SS, Crespi CL, Highly selective inhibition of human CYP3A in vitro by azamulin and evidence that inhibition is irreversible, Drug Metab. Dispos 32 (1) (2004) 105–112. [DOI] [PubMed] [Google Scholar]
- [89].Foster BC, Kearns N, Arnason JT, Saleem A, Ogrodowczyk C, Desjardins S, Comparative study of hop-containing products on human cytochrome p450-mediated metabolism, J. Agric. Food Chem 57 (11) (2009) 5100–5105. [DOI] [PubMed] [Google Scholar]
- [90].Agbonon A, Eklu-Gadegbeku K, Aklikokou K, Gbeassor M, Akpagana K, Tam TW, Arnason JT, Foster BC, In vitro inhibitory effect of West African medicinal and food plants on human cytochrome P450 3A subfamily, J. Ethnopharmacol 128 (2) (2010) 390–394. [DOI] [PubMed] [Google Scholar]
- [91].Foster BC, Cvijovic K, Boon HS, Tam TW, Liu R, Murty M, Vu D, Jaeger W, Tsuyuki RT, Barnes J, Vohra S, Melatonin interaction resulting in severe sedation, J. Pharm. Pharm. Sci 18 (2) (2015) 124–131. [DOI] [PubMed] [Google Scholar]
- [92].Ekins S, Stresser DM, Williams JA, In vitro and pharmacophore insights into CYP3A enzymes, Trends Pharmacol. Sci 24 (4) (2003) 161–166. [DOI] [PubMed] [Google Scholar]
- [93].Atkins WM, Current views on the fundamental mechanisms of cytochrome P450 allosterism, Expert Opin. Drug Metabol. Toxicol 2 (4) (2006) 573–579. [DOI] [PubMed] [Google Scholar]
- [94].Yang J, Atkins WM, Isoherranen N, Paine MF, Thummel KE, Evidence of CYP3A allosterism in vivo: analysis of interaction between fluconazole and midazolam, Clin. Pharmacol. Ther 91 (3) (2012) 442–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Lampe JN, Atkins WM, Time-resolved fluorescence studies of heterotropic ligand binding to cytochrome P450 3A4, Biochemistry 45 (40) (2006) 12204–12215. [DOI] [PubMed] [Google Scholar]
- [96].Davydov DR, Davydova NY, Sineva EV, Kufareva I, Halpert JR, Pivotal role of P450-P450 interactions in CYP3A4 allostery: the case of alpha-naphthoflavone, Biochem. J. 453 (2) (2013) 219–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Ueng YF, Kuwabara T, Chun YJ, Guengerich FP, Cooperativity in oxidations catalyzed by cytochrome P450 3A4, Biochemistry 36 (2) (1997) 370–381. [DOI] [PubMed] [Google Scholar]
- [98].Korzekwa KR, Krishnamachary N, Shou M, Ogai A, Parise RA, Rettie AE, Gonzalez FJ, Tracy TS, Evaluation of atypical cytochrome P450 kinetics with two-substrate models: evidence that multiple substrates can simultaneously bind to cytochrome P450 active sites, Biochemistry 37 (12) (1998) 4137–4147. [DOI] [PubMed] [Google Scholar]
- [99].Fernando H, Rumfeldt JA, Davydova NY, Halpert JR, Davydov DR, Multiple substrate-binding sites are retained in cytochrome P450 3A4 mutants with decreased cooperativity, Xenobiotica 41 (4) (2011) 281–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Davydov DR, Yang Z, Davydova N, Halpert JR, Hubbell WL, Conformational mobility in cytochrome P450 3A4 explored by pressure-perturbation EPR spectroscopy, Biophys. J. 110 (7) (2016) 1485–1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Krusekopf S, Roots I, Kleeberg U, Differential drug-induced mRNA expression of human CYP3A4 compared to CYP3A5, CYP3A7 and CYP3A43, Eur. J. Pharmacol 466 (1–2) (2003) 7–12. [DOI] [PubMed] [Google Scholar]
- [102].Veeriah S, Miene C, Habermann N, Hofmann T, Klenow S, Sauer J, Bohmer F, Wolfl S, Pool-Zobel BL, Apple polyphenols modulate expression of selected genes related to toxicological defence and stress response in human colon adenoma cells, International journal of cancer, J. Int. Cancer 122 (12) (2008) 2647–2655. [DOI] [PubMed] [Google Scholar]
- [103].Matsunaga T, Maruyama M, Matsubara T, Nagata K, Yamazoe Y, Ohmori S, Mechanisms of CYP3A induction by glucocorticoids in human fetal liver cells, Drug Metab. Pharmacokinet 27 (6) (2012) 653–657. [DOI] [PubMed] [Google Scholar]
- [104].Usui T, Saitoh Y, Komada F, Induction of CYP3As in HepG2 cells by several drugs. - association between induction of CYP3A4 and expression of glucocorticoid receptor, Biological & pharmaceutical bulletin 26 (4) (2003) 510–517. [DOI] [PubMed] [Google Scholar]
- [105].Truffin D, Garcon G, Hannothiaux MH, Colein P, Shirali P, Grave-Descampiaux B, Involvement of oxidative stress in the toxicity of 4-monochlorophenol in Hep G2 cells in culture, J. Appl. Toxicol 23 (2) (2003) 109–114. [DOI] [PubMed] [Google Scholar]
- [106].Roberts JK, Moore CD, Ward RM, Yost GS, Reilly CA, Metabolism of beclomethasone dipropionate by cytochrome P450 3A enzymes, J. Pharmacol. Exp. Ther 345 (2) (2013) 308–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Dubois M, Pfohl-Leszkowicz A, De Waziers I, Kremers P, Selective induction of the CYP3A family by endosulfan and DNA-adduct formation in different hepatic and hepatoma cells, Environ. Toxicol. Pharmacol 1 (4) (1996) 249–256. [DOI] [PubMed] [Google Scholar]
- [108].Goldstein I, Rivlin N, Shoshana OY, Ezra O, Madar S, Goldfinger N, Rotter V, Chemotherapeutic agents induce the expression and activity of their clearing enzyme CYP3A4 by activating p53, Carcinogenesis 34 (1) (2013) 190–198. [DOI] [PubMed] [Google Scholar]
- [109].Dubois M, Plaisance H, Thome JP, Kremers P, Hierarchical cluster analysis of environmental pollutants through P450 induction in cultured hepatic cells - indications for a toxicity screening test, Ecotoxicol. Environ. Saf 34 (3) (1996) 205–215. [DOI] [PubMed] [Google Scholar]
- [110].Smutny T, Bitman M, Urban M, Dubecka M, Vrzal R, Dvorak Z, Pavek P, U0126, a mitogen-activated protein kinase kinase 1 and 2 (MEK1 and 2) inhibitor, selectively up-regulates main isoforms of CYP3A subfamily via a pregnane X receptor (PXR) in HepG2 cells, Arch. Toxicol 88 (12) (2014) 2243–2259. [DOI] [PubMed] [Google Scholar]
- [111].O'Shaughnessy PJ, Monteiro A, Bhattacharya S, Fowler PA, Maternal smoking and fetal sex significantly affect metabolic enzyme expression in the human fetal liver, J Clin. Endocrinol. Metab 96 (9) (2011) 2851–2860. [DOI] [PubMed] [Google Scholar]
- [112].Caron-Beaudoin E, Viau R, Hudon-Thibeault AA, Vaillancourt C, Sanderson JT, The use of a unique co-culture model of fetoplacental steroidogenesis as a screening tool for endocrine disrupters: the effects of neonicotinoids on aromatase activity and hormone production, Toxicol. Appl. Pharmacol 332 (2017) 15–24. [DOI] [PubMed] [Google Scholar]
- [113].He H, Wang P, Li S, Chen Y, Kan Q, Zhang L, Effects of pargyline on histone methylation in promoter and enhancer regions and transcription of CYP3A4/3A7, Yaoxue Xuebao 52 (1) (2017) 91–98. [PubMed] [Google Scholar]
- [114].Cartularo L, Laulicht F, Sun H, Kluz T, Freedman JH, Costa M, Gene expression and pathway analysis of human hepatocellular carcinoma cells treated with cadmium, Toxicol. Appl. Pharmacol 288 (3) (2015) 399–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Greuet J, Pichard L, Bonfils C, Domergue J, Maurel P, The fetal specific gene CYP3A7 is inducible by rifampicin in adult human hepatocytes in primary culture, Biochem. Biophys. Res. Commun 225 (2) (1996) 689–694. [DOI] [PubMed] [Google Scholar]
- [116].Schuetz EG, Schuetz JD, Strom SC, Thompson MT, Fisher RA, Molowa DT, Li D, Guzelian PS, Regulation of human liver cytochromes P-450 in family 3A in primary and continuous culture of human hepatocytes, Hepatology 18 (5) (1993) 1254–1262. [PubMed] [Google Scholar]
- [117].Dannenberg LO, Edenberg HJ, Epigenetics of gene expression in human hepatoma cells: expression profiling the response to inhibition of DNA methylation and histone deacetylation, BMC Genomics 7 (2006) 181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Ogino M, Nagata K, Yamazoe Y, Selective suppressions of human CYP3A forms, CYP3A5 and CYP3A7, by troglitazone in HepG2 cells, Drug Metab. Pharmacokinet 17 (1) (2002) 42–46. [DOI] [PubMed] [Google Scholar]
- [119].Maezawa K, Matsunaga T, Takezawa T, Kanai M, Ohira S, Ohmori S, Cytochrome P450 3As gene expression and testosterone 6 beta-hydroxylase activity in human fetal membranes and placenta at full term, Biological & pharmaceutical bulletin 33 (2) (2010) 249–254. [DOI] [PubMed] [Google Scholar]
- [120].Rabe T, Hosch R, Runnebaum B, Diagnosis of intrauterine fetal growth retardation (IUGR) and placental insufficiency by a dehydroepiandrosterone sulfate (DHAS) loading test, Biol. Res. Pregnancy Perinatol 4 (3) (1983) 130–136. [PubMed] [Google Scholar]
- [121].Schulz S, Klann RC, Schonfeld S, Nyce JW, Mechanisms of cell growth inhibition and cell cycle arrest in human colonic adenocarcinoma cells by dehydroepiandrosterone: role of isoprenoid biosynthesis, Cancer Res 52 (5) (1992) 1372–1376. [PubMed] [Google Scholar]
- [122].Mesiano S, Jaffe RB, Developmental and functional biology of the primate fetal adrenal cortex, Endocr. Rev 18 (3) (1997) 378–403. [DOI] [PubMed] [Google Scholar]
- [123].Romano WM, Lukash LA, Challis JR, Mitchell BF, Substrate utilization for estrogen synthesis by human fetal membranes and decidua, Am. J. Obstet. Gynecol 155 (6) (1986) 1170–1175. [DOI] [PubMed] [Google Scholar]
- [124].Hill M, Paskova A, Kanceva R, Velikova M, Kubatova J, Kancheva L, Adamcova K, Mikesova M, Zizka Z, Koucky M, Sarapatkova H, Kacer V, Matucha P, Meloun M, Parizek A, Steroid profiling in pregnancy: a focus on the human fetus, J. Steroid Biochem. Mol. Biol 139 (2014) 201–222. [DOI] [PubMed] [Google Scholar]
- [125].Means AL, Gudas LJ, The roles of retinoids in vertebrate development, Annu. Rev. Biochem 64 (1995) 201–233. [DOI] [PubMed] [Google Scholar]
- [126].Soprano DR, Soprano KJ, Retinoids as teratogens, Annu. Rev. Nutr 15 (1995) 111–132. [DOI] [PubMed] [Google Scholar]
- [127].Doi AM, Patterson PE, Gallagher EP, Variability in aflatoxin B(1)-macromolecular binding and relationship to biotransformation enzyme expression in human prenatal and adult liver, Toxicol. Appl. Pharmacol 181 (1) (2002) 48–59. [DOI] [PubMed] [Google Scholar]
- [128].Eaton DL, Gallagher EP, Mechanisms of aflatoxin carcinogenesis, Annu. Rev. Pharmacol. Toxicol 34 (1994) 135–172. [DOI] [PubMed] [Google Scholar]
- [129].Bahari A, Mehrzad J, Mahmoudi M, Bassami MR, Dehghani H, Cytochrome P450 isoforms are differently up-regulated in aflatoxin B(1)-exposed human lymphocytes and monocytes, Immunopharmacol. Immunotoxicol 36 (1) (2014) 1–10. [DOI] [PubMed] [Google Scholar]
- [130].Hashimoto H, Nakagawa T, Yokoi T, Sawada M, Itoh S, Kamataki T, Fetus-specific Cyp3a7 and adult-specific Cyp3a4 expressed in Chinese-hamster chl cells have similar capacity to activate carcinogenic mycotoxins, Cancer Res 55 (4) (1995) 787–791. [PubMed] [Google Scholar]
- [131].Kitada M, Taneda M, Ohta K, Nagashima K, Itahashi K, Kamataki T, Metabolic activation of aflatoxin B1 and 2-amino-3-methylimidazo [4,5-f]-quinoline by human adult and fetal livers, Cancer Res 50 (9) (1990) 2641–2645. [PubMed] [Google Scholar]
- [132].Li Y, Yokoi T, Kitamura R, Sasaki M, Gunji M, Katsuki M, Kamataki T, Establishment of transgenic mice carrying human fetus-specific CYP3A7, Arch. Biochem. Biophys 329 (2) (1996) 235–240. [DOI] [PubMed] [Google Scholar]
- [133].Li Y, Yokoi T, Katsuki M, Wang JS, Groopman JD, Kamataki T, In vivo activation of aflatoxin B1 in C57BL/6N mice carrying a human fetus-specific CYP3A7 gene, Cancer Res 57 (4) (1997) 641–645. [PubMed] [Google Scholar]
- [134].Yamada A, Fujita K, Yokoi T, Muto S, Suzuki A, Gondo Y, Katsuki M, Kamataki T, In vivo detection of mutations induced by aflatoxin B1 using human CYP3A7/HITEC hybrid mice, Biochem. Biophys. Res. Commun 250 (1) (1998) 150–153. [DOI] [PubMed] [Google Scholar]
- [135].Wells PG, Winn LM, Biochemical toxicology of chemical teratogenesis, Crit. Rev. Biochem. Mol. Biol 31 (1) (1996) 1–40. [DOI] [PubMed] [Google Scholar]
- [136].Moreira RPP, Jorge AAL, Gomes LG, Kaupert LC, Massud J, Mendonca BB, Bachega TASS, Pharmacogenetics of glucocorticoid replacement could optimize the treatment of congenital adrenal hyperplasia due to 21-hydroxylase deficiency, Clinics 66 (8) (2011) 1361–1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Johnson N, De Ieso P, Migliorini G, Orr N, Broderick P, Catovsky D, Matakidou A, Eisen T, Goldsmith C, Dudbridge F, Peto J, Dos-Santos-Silva I, Ashworth A, Ross G, Houlston RS, Fletcher O, Cytochrome P450 allele CYP3A7*1C associates with adverse outcomes in chronic lymphocytic leukemia, breast, and lung cancer, Cancer Res 76 (6) (2016) 1485–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Hevir N, Ribic-Pucelj M, Rizner TL, Disturbed balance between phase I and II metabolizing enzymes in ovarian endometriosis: a source of excessive hydroxyestrogens and ROS? Mol. Cell. Endocrinol 367 (1–2) (2013) 74–84. [DOI] [PubMed] [Google Scholar]
- [139].Goodarzi MO, Xu N, Azziz R, Association of CYP3A7*1C and serum dehydroepiandrosterone sulfate levels in women with polycystic ovary syndrome, J. Clin. Endocrinol. Metab 93 (7) (2008) 2909–2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Haas DM, Lehmann AS, Skaar T, Philips S, McCormick CL, Beagle K, Hebbring SJ, Dantzer J, Li L, Jung JS, The impact of drug metabolizing enzyme polymorphisms on outcomes after antenatal corticosteroid use, Am. J. Obstet. Gynecol 206 (5) (2012) 447.e17–447.e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].Hevir N, Sinkovec J, Rizner TL, Disturbed expression of phase I and phase II estrogen-metabolizing enzymes in endometrial cancer: lower levels of CYP1B1 and increased expression of S-COMT, Mol. Cell. Endocrinol 331 (1) (2011) 158–167. [DOI] [PubMed] [Google Scholar]
- [142].Chen H, Shen ZY, Xu W, Fan TY, Li J, Lu YF, Cheng ML, Liu J, Expression of P450 and nuclear receptors in normal and end-stage Chinese livers, World J. Gastroenterol 20 (26) (2014) 8681–8690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Bacsi K, Kosa JP, Borgulya G, Balla B, Lazary A, Nagy Z, Horvath C, Speer G, Lakatos P, CYP3A7*1C polymorphism, serum dehydroepiandrosterone sulfate level, and bone mineral density in postmenopausal women, Calcif. Tissue Int 80 (3) (2007) 154–159. [DOI] [PubMed] [Google Scholar]
- [144].Smit P, van Schaik RH, van der Werf M, van den Beld AW, Koper JW, Lindemans J, Pols HA, Brinkmann AO, de Jong FH, Lamberts SW, A common polymorphism in the CYP3A7 gene is associated with a nearly 50% reduction in serum dehydroepiandrosterone sulfate levels, J. Clin. Endocrinol. Metab 90 (9) (2005) 5313–5316. [DOI] [PubMed] [Google Scholar]
- [145].Knops N, Herman J, van Dyck M, Ramazani Y, Debbaut E, van Damme-Lombaerts R, Levtchenko E, van den Heuvel LP, Fieuws S, Kuypers D, Tacrolimus dose requirements in paediatric renal allograft recipients are characterized by a biphasic course determined by age and bone maturation, Br. J. Clin. Pharmacol 83 (4) (2017) 863–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [146].Sood D, Johnson N, Jain P, Siskos AP, Bennett M, Gilham C, Busana MC, Peto J, Dos-Santos-Silva I, Keun HC, Fletcher O, CYP3A7*1C allele is associated with reduced levels of 2-hydroxylation pathway oestrogen metabolites, Br. J. Canc 116 (3) (2017) 382–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [147].Johnson N, Walker K, Gibson LJ, Orr N, Folkerd E, Haynes B, Palles C, Coupland B, Schoemaker M, Jones M, Broderick P, Sawyer E, Kerin M, Tomlinson IP, Zvelebil M, Chilcott-Burns S, Tomczyk K, Simpson G, Williamson J, Hillier SG, Ross G, Houlston RS, Swerdlow A, Ashworth A, Dowsett M, Peto J, Dos Santos Silva I, Fletcher O, CYP3A variation, premenopausal estrone levels, and breast cancer risk, J. Natl. Cancer Inst 104 (9) (2012) 657–669. [DOI] [PubMed] [Google Scholar]
- [148].Lazorwitz A, Aquilante CL, Oreschak K, Sheeder J, Guiahi M, Teal S, Influence of genetic variants on steady-state etonogestrel concentrations among contraceptive implant users, Obstet. Gynecol 133 (4) (2019) 783–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Bertilsson G, Berkenstam A, Blomquist P, Functionally conserved xenobiotic responsive enhancer in cytochrome P450 3A7, Biochem. Biophys. Res. Commun 280 (1) (2001) 139–144. [DOI] [PubMed] [Google Scholar]
- [150].Saito T, Takahashi Y, Hashimoto H, Kamataki T, Novel transcriptional regulation of the human CYP3A7 gene by Sp1 and Sp3 through nuclear factor kappa B-like element, J. Biol. Chem 276 (41) (2001) 38010–38022. [DOI] [PubMed] [Google Scholar]
- [151].Kaupert LC, Lemos-Marini SH, De Mello MP, Moreira RP, Brito VN, Jorge AA, Longui CA, Guerra G Jr., Mendonca BB, Bachega TA, The effect of fetal androgen metabolism-related gene variants on external genitalia virilization in congenital adrenal hyperplasia, Clin. Genet 84 (5) (2013) 482–488. [DOI] [PubMed] [Google Scholar]
- [152].Kuehl P, Zhang J, Lin Y, Lamba J, Assem M, Schuetz J, Watkins PB, Daly A, Wrighton SA, Hall SD, Maurel P, Relling M, Brimer C, Yasuda K, Venkataramanan R, Strom S, Thummel K, Boguski MS, Schuetz E, Sequence diversity in CYP3A promoters and characterization of the genetic basis of polymorphic CYP3A5 expression, Nat. Genet 27 (4) (2001) 383–391. [DOI] [PubMed] [Google Scholar]
- [153].Lee SS, Jung HJ, Park JS, Cha IJ, Cho DY, Shin JG, Identification of a null allele of cytochrome P450 3A7: CYP3A7 polymorphism in a Korean population, Mol. Biol. Rep 37 (1) (2010) 213–217. [DOI] [PubMed] [Google Scholar]
- [154].Salameh G, Al Hadidi K, El Khateeb M, Genetic polymorphisms of the CYP3A4, CYP3A5, CYP3A7 and CYP1A2 among the Jordanian population, Environ. Toxicol. Pharmacol 34 (1) (2012) 23–33. [DOI] [PubMed] [Google Scholar]
- [155].Elens L, Capron A, Kerckhove VV, Lerut J, Mourad M, Lison D, Wallemacq P, Haufroid V, 1199G > A and 2677G > T/A polymorphisms of ABCB1 independently affect tacrolimus concentration in hepatic tissue after liver transplantation, Pharmacogenetics Genom 17 (10) (2007) 873–883. [DOI] [PubMed] [Google Scholar]
- [156].Sim SC, Edwards RJ, Boobis AR, Ingelman-Sundberg M, CYP3A7 protein expression is high in a fraction of adult human livers and partially associated with the CYP3A7*1C allele, Pharmacogenetics Genom 15 (9) (2005) 625–631. [DOI] [PubMed] [Google Scholar]
- [157].Kristiansen W, Haugen TB, Witczak O, Andersen JM, Fossa SD, Aschim EL, CYP1A1, CYP3A5 and CYP3A7 polymorphisms and testicular cancer susceptibility, Int. J. Androl 34 (1) (2011) 77–83. [DOI] [PubMed] [Google Scholar]
- [158].Vyhlidal CA, Gaedigk R, Leeder JS, Nuclear receptor expression in fetal and pediatric liver: correlation with CYP3A expression, Drug Metab. Dispos 34 (1) (2006) 131–137. [DOI] [PubMed] [Google Scholar]
- [159].Schuetz JD, Beach DL, Guzelian PS, Selective expression of cytochrome P450 CYP3A mRNAs in embryonic and adult human liver, Pharmacogenetics 4 (1) (1994) 11–20. [DOI] [PubMed] [Google Scholar]
- [160].He H, Nie YL, Li JF, Meng XG, Yang WH, Chen YL, Wang SJ, Ma X, Kan QC, Zhang LR, Developmental regulation of CYP3A4 and CYP3A7 in Chinese Han population, Drug Metab. Pharmacokinet 31 (6) (2016) 433–444. [DOI] [PubMed] [Google Scholar]
- [161].Vyhlidal CA, Bi C, Ye SQ, Leeder JS, Dynamics of cytosine methylation in the proximal promoters of CYP3A4 and CYP3A7 in pediatric and prenatal livers, Drug Metab. Dispos 44 (7) (2016) 1020–1026. [DOI] [PubMed] [Google Scholar]
- [162].Betts S, Bjorkhem-Bergman L, Rane A, Ekstrom L, Expression of CYP3A4 and CYP3A7 in human foetal tissues and its correlation with nuclear receptors, Basic Clin. Pharmacol. Toxicol 117 (4) (2015) 261–266. [DOI] [PubMed] [Google Scholar]
- [163].Chaudhry AZ, Lyons GE, Gronostajski RM, Expression patterns of the four nuclear factor I genes during mouse embryogenesis indicate a potential role in development, Dev. Dynam 208 (3) (1997) 313–325. [DOI] [PubMed] [Google Scholar]
- [164].Ourlin JC, Jounaidi Y, Maurel P, Vilarem MJ, Role of the liver-enriched transcription factors C/EBP alpha and DBP in the expression of human CYP3A4 and CYP3A7, J. Hepatol 26 (Suppl 2) (1997) 54–62. [DOI] [PubMed] [Google Scholar]
- [165].Puzianowska-Kuznicka M, Shi YB, Nuclear factor I as a potential regulator during postembryonic organ development, J. Biol. Chem 271 (11) (1996) 6273–6282. [DOI] [PubMed] [Google Scholar]
- [166].Saffer JD, Jackson SP, Annarella MB, Developmental expression of Sp1 in the mouse, Mol. Cell. Biol 11 (4) (1991) 2189–2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [167].Itoh S, Yanagimoto T, Tagawa S, Hashimoto H, Kitamura R, Nakajima Y, Okochi T, Fujimoto S, Uchino J, Kamataki T, Genomic organization of human fetal specific P-450IIIA7 (cytochrome P-450HFLa)-related gene(s) and interaction of transcriptional regulatory factor with its DNA element in the 5' flanking region, Biochim. Biophys. Acta 1130 (2) (1992) 133–138. [DOI] [PubMed] [Google Scholar]
- [168].Rodriguez-Antona C, Bort R, Jover R, Tindberg N, Ingelman-Sundberg M, Gomez-Lechon MJ, Castell JV, Transcriptional regulation of human CYP3A4 basal expression by CCAAT enhancer-binding protein alpha and hepatocyte nuclear factor-3 gamma, Mol. Pharmacol 63 (5) (2003) 1180–1189. [DOI] [PubMed] [Google Scholar]
- [169].Mueller CR, Maire P, Schibler U, Dbp, a liver-enriched transcriptional activator, is expressed late in ontogeny and its tissue-specificity is determined post-transcriptionally, Cell 61 (2) (1990) 279–291. [DOI] [PubMed] [Google Scholar]
- [170].Pang XY, Cheng J, Kim JH, Matsubara T, Krausz KW, Gonzalez FJ, Expression and regulation of human fetal-specific CYP3A7 in mice, Endocrinology 153 (3) (2012) 1453–1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [171].Matsunaga T, Maruyama M, Harada E, Katsuyama Y, Sugihara N, Ise H, Negishi N, Ikeda U, Ohmori S, Expression and induction of CYP3As in human fetal hepatocytes, Biochem. Biophys. Res. Commun 318 (2) (2004) 428–434. [DOI] [PubMed] [Google Scholar]
- [172].van Ooij C, Snyder RC, Paeper B, Duester G, Temporal expression of the human alcohol dehydrogenase gene family during liver development correlates with differential promoter activation by hepatocyte nuclear factor 1, CCAAT/enhancer-binding protein alpha, liver activator protein, and D-element-binding protein, Mol. Cell. Biol 12 (7) (1992) 3023–3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [173].Goodwin B, Hodgson E, Liddle C, The orphan human pregnane X receptor mediates the transcriptional activation of CYP3A4 by rifampicin through a distal enhancer module, Mol. Pharmacol 56 (6) (1999) 1329–1339. [DOI] [PubMed] [Google Scholar]
- [174].Pascussi JM, Jounaidi Y, Drocourt L, Domergue J, Balabaud C, Maurel P, Vilarem MJ, Evidence for the presence of a functional pregnane X receptor response element in the CYP3A7 promoter gene, Biochem. Biophys. Res. Commun 260 (2) (1999) 377–381. [DOI] [PubMed] [Google Scholar]
- [175].Blumberg B, Sabbagh W Jr., Juguilon H, Bolado J Jr., van Meter CM, Ong ES, Evans RM, SXR, a novel steroid and xenobiotic-sensing nuclear receptor, Genes Dev 12 (20) (1998) 3195–3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [176].Riffel AK, Schuenemann E, Vyhlidal CA, Regulation of the CYP3A4 and CYP3A7 promoters by members of the nuclear factor I transcription factor family, Mol. Pharmacol 76 (5) (2009) 1104–1114. [DOI] [PubMed] [Google Scholar]
- [177].Song X, Xie M, Zhang H, Li Y, Sachdeva K, Yan B, The pregnane X receptor binds to response elements in a genomic context-dependent manner, and PXR activator rifampicin selectively alters the binding among target genes, Drug Metab. Dispos 32 (1) (2004) 35–42. [DOI] [PubMed] [Google Scholar]
- [178].Hara H, Yasunami Y, Adachi T, Loss of CYP3A7 gene induction by 1,25-dihydroxyvitamin D3 is caused by less binding of VDR to the proximal ER6 in CYP3A7 gene, Biochem. Biophys. Res. Commun 321 (4) (2004) 909–915. [DOI] [PubMed] [Google Scholar]
- [179].Timchenko N, Wilson DR, Taylor LR, Abdelsayed S, Wilde M, Sawadogo M, Darlington GJ, Autoregulation of the human C/EBP alpha gene by stimulation of upstream stimulatory factor binding, Mol. Cell. Biol 15 (3) (1995) 1192–1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [180].Friedman AD, Landschulz WH, McKnight SL, CCAAT/enhancer binding protein activates the promoter of the serum albumin gene in cultured hepatoma cells, Genes Dev 3 (9) (1989) 1314–1322. [DOI] [PubMed] [Google Scholar]
- [181].Li Y, Cui Y, Hart SN, Klaassen CD, Zhong XB, Dynamic patterns of histone methylation are associated with ontogenic expression of the Cyp3a genes during mouse liver maturation, Mol. Pharmacol 75 (5) (2009) 1171–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]