Abstract
Owing to the central role of osteoclasts in bone physiology and remodeling, manipulation of their maturation process provides a potential therapeutic strategy for treating bone diseases. To investigate this, we genetically inhibited the Notch signaling pathway in the myeloid lineage, which includes osteoclast precursors, using a dominant negative form of MAML (dnMAML) that inhibits the transcriptional complex required for downstream Notch signaling. Osteoclasts derived from dnMAML mice showed no significant differences in early osteoclastic gene expression compared to the wild type. Further, these demonstrated significantly lowered resorption activity using bone surfaces while retaining their osteoblast stimulating ability using ex vivo techniques. Using in vivo approaches, we detected significantly higher bone formation rates and osteoblast gene expression in dnMAML cohorts. Further, these mice exhibited increased bone/tissue mineral density compared to wild type and larger bony calluses in later stages of fracture healing. These observations suggest that therapeutic suppression of osteoclast Notch signaling could reduce, but not eliminate, osteoclastic resorption without suppression of restorative bone remodeling and, therefore, presents a balanced paradigm for increasing bone formation, regeneration, and healing.
Keywords: Notch Signaling, Osteoclasts, Osteoblasts, bone formation, Regeneration, Fracture healing
Introduction
Fracture healing is a complex multi-phase process. At the early stage of healing, osteoclasts remodel the fracture margin in preparation for callus formation1–3. As the initially cartilaginous soft callus calcifies into a fracture-stabilizing bony callus, osteoclasts return and, alongside osteoblasts, remodel the callus until physiological bone architecture is restored. Should fracture healing proceed inefficiently, however, the discontinuity caused by the fracture may persist or, following apparent healing, there may be persistent structural weakness that can contribute to re-fracture4; 5. This is of concern in aged individuals, where the efficiency of the bone healing process is diminished. Current strategies to improve skeletal healing include local surgical delivery of osteo-inductive materials or proteins and systemic delivery of anti-resorptive or anabolic therapy6. However, none of these therapies accelerate normal fracture healing nor are they recommended for long-term use as re-fracture preventatives7–9. New approaches based on a clear mechanistic understanding of bone biology and regeneration represent opportunities to address these limitations.
Bone resorbing osteoclasts are formed from hematopoietic progenitors of the monocyte/macrophage lineage in a multi-step process which includes progenitor proliferation, expression of functional genes, and fusion into mature, resorption-capable giant cells10; 11. In addition to their role as bone resorbing cells, osteoclasts promote the differentiation of bone-forming osteoblasts12–15. This coupling role is apparent in both genetic and pharmacologic suppression of osteoclast numbers, which result in a coupled reduction in osteoblasts12; 13. Conversely, approaches that inhibit osteoclast activity while preserving osteoclast number preserve osteoblast function16. Thus, understanding of mechanisms of osteoclast function (as opposed to those of initial differentiation or survival) are of interest, as inhibition of osteoclast maturation may reduce resorption while preserving osteoblast activity and physiological remodeling.
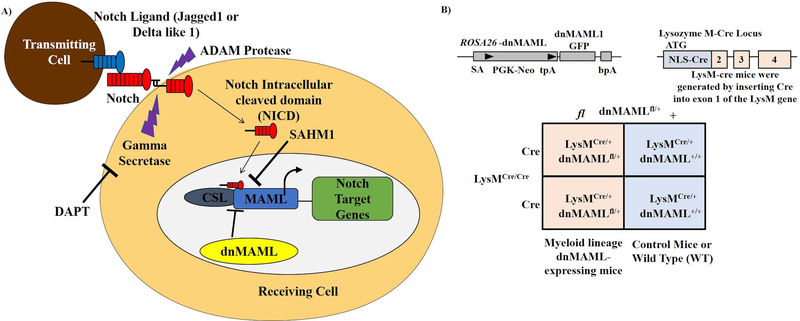
One such osteoclast maturation-controlling system is Notch signaling. We, along with other research groups, have demonstrated that Notch signaling plays a bi-phasic role in osteoclastogenesis, in which, prior to commitment, stimulation of Notch signaling inhibits osteoclast differentiation. After commitment with RANKL, however, Notch signaling is required for final maturation and full functionality17–21. Notch signaling is initiated by binding of one of 5 ligands to one of 4 Notch receptors. This binding releases the intracellular portion of the receptor (NICD), which translocates to the nucleus and binds RBPJκ, a transcriptional regulator, to displace co-repressors and recruit co-activators. Mastermind-like (MAML) is a crucial transcriptional co-activator that serves as an interface between NICD and RBPJκ22–24. In the present study, inhibition of Notch signaling was achieved via expression of dnMAML, which competitively inhibits transcription complex formation by binding NICD and RBPJκ, but no other co-activators (Fig. 1A)22; 25. dnMAML was expressed in the myeloid lineage, which includes osteoclasts and their precursors, via cross between Rosa-dnMAML mice, which carry a LoxP-STOP-LoxP-dnMAML expression cassette, and mice that express Cre recombinase under the control of the Lysozyme M (LysM), promoter.
Figure 1. Genetic manipulation of Notch signaling in osteoclasts.
(A) Trans-activation of Notch signaling in the receiving cell is initiated via interaction between Notch receptor and ligand (Jagged1 or Delta-like1) presented by the transmitting cell. This initiates proteolytic cleavage events that release the Notch intracellular domain (NICD) that translocates to the nucleus and forms a transcriptional complex with MAML and CSL. Notch signaling can be inhibited at the receptor level with DAPT, a γ-secretase inhibitor that inhibits NICD release, or at the transcriptional level by either expressing dominant negative MAML (dnMAML) or treating with SAHM1, a peptide dnMAML mimetic. (B) In this study, we inhibited Notch signaling in osteoclast precursors via Lysozyme M promoter-driven dnMAML expression. Mice with genotypes LysM-Cre/- dnMAML−/− (WT) and LysM-Cre/- dnMAMLMye+/− (dnMAML) were utilized for subsequent studies.
We asked, (1) is osteoclast Notch signaling required for bone remodeling and architecture at physiological baseline, and (2) will inhibition of osteoclast Notch signaling improve fracture healing?
Methods and Materials
Mice breeding
Experimental mice cohorts were generated by mating homozygous LysMCre/Cre (B6.129P2-Lyz2tm1(cre)Ifo/J; Jackson Labs) with heterozygous Rosafl-STOP-fl-dnMAML/+ mice (C57/Bl6 genetic background). The genotypes of the offspring obtained were LysMCre/+/Rosafl-STOP-fldnMAML/+ (dnMAML mice) and LysMCre/+/Rosa+/+ (control or WT) (Fig. 1B). The LysM-Cre26 mouse has been used previously in multiple studies to investigate the roles of specific molecules in osteoclast differentiation without confounding effects during embryonic development27–29.
Osteoclast culture and maintenance
Osteoclast precursors were cultured and differentiated as previously described21; 30. Briefly, bone marrow from femurs and tibias were flushed using α-MEM medium and centrifuged. Cells were later incubated overnight to allow adherent cells to attach to the plate. Next day, non-adherent cells were transferred to new plates with 35ng/mL recombinant mouse Monocyte/Macrophage Colony-Stimulating factor (MCSF) (Biolegend, USA). Upon 70% confluency, adherent osteoclast precursors were harvested using Accutase and later seeded at a density of approximately 26,000 cells per cm2 in α-MEM with 35 ng/mL M-CSF and 100 ng/mL recombinant mouse Receptor Activator of Nuclear Factor kβ Ligand (RANKL) (Shenanandoah Biotechnology). Differentiation medium was replaced every other day. After a period of 3–4 days, osteoclastogenesis was completed and multinuclear cells were observed. The osteoclasts were later tartrate-resistant acid phosphatase (TRAP) stained according to the kit instructions (Sigma Aldrich) to evaluate the enzymatic activity or lysed for RNA extraction.
Fluorescence Microscopy
Osteoclasts were differentiated on cover glass circles #1.5, 12 mm (Electron Microscopy Sciences) and placed into 24-well plates. After a period of 4 days, cells were fixed using 4% paraformaldehyde for 10 min at RT and washed with PBS twice. 0.1% Triton X-100 was added for 5 min with subsequent washing using PBS. Cells were then blocked in 1% BSA for 30 min and stained using Alexa Fluor 594 labelled Phalloidin (Molecular probes) with 5 μl in 200ul PBS/well. The samples were incubated for 30 min in dark. Cells were washed using PBS and then coverslips mounted on clear glass slides after addition of Prolong Gold antifade reagent with DAPI (Invitrogen, Thermo Fischer). Finally, the coverslips were sealed using nail paint and Images were acquired using a Nikon Eclipse fluorescence microscope.
RNA isolation and Quantitative PCR
RNA was extracted using Trizol from Thermo Scientific as per the manufacturer’s instructions21. RNA concentration was determined using a spectrophotometer and 1 μg of RNA was used as starting material for cDNA preparation. Total RNA was reverse transcribed using SuperScript VILO (Thermo Fischer). cDNA was then used as a template for amplification using SYBR Green in a ViiA7 Real time PCR system (Applied Biosciences, Thermo Fischer, CA, USA). Each of the samples were set up in triplicates and the experiments were repeated at least thrice for data integrity and accuracy. Gene expression was normalized to 18S. The list of primers is provided in Table 1.
Table 1:
List of primers used for semi-quantitative RT-PCR (q-PCR).
| S.No. | Name | Forward | Reverse |
|---|---|---|---|
| 1 | TRAP/ACP5 | CAGCTCAGTTGGGTAGCACA | AGCCACAAATCTCAGGGTGG |
| 2 | CSTK | CAGTGTTGGTGGTGGGCTAT | CATGTTGGTAATGCCGCAGG |
| 3 | MMP9 | CAGACGTGGGTCGATTCCAA | TCATCGATCATGTCTCGCGG |
| 4 | Wnt10b | ACCACGACATGGACTTCGGAGA | CCGCTTCAGGTTTTCCGTTACC |
| 5 | Hes1 | GAGGCTGCCAAGGTTTTTGG | ACTTTACGGGTAGCAGTGGC |
| 6 | ALP | CCAGAAAGACACCTTGACTGTGG | TCTTGTCCGTGTCGCTCACCAT |
| 7 | OSX | ATGGCGTCCTCTCTGCTTGA | TTCCCCAGGGTTGTTGAGTC |
| 8 | OCN | GCAATAAGGTAGTGAACAGACTCC | CCATAGATGCGTTTGTAGGCGG |
| 9 | Runx2 | CCTGAACTCTGCACCAAGTCCT | TCATCTGGCTCAGATAGGAGGG |
| 10 | DC-STAMP | TTTGCCGCTGTGGACTATCTGC | GCAGAATCATGGACGACTCCTTG |
| 11 | OC-STAMP | GGCTCAGAAGTTACCCACTGTC | GGAGGTTGGTTGAGGACGAAGA |
| 12 | MAML | CCAGCTTTGATGGCATATCTTCC | CTACAGGGACACTGGAAGGGTT |
| 13 | 18S | GGTAACCCGTTGAACCCCAT | CAACGCAAGCTTATGACCCG |
Jagged 1 stimulation and Hes1 expression
Notch signaling was stimulated using immobilized Jagged1 as previously described30; 31. Goat IgG (Jackson Immunology) and Jagged-1, a Notch ligand (R & D Systems) were made in PBS at a concentration of 10 μg/ml and added to 48 well plates. Wells were then washed using PBS and later 26,000 osteoclast precursors were seeded (24 hr pre-exposure to either MCSF alone or both MCSF and RANKL). Plates were incubated for at least 24 h prior to harvesting the cells for RNA extraction and analysis of Hes1 gene expression.
Mineral dissolution assay
50,000 cells were seeded into a 24-well Osteo-assay plate (Corning life sciences) coated with calcium phosphate. Both WT and dnMAML osteoclast precursors were seeded and later treated with MCSF or MCSF and RANKL for a total of 5 days. Media was replenished alternatively as described earlier. The remaining mineral was Von Kossa stained as per the protocol described21. Cells were washed with PBS and incubated in 10% bleach for 5 min followed by washing twice with deionized water. 300 μl of 5% silver nitrate was added to the plate and incubated at RT for 30 min. Wells were then washed thoroughly using deionized water. Later, 300 μl of 5% Sodium carbonate made in formalin was added and incubated for another 5 minutes at RT. Finally, the solution was removed from the wells and air dried. Plates were then scanned and quantified using Image J software.
Pit formation assay and determination of TRAP activity
Cortical bone slices (Immunodiagnostic Systems, IDS) were purchased for in vitro assessment of osteoclastic bone resorption using pit formation assay as per the procedure described with slight modifications32–34. Bone chips were sterilized under UV for at least 2 h and then washed with media before placing in 96 well plates. 20,000 cells/200 μl (WT and dnMAML) were seeded onto the bone slices containing either MCSF or MCSF and RANKL for a total of 12 days. Conditioned media was collected and replenished with new media every 3rd day. The former was then stored at −20°C for further analysis of TRAP and CTX-1 levels. At day 12th, osteoclasts were removed, and slices were stained with 1% toluidine blue in 1% sodium borate solution. Bone slices were dried and imaged using Nikon Eclipse light microscope. Pits were counted and measured using Image J software. TRAP/TRACP activity was determined35; 36 using TRAP solution buffer consisting of L- Ascorbic acid, di-Sodium tartrate, 4-Nitrophenylphosphate and Reaction buffer (1 M Acetate, 0.5% Triton X-100, 1 M NaCl, 10 mM EDTA pH=5.5). 10 μl of condition medium was dispensed into a new 96-well plate and 90 μl TRAP solution buffer was added. The mix was incubated at 37°C for about 15 min in the dark. The reaction was stopped using 0.3 N NaOH, and absorbance was measured at 405 nm using a Biotek spectrophotometer.
Determination of CTX-1 and Osteocalcin levels
Type I Collagen C-telopeptide (CTX-1) levels were measured in serum collected from 3-month-old female and male (WT and dnMAML mice) and conditioned medium collected from bone slices using CTX-I detection ELISA kit (Chondrex, Inc). It is based on a competitive assay system with a monoclonal antibody which recognizes conserved peptide sequences in mouse37. Levels of osteocalcin, a bone formation marker38 was evaluated using mouse osteocalcin ELISA (Novus Biologicals, NBP2–68151) in serum collected from 3-month old WT and dnMAML mice respectively as per the instructions provided in the kit.
Osteoblast culture maintenance, ALP activity. and Alizarin staining
Adherent bone marrow stromal cells were cultured to 70% confluency. Cells were then harvested and then seeded at a density of 30,000 cells per cm2. WT cells were maintained in maintenance media (α-MEM supplemented with 10% Fetal bovine serum (FBS), 1% Penicillin-streptomycin, 1% L-glutamine, 1% Non-essential amino acids), osteogenic media (maintenance media + 10 mM β-glycerol phosphate + 50 μM ascorbate-2-phosphate + 100 nM Dexamethasone), osteogenic media in combination with conditioned media collected form either WT or dnMMAL osteoclasts (here condition media was first concentrated using 3 KDa cut off concentration filters (Amicon, Millipore) and then mixed in a ratio of 1:10 with osteogenic media), or osteogenic media in combination with 30 ng/mL Bone Morphogenetic Protein 6 (BMP-6) (R & D Systems). Media were refreshed every 3rd days. Alkaline phosphatase (ALP) activity was measured by SensoLyte pNPP Alkaline Phosphatase Assay Kit (AnaSpec, Inc)39 as per the manufacturer’s instructions. Mineralized nodules were stained with alizarin red as per the protocol described earlier40.
Skeletal Phenotyping
WT and dnMAML mice were given a subcutaneous injection of calcein and alizarin red to label bone mineralizing surfaces fluorescent green and red (9 and 2 days prior to euthanasia). Following euthanasia, right femurs were fixed, decalcified and sectioned. Paraffin sections were TRAP stained for quantification of osteoclast numbers. Left femurs were fixed, embedded in polymethylmethacrylate (PMMA), sectioned, and stained using trichrome41; 42 for quantification of osteoblasts per trabecular area and osteoblast surface per bone surface. Unstained sections were imaged fluorescently for quantification of mineralizing surfaces and determination of bone formation rate. Images were acquired using a Nikon Eclipse fluorescence microscope and analyzed using Osteomeasure software. Left tibias were fixed and analyzed via μCT with subsequent mechanical testing. RNA was extracted from right tibias for analysis of osteoclast (TRAP, CSTK, MMP9 and Wnt10B) and osteoblast gene expression (ALP, Osterix, Osteocalcin and Runx2) respectively.
Fracture Protocol and Surgery
Mice were fractured as per the protocol described earlier43. Briefly, a small incision was created around the mouse’s knee and a 27-gauge needle was placed in the tibial tuberosity just above the patella for fracture stabilization. Following pinning and incision, a traumatic 3-point-bend fracture was generated using a modified guillotine (consists of a weight (220 g) sitting 7 cm above the metal lever). The setup of fracture device is outlined in Supplemental Figure 1. Mice were recovered from anesthesia on a warm water pad and received a subcutaneous dose of extended release buprenorphine (0.5 mg/kg) after the surgery, and then once every third day. Animals were humanely euthanized for analysis at 10, 20- and 40-Days post fracture (DPF). Fracture calluses were analyzed using μCT and histology.
MicroCT (μCT) parameters44–47
Samples were imaged in a μCT scanner (model μCT50, Scanco Medical). A Gaussian low-pass filter (σ = 0.8, support = 1) was used for all analyses. Trabecular bone parameters were measured by analyzing 150 slices of the distal metaphysis. Cortical bone parameters were measured by analyzing 50 slices in the mid-diaphysis. A semi-automated contouring method was used to determine the outer cortical bone perimeter. The distal metaphyseal regions were used for evaluation of bone volume, total volume, bone volume/total volume, trabecular number, trabecular thickness, trabecular spacing, bone mineral density, and tissue mineral density while mid-diaphyseal regions for determining cortical thickness, cortical bone mineral density, and cortical tissue mineral density. Fractured tibias were quantified based on ROI (region of interest) of each callus and the following parameters determined: mineralized callus volume (BV), total callus volume (TV), mineralized callus volume fraction (BV/TV), bone mineral density (BMD), tissue mineral density (TMD), and average callus area. The latter was calculated by counting the number of slices multiplied by the size of the slice (0.006 mm/slice) to obtain the total length of the callus, and subsequently, average callus area was determined by dividing bone volume to total callus length.
Mechanical Testing
Following μCT, tibias were placed on the fixed supports of 3-point bending fixture (14 mm span). The actuator of testing system (DaynaMight, Instron Corp.) was centered above the medial region and anterior sides facing forward and down respectively. To keep the bone in place a pre-load of 2N was applied. A load at constant rate of 0.005 mm/s was applied centrally until failure occurred. The force-displacement curve was then recorded for each sample and the structural stiffness determined by measuring the slope of curve at its linear portion. Additionally, ultimate force (force at failure) was determined.
Histological parameters
Femurs from the skeletal phenotyping group were paraffin embedded and sectioned for TRAP staining. Fracture callus sections were TRAP stained for quantification of osteoclast numbers and safranin O stained for quantification of callus area and cartilage area. TRAP staining was performed as per the protocol form University of Rochester. Paraffin sections were first deparaffinized using xylene and rehydrated through graded ethanol (100%, 95% and 80%) to distilled water. Slides were then placed in pre-warmed TRAP staining solution mix (Sodium acetate, tartaric acid, glacial acetic acid, fast Red violet and Naphthol) for about an hour and rinsed in distilled water. Slides were later counterstained with 0.02% Fast Green for 45 sec. and rinsed quickly in distilled water followed by dehydration using ethanol (80%, 95% and 100%) with clearing in xylene. Slides were mounted using Permount (Thermo Fischer) and imaged using Nikon Eclipse light microscope. Osteoclasts were counted using the osteomeasure software. Safranin O staining was performed as per the protocol described earlier48–50 and from IHC world. Briefly, sections were deparaffinized and hydrated. Slides were stained using Weigert’s iron hematoxylin working solution, washed and then counterstained using fast green. Slides were rinsed quickly in 1% acetic acid and then stained using 0.1% safranin O solution. Finally, the slides were dehydrated and mounted as described above. Slides were imaged using Nikon Eclipse light microscope. Cartilage and callus area were determined using the Osteomeasure software.
Biological Variables and Data Analysis
Experiments were carried out in both male and female animals as skeletal problems, fractures and healing issues affect both sexes. Skeletal phenotyping was performed in both 3-month and 6-month males and female C57/BL6 mice cohorts whereas fractures and other ex vivo work in 3-month male and female mice (WT and dnMAML).
Statistics
All experiments were performed at least thrice independently and represented as Mean ± S.D. Student t-test was used for statistical analysis and P<0.05 was considered statistically significant. Graphpad Prism 6.0 software was used for graphical representation of the data. To increase rigor, analyses subject to measurement variability (e.g. qPCR, ex vivo cellular assays, serum measurements) were performed in triplicate and those subject to evaluator variability (e.g. histomorphometry analysis) performed in a blinded fashion.
Study Approval
Mice were housed, and procedures carried out in facilities provided by the University of Pennsylvania and the Philadelphia VA Medical Center both of which are AAALAC accredited. Procedures were approved by the Institutional animal care and use Committee (IACUC) of the University of Pennsylvania.
Results
dnMAML suppresses Jagged1 induction of Hes1 in bone marrow macrophages and pre-osteoclasts
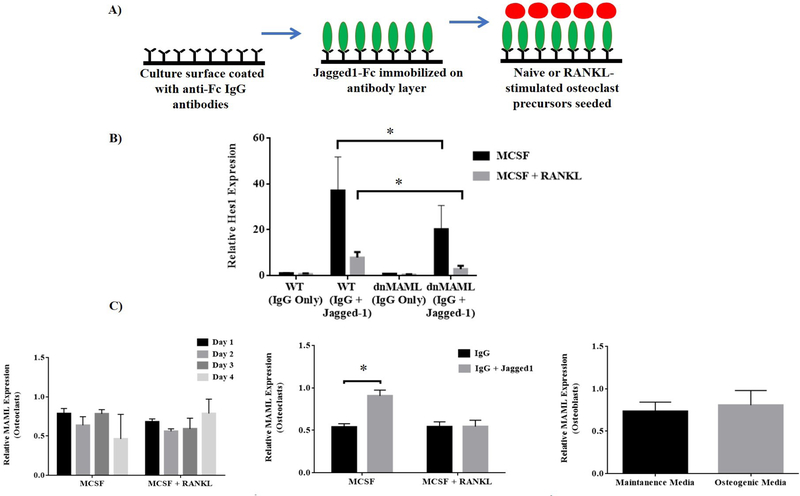
To assess the functionality of dnMAML in osteoclast precursors, cells from both WT and dnMAML mice were subjected to Notch signaling stimulation via immobilized Jagged1 after a 24-hr pre-exposure to either MCSF alone or MCSF + RANKL (Fig. 2A). 24 hr of Jagged1 stimulation produced a robust induction of Hes1, a Notch target gene, in both bone marrow macrophages and pre-osteoclasts, and this induction was blunted by the presence of dnMAML (Fig. 2B). Interestingly, prior stimulation of RANK signaling likewise reduced Hes1 transcription, and this effect was further diminished by the presence of dnMAML (Fig. 2B). We also estimated the levels of MAML during the differenenciation of both osteoclasts and osteoblasts. Levels of MAML were relatively unaltered in both the osteoclasts and osteoblasts (Fig. 2C). However, we observed a significant difference in MAML expression levels upon stimulation with Jagged 1 in MCSF alone group whereas in MCSF +RANKL treated osteoclasts the levels of MAML were unchanged (with or without Jagged 1 stimulation) shown in Fig. 2C.
Figure 2: dnMAML suppresses Jagged1 induction of Hes1.
Ex-vivo generated osteoclasts from both WT and dnMAML mice were subjected to Notch signaling stimulation via Jagged1 following culture with MCSF alone or MCSF + RANKL. After 24 hr of Jagged1 stimulus, relative expression of Hes1 was determined via qRT-PCR. (A) Schematic representation of Notch signaling stimulation via immobilized Jagged1. (B) Relative expression of Hes1 (Notch target gene) following exposure to Jagged1 or IgG demonstrated that Jagged1 stimulation produced a robust increase in Hes1 expression that was blunted by dnMAML. MCSF + RANKL treatment also reduced Jagged1-stimulated Hes1 expression. (C) MAML gene expression analysis in osteoclasts and osteoblasts was performed using qRT-PCR. Relative levels of MAML were unchanged during the differentiation process for both the cell types. Jagged1 treatment, however, significantly increased MAML expression in osteoclast precursors maintained with MCSF, but not MCSF + RANKL. Values represented are Mean ± S.D from at least three independent experiments. P value was calculated using Student t-test (*P<0.05).
Notch signaling is not required for early osteoclast gene expression
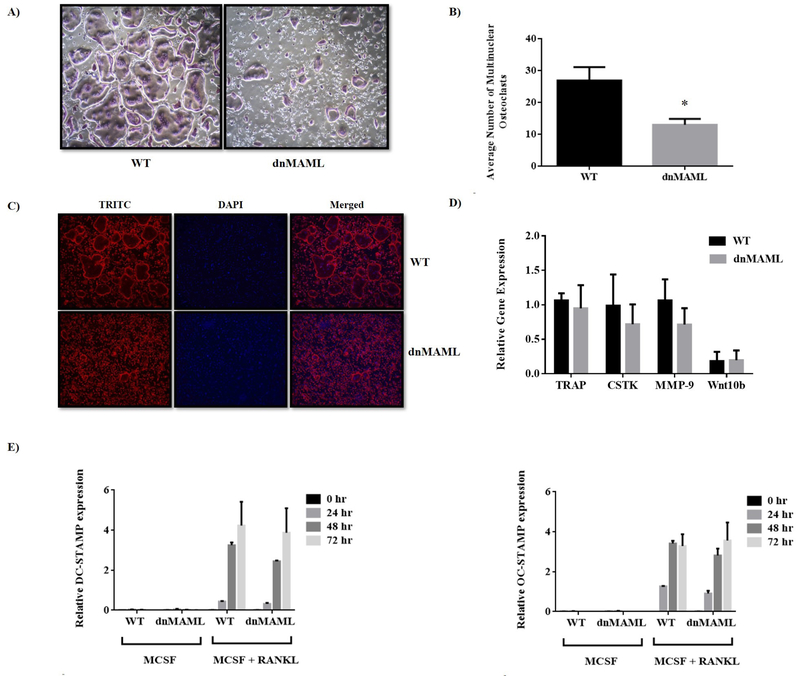
Osteoclasts derived from both WT and dnMAML mice were subjected to TRAP staining. Osteoclasts from WT were giant and fully mature while the osteoclasts from dnMAML mice failed to fuse effectively and remained immature (Fig. 3A). Further, the number of mature osteoclasts were significantly higher in the control group (Fig. 3B). dnMAML cells, nevertheless, remained TRAP positive indicating a failure not in differentiation per se, but fusion and maturation. To delineate cytoskeletal reorganization and actin ring formation, actin filaments were stained with phalloidin. While dnMAML osteoclasts were smaller and fewer, those that did form demonstrated normal podosome belts (Fig. 3C). Gene expression analysis of early osteoclast marker genes (tartrate-resistant acid phosphatase – TRAP, cathepsin k – CTSK, matrix metalloproteinase 9 – MMP9, and Wnt10b) found no significant difference between WT and dnMAML osteoclasts providing further evidence that Notch signaling drives maturation rather than initial differentiation of osteoclasts (Fig. 3D). The smaller and immature osteoclasts observed in dnMAML group indicated a possible defect in fusion process. To address this, we carried out gene expression analysis for fusion markers specific to osteoclasts such as Dendritic cell-specific transmembrane protein – DC-STAMP and Osteoclast stimulatory transmembrane protein – OC-STAMP11. The expression was relatively the same between the 2 groups (Fig. 3E). This suggests that dnMAML expressing osteoclasts do not have a defect in gene expression required for differentiation process. However, we do expect that the fusion markers will be reduced at the protein or surface level, but this needs to be investigated further.
Figure 3. Genetic inhibition of Notch signaling suppresses osteoclast maturation but does not impact early gene expression.
Bone marrow-derived macrophages were collected from both WT and dnMAML mice and differentiated into osteoclasts ex vivo. (A) TRAP stained osteoclasts (purple) in WT group were giant and well-fused while osteoclasts from Notch-inhibited mice failed to fuse effectively and remained immature. (B) Quantification of TRAP stained multinuclear osteoclasts showed a significant reduction in average number of osteoclasts in dnMAML group. (C) To delineate cytoskeletal reorganization, actin filaments were assessed using Phalloidin staining. There were fewer and smaller osteoclasts in dnMAML group, but these cells still demonstrated podosome belts, suggesting normal actin organization. (D) and (E) Expression analysis of osteoclastic marker genes (TRAP, CTSK, MMP9 and Wnt10b) and osteoclast specific fusion markers (DC-STAMP and OC-STAMP) ex vivo showed no significant differences between the dnMAML and WT groups. Values represented are Mean ± S.D from at least three independent experiments. P value was calculated using Student t-test (*P<0.05).
dnMAML expression suppresses osteoclast activity
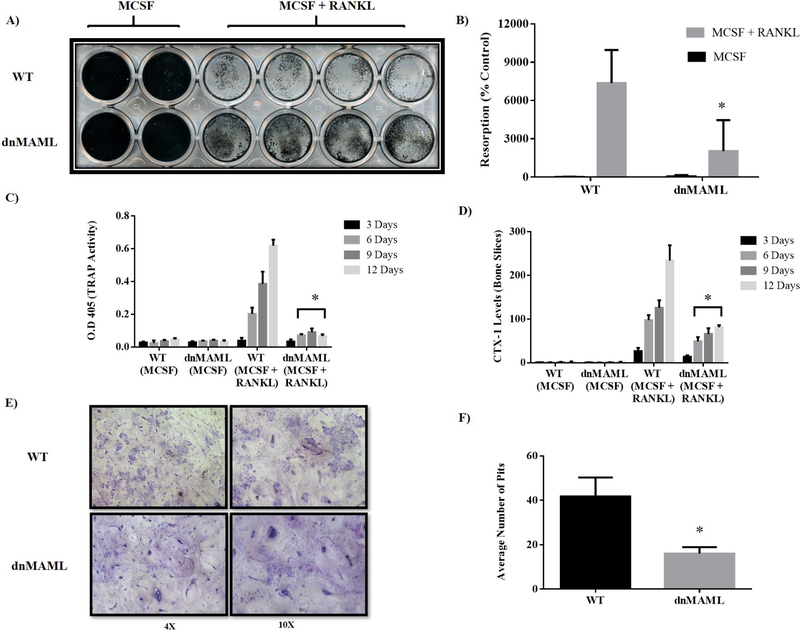
To measure osteoclast activity, WT and dnMAML osteoclasts were differentiated and cultured on calcium phosphate coated plates and bovine cortical bone slices. In both instances, dnMAML osteoclasts demonstrated reduced resorption areas (Fig. 4A, B, E, &F). In addition, assessment of TRAP activity and C-terminal type I collagen fragments (CTX-I) in conditioned media revealed that dnMAML osteoclasts secreted lower amounts of TRAP and digested less collagen than their WT counterparts (Fig. 4C &D).
Figure 4. Genetic inhibition of Notch signaling reduces osteoclast resorptive function.
(A) Osteoclast precursors generated ex vivo from both WT and dnMAML mice were differentiated and cultured on calcium phosphate coated surfaces for a period of 5 days. Following culture, osteoclasts were removed and remaining minerals were von Kossa stained and (B) quantified using ImageJ software. dnMAML osteoclasts demonstrated significantly reduced resorptive activity compared to WT. (C) Osteoclast precursors were seeded on bovine cortical bone slices in 96 well plates and cultured in the presence of MCSF alone or MCSF + RANKL for a period of two weeks. Conditioned media were collected every third day and TRAP enzymatic activity was evaluated using pNPP chromogenic substrate. There was a significant reduction in TRAP activity in the conditioned media of dnMAML osteoclasts cultured with MCSF + RANKL on days 6, 9 and 12. (D) CTX-1, c-terminal fragments of type I collagen, levels were determined in condition media from WT and dnMAML osteoclasts. CTX-1 levels were significantly reduced in dnMAML osteoclasts cultured with MCSF + RANKL on days 6, 9 and 12. (E) At the conclusion of culture, bone slices were toluidine blue stained to reveal resorption pits. (F) Quantification showed a significant reduction in number of resorption pits from dnMAML osteoclast cultures. Values represented are Mean ± S.D from at least three independent experiments. P value was calculated using Student t-test (*P<0.05).
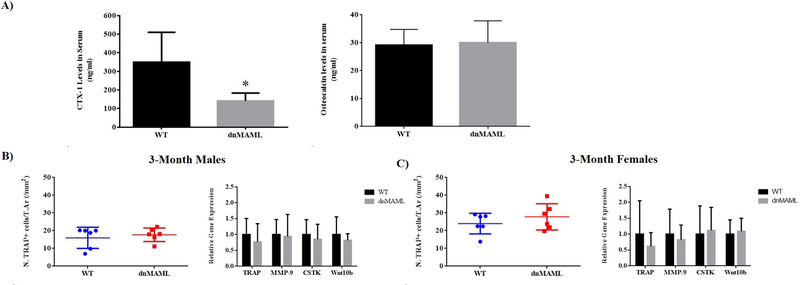
dnMAML suppresses osteoclast activity without impacting TRAP+ cell number or gene expression in vivo
As measured by serum CTX-I ELISA, dnMAML mice (3-month old males and females) demonstrated reduced osteoclast activity compared to WT (Fig. 5A). To assess whether this effect was a result of decreased osteoclast number, TRAP staining was performed. Histomorphometric analysis of TRAP-stained sections and gene expression analysis in whole bone RNA revealed no significant differences in osteoclastic gene expression or TRAP+ cells regardless of sex (Fig. 5B & C). This suggests that, as observed ex vivo, dnMAML reduces osteoclast activity by keeping osteoclasts in an immature, mononuclear state.
Figure 5. Notch inhibition impairs resorptive activity in vivo without impacting TRAP+ cell numbers and gene expression.
profiles. (A) CTX-1 levels in serum obtained from 3-month-old male and female WT and dnMAML mice were significantly reduced in dnMAML mice suggesting reduced resorptive activity. No differences in serum osteocalcin (OCN) levels were observed between the 2 groups suggesting comparable osteoblast activity. TRAP+ cell numbers and expression profile of osteoclastic marker genes (TRAP, CTSK, MMP-9 and Wnt10b) were analyzed in (B) 3-month-old males and (C) 3-month-old females using TRAP staining and qRT-PCR, respectively. There were no significant differences in TRAP+ cell numbers or gene expression profiles between these groups. Values represented are Mean ± S.D and P value was calculated using Student t-test (*P<0.05).
Osteoblast coupling to osteoclasts is maintained in the absence of osteoclast Notch signaling
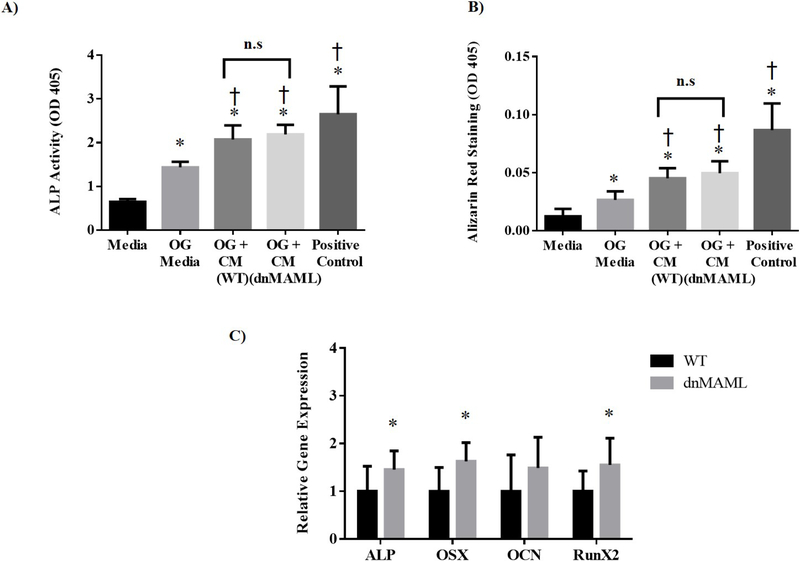
Osteoblast precursors from WT mice were cultured in presence of condition media obtained from either WT or dnMAML osteoclasts or BMP-6 as a positive control. Both alkaline phosphatase (ALP) activity and mineralization were enhanced by treatment with osteoclast-conditioned media regardless of whether the media came from WT or dnMAML osteoclasts (Fig. 6A & B). This indicates that osteoclasts from dnMAML-expressing precursors do not lose their ability to stimulate osteoblasts even though they are deficient in resorptive functionality. However, we cannot rule out the fact that the condition media might have some contribution from monocytes in addition to osteoclasts serving as a limiting factor for data represented in Fig. 6A & B. These observations were further evidenced in vivo where osteoblast gene expression was found to be higher in 6-month female dnMAML mice and suggesting that inhibition of Notch pathway in osteoclasts does not negatively affect osteoblasts (Fig. 6C, Supplemental Fig. 2).
Figure 6. dnMAML osteoclasts stimulate osteogenesis.
Osteoclast and osteoblast precursors were obtained from bone marrow of WT and dnMAML mice. Osteoblast precursors were cultured in the presence of maintenance media, osteogenic (OG) media (maintenance media supplemented with β-glycerol phosphate, ascorbate, and dexamethasone), OG media were combined with conditioned media from WT or dnMAML osteoclasts, or OG media supplemented with BMP-6 as a positive control. After 15 days of differentiation, (A) ALP activity was determined using SensoLyte pNPP Alkaline Phosphatase Assay Kit, and (B) calcium deposits were quantified after alizarin red staining. Both ALP activity and alizarin red staining were significantly increased in all treatment groups compared to maintenance media (*P<0.05). Similarly, ALP activity and alizarin red staining were significantly increased when OG medium was supplemented with conditioned medium from both WT and dnMAML osteoclasts as well as with BMP-6 (†P<0.05). There was no significant difference between cells treated with WT and dnMAML conditioned media. (C) Osteoblast genes (ALP, OSX and RunX2) in 6-month dnMAML females were significantly increased as measured by qRT-PCR. Values represented are Mean ± S.D and P value was calculated using Student t-test (*P<0.05, †P<0.05).
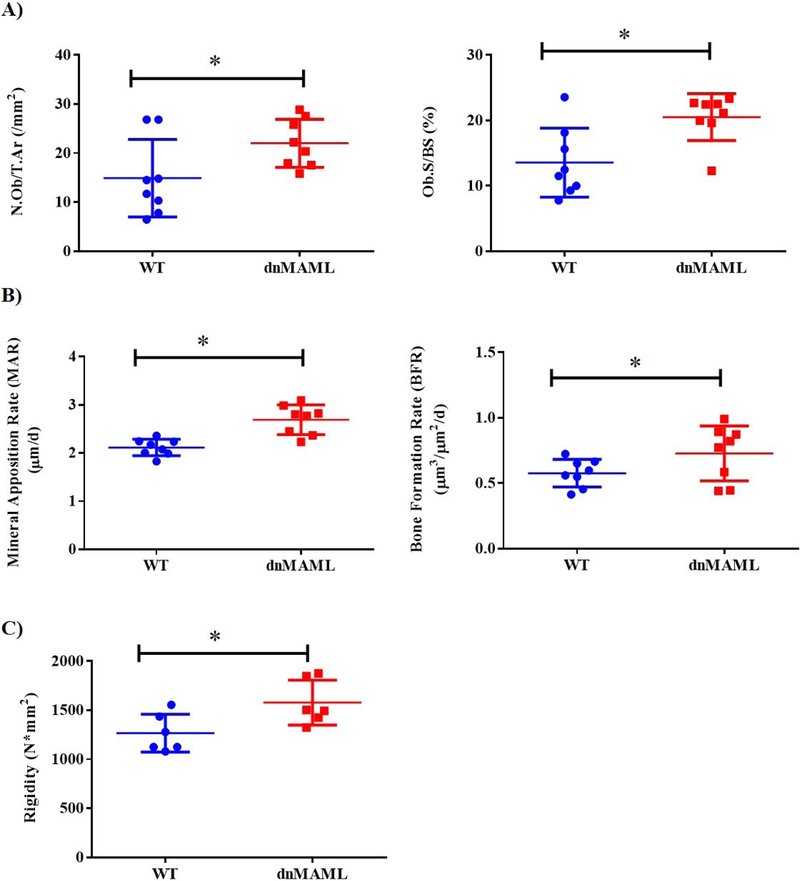
LysM-dnMAML mice have increased osteoblast activity and bone strength, but normal bone architecture
Mice were analyzed for morphology of key skeletal elements and bone architecture using μCT and later tested mechanically using 3-point bending. No changes were observed in any of the parameter evaluated using μCT (Supplemental Figs. 3A–6A and Table S1) while there were significant changes in rigidity in 6-month male dnMAML mice (Fig. 7C, Supplemental Figs. 3B–6B and Table S1). A significantly higher number of osteoblast/trabecular area and osteoblast surface per surface were observed in 6-month females (Fig. 7A, Supplemental Fig. 7A–C and Table S2). There were also significant increases in mineral apposition rate and bone formation rate in 6-month dnMAML females (Fig. 7B, Supplemental Fig. 8A–C and Table S3). These data indicate that inhibition of osteoclast Notch signaling does not produce gross architectural changes at physiological baseline, but, nevertheless, results in stronger bones due to increased osteoblast activity, particularly in older animals.
Figure 7. dnMAML mice demonstrate increased bone formation rates and bone rigidity.
WT and dnMAML mice were given injections of alizarin red and calcein, 8 and 1 days prior to sacrifice, respectively, to label bone mineralizing surfaces. Static and dynamic histomorphometric analyses were later performed on plastic-embedded femur sections. (A) Plastic sections were trichrome stained, imaged, and analyzed. There was a significant increase in osteoblast number/trabecular area and osteoblast surface per bone surface percentage in dnMAML mice. (B) Dynamic histomorphometry using fluorescence microscopy on unstained sections showed significantly higher mineral apposition and bone formation rates in dnMAML group. Representative data for 6-month females are depicted. (C) Mechanical testing using 3-point bending demonstrated bones from dnMAML mice of 6-month males exhibited significantly higher rigidity. Values represented are Mean ± S.D and P value was calculated using Student t-test (*P<0.05).
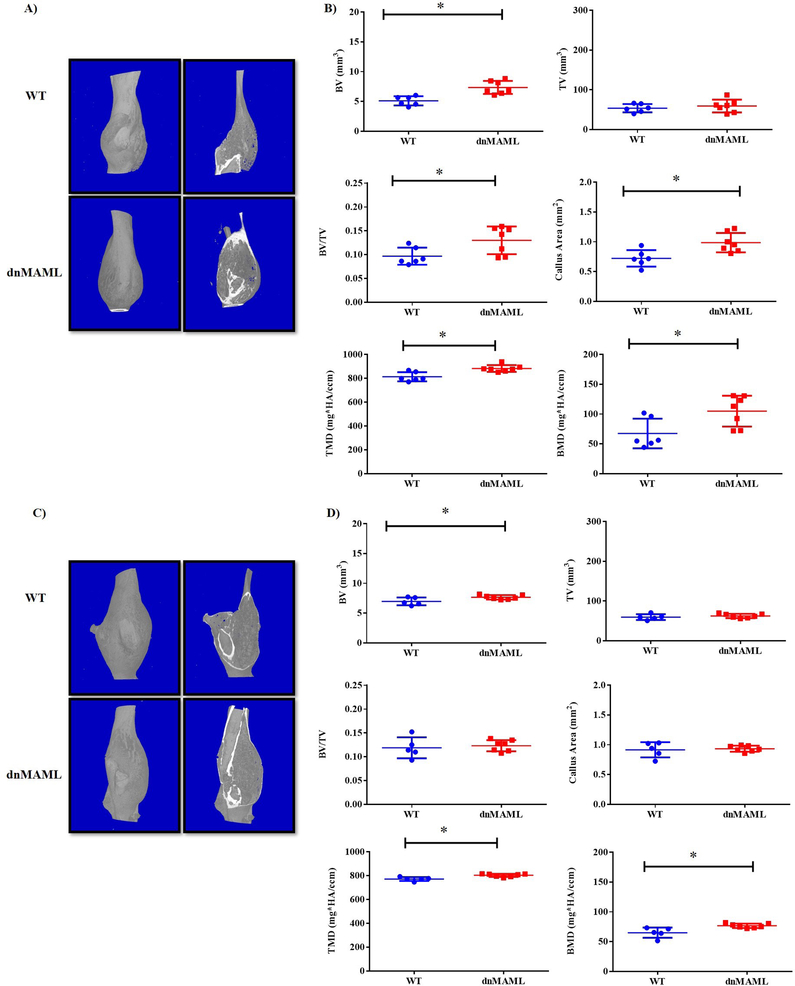
Inhibition of Notch signaling in osteoclasts results in larger bony calluses with increased mineral density following traumatic fracture
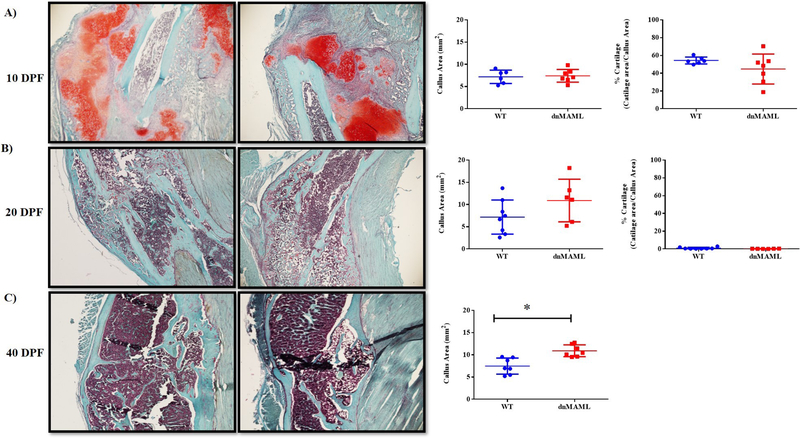
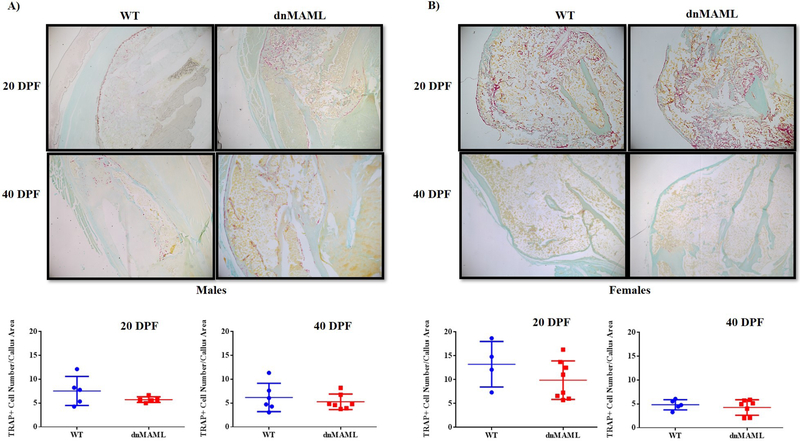
We observed that both 3-month-old male and female dnMAML mice showed significant improvements in fracture healing. These mice displayed significantly higher bone volume, tissue mineral and bone mineral densities respectively at 40 DPF (p<0.05) while no differences were observed at prior time points (Fig.8A–D, Supplemental Fig. 9A–C and Table S4). We also observed significantly higher callus area in 40 DPF 3-month males but not females. To further corroborate these findings, histology was performed using paraffin embedded samples. There was a significant increase in callus area in 40 DPF group (p<0.05) for the males but not for female group (Fig. 9, Supplemental Fig. 10 and Table S4) while no differences in osteoclasts numbers/callus area were observed in TRAP stained sections (Fig. 10). This indicates Notch inhibition in osteoclasts does not affect their cellular number but, rather, their function.
Figure 8. Inhibition of Notch signaling in osteoclasts results in larger bony calluses during fracture healing.
Both 3-month old male and female mice at 40 days post fracture (DPF) showed significantly larger bony calluses. (A) 3D images and sectional cut views obtained from μCT in 3-month males showed (B) significant increases in bone volume, bone volume/total volume, callus area, tissue mineral density, and bone mineral density. (C) In 3-month females, there were (D) significant increases in bone volume, tissue mineral density and bone mineral density. Values represented are Mean ± S.D and P value was calculated using Student t-test (*P<0.05).
Figure 9. Inhibition of Notch signaling results in larger bony callus areas without impacting soft callus formation.
Fracture calluses in 3-month males were Safranin O stained to quantify callus and cartilage areas. Following staining, cartilage-containing areas are orange/red, nuclei are black, and background bone and muscle are blue/green. Representative images at (A) 10 DPF and (B) 20 DPF showed no significant difference in callus or cartilage area between WT and dnMAML. (C) Callus area was significantly higher in dnMAML samples at 40 DPF, at which point, no cartilage was visible in either group. Images were captured using a Nikon Eclipse light microscope at 20X magnification and were analyzed using Osteomeasure software. Values represented are Mean ± S.D and P value was calculated using Student t-test (*P<0.05).
Figure 10. Notch inhibition does not impact TRAP+ cell number in fracture calluses.
Fracture calluses from male and female 3-month-old WT and dnMAML mice were TRAP stained to evaluate the TRAP+ cell numbers. No differences in TRAP+ cell number/callus area were observed in either 20 DPF or 40 DPF samples from (A) 3-month males and (B) 3-month females respectively. Images were captured using a Nikon Eclipse light microscope at 20X magnification and were analyzed using Osteomeasure software.
Discussion
Inhibition of Notch signaling or the genes it regulates in osteoclasts may be a viable method for reducing bone resorption while maintaining bone formation. Systemic inhibition of Notch signaling through global, inducible expression of dnMAML produces a net inhibitory effect on fracture healing due to the deleterious effects of dnMAML expression on bone formation51. Here, we investigated the effects of Notch signaling inhibition in osteoclast precursors on resting bone characteristics and the fracture healing process in vivo. Congruent with prior studies, we found that Notch inhibition via dnMAML suppressed osteoclast function both in vitro and in vivo without significant reduction of early osteoclastic gene expression or coupled osteoblast activity. From our ex vivo experiments, we hypothesize that dnMAML-expressing osteoclast precursors undergo early differentiation toward osteoclasts but fail to fuse and persist as mononuclear TRAP+ cells, though this morphological difference is difficult to ascertain in vivo. Furthermore, where alterations in osteoblast number or function were observed, these parameters were increased, and this resulted in increased bone formation rate and strength. With this increased osteoblast activity, however, we did not observe any significant alterations in bone architecture, which suggests that, while dnMAML-expressing osteoclasts have reduced activity, their resorptive function is sufficient for physiological bone remodeling.
Despite modest changes at physiological baseline, LysM-dnMAML mice showed significant increases in bony callus and mineralization compared to WT. Unaltered TRAP+ cell numbers in the healing fractures suggests that, as with uninjured animals, the increase in bone is more likely due to reduced resorptive activity rather than cell number. Taken with the physiological remodeling data, our work suggests that osteoclast Notch signaling is dispensable with respect to level of osteoclast activity required for normal remodeling. Thus, therapeutic inhibition of osteoclast Notch signaling could result in reduction of osteoclast activity in regions of the skeleton where that activity would have been pathologically elevated while sparing restorative remodeling elsewhere.
While previous clinical studies utilizing bisphosphonates and RANKL inhibition52–54 suggested that osteoclast inhibition does not significantly impact fracture callus parameters, the conclusions of these studies cannot distinguish between osteoclast function and osteoclast number as both interventions reduce osteoclast number by either triggering osteoclast apoptosis (bisphosphonates) or inhibiting osteoclastogenesis from an early stage (RANKL inhibition). The negative findings of these studies may be due to a concomitant decrease in osteoblast activity following reduction in osteoclast number, which would effectively neutralize any positive effect that osteoclast inhibition might have had on callus size and mineralization. Thus, while reduction of osteoclast number can promote increased bone mass and decreased fracture risk in the short term, long term suppression of bone remodeling is a source of concern55; 56. Reduction of osteoclast activity without impacting their coupling to osteoblasts, as we have demonstrated with osteoclast-specific inhibition of Notch signaling, has the potential to alleviate concerns regarding long-term anti-resorptive therapy and produce larger, potentially more stable fracture calluses through the preservation of osteoclast-stimulated osteogenesis in the context of reduced bone resorption.
Supplementary Material
Acknowledgements
We thank the AO foundation (JWA) [Project number for the AO grant is S-16–12A], NIH/NIAMS P30AR069619, and support from Department of Orthopaedic Surgery, Perelman School of Medicine at the University of Pennsylvania, and Corporal Michael J. Crescenz VA Medical Center. We also thank Sarah Teegarden, Ph.D. and Garret FitzGerald, MD of the University of Pennsylvania for providing us with Lysozyme M-Cre mice. J Ahn and KD Hankenson are co-owners of Skelegen, a biologics company.
Grant Supporters: AO foundation (Project number S-16–12A) and NIAMS (P30AR069619) of the NIH.
Footnotes
Conflict of interest: None
Ethical review committee: Procedure accepted by IACUC at University of Pennsylvania (protocol approval attached)
Work performed: VA Medical Centre, Philadelphia and School of Veterinary Medicine, Hill Pavilion at University of Pennsylvania.
References
- 1.Bahney CS, Zondervan RL, Allison P, et al. 2019. Cellular biology of fracture healing. J Orthop Res 37:35–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schell H, Lienau J, Epari DR, et al. 2006. Osteoclastic activity begins early and increases over the course of bone healing. Bone 38:547–554. [DOI] [PubMed] [Google Scholar]
- 3.Takeyama K, Chatani M, Takano Y, et al. 2014. In-vivo imaging of the fracture healing in medaka revealed two types of osteoclasts before and after the callus formation by osteoblasts. Dev Biol 394:292–304. [DOI] [PubMed] [Google Scholar]
- 4.Beraldi R, Masi L, Parri S, et al. 2014. The role of the orthopaedic surgeon in the prevention of refracture in patients treated surgically for fragility hip and vertebral fracture. Clin Cases Miner Bone Metab 11:31–35. [PMC free article] [PubMed] [Google Scholar]
- 5.Hobby B, Lee MA. 2013. Managing atrophic nonunion in the geriatric population: incidence, distribution, and causes. Orthop Clin North Am 44:251–256. [DOI] [PubMed] [Google Scholar]
- 6.Sibai T, Morgan EF, Einhorn TA. 2011. Anabolic agents and bone quality. Clin Orthop Relat Res 469:2215–2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dimitriou R, Jones E, McGonagle D, et al. 2011. Bone regeneration: current concepts and future directions. BMC Med 9:66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Giannotti S, Bottai V, Dell’osso G, et al. 2013. Current medical treatment strategies concerning fracture healing. Clin Cases Miner Bone Metab 10:116–120. [PMC free article] [PubMed] [Google Scholar]
- 9.Kostenuik P, Mirza FM. 2017. Fracture healing physiology and the quest for therapies for delayed healing and nonunion. J Orthop Res 35:213–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Levaot N, Ottolenghi A, Mann M, et al. 2015. Osteoclast fusion is initiated by a small subset of RANKL-stimulated monocyte progenitors, which can fuse to RANKL-unstimulated progenitors. Bone 79:21–28. [DOI] [PubMed] [Google Scholar]
- 11.Soe K, Hobolt-Pedersen AS, Delaisse JM. 2015. The elementary fusion modalities of osteoclasts. Bone 73:181–189. [DOI] [PubMed] [Google Scholar]
- 12.Henriksen K, Karsdal MA, Martin TJ. 2014. Osteoclast-derived coupling factors in bone remodeling. Calcified tissue international 94:88–97. [DOI] [PubMed] [Google Scholar]
- 13.Karsdal MA, Martin TJ, Bollerslev J, et al. 2007. Are nonresorbing osteoclasts sources of bone anabolic activity? Journal of bone and mineral research : the official journal of the American Society for Bone and Mineral Research 22:487–494. [DOI] [PubMed] [Google Scholar]
- 14.Ota K, Quint P, Ruan M, et al. 2013. TGF-beta induces Wnt10b in osteoclasts from female mice to enhance coupling to osteoblasts. Endocrinology 154:3745–3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pederson L, Ruan M, Westendorf JJ, et al. 2008. Regulation of bone formation by osteoclasts involves Wnt/BMP signaling and the chemokine sphingosine-1-phosphate. Proceedings of the National Academy of Sciences of the United States of America 105:20764–20769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Teitelbaum SL. 2016. Therapeutic implications of suppressing osteoclast formation versus function. Rheumatology (Oxford) 55:ii61–ii63. [DOI] [PubMed] [Google Scholar]
- 17.Bai S, Kopan R, Zou W, et al. 2008. NOTCH1 regulates osteoclastogenesis directly in osteoclast precursors and indirectly via osteoblast lineage cells. J Biol Chem 283:6509–6518. [DOI] [PubMed] [Google Scholar]
- 18.Fukushima H, Nakao A, Okamoto F, et al. 2008. The association of Notch2 and NF-kappaB accelerates RANKL-induced osteoclastogenesis. Mol Cell Biol 28:6402–6412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sekine C, Koyanagi A, Koyama N, et al. 2012. Differential regulation of osteoclastogenesis by Notch2/Delta-like 1 and Notch1/Jagged1 axes. Arthritis Res Ther 14:R45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yamada T, Yamazaki H, Yamane T, et al. 2003. Regulation of osteoclast development by Notch signaling directed to osteoclast precursors and through stromal cells. Blood 101:2227–2234. [DOI] [PubMed] [Google Scholar]
- 21.Ashley JW, Ahn J, Hankenson KD. 2015. Notch signaling promotes osteoclast maturation and resorptive activity. J Cell Biochem 116:2598–2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kitagawa M 2016. Notch signalling in the nucleus: roles of Mastermind-like (MAML) transcriptional coactivators. J Biochem 159:287–294. [DOI] [PubMed] [Google Scholar]
- 23.McElhinny AS, Li JL, Wu L. 2008. Mastermind-like transcriptional co-activators: emerging roles in regulating cross talk among multiple signaling pathways. Oncogene 27:5138–5147. [DOI] [PubMed] [Google Scholar]
- 24.Andersson ER, Sandberg R, Lendahl U. 2011. Notch signaling: simplicity in design, versatility in function. Development 138:3593–3612. [DOI] [PubMed] [Google Scholar]
- 25.Maillard I, Tu L, Sambandam A, et al. 2006. The requirement for Notch signaling at the beta-selection checkpoint in vivo is absolute and independent of the pre-T cell receptor. J Exp Med 203:2239–2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Clausen BE, Burkhardt C, Reith W, et al. 1999. Conditional gene targeting in macrophages and granulocytes using LysMcre mice. Transgenic Res 8:265–277. [DOI] [PubMed] [Google Scholar]
- 27.Croke M, Ross FP, Korhonen M, et al. 2011. Rac deletion in osteoclasts causes severe osteopetrosis. J Cell Sci 124:3811–3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qi B, Cong Q, Li P, et al. 2014. Ablation of Tak1 in osteoclast progenitor leads to defects in skeletal growth and bone remodeling in mice. Sci Rep 4:7158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stemig M, Astelford K, Emery A, et al. 2015. Deletion of histone deacetylase 7 in osteoclasts decreases bone mass in mice by interactions with MITF. PLoS One 10:e0123843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kaur G, Ahn J, Hankenson KD, et al. 2017. Stimulation of Notch Signaling in Mouse Osteoclast Precursors. J Vis Exp. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ashley JW, Ahn J, Hankenson KD. 2015. Notch Signaling Promotes Osteoclast Maturation and Resorptive Activity. J Cell Biochem. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vesprey A, Yang W. 2016. Pit Assay to Measure the Bone Resorptive Activity of Bone Marrow-derived Osteoclasts. Bio Protoc 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Merrild DM, Pirapaharan DC, Andreasen CM, et al. 2015. Pit- and trench-forming osteoclasts: a distinction that matters. Bone Res 3:15032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Panwar P, Soe K, Guido RV, et al. 2016. A novel approach to inhibit bone resorption: exosite inhibitors against cathepsin K. Br J Pharmacol 173:396–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boissy P, Andersen TL, Abdallah BM, et al. 2005. Resveratrol inhibits myeloma cell growth, prevents osteoclast formation, and promotes osteoblast differentiation. Cancer Res 65:9943–9952. [DOI] [PubMed] [Google Scholar]
- 36.Soe K, Andersen TL, Hobolt-Pedersen AS, et al. 2011. Involvement of human endogenous retroviral syncytin-1 in human osteoclast fusion. Bone 48:837–846. [DOI] [PubMed] [Google Scholar]
- 37.Srivastava AK, Bhattacharyya S, Castillo G, et al. 2000. Development and evaluation of C-telopeptide enzyme-linked immunoassay for measurement of bone resorption in mouse serum. Bone 27:529–533. [DOI] [PubMed] [Google Scholar]
- 38.Greenblatt MB, Tsai JN, Wein MN. 2017. Bone Turnover Markers in the Diagnosis and Monitoring of Metabolic Bone Disease. Clin Chem 63:464–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li LJ, Kim SN, Cho SA. 2016. Comparison of alkaline phosphatase activity of MC3T3-E1 cells cultured on different Ti surfaces: modified sandblasted with large grit and acid-etched (MSLA), laser-treated, and laser and acid-treated Ti surfaces. J Adv Prosthodont 8:235–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Knight MN, Karuppaiah K, Lowe M, et al. 2018. R-spondin-2 is a Wnt agonist that regulates osteoblast activity and bone mass. Bone Res 6:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rentsch C, Schneiders W, Manthey S, et al. 2014. Comprehensive histological evaluation of bone implants. Biomatter 4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gruber HE. 1992. Adaptations of Goldner’s Masson trichrome stain for the study of undecalcified plastic embedded bone. Biotech Histochem 67:30–34. [DOI] [PubMed] [Google Scholar]
- 43.Zondervan RL, Vorce M, Servadio N, et al. 2018. Fracture Apparatus Design and Protocol Optimization for Closed-stabilized Fractures in Rodents. J Vis Exp. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bouxsein ML, Boyd SK, Christiansen BA, et al. 2010. Guidelines for assessment of bone microstructure in rodents using micro-computed tomography. Journal of bone and mineral research : the official journal of the American Society for Bone and Mineral Research 25:1468–1486. [DOI] [PubMed] [Google Scholar]
- 45.de Bakker CM, Altman AR, Tseng WJ, et al. 2015. muCT-based, in vivo dynamic bone histomorphometry allows 3D evaluation of the early responses of bone resorption and formation to PTH and alendronate combination therapy. Bone 73:198–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.O’Neill KR, Stutz CM, Mignemi NA, et al. 2012. Micro-computed tomography assessment of the progression of fracture healing in mice. Bone 50:1357–1367. [DOI] [PubMed] [Google Scholar]
- 47.Morgan EF, Mason ZD, Chien KB, et al. 2009. Micro-computed tomography assessment of fracture healing: relationships among callus structure, composition, and mechanical function. Bone 44:335–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kahveci Z, Minbay FZ, Cavusoglu L. 2000. Safranin O staining using a microwave oven. Biotech Histochem 75:264–268. [DOI] [PubMed] [Google Scholar]
- 49.Tran D, Golick M, Rabinovitz H, et al. 2000. Hematoxylin and safranin O staining of frozen sections. Dermatol Surg 26:197–199. [DOI] [PubMed] [Google Scholar]
- 50.Camplejohn KL, Allard SA. 1988. Limitations of safranin ‘O’ staining in proteoglycan-depleted cartilage demonstrated with monoclonal antibodies. Histochemistry 89:185–188. [DOI] [PubMed] [Google Scholar]
- 51.Dishowitz MI, Mutyaba PL, Takacs JD, et al. 2013. Systemic inhibition of canonical Notch signaling results in sustained callus inflammation and alters multiple phases of fracture healing. PLoS One 8:e68726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Delos D, Yang X, Ricciardi BF, et al. 2008. The effects of RANKL inhibition on fracture healing and bone strength in a mouse model of osteogenesis imperfecta. J Orthop Res 26:153–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kates SL, Ackert-Bicknell CL. 2016. How do bisphosphonates affect fracture healing? Injury 47 Suppl 1:S65–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xue D, Li F, Chen G, et al. 2014. Do bisphosphonates affect bone healing? A meta-analysis of randomized controlled trials. J Orthop Surg Res 9:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rasmusson L, Abtahi J. 2014. Bisphosphonate associated osteonecrosis of the jaw: an update on pathophysiology, risk factors, and treatment. Int J Dent 2014:471035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sellmeyer DE. 2010. Atypical fractures as a potential complication of long-term bisphosphonate therapy. JAMA 304:1480–1484. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.