Abstract
CCL20 (CC chemokine ligand 20) is emerging as an important regulatory molecule in a pathway common to virus infection, alcoholic hepatitis, and non-alcoholic fatty liver disease (NAFLD) leading to the development of hepatic fibrosis. We previously observed upregulation of CCL20 in patients with NAFLD fibrosis and human hepatic stellate cells (LX-2 cells) in response to lipid loading. To date, the mechanisms mediating the relationship between CCL20 and hepatic fibrogenesis remain unknown. In this study, we sought to characterize the molecular mechanisms by which CCL20 may contribute to fibrogenesis in NAFLD. We observed that CCL20 levels increased with worsening severity of liver histology in NAFLD patients (normal < steatosis <inflammation < fibrosis) and during LX-2 cell activation in a time-dependent manner. We found that treatment of LX-2 cells with CCL20 corresponded with increased levels of CCL20 and ACTA2, and decreased levels of PLAU and SERPINE1, effects mitigated by CCL20 knockdown. We identified a putative binding site for miR-590-5p, which we previously reported to be downregulated in NAFLD fibrosis, in the CCL20 3’ untranslated region (3’UTR), and found that exogenous miR-590-5p functionally interacted with the CCL20 3’UTR to downregulate its expression. Transfection of LX-2 hepatic stellate cells with miR-590-5p mimic or silencing RNA resulted in decreased or increased CCL20 levels, respectively. Our results indicate an association between CCL20 and hepatic stellate cell activation that includes modulation of key ECM components and functional interactions with a miRNA previously implicated in NAFLD fibrosis. Together, these findings support a novel mechanism by which CCL20 may promote fibrogenesis in NAFLD.
Keywords: liver, hepatic stellate cells, LX-2 cells, nonalcoholic steatohepatitis, liver fibrosis, miRNA, chemokine, inflammation, miR-590-5p
Introduction
Nonalcoholic fatty liver disease (NAFLD), the most common cause of chronic liver disease worldwide [1], is characterized by excessive hepatic triglyceride accumulation and encompasses a spectrum of pathologies from steatosis to nonalcoholic steatohepatitis (NASH), which can present with varying degrees of fibrosis [2]. NAFLD progression from simple steatosis to hepatic inflammation and liver fibrosis results from an intricate interplay among a number of cell types including hepatocytes, hepatic stellate cells, sinusoidal endothelial cells, Kupffer cells, and infiltrating immune cells [3]. The molecular mechanisms regulating this complex, multifactorial process remain only partially defined, although our current understanding is that multiple insults that include, but are not limited to, insulin resistance [4], oxidative stress [5], gut dysbiosis [6], and adipose-tissue-derived hormones [4] act in concert to induce specific manifestations of the disease in susceptible individuals [4].
In addition to these factors, excess hepatic fat accumulation is accompanied by increased levels of inflammatory mediators, including cytokines, chemokines, and adipocytokines, that stimulate the development of hepatic steatosis and fibrosis by hepatocytes and hepatic stellate cells, respectively [7]. Chemokines belong to a family of small cytokines with chemotactic functions [8], and are known to induce leukocyte trafficking, growth, and activation in inflammatory sites [9]. In the liver, endothelial cells, hepatocytes, leukocytes, and stellate cells secrete chemokines in response to injury [10]. Experimental studies aimed at elucidating the contributions of chemokine (C-C motif) ligand 2 (CCL2) and chemokine (C-C motif) ligand 5 (CCL5), and their cognate receptors (CCR2 and CCR5), in NAFLD pathogenesis and disease progression have been ongoing for at least a decade [11–14]. More recently, chemokine (C-C motif) ligand 20 (CCL20) has emerged as an important proinflammatory and profibrogenic hepatic chemokine [15]. CCL20 levels are elevated in a number of chronic liver diseases, including alcoholic hepatitis [15] and hepatocellular carcinoma [16–19]. Serum [20, 21] and hepatic [20] levels of CCL20 have also been observed in NAFLD patients with coincident fibrosis compared to those without fibrosis or with normal liver histology. In patients with hepatitis C virus infection [19] and alcoholic hepatitis [15], CCL20 levels have been correlated with disease severity, although the correspondence of hepatic CCL20 levels with histological findings of steatosis, lobular inflammation, and hepatic fibrosis in NAFLD patients has not yet been explored.
In the liver, hepatic stellate cells are one of the main CCL20-producing cell types [15]. In these cells, CCL20 expression is induced by lipopolysaccharide (LPS), tumor necrosis factor alpha, and interleukin 1β [15]. In a recent study, we demonstrated that CCL20 expression is upregulated by fatty acid loading in LX-2 cells, an immortalized human hepatic stellate cell line that retains key features of primary cells [20]. Hepatic stellate cells are pivotal in fibrogenesis, existing in a biologically quiescent state under physiologic conditions, and transforming to myofibroblasts in response to hepatic injury [22]. Increased production of alpha-smooth muscle actin (ACTA2), collagen type 1, alpha 1 (COL1A1), and other components of the extracellular matrix (ECM) accompanies this phenotypic transition, and in the presence of chronic hepatic inflammation, activated hepatic stellate cells produce excessive ECM, which can eventually lead to hepatic scarring [23]. Taken together, these findings support a relationship among CCL20, hepatic stellate cell function, and biological mechanisms relevant to the pathogenesis of NAFLD fibrosis.
In the present study, we used a combination of human liver samples and LX-2 cells to extend our previous work on the regulation of CCL20 in NAFLD fibrosis. We show that hepatic CCL20 expression increases incrementally across the histological spectrum of NAFLD (steatosis, inflammation, and fibrosis) and in LX-2 cells grown under conditions to induce myofibroblastic activation. We found evidence that CCL20 is associated with ACTA2, COL1A1, endothelial plasminogen activator inhibitor 1 (PAI-1, encoded by the SERPINE1 gene), and plasminogen activator, urokinase (PLAU), which are important ECM components or regulators of ECM maintenance. In analyses aimed at characterizing CCL20 regulation, we identified a potential binding site for miR-590-5p in the CCL20 3’-untranslated region, confirmed that the miRNA functionally interacts with this sequence, and demonstrated that miR-590-5p regulates CCL20 expression in LX-2 cells. These results point to a novel mechanism by which CCL20 participates in hepatic fibrogenesis in NAFLD, including an apparent upregulation as early as the development of simple steatosis.
Materials and Methods
RNA sequencing data analysis
Details of the patient samples, RNA-sequencing experiments, and raw data processing were previously described [24]. For the analysis here, the raw count table data was filtered including genes with an average count > 5 across all samples. To make the data suitable for regression analysis, we normalized the counts using the voom algorithm [25] and adjusted for batch effects using the ComBat algorithm [26]. Following removal of outliers, the final sample size across the four histological grades was: normal (n = 36), steatosis (n = 50), lobular inflammation (n = 52), and severe fibrosis (n = 53). To investigate the relationship between CCL20 gene expression and histological grade, we modeled an ordinal logistic regression considering grade as dependent variable, adjusting for sex and age. Finally, we conducted pairwise comparison of the expression level of normal samples versus the other classes using a logistic regression considering increasing histological severity as dependent variable, and adjusting for sex and age. P values were adjusted for multiple comparisons using the Bonferroni method. All the analyses were conducted using R software.
RNA extraction from liver wedge biopsies
Liver wedge biopsies were obtained from individuals enrolled in the Bariatric Surgery Program at the Geisinger Clinic Center for Nutrition and Weight Management [27]. Details of the study population can be found elsewhere [28–30]. All study participants provided written informed consent for research, which was conducted according to The Code of Ethics of the World Medical Association (Declaration of Helsinki). The Institutional Review Boards of Geisinger Health System, Translational Genomics Research Institute, and Temple University School of Medicine approved the research protocol. Approximately 5–10 mg of biopsied liver tissue was placed in RINO tubes (Next Advance; Troy, NY) containing Buffer RLT Plus with 2- mercaptoethanol (Qiagen; Germantown, MD) and processed in a Bullet Blender Tissue Homogenizer (Next Advance) for 15 minutes. Total RNA was extracted from homogenized lysate using the RNeasy Mini Kit (Qiagen) and quantified by the NanoDrop One spectrophotometer (Applied Biosystems; Foster City, CA). RealTime quantitative PCR (qPCR) was performed as described below.
Cell culture
LX-2 cells (Merck Millipore; Billerica, MA) were cultured in Dulbecco’s Modified Eagle Medium (DMEM) (Thermo Fisher Scientific; Waltham, MA) supplemented with 2% fetal bovine serum (FBS) and 1% Pen/Strep (Omega Scientific; Tarzana, CA). Approximately 1 × 106 cells were thawed in T-75 flasks (Corning Life Sciences; Corning, NY) containing 12 mL cell culture medium and placed at 37°C in a Hera Cell 5% CO2 incubator (Thermo Fisher Scientific). Culture medium was replaced the first day after thawing, and then every 72 hours until 80% confluent. In preparation for plating on Matrigel, LX-2 cells were seeded at 0.5 × 106 cell/well on 6-well culture dishes (VWR International; Radnor, PA) and serum-starved overnight. Matrigel Growth Factor Reduced (GFR) Basement Membrane Matrix (Corning) was thawed by submerging the unopened bottle in ice at 4°C for four hours. Using chilled pipet tips, thawed Matrigel was transferred to a conical tube containing cold serum-free media to achieve a final concentration of 1 mg/mL, and 350 μL of this mixture was added to chilled 6-well plates (VWR International) and rocked to ensure even coating. Plates were incubated overnight at 37°C to allow matrix solidification. LX-2 cells were seeded onto Matrigel-coated plates at a density of 1×105 cells/well and incubated at 37°C for up to 72 hours. General morphology was observed with a light microscope and cell state was assessed by levels of Oil Red O staining and measurement of alpha smooth muscle actin (ACTA2) and collagen 1 alpha 1 (COL1A1).
HepG2 and HEK293 cells (ATCC; Manassas, VA) were cultured in Dulbecco’s Modified Eagle Medium (DMEM) (Thermo Fisher Scientific) supplemented with 10% fetal bovine serum (FBS) and 1% Pen/Strep (Omega Scientific). Culture medium was replaced the first day after thawing, and then every 48 hours until 80% confluent. Primary human hepatocytes (Thermo Fisher Scientific) were thawed in 50 mL Cryopreserved Hepatocytes Recovery Medium (Thermo Fisher Scientific) and plated in 500 μL William’s E Medium supplemented with Hepatocyte Plating Supplement Pack on collagen-coated 24-well plates (Thermo Fisher Scientific). Culture medium was replaced the first day after thawing with William’s E Medium supplemented with Hepatocyte Maintenance Supplement Pack (Thermo Fisher Scientific).
Cell treatments
For the CCL20 experiments, LX-2 cells were seeded at 2×105 cells/well in 6-well culture dishes (VWR International) containing 2.0 mL cell culture medium and placed at 37°C in a Hera Cell 5% CO2 incubator (Thermo Fisher Scientific). Prior to all treatments, cells were serum-starved overnight. CCL20 stock solution was prepared by dissolving lyophilized human recombinant CCL20 (Sigma-Aldrich; St. Louis, MO) in water to a concentration of 1 mg/mL. Serum-free media was aspirated from cells and replaced with media containing 250 ng/mL or 1 μg/mL CCL20.
For treatments with primary human hepatocytes, palmitate (Sigma-Aldrich) was first conjugated to bovine serum albumin (BSA; [Omega Scientific]) at a final concentration of 1 mM palmitate and 1% BSA by heating at 37°C for 30 minutes prior to cell treatments. Cells were serum-starved overnight, and then treated with 1% BSA (as the negative control), 20 mM fructose, 1 mM BSA-conjugated palmitate, or a combination of the two for 48 hours. Oil Red O staining was used to assess lipid uptake. For the glucose treatments, LX-2 and HepG2 cells were seeded at 1×105 cells/well in 6-well plates and serum-starved overnight. Cells were grown in 5 mM glucose (normal glucose, NG), 25 mM glucose (high glucose, HG), or 25 mM mannitol as an osmotic control. CCL20 fold-change relative to normal glucose was measured at 0, 4, 24, and 48 hours in LX-2 cells and 0, 24, and 48 hours in HepG2 cells.
Total RNA extraction and quantification from cells
Total RNA was extracted from cells using the RNeasy mini kit (Qiagen) according to the manufacturer’s protocol. RNA quality and concentration was determined by absorbance at 260 nm using the NanoDrop One spectrophotometer (Thermo Fisher Scientific). MicroRNA was extracted from cells using the miRNeasy mini kit (Qiagen) according to the manufacturer’s protocol. The miRNA quantity and quality were assessed using the small RNA chip on the 2100 Bioanalyzer System (Agilent Technologies; Santa Cruz, CA).
Quantitative real-time PCR (qPCR)
The TaqMan RNA-to-Ct 1-Step kit (Thermo Fisher Scientific) in conjunction with the QuantStudio 6 Flex Real-Time PCR system (Thermo Fisher Scientific) and TaqMan commercial primers (information available upon request) were used to measure mRNA levels. MiRNA was processed using the Taqman MicroRNA Reverse Transcription Kit (Applied Biosystems), followed by amplification using commercial probes (Applied Biosystems). Cycle threshold levels were generated using QuantStudio Real-Time PCR Software 1.0. Messenger RNA and miRNA data were normalized using glyceraldehyde 3-phosphate dehydrogenase (GAPDH) or a combination of RNU48 and RNU6B (which showed invariant expression levels), respectively. The −ΔΔCt method was used to determine fold-change of gene expression. All experiments were performed in triplicate. Primer sequences are available upon request.
Transfection with CCL20 siRNA
Approximately 2×105 LX-2 cells/well were seeded in a 24-well plate with 6 μL Lipofectamine RNAiMAX transfection reagent (Thermo Fisher Scientific) containing either 50 nM or 100 nM Silencer Select Pre-Designed siRNA directed against CCL20 or Silencer Select Negative Control Number 1 siRNA (both Thermo Fisher Scientific) and incubated at 37°C. Cells were harvested after 48 or 72 hours. Total RNA was extracted and expression of CCL20, ACTA2, COL1A1, SERPINE1, and PLAU was assessed as described.
Prediction of miRNAs targeting the 3’ untranslated region (3’UTR) of CCL20
Potential CCL20-targeting miRNAs were identified using six different prediction algorithms (miRWalk, Microt4, miRanda, miRDB, RNA22, and Targetscan) as implemented in miRWalk 2.0 (http://zmf.umm.uni-heidelberg.de/apps/zmf/mirwalk2/). The miRmap tool [31] was used to predict miRNA target repression strength.
3’UTR dual-luciferase reporter assay
Reporters were constructed using the pmirGLO Dual-Luciferase miRNA Target Expression Vector (Promega Corp; Madison, WI). The 3’ untranslated region (3’UTR) of CCL20 or a scrambled sequence insert were generated by PCR and cloned into the multiple cloning site (MCS) of the pmirGLO vector by the Emory Integrated Genomics Core (Atlanta, GA). The resulting (pGLO-CCL20 3’UTR) and (pGLO-Scramble) reporter plasmids were sequence-verified. Approximately 2×105 HEK293 cells/well were seeded in a 24-well plate, co-transfected with 2 μg of the indicated vector and 10 pM miR-590-5p mimic or negative control mimic using 4 μl Lipofectamine 2000 per well (Thermo Fisher Scientific), and then incubated at 37°C for 48 hours. Luciferase activity was measured using the Dual-Glo Luciferase Assay System (Promega). Luminescence was detected using the EnVision 2105 Multimode Plate Reader (PerkinElmer; Waltham, MA). The ratio of firefly luciferase activity to Renilla luciferase activity was used to calculate normalized luciferase activity. The effect of miR-590-5p on gene expression was determined by setting the normalized luciferase activity of the negative mimic control + pGLO-CCL20 3’UTR vector to 100% and showing the normalized luciferase activity of miR-590-5p mimic + CCL20 3’UTR vector as a percentage. For the control conditions, relative luciferase activity was determined by showing the normalized luciferase activity of miR-590-5p + pGLO-Scramble vector as a percentage of the negative mimic control + vector control. All experiments were performed in triplicate. The Mann-Whitney U test was used to assess differences between conditions. A P-value <0.05 was considered statistically significant.
Transfection with miR-590-5p mimic and anti-miR-590-5p inhibitor
Approximately 2×105 LX-2 cells/well were seeded in a 24-well plate with 6 μL Lipofectamine RNAiMAX transfection reagent (Thermo Fisher Scientific) containing either 75 nM mirVana miRNA-590-5p mimic, 50 nM anti-miR-590-5p inhibitor, or 50 nM scrambled sequence control (mirVana Negative Control #1 [Thermo Fisher Scientific]) and incubated for 48 hours at 37°C. Total RNA was extracted using the miRNeasy kit (Qiagen; Germantown, MD) and miR-590-5p over-expression and knockdown was verified using qPCR as described.
Statistical analysis
All statistical analyses were performed using GraphPad Prism 8 (GraphPad Software; La Jolla, CA). Data were analyzed as the mean ± standard deviation (SD) from at least three independent assays. The Kruskal-Wallis one-way analysis of variance test with a Dunn post-hoc test were used to assess differences between conditions, unless otherwise indicated. A P-value <0.05 was considered statistically significant.
Results
CCL20 expression increases with worsening liver histology in NAFLD patients
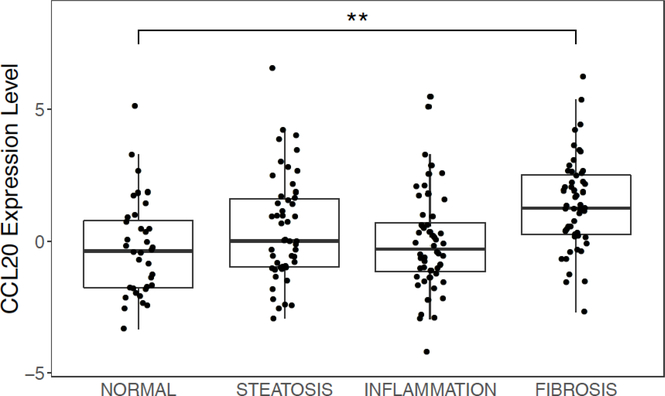
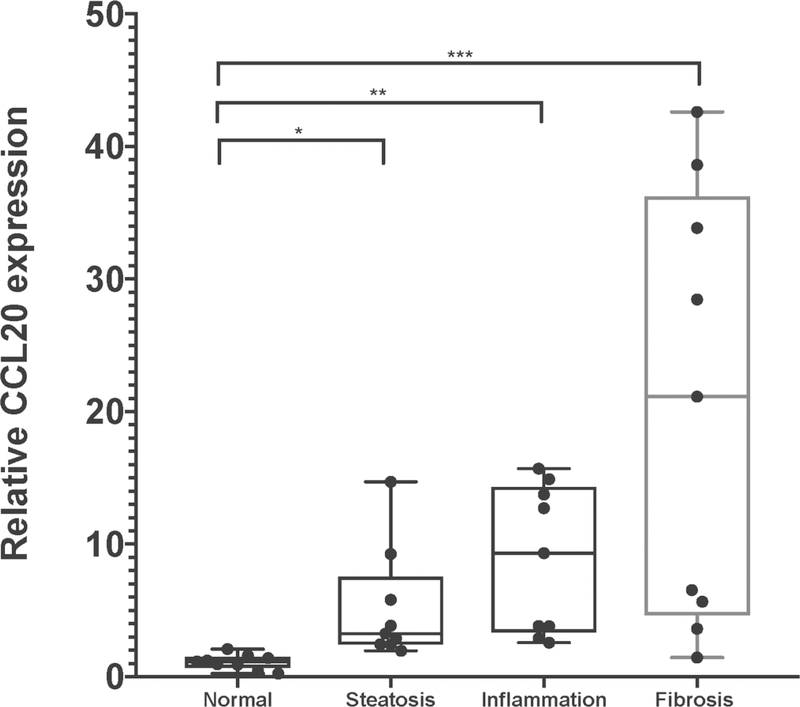
We previously observed an increase in circulating CCL20 protein levels that paralleled severity of liver fibrosis in NAFLD patients [20], a finding consistent with earlier work showing higher levels of CCL20 in hepatitis C virus-infected individuals with cirrhosis compared to those without cirrhosis [19] and correlation of hepatic CCL20 gene expression with prognostic scores (i.e., Model for End-stage Liver Disease) and hepatic cirrhosis in alcoholic hepatitis [15]. To evaluate hepatic CCL20 patterns across the histological spectrum of NAFLD, we first compared normalized counts of CCL20 derived from RNA-sequencing in patients with biopsy-proven steatosis, inflammation, and fibrosis (Supplementary Table 1). As shown in Figure 1A, CCL20 expression increased with worsening liver histology in NAFLD (ordinal logistic regression: β =0.244; Padj =4.8E-04). However, the comparison between normal liver histology and the other histological stages revealed that CCL20 levels were significantly different only between normal and severe fibrosis (β = 0.512; Padj = 1.4E-03). To validate these findings, we analyzed CCL20 expression using quantitative RT-PCR in a subset of NAFLD patients and found 5.2-fold, 8.8-fold, and 20.2fold upregulation of CCL20 expression in samples with biopsy-proven steatosis, inflammation, and severe fibrosis, respectively, compared to normal liver histology (Figure 1B), consistent with CCL20 expression increasing with progression across NAFLD histological stages.
Figure 1. CCL20 expression in liver samples from individuals spanning the histological spectrum of NAFLD.
A) Using RNA-sequencing data [24], we determined average CCL20 transcript abundance in NAFLD samples across a spectrum of liver histology (36 normal, 50 steatosis, 52 inflammation, and 53 fibrosis). The ordinal logistic regression showed a significant positive trend (β = 0.244; P = 4.8E-4), whereas the pairwise comparisons conducted with logistic regression showed a significant difference only between normal and fibrosis histological grades (Padj =1.3E-03). B) CCL20 gene expression was measured in a subset of NAFLD patients with steatosis (n=10), lobular inflammation (n=10), advanced fibrosis (n=10), and normal liver histology (n=10). Real time qPCR was performed using Taqman gene expression assays for CCL20 and GAPDH. The Kruskal-Wallis one-way analysis of variance test with a Dunn post-hoc test were used to assess differences in expression between each histological grade of NAFLD and normal histology. *P≤0.05, **P≤0.001, and *** P≤ 0.0001.
CCL20 expression is upregulated in activated LX-2 cells and lipid-loaded PHH
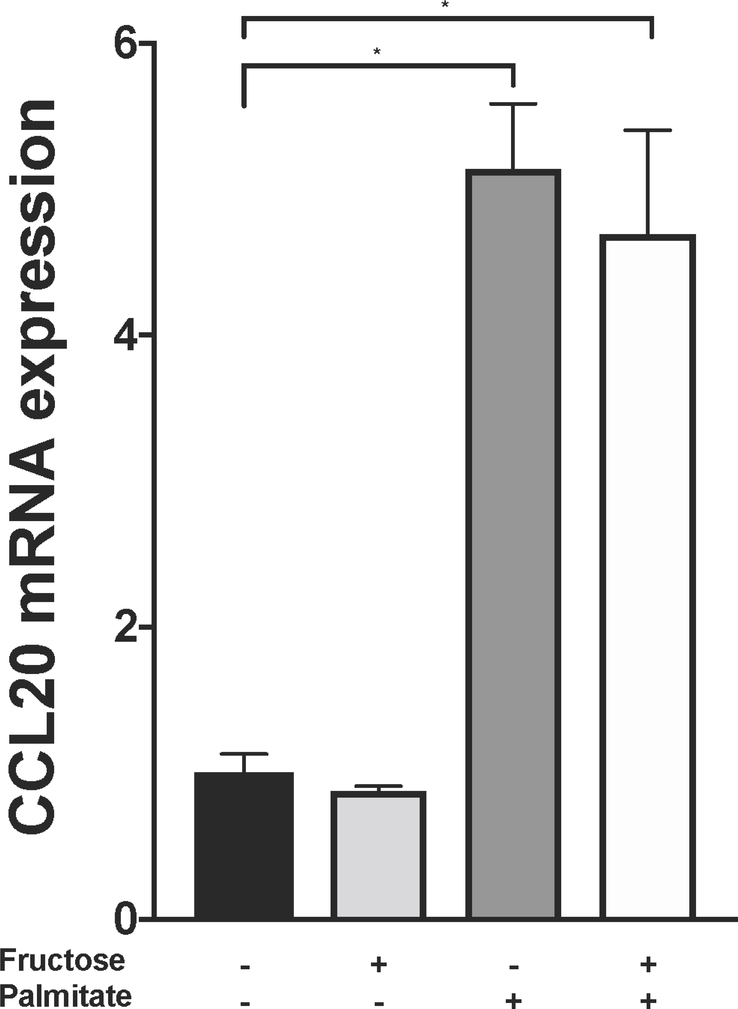
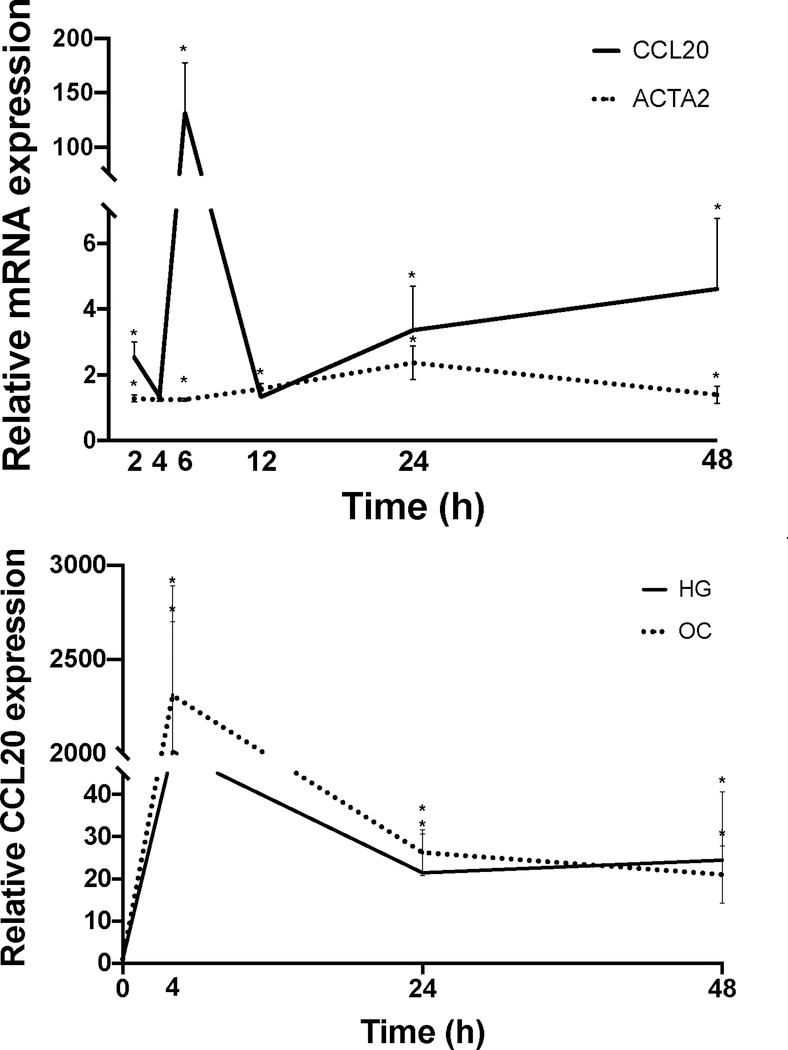
In LX-2 cells, CCL20 expression was upregulated in response to treatment with palmitic acid, oleic acid, or a palmitic acid/oleic acid (50/50) mixture, suggesting that hepatic stellate cells may play a participatory role during hepatic lipid-loading [20]. Similar results were observed in HepG2 cells, although to a lesser extent. To extend these findings, we first measured CCL20 expression in primary human hepatocytes in response to treatment with palmitate and fructose (Figure 2A). In the presence of 20 mM fructose, CCL20 levels were similar to those in untreated cells. However, in the presence of 1 mM palmitate, CCL20 expression was over five-fold higher compared to untreated cells; this finding was similar in cells treated with both palmitate and fructose, suggesting that the palmitate-induced upregulation of CCL20 was not affected by the presence of fructose. In light of the changes in expression that occurred during lipid loading of LX-2 cells [20], we next asked whether CCL20 levels were different between “activated” and “quiescent” LX-2 cells [32]. LX-2 cells plated on plastic quickly transform to a myofibroblastic phenotype; in these cells, we saw CCL20 expression increase 132-fold at six hours compared to time of plating (Figure 2B). After this, CCL20 expression decreased to levels observed in cells plated on Matrigel, which resemble a “quiescent” phenotype.
Figure 2. CCL20 expression patterns in hepatic cells.
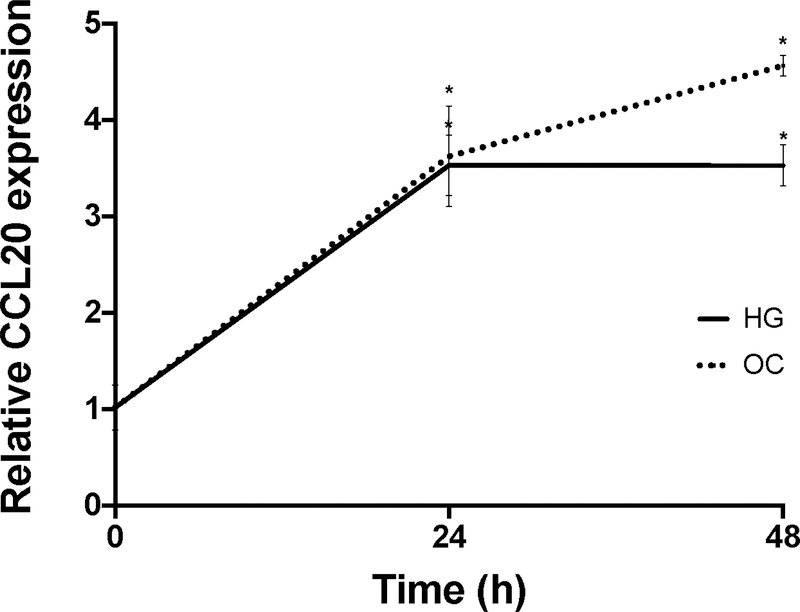
A) Primary human hepatocytes (PHHs) were treated with 1 mM BSA-conjugated palmitate, 20 mM fructose, or a combination of palmitate and fructose for 48 hours. The black bar represents BSA-only treatment conditions. B) LX-2 cells were either grown on plastic to achieve a myofibroblastic phenotype or on Matrigel (MG) to induce a state resembling biological quiescence. Expression levels for CCL20 and ACTA2 were normalized against GAPDH. C) LX-2 cells and D) HepG2 cells grown under normal (5mM), high (25 mM) glucose concentration, or mannitol (25 mM), as an osmotic control. CCL20 transcript abundance was measured by qPCR. CCL20 fold-change relative to normal glucose was measured at 4, 24, and 48 hours in LX-2 cells and 0, 24, and 48 hours in HepG2 cells. Differences in expression were assessed using the Mann-Whitney test. *P <0.05.
Insulin resistance is thought to be a central process in NAFLD [33]. CCL20 expression increases in renal proximal tubule cells exposed to high glucose (25 mM) and in the kidneys of diabetic rats [34]. To determine whether high glucose conditions affect CCL20 in liver cells, we treated LX-2 cells and HepG2 cells with 25 mM glucose for 48 hours. In LX-2 cells, CCL20 expression increased markedly at four hours, and then decreased, but remained at levels 20-fold higher than untreated cells (Figure 2C). In contrast, CCL20 levels in HepG2 cells steadily rose over the first 24 hours, and then plateaued at 3.5-fold induction (Figure 2D). In both cell types, CCL20 levels increased in response to treatment with the osmotic control (25 mM mannitol), suggesting that elevated CCL20 expression is due to osmotic stress and not high extracellular glucose concentration in liver cells.
CCL20 treatment increases levels of extracellular matrix components
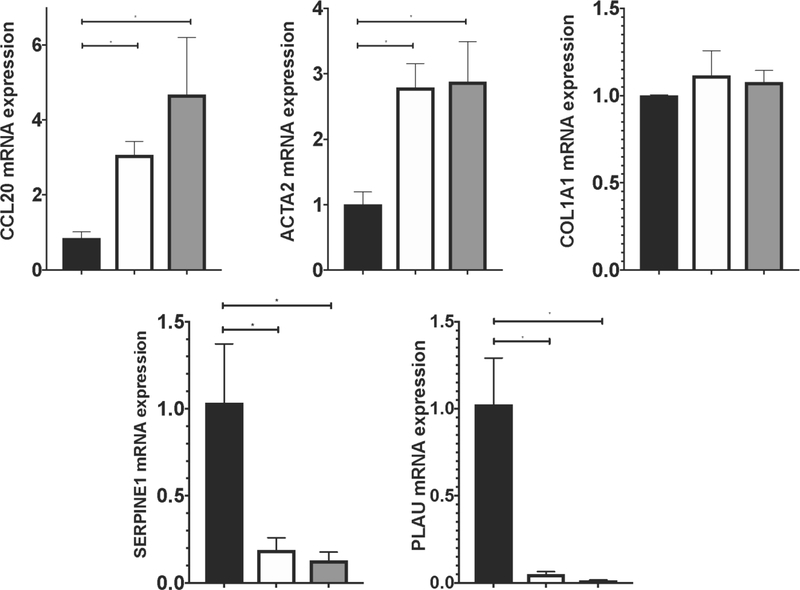
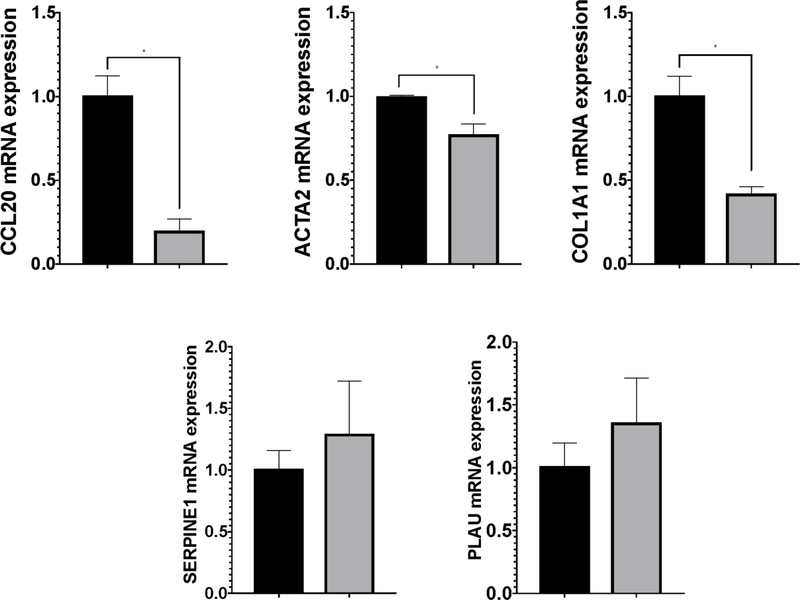
The presence of increased CCL20 levels in fibrotic liver samples from NAFLD patients suggests a potential relationship between CCL20 and expression of ECM-related components in hepatic stellate cells, the key fibrogenic cells in the liver. To address this question, we treated LX-2 cells with 250 ng/mL and 1 μg/mL of CCL20 for 24 hours. In the presence of CCL20, we observed upregulation of CCL20 and ACTA2 levels, and downregulation of SERPINE1 (serine protease inhibitor family E, member 1) and PLAU (plasminogen activator, urokinase, also known as urokinase plasminogen activator [uPA]) (Figure 3A). In contrast to results reported for primary human HSCs [15], we did not observe a statistically significant change in COL1A1 levels with CCL20 treatment, although knockdown of CCL20 resulted in a significant reduction of COL1A1 levels. CCL20 knockdown also corresponded with decreased levels of CCL20, and ACTA2, and a trend toward increased levels of SERPINE1 and PLAU (Figure 3B).
Figure 3. Effects of CCL20 on ECM components.
A) LX-2 cells were treated with 250 ng/mL (white bars), 1 μg/mL CCL20 (gray bars), or no treatment (NTC: black bars) for 48 hours and levels of CCL20, ACTA2, COL1A1, SERPINE1, and PLAU were measured by qPCR. B) LX-2 cells were transfected with 100 nM CCL20 siRNA for 72 hours. Black bars represent NTC and gray bars reflect cells treated with CCL20 siRNA. All experiments were performed in triplicate and data represent means ±SD. The Mann-Whitney U test was used to assess differences between conditions. *P<0.05.
CCL20 is regulated by miR-590-5p
In colorectal cancer cells, CCL20 is regulated by miR-21 [35], indicating an additional level of regulation with potential importance. Combined with our previous work showing dysregulated hepatic miRNA expression in NAFLD fibrosis [36], we sought to determine whether miRNA-mediated regulation of CCL20 occurred in NAFLD fibrosis. We first analyzed the CCL20 sequence to identify putative miRNA binding sites using a comparative platform comprised of six different algorithms (see METHODS section). Although 768 miRNA binding site predictions were identified using one or more of algorithms, the combination of all six algorithms together predicted binding sites for only three miRNAs: miR-590-5p, miR-557, and miR-507. Using the integrated miRmap tool [31], miRmap scores for the three miRNAs were 91.69, 95.91, and 92.99 for miR-590-5p, miR-557, and miR-507, respectively. Based on our previous work showing a 2.3-fold reduction (Padj =0.01) in miR-590-5p expression in NAFLD patients with fibrosis compared to those with normal histology (but no differences in miR-557 and miR-507 levels) [28], we sought to ascertain whether miR-590-5p might regulate CCL20 expression in liver cells.
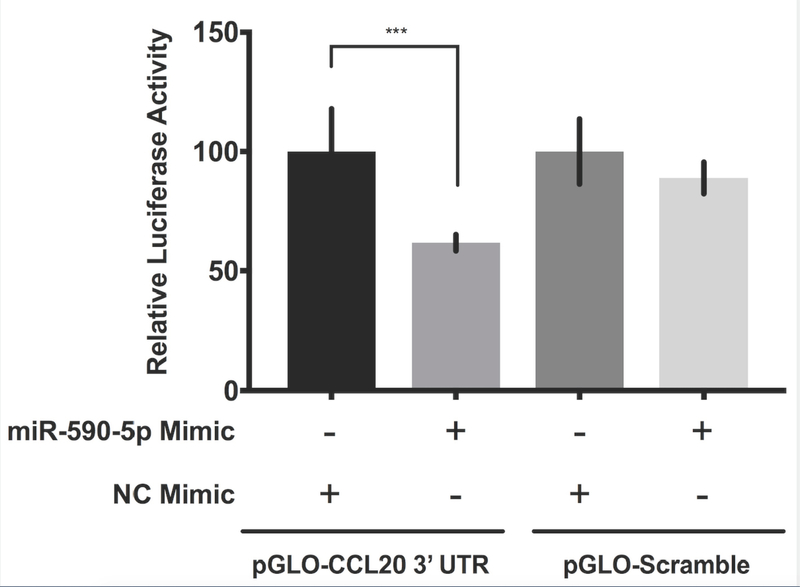
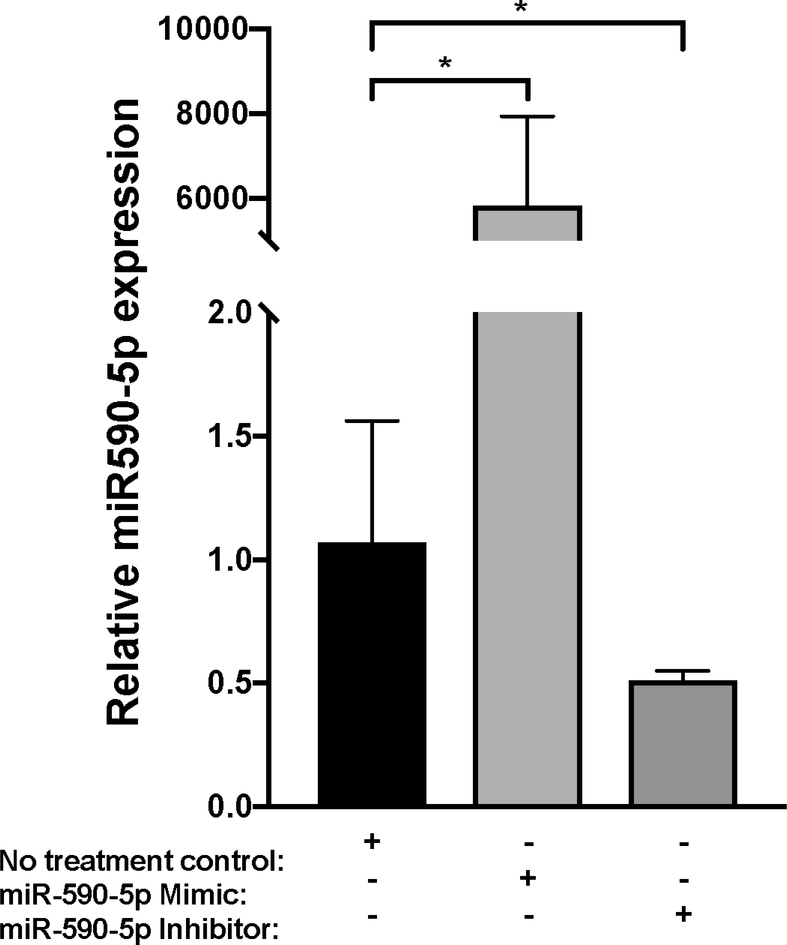
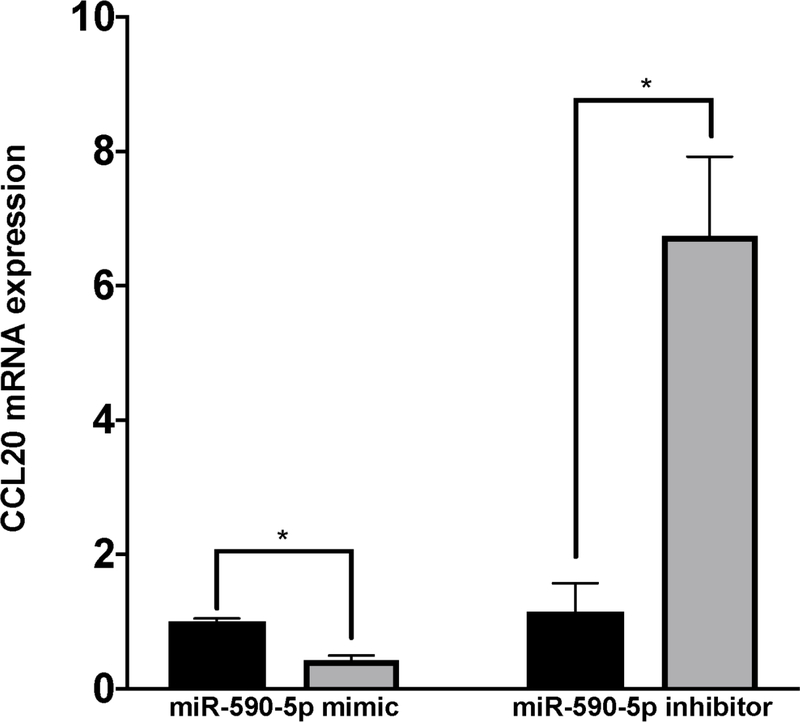
According to miRmap [31], the predicted miR-590-5p binding site aligns to positions 261–267 in the CCL20 3’UTR (Figure 4A). To validate this interaction, we used a dual luciferase reporter assay to measure direct binding between miR-590-5p and the CCL20 3’UTR. Co-transfection of a construct containing the CCL20 3’UTR and exogenous miR-590-5p resulted in ~40% decrease in relative luciferase activity compared to control conditions (Figure 4B). We next transfected LX-2 cells with miR-590-5p mimic or siRNA to determine possible functional consequences of miR-590-5p on CCL20 expression. We observed the expected increased or decreased miR-590-5p expression in the presence of mimic and inhibitor, respectively (Figure 4C). In the presence of exogenous miR-590-5p, CCL20 levels decreased 2.4-fold compared to untreated cells, and increased 6.7-fold in response to treatment with miR-590-5p siRNA (Figure 4D), suggesting that this miRNA directly downregulates expression of CCL20.
Figure 4. miR-590-5p functionally interacts with CCL20 to regulate its expression.
A) The miR-590-5p target site was predicted to align to positions 261–267 in the CCL20 3’UTR (GenBank Accession NM_004591). Vertical lines indicate paired alignment between the miRNA seed region and CCL20 3’UTR sequence. B) Approximately 2 × 105 human embryonic kidney 293 (HEK293) cells were co-transfected with the pGLO-CCL20 3’UTR vector or scrambled control vector (pGLO-Scramble) and miR-590-5p mimic or scrambled sequence control miRNA (negative control [NC] mimic), and luciferase activity was measured 48 hours post-transfection using the Dual-Glo Luciferase Assay System (Promega). The ratio of firefly luciferase activity to renilla luciferase activity was determined for each sample and the firefly/renilla relative luminometer units (RLU) in HEK293 cells co-transfected with miR-590-5p mimic and the pGLO-CCL20 3’UTR vector were compared with those from cells co-transfected with pGLO-Scramble and NC mimic. The Mann-Whitney U test was used to assess differences between conditions; differences between the negative controls were not statistically significant. Activated LX-2 cells were transfected with 75 nM mirVana miRNA-590-5p mimic, 50 nM anti-miR-590-5p inhibitor, or 50 nM scrambled sequence control. After 48 hours cells were harvested and total RNA was extracted.
The TaqMan RNA-to-Ct 1-Step kit (Thermo Fisher Scientific) and TaqMan commercial primers were used to measure C) miR-590-5p and D) CCL20 transcript levels in the presence of miR-590-5p mimic and miR-590-5p siRNA. Cycle threshold levels were generated using QuantStudio Real-Time PCR Software 1.0 and mRNA data were normalized using GAPDH. Black bars represent the scrambled miRNA negative control, while the gray bars represent treatment conditions. All experiments were performed in triplicate. Data are means ±SD. *P<0.05; **P<0.001; ***P<0.0001.
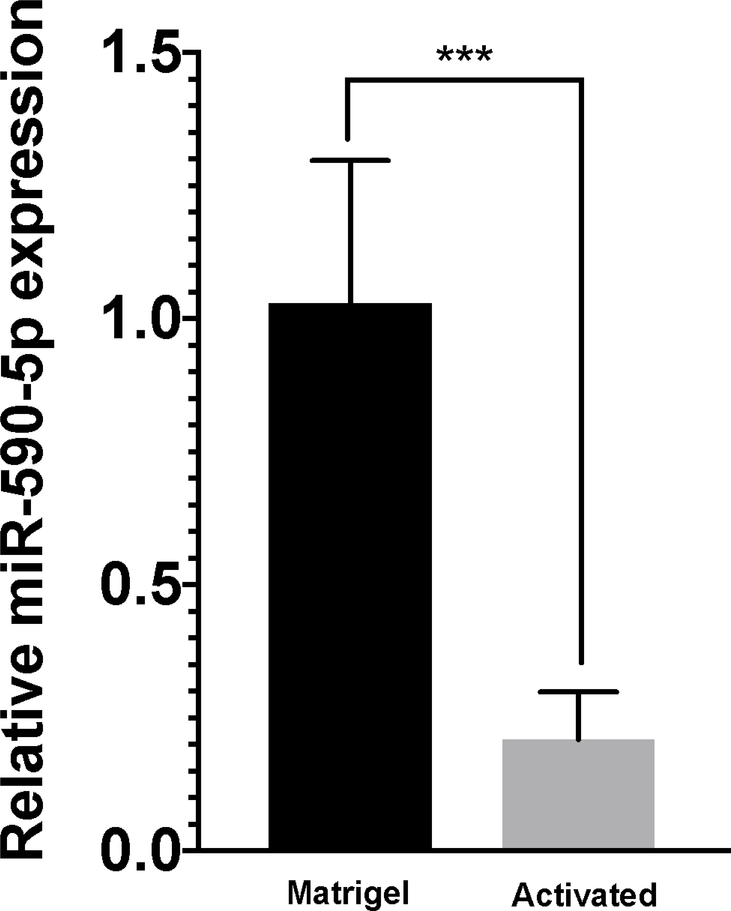
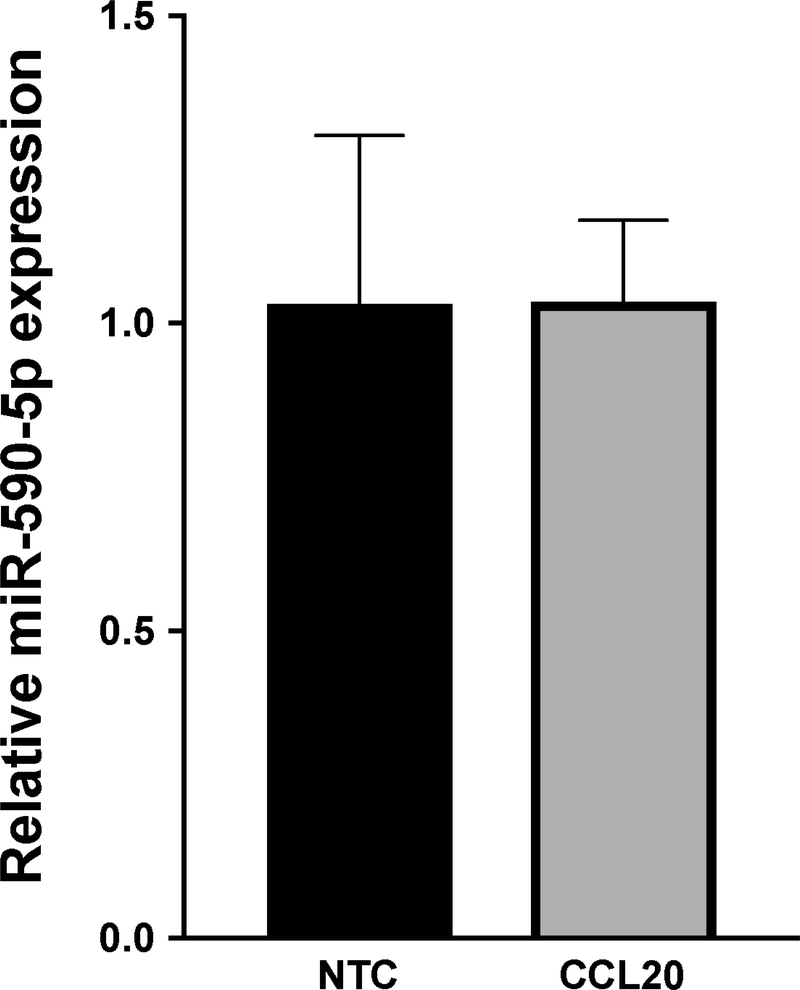
Based on our observation that miR-590-5p levels were reduced in NAFLD patients with fibrosis, we hypothesized that expression patterns would be mirrored in hepatic stellate cells following myofibroblastic transition. As shown in Figure 5A, levels of miR-590-5p were reduced 4.8-fold in activated LX-2 cells compared to cells grown on MG, consistent with increased CCL20 expression in these cells. To determine whether CCL20 conversely affects miR-590-5p expression, we measured miR-590-5p levels in response to treatment with 250 ng/mL CCL20 for six hours. We observed no difference in miR-590-5p expression between CCL20-treated and untreated cells (Figure 5B).
Figure 5. miR-590-5p expression in LX-2 cells.
A) miR-590–5 expression levels were measured in activated (gray bar) LX-2 cells and cells grown on Matrigel (black bar). B) LX-2 cells were treated with 250 ng/mL CCL20 (CCL20: gray bar) or untreated (NTC: black bar) for six hours. Data were normalized using RNU48 and RNU6B. The −ΔΔCt method was used to determine fold-change of miR-590-5p expression. All experiments were performed in triplicate. *P<0.05.
Discussion
The cellular and molecular mechanisms underlying the development and progression of NAFLD are complex and multifactorial [4]. Up until recently, this process was postulated to follow a “two-hit” mechanism, by which factors such as obesity, insulin resistance, diet, and lifestyle result in hepatic fat accumulation that sensitize the liver to further insult (comprising the “first hit”), with a “second hit” leading to the consequent cascade of inflammation and fibrogenesis [4]. While this mechanism was supported by findings from animal models of obesity [37], it failed to recapitulate the complexity of NAFLD development and progression in humans. A number of pathways have now been implicated in the progression from steatosis to hepatic inflammation and fibrogenesis in NAFLD, one of which includes the activation of proinflammatory cytokines, including chemokines, which participate in complex crosstalk among a variety of different cells. The chemokine CCL20 has recently emerged as a potential mediator of hepatic inflammation, injury, and fibrosis in alcoholic hepatitis [15], while in NAFLD fibrosis, serum and hepatic CCL20 levels are elevated relative to NAFLD patients with normal liver histology [20]. Here we provide evidence that regulation of CCL20 via a miRNA mediator in hepatic stellate cells may be a candidate molecular pathway in NAFLD fibrosis through mechanisms involving ECM.
Abundant evidence implicates CCL20 in liver injury. In mice, CCL20 mediates hepatocellular damage resulting from LPS administration [15]. In patients with alcoholic hepatitis (AH), CCL20 is the most upregulated CC chemokine [38] and hepatic CCL20 levels were positively correlated with scores used to assess prognosis [15]. Circulating CCL20 levels were correlated with serum LPS concentration in AH and higher levels of serum CCL20 were associated with more severe hepatic infiltration of polymorphonuclear cells compared to a mild grade of infiltration [15]. Levels of hepatic CCL20 mRNA and serum CCL20 protein were also higher in AH patients who died within 90 days of admission compared to those who survived [15]. In patients with NAFLD fibrosis, CCL20 was one of the most upregulated transcripts relative to normal liver histology and median serum CCL20 protein levels were increased in all grades of fibrosis [20]. In the current study, we observed increasing upregulation of CCL20 transcript levels in NAFLD patients with steatosis < lobular inflammation < fibrosis, demonstrating that activation of this chemokine occurs even in the early stages of simple steatosis and continues to increase with worsening liver histology. Together, these results suggest that CCL20 may mediate inflammation and fibrogenesis in chronic liver disease independent of underlying pathophysiology and may therefore serve as a useful biomarker for early detection of liver damage.
Our finding that CCL20 expression increased during the early activation stages in LX-2 cells, but remained stable in cells grown under conditions to resemble a “quiescent” phenotype prompted us to speculate that CCL20 may exert measurable autocrine effects in LX-2 cells. CCL20 treatment of LX-2 cells did result in increased levels of CCL20 and ACTA2 and decreased expression of SERPINE1 and PLAU, while knockdown of CCL20 corresponded with decreased CCL20, ACTA2, and COL1A1. ACTA2 and COL1A1 are well-characterized components of the ECM and are known to increase during activation of hepatic stellate cells [23]; regulation of these components by CCL20 therefore suggests a role in ECM composition, consistent with our findings of CCL20 upregulation in activated LX-2 cells. PLAU is a secreted serine protease that binds with its receptor (uPAR) to convert plasminogen to plasmin. In turn, plasmin plays key roles in extracellular matrix degradation, chemotaxis, and growth factor release [39]. Here, we observed downregulation of PLAU expression by CCL20 treatment, which was reversed with siRNA-mediated knockdown of CCL20, supporting the idea that this chemokine impacts fibrogenesis via regulation of ECM in hepatic stellate cells. We also observed downregulation of SERPINE1, which encodes PAI-1 (plasminogen activator inhibitor-1), by CCL20. PAI-1 functions to protect ECM proteins from proteolytic degradation [40]: PAI-1 binds PLAU, and the resulting PLAU-PAI-1-uPAR structure is internalized into the cytosol, whereupon, PLAU and PAI-1 are degraded and uPAR is recycled back to the cell membrane [39]. These unexpected findings suggest additional levels of ECM regulation in activated LX-2 cells. In primary human hepatic stellate cells, CCL20 induced the expression of CCL2, CCL5, and intracellular adhesion molecule 1, as well as COL1A1 and transforming growth factor beta 1 [15]. Taken together, the findings demonstrate that CCL20 has both profibrogenic and proinflammatory effects on hepatic stellate cells.
Our results further suggest that hepatic stellate cells may also be a key cell type in cellular response to steatosis. In LX-2 cells, CCL20 transcript expression was upregulated ~15-fold by palmitate, while in HepG2 cells, both palmitate and oleate treatment increased expression, but to a lesser degree (~5-fold) [20]. We observed a similar upregulation of CCL20 by palmitate in primary human hepatocytes, confirming the results obtained in HepG2 cells. In contrast to fatty acid loading, increased CCL20 levels observed following exposure to elevated glucose paralleled levels were caused by osmotic stress in both LX-2 and HepG2 cells. Given the complex, multicellular nature of NAFLD progression, investigation of the production of CCL20 in individual cell types due to multiple stimuli and mechanisms of paracrine and autocrine signaling among these cell types is warranted.
While the evidence supporting a role for CCL20 in the development and progression of NAFLD fibrosis is only now emerging, evidence supporting a role for chemokines has been accumulating over the past several years. For example, CCL2 expression in hepatocytes is upregulated in mice fed a high fat diet [41], and the pathway involving CCL2 and its receptor, C-C chemokine receptor 2 (CCR2), is also upregulated in the livers of animals with diet-induced NASH [7]. CCL2-CCR2 signaling promotes the migration of hepatic stellate cells, which impacts the development of fibrosis [13]. In contrast, inactivation of hepatic CCR2 inhibits the development of NASH [7] and deletion of CCL2 and CCR2 ameliorates hepatic steatosis [11, 14]. Likewise, expression of CCL5, which is primarily involved in the migration of T-cells, monocytes, neutrophils, and dendritic cells, is upregulated in diet-induced obese mice [42] and obese patients [43]. Inactivation of the CCL5 receptor, CCR5, protects mice from insulin resistance and hepatic fatty infiltration, and corresponds with a shift in macrophage polarization toward the alternative M2 activation [42]. CCL5 is also involved in the progression of hepatic fibrosis in mice [12]. A phase 2b randomized double blind clinical trial of a CCR2 and CCR5 dual antagonist revealed a significant anti-fibrotic effect at year 1 that was well tolerated, although the study did not meet its primary endpoint [44].
Our finding that miR-590-5p regulates CCL20 expression through direct interaction with the CCL20 3’UTR reveals potential further complexity in the pathogenesis of NAFLD. Early work demonstrated a functional interaction between miR-21 and CCL20 in colorectal cancer cells [35], and miR-150 inhibits tumor invasion and metastasis by targeting CCR6, the sole receptor for CCL20, in advanced cutaneous T-cell lymphoma [45]. However, this is the first evidence showing a role for miRNA-mediated regulation of CCL20 in liver cells, Interestingly, in a miRNA-profiling study conducted in NAFLD patients, we found that miR-590-5p was significantly downregulated in samples with biopsy-proven fibrosis compared to normal liver histology [36]. CCL20 treatment did not affect miR-590-5p levels, indicating that other factors are responsible for the downregulation of this miRNA. We postulate that downregulated miR-590-5p expression in NAFLD fibrosis may lead to disinhibited regulation of CCL20 expression that is observed with progressive NAFLD stages. Additional experiments are necessary to fully address this possibility.
Our data indicate that CCL20 expression increases in parallel with worsening liver histology in NAFLD patients and with transition to a myofibroblastic phenotype in hepatic stellate cells. CCL20 effects on components and regulators of ECM confirm a profibrogenic role for this chemokine, and evidence that miR-590-5p regulates CCL20 expression suggests a novel pathway for NAFLD progression. These results highlight the importance of further investigation of CCL20 as a key player in NAFLD-related fibrogenesis.
Supplementary Material
Acknowledgements
This study was funded in part by NIDDK DK088231. Vector cloning was supported by the Emory Integrated Genomics Core (EIGC), which is subsidized by the Emory University School of Medicine and is one of the Emory Integrated Core Facilities. Additional support was provided by the National Center for Advancing Translational Sciences of the National Institutes of Health under Award Number UL1TR000454. The content is solely the responsibility of the authors and does not necessarily reflect the official views of the National Institutes of Health.
Footnotes
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Rinella ME, Nonalcoholic fatty liver disease: a systematic review. JAMA, 2015. 313(22): p. 2263–73. [DOI] [PubMed] [Google Scholar]
- 2.McCullough AJ, The clinical features, diagnosis and natural history of nonalcoholic fatty liver disease. Clin Liver Dis, 2004. 8(3): p. 521–33, viii. [DOI] [PubMed] [Google Scholar]
- 3.Younossi ZM, Review article: current management of non-alcoholic fatty liver disease and non-alcoholic steatohepatitis. Aliment Pharmacol Ther, 2008. 28(1): p. 2–12. [DOI] [PubMed] [Google Scholar]
- 4.Buzzetti E, Pinzani M, and Tsochatzis EA, The multiple-hit pathogenesis of non-alcoholic fatty liver disease (NAFLD). Metabolism, 2016. 65(8): p. 1038–48. [DOI] [PubMed] [Google Scholar]
- 5.Tariq Z, Green CJ, and Hodson L, Are oxidative stress mechanisms the common denominator in the progression from hepatic steatosis towards non-alcoholic steatohepatitis (NASH)? Liver Int, 2014. 34(7): p. e180–90. [DOI] [PubMed] [Google Scholar]
- 6.Wieland A, et al. , Systematic review: microbial dysbiosis and nonalcoholic fatty liver disease. Aliment Pharmacol Ther, 2015. 42(9): p. 1051–63. [DOI] [PubMed] [Google Scholar]
- 7.Miura K, et al. , Hepatic recruitment of macrophages promotes nonalcoholic steatohepatitis through CCR2. Am J Physiol Gastrointest Liver Physiol, 2012. 302(11): p. G1310–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Griffith JW, Sokol CL, and Luster AD, Chemokines and chemokine receptors: positioning cells for host defense and immunity. Annu Rev Immunol, 2014. 32: p. 659–702. [DOI] [PubMed] [Google Scholar]
- 9.Gerard C and Rollins BJ, Chemokines and disease. Nat Immunol, 2001. 2(2): p. 108–15. [DOI] [PubMed] [Google Scholar]
- 10.Wasmuth HE, Tacke F, and Trautwein C, Chemokines in liver inflammation and fibrosis. Semin Liver Dis, 2010. 30(3): p. 215–25. [DOI] [PubMed] [Google Scholar]
- 11.Kanda H, et al. , MCP-1 contributes to macrophage infiltration into adipose tissue, insulin resistance, and hepatic steatosis in obesity. J Clin Invest, 2006. 116(6): p. 1494–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seki E, et al. , CCR1 and CCR5 promote hepatic fibrosis in mice. J Clin Invest, 2009. 119(7): p. 1858–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Seki E, et al. , CCR2 promotes hepatic fibrosis in mice. Hepatology, 2009. 50(1): p. 185–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weisberg SP, et al. , CCR2 modulates inflammatory and metabolic effects of high-fat feeding. J Clin Invest, 2006. 116(1): p. 115–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Affo S, et al. , CCL20 mediates lipopolysaccharide induced liver injury and is a potential driver of inflammation and fibrosis in alcoholic hepatitis. Gut, 2014. 63(11): p. 1782–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hou KZ, Fu ZQ, and Gong H, Chemokine ligand 20 enhances progression of hepatocellular carcinoma via epithelial-mesenchymal transition. World J Gastroenterol, 2015. 21(2): p. 475–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li WM and Liu HR, CCL20-CCR6 Cytokine Network Facilitate Treg Activity in Advanced Grades and Metastatic Variants of Hepatocellular Carcinoma. Scand J Immunol, 2016. 83(1): p. 33–7. [DOI] [PubMed] [Google Scholar]
- 18.Rubie C, et al. , Enhanced expression and clinical significance of CC-chemokine MIP-3 alpha in hepatocellular carcinoma. Scand J Immunol, 2006. 63(6): p. 468–77. [DOI] [PubMed] [Google Scholar]
- 19.Soliman HH, et al. , The role of chemokine CC ligand 20 in patients with liver cirrhosis and hepatocellular carcinoma. Int J Biol Markers, 2012. 27(2): p. e125–31. [DOI] [PubMed] [Google Scholar]
- 20.Chu X, et al. , CCL20 is up-regulated in non-alcoholic fatty liver disease fibrosis and is produced by hepatic stellate cells in response to fatty acid loading. J Transl Med, 2018. 16(1): p. 108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Serhal R, et al. , Nonalcoholic Steatohepatitis: Involvement of the Telomerase and Proinflammatory Mediators. Biomed Res Int, 2015. 2015: p. 850246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hendriks HF, et al. , Perisinusoidal fat-storing cells are the main vitamin A storage sites in rat liver. Exp Cell Res, 1985. 160(1): p. 138–49. [DOI] [PubMed] [Google Scholar]
- 23.Friedman SL, Hepatic stellate cells: protean, multifunctional, and enigmatic cells of the liver. Physiol Rev, 2008. 88(1): p. 125–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gerhard GS, et al. , Transcriptomic Profiling of Obesity-Related Nonalcoholic Steatohepatitis Reveals a Core Set of Fibrosis-Specific Genes. J Endocr Soc, 2018. 2(7): p. 710–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Law CW, et al. , voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol, 2014. 15(2): p. R29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leek JT, et al. , The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics, 2012. 28(6): p. 882–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wood GC, et al. , An electronic health record-enabled obesity database. BMC Med Inform Decis Mak, 2012. 12(1): p. 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leti F, et al. , High-throughput sequencing reveals altered expression of hepatic microRNAs in nonalcoholic fatty liver disease-related fibrosis. Transl Res, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.DiStefano JK, et al. , Genome-wide analysis of hepatic lipid content in extreme obesity. Acta Diabetol, 2015. 52(2): p. 373–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gerhard GS, et al. , Identification of novel clinical factors associated with hepatic fat accumulation in extreme obesity. J Obes, 2014. 2014: p. 368210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vejnar CE and Zdobnov EM, MiRmap: comprehensive prediction of microRNA target repression strength. Nucleic Acids Res, 2012. 40(22): p. 11673–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu L, et al. , Human hepatic stellate cell lines, LX-1 and LX-2: new tools for analysis of hepatic fibrosis. Gut, 2005. 54(1): p. 142–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bessone F, Razori MV, and Roma MG, Molecular pathways of nonalcoholic fatty liver disease development and progression. Cell Mol Life Sci, 2019. 76(1): p. 99–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qi W, et al. , High glucose induces macrophage inflammatory protein-3 alpha in renal proximal tubule cells via a transforming growth factor-beta 1 dependent mechanism. Nephrol Dial Transplant, 2007. 22(11): p. 3147–53. [DOI] [PubMed] [Google Scholar]
- 35.Vicinus B, et al. , miR-21 functionally interacts with the 3’UTR of chemokine CCL20 and down-regulates CCL20 expression in miR-21 transfected colorectal cancer cells. Cancer Lett, 2012. 316(1): p. 105–12. [DOI] [PubMed] [Google Scholar]
- 36.Leti F, et al. , High-throughput sequencing reveals altered expression of hepatic microRNAs in nonalcoholic fatty liver disease-related fibrosis. Transl Res, 2015. 166(3): p. 304–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leamy AK, Egnatchik RA, and Young JD, Molecular mechanisms and the role of saturated fatty acids in the progression of non-alcoholic fatty liver disease. Prog Lipid Res, 2013. 52(1): p. 165–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Affo S, et al. , Transcriptome analysis identifies TNF superfamily receptors as potential therapeutic targets in alcoholic hepatitis. Gut, 2013. 62(3): p. 452–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Choong PF and Nadesapillai AP, Urokinase plasminogen activator system: a multifunctional role in tumor progression and metastasis. Clin Orthop Relat Res, 2003(415 Suppl): p. S46–58. [DOI] [PubMed] [Google Scholar]
- 40.Ghosh AK and Vaughan DE, PAI-1 in tissue fibrosis. J Cell Physiol, 2012. 227(2): p. 493–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Obstfeld AE, et al. , C-C chemokine receptor 2 (CCR2) regulates the hepatic recruitment of myeloid cells that promote obesity-induced hepatic steatosis. Diabetes, 2010. 59(4): p. 916–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kitade H, et al. , CCR5 plays a critical role in obesity-induced adipose tissue inflammation and insulin resistance by regulating both macrophage recruitment and M1/M2 status. Diabetes, 2012. 61(7): p. 1680–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu H, et al. , T-cell accumulation and regulated on activation, normal T cell expressed and secreted upregulation in adipose tissue in obesity. Circulation, 2007. 115(8): p. 1029–38. [DOI] [PubMed] [Google Scholar]
- 44.Friedman SL, et al. , A randomized, placebo-controlled trial of cenicriviroc for treatment of nonalcoholic steatohepatitis with fibrosis. Hepatology, 2018. 67(5): p. 1754–1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ito M, et al. , MicroRNA-150 inhibits tumor invasion and metastasis by targeting the chemokine receptor CCR6, in advanced cutaneous T-cell lymphoma. Blood, 2014. 123(10): p. 1499–511. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.