Abstract
Background:
The identification of meaningful biomarkers of tuberculosis (TB) has potential to improve diagnosis, disease staging and prediction of treatment outcomes. It has been shown that active pulmonary TB (PTB) is associated with qualitative and quantitative changes in systemic immune profile, suggesting a chronic inflammatory imbalance. Here we characterized the profile of PTB and extrapulmonary TB (EPTB) in a prospective cohort study.
Methods:
We measured a panel of 27 inflammatory cytokines, soluble receptors, and lipid mediators in peripheral blood from patients with PTB or EPTB from a prospective clinical study in China. Multidimensional analyses were performed to describe associations between plasma levels of biomarkers and different TB disease presentation profiles.
Results:
Mycobacterium tuberculosis infection induced changes in both the expression and correlation profiles of plasma mediators of inflammation in patients with PTB compared to those with EPTB. Increases in mycobacterial loads in sputum smears were associated with rises in concentrations of several molecules involved in TB pathogenesis, such as IL-1β, IFN-α, IL-10 and PGF2α. Moreover, PTB patients presenting with severe disease exhibited a distinct inflammatory profile hallmarked by heightened levels of TNF-α, IL-1β, IL17, IL-18 and IL-27. Interestingly, while antitubercular treatment (ATT) resulted in early changes of plasma concentrations of markers in PTB, changes were delayed in EPTB patients. Exploratory analyses of the molecular degree of perturbation (MDP) of the inflammatory mediators before and during ATT suggested the occurrence of infection and/or treatment-induced long lasting “inflammatory imprinting” of biomarker profiles in TB. At 24 weeks post ATT commencement, markers underlying the observed increases in MDP scores were IL-27 in PTB and IL-1β in EPTB patients.
Conclusion:
Our findings describe systemic and durable changes in the concentrations of inflammatory cytokines and lipid mediators in both PTB and EPTB and emphasize the role of M. tuberculosis bacterial burden and site of disease in modulating patient immune biomarkers.
Keywords: Cytokines, lipid mediator, inflammation, Mycobacterium tuberculosis, acid-fast bacilli, extrapulmonary tuberculosis
1. Introduction
Tuberculosis (TB) is a devastating disease and infects 10 million people worldwide every year and is among the top ten causes of death, particularly within low-income countries, according to the World Health Organization [1], M. tuberculosis is among the most successful human pathogens, with up to 1/2 of the world’s population having been exposed to the bacterium, functioning similar to a commensal or parasite, as it exploits human cells for proliferation while simultaneously engineering a multi-faceted interaction with the host immune system [2]. A recent concern is the emergence of new multidrug resistant (MDR) M. tuberculosis strains, which have already reached 25% of cases in China and the emergence of totally resistant drug strains (TDR) in India and many other countries [2].
The immunological host response against M. tuberculosis is a complex interplay between innate and adaptive immune responses [3, 4]. Initially, infection with M. tuberculosis triggers activation of innate immune cells such as alveolar macrophages and dendritic cells, leading to secretion of several pro-inflammatory cytokines, such as interferons, TNF, IL-1 and eicosanoid lipid mediators [5-8]. This scenario culminates in antigen presentation to lymphocytes in the lung draining lymph nodes and formation of pulmonary granulomas to promote containment of further bacillary growth and dissemination [7, 9, 10]. Understanding the inflammatory nuances of these immune interactions is important to drive the development of new therapeutic and preventative treatment strategies that are based on optimization anti-M. tuberculosis host responses.
We have recently described that active TB is associated with distinct expression of surrogates of oxidative stress, pro-inflammatory cytokines and lipid mediators [6, 11]. Moreover, we have shown that cross-regulation of inflammatory pathways linking lipid mediators, IL-1 and type I interferons (IFN) can distinguish hallmarks of TB disease in patients and that manipulation of these pathways can lead to novel host-directed therapies [6]. Our studies demonstrated that prostaglandin E2 (PGE2) dampens uncontrolled type I IFN driven inflammation, suggesting that balance between eicosanoids and inflammatory cytokines was associated with control of bacillary growth and lung pathology [6]. However, when this balance is disturbed, as is the case in severe TB, suppression of PGE2 and/or high levels of type I IFN may occur, resulting in mycobacterial replication within the caseous lesions followed by an exuberant host immune activation and the chance of affecting sites other than lung [6]. Additionally, the lack of IL-1 and PGE2 leads to increased type I IFN production, which in turn increases IL-10 and IL-1RA levels, further reducing the concentration of IL-1 and PGE2 [5, 6]. Most importantly, there is evidence that an inflammatory imbalance involving type I IFNs is likely an important component of TB pathogenesis [6, 12-14].
Here, we measured and analyzed cytokines, chemokines, and lipid mediators of the IL-1, type IFN and eicosanoid axis in a prospective cohort of Chinese individuals infected with M. tuberculosis [6]. We explored the relationship between inflammatory immune responses in circulation of pulmonary versus extrapulmonary TB and bacillary sputum loads before and after antitubercular treatment (ATT). We found that both clinical forms of TB exhibited durable imprinting of specific biomarker profiles that persisted for up to 24 weeks of ATT.
2. Materials and Methods
2.1. Ethics statement
The original study is registered on the platform ClinicalTrials.gov () and has been approved and reviewed by the Institutional Review Board (IRB) from Henan Chest Hospital (HCH), China, and US National Institute for Allergy and Infectious Disease (NIAID), National Institutes of Health (NIH), Bethesda, Maryland. Written informed consent was obtained from all participants or their legally responsible guardians, and all clinical investigations were conducted according to the principles expressed in the Declaration of Helsinki and local and national Chinese regulations.
2.2. Study design and participants
The cohort study was performed in Zhengzhou, China, between 2010 and 2012 [6, 15, 16]. Plasma samples were collected from 100 patients with active PTB, 50 patients with EPTB, and 11 age and gender-matched healthy controls, who served as reference for the laboratory measurements in an exploratory analysis. For the present study, 6 patients from the PTB group were excluded due to lack of sample available for the laboratory assays, thus the analyses shown here are from 94 individuals with pulmonary disease. Plasma was collected at baseline (first visit), 2 weeks, 8 weeks and 24 weeks after onset of treatment.
Persons with signs and symptoms indicative of active TB and who were HIV-unexposed and administered less than two weeks of anti-tubercular chemotherapy or community controls were enrolled into a prospective clinical protocol to assess response to chemotherapy () conducted at the Henan Chest Hospital. For the present study, only treatment-naive patients at study enrollment were included. Exclusion criteria were HIV infection, diabetes, cancer and other lung diseases identified at the first clinical visit and reported resistance to anti-TB drugs. The enrolled subjects all underwent a chest Computer Tomography (CT) scan, provided three sputum samples for AFB smear and culture by both the BACTEC MGIT 960 system (Becton, Dickenson and Company) and Lowenstein-Jensen medium (Chuang Xin Company; Hangzhou, China), and had blood drawn for routine blood chemistry and cytology as well as several experimental assays at enrolment. The HCH radiology department provided a scored assessment of the chest tomography (CT) scans that included locations of disease by lobe, type of abnormalities and number of cavities for each CT. The sputum smears were prepared by the Ziel–Nielsen method using 1% carbol-fuchsin and scored using the International Union Against Tuberculosis and Lung Disease (IUATLD) scale. The 94 subjects (347 samples) included in this analysis of the PTB were confirmed to match their cohort definition, and to have abnormal chest CT scans consistent with active PTB. The 50 patients (169 plasma samples) with EPTB were diagnosed on the basis of AFB staining and/ or culture positivity of fine-needle aspiration biopsies of lymph nodes or pleural effusions. Of the 50 EPTB cases examined in the present study, 42 had pleural TB (n=42) whereas 4 had TB lymphadenitis (n=4) and 4 had miliary TB (n=4). Healthy controls were recruited from Zhengzhou, China and included hospital personnel but excluded persons known to have a history of TB or HIV infection. Each enrolled individual was given a physical examination, a chest CT, and the same microbiologic, biochemical and cytology tests. Healthy controls (HC) participants presented in this analysis lacked radiologic and microbiological signs of active M. tuberculosis infection had no pulmonary symptoms of tuberculosis and had a negative result in the Quantiferon Gold in-the-tube test. BCG vaccination is administered in China and the majority of trial participants had a scar from vaccination and accordingly TST was not administered.
2.3. Immunoassays
Hematologic evaluation was performed using the hospital's clinical laboratory. We evaluated a panel of 27 cytokines, lipid mediators and soluble receptors to examine inflammation and immune activation using different immunoassays. The selection of variables was based on potential role in TB pathogenesis demonstrated by previous work from our group [6] and others [8, 11]. We measured the biomarkers with commercial ELISA kits (for the soluble receptors CD40L, sIL-1R1, SIL-1R2, as well as MPO) (R&D Systems, Minneapolis, MN). The cytokines IL-1α, IL-Aβ, IL-1RA, IL-2, IL-5, IL-6, IL-8, IL10, IL-12p70, IL-17A, IL-18, IL-27, IFN-α, IFN-β, IFN-γ, TNF-α, TNF-β and resistin were measured using a Flowcytomix Multiplex Array kit (eBioscience, San Diego, CA) according to the manufacturer’s instructions. FlowCytomix is a bead-based ELISA like assay allowing for the results of several conventional ELISAs in a single assay [17]. This assay platform reduces sample volume requirements (<20µL of sample) and assay times, all performed using a standard flow cytometer. Levels of lipid mediators (PGE2, PGF2α, LXA4 and 15-EPI-LXA4) were assessed using EIA kits according to the manufacturer’s directions (Oxford Biomedical Research, Oxford, MI).
2.4. Data analysis
The median values with interquartile ranges (IQR) were used as measures of central tendency and dispersion. Cytokines levels were compared between the study groups using the Mann-Whitney U (when 2 groups were compared) or the Kruskal-Wallis test with Dunn’s multiple comparisons (when more than 2 groups were compared), to identify the biomarkers that were statistically significant between groups, or non-parametric linear trend post-test, to evaluate the tendency of linear increase or decrease in biomarkers values according to variations in AFB smear grade. Categorical variables were compared using the Fisher’s exact test (2×2 contingence tables) or Pearson’s qui-square test (contingence table with more variables) with Yates’s correction for continuity. Hierarchical cluster analyses (Ward’s method), with 100X bootstrap [18] of z-score normalized data were employed to depict the overall expression profile of indicated biomarkers in the study groups. Dendograms represent Euclidean distance. Venn diagrams were used to illustrate differentially expressed markers as described in [19]. P-values were adjusted for multiple measurements using Holm-Bonferroni’s method. All univariate comparisons with P-values <0.05 after adjustments were considered statistically significant. Only adjusted P-values were considered.
Profiles of correlations between biomarkers in different clinical groups were examined using network analysis of the Spearman correlation matrices (with 100X bootstrap). In indicated analyses, only strong correlations, defined herein by Spearman rank values (rho) >0.4 or <−0.4 and P<0.05 after Bonferroni adjustment (pre-specified arbitrary definition) were included in the network visualization. In such analyses, markers that exhibited similar correlation profiles were clustered based on a modularity [20], which infers a sub-networks inside the of the correlation network profiles and depicted using Fruchterman-Reingold algorithm (force-directed graph drawing) [21] (R scripts: http://sachaepskamp.com/files/Cookbook.html#installing-r-packages and https://jokergoo.github.io/circlize_book/book/index.html).
Sparse canonical correlation analysis (CCA) modelling was employed to assess whether combinations of circulating biomarkers could discriminate between subgroups of PTB patients stratified by AFB sputum smear status and radiographic extension of lung lesions (unilateral or bilateral) as previously described [6, 22] (R script: https://www.jstatsoft.org/article/view/v023i12). The CCA algorithm was chosen because many variables were studied. This approach reduces dimensionality for two co-dependent data sets (biomarker profile and baseline characteristics profile, which were age and gender) simultaneously so that the discrimination of the clinical endpoints represents a combination of variables that are maximally correlated. Thus, trends of correlations between parameters in different clinical groups rather than their respective distribution within each group are the key components driving the discrimination outcome. In our CCA algorithm, simplified and adapted from previously reported investigations of biomarkers for diagnosis of infectious diseases [6, 23], linear regression graphs represent coefficients from different combinations of plasma factors and baseline characteristics. In the biomarker profile dataset, we included values of all the inflammatory marker variables that exhibited significant differences (P<0.05) in the univariate analysis (MPO, resistin, IL-1;β, IL-10, IL-17, IL-4, IFN-α, IL-27, TNF-α, IL-1RA and IL-6). We pre-selected variables to include in the CCA algorithm to test whether the combination of the markers, rather than each one individually, could distinguish the groups. In addition, to our knowledge, investigating statistical relationships between the markers rather than just concentrations allow us to infer about regulatory immune networks [6].
The molecular degree of perturbation (MDP) was calculated to infer the overall inflammatory profile of TB over the course of antitubercular treatment. The MDP method used for the present study has been described previously [19] and is an adaptation of the molecular distance to health described by Pankla et al. [24] and employed to non-genomic measurements [25]. Healthy uninfected controls were defined as the “reference” group, and the average level and standard deviation of this reference group were calculated for the plasma concentrations of each inflammatory marker. The MDP score of an individual marker in a given sample “s” was defined by taking the difference in concentration level in sample “s” from the average of the marker in reference group divided by the corresponding standard deviation. The MDP score represents the number of standard deviations from the reference. Individuals who had MDP values above 2 standard deviations from mean value of controls were considered molecularly perturbed. The statistical analyses were performed using JMP 14 (SAS, Cary, NC) and Prism 7.0 (GraphPad Software, San Diego, CA) and R statistical software.
3. Results
3.1. Characteristics of the study participants
Baseline and post treatment plasma samples originated from persons with signs and symptoms indicative of active TB and who were HIV-uninfected and who were not undertaking antitubercular therapy or healthy controls that were enrolled into a prospective clinical protocol to assess response to chemotherapy () conducted at the Henan Chest Hospital (HCH) in Zhengzhou, China from 2010 to 2012, as described previously [6, 15, 16]. Individuals with pulmonary tuberculosis (PTB) were similar to those presenting with extra pulmonary tuberculosis (EPTB) with regard to age (median [IQR] in years: 27 [23-44.7] vs. 27 [22-36], respectively; P= 0.1933; Supplemental Table 1) and gender (P=0.4916, Table S1), with higher frequency of male individuals (61.7% vs. 52% vs. respectively).
3.2. Pulmonary and extrapulmonary TB patients display distinct expression profiles of inflammatory biomarkers in peripheral blood
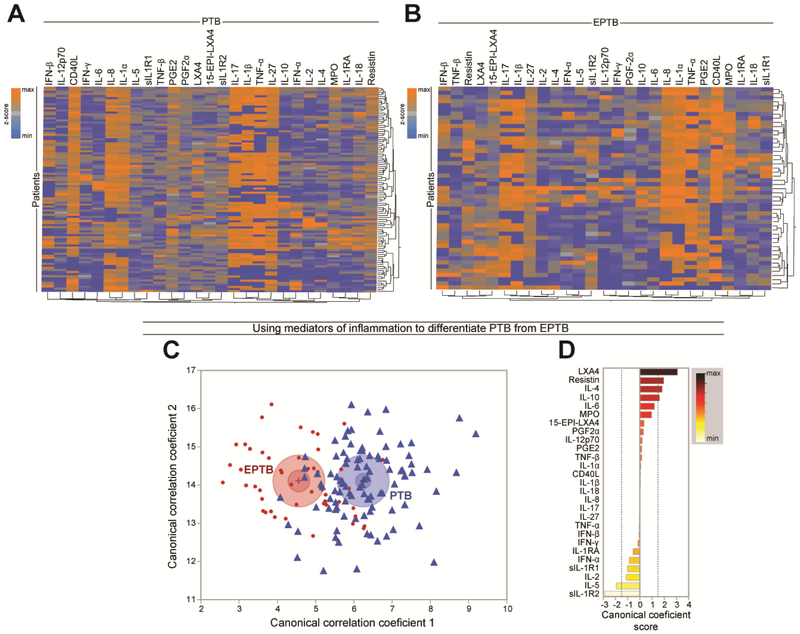
We examined the expression profile of 27 soluble inflammatory proteins and eicosanoid lipid mediators related to IL-1 and type I IFN driven inflammation in plasma from the study population, to test whether active TB in PTB or EPTB is associated with specific changes in these pathways. Preliminary analyses revealed that when z-score normalized values from all markers were considered, there was no discrimination between individuals from the HC, PTB and EPTB groups using unsupervised hierarchical cluster analysis (Supplemental Figure 1). When the subtypes of EPTB were compared (pleural vs. miliary vs. lymph node TB), we found that the EPTB subgroups could not be clustered separately when concentration values from all markers were considered (Supplemental Figure 2A). Although the sample size was small, we compared individual markers among the EPTB subtypes and found very few statistically significant markers (adjusted P <0.05) with small fold-difference values (all below 2.0-fold difference) (Supplemental Figure 2B). When EPTB and PTB were evaluated separately using the same statistical approach, we observed a heterogeneity of expression within each group (Figure 1A and Figure 1B). In both PTB and EPTB groups, the markers with more homogeneously high expression were CD40L, IL-1α, IL-1β, IL-8, IL-17, IL-27 and TNF-α (Figure 1A and Figure 1B), highlighting similarities between the different clinical forms of TB. Indeed, among all the markers, concentrations of only 4 were statistically different between the groups (Supplemental Table 2). We found that concentrations of IL-10, IL-12p70 and resistin were higher in PTB compared to EPTB whereas levels of SIL-1R2 were lower (Supplemental Table 2). Importantly, when all markers were included in a discriminant analysis model based on canonical correlation, we found that PTB patients could be distinguished from those with EPTB (Figure 1C). The markers with the strongest contributions to the canonical model were LXA4, resistin, IL-4, IL-10, SIL-1R2 and IL-5 (Figure 1D). These findings indicate that the overall inflammatory status, as determined here by correlations between inflammatory plasma mediators, is qualitatively different between PTB and EPTB groups and likely underscores differences in the underlying immunopathology of different clinical forms of TB disease.
Figure 1: Plasma levels of inflammatory mediators distinguish PTB from EPTB in Chinese patients.
(A-B) Plasma levels of cytokines, soluble receptors and eicosanoids were assessed in samples from patients with pulmonary TB (n=94) or extrapulmonary TB (n=50). Data were log10 transformed and z-score normalized. (A) A hierarchical cluster analysis (Ward’s method with 100X bootstrap) was employed to test whether the overall expression profile of the biomarkers could separate the study groups. Dendrograms represent Euclidean distance. (C) Discrimination of groups using combination of plasma biomarkers. In an exploratory approach, a sparse canonical correlation analysis (sCCA) was employed to test whether patient groups could be distinguished. (D) Canonical discriminant was performed to evaluate the biomarkers responsible for the difference between groups in model.
3.3. Unique Correlation profiles of inflammatory biomarkers in peripheral blood between pulmonary and extrapulmonary TB patients.
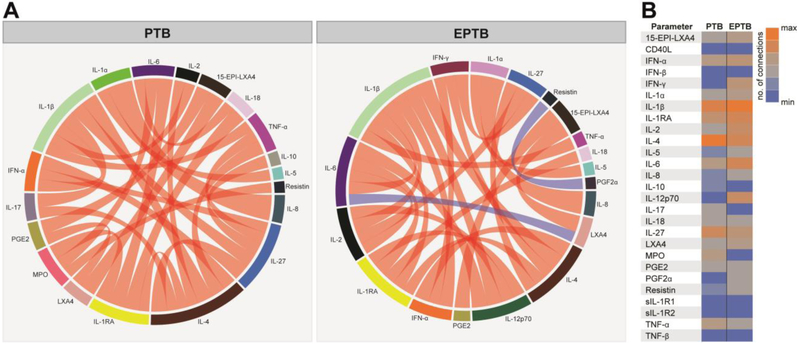
We have previously shown that using analysis of Spearman correlation matrices can provide insights on the degree and quality of inflammation in TB-HIV as well as in other diseases [26-28]. The results obtained from the canonical correlation model described above indicated that there were differences in the correlation profiles between the inflammatory markers from PTB vs. EPTB patients and we next utilized Circos plots to visualize such distinctions. Interestingly, we found that the presence of pulmonary infection was associated with a marked absence of negative correlations (Figure 2A), suggesting that pulmonary infection triggers a coordinated response involving canonical induction of many inflammatory markers. Node analysis of the PTB network indicated that IL-1β, IL-4 and IL-27, followed by IL-1RA were the most highly connected markers (Figure 2), implying that their regulation may be preferentially more susceptible to modulation by pulmonary infection. Importantly, the correlation profile found in the EPTB group was distinct from the PTB group, with appearance of negative correlations involving resistin vs. PGF2α and IL-6 vs. LXA4 (Figure 2A). In addition, extrapulmonary infection was associated with a predominance of IL-1β followed by IL-6, IL-4, IL-1RA and IL-2 (Figure 2B). IL-12p70 and IFN-γ were not relevant in the PTB correlation plot but were relevant nodes in the EPTB group. Differences in correlation profiles between PTB and EPTB, including evidence that IL-12p70 has more statistical relationships with other markers in EPTB but not in PTB further underline potential inflammatory distinctions for extrapulmonary disease pathogenesis.
Figure 2: Network analysis of inflammatory biomarker correlation matrices in pulmonary and extra pulmonary TB patients.
(A)Spearman correlation matrices of the biomarker expression levels in each study group were build and Circos plots illustrate the correlation networks. Each bar represents a different plasma parameter. The length of each bar is proportional to the number of significant correlations. The connecting lines represent statistically significant correlations (p<0.05). Red connecting lines represent positive correlations while blue lines infer negative correlations. The thickness of the connecting lines is proportional to the Spearman correlation coefficient value. Markers which did not exhibit statistically significant correlations are now shown in the Circos plots. (B)Node analysis: heatmap shows the number of statistically significant correlations involving each marker per clinical group.
3.4. The inflammatory signature of pulmonary tuberculosis is substantially associated with changes in acid-fast bacilli smear grades in sputum.
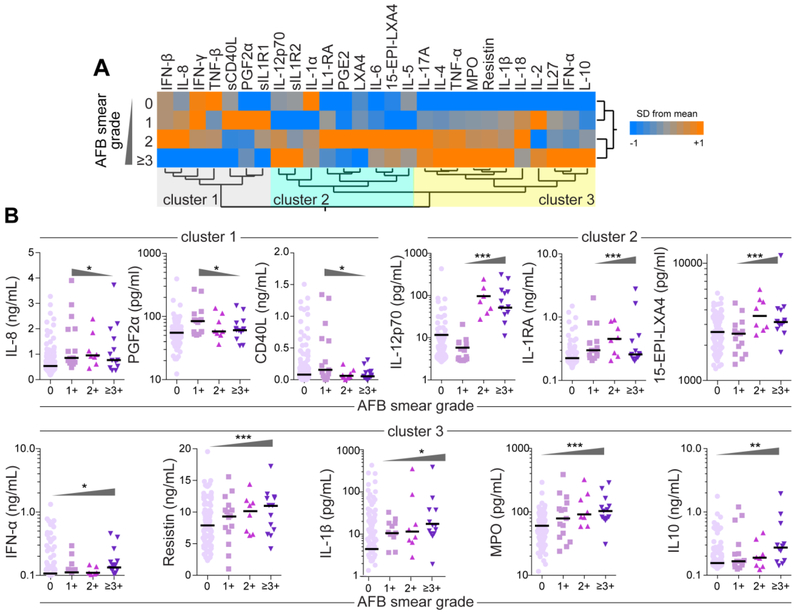
We have previously suggested that M. tuberculosis loads in sputum are directly associated with the status of systemic inflammation and potentially impact the immune profile of patients from Brazil [26] and India [11]. To test whether this phenomenon would also be present in our Chinese patient population, we compared the levels of all the biomarkers between the subgroups of PTB patients presenting with different AFB grades in sputum smears (Supplemental Table 3 and Figure 3). A hierarchical analysis identified three main groups of biomarkers based upon the expression profile between the study groups (Figure 3A). While the first cluster included the markers that displayed higher values in the groups of PTB patients presenting with lower AFB grades (0 and 1+), the other two clusters delineated parameters with higher concentrations that associated with increased mycobacterial loads (AFB of +2 and ≥+3) (Figure 3 A). Linear trend ad hoc analyses of biomarker levels revealed that average levels of IL-8, PGF2α and CD40L were gradually decreased with increased AFB grades (Figure 3B). Average plasma concentrations of IL-12p70, IL-1RA and 15-EPI-LXA4 tended to follow increased mycobacterial sputum loads and were higher in individuals with AFB grade of 2+ (Figure 3B). Other biomarkers such as IFN-α, resistin, IL-1β, MPO and IL-10 consistently displayed higher average concentrations with increasing AFB smear grade values (Figure 3B, Supplemental Table 3). Thus, the above analytical approaches revealed biomarkers for which their concentrations correlated both positively and negatively with sputum mycobacterial loads as measured via AFB grade.
Figure 3: Correlation of sputum AFB smear grade with concentrations of plasma biomarkers in PTB patients.
(A)A hierarchical cluster analysis (Ward’s method with 100X bootstrap) was employed to test whether the overall expression profile could separate the groups of PTB patients based on AFB sputum grades. Dendrograms represent Euclidean distance. Using this approach, 3 clusters of biomarkers were detected. (B) Scatterplots of concentrations of indicated biomarkers from each cluster shown in (A), which values presented statistically significant differences between the study groups using the Kruskal-Wallis test with linear trend ad hoc test, are displayed. Bars represent median values whereas triangles indicate the trends of data variation. *p<0.05; **p<0.01; ***p<0.0001.
3.5. Differences in systemic inflammation can further distinguish mild pulmonary TB patients from those with more severe disease presentation
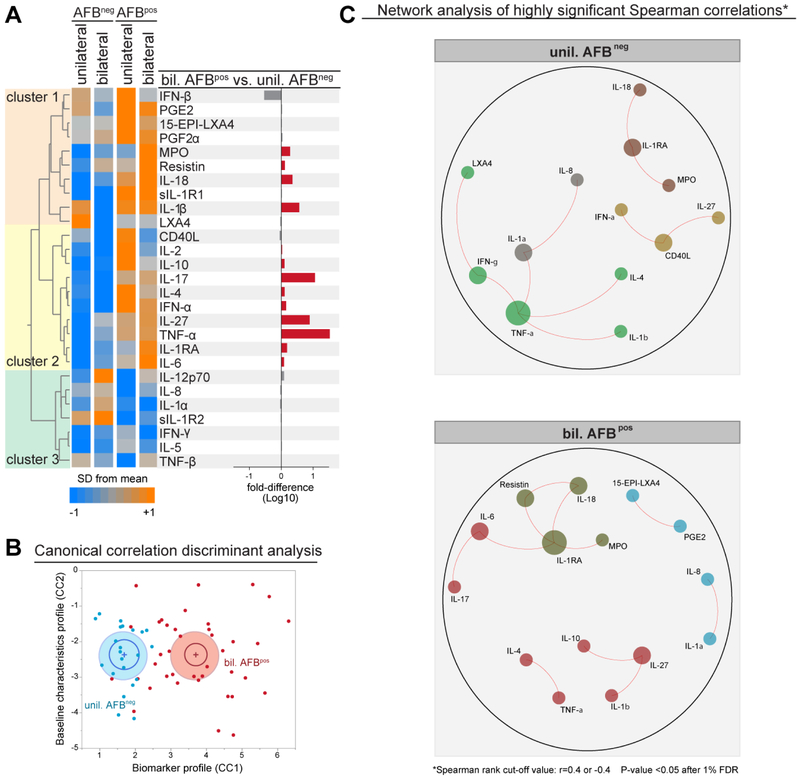
In addition to mycobacterial loads in sputum, radiographic disease extension has been used by our group and others as a potential parameter of TB disease severity [19, 26, 29, 30]. We next stratified the PTB patients into mild and severe clinical subgroups based on AFB smear status (positive vs. negative) and on radiographic extension of lung lesions (unilateral vs. bilateral) and compared the inflammatory signatures (Figure 4). We found that, in general, plasma concentration of most of the markers examined were higher in patients with positive AFB sputum smears, independent of radiographic disease extension, except for IL-1α, IL-5, IL-8, IL-12p70, IFN-γ, sIL-1R2 and TNF-β (cluster 3, Figure 4A). The highest change in fold-differences in biomarker concentrations were found in patients presenting simultaneously with positive AFB in smears and bilateral lung disease (more severe TB disease) when compared with patients with negative AFB smears and unilateral lesions (mild disease) (Figure 4A and Supplemental Table 4). The cytokines TNF-α, IL-17, IL-27 and IL-1β were ranked as top markers differentially expressed between these subgroups of PTB patients, all of them found at higher levels in individuals with more severe disease (Figure 4A). We next asked whether correlations between plasma levels of the mediators could distinguish mild from severe PTB subgroups. A discriminant analysis using a model of canonical correlation with plasma levels of all the markers which were differentially expressed between mild and severe TB could discriminate between mild and severe TB (Figure 4B). Network analysis of highly significant correlations confirmed the hypothesis that there are differences in the profiles between mild and severe PTB groups (Figure 4C). In mild PTB, TNF-α was the most highly connected node, being linked to IL-1β, IFN-γ, IL-1α and IL-4, creating a correlation chain that defined this group (Figure 4C). In severe PTB, a completely distinct pattern was detected and included IL-1RA as a central marker connected to MPO, IL-18, resistin and IL-6. These observations suggest that both the levels of inflammatory mediators and the relationships between such markers can identify PTB patients with more severe forms of TB based on AFB smear grade combined with radiographic extent of lung lesions.
Figure 4: Difference in AFB status and extent of radiographic disease associated with specific changes in plasma inflammatory biomarker profiles in PTB.
(A)Hierarchical cluster analysis (Ward’s method with 100X bootstrap) was employed to illustrate the overall expression profile of the biomarkers in PTB patients stratified per AFB smear status and lung disease extension (AFBneg + unilateral disease, n=22; AFBneg + bilateral disease, n=21; AFBpos + unilateral disease, n=09; AFBpos + bilateral disease, n=34). Dendrograms represent Euclidean distance. The bar graph on the right panel indicate the fold-difference variation from the group of PTB patients with mild disease (AFBneg + unilateral disease) to that with severe disease (AFBpos + bilateral disease). (B) Sparse canonical correlation analysis (sCCA) was employed using the markers indicated in (A) which were statistically different between the groups. (C) Networks represent strong Spearman correlations (p<0.05 after 1% FDR adjustment; Spearman rank value >0.4 or <−0.4). Markers were clustered based on a similarity index of the correlation profiles using a modularity algorithm and depicted with Fruchterman Reingold (force-directed graph drawing). Only markers which had strong correlations were plotted to reduce visual pollution. Size of the circles is proportional to number of connections.
3.6. Longitudinal changes in inflammatory profiles in patients with pulmonary or extrapulmonary tuberculosis undergoing antitubercular treatment.
The results above strongly support our idea that the degree of bacterial burden during active TB disease drives major changes in systemic inflammation and suggest that these changes occur for both pulmonary and extrapulmonary forms of disease. To further explore our hypothesis and ask whether the inflammatory profiles identified above are equally affected by chemotherapy in PTB and EPTB patients, we measured the same parameters at different time points after onset of antitubercular treatment (ATT), when bacterial burdens in patients are actively being reduced (Supplemental Table 5, Supplemental Table 6 and Figure 5). We observed that in PTB patients, levels of TNF-β were significantly increased as early as 2 weeks after ATT initiation (Figure 5A, Figure 5C, Supplemental Table 5). At the week 2 study time point, IL-1β, IL-4, IL-6, IL-10, IL-18 and IL-27 displayed significant reduction in plasma levels. Intriguingly, no early change in biomarker concentrations was found in the group of EPTB patients (Figure 5B, Figure 5C, Supplemental Table 6). After 8 weeks of ATT, several changes in plasma markers were noted in both TB disease groups, with most the parameters presenting with reduced concentrations compared to pre-treatment values, except for TNF-β which increased in EPTB patients (Figures 5A-C).
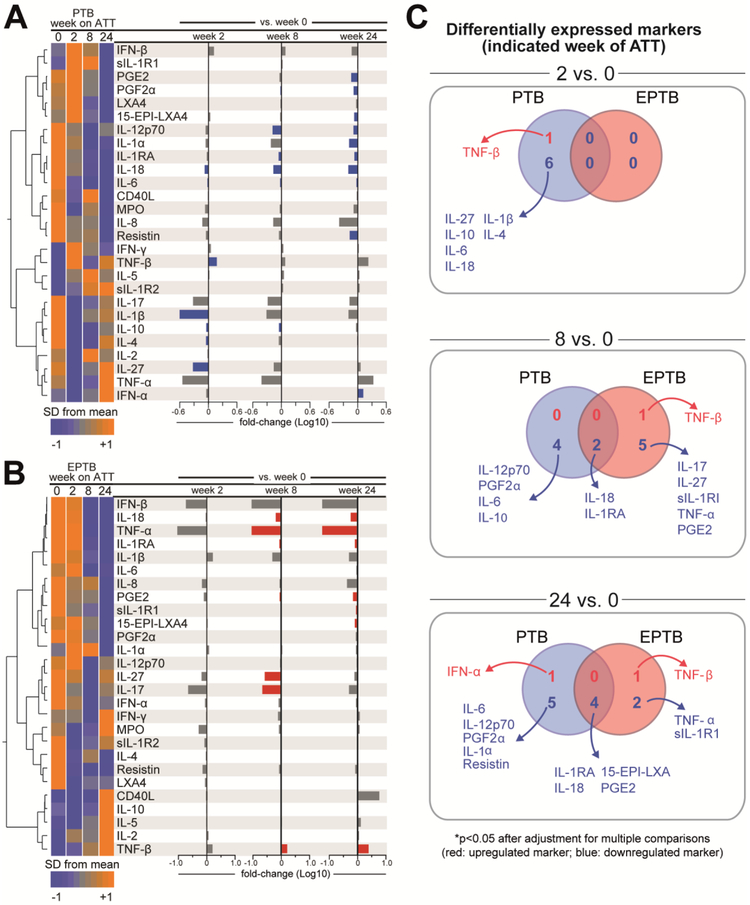
Figure 5: Impact of antitubercular treatment on inflammatory balance and duration of changes observed in inflammatory biomarker profiles in PTB and EPTB patients.
(A-B) Hierarchical cluster analysis (Ward’s method with 100X bootstrap) was employed to illustrate the overall expression profile of the biomarkers in the groups of patients with PTB (n=94) or EPTB (n=50). Dendrograms represent Euclidean distance. The bar graphs on the right panel indicate the fold-change variation in biomarker values between the indicated time points. Differences which reached statistical significance after adjustment for multiple comparisons (adjusted P < 0.05) are represented in colored bars. (C)Venn Diagrams describe the number of markers which values were statistically different (p<0.05) between the indicated study time points in patients with pulmonary TB or extrapulmonary TB. Markers which were upregulated are illustrated in red whereas downregulated markers are shown in blue.
At week 24 of ATT, we observed a reduction in a number of parameters that were significantly different from pre-treatment timepoints in both groups, including IL-1RA, IL-18, 15-EPI-LXA4 and PGE2 (Figures 6A-C). At this post treatment period, IFN-a levels were increased whereas concentrations of IL-1α, IL-6, IL-12p70, PFG2α and resistin were decreased exclusively in PTB patients (Figure 5C). In the EPTB group, TNF-β levels were once again found increased and values of TNF-a and sIL-1R1 were decreased (Figure 5C). These findings demonstrate that ATT induced important global reductions in peripheral blood levels of inflammatory biomarkers, with some differences between PTB and EPTB, specifically with regard to timing of TNF-β increases in plasma concentrations.
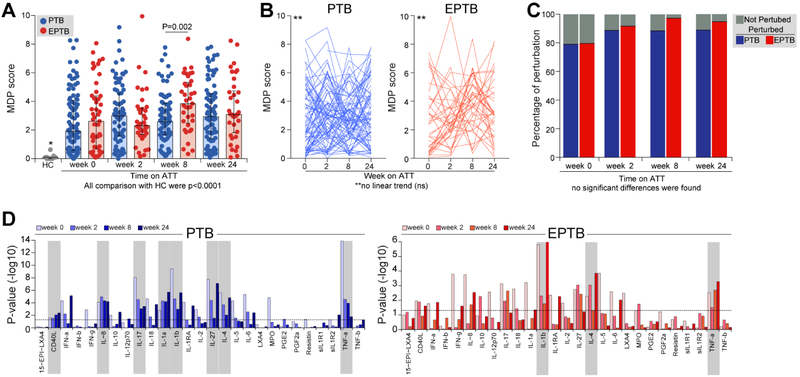
Figure 6. Durable molecular degree of perturbation in TB patients undergoing antitubercular treatment.
(A)The molecular degree of perturbation (MDP) score relative to healthy uninfected controls (HC) was calculated as described in Methods. Individual values with median and IQR per group are shown. Data between PTB and EPTB groups in each time point were compared using the Mann Whitney U test.The Kruskal Wallis test with Dunn’s multiple comparisons was used to compare MDP values between each clinical group and time point with HC. (B) Variations of MDP individual values for each patient over time of treatment were analyzed using the Friedman matched test with non-parametric linear tend ad hoc test. (C) Frequencies of molecularly perturbed TB patients between PTB and EPTB groups in each time point were compared using the Fisher’s exact test (no statistically significant differences were found). (D) Bar graphs display −log10 transformed p-values of the correlations between plasma levels of each biomarker and MDP score values in each disease groups and indicated time points. Shaded rectangles highlight the most relevant markers associated with MDP score values. ns, nonsignificant.
3.7. Persistent molecular perturbation in PTB and EPTB patients during antitubercular treatment
Originally, the molecular degree of perturbation (MDP) is a numerical score of transcriptional perturbation made by summing the differences in the expression levels of preselected genes between a given sample group and healthy control participants [19] (see Methods for details). When applied to the blood transcriptome, MDP largely reflects immunological activation status [12, 19]. Here, we employed this statistical approach in plasma concentrations of biomarkers [25] to assess global effects of PTB or EPTB on the blood inflammatory status that significantly differs from that detected in healthy controls (raw data is found in Supplemental File 1). To answer this question, we performed an exploratory analysis comparing the PTB and EPTB groups with healthy controls (n=11) who were matched by age and sex (median age: 35 years, IQR: 23-40; 45.4% of male individuals). We found that, before ATT initiation, both PTB and EPTB were associated with substantial increases in MDP scores compared to uninfected controls (both P<0.0001; Figure 6A), and no statistical difference was observed in median values between the clinical groups. There was an important spread of MDP values in the TB groups, highlighting the heterogeneity in systemic inflammation and possibly disease activity as suggested previously [31]. The only difference in MDP values between the clinical groups was found at week 8 of treatment, with EPTB exhibiting higher scores than those with PTB (P=0.002, Fig 6A). Strikingly, the median MDP scores within each clinical group (PTB or EPTB) did not significantly change over time on treatment (Figure 6B), demonstrating a persistent molecular perturbation even after 6 months of ATT. Indeed, at week 24, 89.7% individuals with PTB and 94.7% of those with EPTB were still molecularly perturbed (Figure 6C). In addition, while most of the markers contributed to increased MDP values at treatment baseline in both PTB and EPTB patients, at week 24 of follow up, fewer parameters were driving changes in the MDP score (Figure 6D). Importantly, the most influential markers to MDP score values during all the time points in both disease groups included IL-1β, IL-4, and TNF-α (Figure 6D). In PTB patients, additional relevant markers associated with increased MDP values included IL-1α, IL-8, IL-27 and CD40L (Figure 6D). Importantly, these data suggest that M. tuberculosis infection or ATT may have durable long-lasting systemic effects on inflammatory responses in TB patients compared to uninfected and treatment naive individuals.
4. Discussion
Infection with M. tuberculosis is known to cause profound stimulation of both innate and adaptive immune responses both in vitro and in vivo. M. tuberculosis subverts the inflammatory milieu to persist and proliferate in necrotic tissues and disseminate in susceptible hosts [2, 3]. Such immunological events can be read by detecting expression of genes, proteins, lipid mediators and other molecules in the tissue [32] as well and in peripheral blood of patients [4, 33]. While this should be ideally studied at the site of infection, sampling bronchoalveolar lavage or lung tissue is highly invasive, represents a significant exposure risk for health care professionals and is unfeasible for most studies. Studying plasma biomarkers in the context of TB may provide important insights into diagnosis, treatment efficacy and also disease pathogenesis [33]. Several prior studies have reported concentrations of cytokines, chemokines, acute phase proteins and lipid mediators in populations of pulmonary TB patients [30, 34-41], but less is known about extrapulmonary infection [42-44]. Here, we performed multiparametric analyses of plasma concentrations of several inflammatory cytokines, soluble cytokine receptors, receptor ligands, alongside with lipid mediators in a large set of active TB patients with pulmonary or extrapulmonary disease to characterize the systemic inflammatory profile of this disease.
Our primary exploratory results indicate that both pulmonary and extrapulmonary TB states are associated with induction of changes in the systemic inflammatory milieu. At first glance, these findings may sound not surprising, considering that both clinical forms of active TB derive from M. tuberculosis infection of immune myeloid cells. Nevertheless, we were able to find exclusive qualitative and quantitative differences between PTB and EPTB groups in immune markers, such as IL-10, IL-12p70, resistin and sIL-1R2. The distinct inflammatory profiles of PTB versus EPTB could simply be the consequence of the underlying pathology that leads to disseminated versus more localized infection. Alternatively, altered production of these cytokines may be involved in the dissemination of mycobacteria to extrapulmonary compartments. Future experimental studies are warranted to directly test these possibilities.
Perhaps a more important finding was the identification of eicosanoid lipid mediators differentially modulated in the distinct forms of active TB, which included PGE2, PGF2α, LXA4 and 15-EPI-LXA4. The role of lipid mediators derived from cyclooxygenases or lipoxygenases has been extensively studied. We and others have shown that PGE2 promotes relative protection against disease associated with necrotic cell death, resulting in reduced disease burden whereas lipoxins can have detrimental effects on host containment of mycobacterial growth [6, 36, 45]. Here we find that both prostaglandins and lipoxins are induced in the majority of individuals during M. tuberculosis infection. Whether such systemic profile changes represent in situ modulation of lipid mediators at the phagocyte level, the local lung airways and tissue or systemic changes requires further investigation.
Immune homeostasis is a coordinated process that involves cross-regulation of production of several pro and anti-inflammatory mediators. These intricate relationships between the immune and inflammatory markers can be interrogated through analysis of correlation matrices, which have been used in several clinical scenarios [26, 27]. In the present study, we found that PTB and EPTB displayed important differences in correlation profiles, involving the number of correlations, direction of the association as well as the main markers representing nodes with the highest connectivity index in Circos plots. It is possible that the inflammatory response is modulated in such a way during active EPTB disease to results in an unfettered, less organized immune activation. Furthermore, some of the most relevant markers sustaining the density of the correlation matrices in active TB, IL-12p70 and IFN-γ, were different between pulmonary and extrapulmonary disease presentations. Noteworthy, IL-1β was shown to be relevant in matrices networks from both PTB and EPTB groups, underscoring a possible central importance of this marker in immune responses to TB, which has been previously suggested [5, 6, 46].
Our next exploratory analysis tried to delineate the role of mycobacterial loads in driving specific changes in systemic inflammation in pulmonary infection. We did this twofold; first, we show that presence of positive AFB screening in sputum smears was strongly linked to meaningful increases in concentrations of several inflammatory parameters. Secondly, we compared the inflammatory profiles after bacterial burdens were being reduced through antitubercular drug treatment. In the first scenario, we found that many of the induced markers are related to innate immune activation, such as IL-1β, IL-6, and TNF-α. In addition, we found increased levels of IL-1RA, IL-27, sIL1R1, IFN-α and IL-10, which have been described to dampen effective anti-mycobacterial responses in both mice and humans [5, 14, 47, 48]. It is possible that an environment dominated by hyperactivation of myeloid cells and simultaneous interference of effective microbial killing induced by IFN-α and/or IL-10 results in unrestrained M. tuberculosis growth, leading to increased risk of disease transmission trough sputum dissemination. Consistent with this hypothesis, concentrations of both IFN-α and IL-10 gradually increased following the mycobacterial loads in sputum and were part of the bio-signature that defined patients with very high AFB sputum grades.
A number of prospective studies have previously reported changes in biomarker levels during antitubercular treatment in either pulmonary or extrapulmonary TB cases [15, 30, 33, 35, 37, 39-41, 49-51]. Our study adds to the current knowledge in the field by directly comparing these changes between PTB and EPTB disease forms while focusing on both lipid mediators and inflammatory cytokines. Our analyses demonstrated that only in pulmonary, but not EPTB disease, chemotherapy lead to very early effects on biomarker levels. It is thought that the occurrence of EPTB is linked to dissemination of mycobacteria via hematogenous and/or lymphatic routes [52], implying a potentially higher relative microbial load in these patients. It is also possible that there is a limited uptake/availability of the TB drugs at extrapulmonary sites, resulting in delayed microbial clearance. Interestingly, the dynamics of TNF-β levels during treatment illustrate well the differences between the disease groups. Indeed, appearance of higher levels of TNF-β were observed as early as 2 weeks of ATT in PTB, and levels were no longer statistically significant later on during treatment. On the other hand, increases in TNF-β concentrations in EPTB were substantial only at week 8 of treatment, but persisted at high levels for up to week 24. TNF-β is also known as lymphotoxin A and shares receptors with TNF-aα, such as TNF receptors 1 and 2, and many of their effects are similar [53]. This cytokine has been described to contribute to control of chronic M. tuberculosis infection [54]. Thus, it is possible that the expression levels of this cytokine in peripheral blood could serve as a surrogate of mycobacterial loads in TB patients undergoing treatment. A higher or more systemically availability of M. tuberculosis antigens in EPTB patients undergoing therapy may potentially explain a delayed modulation of immune activation. Although interesting, this hypothesis is difficult to test in patients, but could be addressed in appropriate animal models.
Tuberculosis can cause substantial tissue damage and the degree of lesion extension in the lungs is usually employed as a measure of disease activity and/or severity [19, 26, 29, 30]. Here, additional analyses were performed comparing the biomarker signatures between subgroups of PTB stratified by AFB smear status and by extent of radiographic disease. The results demonstrated that more severe PTB was associated with higher levels of several inflammatory markers, including TNF-α, IL-17, IL-27 and IL-1β, as well as changes in relationships between circulating biomarkers, with a central role for IL-1RA. We hypothesize that immune activation driven by direct stimulation with mycobacterial products linked to increased sputum grade and bilateral disease could drive robust inflammatory signals which may hallmark severe PTB. Prospective studies performing simultaneous radiographic and immunologic assessments in PTB patients upon ATT initiation are warranted to test this hypothesis.
A more interesting result was the finding of a potential systemic durable “inflammatory profile imprinting” of specific immune biomarkers in both PTB and EPTB patients at the end of ATT. After 6 months undergoing anti-TB chemotherapy, patients with active TB exhibited significant differences in molecular degree of perturbation score values compared to age and gender-matched healthy controls. The common bio-signature driving increased molecular perturbation in blood was composed by key mediators of anti-M. tuberculosis innate immune response, such as IL-1β and TNF-α, as well as IL-4 [36, 55]. Previous studies have also reported abnormal immune responses in individuals with prior EPTB, supporting our findings of long lasting changes in immune profile [42-44]. Importantly, our findings have multiple possible implications. For one, it is possible that these biomarker differences in the patient cohorts represent an underlying immunological distinction, which may have been present even before exposure to M. tuberculosis and is therefore associated with outcome of PTB or EPTB, respectively. If so, this particular scenario may be of interest for diagnostics and disease outcome prediction. More targeted studies investigating similar cohorts for longer periods, with a more unbiased biomarker or transcriptional profiling approach, in areas with different TB epidemiology could provide valuable insight into the inflammatory basis of persistent profile “imprinting” for systemic biomarker assays. Our observation of a durable systemic inflammatory profile “imprinting” during M. tuberculosis infection and/or during drug-treatment in patients could be tested and explored more definitively in appropriate animal models. Specifically, animal models would make it possible to discern whether the persistent profile imprinting is the result of ongoing or previous M. tuberculosis infection or due to the extensive antibiotic drug treatment regimens themselves, which have been shown to affect microbiota-immune axis and possible related immune functions [56, 57]. Finally, the use of an appropriate animal model could provide insight whether and how the observed “imprinting” of inflammatory biomarker profiles is affecting the host’s ability to fight concomitant infections, particular re-infections and other co-morbidities.
Our study has some clear limitations. This investigation was exploratory and although the disease groups had relatively high number of individuals, the control group was small, but homogeneous. In addition, we investigated easily accessible, systemic circulating markers of inflammation and we have not examined tissue specimens. It is likely that the inflammatory biomarker profile at the site of disease differs from that described in peripheral blood. We also employed only a crude approximation of radiographic extent of disease, more sophisticated analysis to quantitate specific type of abnormalities may reveal more interesting associations. Lastly, we have not followed up patients for longer periods after termination of antitubercular treatment to evaluate TB recurrence, other late outcomes or determined the extent of the duration of the “inflammatory profile imprinting” beyond 6 months after TB chemotherapy.
5. Conclusions
The findings presented here demonstrate a perturbed systemic inflammatory response in active TB linking cytokines and eicosanoid lipid mediators. Our results highlight an active role of M. tuberculosis and its bacterial burden in subversion of the host immune responses as read out by durable changes in the inflammatory biomarker profiles dependent on pulmonary or extrapulmonary manifestations after successful antitubercular treatment.
Supplementary Material
Highlights.
Systemic inflammation is distinct between pulmonary and extrapulmonary tuberculosis
Concentrations of biomarkers correlate with mycobacterial loads in sputum
Levels of biomarkers and their relationships can identify severe tuberculosis
Molecular degree of perturbation persists after antitubercular treatment
Acknowledgments
The authors thank the study participants. We also thank Dr. Artur Queiroz (Biostatistics, Instituto Gonçalo Moniz, FIOCRUZ, Salvador, Brazil) for consulting as external statistician and review of all the analyses.
Funding Sources
This project was supported by the Intramural Research Program of the NIAID to K.D.M.B. and C.E.B. The work of B.B.A. was supported by grants from the NIH (U01AI115940, R01AI069923-08, R01AI20790-02), by Intramural Program of Fundação Oswaldo Cruz, Fundação José Silveira and by the Brazilian National Council for Scientific and Technological Development (CNPq). P.S.S.M. was supported by a PhD fellowship from the Fundação de Amparo à Pesquisa do Estado da Bahia (FAPESB). K.F.F. received a fellowship from the Programa Nacional de Pós-Doutorado, Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) (Finance Code 001). C.L.V. and D.O.S. are supported by CNPq and FAPESB respectively. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Abbreviations:
- AFB
acid fast bacilli
- ATT
antitubercular treatment
- BCG
bacillus Calmette-guérin
- CCA
canonical correlation analysis
- LXA
lipoxin
- EPTB
extrapulmonary tuberculosis
- HC
healthy control
- IFN
interferon
- IL
interleukin
- IQR
interquartile range
- LXA
lipoxin
- MDP
molecular degree of perturbation
- MPO
myeloperoxidase
- PG
prostaglandin
- PTB
pulmonary tuberculosis
- TB
tuberculosis
- TNF
tumor necrosis factor
Footnotes
Disclosure of conflict of interest
Declarations of interest: none.
Conflict of Interest Statement
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication. The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
6. References
- [1].WHO, Global Tuberculosis Report, 2017. http://www.who.int/tb/publications/global_report/en/. (Accessed 07/02/2018 2018).
- [2].Pai M, Behr MA, Dowdy D, Dheda K, Divangahi M, Boehme CC, Ginsberg A, Swaminathan S, Spigelman M, Getahun H, Menzies D, Raviglione M, Tuberculosis, Nat Rev Dis Primers 2 (2016) 16076. [DOI] [PubMed] [Google Scholar]
- [3].Scriba TJ, Coussens AK, Fletcher HA, Human Immunology of Tuberculosis, Microbiol Spectr 4(5) (2016). [DOI] [PubMed] [Google Scholar]
- [4].Cliff JM, Kaufmann SH, McShane H, van Helden P, O'Garra A, The human immune response to tuberculosis and its treatment: a view from the blood, Immunol Rev 264(1) (2015) 88–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Mayer-Barber KD, Andrade BB, Barber DL, Hieny S, Feng CG, Caspar P, Oland S, Gordon S, Sher A, Innate and adaptive interferons suppress IL-1alpha and IL-1beta production by distinct pulmonary myeloid subsets during Mycobacterium tuberculosis infection, Immunity 35(6) (2011) 1023–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Mayer-Barber KD, Andrade BB, Oland SD, Amaral EP, Barber DL, Gonzales J, Derrick SC, Shi R, Kumar NP, Wei W, Yuan X, Zhang G, Cai Y, Babu S, Catalfamo M, Salazar AM, Via LE, Barry CE 3rd, Sher A, Host-directed therapy of tuberculosis based on interleukin-1 and type I interferon crosstalk, Nature 511(7507) (2014) 99–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Mayer-Barber KD, Barber DL, Innate and Adaptive Cellular Immune Responses to Mycobacterium tuberculosis Infection, Cold Spring Harb Perspect Med 5(12) (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Tobin DM, Roca FJ, Oh SF, McFarland R, Vickery TW, Ray JP, Ko DC, Zou Y, Bang ND, Chau TT, Vary JC, Hawn TR, Dunstan SJ, Farrar JJ, Thwaites GE, King MC, Serhan CN, Ramakrishnan L, Host genotype-specific therapies can optimize the inflammatory response to mycobacterial infections, Cell 148(3) (2012) 434–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].O'Garra A, Redford PS, McNab FW, Bloom CI, Wilkinson RJ, Berry MP, The immune response in tuberculosis, Annu Rev Immunol 31 (2013) 475–527. [DOI] [PubMed] [Google Scholar]
- [10].Ernst JD, The immunological life cycle of tuberculosis, Nat Rev Immunol 12(8) (2012) 581–91. [DOI] [PubMed] [Google Scholar]
- [11].Andrade BB, Pavan Kumar N, Amaral EP, Riteau N, Mayer-Barber KD, Tosh KW, Maier N, Conceicao EL, Kubler A, Sridhar R, Banurekha VV, Jawahar MS, Barbosa T, Manganiello VC, Moss J, Fontana JR, Marciano BE, Sampaio EP, Olivier KN, Holland SM, Jackson SH, Moayeri M, Leppla S, Sereti I, Barber DL, Nutman TB, Babu S, Sher A, Heme Oxygenase-1 Regulation of Matrix Metalloproteinase-1 Expression Underlies Distinct Disease Profiles in Tuberculosis, J Immunol 195(6) (2015) 2763–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Berry MP, Graham CM, McNab FW, Xu Z, Bloch SA, Oni T, Wilkinson KA, Banchereau R, Skinner J, Wilkinson RJ, Quinn C, Blankenship D, Dhawan R, Cush JJ, Mejias A, Ramilo O, Kon OM, Pascual V, Banchereau J, Chaussabel D, O'Garra A, An interferon-inducible neutrophil-driven blood transcriptional signature in human tuberculosis, Nature 466(7309) (2010) 973–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Zak DE, Penn-Nicholson A, Scriba TJ, Thompson E, Suliman S, Amon LM, Mahomed H, Erasmus M, Whatney W, Hussey GD, Abrahams D, Kafaar F, Hawkridge T, Verver S, Hughes EJ, Ota M, Sutherland J, Howe R, Dockrell HM, Boom WH, Thiel B, Ottenhoff THM, Mayanja-Kizza H, Crampin AC, Downing K, Hatherill M, Valvo J, Shankar S, Parida SK, Kaufmann SHE, Walzl G, Aderem A, Hanekom WA, Acs, G.C.c.s. groups, A blood RNA signature for tuberculosis disease risk: a prospective cohort study, Lancet 387(10035) (2016) 2312–2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Zhang G, deWeerd NA, Stifter SA, Liu L, Zhou B, Wang W, Zhou Y, Ying B, Hu X, Matthews AY, Ellis M, Triccas JA, Hertzog PJ, Britton WJ, Chen X, Feng CG, A proline deletion in IFNAR1 impairs IFN-signaling and underlies increased resistance to tuberculosis in humans, Nat Commun 9(1) (2018) 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Liang L, Shi R, Liu X, Yuan X, Zheng S, Zhang G, Wang W, Wang J, England K, Via LE, Cai Y, Goldfeder LC, Dodd LE, Barry CE, Chen RY, Interferon-gamma response to the treatment of active pulmonary and extra-pulmonary tuberculosis, Int J Tuberc Lung Dis 21(10) (2017) 1145–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Trauner A, Liu Q, Via LE, Liu X, Ruan X, Liang L, Shi H, Chen Y, Wang Z, Liang R, Zhang W, Wei W, Gao J, Sun G, Brites D, England K, Zhang G, Gagneux S, Barry CE 3rd, Gao Q, The within-host population dynamics of Mycobacterium tuberculosis vary with treatment efficacy, Genome Biol 18(1) (2017) 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Wang X, Dong L, Liang Y, Ni H, Tang J, Xu C, Zhou Y, Su Y, Wang J, Chen D, Mao C, Performance evaluation of FlowCytomix assays to quantify cytokines in patients with rheumatoid arthritis, Int J Clin Exp Med 8(9) (2015) 16158–66. [PMC free article] [PubMed] [Google Scholar]
- [18].Friedman NGMG, M.W.A, Data Analysis with bayesian networks: a bootstrap approach, Elsevier Science B.V (1999) 196–205. [Google Scholar]
- [19].Prada-Medina CA, Fukutani KF, Pavan Kumar N, Gil-Santana L, Babu S, Lichtenstein F, West K, Sivakumar S, Menon PA, Viswanathan V, Andrade BB, Nakaya HI, Kornfeld H, Systems Immunology of Diabetes-Tuberculosis Comorbidity Reveals Signatures of Disease Complications, Sci Rep 7(1) (2017) 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Blondel VD, Guillaueme J, Lamblotte R, Lefebvre E, Fast unfolding of communities in large networks, J. Stat. Meth (2008). [Google Scholar]
- [21].Bastian M. HS, Jacomy M, Gephi: an open source software for exploring and manipulating networks, International AAAI Conference on Weblogs and Social Media, 2009. [Google Scholar]
- [22].Manion M, Andrade BB, DerSimonian R, Gu W, Rupert A, Musselwhite LW, Sierra-Madero JG, Belaunzaran-Zamudio PF, Sanne I, Lederman MM, Sereti I, Country of residence is associated with distinct inflammatory biomarker signatures in HIV-infected patients, J Virus Erad 3(1) (2017) 24–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Rousu J, Agranoff DD, Sodeinde O, Shawe-Taylor J, Fernandez-Reyes D, Biomarker discovery by sparse canonical correlation analysis of complex clinical phenotypes of tuberculosis and malaria, PLoS Comput Biol 9(4) (2013) e1003018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Pankla R, Buddhisa S, Berry M, Blankenship DM, Bancroft GJ, Banchereau J, Lertmemongkolchai G, Chaussabel D, Genomic transcriptional profiling identifies a candidate blood biomarker signature for the diagnosis of septicemic melioidosis, Genome Biol 10(11) (2009) R127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Oliveira-de-Souza D, Vinhaes CL, Arriaga MB, Kumar PK, Cubillos-Angulo JM, Shi R, Wang W, Yuan X, Zhang G, Ying C, Barry CE, Via LE, Sher A, Babu S, Mayer-Barber KD, Nakaya HI, Fukutani KF, Andrade BB, Molecular degree of perturbation of plasma inflammatory markers associated with tuberculosis reveals distinct disease profiles between Indian and Chinese populations, Sci RepIn Press (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Mesquita ED, Gil-Santana L, Ramalho D, Tonomura E, Silva EC, Oliveira MM, Andrade BB, Kritski A, T.B.S.g. Rede, Associations between systemic inflammation, mycobacterial loads in sputum and radiological improvement after treatment initiation in pulmonary TB patients from Brazil: a prospective cohort study, BMC Infect Dis 16 (2016) 368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Andrade BB, Singh A, Narendran G, Schechter ME, Nayak K, Subramanian S, Anbalagan S, Jensen SM, Porter BO, Antonelli LR, Wilkinson KA, Wilkinson RJ, Meintjes G, van der Plas H, Follmann D, Barber DL, Swaminathan S, Sher A, Sereti I, Mycobacterial antigen driven activation of CD14++CD16− monocytes is a predictor of tuberculosis-associated immune reconstitution inflammatory syndrome, PLoS Pathog 10(10) (2014) e1004433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Hsu DC, Breglio KF, Pei L, Wong CS, Andrade BB, Sheikh V, Smelkinson M, Petrovas C, Rupert A, Gil-Santana L, Zelazny A, Holland SM, Olivier K, Barber D, Sereti I, Emergence of Polyfunctional Cytotoxic CD4+ T Cells in Mycobacterium avium Immune Reconstitution Inflammatory Syndrome in Human Immunodeficiency Virus-Infected Patients, Clin Infect Dis (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Ralph AP, Ardian M, Wiguna A, Maguire GP, Becker NG, Drogumuller G, Wilks MJ, Waramori G, Tjitra E, Sandjaja, Kenagalem E, Pontororing GJ, Anstey NM, Kelly PM, A simple, valid, numerical score for grading chest x-ray severity in adult smear-positive pulmonary tuberculosis, Thorax 65(10) (2010) 863–9. [DOI] [PubMed] [Google Scholar]
- [30].Rockwood N, Costa DL, Amaral EP, Du Bruyn E, Kubler A, Gil-Santana L, Fukutani KF, Scanga CA, Flynn JL, Jackson SH, Wilkinson KA, Bishai WR, Sher A, Wilkinson RJ, Andrade BB, Mycobacterium tuberculosis Induction of Heme Oxygenase-1 Expression Is Dependent on Oxidative Stress and Reflects Treatment Outcomes, Front Immunol 8 (2017) 542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Singhania A, Verma R, Graham CM, Lee J, Tran T, Richardson M, Lecine P, Leissner P, Berry MPR, Wilkinson RJ, Kaiser K, Rodrigue M, Woltmann G, Haldar P, O'Garra A, A modular transcriptional signature identifies phenotypic heterogeneity of human tuberculosis infection, Nat Commun 9(1) (2018) 2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Marakalala MJ, Raju RM, Sharma K, Zhang YJ, Eugenin EA, Prideaux B, Daudelin IB, Chen PY, Booty MG, Kim JH, Eum SY, Via LE, Behar SM, Barry CE 3rd, Mann M, Dartois V, Rubin EJ, Inflammatory signaling in human tuberculosis granulomas is spatially organized, Nat Med 22(5) (2016) 531–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Wallis RS, Kim P, Cole S, Hanna D, Andrade BB, Maeurer M, Schito M, Zumla A, Tuberculosis biomarkers discovery: developments, needs, and challenges, Lancet Infect Dis 13(4) (2013) 362–72. [DOI] [PubMed] [Google Scholar]
- [34].Andrade BB, Pavan Kumar N, Mayer-Barber KD, Barber DL, Sridhar R, Rekha VV, Jawahar MS, Nutman TB, Sher A, Babu S, Plasma heme oxygenase-1 levels distinguish latent or successfully treated human tuberculosis from active disease, PLoS One 8(5) (2013) e62618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Bai XJ, Li HM, Yang YR, Zhang JX, Liang Y, Wu XQ, Cytokine and soluble adhesion molecule profiles and biomarkers for treatment monitoring in Re-treated smear-positive patients with pulmonary tuberculosis, Cytokine 108 (2018) 9–16. [DOI] [PubMed] [Google Scholar]
- [36].Mayer-Barber KD, Sher A, Cytokine and lipid mediator networks in tuberculosis, Immunol Rev 264(1) (2015) 264–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Miranda P, Gil-Santana L, Oliveira MG, Mesquita ED, Silva E, Rauwerdink A, Cobelens F, Oliveira MM, Andrade BB, Kritski A, Sustained elevated levels of C-reactive protein and ferritin in pulmonary tuberculosis patients remaining culture positive upon treatment initiation, PLoS One 12(4) (2017) e0175278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Scriba TJ, Penn-Nicholson A, Shankar S, Hraha T, Thompson EG, Sterling D, Nemes E, Darboe F, Suliman S, Amon LM, Mahomed H, Erasmus M, Whatney W, Johnson JL, Boom WH, Hatherill M, Valvo J, De Groote MA, Ochsner UA, Aderem A, Hanekom WA, Zak DE, A.C.S.c.s.t. other members of the, Sequential inflammatory processes define human progression from M. tuberculosis infection to tuberculosis disease, PLoS Pathog 13(11) (2017) e1006687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Sigal GB, Segal MR, Mathew A, Jarlsberg L, Wang M, Barbero S, Small N, Haynesworth K, Davis JL, Weiner M, Whitworth WC, Jacobs J, Schorey J, Lewinsohn DM, Nahid P, Biomarkers of Tuberculosis Severity and Treatment Effect: A Directed Screen of 70 Host Markers in a Randomized Clinical Trial, EBioMedicine 25 (2017) 112–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Singanayagam A, Manalan K, Connell DW, Chalmers JD, Sridhar S, Ritchie AI, Lalvani A, Wickremasinghe M, Kon OM, Evaluation of serum inflammatory biomarkers as predictors of treatment outcome in pulmonary tuberculosis, Int J Tuberc Lung Dis 20(12) (2016) 1653–1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Thompson EG, Du Y, Malherbe ST, Shankar S, Braun J, Valvo J, Ronacher K, Tromp G, Tabb DL, Alland D, Shenai S, Via LE, Warwick J, Aderem A, Scriba TJ, Winter J, Walzl G, Zak DE, T.B.B.C. Catalysis, Host blood RNA signatures predict the outcome of tuberculosis treatment, Tuberculosis (Edinb) 107 (2017) 48–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Fiske CT, de Almeida AS, Shintani AK, Kalams SA, Sterling TR, Abnormal immune responses in persons with previous extrapulmonary tuberculosis in an in vitro model that simulates in vivo infection with Mycobacterium tuberculosis, Clin Vaccine Immunol 19(8) (2012) 1142–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Huaman MA, Sterling TR, Shepherd BE, Fiske CT, 25-Hydroxyvitamin D levels after recovery from tuberculosis: insights into pathogenesis, Tuberculosis (Edinb) 94(1) (2014) 51–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].de Almeida AS, Fiske CT, Sterling TR, Kalams SA, Increased frequency of regulatory T cells and T lymphocyte activation in persons with previously treated extrapulmonary tuberculosis, Clin Vaccine Immunol 19(1) (2012) 45–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Bafica A, Scanga CA, Serhan C, Machado F, White S, Sher A, Aliberti J, Host control of Mycobacterium tuberculosis is regulated by 5-lipoxygenase-dependent lipoxin production, J Clin Invest 115(6) (2005) 1601–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Novikov A, Cardone M, Thompson R, Shenderov K, Kirschman KD, Mayer-Barber KD, Myers TG, Rabin RL, Trinchieri G, Sher A, Feng CG, Mycobacterium tuberculosis triggers host type I IFN signaling to regulate IL-1beta production in human macrophages, J Immunol 187(5) (2011) 2540–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].McNab FW, Ewbank J, Howes A, Moreira-Teixeira L, Martirosyan A, Ghilardi N, Saraiva M, O'Garra A, Type I IFN induces IL-10 production in an IL-27-independent manner and blocks responsiveness to IFN-gamma for production of IL-12 and bacterial killing in Mycobacterium tuberculosis-infected macrophages, J Immunol 193(7) (2014) 3600–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Moreira-Teixeira L, Redford PS, Stavropoulos E, Ghilardi N, Maynard CL, Weaver CT, Freitas do Rosario AP, Wu X, Langhorne J, O'Garra A, T Cell-Derived IL-10 Impairs Host Resistance to Mycobacterium tuberculosis Infection, J Immunol 199(2) (2017) 613–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Ferrian S, Manca C, Lubbe S, Conradie F, Ismail N, Kaplan G, Gray CM, Fallows D, A combination of baseline plasma immune markers can predict therapeutic response in multidrug resistant tuberculosis, PLoS One 12(5) (2017) e0176660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Nouhin J, Pean P, Madec Y, Chevalier MF, Didier C, Borand L, Blanc FX, Scott-Algara D, Laureillard D, Weiss L, Interleukin-1 receptor antagonist, a biomarker of response to anti-TB treatment in HIV/TB co-infected patients, J Infect 74(5) (2017) 456–465. [DOI] [PubMed] [Google Scholar]
- [51].Kathamuthu GR, Moideen K, Baskaran D, Sekar G, Rathinam S, Bharathi VJ, Ganeshan GR, Babu S, Tuberculous lymphadenitis is associated with altered levels of circulating angiogenic factors, Int J Tuberc Lung Dis 22(5) (2018) 557–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Elder NC, Extrapulmonary tuberculosis. A review, Arch Fam Med 1(1) (1992) 91–8. [DOI] [PubMed] [Google Scholar]
- [53].Smith CA, Farrah T, Goodwin RG, The TNF receptor superfamily of cellular and viral proteins: activation, costimulation, and death, Cell 76(6) (1994) 959–62. [DOI] [PubMed] [Google Scholar]
- [54].Jacobs M, Togbe D, Fremond C, Samarina A, Allie N, Botha T, Carlos D, Parida SK, Grivennikov S, Nedospasov S, Monteiro A, Le Bert M, Quesniaux V, Ryffel B, Tumor necrosis factor is critical to control tuberculosis infection, Microbes Infect 9(5) (2007) 623–8. [DOI] [PubMed] [Google Scholar]
- [55].Mayer-Barber KD, Yan B, Clash of the Cytokine Titans: counter-regulation of interleukin-1 and type I interferon-mediated inflammatory responses, Cell Mol Immunol 14(1) (2017) 22–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Namasivayam S, Maiga M, Yuan W, Thovarai V, Costa DL, Mittereder LR, Wipperman MF, Glickman MS, Dzutsev A, Trinchieri G, Sher A, Longitudinal profiling reveals a persistent intestinal dysbiosis triggered by conventional anti-tuberculosis therapy, Microbiome 5(1) (2017) 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Wipperman MF, Fitzgerald DW, Juste MAJ, Taur Y, Namasivayam S, Sher A, Bean JM, Bucci V, Glickman MS, Antibiotic treatment for Tuberculosis induces a profound dysbiosis of the microbiome that persists long after therapy is completed, Sci Rep 7(1) (2017) 10767. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.