Abstract
Although Cyberlindnera fabinaii is a rare opportunist yeast species, its ability to cause septicemia, produce biofilm, and rapid acquisition of resistance to fluconazole and voriconazole, reinforced the urge for its identification from its closely related species. Widely used biochemical assays mainly identify Cyberlindnera fabinaii as Cyberlindnera jadinii and Wickerhamomyces anomalus, resulting in underestimation of this yeast in clinical settings. Moreover, the urge for a reliable molecular means of identification remains unsolved for 28 years. In order to unequivocally differentiate Cy. fabianii, Cy. mississipiensis, Cy. jadinii, and W. anomalus, we designed a dual-function multiplex polymerase chain reaction (PCR) assay. Challenging our dual-function multiplex PCR assay with 30 most clinically important yeast species, proved its specificity. Although conventional PCR could differentiate four target species, the real-time PCR counterpart due to Tm overlap misidentified Cy. mississipiensis as Cy. jadinii. Alongside of presenting a comprehensive literature review of published cases of Cy. fabianii from 1990 to 2018, we collected various clinical isolates from Tehran, Shiraz, and Fasa (July 1, 2017, to December 31, 2017) to find a passive relative distribution of these closely-related species in Iran. Subjecting our Iranian collection of yeast isolates to matrix-assisted laser desorption/ionization–time of flight (MALDI-TOF) MS and LSU and ITS rDNA sequencing revealed six isolates of Cy. fabianii (central venous catheter n = 2 and vaginal swabs n = 4) and one isolate of Cy. jadinii (vaginal swabs). Due to the use of biochemical assays in global ARTEMIS study, we encourage reidentification of clinical isolates of Cy. jadinii and Cy. jadinii using MALDI-TOF or Sanger sequencing that might lead to correcting the distribution of this fungus.
Keywords: Cy. fabianii, Cy. jadinii, Cy. mississipiensis, W. anomalus, biochemical assays, multiplex PCR
Introduction
Candida species are associated with 10–15% causes of nosocomial candiduria1 and considered as the fourth common cause of nosocomial bloodstream infections.2 Although C. albicans is still the most frequently isolated opportunistic yeast species, the dynamic nature of epidemiology shows a shift toward a growing number of rare yeast species.3
Candida fabianii (Cyberlindnera fabianii) is an uncommon and rare ascomycetous opportunistic yeast species with low virulence attributes.4 However, it has been shown that this emerging yeast species can cause invasive bloodstream infections.5–7 Another significant microbiological factor of Cy. fabianii is its ability to form antifungal resistant biofilms.7 Alarmingly, previous studies reported that Cy. fabianii has the potential to develop resistance to different antifungals, including fluconazole, voriconazole, caspofungin, and amphotericin B.4,7,8 Moreover, fluconazole prophylactic treatment of infection caused by Cy. fabianii can lead to a 50% failure outcome8 and may cause fatal septicemia.5,9,10
When subjected to commercially available biochemical assays (ID-32 C, API 20 C Aux, Vitek2 YST Card, and Vitek2 Compact), Cy. fabianii is usually misidentified as Cy. jadinii or Wickerhamomyces anomalus,4,5,11–14 leading to underestimation of this yeast in clinical settings. However, using molecular identification, it was revealed that the prevalence of Cy. fabianii was 10 times higher than W. anomalus and approximately 37 higher than Cy. jadinii.14 Furthermore, susceptibility testing seems to be different between Cy. fabianii, Cy. jadinii, and W. anomalus species,14 hence, developing a specific multiplex polymerase chain reaction (PCR) assays for accurate identification to the species level is important. Here, we described the first Iranian cases of Cy. fabianii from central venous catheter (CVC) and vagina, performed a systematic literature review to identify clinical and mycological features associated with Cy. fabainii, and designed a dual-function multiplex PCR that can be utilized in both conventional and real-time PCR platforms. Using SYBR Green I-based real-time PCR assay, Cy. fabianii, Cy. jadinii, and W. anomalus, and using conventional PCR aforementioned species and Cy. mississipiensis were unequivocally identified.
Methods
Epidemiology and clinical profile
Case reports and epidemiological studies reported Cy. fabianii were systematically searched from Pubmed and Google scholar (searching date: 8 August 2008). The key words for our search were “fabianii” and “infection,” without any limitation in language or date. All the titles and abstracts were checked, and full-text of related papers were studied for further evaluation. All cases have been confirmed by culture, and appropriate diagnostic tools such as matrix-assisted laser desorption/ionization–time of flight (MALDI-TOF) MS or DNA sequencing for species identification (also for epidemiological studies), except for the first clinical isolate of Cy. fabianii in 1990, as it was identified by API 20 C and appropriate complementary mating type assays.
Isolates and conditions
During 6 months (July 1, 2017, to December 31, 2017), six isolates of Cy. fabianii (two from central venous catheter and four from vaginal swabs) and one isolate of Cy. jadinii were collected from Iranian patients in Tehran, Shiraz, and Fasa. In order to check the specificity and sensitivity of our multiplex PCR assay, a panel of 70 strains comprising of 30 clinically important yeast species (40 target and 30 nontarget species) collection were received from the collection of Westerdijk Fungal Biodiversity Institute (formerly known as CBS) (Supplementary Table S1). Moreover, clinical isolates of Cy. fabianii (n = 6) and Cy. jadinii (n = 1) obtained from this study were included in the sensitivity and specificity testing of our PCR assays. CBS reference and type strains were cultured on GYPA plates for 48 hours at 25°C. Central venous catheters and vaginal swabs were struck on Sabouraud dextrose agar and CHROMagar media, followed by incubation at 37°C for 24–48 hours.
MALDI-TOF MS
The initial identification of clinical isolates were carried out by MALDI-TOF MS.15 Scores ≥2 were considered as reliable identification.
Sequencing
Definitive identification of target species was carried out by Sanger sequencing of ITS and LSU of rDNA domain using (ITS1 and ITS4) and (LR5 and LROR) primers.16
DNA extraction, quality and quantity assessments
One full loop of pure colonies (10 μl) were subjected to the DNeasy Blood and Tissue kit and DNA extraction was performed according to manufacturer's protocol. In the last step of DNA extraction, 70 μl of elution buffer (AE buffer) was added to the columns, and they were incubated at room temperature for 15 minutes followed by centrifugation at 10 000 rpm for 1 minute.
NanoDrop™ 2000 (Thermo Fisher Scientific, Waltham, MA, USA) and QuBit dsDNA BR Assay Kit (Thermo Fisher Scientific) were used for quality and quantity assessments, respectively. DNA samples were adjusted at one nanogram/μl and utilized as template for PCR reactions.
Primer design
In order to design specific primers for Cy. fabianii, Cy. mississipiensis, W. anomalus, and Cy. jadinii, ITS1-5.8s-ITS2 rDNA loci of target (along with sequences derived from our isolates) and nontarget species were obtained from NCBI database (https://blast.ncbi.nlm.nih.gov/Blast.cgi), sequences were aligned by MEGA software v7.0, and primers were positioned in the most stable and specific regions to ensure successful amplification of target species from various genotypes and to minimize the risk of cross-reaction with the other species. One universal primer and five species-specific primers were designed (Table 1). The primer properties were evaluated by the IDT Oligo analyzer on-line software (https://eu.idtdna.com/calc/analyzer). Primers were manufactured by the IDT Company (Integrated DNA Technology, Leuven, Belgium).
Table 1.
Information of the primers utilized in this study.
| Primer name | Primer sequence | Target loci | Target species | Melting temperature | PCR product size |
|---|---|---|---|---|---|
| PR-Missi | CCATGCTTCAACACACTACTGC | ITS2 rDNA | Cy. mississipiensis | 81.56 ± 0.38 | 320 bps |
| PR-Ano | GCTTATTAGTACACTCTTGCTAAGTCAA | ITS2 rDNA | W. anomalus | 80.48 ± 0.2 | 235 bps |
| PR-Jad | ACCAAGTCCCCTAGAGGATC | ITS2 rDNA | Cy. jadinii | 81.38 ± 0.23 | 176 bps |
| PF-Universal | CAACGGATCTCTTGGTTCTCG | 5.8 s rDNA | All except for Cy. fabianii | NA | NA |
| PF-Fab | ACTAGCGCGGCGACTAAAAC | ITS1 rDNA | Cy. fabianii | 77.62 ± 0.19 | 84 bps |
| PR-Fab | CGCAGAAAAGCTAGGCTTATTCC | ITS1 rDNA | Cy. fabianii | 77.62 ± 0.19 | 84 bps |
NA, not applicable; PCR, polymerase chain reaction.
Conventional PCR reactions
The multiplex PCR assay was utilized in both conventional and real-time PCR machines. For the conventional PCR, reactions were adjusted at 50 μl as follows: 39 μl MiliQ water (Merck Millipore, Billerica, MA, USA), 5 μl 10 × buffer, 1.5 mM MgCl2, 2.5 units of Taq enzyme (Bio Taq DNA Polymerase, Biolab), 0.2 mM of mixed dNTP (dNTP mix, 100Mm, Biolab), 5 picomoles of the PR-Ano, PR-Jad, PR-Missi, and 10 pM of PF-Universal, PF-Fab and PR-Fab primers, and 1 μl of DNA template.
PCR (2720 Thermal Cycler, Applied Biosystems, Waltham, MA, USA) used the following program, pre-denaturation for 5 minutes at 94°C, 35 cycles of 94°C for 30 seconds, 65°C for 30 seconds, 72°C for 30 seconds, and final extension at 72°C for 8 minutes. PCR products were run on 2% agarose gel for 45 minutes (8 Volt/cm), stained with GelRed (BioTium Corporation, USA) and visualized using gel documentation with exposure time of 4 seconds (Gel Doc XR+, BioRad, CA, USA).
Real-time PCR reactions
For real-time PCR (Applied Biosystems® 7500 fast, Thermo Fisher Scientific), the following PCR conditions were used: 10 μl of Power Up SYBR Green Master Mix (Thermo Fisher Scientific) five pM of the PR-Fab, PF-Fab, PR-Ano, PR-Jad, PR-Missi and 10 pM of PF-Universal primer, 1 μl of DNA template, and Mili-Q water (Merck Millipore, Darmstadt, Germany) to reach the final volume of 20 μl. The PCR program was as following: 50°C for 2 minutes, 95°C for 3 minutes, followed by 40 cycles of 95°C for 15 seconds and 64°C for 30 seconds. Upon termination of the PCR program, the PCR products underwent melting curve analysis with the increment of 0.5°C/second from 65°C to 95°C.Data analysis was carried out by 7500 software version 2.3 (Thermo Fisher Scientific).
Specificity and sensitivity testing
A panel of 70 CBS reference strains comprising 30 clinically related yeast species were used for the specificity testing (Supplementary Table S1).
In order to draw the standard curve, 10 log serial dilutions of DNA samples starting from 10 000 cfu/μl and ending at 10 cfu/μl were prepared in duplicate for each target species. Sensitivity, reproducibility, and the efficiency of the multiplex PCR were obtained from the standard curves.
Antifungal susceptibility testing
Antifungal susceptibility patterns of Cy. fabianii and one isolate of Cy. jadinii were determined by recommended protocol of Clinical and Laboratory Standards Institute (CLSI) M27-A3 and M27-S4 documents).17 The following drugs were included: itraconazole (Janssen Research Foundation, Beerse, Belgium), voriconazole (Pfizer, Central Research, Sandwich, UK), fluconazole (Pfizer, Groton, CT, USA), anidulafungin (Pfizer, Central Research, Sandwich, UK), and amphotericin B (Sigma, St. Louis, MO, USA). The minimum inhibitory concentration (MIC) values for amphotericin B and the rest of antifungal agents were determined by 100 and 50% inhibition compared to control strains of Candida krusei ATCC 6258 and Candida parapsilosis ATCC 22019, respectively. MIC values were interpreted based on CLSI-M27/S4, where amphotericin B (≥2 μg/ml), itraconazole and voriconazole (≥1 μg/ml), fluconazole (≥8 μg/ml) and caspofungin (≥1 μg/ml)18 were considered as resistant.
Utilized software
Geneious (version 10.2.3) software was used to design the primers. SPSS (version 21, International Business Machines Corporation, Armonk, NY, USA) and Graph Pad Prism (version 5, GraphPad Software, Inc., San Diego, CA, USA) were used for statistical analysis. Mean ± standard deviation (SD) was used for data with normal distribution.
Accession numbers
D1/D2 LSU and ITS rDNA sequences obtained for Cy. fabianii and Cy. jadinii with the accession numbers of MH236225, MH236226, MH236227, MH236228, MH236229, MH236230, MH236231, MH236232, MH236233, MH236234, MH236235, MH236236, and MH236237 were deposited in the GenBank database (https://www.ncbi.nlm.nih.gov/genbank/). Isolates of Cy. fabianii were deposited in the culture collection of Westerdijk Fungal Biodiversity Institute and they were given the following CBS reference numbers, 15348, 15349, 15352–15355.
Results
Literature review of cases
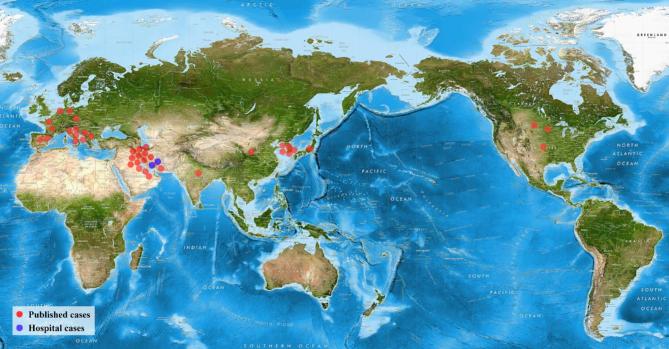
Using the literature review we identified 39 detailed cases (supplementary Tables S2 and S3). The global distribution of infectious caused by Cy. fabianii is shown in Fig. 1. The majority of the cases clustered in Western Europe and the Middle East (Fig. 1). Patient characteristics including demographic, clinical information, and microbiological findings are presented in supplementary Tables S2 and S3. The majority of the cases (35/39; 89.7%) were published after 2010. Small differences in terms of sex were observed (male/female = 16/21). A high proportion of patients were neonates (<1 month, n = 17), followed by adults (≥17 years, n = 15), infant (>1 month and <1 year, n = 2), and children (>1 year and <18 years, n = 4). Fungemia is the most common infection caused by Cy. fabianii (23/39, 59.0%), followed by funguria (n = 4), prostatitis (n = 1), endocarditis (n = 1), meningitis (n = 1) and peritonitis (n = 1). Catheter insertion was performed in 18 cases, from which seven of them showed growth of Cy. fabianii. From the 17 cases of the neonates, 13 of them were preterms. 13 cases had a history of exposure to antibiotics before the manifestation of infections. Post-surgery histories were observed for 12 cases. Five cases were reported with cancer. Other risk factors included steroids exposure, dialysis, leukopenia, mechanical ventilation, posterior urethral valve, and malnutrition.
Figure 1.
Global distribution of published cases of Cy. fabianii. This Figure is reproduced in color in the online version of Medical Mycology.
The most frequently used identification kits were API kits (n = 21) or the VITEK system (n = 14) but in all cases failed to correctly identify Cy. fabianii. However, subjecting misidentified cases to either MALDI-TOF or sequencing resulted in accurate identification.
Antifungal susceptibility testing showed 16.7% (3/21), 12.5% (1/11), 8.3% (1/15), and 12.5% (2/19) resistant rates for fluconazole, itraconazole, voriconazole, and amphotericin B, respectively. No resistance to anidulafungin and flucytosine, were observed. The overall mortality for all included cases was 23.1% (9/39).
As Cy. fabianii, Cy. jadinii, and W. anomalus are easily misidentified by phenotypic and biochemical assays and in order to obtain a clear image about the epidemiology and distribution of Cy. fabianii, papers that utilized MALDI-TOF or sequencing for identification were further investigated (Supplementary Table S4).
MALDI-TOF and Sanger sequencing results
From July 1 2017 to December 31, 2017, six isolates of Cy. fabianii and one isolate of Cy. jadinii were recovered from central venous catheters and vaginal samples. Sanger sequencing using D1/D2 LSU and ITS rDNA domains and MALDI-TOF MS were 100% in agreement with each other. Isolates of Cy. fabianii were recovered from central venous catheter (n = 2) and vaginal swabs (n = 4), and the isolate of Cy. jadinii was obtained from vaginal swab.
Antifungal susceptibility testing
Supplementary Table S5 summarizes the MIC values of the tested antifungal drugs against six clinical strains of Cy. fabianii and one isolate of Cy. jadinii. All strains of Cy. fabiani had low MICs for anidulafungin, followed by itraconazole and voriconazole, whereas the least active drug was fluconazole. Among the antifungal drugs, fluconazole exhibited the highest MIC value that ranged from 1 to 8 μg/ml, followed by amphotericin B (MIC range 0.016–2 μg/ ml), voriconazole (MIC range 0.016–1 μg/ml), and itraconazole (MIC range 0.063–1 μg/ml. All Cy. fabianii and Cy. jadinii isolates were susceptible to anidulafungin (MIC = 0.006 μg/ ml). The MIC value of fluconazole against three isolates of Cy. fabianii (two from CVC and one from vagina) was >3 log3 higher than that of the other isolates.
Conventional multiplex PCR results
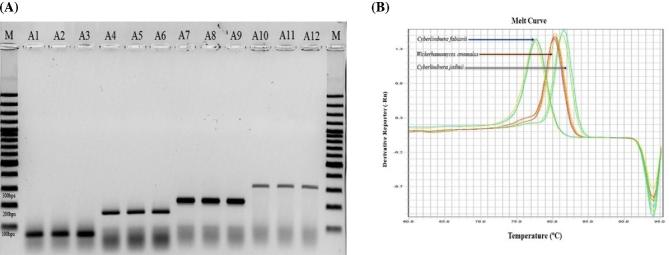
Subjecting genomic DNA samples of Cy. fabianii (n = 14), Cy. jadinii (n = 11), W. anomalus (n = 13), Cy. mississipiensis (n = 2) to our multiplex PCR resulted in amplicons of 84 bps, 176 bps, 235 bps, and 320 bps, respectively (Fig. 2A). All four target species could be distinguished and identified unequivocally. Subjecting CBS 7232 (reference strain of Cy. jadinii) to our multiplex PCR resulted in negative amplification. Further identification by MALDI-TOF and Sanger sequencing identified this strain as Meyerozyma guilliermondii. As a result, designation of CBS 7232 in the collection department of Westerdijk Institute was corrected. Challenging our conventional multiplex PCR with DNA samples of 30 clinically important yeast species, showed 100% specificity for our conventional PCR assay.
Figure 2.
Differentiation of target species using (A) conventional PCR through amplicon size polymorphism (M, 100 bps molecular weight, A1-A3, CBS14917, CBS 5481, CBS 5482, A4-A6, CBS1517, CBS1726, CBS 621, A7-A9, CBS 6417, CBS 110, CBS 2870, A10-A12, CBS 7023, CBS 5837, CBS 5837) and (B) qPCR through melting temperatures. This Figure is reproduced in color in the online version of Medical Mycology.
Multiplex qPCR results
Primers utilized for the conventional multiplex PCR were subjected to a SYBR Green I-based qPCR assay. Distinctive melt temperatures of 77.62 ± 0.19°C, 80.48±0.2°C, 81.38 ± 0.23°C, and 81.56 ± 0.38°C distinguished Cy. fabianii (n = 14), W. anomalus (n = 13), Cy. jadinii (n = 11), Cy. mississipiensis (n = 2) (Fig. 2B). However, the melting Tm of Cy. jadinii showed an overlap with that of Cy. mississipiensis. As a result, the specific primer of PR-Missi was withdrawn from the multiplex reaction to unequivocally identify Cy. jadinii (as the former species is not clinically important). Challenging our multiplex PCR with a specificity test set, resulted in minor cross-reaction at higher Ct values of 25 and 30 with the DNA of Pichia kudriavzevii (Candida krusei) and M. guilliermondii (Candida guilliermondii), respectively (Supplementary Fig. S1). However, by adjusting the threshold at a Ct of 22, target species of Cy. fabianii, W. anomalus, Cy. jadinii were reliably identified (100 cfu), and cross-reaction with other clinically important yeast pathogens was not observed. Subjecting 1 ng DNA of the mistakenly designated reference strain of CBS 7232 to our multiplex qPCR resulted in amplification at a Ct value of 30, while the rest of DNA of Cy. jadinii were amplified at a Ct value of 14, which was in concordance with results obtained from the conventional multiplex PCR, MALDI-TOF MS, and Sanger sequencing. Regarding the reproducibility, five runs in three consecutive days generated an average R2 value of 0.99 for all target species, demonstrating a high reproducibility of our qPCR assay (Supplementary Fig. S2).
Discussion
Using MALDI-TOF MS and sequencing of ITS and LSU rDNA, we identified six isolates of Cy. fabianii (n = 4 from vagina and n = 2 from CVC). One of our patients was suffering from acute myeloid leukemia (AML), end-stage renal diseases, hypertension, and diabetes mellitus, and the other one had severe trauma and subsequently underwent surgery (both CVC isolates). Concordant with previous studies, AML, severe trauma and surgical interventions are risk factors for development of invasive candidiasis.19 Investigating published case-reports showed that the main risk factors associated with the acquirement of Cy. fabianii infection are attributable to previous exposure with antibiotic therapy followed by central venous catheter insertion, low birth weight, surgery, cancer, neutropenia, chemotherapy, and renal failure. In our study, four patients with vaginitis did not have any underlying diseases, and they were all immunocompetent. Review of cases revealed that there were four cases of funguria with Cy. fabianii, and all of them suffered from underlying conditions.4,8 Data mined from literature review in agreement with the presumption that infections pertained to Cy. fabianii are mainly linked to neonates,20 revealed that the majority of the infections were acquired by neonate (< 1 month), followed by adults with the age of ≥17 years and the range of 17–87 years, and infants with the age of <1 year (8%). On the contrary, all of our cases were >18 years, which is in agreement with the Cy. fabianii isolate (obtained from sputum) previously reported from Iran, from a 69 year-old man.21 Although Cy. fabianii features a wide range of clinical syndromes, ranging from fungemia and funguria to meningitis, fungemia with the highest proportion (59%), is considered as the most prevalent clinical complication caused by this opportunistic yeast species.
Antifungal susceptibility pattern of our isolates exhibited the highest MICs for fluconazole, followed by amphotericin B, which is in agreement with previous reports.4 In agreement with previous study (12), 100% of our isolates of Cy. fabianii (n = 6) were susceptible to anidulafungin.
For treatment of Cy. fabianii, fluconazole was the most widely utilized drug followed by different formulation of amphotericin B, caspofungin, and flucytosine. Regarding the efficacy of antifungal drugs for clearance of infection, studies suggested a combination of fluconazole with liposomal AMB or caspofungin or removal of central venous and urinary tract catheter.8 In addition, treatment with anidulafungin led to successful outcome for all patients infected with Cy. fabiani.12 In our cases, one of the patients died despite of treatment with AMB. However, death could not be linked to failure in treatment as the patient suffered from other serious background diseases. Treatment of the second patient (with trauma and surgery) with AMB resulted in successful outcome. Patients suffering from vaginitis (n = 4) were all successfully treated with topical clotrimazole and fluconazole.
In order to overcome the persisting challenge of misidentification of Cy. fabianii, we developed a dual-function multiplex PCR. Our multiplex PCR showed 100% agreement with sequencing and MALDI-TOF MS. Although a previous study described a singleplex PCR that could only detect Cy. fabianii, authors utilized limited number of yeast species for specificity testing.8 However, our assay first tested with a broad range of yeast species and second is a multiplex PCR targeting Cy. fabianii, Cy. mississipiensis, Cy. jadinii, and W. anomalus. MALDI-TOF and Sanger sequencing can definitively identify these three clinically important yeast species, but these devices are usually not available in routine clinical microbiology laboratories, and they are either expensive and/or require highly trained personnel to perform the experiments.22 Although MALDI-TOF MS compared to PCR requires less expenses per reaction, high costs with the initial purchase along with the maintenance cost prevent installation of this platform in routine laboratories with financial constraints.23 Currently, PCR as an affordable and reproducible device has been exploited in the form of a supplementary identification tool in routine laboratories of developing countries.24 Moreover, advances in PCR machinery allowed investigators of low-resourced countries to construct their own home-made PCR machines using the most basic off-the-shelf appliances with an exceptional efficiency that costs only 150 US dollars.25 Our multiplex PCR Compared to time-consuming biochemical assays takes only 2–3.5 hours to report the identification results. Although biochemical assays are used frequently for identification, especially in developing countries,26 they do not have enough discriminatory power and frequently misidentify Cy. fabianii as Cy. jadinii or W. anomalus.13,14 Consequently, important worldwide studies such as the ARTEMIS DISK global antifungal surveillance over a period of 10.5 years did not identify a single isolate of Cy. fabianii, while they could identify 88 and 6 isolates of W. anomalus and Cy. jadinii, respectively.27 In contrast, MALDI-TOF MS correctly identified 163 and 222 isolates of Cy. fabianii that previously using API ID 32C kit were misidentified as W. anomalus and Cy. jadinii.13,14
Obtaining these erroneous results can lead to a concealed image of epidemiology for Cy. fabianii.13 Different epidemiological studies that used either MALDI-TOF MS or sequencing for identification, showed that Cy. fabianii, although with a low rate, but consistently is isolated, while W. anomalus and Cy. jadinii were identified in three and one of the studies, respectively.9,21,27–31 In our study, we found six isolates of Cy. fabianii, one isolate of Cy. jadinii, and no W. anomalus.
Due to the growing number of infections caused by non-Candida albicans yeast species, establishing definitive breakpoints and the development of specific molecular means of identification can contribute to both successful treatment and a better knowledge on the clinical occurrence of emerging and uncommon opportunistic yeast species.
The limitation of our study was that we utilized only 40 reference strains of Cy. fabianii, W. anomalus, Cy. jadinii, and Cy. mississipiensis, and therefore further experiments in different setups are required.
Supplementary Material
Acknowledgements
Author Contributions Statement: A.A., F.D., and W.J.F. participated in primer design, PCR optimization, data collection, and drafted the manuscript. W.H.P., K.Z., and T.B. participated in designing this study and revising the manuscript. A.M.H., W.L., and F.H. actively participated in paper revision. H.B., S.K., M.H.A, M.B., and S.K.A. participated collecting Iranian clinical isolates and associated clinical data. All authors contributed to the revision of the final manuscript.
Funding
This work was supported by the Major National R&D Projects of the National Health Department (2018ZX10101003), European Union's Horizon 2020 research and innovation program under the Marie Sklodowska-Curie grant agreement no. 642095, National Natural Science Foundation of China (31770161), Shanghai Science and Technology Committee (grant numbers 14DZ2272900 and 14495800500)
Declaration of interest
The authors report no conflicts of interest. The authors alone are responsible for the content and the writing of the paper.
References
- 1. Sobel JD, Fisher JF, Kauffman CA, Newman CA. Candida urinary tract infections epidemiology. Clin Infect Dis. 2011; 6: 433–436. [DOI] [PubMed] [Google Scholar]
- 2. Pfaller MA, Jones RN, Messer SA, Edmond MB, Wenzel RP. National surveillance of nosocomial blood stream infection due to species of Candida other than Candida albicans: frequency of occurrence and antifungal susceptibility in the SCOPE Program. Diagn Microbiol Infect Dis. 1998; 30: 121–129. [DOI] [PubMed] [Google Scholar]
- 3. Miceli MH, Díaz JA, Lee SA. Emerging opportunistic yeast infections. Lancet Infect Dis. 2011; 11: 142–151. [DOI] [PubMed] [Google Scholar]
- 4. Jindal N, Arora S, Dhuria N, Arora D. Cyberlindnera (Pichia) fabianii infection in a neutropenic child: importance of molecular identification. JMM Case Reports 2015; 1: 2015–2017. [Google Scholar]
- 5. Valenza G, Valenza R, Brederlau J, Frosch M, Kurzai O. Identification of Candida fabianii as a cause of lethal septicaemia. Mycoses. 2006; 49: 331–334. [DOI] [PubMed] [Google Scholar]
- 6. Bhally HS, Jain S, Shields C, Halsey N, Cristofalo E, Merz WG. Infection in a neonate caused by Pichia fabianii: Importance of molecular identification. Med Mycol. 2006; 44: 185–187. [DOI] [PubMed] [Google Scholar]
- 7. Hamal P, Ostransky J, Dendis M et al.. A case of endocarditis caused by the yeast Pichia fabianii with biofilm production and developed in vitro resistance to azoles in the course of antifungal treatment. Med Mycol. 2008; 46: 601–605. [DOI] [PubMed] [Google Scholar]
- 8. Vágvölgyi C, Mlinarić-Missoni E, Kocsubé S, Hatvani L, Škarić I, Lukić-Grlić A. Cyberlindnera fabianii in the neonatal and paediatric intensive care unit: case reports. JMM Case Reports. 2015; 2: 1–7. [Google Scholar]
- 9. Taj-Aldeen SJ, AbdulWahab A, Kolecka A, Deshmukh A, Meis JF, Boekhout T. Uncommon opportunistic yeast bloodstream infections from Qatar. Med Mycol. 2014; 52: 552–556. [DOI] [PubMed] [Google Scholar]
- 10. Yun JW, Park KS, Ki CS, Lee NY. Catheter-related bloodstream infection by Lindnera fabianii in a neutropenic patient. J Med Microbiol. 2013; 62: 922–925. [DOI] [PubMed] [Google Scholar]
- 11. Baghdadi J, Hemarajata P, Humphries R, Kelesidis T. First report of ventriculoperitoneal shunt infection due to Cyberlindnera fabianii. Case Rep Infect Dis. 2015; 2015: 630816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lee JI, Yu S, Park JS, Joo E, Shin JH, Kwon M. Successful treatment of fungemia caused by Cyberlindnera fabianii with anidulafungin : a case report. Ann Clin Microbiol. 2015; 18: 94–97. [Google Scholar]
- 13. Svobodova L, Bednarova D, Hamal P. The prevalence of Candida pelliculosa, Candida utilis, and Candida fabianii in the Olomouc University Hospital: epidemiological study. Epidemiol Mikrobiol Imunol. 2016; 65: 34–38. [PubMed] [Google Scholar]
- 14. Svobodova L, Bednarova D, Ruzicka F et al.. High frequency of Candida fabianii among clinical isolates biochemically identified as Candida pelliculosa and Candida utilis. Mycoses. 2016; 59: 241–246. [DOI] [PubMed] [Google Scholar]
- 15. Marklein G, Josten M, Klanke U et al.. Matrix-assisted laser desorption ionization-time of flight mass spectrometry for fast and reliable identification of clinical yeast isolates. J Clin Microbiol. 2009; 47: 2912–2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Stielow JB, Lévesque CA, Seifert KA et al.. One fungus, which genes? Development and assessment of universal primers for potential secondary fungal DNA barcodes. Persoonia. 2015; 35: 242–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rex JH, Alexander BD, Andes D et al.. Reference method for broth dilution antifungal susceptibility testing of yeasts: approved standard – 3rd edn. Clin Lab Stand Inst. 2008; 1–25. [Google Scholar]
- 18. Pfaller MA, Diekema DJ. Progress in antifungal susceptibility testing of Candida spp. by use of Clinical and Laboratory Standards Institute broth microdilution methods, 2010 to 2012. J Clin Microbiol. 2012; 50: 2846–2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yapar N. 2014. Epidemiology and risk factors for invasive candidiasis. Ther Clin Risk Manag. 2014; 10: 95–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hof H, Amann V, Tauber C, Paulun A. Peritonitis in a neonate due to Cyberlindnera fabianii, an ascomycetic yeast. Infection. 2017; 45: 921–924. [DOI] [PubMed] [Google Scholar]
- 21. Karimi L, Mirhendi H, Khodadadi H, Mohammadi R. Molecular identification of uncommon clinical yeast species in Iran. Curr Med Mycol. 2015; 1: 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Criseo G, Scordino F, Romeo O. Current methods for identifying clinically important cryptic Candida species. J Microbiol Methods. 2015; 111: 50–56. [DOI] [PubMed] [Google Scholar]
- 23. Singhal N, Kumar M, Kanaujia PK, Virdi JS. MALDI-TOF mass spectrometry: an emerging technology for microbial identification and diagnosis. Front. Microbiol. 2015; 6: 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. World Health Organization. Establishment of PCR Laboratory in Developing Countries. 2016; Geneva: WHO. [Google Scholar]
- 25. Wong G, Wong I, Chan K, Hsieh Y, Wong S. A rapid and low-cost PCR thermal cycler for infectious disease diagnostics. PLoS One. 2015; 10: 1–20. [Google Scholar]
- 26. Kathuria S, Singh PK, Sharma C et al.. Multidrug-resistant Candida auris misidentified as Candida haemulonii: characterization by matrix-assisted laser desorption ionization-time of flight mass spectrometry and DNA sequencing and its antifungal susceptibility profile variability by vitek 2, CLSI broth microdilution and Etest method. J Clin Microbiol. 2015; 53: 1823–1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pfaller MA, Diekema DJ, Gibbs DL et al.. Results from the artemis disk global antifungal surveillance study, 1997 to 2007: a 10.5-year analysis of susceptibilities of Candida species to fluconazole and voriconazole as determined by CLSI standardized disk diffusion. J Clin Microbiol. 2010; 48: 1366–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Won EJ, Shin JH, Lee K et al.. Accuracy of species-level identification of yeast isolates from blood cultures from 10 university hospitals in south Korea by use of the matrix-assisted laser desorption ionization-time of flight mass spectrometry-based vitek MS system. J Clin Microbiol. 2013; 51: 3063–3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pongrácz J, Juhász E, Iván M, Kristóf K. Significance of yeasts in bloodstream infection: Epidemiology and predisposing factors of candidaemia in adult patients at a university hospital (2010–2014). Acta Microbiol Immunol Hung. 2015; 62: 317–329. [DOI] [PubMed] [Google Scholar]
- 30. Fernández-Ruiz M, Guinea J, Puig-Asensio M et al.. Fungemia due to rare opportunistic yeasts: data from a population-based surveillance in Spain. Med Mycol. 2017; 55: 125–136. [DOI] [PubMed] [Google Scholar]
- 31. Bretagne S, Renaudat C, Desnos-Ollivier M, Sitbon K, Lortholary O, Dromer F. French Mycosis Study Group. Predisposing factors and outcome of uncommon yeast species-related fungaemia based on an exhaustive surveillance programme (2002–14). J Antimicrob Chemother. 2017; 72: 1784–1793. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.