Abstract
The present study aimed to systematically evaluate the genetic diversity of Chinese domestic duck breeds and ensure the most effective allocation and usage of conservation funds. We first performed an analysis of DNA genetic distance in 21 duck breeds by measuring short tandem repeats. Then, we calculated the extinction probability, contribution rate, and marginal diversity for each breed. The results showed that the extinction rate of the Zhongshan duck, Guangxi duck, and Ji’an duck were the highest at 0.67, 0.59, and 0.59, respectively, and that of the Linwu duck, Jinding duck, and Gaoyou duck were the lowest at 0.15, 0.18, and 0.19, respectively. The current diversity of populations was 7.72 and the expected diversity in five hundred years is 5.14 ± 1.15. The marginal diversity of the Chinese Muscovy duck was the largest (−2.20), accounting for 42.61% of the expected diversity, followed by the Guangxi duck (−0.49, 9.44%), whereas the Jinding duck was the smallest (−0.12; 2.32%). The protection potency of the Chinese Muscovy duck was the largest (0.61), followed by Guangxi duck (0.29), whereas the Jinding duck was the smallest (0.02). This study provides a reference for determining the conservation priority of Chinese domestic duck breeds or genetic resources.
Subject terms: Genetics, Predictive markers
Introduction
Animal genetic resources are the basis of the sustainable development of animal husbandry1,2, and China is one of the foremost countries in this respect, accounting for one-sixth of the world’s animal genetic resources3,4. Two systematic and comprehensive surveys were conducted on animal genetic resources: one from 1970 to 1980, and the other in 2004. According to the Report on Domestic Animal Genetic Resource in China (edited in 2012)5, 777 breeds of animal genetic resources have been formally named in China, including 556 local breeds, 109 cultivated breeds, 104 introduced breeds, and eight other breeds. With respect to breeds of poultry, there are 116 chickens, 34 ducks, 31 geese, three turkeys, three pigeons, and two species of quail.
However, animal genetic resources have shown an overall decline in China since 1970s, due to unknown resources in some areas, low in vivo conservation, loss of animal genetic resources, and large-scale adoption of breeding and intensification processes6. In particular, the large number of imported breeds, and their wide promotion, have greatly threatened Chinese domestic animal genetic resources7. As examples, the Guping chicken, Lintao chicken, Wenshan goose, and Simao goose have all become extinct and, in total, 44 breeds are on the edge of extinction and 15 are endangered8,9. In response to this crisis, the departments responsible for managing Chinese animal genetic resources have allocated yearly funds toward conservation efforts. Since these funds are limited, both conservation strategy and fund allocation are determined by the economic value and population size of a breed10. However, the subjectivity of this system could result in ineffective conservation of precious and endangered genetic resources. Therefore, a better system is needed to determine priority in these conservation efforts and achieve optimal allocation of funds11,12.
One option is through marginal diversity, which was defined by Weitzman in 199213 as a mechanism for measuring genetic diversity. The concept uses genetic and non-genetic factors to calculate a “maximum-likelihood tree”14 and the current diversity of breeds, and estimates the expected variations in diversity over a certain time. This approach defines criteria of diversity and relies on quantitative assessments of different strategies, providing concrete reasoning for breed conservation. At present, marginal diversity has been applied in studies on European pigs15 and cows16; however, no systematic assessment with this approach has been conducted in Chinese domestic duck breeds and the managers also do not know how to allocate funds for breed insurance. Here, we use short tandem repeat profiling to perform a marginal diversity analysis of 21 Chinese domestic duck breeds or genetic resources, which can be used to determine conservation priority.
Materials and Methods
All animal experiments were performed in accordance with the Regulations for the Administration of Experimental Animals issued by the Ministry of Science and Technology (Beijing, China). All experiments were approved by the Animal Care and Use Committee of Yangzhou University.
Breed and genetic distance measurements
The objects of the study were Chinese domestic duck breeds or their genetic resources. Their name, sample size, and origin are shown in Table 1. Blood was collected according to pedigree, to ensure that samples were from unrelated individuals. The samples (0.5 mL) obtained from the vein of the ducks wings were carefully mixed with lysis solution and kept at 4 °C for subsequent DNA extraction. DNA extraction was performed according to the method described by Huang et al.17.
Table 1.
Name, sample size, and origin of 21 Chinese domestic duck breeds.
| Breed | Abbreviation | Sample size | Economic use | Feather color | Existing quantity | Origin |
|---|---|---|---|---|---|---|
| Beijing duck | BJ | 96 | meat | white | 49,900,000 | Jade Spring Hill, Beijing |
| Chaohu duck | CH | 80 | meat/egg | hemp | 2,000,000 | Lujiang, Chaohu, Anhui |
| Dayu duck | DY | 96 | meat | hemp | 110,000 | Dayu, Ganzhou, Jiangxi |
| Chinese Muscovy duck (Chinese Fanya) | FY | 96 | meat | white/black | 1,200,000 | Honduras |
| Guangxi small sheldrake | GX | 72 | egg/meat | hemp | 10,000,000 | Xilin, Baise, Guangxi |
| Gaoyou duck | GY | 66 | egg/meat | hemp | 2,000,000 | Gaoyou, Jiangxi |
| Ji’an red duck | JA | 80 | meat/egg | brown red | 10,000,000 | Suichuan, Ji’an, Jiangxi |
| JIanchang duck | JC | 96 | meat/egg | hemp | 530,000 | Xichang City and Dechang County, Sichuan |
| Jinding duck | JD | 80 | egg | hemp | 12,000 | Zini, Longhai, Fujian |
| Jingjiang sheldrake | JJ | 80 | meat | hemp | 136,000 | Jingzhou, Hubei |
| Jianshui brown duck | JS | 96 | meat/egg | brown | 12,000 | Jianshui, Lin’an, etc., Yunnan |
| Jingxi large sheldrake | JX | 72 | egg/meat | hemp | 400,000 | Jingxi, Baise, Guangxi |
| Liancheng white duck | LC | 96 | fancy | white | 1,500,000 | Liancheng, Longyan, Fujian |
| Linwu duck | LW | 72 | egg | light gray hemp | 6,510,000 | Linwu, Chenzhou, Hunan |
| Mawang duck | MW | 96 | egg | light gray hemp | 466,000 | Youyang, Chongqing |
| Putian black duck/coot | PT | 96 | meat/egg | black | 150 000 | Lingchuan, Putian, Fujian |
| Shanma (Mountain) duck | SM | 72 | egg | light gray hemp | 25,000 | Longyan, Fujian |
| Sansui duck | SS | 96 | egg/meat | hemp | 10,000 | Sansui, Guizhou |
| Taiwan duck | TW | 96 | egg/meat | dun | 2,400,000 | Yilan, Dalin, etc., Taiwan |
| Youxian County sheldrake | YX | 72 | egg | light gray hemp | 5,800,000 | Youxian, Zhuzhou, Hunan |
| Zhongshan sheldrake | ZS | 96 | meat/egg | hemp | None | Zhongshan, Guangdong |
Twelve pairs of microsatellite primers with rich polymorphism were selected as follows: APH01, APL2, AJ272579, AJ272578, AJ272577, AJ415887, AJ515884, AJ515893, AY493256, AY493289, AY493313, and CMO11. The Sequences, combination, and optimal reaction condition have been reported previously18. A total of 1802 ducks were genotyped and the population genetic parameters calculated were described in a published paper18. The standard genetic distance between populations was calculated with Microsatellite-Toolkit19 and Dispan (http://www.softpedia.com/get/Science-CAD/DISPAN.shtml).
PCA and population structure analysis for all breeds
In this study, SPSS13.0 software was used for principal component analysis (PCA) of all the detected alleles20, and Structure 2.0 (http://rosenberglab.bioinformatics.med.umich.edu/distruct.htm) software was used for genetic Structure analysis of 21 populations.
Extinction probability
Extinction probability is an important index for genetic resource diversity. Future changes in the diversity of local breeds or genetic resources can be measured as the extinction probability over time (500 years)21. In Weitzman’s approach, the extinction probability (Zi) of each set is a variable that needs special attention. There are various methods for calculating Zi; however, we adopted the method proposed by Reust-Marti11. This method uses seven variables: the total population size (POS), its change over the past 10 years (CHA), distribution of the breed (DIS), risk of indiscriminate crossing (CRO), organization and conservation measures of breeding (ORG), special traits (SPE), and threat of production transition (PRO) (Table 2). Different weights (wi) were given to different variables to estimate Zi in the future 500 years. The estimate formula and correction formula are as follows:
| 1 |
where, wi is the weight of each variable (w1 = 0.35, w2 = 0.15, w3 = 0.14, w4 = 0.10, w5 = 0.10, w6 = 0.06 and w7 = 0.10) and xi is the estimate of the ith indicator. The seven parameters for this analysis were attained by on-site observation, literature review, and estimation, in order to calculate the Zi of each breed and genetic resource in the next 500 years. For the convenience of calculation, Zia of each breed or genetic resource was corrected to 0.1–0.9, according to the formula below22.
| 2 |
Table 2.
Influencing factors and criteria of extinction probability.
| Influencing factor | Abbreviation | Grading standard |
|---|---|---|
| Total population size | POS | 0.3 < ten thousand; 0.2 = ten thousand to one hundred thousand; 0.1 = one hundred thousand to one million; 0 = one million |
| Change of total population size over the past 10 years | CHA | 0.1 = decreasing (>20%); 0 = increasing or maintaining stability |
| Distribution of the breed | DIS | 0.2 = county; 0.1 = city; 0 = trans-regional and trans-provincial areas |
| Risk of indiscriminate crossing | CRO | 0.2 = high degree; 0.1 = moderate degree; 0.05 = low degree; 0 = No |
| Organization and conservation measures of breeding | ORG | 0.2 = No; 0 = Yes |
| Special traits | SPE | 0.1 = None; 0 = Yes |
| Threat of production transition | TRA | 0.3 = high degree; 0.2 = moderate degree; 0.1 = low degree |
For the set (S) containing a certain number (N) of breeds and genetic resources, and a breed i, the distance of j ∈ S can be expressed as dij. According to Weitzman’s recursive algorithm, the diversity variable D (S) can be calculated by an N × N distance matrix. The probability of a breed’s existence in 500 years is 1-Zi, if Z is an N-dimensional vector containing Zi of N sets. K is an N-dimensional vector containing the indicator variable Ki (i = 1, 2, … N). Ki = 1 if the set i exists, whereas Ki = 0 if the set i is extinct. Therefore, K represents an overview of the status in which a subset of breeds exists and its complementary subset is extinct. The formula of the existence probability of a subset of breeds is as follows:
| 3 |
DK is the diversity of the subsets safe from extinction. The expected diversity at the end of the time horizon (500 years) is calculated as:
| 4 |
The variance of the expected diversity is:
| 5 |
The marginal diversity of a breed or genetic resource reflects the variation of the expected diversity when the extinction probability is increased by one unit. The marginal diversity is calculated as follows:
| 6 |
Based on the extinction probability and expected diversity of a breed or genetic resource, Weitzman suggested conservation potency as the optimal parameter to assess the genetic diversity over a given time horizon. The conservation potency (CP) is calculated as follows:
| 7 |
CPi represents a possible increase in the expected diversity of a breed or genetic resource when the threat is completely removed. According to previous work by Simianer et al.10, CPi is the optimal parameter for determining conservation schemes, with the highest CPi requiring a minimum amount of capital required for the protection scheme22. The breed or genetic resource with the highest CPi should be allocated the least funds in breeding conservation efforts10,23.
Results
Genetic distance
Twelve simple sequence repeats were detected in the 21 duck breeds. Nei’s standard genetic distance was estimated using Microsatellite-Toolkit and Dispan24. According to the genetic distance matrix (Table 3) and cluster analysis by the unweighted pair-group method with arithmetic means (Fig. 1)25, FY belonging to the Cairina breed forms a single set, whereas the other 20 duck breeds (Anas) form three large sets. The distance between GY and JA was the shortest, and the distance between FY and GX was the longest First bullet.
Table 3.
Nei’s standard genetic distance between 21 Chinese domestic duck breeds.
| BJ | CH | DY | FY | GX | GY | JA | JC | JD | JJ | JS | JX | LC | LW | MW | PT | SM | SS | TW | YX | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CH | 0.3849 | |||||||||||||||||||
| DY | 0.3552 | 0.2093 | ||||||||||||||||||
| FY | 1.4498 | 1.5928 | 1.7169 | |||||||||||||||||
| GX | 0.5577 | 0.5698 | 0.2720 | 2.3350 | ||||||||||||||||
| GY | 0.4749 | 0.3001 | 0.2800 | 2.0529 | 0.1984 | |||||||||||||||
| JA | 0.5655 | 0.4483 | 0.3074 | 2.1508 | 0.1557 | 0.0700 | ||||||||||||||
| JC | 0.1875 | 0.4740 | 0.3797 | 1.4489 | 0.5702 | 0.6435 | 0.7541 | |||||||||||||
| JD | 0.5225 | 0.3202 | 0.2559 | 1.9372 | 0.1793 | 0.1212 | 0.1301 | 0.6755 | ||||||||||||
| JJ | 0.3738 | 0.2097 | 0.1989 | 1.7176 | 0.2734 | 0.1910 | 0.2287 | 0.4072 | 0.1355 | |||||||||||
| JS | 0.2040 | 0.3825 | 0.4276 | 1.3069 | 0.6836 | 0.5397 | 0.6659 | 0.2801 | 0.5232 | 0.4767 | ||||||||||
| JX | 0.3935 | 0.4419 | 0.1989 | 2.0945 | 0.2999 | 0.4227 | 0.4163 | 0.3683 | 0.3702 | 0.2440 | 0.4795 | |||||||||
| LC | 0.5355 | 0.5450 | 0.4381 | 1.2576 | 0.7158 | 0.6798 | 0.7618 | 0.4471 | 0.7652 | 0.5228 | 0.5866 | 0.4956 | ||||||||
| LW | 0.4433 | 0.3063 | 0.2631 | 2.1002 | 0.3364 | 0.3405 | 0.3244 | 0.4824 | 0.2751 | 0.2529 | 0.5010 | 0.3217 | 0.7047 | |||||||
| MW | 0.2260 | 0.3622 | 0.3898 | 1.2259 | 0.6588 | 0.5169 | 0.6061 | 0.3178 | 0.4908 | 0.4347 | 0.2043 | 0.4445 | 0.3674 | 0.4905 | ||||||
| PT | 0.6262 | 0.6331 | 0.5018 | 1.2362 | 0.7920 | 0.7392 | 0.8338 | 0.5651 | 0.6991 | 0.5654 | 0.5596 | 0.4819 | 0.1791 | 0.5982 | 0.4644 | |||||
| SM | 0.4500 | 0.3127 | 0.2506 | 1.7219 | 0.3043 | 0.2494 | 0.2266 | 0.6413 | 0.1110 | 0.2022 | 0.4304 | 0.4134 | 0.5293 | 0.4080 | 0.3766 | 0.6702 | ||||
| SS | 0.2863 | 0.2854 | 0.2604 | 1.4692 | 0.4835 | 0.3242 | 0.4179 | 0.3951 | 0.3245 | 0.2849 | 0.3223 | 0.3351 | 0.2986 | 0.3026 | 0.1489 | 0.3283 | 0.3262 | |||
| TW | 0.3868 | 0.6315 | 0.5551 | 1.2674 | 0.8891 | 0.8208 | 0.8472 | 0.5227 | 0.7868 | 0.6760 | 0.4128 | 0.6160 | 0.4060 | 0.7507 | 0.1730 | 0.3802 | 0.5840 | 0.3922 | ||
| YX | 0.4048 | 0.2970 | 0.1567 | 1.8717 | 0.3296 | 0.3464 | 0.3301 | 0.4498 | 0.3180 | 0.3098 | 0.4438 | 0.3354 | 0.5949 | 0.1912 | 0.4687 | 0.5947 | 0.2805 | 0.3386 | 0.6838 | |
| ZS | 0.1228 | 0.3896 | 0.3505 | 1.4752 | 0.5732 | 0.5210 | 0.5913 | 0.1998 | 0.4878 | 0.3876 | 0.2101 | 0.3239 | 0.4621 | 0.4121 | 0.1372 | 0.5241 | 0.4182 | 0.2113 | 0.3126 | 0.4576 |
Note: Breed abbreviations are defined in Table 1.
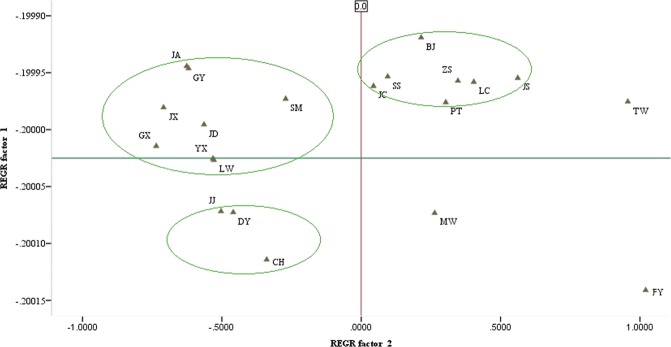
Figure 1.
Two-dimensional scatter plot of the first and second factors for 21 duck populations. Note: Breed abbreviations are defined in Table 1.
PCA and population structure analysis for all breeds
PCA was performed on the gene frequencies of all alleles detected in 12 SSR seats in 21 populations. The plane distance graph constructed according to the first two principal components was shown in Fig. 1. 21 duck breeds were divided into three large groups. Among them, BJ, ZS, SS, JC, PT, JS, LC were relatively close to each other. JJ, DY and CH were close to each other, forming another group. In addition, 9 breeds including JA, GY, SM, JX, JD, GX, YX, LW constituted a group. The distances between FY and other 20 domestic duck breeds were relatively large.
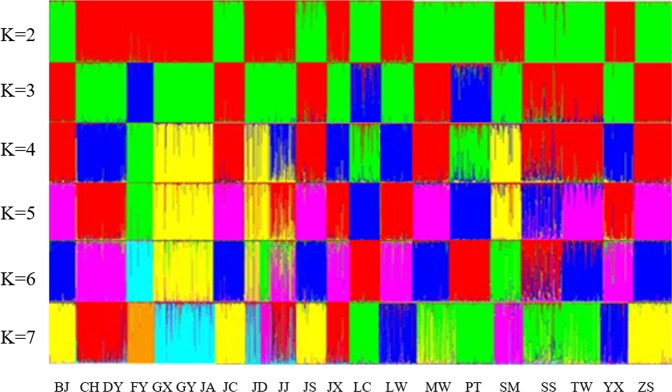
Structure 2.0 program can group individuals with similar genotypes on multiple SSR seats without prior knowledge of the fusion or evolutionary history of the populations (Fig. 2). When K = 2, no single population was isolated; Of these, 9 breeds such as BJ, JC, JS, LC, MW, PT, SS, TW, ZS were grouped together, and the rest of 12 breeds were clustered into one group. When K = 3 and K = 4, no single group was isolated. FY, LC, PT were clustered into one group. FY ducks were isolated as a single group until K = 5. When K = 7, BJ, JC, JS and ZS still formed a group.CH, DY and JX still gathered into one group: GX, GY, JA, JD CLUSTERED into the last one group.
Figure 2.
Population structure of 21 populations by the individual Q matrix structure. (Running Structure 1000 times from K = 2 to 7). Note: Breed abbreviations are defined in Table 1.
Extinction probability
The average extinction probability for the 21 Chinese domestic duck breeds and genetic resources was 0.38% (Table 4). ZS, GX, and JA had the largest extinction probabilities, whereas LW, JD, and GY had the smallest.
Table 4.
Extinction factor weighting and extinction probability correction of each population.
| Breed name | POS | CHA | DIS | CRO | ORG | SPE | TRA | Zi | Correction |
|---|---|---|---|---|---|---|---|---|---|
| Weight | 0.35 | 0.15 | 0.14 | 0.10 | 0.10 | 0.06 | 0.10 | 1.00 | |
| BJ | 0.00 | 0.00 | 0.00 | 0.20 | 0.00 | 0.00 | 0.30 | 0.05 | 0.23 |
| CH | 0.20 | 0.00 | 0.10 | 0.20 | 0.20 | 0.10 | 0.20 | 0.15 | 0.48 |
| DY | 0.30 | 0.00 | 0.20 | 0.10 | 0.20 | 0.10 | 0.20 | 0.19 | 0.58 |
| FY | 0.10 | 0.00 | 0.00 | 0.00 | 0.20 | 0.10 | 0.10 | 0.07 | 0.28 |
| GX | 0.30 | 0.10 | 0.10 | 0.05 | 0.20 | 0.10 | 0.30 | 0.20 | 0.59 |
| GY | 0.00 | 0.00 | 0.10 | 0.10 | 0.00 | 0.00 | 0.10 | 0.03 | 0.19 |
| JA | 0.30 | 0.10 | 0.20 | 0.20 | 0.00 | 0.10 | 0.20 | 0.19 | 0.59 |
| JC | 0.30 | 0.00 | 0.10 | 0.20 | 0.00 | 0.00 | 0.10 | 0.15 | 0.48 |
| JD | 0.00 | 0.00 | 0.00 | 0.20 | 0.00 | 0.00 | 0.10 | 0.03 | 0.18 |
| JJ | 0.20 | 0.10 | 0.10 | 0.10 | 0.00 | 0.10 | 0.20 | 0.14 | 0.44 |
| JS | 0.20 | 0.10 | 0.10 | 0.20 | 0.20 | 0.10 | 0.30 | 0.18 | 0.54 |
| JX | 0.10 | 0.00 | 0.10 | 0.20 | 0.20 | 0.10 | 0.30 | 0.13 | 0.42 |
| LC | 0.20 | 0.00 | 0.00 | 0.20 | 0.00 | 0.00 | 0.10 | 0.10 | 0.35 |
| LW | 0.00 | 0.00 | 0.00 | 0.10 | 0.00 | 0.00 | 0.10 | 0.02 | 0.15 |
| MW | 0.10 | 0.00 | 0.00 | 0.20 | 0.20 | 0.10 | 0.10 | 0.09 | 0.33 |
| PT | 0.10 | 0.10 | 0.20 | 0.10 | 0.00 | 0.00 | 0.30 | 0.12 | 0.40 |
| SM | 0.00 | 0.00 | 0.00 | 0.20 | 0.20 | 0.10 | 0.20 | 0.07 | 0.27 |
| SS | 0.10 | 0.10 | 0.10 | 0.20 | 0.00 | 0.10 | 0.20 | 0.11 | 0.38 |
| TW | 0.00 | 0.00 | 0.10 | 0.05 | 0.20 | 0.10 | 0.10 | 0.06 | 0.24 |
| YX | 0.00 | 0.00 | 0.00 | 0.10 | 0.00 | 0.10 | 0.20 | 0.04 | 0.19 |
| ZS | 0.30 | 0.10 | 0.20 | 0.20 | 0.20 | 0.10 | 0.30 | 0.22 | 0.67 |
Breed abbreviations are defined in Table 1.
Current and expected future diversity
The current diversity of the 21 breeds and genetic resources was determined to be 7.72, and the expected diversity of all sets in 500 years is 5.14 ± 1.15. Therefore, an overall decrease of 2.58 (33.43%) is anticipated.
Contributions and marginal diversities of each breed
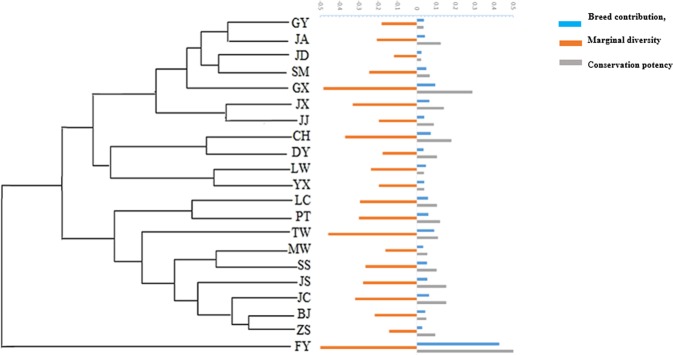
The term “contributions” is defined as the percentage of contribution of each breed to overall diversity. The contributions and marginal diversities of each of the 21 breeds are shown in Table 5 and Fig. 3. The contribution of FY was the largest (Table 3) followed by GX, whereas JD had the smallest contribution. Similarly, in terms of marginal diversity, FY showed the largest (Table 5), followed by GX, whereas JD has the smallest (−0.12, 2.32). Finally, FY had the highest conservation potency, followed by GX, and JD had the lowest.
Table 5.
Marginal diversity of 21 Chinese domestic duck breeds.
| Breed name | Extinction probability | Contribution (%) | Marginal diversity | Conservation potency |
|---|---|---|---|---|
| BJ | 0.23 | 4.25 | −0.2183 | 0.0494 |
| CH | 0.48 | 7.26 | −0.3732 | 0.1789 |
| DY | 0.58 | 3.47 | −0.1783 | 0.1031 |
| FY | 0.28 | 42.61 | −2.1896 | 0.6120 |
| GX | 0.59 | 9.44 | −0.4849 | 0.2876 |
| GY | 0.19 | 3.57 | −0.1834 | 0.0341 |
| JA | 0.59 | 4.07 | −0.2091 | 0.1235 |
| JC | 0.48 | 6.23 | −0.3202 | 0.1527 |
| JD | 0.18 | 2.32 | −0.1192 | 0.0210 |
| JJ | 0.44 | 3.85 | −0.1977 | 0.0872 |
| JS | 0.54 | 5.43 | −0.2790 | 0.1513 |
| JX | 0.42 | 6.49 | −0.3335 | 0.1388 |
| LC | 0.35 | 5.75 | −0.2955 | 0.1043 |
| LW | 0.15 | 4.63 | −0.2381 | 0.0359 |
| MW | 0.33 | 3.18 | −0.1633 | 0.0539 |
| PT | 0.40 | 5.87 | −0.3015 | 0.1201 |
| SM | 0.27 | 4.80 | −0.2469 | 0.0659 |
| SS | 0.38 | 5.22 | −0.2682 | 0.1014 |
| TW | 0.24 | 8.95 | −0.4597 | 0.1099 |
| YX | 0.19 | 3.85 | −0.1977 | 0.0378 |
| ZS | 0.67 | 2.79 | −0.1434 | 0.0955 |
Breed abbreviations are defined in Table 1.
Figure 3.
A maximum-likelihood tree showing the marginal diversity, contribution, and conservation potency of each breed. Note: Breed abbreviations are defined in Table 1.
Discussion
There are numerous domestic duck breeds in China including 27 indigenous breeds, two introduced breeds, and a few developing breeds. However, with the introduction and promotion of cherry valley duck, the number of local duck species in China has dropped sharply, and many species are facing the danger of extinction26. Conservation of genetic diversity plays an important role in sustaining the livestock breeds27. At present, Weitzman marginal diversity method has attracted more and more attention in the research on rational allocation of livestock and poultry resources protection funds, and has become one of the most dynamic theories in the field of livestock genetic resources protection and utilization28,29. There were many researches on animal genetic diversity in the world30–32, but few of them analyze the application of marginal diversity method To our knowledge, Reist-Marti et al.(2003) have estimated extinction probability in livestock breeds11. Bennewitz (2005) estimated the extinction probabilities of 5 German dual-purpose cattle breeds by population viability analysis33. And then (2006) he analyzed 44 North Eurasian cattle breeds using simplified determined extinction probabilities. The results show that the expected loss of diversity within the next 50 years is between 1 and 3% of the actual diversity34. The marginal diversity analysis of goat29, sheep35, cattle36, pigs37 has been completed in China, which provides a reliable data reference for the division of conservation funds. After years of investigation of domestic duck resources and collection of blood samples, this paper analyzed the marginal diversity and extinction probability of local duck breeds in China for the first time.
In this paper, the PCA was used to explain the molecular genetic relationships among the populations and a plane distance map was constructed, reflecting the real genetic information and genetic relationships of the 21 populations. Structure cluster analysis use allelic and genotype data from multiple loci, such as SSR loci, to construct a cluster model. Structure 2.0 program was based on Bayesian probability theory, adopt Markov-Monte Carlo simulation algorithm, and used mixed model when running the program to reveal the unknown population genetic relationship and potential population Structure from all population levels38. The expected number of classification (K value) of the detected group was set at runtime, which can be used to divide all individuals and reflect the genetic structure of the group. It is especially suitable for the study of the genetic structure, the differentiation and migration of individuals. In this paper, the population Structure diagram and the maximum-likelihood tree39 obtained based on Structure 2.0 program were consistent with the results from PCA, verifying the accuracy of population Structure inference.
The calculation for extinction probability considers all factors that might cause change in a breed or genetic resource, making it an accurate and reliable estimation40. However, due to the political and economic situation in China, as well as the distribution of indigenous duck breeds and resources, some factors were not considered in this study, such as natural disasters, reliability of the information source, and development of reasonable storage approaches. For this measurement, seven variables (POS, CHA, DIS, CRO, ORG, SPE, and TRA) were assigned to different weights as major factors. These variables have been proved to be important factors reflecting population diversity. Here, we calculated the current and expected population diversities of a total of 21 Chinese duck breeds, respectively. Importantly, we found that the expected diversity (within 500 years) were 33.43% lower than current diversity.
Some variables for calculating the extinction probability, such as CHA and CRO, only consider the conservation of a single breed or genetic resource, and do not account for the effect of the conserved breed on the genetic diversity of the entire population41. If limited breeding conservation funding is allocated based on extinction probability parameters, it may not be the most beneficial solution for the entire population, especially if that population includes numerous breeds and strains. Instead, the breed with the largest contribution should be given the highest priority42,43. In this study, the largest contributor was FY, followed by GX.
However, breed contribution is not the only consideration for conservation planning efforts, and its calculation does not consider extinction probability. In contrast, marginal diversity considers both contribution and the extinction probability, and can therefore act as a comprehensive measurement of the importance of each breed. According to Weitzman, marginal diversity parameters should be considered the preferred reference during breeding conversation planning44. Conserving the breed with the largest conservation potency is the most effective way to maintain overall genetic diversity. Therefore, the first two breeds prioritized should be FY and GX, followed by CH, JC, JS, JX, JA, PT, TW, LC, DY, SS, ZS, JJ, SM, MW, BJ, YX, LW, GY, and JD.
Based on the marginal diversity parameters, we identified the conservation priority of 21 local duck breeds and genetic resources. FY and GX are the first two breeds that should be protected. The conservation priority in this study can provide a reference for breed conservation planning.
Acknowledgements
We are grateful to Professor Y.J. Mao at Yangzhou University for his participation in and guidance to calculation and analysis of the standard genetic distance and calculation of the marginal diversity parameters. This research work was supported by the National Natural Science Foundation of China (31601931), Jiangsu modern Agricultural Industrial Technology system construction project (JATS[2018]301).
Author Contributions
Q.X. and G.H.C. conceived and designed the experiments; Y.Z. and L.D.W. performed the experiments and wrote the paper; Y.Q.B. analyzed the data; Z.S.W. contributed materials; G.B.C. reviewed and edited the manuscript. All authors read and approved the manuscript.
Data Availability
The data generated and analyzed as a part of this study are included within this article.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Anderson S. Animal genetic resources and sustainable livelihoods. Ecol Econ. 2003;45:331–339. doi: 10.1016/S0921-8009(03)00088-0. [DOI] [Google Scholar]
- 2.Rege JEO, Gibson JP. Animal genetic resources and economic development: issues in relation to economic valuation. Ecol Econ. 2003;45:319–330. doi: 10.1016/S0921-8009(03)00087-9. [DOI] [Google Scholar]
- 3.Lu GB, Wang XH, Chang WK, Zhang H. Analysis of Conservation Status of Chinese Livestock and Poultry Genetic Resources. Acta Ecol Anim Dom. 2014;35:6. [Google Scholar]
- 4.Feng W, Ma Y, Chen Y, Fu B. General viewson status of domestic animal genetic resources in china. Acta Veterinaria et Zootechnica Sinica. 1997;28:4. [Google Scholar]
- 5.Resources, N. C. f. A. G. Report on Domestic Animal Genetic Resources in China. (China Agriculture Press, 2012).
- 6.Ma Y, Xu G, Wang D, Liu H. Study onDynamic Information of Animal Genetic Resources in China. Scientia Agricultura Sinica. 2002;35:4. [Google Scholar]
- 7.Liu Fang, Long Huaping, Yang Yuze, He Zhongwei. Proceedings of 2013 World Agricultural Outlook Conference. Berlin, Heidelberg: Springer Berlin Heidelberg; 2014. Temporal Changes and the Influencing Factor of China Importing Genetic Resources of Livestock and Poultry; pp. 331–339. [Google Scholar]
- 8.Ma Y, Wu C. Evaluation of Threatening Degree of the Animal Genetic Resources in China. Ecol Dom Anim. 2001;22:6. [Google Scholar]
- 9.Li J, Li M, Wang Y. The problem and protection strategy of the genetic resources of livestock and poultry in China. J. NU Nationalities.(Natura1 Science) 2013;34:5. doi: 10.14084/j.cnki.cn62-1188/n.2013.04.007. [DOI] [Google Scholar]
- 10.Simianer H, Marti SB, Gibson J, Hanotte O, Rege JEO. An approach to the optimal allocation of conservation funds to minimize loss of genetic diversity between livestock breeds. Ecol Econ. 2003;45:377–392. doi: 10.1016/S0921-8009(03)00092-2. [DOI] [Google Scholar]
- 11.Reist-Marti SB, Simianer H, Gibson J, Hanotte O, Rege JEO. Weitzman’s approach and conservation of breed diversity: an application to African cattle breeds. Conserv Biol. 2003;17:1299–1311. doi: 10.1046/j.1523-1739.2003.01587.x. [DOI] [Google Scholar]
- 12.Knapp SM, Russell RE, Swihart RK. Setting priorities for conservation: the influence of uncertainty on species rankings of Indiana mammals. Biol Conserv. 2003;111:223–234. doi: 10.1016/S0006-3207(02)00278-1. [DOI] [Google Scholar]
- 13.Weitzman, M. What to preserve? An application of diversity theory to crane conservation. Quar J. Eco. 26 (1993).
- 14.Zoller S, Boskova V, Anisimova M. Maximum-Likelihood Tree Estimation Using Codon Substitution Models with Multiple Partitions. Mol Biol Evol. 2015;32:2208–2216. doi: 10.1093/molbev/msv097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Laval G, et al. Genetic diversity of eleven European pig breeds. Genet Sel Evol. 2000;32:187–203. doi: 10.1186/1297-9686-32-2-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Canon J, et al. Genetic diversity measures of local European beef cattle breeds for conservation purposes. Genet Sel Evol. 2001;33:311–332. doi: 10.1186/1297-9686-33-3-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang YH, et al. A genetic and cytogenetic map for the duck (Anas platyrhynchos) Genetics. 2006;173:287–296. doi: 10.1534/genetics.105.053256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Y, et al. Analysis of the Genetic Diversity and Origin of Some Chinese Domestic Duck Breeds. J. Integr Agr. 2014;13:849–857. doi: 10.1016/S2095-3119(13)60447-5. [DOI] [Google Scholar]
- 19.Berardi G, Toscanini U, Raimondi E. STR data for PowerPlex (R) 16 system from Buenos Aires population, Argentina. Forensic Sci Int. 2003;134:222–224. doi: 10.1016/S0379-0738(03)00141-5. [DOI] [PubMed] [Google Scholar]
- 20.Gauch HG, Jr., Qian S, Piepho HP, Zhou L, Chen R. Consequences of PCA graphs, SNP codings, and PCA variants for elucidating population structure. PloS one. 2019;14:e0218306. doi: 10.1371/journal.pone.0218306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li., P. Marginal diversity and conservation priority of the local goat populations in Shandong phD thesis, Shandong Agricultural University, (2008).
- 22.Ollivier L, et al. An assessment of European pig diversity using molecular markers: Partitioning of diversity among breeds. Conserv Genet. 2005;6:729–741. doi: 10.1007/s10592-005-9032-6. [DOI] [Google Scholar]
- 23.Pham MH, et al. Genetic diversity of Vietnamese domestic chicken populations as decision-making support for conservation strategies. Anim Genet. 2013;44:509–521. doi: 10.1111/age.12045. [DOI] [PubMed] [Google Scholar]
- 24.Liu JB, et al. Analysis of geographic and pairwise distances among sheep populations. Genet Mo Res. 2014;13:4177–4186. doi: 10.4238/2014.June.9.4. [DOI] [PubMed] [Google Scholar]
- 25.Zhang Y, et al. Genetic diversity of endangered Polyporus umbellatus from China assessed using a sequence-related amplified polymorphism technique. Genet Mol Res. 2012;11:4121–4129. doi: 10.4238/2012.December.3.1. [DOI] [PubMed] [Google Scholar]
- 26.He DQ, et al. A homogenous nature of native Chinese duck matrilineal pool. BMC Evol Biol. 2008;8:298. doi: 10.1186/1471-2148-8-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.(Organization), F. F. a. A. Coping with climate changethe roles of genetic resources for food and agriculture, (Italy: FAO, 2015).
- 28.Zerabruk M, Bennewitz J, Kantanen J, Olsaker I, Vangen O. Analysis of genetic diversity and conservation priorities for six north Ethiopian cattle breeds. J. Anim Breed Genet. 2007;124:236–241. doi: 10.1111/j.1439-0388.2007.00660.x. [DOI] [PubMed] [Google Scholar]
- 29.Ling YH, et al. Assessment of Genetic Diversity among the Twelve Chinese Meat Goat Breeds Using Weitzman Approach. J. Anim Plant Sci. 2014;24:986–990. [Google Scholar]
- 30.Souza CA, et al. Genetic diversity and assessment of 23 microsatellite markers for parentage testing of Santa Ines hair sheep in Brazil. Genet Mol Res. 2012;11:1217–1229. doi: 10.4238/2012.May.8.4. [DOI] [PubMed] [Google Scholar]
- 31.Curkovic M, et al. The genetic diversity and structure of 18 sheep breeds exposed to isolation and selection. J. Anim Breed Genet. 2016;133:71–80. doi: 10.1111/jbg.12160. [DOI] [PubMed] [Google Scholar]
- 32.Hariyono DNH, et al. Genetic diversity and phylogenetic relationship analyzed by microsatellite markers in eight Indonesian local duck populations. Asian-Australas J Anim Sci. 2019;32:31–37. doi: 10.5713/ajas.18.0055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bennewitz J, Meuwissen THE. Estimation of extinction probabilities of five German cattle breeds by population viability analysis. J. Dairy Sci. 2005;88:2949–2961. doi: 10.3168/jds.S0022-0302(05)72975-1. [DOI] [PubMed] [Google Scholar]
- 34.Bennewitz J, et al. Estimation of breed contributions to present and future genetic diversity of 44 North Eurasian cattle breeds using core set diversity measures. Genet Sel Evol. 2006;38:201–220. doi: 10.1051/gse:2005036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yuehui M. Marginal diversity and its application to Chinese sheep breeds conservation. China Biod Sci. 2005;13:5. [Google Scholar]
- 36.Yongjiang M, Hong C, Zhangping Y, Pinent T, Simianer H. The application of marginal diversity on yellow cattle breeds conservation in China. China Cattle Sci. 2006;32:8. [Google Scholar]
- 37.Qianjun Z, Ma Y. Optimal allocation of funds for conservation of Chinese pig breeds using marginal diversity estimates. China Biod Sci. 2007;15:7. doi: 10.1360/biodiv.060166. [DOI] [Google Scholar]
- 38.Moore BR, Donoghue MJ. A Bayesian approach for evaluating the impact of historical events on rates of diversification. Proc Natl Acad Sci USA. 2009;106:4307–4312. doi: 10.1073/pnas.0807230106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marshall TC, Slate J, Kruuk LE, Pemberton JM. Statistical confidence for likelihoodbased paternity inference in natural populations. Mol Ecol. 1998;7:639–55. doi: 10.1046/j.1365-294x.1998.00374.x. [DOI] [PubMed] [Google Scholar]
- 40.Bokma F. Bayesian estimation of speciation and extinction probabilities from (in)complete phylogenies. Evolution. 2008;62:2441–2445. doi: 10.1111/j.1558-5646.2008.00455.x. [DOI] [PubMed] [Google Scholar]
- 41.Garcia D, Corral N, Canon J. Combining inter- and intrapopulation information with the Weitzman approach to diversity conservation. J. Heredity. 2005;96:704–712. doi: 10.1093/jhered/esi103. [DOI] [PubMed] [Google Scholar]
- 42.Barker JSF. Animal breeding and conservation genetics. Conserv Genet. 1994;68:15. doi: 10.1007/978-3-0348-8510-2_30. [DOI] [PubMed] [Google Scholar]
- 43.Zhao, Q. Origin,genetic diversity and eonservation of Chinese sheep populaions Ph.D thesis, Chinese Academy of Agricultural Science, (2007).
- 44.Weitzman M. On diversity. The Quarterly Journal of Economics. 1992;107:43. doi: 10.2307/2118476. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data generated and analyzed as a part of this study are included within this article.