Abstract
It was reported that oral administration of Bacillus favored the growth of Lactobacillus in the intestinal tract. Here, this phenomenon was confirmed by co-cultivation of Bacillus subtilis 168 and Lactobacillus plantarum SDMCC050204-pL157 in vitro. To explain the possible molecular mechanisms, B. subtilis 168 cells were incubated in simulated intestinal fluid at 37 °C for 24 h, and up to 90% of cells autolysed in the presence of bile salts. Addition of the autolysate to medium inoculated with Lb. plantarum SDMCC050204 decreased the concentration of H2O2 in the culture, alleviated DNA damage and increased the survival of Lb. plantarum, as like the results of exogenous heme addition. These results suggested that the autolysate provided heme, which activated the heme-dependent catalase KatA in Lb. plantarum SDMCC050204. HPLC confirmed the presence of heme in the autolysate. Disruption of the Lb. plantarum SDMCC050204 katA gene abolished the protective effect of the B. subtilis 168 autolysate against H2O2 stress. We thus hypothesized that the beneficial effect of Bacillus toward Lactobacillus was established through activation of the heme-dependent catalase and remission of the damage of reactive oxygen species against Lactobacillus. This study raised new crosstalk between the two frequently-used probiotics, highlighting heme-dependent catalase as the key mediator.
Subject terms: Symbiosis, Applied microbiology
Introduction
Numerous microorganisms that inhabit the human gastrointestinal tract form a dynamic and diverse community, referred to as the gut microbiota1. An ever-increasing number of studies have pointed out the relationship between gut microbiota and body health, showing that these microbial populations impact an array of physiological functions in the gastrointestinal tract, including digestion, immune response and disease prevention2–4. Bacillus and Lactobacillus strains are bacterial members of the gut microbiota and are widely used as probiotics1,5–7. In China, B. subtilis, B. licheniformis and B. cereus are ingested by adults, infants and newborns to relieve diarrhea and build health intestinal microbiota. Bacillus strains are considered to efficiently antagonize pathogenic bacteria, while promote the growth of Lactobacillus to balance the microbiota8, achieving therapeutic purposes. The question is then, how does Bacillus crosstalk with Lactobacillus?
Previously, it was reported that co-cultivation with Bacillus strains could facilitate the growth of Lactobacillus in animal intestinal tracts9–12. This beneficial effect may be interpreted by the “Biological Oxygen-Capturing Theory”. Briefly, according to this theory, Bacillus spores are orally administered because of their resistance to the gastrointestinal tract. When arriving in the small intestine, where is rich in nutrients, the spores are induced to germinate, proliferate, and then resporulate13. This process consumes oxygen in the intestinal tract, and generates an anaerobic environment for the proliferation of Lactobacillus14. Moreover, Bacillus strains produce bioactive molecules including hydrolytic enzymes (amylase and protease), antioxidative enzymes (catalase and superoxide dismutase) and surface proteins that could favor the growth and viability of Lactobacillus15,16.
Normally, lactic acid bacteria are sensitive to reactive oxygen species (ROS) because they do not equip with perfect antioxidant systems found in aerobic microorganisms17. In fact, several Lactobacillus strains, including Lb. plantarum, Lb. brevis and Lb. casei, have the heme-dependent catalase coding gene in their genomes18–20. Catalase catalyzes the decomposition of the ROS hydrogen peroxide. However, owing to their lack of the heme biosynthesis ability, these Lactobacillus strains are defined as catalase-negative strains. When these Lactobacillus cells are supplemented with exogenous heme, the heme-dependent catalase in these cells could be activated, which increase survival and biomass of the Lactobacillus strains20. However, whether Bacillus is involved in this process in the intestinal tract is yet to be uncovered besides the “Biological Oxygen-Capturing Theory”.
In this study, we performed co-cultivation of B. subtilis 168 and Lb. plantarum SDMCC050204-pL157, analyzed the presence of heme in autolysate of B. subtilis 168, and confirmed the role of the autolysate in activation of catalase (KatA) in Lb.plantarum SDMCC050204. The aim of this study was to propose a new crosstalk model between Bacillus and Lactobacillus strains in the intestinal tract after oral administration of Bacillus.
Materials and Methods
Bacterial strains, culture media, and growth conditions
Bacterial strains and plasmids used in this study are listed in Table 1. B. subtilis 168 and Escherichia coli DH5α were grown aerobically in Luria-Bertani (LB) medium at 37 °C. Lb. plantarum strains were grown in de Man, Rogosa and Sharpe (MRS) broth containing 0.5% glucose at 37 °C in two different conditions, either (i) static cultivation in 100-mL Erlenmeyer flasks containing 50 mL medium, or (ii) aerated cultivation in 300-mL Erlenmeyer flasks containing 50 mL medium with agitation on a rotary shaker at 200 rpm. When appropriate, the following antibiotics were added to the medium: ampicillin (100 μg/mL for E. coli), chloramphenicol (5 μg/mL for Lb. plantarum) and erythromycin (5 μg/mL for Lb. plantarum). When required, 20 μM heme (Sigma, USA) was added to medium. Cell turbidity was monitored by the optical density at 600 nm (OD600).
Table 1.
Bacterial strains and plasmids used in this study.
| Strain, plasmid or primer | Feature(s) or sequence | Reference |
|---|---|---|
| Strain | ||
| B. subtilis 168 | Standard strain | CGMCC |
| Lb. plantarum SDMCC050204 | Wild-type strain isolated from Chinese artisanal cheese; Whole genome sequencing (unpublished data) | Our laboratory |
| Lb. plantarum SDMCC050276 | Wild-type strain isolated from silage | Our laboratory |
| Lb. plantarum SDMCC050277 | Wild-type strain isolated from silage | Our laboratory |
| Lb. plantarum SDMCC050204-pL157 | Lb. plantarum SDMCC050204 containing the plasmid pL157; Cmr | This work |
| Lb. plantarum SDMCC050276-pL157 | Lb. plantarum SDMCC050276 containing the plasmid pL157; Cmr | This work |
| Lb. plantarum SDMCC050277-pL157 | Lb. plantarum SDMCC050277 containing the plasmid pL157; Cmr | This work |
| Lb. plantarum SDMCC050204ΔkatA | Lb. plantarum SDMCC050204 with the heme-dependent catalase gene (katA) inactivation in the genome | This work |
| E. coli DH5α | Cloning host | Novagen |
| Plasmid | ||
| pL157 | Cmr; shuttle vector in lactobacilli and E. coli | 21 |
| pUC-erm | Ampr, Ermr; the erythromycin resistance gene from the vector pMG36e was cloned into the BamHI site of the vector pUC19 | 27 |
Co-cultivation of Lb. plantarum SDMCC050204-pL157 with B. subtilis 168
To easily distinguish and enumerate Lb. plantarum easily in co-culture with B. subtilis 168 (which is chloramphenicol sensitive), Lb. plantarum SDMCC050204 was conferred with a chloramphenicol resistance phenotype by transformation of plasmid pL157 by electroporation21,22, generating Lb. plantarum SDMCC050204-pL157. Since previous report showed the physical stability of pL157 in host cells without selective pressure21, chloramphenicol was not added into the co-culture of Lb. plantarum SDMCC050204-pL157 and B. subtilis 168.
Lb. plantarum SDMCC050204-pL157 and B. subtilis 168 were individually cultivated to reach early stationary phase. The cells were collected by centrifugation at 5,000 × g for 5 min, washed three times with sterile saline solution and resuspended in MRS broth. Then, 5.6 × 108 CFU/mL of Lb. plantarum SDMCC050204-pL157 and 6.0 × 107 CFU/mL of B. subtilis 168 were inoculated into MRS broth, and cultivated aerobically for 96 h. Viable cell counts of Lb. plantarum SDMCC050204-pL157 were determined on MRS agar supplemented with chloramphenicol.
Autolysis of B. subtilis in simulated intestinal fluid
B. subtilis 168 spores (2.0 × 107 CFU/mL) were incubated in LB medium for 9 h to reach late logarithmic phase to early stationary phase. The vegetative cells were collected, washed and resuspended in simulated intestinal fluid (SIF) at an OD600 of 17.00 ± 0.46. The bacterial suspensions were incubated at 37 °C with agitation (200 rpm) to simulate peristalsis, and aliquots were taken to determine the OD600 at 0, 12 and 24 h. The SIF contained 0.5% NaCl, 1 g/L pancreatin (Sigma, USA), and 0%, 0.05%, 0.1% or 0.3% pig bile salts (Sigma, USA), pH 8.023.
Cultivation of Lb. plantarum with the autolysate of B. subtilis 168
To prepare autolysate of B. subtilis 168, vegetative cells were resuspended in 0.1% NaCl to one-tenth of the initial culture volume and stored at 4 °C for 7 days. Then, the intact cells and cell debris were removed by centrifugation at 13,000 × g for 20 min. The resultant supernatant was filtered with a 0.22 μm membrane and then heated at 100 °C for 15 min to inactivate any proteinic enzymes to obtain the autolysate. Culture medium was prepared by mixing an equal volume of two-fold concentrated MRS broth with the autolysate, or with 0.1% NaCl as a control. Then, 5.6 × 108 CFU/mL Lb. plantarum SDMCC050204 were inoculated into the culture medium and incubated aerobically for 60 h.
Analysis of catalase activity and H2O2 concentration
Cell pellets from 2 mL of Lb. plantarum SDMCC050204 culture were collected, washed three times with sterile saline solution, and resuspended in 50 μL saline solution. The cell suspensions were mixed with 20 μL of 30% H2O2 solution, and the air bubble formation was determined24. H2O2 concentrations in the culture were measured using a H2O2 Quantified Analysis Kit (Sangon Biotech, China) as stated by standard procedures.
Detection of heme in the autolysate of B. subtilis
Heme was extracted from the autolysate of B. subtilis 168 according to the Weinstein method with some modifications25. Specifically, 100 mL of autolysate was concentrated to 10 mL by lyophilization (Thermo Savant, USA). Then, 30 mL of 90% aqueous acetone containing 5% HCl (v/v) were added to the autolysate. The mixture was vortexed for 10 min at room temperature, and then centrifuged at 13,000 × g for 10 min. The heme-containing supernatant was recovered, while the pellets were extracted with another 10 mL of acidic acetone. After centrifugation, the two fractions of supernatant were combined followed by evaporation in a vacuum rotary evaporator (Thermo Savant, USA) to remove the organic phase and a vacuum freeze-dryer to remove the water phase. The dry residue was dissolved in 3 mL of distilled water and the pH was adjusted to 12.0 for the transformation of heme to soluble hematin. The solution was filtered with a 0.22 μm membrane for the further detection.
Hematin was detected by high-performance liquid chromatography (HPLC; Shimadzu, Japan) using an XBridge BEH300 C18 reverse phase column (150 × 4.6 mm; Waters, USA) with a flow rate of 0.6 mL/min. The column was equilibrated with solvent A. Separation of the hematin was effected with a gradient of 20% to 70% solvent B over 40 min. The column effluent was monitored by photo-diode array detection at 398 nm. Solvent A was 0.1% trifluoroacetic acid (TFA) (v/v) in water; solvent B was 0.1% TFA (v/v) in acetonitrile. Hematin sample as prepared above (50 μL) was injected into the column for analysis. 10 μL of 30 μg/mL hematin (Sigma, USA) was used as the standard sample.
Disruption of the heme-dependent catalase gene (katA) in Lb. plantarum SDMCC050204
Molecular cloning techniques were performed essentially as described previously26. Taq polymerase, restriction enzymes and T4 DNA ligase were used according to the manufacturer’s instructions (TaKaRa, Japan).
Disruption of the katA gene was carried out using a single-crossover integration strategy27. A DNA fragment containing the katAk was PCR amplified from Lb. plantarum SDMCC050204 genomic DNA with primers laX-1 (5′-ACCTCTAGAGACGTGCGCGGTTTT-3′; the underlined bases indicate an XbaI site) and laX-2 (5′-CAGAAGCTTAGGCTCGTAGTTGACC-3′; the underlined bases indicate a HindIII site). The PCR products were digested with XbaI/HindIII and then ligated into the compatible end of pUC-erm, yielding pUC-erm-kat. pUC-erm-kat (200 ng) was introduced into competent Lb. plantarum SDMCC050204 cells by electroporation. Transformants were selected on MRS agar containing erythromycin at a final concentration of 5 μg/mL, generating the mutant Lb. plantarum SDMCC050204ΔkatA.
Evaluation of DNA damage due to H2O2 stress
Evaluation of DNA damage was conducted according to the methods of Rezaïki with some modifications28. Briefly, Lb. plantarum SDMCC050204 and Lb. plantarum SDMCC050204ΔkatA were incubated with or without the autolysate of B. subtilis 168 or heme for 12, 36 and 60 h. After cultivation, the genomic DNA was extracted using a TIANamp Bacteria DNA Kit (TIANGEN, China) and electrophoresed on Tris-acetate-EDTA agarose (1.0%) gels to compare the extent of DNA degradation in each sample.
Statistical analysis
Statistical analysis was performed using data from three technical replicates by unpaired two-tailed Student’s t-test. P values of <0.05 were considered statistically significant; P values of <0.01 were considered statistically highly significant.
Results
Detection of survival of Lb. plantarum SDMCC050204-pL157 co-cultured with B. subtilis 168 in vitro
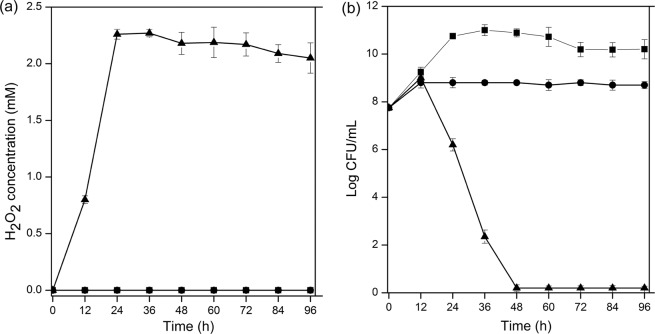
It was reported that oral administration of Bacillus favored the growth of Lactobacillus in the intestinal tract29,30. To confirm this result in vitro, Lb. plantarum SDMCC050204-pL157 was incubated with or without B. subtilis 168. The H2O2 concentrations and cell viability of Lb. plantarum SDMCC050204-pL157 were detected. As shown in Fig. 1a,b, when Lb. plantarum SDMCC050204-pL157 was cultivated alone under aerobic conditions for 96 h, about 2 mM H2O2 was detected in the cultures, and the viable cell numbers showed a significant decrease, from 5.62 ± 0.2 × 107 CFU/mL at 0 h to 0 CFU/mL at 96 h. In contrast, in the static culture, very little H2O2 was detected, and the viable cell counts were 1.0 ± 0.4 × 109 CFU/mL at 96 h, suggesting that Lb. plantarum SDMCC050204-pL157 suffered from H2O2 stress under aerobic conditions, causing cellular damage.
Figure 1.
H2O2 accumulation (a) and viable cell counts (b) of Lb. plantarum SDMCC050204-pL157 when statically cultivated (■), and aerobically cultivated with (●) or without (▲) B. subtilis 168. In all panels, the values are means ± standard deviations of three independent experiments. The curves with symbols ■ and ● overlap in panel (a).
To test whether B. subtilis could decrease the H2O2 level in aerobic culture of Lb. plantarum, Lb. plantarum SDMCC050204-pL157 as well as two other Lb. plantarum strains SDMCC050276-pL157 and SDMCC050277-pL157 were respectively co-cultured with B. subtilis 168. The results showed that the H2O2 was not detectable in the co-cultures (Fig. 1a), as that of the static culture, and the viable cell counts of SDMCC050204-pL157 was 8.4 ± 0.1 × 108 CFU/mL after 96 h incubation, close to that in the static culture (Fig. 1b). Similar phenomena were observed for co-cultivation of B. subtilis 168 with strains SDMCC050276-pL157 and SDMCC050277-pL157 (data not shown). These results confirmed that B. subtilis 168 could provide bioactive molecules to protect Lb. plantarum strains from H2O2 stress.
Autolysis of B. subtilis 168 in SIF
B. subtilis is prone to autolysis due to environmental stressors or the regulated processes that occur at different stages of the cell life31,32. To find out the bioactive molecules, B. subtilis 168 cells were suspended in SIF, and the OD600 was determined at 12 and 24 h, respectively. As shown in Fig. 2, autolysis of B. subtilis 168 cells was observed, and lysis extent reached up to 60% of the cells after 24 h. Moreover, the autolysis was significantly boosted up to 90% of the cells in the presence of 0.05% bile salts, revealing that B. subtilis 168 cells could autolyse in the intestinal tract.
Figure 2.
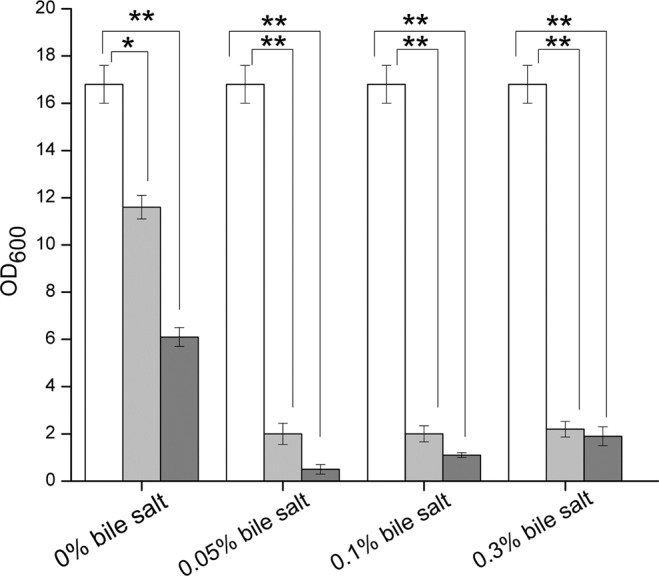
Autolysis of B. subtilis 168 cells in simulated intestinal fluid containing 0%, 0.05%, 0.1% and 0.3% pig bile salts determined by the decline of OD600 of cell suspensions. White column, 0 h incubation; light grey column, 12 h incubation; dark grey column, 24 h incubation. The values are means ± standard deviations of three independent experiments. *P < 0.05; **P < 0.01.
Activation of heme-dependent catalase by the autolysate
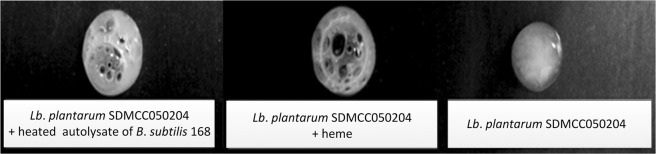
B. subtilis cells could autolyse, accompanied by the release of intracellular solutes. Here, solution containing the intracellular solutes of B. subtilis 168 without intact cells or cell debris was termed the autolysate. To identify the role(s) of the autolysate in the elimination of H2O2 from Lb. plantarum culture, the germfree filtering lysate of B. subtilis 168 cells was heated at 100 °C for 15 min to inactivate any proteinic enzymes, including catalase, peroxidase, superoxide dismutase, and so on. Then, Lb. plantarum SDMCC050204 was incubated in MRS medium supplemented with the heated autolysate for 24 h. As a result, H2O2 was undetectable in the cultures (data not shown), while air bubble formation was observed from cells of Lb. plantarum SDMCC050204 when treated with H2O2 (Fig. 3), suggesting that the heated autolysate contributed biomolecules that activated catalase activity in Lb. plantarum SDMCC050204. The same phenomenon was obtained when addition of exogenous heme to the medium instead of the heated autolysate. However, nor did these results when Lb. plantarum SDMCC050204 was cultivated in the medium without the heated autolysate or heme (Fig. 3). Thus, we concluded that the autolysate of B. subtilis 168 supplied heme, which in turn resulted in activity of the heme-dependent catalase KatA in Lb. plantarum SDMCC050204.
Figure 3.
Qualitative analysis of catalase activity of Lb. plantarum SDMCC050204 in medium supplemented with the heated autolysate of B. subtilis 168, heme, or without supplementation. Catalase activity was indicated by bubble formation due to liberation of O2.
Detection of heme in the autolysate of B. subtilis 168
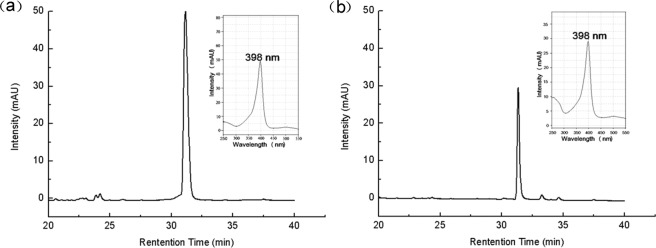
To confirm the presence of heme in the autolysate, HPLC analysis was performed. As shown in Fig. 4, a specific absorption peak (retention time = 31.36 min) at 398 nm appeared, close to that for a standard hematin sample (retention time = 31.41 min), indicating that heme could be quantitatively provided by the autolysis of B. subitlis 168 cells.
Figure 4.
Detection of hematin in the autolysate of B. subtilis 168 by HPLC analysis. (a) Standard sample of hematin. (b) Sample extracted from the autolysate of B. subtilis 168. In the inset panels, the Soret band region of the spectrum of hematin is shown. Hematin is a soluble transforming product of heme.
Functional analysis of heme-dependent catalase in Lb. plantarum SDMCC050204
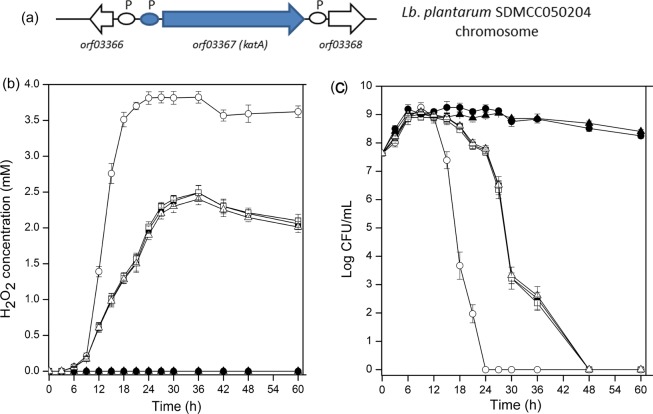
Genomic sequencing indicated that a heme-dependent catalase coding gene, katA, is present in the genome of Lb. plantarum SDMCC050204 (unpublished data). Figure 5a shows the genetic organization of katA in Lb. plantarum SDMCC050204. To determine whether the positive response of H2O2 decomposition in cultures of Lb. plantarum supplemented with heme or B. subtilis 168 autolysate was catalyzed by KatA, the katA gene was disrupted. Cells of the mutant SDMCC050204ΔkatA could not degrade H2O2 to yield detectable air bubbles, even in the presence of either the autolysate of B. subtilis 168 or heme (data not shown).
Figure 5.
The genetic organization of katA in Lb. plantarum SDMCC050204 (a), H2O2 accumulation (b) and viable cell counts (c) of Lb. plantarum SDMCC050204 (solid symbols) or Lb. plantarum SDMCC050204ΔkatA mutant (open symbols) when cultivated aerobically alone (■, □), with the heated autolysate of B. subtilis 168 (●, ○), or with heme (▲, △). In panel (a), P indicates putative promoter. In panels (b,c) the values are means ± standard deviations of three independent experiments. The curves with symbols ■, □ and △ partially overlap.
The H2O2 concentrations in cultures of mutant SDMCC050204ΔkatA and the wild type SDMCC050204 were compared after supplementation with the autolysate or heme. As stated above, about 2 mM H2O2 was detected in the culture of strain SDMCC050204 in MRS medium, the same was observed in culture of the mutant SDMCC050204ΔkatA. When the autolysate was added, no H2O2 was detected in culture of strain SDMCC050204, while as much as 3.6 mM H2O2 was present in culture of the mutant SDMCC050204ΔkatA (Fig. 5b). Heme addition resulted in similar comparison of H2O2 levels in the cultures of the wild type and the mutant. Moreover, the mutant strain exhibited an earlier and sharper decline in viable cell numbers (Fig. 5c). These results indicated that SDMCC050204ΔkatA lost the ability to produce active catalase even in the presence of the B. subtilis autolysate or heme, highlighting the heme-dependent catalase as the key component mediating interaction between the Bacillus sp. and the Lactobacillus. No significant differences were observed between the addition of the autolysate and exogenous heme in respect to H2O2 concentration or cell viability (Fig. 5b,c).
Detection of DNA damage under oxidative stress
ROS have long been viewed as dangerous, highly-reactive molecules that cause cellular damage17,28. To examine DNA integrity, chromosomal DNA was extracted from cells of Lb. plantarum SDMCC050204 and SDMCC050204ΔkatA cultivated aerobically for 12, 36 and 60 h (Fig. 6). DNA damage was obviously alleviated in strain SDMCC050204 by the addition of heme or the B. subtilis 168 autolysate, but not in strain SDMCC050204ΔkatA. These results demonstrated that the activation of the heme-dependent catalase KatA efficiently protected Lb. plantarum strains against oxidative stress, and consequently helped maintain the DNA integrity.
Figure 6.

Evaluation of DNA damage of Lb. plantarum SDMCC050204 and the SDMCC050204ΔkatA mutant from aeration, aeration in the presence of heme, and aeration in the presence of the autolysate of B. subtilis. Chromosomal DNA was prepared from cells taken at 12, 36 and 60 h, and separated by electrophoresis on a 1% agarose gel to observe the extent of DNA degradation. Distance of migration reflects the degree of degradation, as indicated.
Discussion
It is generally considered that Bacillus strains are beneficial for the survival and growth of Lactobacillus in animal intestinal tracts on the basis of the “Biological Oxygen-Capturing Theory9–12,33”. According to this theory, the anaerobic environment generated by the growth of Bacillus strains plays major role in promotion of the growth of Lactobacillus14. Others have stated that catalase, subtilisin and surface proteins produced by Bacillus helped Lactobacillus inhabit the same niche15,16. In this study, we found that B. subtilis was prone to autolysis, particularly in the presence of bile salts, which offered the possibility to release heme that could be beneficial to other microbiota, including the core Lactobacillus in the intestinal tract. Our work focused on exploring the key factors linking the B. subtilis autolysate and Lb. plantarum, and the critical roles of heme and KatA, a heme-dependent catalase, were consequently demonstrated. This work thus proposed a novel crosstalk model between Bacillus and Lactobacillus in the intestinal tract, which would shed new light on the complicated interactions of different bacterial species in the gut microbiota.
ROS, including , H2O2 and , are generated as by-products of normal human cellular metabolic activities34. Alcohol, chronic infections and inflammatory disorders stimulate the production of ROS, and thus the intestinal tract is a key source of ROS34,35. Excessive accumulation of ROS results in oxidative stress, leading to intracellular biological macromolecular damage34. Meanwhile, the metabolism of Lb. plantarum is impacted by oxidative stress. H2O2 could induce the activity of pyruvate oxidase (POX), which converts pyruvate into acetate, accompanied by the production of extra H2O236. Here, to imitate the ROS pool in the animal intestinal environment, aerobic cultivation was carried out to subject Lb. plantarum cells to oxidative stress, as 2 mM H2O2 was detected in the aerobic culture of Lb. plantarum SDMCC050204-pL157. We also found that Lb. plantarum was sensitive to oxidative damage from H2O2, agreeing with previous reports37. H2O2 damage to the Lb. plantarum cells mainly resulted from the lack of effective antioxidant systems. Although ROS erasers, including catalase, superoxide dismutase and NADH peroxidase, can be produced in some Lactobacillus strains by genetic and physiological analysis, the enzymatic activities are low38,39. In particular, catalase is commonly inactive in Lactobacillus, because the main cofactor heme is absent18,40–42. Therefore, mechanisms to cope with oxidative stress from H2O2 in the intestinal tract are of great importance for Lactobacillus. Our results here demonstrated that co-cultivation with B. subtilis significantly decreased the level of H2O2 and enhanced the survival of Lb. plantarum cells (Fig. 1).
Bacillus are complex organisms that exist as vegetative cells or metabolically inert spores or as part of a multicellular biofilm when encountering extreme environments43. When the environment is deficient in nutrients, rich in growth inhibitors and the pH or osmotic pressure is unfavorable, the vegetative cells of Bacillus are prone to autolysis, releasing resistant spores31,32. The above extreme conditions, to a large extent, can be provided by the gastrointestinal tract. Our results confirmed that most B. subtilis 168 vegetative cells spontaneously lysed in SIF (Fig. 2). The high-level autolysis released not only spores but also intercellular substances, making them candidate bioactive molecules to help Lb. plantarum resist H2O2 stress.
When exogenous heme or the autolysate of B. subtilis 168 devoid of catalase was added to the culture medium of Lb. plantarum SDMCC050204, the grown cell suspensions showed air bubble formation in the presence of H2O2. This result provided an indication of the presence of active catalase in cell suspensions of Lb. plantarum25. According to the genomic sequence, a heme-dependent catalase coding gene (katA) was in the genome of Lb. plantarum SDMCC050204. Thus, one hypothesis was that the autolysate of B. subtilis 168 offered heme to Lb. plantarum SDMCC050204 for activation of the heme-dependent catalase KatA. Then, heme was confirmed to be present in the autolysate of B. subtilis 168 (Fig. 4). Moreover, after disruption of the katA gene in the Lb. plantarum SDMCC050204 genome, the autolysate could not help Lb. plantarum SDMCC050204 to decrease the H2O2 concentration, avoid DNA damage and survive H2O2 stress (Figs 5 and 6). Thus, KatA and the heme provided by the autolysis of B. subtilis, were identified as important elements involved in the communication between Lb. plantarum and B. subtilis. The observation that strain SDMCC050204ΔkatA exhibited an earlier and sharper decline in viable cells than the wild type suggested that there were also other substances in the autolysate that promoted the growth of Lb. plantarum (Fig. 5c). In keeping with this speculation, small peaks in addition to the major one (hematin) were visible during HPLC analysis of the autolysate of B. subtilis 168. Our future work will focus on characterization of these unknown bioactive compounds produced by.
In conclusion, this study explored the molecular mechanisms by which Lb. plantarum benefits from co-culture with B. subtilis. It is suggested that the heme-dependent catalase KatA was activated, and this change significantly improved the survival of Lb. plantarum under H2O2 stress. Heme and KatA were the key linkage, whose functions were highlighted during the interaction of the two species. Furthermore, a crosstalk model in the intestinal tract was proposed: the life cycle of Bacillus strains led to the release of sufficient heme into the environment; the activity of Lactobacillus catalase was stimulated by the heme; H2O2 was degraded and oxidative stress was relieved; thus, the survival of Lactobacillus was promoted. Our finding will promote better and rational use of the two probiotics.
Acknowledgements
This research was funded by National Key Research and Development Program of China (2017YFD0400301) and the Public Service Sectors (Agriculture) Special and Scientific Research Projects (201503134).
Author Contributions
J.K., L.Z. and T.-T.G. contributed to developing the study concept, design, and supervision. T.Y., J.K., L.Z., X.-Y.G. and M.-Y.W. conducted the experiments. J.K., L.Z. and T.-T.G. contributed to the writing and reviewing of the manuscript. T.-T.G. did the final check and submitted the manuscript. All the authors approved the final manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Kaoutari AEI, Armougom F, Gordon JI, Raoult D, Henrissat B. The abundance and variety of carbohydrate-active enzymes in the human gut microbiota. Nat. Rev. Microbiol. 2013;11:497–504. doi: 10.1038/nrmicro3050. [DOI] [PubMed] [Google Scholar]
- 2.Krishnan S, Alden N, Lee K. Pathways and functions of gut microbiota metabolism impacting host physiology. Curr. Opin. Biotech. 2015;36:137–145. doi: 10.1016/j.copbio.2015.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ni J, et al. A role for bacterial urease in gut dysbiosis and Crohn’s disease. Sci. Transl. Med. 2017;9(416):eaah6888. doi: 10.1126/scitranslmed.aah6888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wilck N, et al. Salt-responsive gut commensal modulates TH17 axis and disease. Nature. 2017;551(7682):585–589. doi: 10.1038/nature24628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kechaou N, et al. Identification of one novel candidate probiotic Lactobacillus plantarum strain active against influenza virus infection in mice by a large-scale screening. Appl. Environ. Microbiol. 2013;79:1491–1499. doi: 10.1128/AEM.03075-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Askelson TE, Campasino A, Lee JT, Duong T. Evaluation of phytate-degrading Lactobacillus culture administration to broiler chickens. Appl. Environ. Microbiol. 2014;80:943–950. doi: 10.1128/AEM.03155-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elshaghabee FM, Rokana N, Gulhane RD, Sharma C, Panwar H. Bacillus as potential probiotics: status, concerns, and future perspectives. Front. Microbiol. 2017;8:1490. doi: 10.3389/fmicb.2017.01490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mingmongkolchai S, Panbangred W. Bacillus probiotics: an alternative to antibiotics for livestock production. J. Appl. Microbiol. 2018;124:1334–1346. doi: 10.1111/jam.13690. [DOI] [PubMed] [Google Scholar]
- 9.Endo T, Nakano M, Shimizu S, Fukushima M, Miyoshi S. Effects of a probiotic on the lipid metabolism of cocks fed on a cholesterol-enriched diet. Biosci. Biotech. Bioch. 1999;63:1569–1575. doi: 10.1271/bbb.63.1569. [DOI] [PubMed] [Google Scholar]
- 10.Hosoi T, Ametani A, Kiuchi K, Kaminogawa S. Changes in fecal microflora induced by intubation of mice with Bacillus subtilis (natto) spores are dependent upon dietary components. Can. J. Microbiol. 1999;45:59–66. doi: 10.1139/w98-206. [DOI] [PubMed] [Google Scholar]
- 11.Guo X, Li D, Lu W, Piao X, Chen X. Screening of Bacillus strains as potential probiotics and subsequent confirmation of the in vivo effectiveness of Bacillus subtilis MA139 in pigs. Antonie van leeuwenhoek. 2006;90:139–146. doi: 10.1007/s10482-006-9067-9. [DOI] [PubMed] [Google Scholar]
- 12.Fukushima M, Nakano M. The effect of a probiotic on faecal and liver lipid classes in rats. Brit. J. Nutr. 2007;73:701–710. doi: 10.1079/BJN19950074. [DOI] [PubMed] [Google Scholar]
- 13.Tam NK, et al. The intestinal life cycle of Bacillus subtilis and close relatives. J. Bacteriol. 2006;188(7):2692–2700. doi: 10.1128/JB.188.7.2692-2700.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang Y, Xiong H, Guo X. Enhanced viability of Lactobacillus reuteri for probiotics production in mixed solid-state fermentation in the presence of Bacillus subtilis. Folia. Microbiol. 2014;59:31–36. doi: 10.1007/s12223-013-0264-4. [DOI] [PubMed] [Google Scholar]
- 15.Hosoi T, Ametani A, Kiuchi K, Kaminogawa S. Improved growth and viability of lactobacilli in the presence of Bacillus subtilis (natto), catalase, or subtilisin. Can. J. Microbiol. 2000;46:892–897. doi: 10.1139/w00-070. [DOI] [PubMed] [Google Scholar]
- 16.Horie M, et al. Evaluation of probiotic and prebiotic-like effects of Bacillus subtilis BN on growth of lactobacilli. J. Gen. Appl. Microbiol. 2018;64:26–33. doi: 10.2323/jgam.2017.03.002. [DOI] [PubMed] [Google Scholar]
- 17.Papadimitriou K, et al. Stress physiology of lactic acid bacteria. Microbiol. Mol. Biol. Rev. 2016;80:837–890. doi: 10.1128/MMBR.00076-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Abriouel H, et al. Cloning and heterologous expression of hematin-dependent catalase produced by Lactobacillus plantarum CNRZ1228. Appl. Environ. Microbiol. 2004;70:603–606. doi: 10.1128/AEM.70.1.603-606.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fu L, et al. Enhancing the oxidative resistance of yoghurt starter bacteria with heterologous catalase expression in Streptococcus thermophilus. Int. Dairy J. 2013;30:68–72. doi: 10.1016/j.idairyj.2012.11.012. [DOI] [Google Scholar]
- 20.Ricciardi A, Ianniello RG, Parente E, Zotta T. Factors affecting gene expression and activity of heme- and manganese-dependent catalases in Lactobacillus casei strains. Int. J. Food Microbiol. 2018;280:66–77. doi: 10.1016/j.ijfoodmicro.2018.05.004. [DOI] [PubMed] [Google Scholar]
- 21.Sudhamani M, et al. Characterisation of pSMA23, a 3.5 kbp plasmid of Lactobacillus casei, and application for heterologous expression in Lactobacillus. Plasmid. 2008;59:11–19. doi: 10.1016/j.plasmid.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 22.Aukrust TW, Brurberg MB, Nes IF. Transformation of Lactobacillus by electroporation. Methods Mol. Biol. 1995;47:201–208. doi: 10.1385/0-89603-310-4:201. [DOI] [PubMed] [Google Scholar]
- 23.Frece J, et al. Importance of S-proteins in probiotic activity of Lactobacillus acidophilus M92. J. Appl. Microbiol. 2005;98:285–292. doi: 10.1111/j.1365-2672.2004.02473.x. [DOI] [PubMed] [Google Scholar]
- 24.Zotta T, et al. Assessment of aerobic and respiratory growth in the Lactobacillus casei group. PLoS One. 2014;9(6):e99189. doi: 10.1371/journal.pone.0099189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weinstein JD, Beale SI. Separate Physiological Roles and Subcellular Compartments for Two Tetrapyrrole Biosynthetic Pathways in Euglena gracilis. J. Biol. Chem. 1983;258:6799–6807. [PubMed] [Google Scholar]
- 26.Sambrook, J., Fritsch, E. F. & Maniatis, T. Molecular Cloning: A Laboratory Manual, 2nd ed. Cold Spring Harbor Laboratory Press: Cold Spring Harbor, New York (1989).
- 27.Guo TT, et al. Oxygen-inducible conversion of lactate to acetate in heterofermentative Lactobacillus brevis ATCC 367. Appl. Environ. Microbiol. 2017;83:e01659–17. doi: 10.1128/AEM.01659-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rezaïki L, et al. Respiration metabolism reduces oxidative and acid stress to improve long-term survival of Lactococcus lactis. Mol. Microbiol. 2004;53:1331–1342. doi: 10.1111/j.1365-2958.2004.04217.x. [DOI] [PubMed] [Google Scholar]
- 29.Jeong JS, Kim IH. Effect of Bacillus subtilis C-3102 spores as a probiotic feed supplement on growth performance, noxious gas emission, and intestinal microflora in broilers. Poultry Sci. 2014;93:3097–3103. doi: 10.3382/ps.2014-04086. [DOI] [PubMed] [Google Scholar]
- 30.Tang Wenjie, Qian Ye, Yu Bing, Zhang Tao, Gao Jun, He Jun, Huang Zhiqing, Zheng Ping, Mao Xiangbing, Luo Junqiu, Yu Jie, Chen Daiwen. Effects of Bacillus subtilis DSM32315 supplementation and dietary crude protein level on performance, gut barrier function and microbiota profile in weaned piglets1. Journal of Animal Science. 2019;97(5):2125–2138. doi: 10.1093/jas/skz090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smith TJ, Blackman SA, Foster SJ. Autolysins of Bacillus subtilis: multiple enzymes with multiple functions. Microbiology. 2000;146:249–262. doi: 10.1099/00221287-146-2-249. [DOI] [PubMed] [Google Scholar]
- 32.Chung JK, et al. Induction of growth phase-specific autolysis in Bacillus subtilis 168 by growth inhibitors. J. Microbiol. 2009;47:50–59. doi: 10.1007/s12275-008-0256-2. [DOI] [PubMed] [Google Scholar]
- 33.Chen W, et al. Effects of Bacillus subtilis var. natto and Saccharomyces cerevisiae fermented liquid feed on growth performance, relative organ weight, intestinal microflora, and organ antioxidant status in Landes geese. J. Anim. Sci. 2013;91:978–985. doi: 10.2527/jas.2012-5148. [DOI] [PubMed] [Google Scholar]
- 34.Bhattacharyya A, Chattopadhyay R, Mitra S, Crowe SE. Oxidative stress: an essential factor in the pathogenesis of gastrointestinal mucosal diseases. Physiol. Rev. 2014;94:329–354. doi: 10.1152/physrev.00040.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.EI-Kenawi A, Ruffell B. Inflammation, ROS, and Mutagenesis. Cancer Cell. 2017;32(6):727–729. doi: 10.1016/j.ccell.2017.11.015. [DOI] [PubMed] [Google Scholar]
- 36.Lorquet F, et al. Characterization and functional analysis of the poxB gene, which encodes pyruvate oxidase in Lactobacillus plantarum. J. Bacteriol. 2004;186:3749–3759. doi: 10.1128/JB.186.12.3749-3759.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zotta T, Parente E, Ricciardi A. Aerobic metabolism in the genus Lactobacillus: impact on stress response and potential applications in the food industry. J.Appl. Microbiol. 2017;122:857–869. doi: 10.1111/jam.13399. [DOI] [PubMed] [Google Scholar]
- 38.Ianniello RG, et al. Biochemical analysis of respiratory metabolism in the heterofermentative Lactobacillus spicheri and Lactobacillus reuteri. J. Appl. Microbiol. 2015;119:763–775. doi: 10.1111/jam.12853. [DOI] [PubMed] [Google Scholar]
- 39.Chooruk A, Piwat S, Teanpaisan R. Antoxidant activity of various oral Lactobacillus strains. J. Appl. Microbiol. 2017;123:271–279. doi: 10.1111/jam.13482. [DOI] [PubMed] [Google Scholar]
- 40.An H, et al. High-level expression of heme-dependent catalase gene katA from Lactobacillus sakei protects Lactobacillus rhamnosus from oxidative stress. Mol. Biotech. 2010;45:155–160. doi: 10.1007/s12033-010-9254-9. [DOI] [PubMed] [Google Scholar]
- 41.Ianniello RG, et al. Investigation of factors affecting aerobic and respiratory growth in the oxygten-tolerant strain Lactobacillus casei N87. PLoS One. 2016;11:e0164064. doi: 10.1371/journal.pone.0164065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brooijmans R, et al. Heme and menaquinone induced electron transport in lactic acid bacteria. Microb. Cell Fact. 2009;8:28. doi: 10.1186/1475-2859-8-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jones SE, Knight KL. Bacillus subtilis-mediated protection from Citrobacter rodentium-associated enteric disease requires espH and functional flagella. Infect. Immun. 2012;80:710–719. doi: 10.1128/IAI.05843-11. [DOI] [PMC free article] [PubMed] [Google Scholar]