Abstract
Non-small-cell lung cancer (NSCLC) is a major cause for cancer-related deaths around the globe, partially due to the frequent recurrence and metastasis. Leucine-rich-alpha2-glycoprotein 1 (LRG1) is reportedly upregulated in several cancers including NSCLC; however, its functions in NSCLC remain elusive. We used quantitative real-time PCR and western blot assays to evaluate the expression patterns of LRG1 in tumor tissues collected from NSCLC patients, as well as NSCLC cell lines, and examined the effects of LRG1 on the proliferation, migration, and invasion of NSCLC cells. Further, we isolated exosomes from the blood of NSCLC patients, as well as NSCLC cell cultures, and assessed the impact of exosome exposure on the angiogenic capacities of human umbilical vein endothelial cells. LRG1 was upregulated in NSCLC tissues and cells and induced an enhancement of NSCLC cell proliferation, migration, and invasion. In addition, LRG1 was enriched in the exosomes derived from NSCLC tissue and cells, and mediated a proangiogenic effect via the activation of transforming growth factor β (TGF-β) pathway. Exosomal LRG1 derived from NSCLC cells promotes angiogenesis via TGF-β signaling and possesses the potential of a therapeutic target in NSCLC treatment.
Introduction
Lung cancer is one of the leading causes for cancer-related casualties around the global, and non-small-cell lung cancer (NSCLC) represents approximately 80% of total lung cancer incidences.1, 2 During the past few decades, substantial progresses have been achieved in the diagnostic and therapeutic strategies for NSCLS, including advancement of treatment options such as surgical, radio- or chemotherapy, and targeted therapies. However, the 5-year survival rate of NSCLC patients remains poor due to frequent recurrence, metastasis, and the fact that the majority of the patients present at an advanced stage.3 Therefore, a better understanding of the pathological mechanisms involved in the proliferation, invasion, and migration of tumor cells is critical for the development of effective approaches to cure NSCLC.
Cancer metastasis is a coordinated and complex process involving the proliferation and invasion of cancer cells at the primary site, migration through the circulation, and adaptation to the distal organ or tissue to form metastases,4 during which tumor cells constantly and actively interact with their microenvironment.5 There are multiple means for intercellular communication that tumor cells utilize to support a pro-tumorigenic microenvironment, including the production and exchange of exosomes.6 Exosomes are a form of extracellular vesicles, around 100 nm in diameter, secreted by all cell types.7, 8 Exosomes are believed to carry cellular contents including protein, lipids, and microRNAs that reflect the identity and the state of the cells of origin. Once transported to the distal site, exosomes can fuse with the recipient cells and release their contents; therefore, exosomes have attracted increasing interests for their prominent roles in long-range cell-cell communications.9 Accumulating evidence suggested that exosomes exert critical functions in the progression of several cancers, promoting tumor growth, angiogenesis, and metastasis.10, 11 A recent report showed that high levels of exosomal proteins were positively correlated with several malignant parameters in NSCLC, raising the possibility that exosomes could serve as a therapeutic target or biomarker in the treatment of NSCLC.
Leucine-rich-alpha2-glycoprotein 1 (LRG1) was first isolated and characterized in 1977 by Haupt and colleagues.12 It is the founding member of the leucine-rich repeat (LRR) protein family, consisting of eight repeating sequences.13 LRG1 has been implicated in various types of cancers, including pancreatic cancer, hepatocellular carcinoma, bladder cancer, gastric cancer, and NSCLC.14, 15, 16, 17, 18 It was demonstrated that in colorectal cancer LRG1 was overexpressed and promoted angiogenesis, a crucial process during cancer metastasis, through activation of hypoxia-inducible factor (HIF)-1α pathway.19, 20 However, the mechanistic details regarding the roles of LRG1 in NSCLC remain largely unknown. Thus, in the current study, we aimed to investigate the expression pattern and the effects on angiogenesis of LRG1 in NSCLC, as well as to reveal the underlying cellular mechanisms. With a combinatorial approach using molecular, cellular, and biochemical techniques, we found that LRG1 was upregulated in NSCLC tissues and responsible for the enhanced proliferation, migration, and invasion capabilities of the cancer cells. Further, LRG1 was enriched in the exosomes derived from NSCLC tissues and cell cultures to promote angiogenesis, likely through the activation of transforming growth factor β (TGF-β) pathway.
Results
LRG1 Was Upregulated in NSCLC
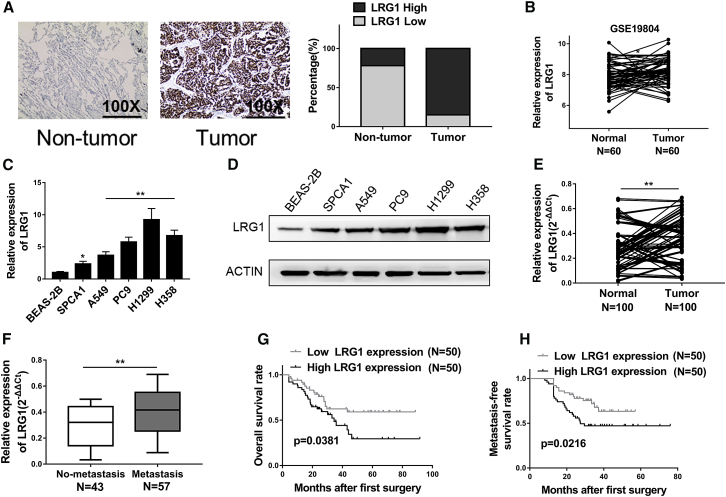
In order to evaluate the physiological relevance of LRG1 in NSCLC, we performed immunohistochemical analysis of NSCLC tissues with corresponding adjacent non-tumor tissues collected from 100 NSCLC patients. Close examination of the staining revealed that LRG1 was highly expressed in the NSCLC tumor tissues compared to the benign control tissues (Figure 1A). This note was further supported by the data from GSE 19804 dataset, in which LRG1 exhibited a significantly higher expression level in NSCLC tissues in comparison to the adjacent normal tissues (Figure 1B). We then assessed the levels of LRG1 mRNA in several NSCLC cell lines, namely SPCA1, A549, PC9, H1299, and H358, and found that LRG1 was upregulated in all five tumor cell lines compared to the expression level in normal human bronchial epithelial (BEAS-2B) cells (Figure 1C). Consistently, western blot assays demonstrated that the protein levels of LRG1 were also elevated in NSCLC cells compared to BEAS-2B cells (Figure 1D), suggesting an association between LRG1 and NSCLC. To further assess the involvement of LRG1 in NSCLC, we examined the tumor tissues from 100 NSCLC patients (Table S1) and again observed a significantly higher transcript level of LRG1 in tumor tissues compared to the normal adjacent tissues (Figure 1E). In addition, LRG1 expression level seemed to be linked with metastasis because it was notably higher in the 57 cases with lymph node metastasis (Figure 1F). Kaplan-Meier survival curves also revealed that poor survival, both overall (Figure 1G) and metastasis-free survival (Figure 1H), was observed in patients with higher levels of LRG1, indicating a likely role of LRG1 in the progression of NSCLC.
Figure 1.
LRG1 Was Highly Upregulated in NSCLC, and Increased LRG1 Expression Predicted Poor Prognosis in NSCLC Patients
(A) IHC analysis of LRG1 expression in NSCLC tissue samples, as well as non-tumor adjacent tissues, 100×. (B) The LRG1 expression levels in 60 NSCLC tissues (tumor) and 60 adjacent normal tissues (normal) from GSE19804. (C and D) The expression levels of LRG1 in NSCLC cells (SPCA1, A549, PC9, H1299, and H358) and BEAS-2B were detected by qRT-PCR (C) and western blotting (D). (E) The LRG1 expression levels in 100 NSCLC tissues (tumor) and 100 adjacent normal tissues (normal) were detected by qRT-PCR. (F) The LRG1 expression in NSCLC tissues with lymph node metastasis (metastasis) was significantly higher than that in tissues without lymph node metastasis (no-metastasis). (G and H) Kaplan-Meier survival curves revealed that higher levels of LRG1 were associated with poor overall survival rate (G) and metastasis-free survival rate (H). *p < 0.05; **p < 0.01.
LRG1 Regulated the Proliferation, Migration, and Invasion of NSCLC Cells
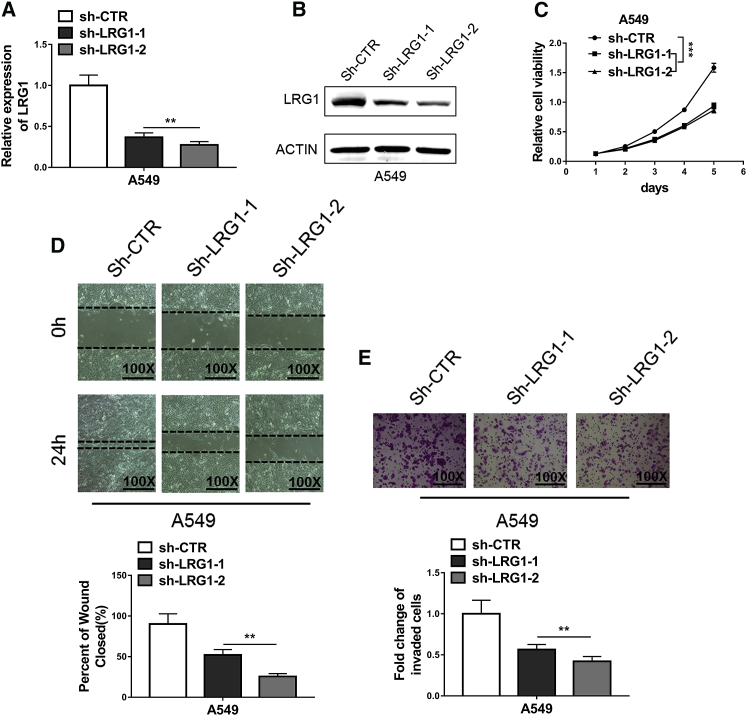
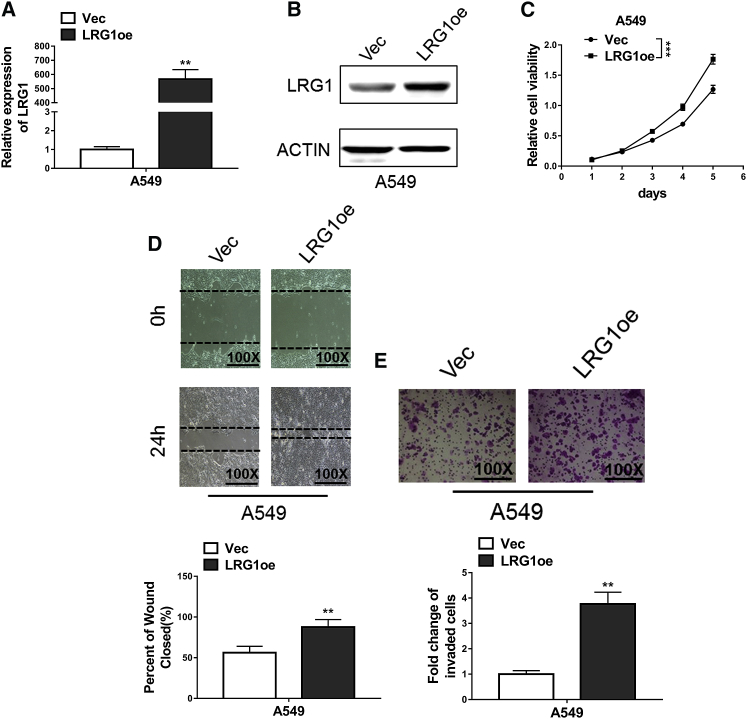
The exact functions of LRG1 in NSCLC were assessed through short hairpin RNA (shRNA)-mediated knockdown experiments. Our qRT-PCR results (Figure 1C) demonstrated that A549 cells, a common NSCLC cell line and a most widely used model, expressed an intermedium level of LRG1 and were therefore perfectly suitable for knockdown, as well as overexpression manipulations. The efficiency of knockdown at both mRNA and protein levels was confirmed using quantitative PCR and western blot assay, respectively (Figures 2A and 2B). The attenuated expression of LRG1 mediated by either sh-LRG1 or sh-LRG2, but not the negative control shRNA-control (sh-CTR), resulted in suppressed proliferative activities of A549 cells, as indicated by the reduced proliferation rate in 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay (Figure 2C). Similarly, decreased migration capacities in the wound-healing assay were observed in A549 cells with LRG1 knockdown (Figure 2D), which also exhibited inhibited invasion capabilities in the Transwell assay (Figure 2E). We then examined the impact of LRG1 overexpression (Figures 3A and 3B) on the proliferation, migration, and invasion of A549 cells using aforementioned assays, and we found that A549 cells with LRG1 overexpression indeed displayed enhanced capabilities of proliferation (Figure 3C), migration (Figure 3D), and invasion (Figure 3E). These findings suggested that LRG1 exerted critical functions to promote the proliferation, migration, and the invasion of NSCLC cells.
Figure 2.
Knockdown of LRG1 Inhibited Proliferation, Migration, and Invasion in NSCLC Cells
(A and B) Knockdown efficiency of LRG1 in A549 cells transfected with shRNAs targeting LRG1 (sh-LRG1-1 and sh-LRG1-2) or negative control (sh-CTR) was confirmed by qRT-PCR (A) and western blotting (B). (C) MTT assays showed that knockdown of LRG1 significantly suppressed proliferation of A549 cells. (D) Wound-healing assays showed the repression of cell migration of A549 due to knockdown of LRG1. (E) Transwell assays showed the repression of cell invasion of A549 due to knockdown of LRG1. **p < 0.01; ***p < 0.001 (two-way ANOVA for C, Student’s t test for others).
Figure 3.
LRG1 Overexpression Promoted Proliferation, Migration, and Invasion in NSCLC Cells
(A and B) The expression levels of LRG1 in A549 cells transfected with LRG1 overexpression plasmid (LRG1oe) or empty vector (Vec) were confirmed using qRT-PCR (A) and western blotting (B). (C) MTT assays showed LRG1 overexpression significantly enhanced proliferation of A549 cells. (D) Wound-healing assays showed the enhancement of cell migration of A549 due to overexpression of LRG1. (E) Transwell assays showed the increase of cell invasion of A549 due to overexpression of LRG1. **p < 0.01; ***p < 0.001 (two-way ANOVA for C, Student’s t test for others).
Exosomes Derived from NSCLC Cells Promoted Angiogenesis
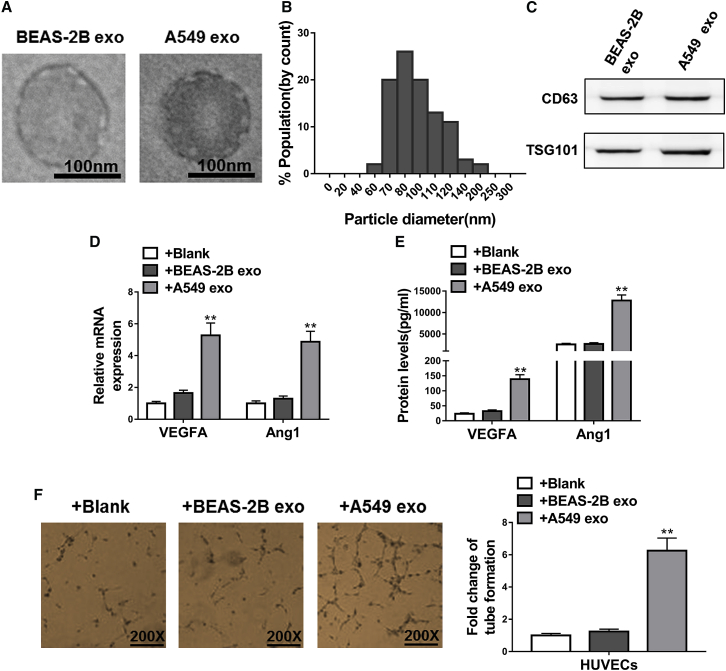
As cancer progresses and tumors grow, the lack of sufficient supply of oxygen and nutrients triggers the process of angiogenesis, which is considered a key feature of cancer pathology.21 Because exosomes have been previously implicated in pathological angiogenesis,10 we next sought to investigate the role of exosomes in the progression of NSCLC. Particles of the typical size and morphological features of exosomes were enriched in A549 cells compared to BEAS-2B cells (Figures 4A and 4B), and their exosomal identity was further verified by the expression of known exosome markers CD63 and tumor susceptibility gene (TSG) 101 (Figure 4C). We found that exosomes derived from NSCLC cells possessed the capacity to promote angiogenesis, as human umbilical vein endothelial cells (HUVECs) exposed to the A549-derived exosomes expressed markedly higher levels of proangiogenic markers, namely vascular endothelial growth factor A (VEGFA) and angiopoietin-1 (Ang1), at both mRNA and protein levels, whereas exosomes derived from normal BEAS-2B cells had no effect (Figures 4D and 4E, respectively). It was found that there was no difference in exosome uptake for the two exosome groups, as evidenced by the comparable fluorescence intensity in the Exo-Red-labeled exosome-treated cells (data not shown). In agreement with these findings, HUVECs exposed to A549-derived but not BEAS-2B-derived exosomes exhibited increased tube formation, indicating a greater angiogenic tendency induced by exosomes derived from NSCLC cells (Figure 4F). Another NSCLC cell line, H1299, in which LRG1 was expressed at the highest level among cell lines examined (Figure 1C), was subjected to the same angiogenesis assessments. Similar to what was found with A549 cells, H1299 cell-derived exosomes also promoted angiogenesis (Figure S1), suggesting a possible involvement of LRG1 in such pro-angiogenic effects.
Figure 4.
Exosome Derived from NSCLC Cells Promoted Angiogenesis
(A) Representative electron micrographs of exosomes of the typical size isolated from BEAS-2B cells conditioned and A549 cells conditioned medium exhibiting exosomal morphological traits. Scale bars, 100 nm. (B) Histogram showing the particle diameter (nm) of the small vesicles harvested from the medium of the A549 cells using the standard exosome isolation protocol. The amount of exosomes released into the conditioned media was normalized to the number of cells in the culture for conditioned media collection. (C) Western blotting revealed substantial expression of TSG101 and CD63 in exosomes isolated from the medium of BEAS-2B cells (BEAS-2B exo) and A549 cells (A549 exo). (D) The mRNA levels of proangiogenic factors VEGFA and Ang1 in HUVECs were determined by qRT-PCR. HUVECs were exposed to BEAS-2B cell-derived exosomes (+BEAS-2B exo), A549 cell-derived exosomes (+A549 exo), or alone (blank) for 48 h. (E) The protein levels of proangiogenic factors VEGFA and Ang1 in conditioned media of HUVECs were determined by ELISA. HUVECs were exposed to BEAS-2B cell-derived exosomes (+BEAS-2B exo), A549 cell-derived exosomes (+A549 exo), or alone (blank) for 48 h. (F) Representative images show the tube formation of HUVECs after the same exposure of corresponding exosomes. *p < 0.05; **p < 0.01.
LRG1 Was Enriched in Exosomes of NSCLC and Promoted Angiogenesis via TGF-β Signal Pathway
Given the enhanced proliferation, migration, and invasion of NSCLC cells we previously observed with LRG1 expression, we next examined whether LRG1 was involved in the proangiogenic activities of A549-derived exosomes. We isolated exosomes from the blood of NSCLC patients (tumor), as well as healthy controls (normal), and discovered a higher plasma exosomal level of LRG1 in the tumor samples (Figure 5A), particularly in the samples collected from patients with lymph node metastasis (Figure 5B). LRG1 was also enriched in exosomes derived from NSCLC cell lines in comparison to that of the BEAS-2B cells, at both transcript and protein levels (Figures 5C and 5D, respectively). Moreover, the expression of both LRG transcript and protein by HUVECs was increase when challenged with A549-derived exosomes (Figures 5E and 5F), which again implied a link between upregulated LRG1 and the progression of NSCLC. Finally, we assessed the impacts of LRG-1 knockdown on the proangiogenic activities of NSCLC-derived exosomes. The knockdown efficiency was evaluated using qRT-PCR, as well as western blot assay (Figures 5G and 5H). Marked decreases of exosomal LRG1 at both transcript and protein levels were consistently observed in A549 cells transfected with LRG1 small interfering RNAs (LRG1-KD) when compared with the negative control (control). The expression of proangiogenic markers by HUVECs induced by the exposure to A549-derived exosomes, as well as the tube formation of exosome-exposed HUVECs, were both reversed or attenuated by LRG1 knockdown in A549 cells (Figures 5I–5K). These results presented evidence for a critical role of exosomal LRG1 in the pathological angiogenesis in NSCLC.
Figure 5.
LRG1 Was Enriched in Exosome of NSCLC and Can Be Transferred to HUVECs to Promote Angiogenesis
(A) Quantitative analysis of exosomal LRG1 levels in plasma of 100 NSCLC patients, as well as 60 healthy donors by ELISA. (B) Quantitative analysis of exosomal LRG1 levels in plasma of 43 NSCLC patients without lymph node metastasis (no-metastasis) and 57 NSCLC patients with lymph node metastasis (metastasis) by ELISA. (C and D) The expression levels of exosomal LRG1 derived from NSCLC cells (SPCA1, A549, PC9, H1299, and H358) and BEAS-2B were detected by qRT-PCR (C) and western blotting (D). (E and F) LRG1 levels in HUVECs exposed to exosomes following various treatments were measured using qRT-PCR (E) and western blotting (F). (G and H) The expression levels of exosomal LRG1 derived from A549 cells transfected with LRG1 si-RNAs (LRG1-KD) or negative control (control) were measured using qRT-PCR (G) and western blotting (H). (I) The mRNA levels of proangiogenic factors VEGF and Ang1 in HUVECs were determined by qRT-PCR. HUVECs were exposed to exosomes derived from A549 cells (+A549 exo) or LRG1 knockdown-A549 cells (+A549 LRG1-KD exo) for 48 h. (J) The protein levels of proangiogenic factors VEGFA and Ang1 in conditioned media of HUVECs were determined by ELISA. HUVECs were exposed to exosomes derived from A549 cells (+A549 exo) or LRG1 knockdown-A549 cells (+A549 LRG1-KD exo) for 48 h. (K) Tube formation of HUVECs after the same exposure of corresponding exosomes. *p < 0.05; **p < 0.01; ***p < 0.001, Student’s t test.
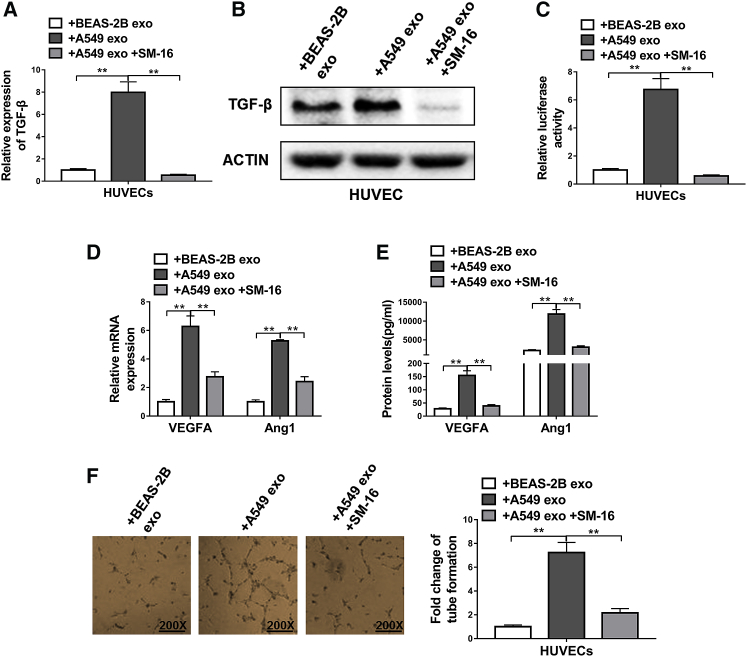
Lastly, we explored the molecular mechanisms underlying the promotion of angiogenesis mediated by exosomal LRG1. A prior report has shown that LRG1 promoted angiogenesis through TGF-β pathway in a retinal degenerative disease model.22 In light of this, we evaluated the role of TGF-β pathway in the proangiogenic events in NSCLC. The TGF-β expression in HUVECs, at both mRNA and protein levels, was examined following exposure to A549-derived exosomes with or without SM-16, a specific blocker for TGF-β receptors. We found that the expression level of TGF-β (Figures 6A and 6B), as well as its signaling activities (Figure 6C), were strongly elevated after exposure to exosomes derived from NSCLC cells in a SM-16-sensitive manner. Furthermore, blockage of the TGF-β pathway using SM-16 greatly dampened the proangiogenic effects of A549-derive exosomes, as evidenced by the suppressed expression of VEGFA and Ang1 (Figures 6D and 6E), as well as the repressed tube-forming capabilities of exosome-exposed HUVECs (Figure 6F). Taken together, these data demonstrated that the proangiogenic properties of exosomal LRG1 depended on the activities of TGF-β pathway.
Figure 6.
Exosomal LRG1-Mediated Promotion of Angiogenesis Was Dependent on TGF-β Signal Pathway
(A and B) The expression levels of TGF-β in HUVECs were determined by qRT-PCR (A) and western blotting (B). HUVECs were exposed to BEAS-2B cell-derived exosomes (+BEAS-2B exo) or A549 cell-derived exosomes (+A549 exo) or A549 cell-derived exosomes plus TGF-β inhibitor SM-16 (10 μg/mL; +A549 exo+SM-16). (C) TMLC assay for active TGF-β from conditioned media of HUVECs following the same exposure. (D) The mRNA levels of proangiogenic factors VEGF and Ang1 in HUVECs were determined by qRT-PCR. HUVECs were exposed to BEAS-2B cell-derived exosomes (+BEAS-2B exo), A549 cell-derived exosomes (+A549 exo), or A549 cell-derived exosomes plus TGF-β inhibitor SM-16 (+A549 exo+SM-16). (E) The protein levels of proangiogenic factors VEGFA and Ang1 in conditioned media of HUVECs were determined by ELISA. HUVECs were exposed to BEAS-2B cell-derived exosomes (+BEAS-2B exo), A549 cell-derived exosomes (+A549 exo), or A549 cell-derived exosomes plus TGF-β inhibitor SM-16 (+A549 exo+SM-16). (F) The tube formation of HUVECs following the same exposure. **p < 0.01, Student’s t test.
Discussion
NSCLC is a major cause for cancer-related deaths around the globe, due to the frequent recurrence and metastasis. Therefore, in-depth knowledge regarding the cellular mechanisms that are responsible for the progression and metastasis of NSCLC will provide valuable information for the clinical management of NSCLC. In the present study, we focused on the functions of exosomal LRG1 in the progression of NSCLC. We collected tumor tissues from NSCLC patients and discovered an upregulation of LRG1 in NSCLC tissues that was correlated with metastasis, as well as poor survival. Using NSCLC cell lines, we provided evidence that higher levels of LRG1 were associated with better capacities of proliferation, migration, and invasion of NSCLC cells. Further, we discovered that LRG1 was enriched in the exosomes derived from NSCLC tissue or cells and mediated the proangiogenic effects of NSCLC-origin exosomes, a process depending on the activities of the TGF-β pathway. Our findings revealed for the first time that exosomal LRG1 promoted angiogenesis in NSCLC and raised the possibility of exosomal LRG1 serving as a novel therapeutic target or biomarker in the treatment against NSCLC.
A growing body of literature has implicated a key role of LRG1 in various forms of carcinomas. For instance, Ladd and colleagues reported a higher level of LRG1 in the colorectal cancer samples collected from female patients, even before their diagnosis.19 Linder et al. used combined techniques of mass spectrometry and western blot assays to screen the urinary samples harvested from individuals diagnosed with non-muscle bladder cancer and discovered a greater abundance of LRG1 in the urine of cancer patients.16 Similarly, it has been shown that the expression of LRG1 was upregulated by more than 2-fold in gastric cancer samples through comparative proteomics analysis17 and the serum level of LRG1 increased with progressive stages of the pancreatic cancer.14 Of particular relevance to our study, it was previously reported that LRG1 was overexpressed in both the blood and tumor tissues harvested from NSCLC patients and therefore possessed the potential of a biomarker. However, the precise functions of LRG1 in NSCLC were poorly understood. Our data presented in the current study showed that LRG1 not only enhanced the capabilities of NSCLC cells to proliferate, migrate, and invade, consistent with prior findings,23 but also promoted angiogenesis through exosomal activities. These findings were in agreement with prior reports describing the modulation of angiogenesis by LRG1 in other pathological models20, 22 and shone light on the cellular events underlying the elevated expression of LRG1 in NSCLC.
It is also noteworthy that our data demonstrated that LRG1 was enriched in exosomes and possibly modulated angiogenesis through exosome secretion and modification of the microenvironment, as well as the targeted endothelial cells. Exosomes are virus-size extracellular particles, which were initially regarded as mere cellular garbage bags but are now gaining increasing attention for their unraveled roles in intracellular communications.7, 8 Exosomes carry cellular contents reflecting the information of the parent cell and may be utilized by tumor cells to sculpt and modify their microenvironment.5, 9, 10, 24 For example, when a tumor reaches a certain size, there is a great drive to grow new blood vessels due to hypoxia and nutrients deprivation, in which exosomes originated from tumor cells may play essential roles to facilitate such pathological angiogenesis.21 Indeed, in our study we found that exosomes derived from either NSCLC patient samples or NSCLC cells were capable of increasing the expression of proangiogenic markers VEGFA and Ang1 in endothelial cells and enhanced the tube formation of HUVECs. Furthermore, such an effect of NSCLC-derived exosomes was dependent upon the expression of LRG1 and the activities of TGF-β pathway, the latter of which has also been implicated in the progression of several cancers such as hepatocellular carcinoma.25
In summary, we hereby provided the first evidence that LRG1 not only promoted the progression, migration, and invasion of NSCLC cells but also further induced angiogenesis through exosomal enrichment, via the activation of TGF-β pathway. Our findings proved that LRG1 likely contributes to the progression and metastasis of NSCLC and therefore supported the possibility of targeting exosomal LRG1 in the treatment of NSCLC.
Materials and Methods
Collection of Tumor Samples
A total of 100 NSCLC patients were recruited for the collection of tumor samples with corresponding adjacent benign tissues in Peking University Shenzhen Hospital. The procedures and protocols involved in the current study were approved by the Ethics Committee of Peking University Shenzhen Hospital. All patients provided written informed consent prior to recruitment.
Immunohistochemistry
NSCLC tumor tissue and corresponding non-tumor tissue samples were examined using immunohistochemistry. Paraffin-embedded sample sections were subjected to antigen retrieval in citrate buffer (pH 6.0; 10 mM) followed by incubation with anti-LRG1 primary antibody (#ab178698, 1:100, Abcam, Cambridge, MA, USA) overnight at 4°C, and then incubated with HRP-conjugated secondary antibody at room temperature.
Quantitative Real-Time PCR
Extraction of total RNA from tissue or cell samples was conducted using Trizol (Invitrogen, Waltham, MA, USA), followed by reverse transcription into cDNA with the use of the PrimeScript RT Reagent Kit (Perfect Real Time, TaKaRa, Dalian, China). Quantitative real-time PCR was conducted using the SYBR Green Kit (TaKaRa, Dalian, China), and the relative expression levels were calculated using the 2−ΔΔCt method with GAPDH as the internal control. The primers used in the current study were as follows: LRG1, F: 5′-GTTGGAGACCTTGCCACCT-3′, R: 5′-GCTTGTTGCCGTTCAGGA-3′; VEGFA, F: 5′-CTTTCTGCTGTCTTGGGTG-3′, R: 5′-ACTTCGTGATGATTCTGCC-3′; Ang1, F: 5′-CCAGTACAACACAAACGCTCT-3′, R: 5′-TCTCCGACTTCATGTTTTCCAC-3′; TGF-β, F: 5′-CCCTACATTTGGAGCCTGGA-3′, R: 5′-CCGGGTTATGCTGGTTGTAC-3′; GAPDH: F: 5′-ACAACTTTGGTATCGTGGAAGG-3′, R: 5′-GCCATCACGCCACAGTTTC-3′.
Western Blot Assay
Cell samples were lysed in standard radioimmunoprecipitation buffer for protein purification and the protein concentrations were examined using the bicinchoninic acid assay. The extracted total proteins of equal amount were run and separated on a sodium dodecyl sulfate-polyacrylamide gel and subsequently transferred to a polyvinylidene fluoride membrane. Then the membrane was blocked with 5% skim milk and incubated the primary antibodies overnight at 4°C, followed by 2 h incubation with the appropriate secondary antibody at room temperature. Primary antibodies used in the current study were as follows: LRG1 (#ab178698, 1:500, Abcam, Cambridge, MA, USA); CD63 (#ab59479, 1:100, Abcam); TSG101 (#ab125011, 1:1,000, Abcam); TGF-β (#3711, 1:1,000, CST, Danvers, MA, USA). The final blots were visualized using the chemiluminescence method (Thermo Fisher, Waltham, MA, USA) and quantified with the ImageJ software.
Cell Culture and Transfection
NSCLC cells lines (SPCA1, A549, PC9, H1299, and H358) were cultured in RPMI 1640 medium containing 10% fetal bovine serum (FBS, Invitrogen, Pleasanton, CA, USA). HUVECs were cultured in DMEM containing 10% FBS. All cell cultures were maintained at 37°C with 5% CO2 in a humidified incubator. For LRG1 knockdown experiments, two different shRNAs were designed to target LRG1 (sh-LRG-1: F: 5′-ccggGCAATTAGAACGGCTACATCTggatccAGATGTAGCCGTTCTAATTGCtttttg-3′, R: 5′-aattcaaaaaGCAATTAGAACGGCTACATCTggatccAGATGTAGCCGTTCTAATTGC-3′; sh-LRG2: F: 5′-ccggCAGCCGACACCGTGCACCTGGggatccCCAGGTGCACGGTGTCGGCTGtttttg, R: 5′-aattcaaaaaCAGCCGACACCGTGCACCTGGggatccCCAGGTGCACGGTGTCGGCTG-3′). For the construction of LRG1 overexpression plasmid, LRG1 cDNA was cloned using primers as follows: F: 5′-CTAGAATTCATGTCCTCTTGGAG-3′, R: 5′-CTAGGATCCTCACTGGGACTTGGCCAC-3′.
Cell Proliferation Assay
The proliferative capacities of cells were assessed using the MTT assay. In brief, cells were placed in 96-well plates at the density of 1 × 103 cells per well, and transfections were performed 24 h later. Cell viability was examined every 24 h for 4 days with the cell counting kit-8 (CCK-8; Dojindo, Kumamoto, Japan). The rate of proliferation was measured as OD450 at each sampling point divided by that of cell samples immediately after transfection.
Wound-Healing Assay
The migration capacity of NSCLC cells was evaluated using a wound-healing assay. In brief, a linear cut was generated across a confluent cell monolayer using the tip of a 200 μL pipette. The edge of the wound was monitored and recorded at 0, 24, and 48 h. The distance of cell migration into the wound was quantified using Image-Pro Plus 6.0 software.
Transwell Assays
Cells maintained in serum-free RPMI 1640 medium for 12–24 h were seeded at the density of 5 × 104 or 1 × 105 cells per well into the upper chamber of 24-well transwell inserts (Corning, 8.0 μm pores) coated with Matrigel (BD Biosciences, Franklin Lakes, NJ, USA), then allowed for 12–24 hours to invade toward serum-containing medium within the lower compartment. After the testing period, the cells on the bottom side of the chamber were stained and counted. The relative invasion capabilities of the cells were quantified as the fold increase of invaded cells from three random views.
Exosome Isolation
For exosome isolation from NSCLC cell lines, ExoQuick exosome precipitation solution was added to the cell culture media, and the isolation of exosomes was conducted following the manufacturer’s instructions (SBI System Biosciences, San Francisco, CA, USA). BCA protein quantization was utilized to quantify isolated exosomes.
For exosome isolation from blood samples of NSCLC patients, all samples were subjected to centrifugation at 3,000 g for 15 min to separate plasma from cells or debris. Then an additional centrifuge at 12,000 g for 20 min was performed and the supernatant was further filtered through a sterile syringe filter (0.22 mm, Millipore, Billerica, MA, USA) into individual aliquots. Each plasma sample was mixed with an Exosome Precipitation Solution (3D Medicines biotechnology, Shanghai, China) at 4:1 v/v. The mixture was incubated for 20 min at 20°C and then centrifuged at 6,500 g for 20 min at 4°C. The liquid phase was discarded and the remaining content was further centrifuged at 1,500 g for 5 min at 4°C to remove residual liquid. Finally, the exosome pellet was obtained for immediate use.
Enzyme-Linked Immunosorbent Assay
The protein levels of proangiogenic factors VEGFA and Ang1 in conditioned media of HUVECs were examined using respective ELISA kits (R&D Systems, Minneapolis, MN, USA) according to the provided guides. The plate was read using Infinite 200 PRO plate reader (Tecan, Männedorf, Switzerland) at 450 nm wavelength with wavelength correction set to 540 nm.
Exosomal LRG1 levels in the plasma of NSCLC patients were assessed using abovementioned methods with commercial ELISA kits purchased from Uscn Life Science (Wuhan, China).
Endothelial Cell Tube Formation Assay
A 48-well plate was coated with Matrigel (BD Biosciences, Franklin Lakes, NJ, USA) and allowed 30 min at 37°C for polymerization. Then, HUVECs at the density of 3 × 104 cells per well were placed into Matrigel-treated 48-well plate and cultured in conditioned medium. After 8 h, capillary-like tubes were photographed and quantified from four random fields.
Transformed Mink Lung Epithelial Cell TGF-β Assay
Transformed mink lung epithelial cells (TMLCs) containing a plasminogen activator inhibitor-1 promoter-driven luciferase reporter system were digested, washed twice, resuspended in serum-free DMEM, and then seeded at the density of 2.5 × 104 cells per well into a 96-well plate. The cells were allowed 4–6 h to adhere and then the media were replaced with the conditioned media of HUVECs following various treatments. After an 18–20 h incubation at 37°C, the cells were washed with PBS 2–3 times and then lysed using lysis buffer (50 μL). The resulted cell lysate (20 μl) was added to 90 μL of luciferase assay reagent (Promega, Madison, WI, USA), and the luciferase activities were quantified.
Statistics
All experiments were performed independently at least three times. Data were analyzed using Statistical Product and Service Solutions (SPSS) 23.0 software and presented as mean ± SD unless otherwise indicated. The differences between treatment groups were determined using two-tailed Student’s t test or two-way ANOVA, wherever appropriate. p < 0.05 was considered to indicate statistical significance.
Author Contributions
Z.L., C.Z., Q.N., F.L., J.L., Z.M., B.C., and D.W. conducted the experiments. Z.L. and C.Z. collected and analyzed the data. H.W. conceived this study. Z.L., C.Z., and H.W. wrote the manuscript.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This study was funded by the Science and Technology Development Fund Project of Shenzhen (grant number: JCYJ 20150403091443310).
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omto.2019.08.001.
Supplemental Information
References
- 1.Miller K.D., Siegel R.L., Lin C.C., Mariotto A.B., Kramer J.L., Rowland J.H., Stein K.D., Alteri R., Jemal A. Cancer treatment and survivorship statistics, 2016. CA Cancer J. Clin. 2016;66:271–289. doi: 10.3322/caac.21349. [DOI] [PubMed] [Google Scholar]
- 2.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2018. CA Cancer J. Clin. 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 3.Wang T., Nelson R.A., Bogardus A., Grannis F.W., Jr. Five-year lung cancer survival: which advanced stage nonsmall cell lung cancer patients attain long-term survival? Cancer. 2010;116:1518–1525. doi: 10.1002/cncr.24871. [DOI] [PubMed] [Google Scholar]
- 4.Gupta G.P., Massagué J. Cancer metastasis: building a framework. Cell. 2006;127:679–695. doi: 10.1016/j.cell.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 5.Zhang Y., Yang P., Wang X.F. Microenvironmental regulation of cancer metastasis by miRNAs. Trends Cell Biol. 2014;24:153–160. doi: 10.1016/j.tcb.2013.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Penfornis P., Vallabhaneni K.C., Whitt J., Pochampally R. Extracellular vesicles as carriers of microRNA, proteins and lipids in tumor microenvironment. Int. J. Cancer. 2016;138:14–21. doi: 10.1002/ijc.29417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Théry C. Exosomes: secreted vesicles and intercellular communications. F1000 Biol. Rep. 2011;3:15. doi: 10.3410/B3-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tkach M., Théry C. Communication by Extracellular Vesicles: Where We Are and Where We Need to Go. Cell. 2016;164:1226–1232. doi: 10.1016/j.cell.2016.01.043. [DOI] [PubMed] [Google Scholar]
- 9.Mathivanan S., Ji H., Simpson R.J. Exosomes: extracellular organelles important in intercellular communication. J. Proteomics. 2010;73:1907–1920. doi: 10.1016/j.jprot.2010.06.006. [DOI] [PubMed] [Google Scholar]
- 10.Kalluri R. The biology and function of exosomes in cancer. J. Clin. Invest. 2016;126:1208–1215. doi: 10.1172/JCI81135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Skog J., Würdinger T., van Rijn S., Meijer D.H., Gainche L., Sena-Esteves M., Curry W.T., Jr., Carter B.S., Krichevsky A.M., Breakefield X.O. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat. Cell Biol. 2008;10:1470–1476. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haupt H., Baudner S. [Isolation and characterization of an unknown, leucine-rich 3.1-S-alpha2-glycoprotein from human serum (author’s transl)] Hoppe Seylers Z. Physiol. Chem. 1977;358:639–646. [PubMed] [Google Scholar]
- 13.Takahashi N., Takahashi Y., Putnam F.W. Periodicity of leucine and tandem repetition of a 24-amino acid segment in the primary structure of leucine-rich alpha 2-glycoprotein of human serum. Proc. Natl. Acad. Sci. USA. 1985;82:1906–1910. doi: 10.1073/pnas.82.7.1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Furukawa K., Kawamoto K., Eguchi H., Tanemura M., Tanida T., Tomimaru Y., Akita H., Hama N., Wada H., Kobayashi S. Clinicopathological Significance of Leucine-Rich α2-Glycoprotein-1 in Sera of Patients With Pancreatic Cancer. Pancreas. 2015;44:93–98. doi: 10.1097/MPA.0000000000000205. [DOI] [PubMed] [Google Scholar]
- 15.He X., Wang Y., Zhang W., Li H., Luo R., Zhou Y., Liao C.L., Huang H., Lv X., Xie Z., He M. Screening differential expression of serum proteins in AFP-negative HBV-related hepatocellular carcinoma using iTRAQ -MALDI-MS/MS. Neoplasma. 2014;61:17–26. [PubMed] [Google Scholar]
- 16.Lindén M., Lind S.B., Mayrhofer C., Segersten U., Wester K., Lyutvinskiy Y., Zubarev R., Malmström P.U., Pettersson U. Proteomic analysis of urinary biomarker candidates for nonmuscle invasive bladder cancer. Proteomics. 2012;12:135–144. doi: 10.1002/pmic.201000810. [DOI] [PubMed] [Google Scholar]
- 17.Uen Y.H., Lin K.Y., Sun D.P., Liao C.C., Hsieh M.S., Huang Y.K., Chen Y.W., Huang P.H., Chen W.J., Tai C.C. Comparative proteomics, network analysis and post-translational modification identification reveal differential profiles of plasma Con A-bound glycoprotein biomarkers in gastric cancer. J. Proteomics. 2013;83:197–213. doi: 10.1016/j.jprot.2013.03.007. [DOI] [PubMed] [Google Scholar]
- 18.Liu Y., Luo X., Hu H., Wang R., Sun Y., Zeng R., Chen H. Integrative proteomics and tissue microarray profiling indicate the association between overexpressed serum proteins and non-small cell lung cancer. PLoS ONE. 2012;7:e51748. doi: 10.1371/journal.pone.0051748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ladd J.J., Busald T., Johnson M.M., Zhang Q., Pitteri S.J., Wang H., Brenner D.E., Lampe P.D., Kucherlapati R., Feng Z. Increased plasma levels of the APC-interacting protein MAPRE1, LRG1, and IGFBP2 preceding a diagnosis of colorectal cancer in women. Cancer Prev. Res. (Phila.) 2012;5:655–664. doi: 10.1158/1940-6207.CAPR-11-0412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang J., Zhu L., Fang J., Ge Z., Li X. LRG1 modulates epithelial-mesenchymal transition and angiogenesis in colorectal cancer via HIF-1α activation. J. Exp. Clin. Cancer Res. 2016;35:29. doi: 10.1186/s13046-016-0306-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weis S.M., Cheresh D.A. Tumor angiogenesis: molecular pathways and therapeutic targets. Nat. Med. 2011;17:1359–1370. doi: 10.1038/nm.2537. [DOI] [PubMed] [Google Scholar]
- 22.Wang X., Abraham S., McKenzie J.A.G., Jeffs N., Swire M., Tripathi V.B., Luhmann U.F.O., Lange C.A.K., Zhai Z., Arthur H.M. LRG1 promotes angiogenesis by modulating endothelial TGF-β signalling. Nature. 2013;499:306–311. doi: 10.1038/nature12345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou Y., Zhang X., Zhang J., Fang J., Ge Z., Li X. LRG1 promotes proliferation and inhibits apoptosis in colorectal cancer cells via RUNX1 activation. PLoS ONE. 2017;12:e0175122. doi: 10.1371/journal.pone.0175122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Abd Elmageed Z.Y., Yang Y., Thomas R., Ranjan M., Mondal D., Moroz K., Fang Z., Rezk B.M., Moparty K., Sikka S.C. Neoplastic reprogramming of patient-derived adipose stem cells by prostate cancer cell-associated exosomes. Stem Cells. 2014;32:983–997. doi: 10.1002/stem.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shen Y., Wei Y., Wang Z., Jing Y., He H., Yuan J., Li R., Zhao Q., Wei L., Yang T., Lu J. TGF-β regulates hepatocellular carcinoma progression by inducing Treg cell polarization. Cell. Physiol. Biochem. 2015;35:1623–1632. doi: 10.1159/000373976. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.