These results from whole-genome transcriptional profiling provide further insights into the biological activity and mode of action of a small-molecule inhibitor of C. albicans filamentation. This information will assist in the development of novel antivirulence strategies against C. albicans infections.
Keywords: Candida albicans, candidiasis, filamentation, antivirulence
ABSTRACT
The opportunistic pathogenic fungus Candida albicans can cause devastating infections in immunocompromised patients. Its ability to undergo a morphogenetic transition from yeast to filamentous forms allows it to penetrate tissues and damage tissues, and the expression of genes associated with a number of pathogenetic mechanisms is also coordinately regulated with the yeast-to-hypha conversion. Therefore, it is widely considered that filamentation represents one of the main virulence factors of C. albicans. We have previously identified N-[3-(allyloxy)-phenyl]-4-methoxybenzamide (compound 9029936) as the lead compound in a series of small-molecule inhibitors of C. albicans filamentation and characterized its activity both in vitro and in vivo. This compound appears to be a promising candidate for the development of alternative antivirulence strategies for the treatment of C. albicans infections. In this study, we performed RNA sequencing analysis of samples obtained from C. albicans cells grown under filament-inducing conditions in the presence or absence of this compound. Overall, treatment with compound 9029936 resulted in 618 upregulated and 702 downregulated genes. Not surprisingly, some of the most downregulated genes included well-characterized genes associated with filamentation and virulence such as SAP5, ECE1 (candidalysin), and ALS3, as well as genes that impact metal chelation and utilization. Gene ontology analysis revealed an overrepresentation of cell adhesion, iron transport, filamentation, biofilm formation, and pathogenesis processes among the genes downregulated during treatment with this leading compound. Interestingly, the top upregulated genes suggested an enhancement of vesicular transport pathways, particularly those involving SNARE interactions.
IMPORTANCE These results from whole-genome transcriptional profiling provide further insights into the biological activity and mode of action of a small-molecule inhibitor of C. albicans filamentation. This information will assist in the development of novel antivirulence strategies against C. albicans infections.
INTRODUCTION
The opportunistic pathogenic fungus Candida albicans is a common member of the human microbiota (1, 2). However, this otherwise normal commensal of humans is also capable of causing a range of diseases in immunocompromised and medically compromised individuals, as well as those on heavy doses of antibiotics. Candidiasis carries mortality rates of 40 to 60% (3) and current therapeutic options are few, mostly restricted to polyenes, azoles, and echinocandins. Use of these antifungals is limited by toxicity, drug-drug interactions, and the emergence of resistance (4–6). Clearly, novel approaches to antifungal drug development are urgently needed (7, 8).
C. albicans is able to undergo reversible morphogenetic conversions between yeast and filamentous morphologies, which are intimately linked to the pathogenesis of candidiasis, as filaments can invade tissues and cause damage (9–12), and filamentation also plays a central role in biofilm formation (13–18). Moreover, a number of other pathogenetic properties of C. albicans, such as adhesive interactions and production of proteolytic enzymes and toxins, are also coordinately regulated with the morphological conversion to hyphae (1, 19–26). Altogether, it is now widely accepted that filamentation constitutes one of the main virulence factors associated with the pathogenesis of C. albicans, thereby representing an attractive, yet unexploited, target for the development of a novel antivirulence strategy for the treatment of candidiasis (7, 27–29). We have previously reported on the in vitro and in vivo activities of compound 9029936 (30, 31). This compound, with a biaryl amide core structure, was originally identified as one of the top hits in a screen of 30,000 small-molecule compounds from a commercially available chemical library in a search for inhibitors of C. albicans filamentation (30, 31). Interestingly, compound 9029936 inhibits filamentation of all C. albicans strains tested (including collection strains and clinical isolates) and in all different media used to induce the yeast-to-hypha transition, indicating that it probably impacts a common node of the multiple signaling pathways that control hypha formation under different environmental conditions (30, 31). It displays a good safety profile and potent in vivo activity in different animal models of C. albicans infections, and therefore, it represents a promising candidate for the development of novel antifungal approaches targeting virulence instead of cell proliferation or viability (as current fungistatic or fungicidal drugs do). In this study, we used RNA sequencing (RNA-seq) to investigate the impact of treatment with compound 9029936 on the whole transcriptome of C. albicans. Our results indicate that exposure of cells to the compound leads to a downregulation of genes and pathways associated with C. albicans virulence, providing further insights into the antivirulence mode of action of this small-molecule compound.
RESULTS AND DISCUSSION
Global transcriptomic changes of C. albicans in response to treatment with compound 9029936.
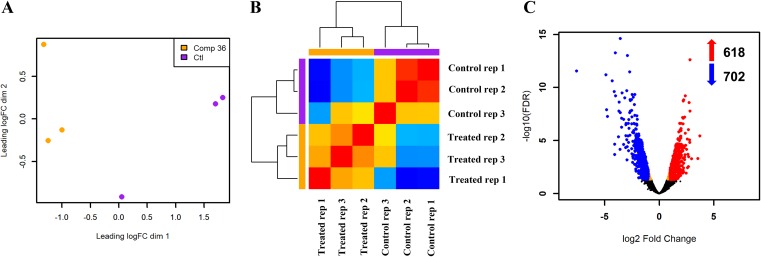
We investigated changes in C. albicans gene expression under filament-inducing conditions as a result of treatment with compound 9029936 through the use of RNA sequencing. To that end, C. albicans SC5314 was grown under strong filament-inducing conditions (yeast extract-peptone-dextrose [YPD] containing 10% fetal bovine serum [FBS] at 37°C) in the presence or absence of compound 9029936 at 5 μM. Cells were harvested after 90 min, since prior experiments have indicated that compound 9029936 exerts its inhibitory effects during the early stages of the morphological transition (30). The efficacy of treatment with this leading compound was monitored by microscopic examination, which corroborated the fact that compound 9029936 was able to block filamentation, while the untreated samples filamented normally (data not shown). Experiments were conducted in triplicate and RNA was extracted from the different cultures. Samples were then subjected to RNA sequencing using an Illumina HiSeq 2000 platform (as described in Materials and Methods). Table S1 in the supplemental material shows the sequence reads produced from all samples, with an average of 38,090,680 reads produced per sample and with 90% or more successfully mapped to the C. albicans strain SC5314 reference genome (assembly 21, downloaded from the Candida Genome Database [http://www.candidagenome.org/]). Principal-component analysis (PCA) and hierarchical clustering were applied to provide a visual representation of the transcriptomic similarities between samples treated with compound 9029936 and the untreated controls. Samples from different conditions (presence or absence of compound 9029936) clustered separately, while those from the same conditions clustered together, indicating a high level of correlation among samples, as well as distinctive transcriptome profiles (Fig. 1A and B). Analyses of the RNA sequencing data clearly indicate that compound 9029936 has a profound effect on C. albicans gene expression leading to vast alterations in the transcriptome. For this analysis, genes that showed greater than a 2-fold (up or down) change in their level of expression were considered differentially expressed, and the cutoff for statistical significance used a Benjamini-Hochberg adjusted P value of <0.05. A total of 1,320 genes showed a significant difference in expression between samples treated with 5 μM compound 9029936 and those left untreated under filament-inducing conditions (Fig. 1C). Among these differentially expressed genes (DEGs), 618 were upregulated and 702 were downregulated in the compound-treated samples (relative to the untreated controls).
FIG 1.
Overall transcriptomic changes of C. albicans growing under filament-inducing conditions in response to treatment with compound 9029936. (A) Principal-component analysis (PCA) plot showing the level of correlation and reproducibility among control untreated samples (purple) and samples treated with 5 μM compound 9029936 (orange). (B) Hierarchical-clustering heat map of gene expression data. The color scale indicates the degree of correlation (red, high correlation; blue, low correlation), while the height of the dendrogram branches represents the variability in gene expression between samples. (C) Volcano plot showing the significantly upregulated (red) and downregulated (blue) genes in samples treated with compound 9029936. A cutoff absolute value of log fold change >1 (2-fold change) was used. Adjusted P value < 0.05.
Sequenced reads obtained from the sample set from RNA sequencing experiments. Download Table S1, PDF file, 0.2 MB (252.1KB, pdf) .
Copyright © 2019 Romo et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Treatment with compound 9029936 results in downregulation of genes associated with C. albicans pathogenetic processes.
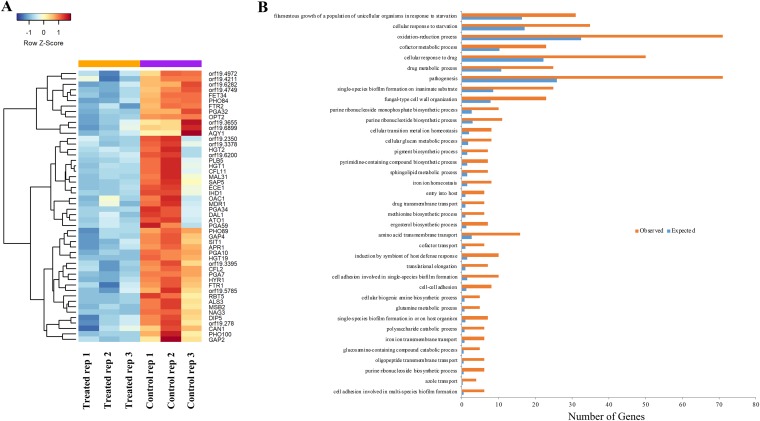
We first focused our attention on those genes whose expression was downregulated during treatment with our leading compound. Figure 2A shows a heat map of the top 50 downregulated genes. This list of most downregulated genes includes some uncharacterized open reading frames (ORFs), such as orf19.4749 and orf19.6282, whereas orf19.4972 (OFI1) encodes a putative transcription factor that has been shown to be involved in the regulation of white-opaque switching and filamentous growth (32, 33). Two genes (FET31 and FET34), encoding the major multicopper oxidases (MCOs) that form part of the high-affinity iron uptake system, are normally activated in response to iron-limited conditions inside the host and have been shown to play important roles in hyphal development and virulence (34, 35). Interestingly, FTR2 and, to a lesser extent, FTR1, encoding high-affinity iron permeases with essential roles in virulence (34, 36–38), were also in this list of most-downregulated genes, as was RBT5 (PGA1), encoding a glycosylphosphatidylinositol (GPI)-anchored cell wall protein involved in the utilization of hemin and hemoglobin for iron in the host (36, 39–41).
FIG 2.
The top 50 downregulated genes and overrepresented GO terms for biological processes downregulated in response to treatment with compound 9029936 under filament-inducing conditions. (A) Heat map displaying the top 50 downregulated genes in C. albicans cells treated with 5 μM compound 9029936 versus untreated control cultures. (B) Bar graph representation of significantly overrepresented GO terms for biological processes with downregulated genes in RNA sequencing analysis in response to treatment with 5 μM compound 9029936.
Besides iron uptake, both morphogenetic conversions and invasion of tissues by C. albicans require phosphate transporters (42), and of note, the eighth most downregulated gene was PHO84, which encodes a high-affinity phosphate transporter that intersects with the TOR pathway and was recently suggested as a potential antifungal target (43, 44). PHO84 and two other PHO genes (PHO89 and PHO100) in the 50 most significantly downregulated genes are induced during phosphate starvation (45), indicating that treatment with compound 9029936 leads to downregulation of genes associated with phosphate acquisition with important roles in oxidative stress and virulence.
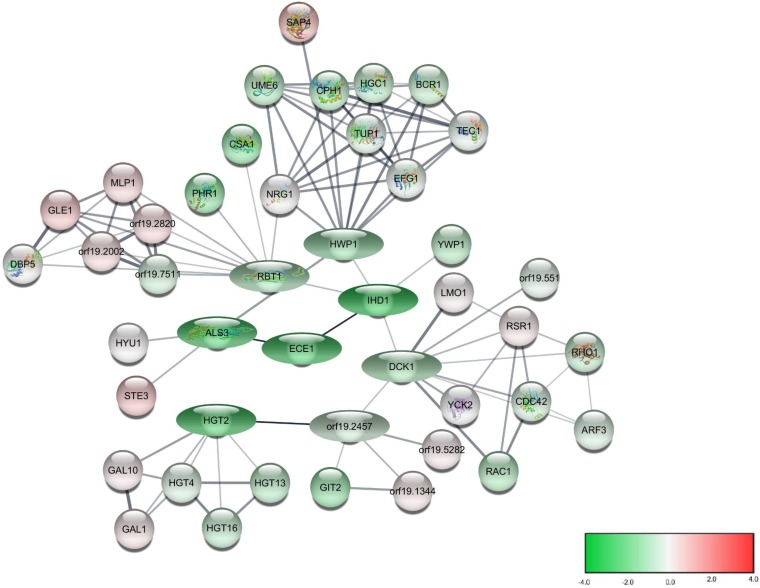
As expected from a filamentation inhibitor, featured prominently in this group of top 50 genes downregulated during treatment with compound 9029936 are well-characterized genes associated with filamentation and pathogenetic mechanisms of C. albicans (i.e., adhesion and production of proteolytic enzymes and toxins), including SAP5, and a majority of genes that form part of the C. albicans core filamentation response network (46), such as ECE1, ALS3, IHD1, and HGT2. Sap5 is a well-known secreted aspartyl proteinase important for virulence and pathogenesis during filamentation and biofilm formation by C. albicans (47–50). Ece1p, long associated with C. albicans hypha formation, with its internal small peptide candidalysin, is a vital virulence factor via which C. albicans permeabilizes host epithelial membranes, enabling tissue invasion (21). Als3 (a member of the agglutinin-like sequence adhesins) is utilized by C. albicans during filamentation for epithelial and endothelial adhesion, and null mutants are defective in these adhesive properties (51). Furthermore, Als3p is also known to play a role in iron acquisition, which, as mentioned previously, is crucial for fungal pathogenesis (19). Interestingly, although not in the top 50 list, levels of expression of all other members of the ALS gene family were significantly downregulated too. IHD1 encodes a relatively poorly characterized GPI-anchored protein induced during hypha formation (52, 53). Little information is available for HGT2, except that it constitutes part of the core filamentation response network in C. albicans (46); it is homologous to the high-affinity glucose transporter HGT1, which also plays multiple roles in virulence and in the evasion of immune defenses (54) and, interestingly, was also among the top 50 genes most downregulated by treatment with our compound. Also in this list of top 50 downregulated genes were PLB5, encoding a phospholipase with a role in virulence (55), and HYR1, encoding a prototypical hypha-specific cell wall protein (56). Other genes encoding surface adhesins, such as PGA32 (also induced in high iron), PGA59, and MSB2, were also downregulated, pointing to potential secondary effects on the cell surface as a consequence of the inhibition of hypha formation. Of note, although absent from the top 50 downregulated genes, expression of all other genes coding for the other members of the core filamentation response network was also significantly downregulated during treatment with compound 9029936. These included HWP1, encoding a hypha-specific adhesin complementary to Als3p (57), RBT1, encoding a hyphal adhesin related to Hwp1p involved in mating and filamentation (58), and DCK1, coding for a putative guanine nucleotide exchange factor required for embedded filamentous growth (59), as well as the uncharacterized orf19.2457. The data were also used to generate a protein interaction network using the STRING (Search Tool for the Retrieval of Interacting Genes) database; results are shown in Fig. 3. This figure depicts protein-protein associations for genes in the C. albicans core filamentation response, based on both known and predicted interactions of their corresponding protein products. The gradient of colors indicates differential levels of expression from RNA sequencing results for each different gene in cells treated with compound 9029936.
FIG 3.
Network visualization for predicted protein-protein interactions and expression levels of genes involved in the core filamentation response network. The STRING database is a curated knowledge database of known and predicted protein-protein associations. The genes whose protein products are known or predicted to have direct protein-protein interaction are highlighted. The eight genes identified as forming part of the C. albicans core filamentation response network are represented by ovals. All other interacting genes are represented by circles. The relative expression levels of genes in C. albicans cells treated with 5 μM compound 9029936 versus untreated control cultures are represented by a gradient of colors from green (downregulation) to red (upregulation) indicating log2 fold changes. The input genes are drawn as filled nodes when their protein structures are known or predicted, while unfilled nodes are used for the additional genes with unknown protein structures.
Importantly, the set of genes that is downregulated in response to treatment with compound 9029936 shows significant overlap with the set of genes previously shown to be strongly upregulated by C. albicans in response to serum and temperature (60), one of the strongest filament-inducing conditions. Many of these genes are involved in a diverse array of processes important for pathogenicity. These findings confirm our previous results (30, 31) suggesting that compound 9029936 may target an upstream regulator (i.e., transcription factor) that controls expression of the C. albicans filamentous growth program.
To identify larger patterns in differential gene expression and obtain overall insight into the impact of compound 9029936, Gene Ontology (GO) terms were assigned to all of the genes in the C. albicans genome and we then compared terms for the downregulated genes to a background of all terms. We found a total of 37 GO terms that were overrepresented (enriched) in this analysis (Fig. 2B). Somewhat unsurprisingly due to the antivirulence nature of the compound, filamentation, biofilm formation, cell adhesion, and pathogenesis were among the most enriched gene classes found to be downregulated. These results further support the finding that the morphogenetic transition is the main biological process affected by treatment with this compound. Other notable overrepresented GO terms for downregulated genes were those involved in response to starvation, oxidation reduction, and iron homeostasis and transport, all of which have been linked to C. albicans pathogenesis (Fig. 2B).
C. albicans displays a high degree of metabolic plasticity that greatly contributes to virulence by allowing it to rapidly adapt to different niches within the host where nutrient availability may be limited. This capacity to sense its surroundings and adapt to changing microenvironments in the human host is critical for both C. albicans survival as a commensal and as an opportunistic pathogen (61, 62). Moreover, morphological transitions in C. albicans are accompanied by changes in metabolism, giving each morphological state its own metabolic fingerprint (63–66). Thus, we also performed Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis based on the sequencing data in order to detect which metabolic pathways were impacted by treatment with compound 9029936. This analysis detected a total of 20 KEGG pathways that were significantly downregulated by treatment with our leading compound under filament-inducing conditions (Table S2). These overwhelmingly included pathways involved in carbon metabolism, biosynthesis and metabolism of amino acids, and biosynthesis of secondary metabolites. Thus, it would seem that treatment with compound 9029936 leads to the downregulation of a multitude of metabolic and biosynthetic pathways associated not only with the morphogenetic yeast-to-hypha transition but also with the pathogenesis of candidiasis, since induction of these pathways is required for C. albicans to adapt to different environments and display its full virulence potential within the host (60, 67–74). Interestingly, we did not observe an overall downregulation of genes involved in endocytosis pathways, indicating that compound 9029936 functions differently from or has a mechanism of action different from those of some previously described inhibitors of C. albicans filamentation (75).
Significantly overrepresented KEGG pathways downregulated in C. albicans cells during treatment with compound 9029936. Download Table S2, XLSX file, 0.01 MB (11.9KB, xlsx) .
Copyright © 2019 Romo et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Treatment with compound 9029936 upregulates the expression of C. albicans genes required for vesicular transport.
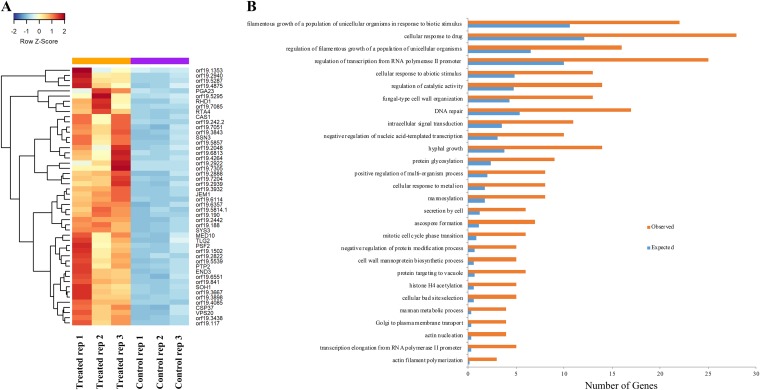
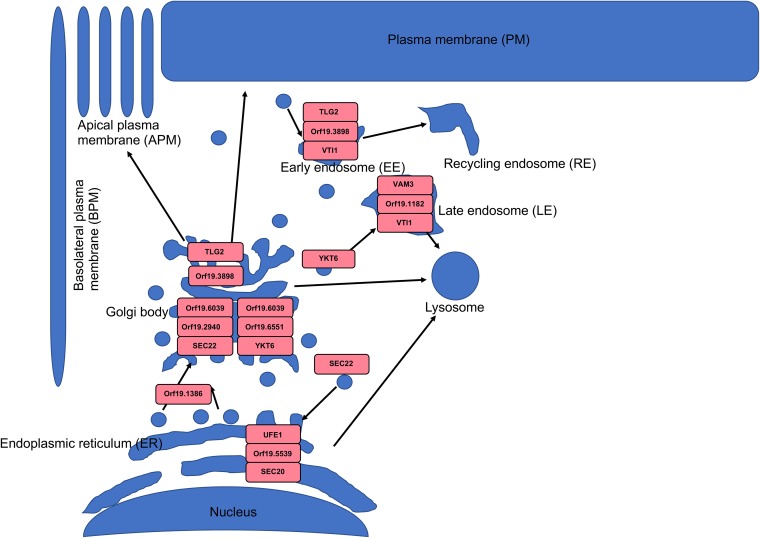
In the analysis of the top 50 genes upregulated in response to treatment with compound 9029936 under filament-inducing conditions (Fig. 4A), a large number of uncharacterized genes (indicated by open reading frame numbers) with unknown functions emerged. In fact, of the top most upregulated genes, two are completely uncharacterized (orf19.1353 and orf19.5287), while one is predicted to play a role in mRNA splicing (orf19.4875) and one is predicted to be a putative v-SNARE of the endoplasmic reticulum (ER) membrane (orf19.2940/BOS1). Interestingly, several of the products encoded by upregulated genes are involved in vesicular transport within the cell. These included Bos1p (described above), Orf19.5875p, and Tlg1p/Tlg2p, all of which are predicted to encode syntaxin-like vacuolar t-SNAREs involved in vacuolar inheritance, and Orf19.2888p, which is predicted to play a role in protein complex assembly as well as early endosome-to-Golgi transport. Additionally, Orf19.5539p is also predicted to be involved in retrograde vesicle-mediated transport as well as Golgi-to-ER transport and SNARE complexes, whereas Gos1p is predicted to play a role in vesicle transport and fusion as well as SNARE complex localization. Orf19.841p is predicted to be involved in Golgi vesicle transport and Golgi membrane localization, while Sys3p plays a role in Golgi vesicle docking. Among the others, Kip99p is predicted to have microtubule motor activity and protein homodimerization activity and plays a role in kinesin complex localization. Vps20p is involved in multivesicular body trafficking, and Sec9p is a t-SNARE protein required for secretory vesicle-membrane fusion. The theme that emerged during this analysis points to the upregulation of genes whose products play a role in vesicle transport, localization, and fusion, all of which are required for hyphal tip elongation since cell membrane and wall components are transported via motor proteins to the growing tip to meet the demands of filamentation (76, 77). This could indicate a compensatory mechanism being utilized by C. albicans in an attempt to overcome the potent inhibition of filamentation exerted by compound 9029936 or, alternatively, an attempt to remove the compound from the cell.
FIG 4.
The top 50 upregulated genes and overrepresented GO terms for biological processes upregulated in response to treatment with compound 9029936 under filament-inducing conditions. (A) Heat map displaying the top 50 upregulated genes in C. albicans cells treated with 5 μM compound 9029936 versus untreated control cultures. (B) Bar graph representation of significantly overrepresented GO terms for biological processes with upregulated genes in RNA sequencing analysis in response to treatment with 5 μM compound 9029936.
GO analysis identified 28 GO terms enriched (overrepresented) in the upregulated gene set in the presence of compound 9029936 under filament-inducing conditions. The most enriched terms included processes such as hyphal growth and filamentation in response to abiotic stimuli (most likely due to the inclusion of genes involved in the negative regulation of filamentation), cellular response to drug, regulation of transcription, and DNA repair (Fig. 4B).
As with the downregulated data set, KEGG pathway impact analysis was also performed on the list of genes upregulated by treatment with our leading compound. This analysis detected eight KEGG pathways that were significantly impacted (Table S3). Of these, SNARE interactions in vesicular transport proved to be the pathway most significantly affected. These results support our initial observation based on the heat map of upregulated DEGs (Fig. 4A). Indeed, the morphological transition from yeast to hyphae requires the constant transport and fusion of vesicles into the growing hyphal tip to deliver the components required for the expansion of the membrane as well as the necessary enzymes for cell wall synthesis (76). Furthermore, these results support our previous findings from cytological profiling experiments, in which we observed that the vacuole integrity was compromised by treatment with compound 9029936 (31). A map of SNARE interactions in vesicular transport was generated from the KEGG analysis and is shown in Fig. 5. Genes whose expression is upregulated by treatment with compound 9029936 are shown in red. It is apparent that a large proportion of the components of this pathway show enhanced gene expression in response to this treatment, which again suggests a possible compensatory mechanism used by C. albicans in an attempt to overcome the inhibitory effects of compound 9029936 on filamentation/hypha formation.
FIG 5.
Upregulation of genes in the SNARE interactions in vesicular transport pathway during treatment with compound 9029936. KEGG analysis indicated that a total of 20 genes involved in the SNARE interactions in vesicular transport pathway, indicated in red in the map, were significantly upregulated in C. albicans cells treated with 5 μM compound 9029936 versus untreated control cultures.
Significantly overrepresented KEGG pathways upregulated in C. albicans cells during treatment with compound 9029936. Download Table S3, XLSX file, 0.01 MB (11.3KB, xlsx) .
Copyright © 2019 Romo et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
In summary, this study provides a comprehensive view of transcriptomic changes associated with the treatment of C. albicans with a novel small-molecule inhibitor of filamentation. Major changes in the expression of key genes and pathways associated with C. albicans pathogenesis processes, including filamentation, iron and phosphate acquisition, metabolic processes, and vesicular transport, among others, provide further information and insight into the antivirulence mode of action of compound 9029936.
MATERIALS AND METHODS
Strains, media, and culture conditions.
The wild-type C. albicans strain SC5314 was utilized for these studies. Cell stocks were stored at –80°C, propagated by streaking onto yeast extract-peptone-dextrose (YPD) agar plates (1% [wt/vol] yeast extract, 2% [wt/vol] Bacto peptone, 2% [wt/vol] dextrose, and 1.5% agar), and incubated overnight at 30°C. From these, a loopful of cells was inoculated into flasks (150 ml) containing 25 ml of YPD liquid medium in an orbital shaker at 180 rpm and grown overnight for 14 to 16 h at 30°C. Under these conditions, C. albicans grows as a budding yeast.
Drugs.
Milligram quantities of the lead small-molecule compound 9029936 were obtained from hit resupply stocks available at Chembridge Corporation (San Diego, CA). Concentrated stock solutions were prepared in dimethyl sulfoxide (DMSO) and stored at −20°C. Working dilutions of the compound at the appropriate final concentration were prepared fresh before each experiment.
RNA isolation, purification, and sequencing.
The C. albicans SC5314 strain was grown overnight as described above, washed with phosphate-buffered saline (PBS), used to inoculate YPD containing 10% fetal bovine serum (FBS) at a 1:30 dilution, and incubated at 37°C for 90 min in the presence or absence of compound 9029936 at 5 μM as previously described (30). RNA was extracted by using a hot-acid-phenol protocol (78). Three biological replicates were obtained for each condition (treated and untreated). To determine the final RNA concentration and quality, samples were analyzed using a 2100 series bioanalyzer (Agilent Technologies, CA).
RNA sequencing was performed at the Genome Sequencing Facility at the Greehey Children’s Cancer Research Institute at the University of Texas Health Science Center at San Antonio. Briefly, cDNA libraries for RNA-seq analysis were prepared from total RNA samples using an Illumina TruSeq stranded mRNA-seq kit. RNA sequencing was performed using an Illumina HiSeq 2000 machine (San Diego, CA) to obtain 100-bp paired-end reads. After the sequencing run, demultiplexing with CASAVA was employed to generate a fastq file for each sample.
Transcriptomic analysis.
For analyses of data, the RNA sequencing reads were processed using CLC Genomics Workbench 10.0 (Qiagen). Quality trimming and adapter trimming were performed using default parameters. Reads were mapped to the C. albicans strain SC5314 reference genome (assembly 21) (http://www.candidagenome.org/). Only the uniquely mapped reads were used as the raw expression value, followed by the trimmed mean of M-values (TMM) normalization (79) to eliminate RNA composition bias. Differential gene expression profiling was carried out using the edgeR package implemented in R (79). The Benjamini and Hochberg false-discovery rate (FDR) procedure was used for multiple-hypothesis testing correction (80). Genes with FDR-adjusted P value (<0.05) and expression fold changes of more than 2 or less than −2 were considered to be differentially expressed. To validate the RNA sequencing data, we performed quantitative reverse transcription-PCR (qRT-PCR) to measure changes in the amount of mRNA of selected genes between treated and untreated samples (Fig. S1).
Confirmatory qRT-PCR analysis of hypha-specific genes differentially expressed during treatment with compound 9029936. Levels of expression of PHO84, ALS3, and HWP1 were measured by qRT-PCR in order to confirm results from RNA-seq experiments. The primers used were TTTGTTGGGTTTGTTCGTCA (forward) and GCAATAATGGCACCGACTTT (reverse) for PHO84, CAACTTGGGTTATTGAAACAAAAACA (forward) and AGAAACAGAAACCCAAGAACAACCT (reverse) for ALS3, and TCAGCCTGATGACAATCCTC (forward) and GCTGGAGTTGTTGGCTTTTC (reverse) for HWP1. **, P < 0.0015; ***, P < 0.0008; ****, P < 0.0001. Download FIG S1, PDF file, 0.2 MB (253.3KB, pdf) .
Copyright © 2019 Romo et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Functional enrichment analysis was performed on the differentially expressed genes to identify overrepresented Gene Ontology (GO) biological processes using the Panther classification system (81). Kyoto Encyclopedia of Genes and Genomes (KEGG) (82) pathway enrichment analysis was performed using KOBAS 3.0 software (83). The Benjamini and Hochberg procedure was used for multiple-testing correction for both GO and KEGG pathway analyses, with the cutoff criterion of an FDR of <0.05 (80). Protein-protein association data for Candida albicans (taxon identifier [ID] 5476) were extracted from the STRING (Search Tool for the Retrieval of Interacting Genes) database (84). STRING uses Bayesian models to integrate various sources of data, including genomic context (sequence similarity, genome organization, chromosome synteny, and phylogenetic reconstruction), gene coexpression, biochemical and genetic experimental data, pathway analysis, computational predictions, and literature text mining, to infer protein-protein associations. From a functional perspective, “association” can mean direct physical binding, but it can also mean indirect interaction, such as participation in the same cellular process. A confidence score (S) ranging from 0 to 1 was assigned to each predicted association. Cytoscape 3.7.1 (85) and StringApp (86) were used for the interactome network visualization. The relative expression levels of genes in C. albicans cells treated with 5 μM compound 9029936 versus untreated control cultures are represented in Fig. 3 by a gradient of colors from green (downregulation) to red (upregulation) for log2 fold change.
Data availability.
The RNA sequencing data were deposited into the GEO database under accession number GSE136116.
ACKNOWLEDGMENTS
This work was supported by grant R01AI119554 from the National Institute of Allergy and Infectious Diseases (to J.L.L.-R.). Additional support was provided by the Margaret Batts Tobin Foundation, San Antonio, TX. D.K. was supported by grants R01AI127692 and R21AI142560 from the National Institute of Allergy and Infectious Diseases. Y.W. was supported by grant GM100806 from the National Institute of General Medical Sciences. J.A.R. was supported by the UTSA RISE-PhD Trainee Program (NIH/NIGMS RISE GM60655). The Genome Sequencing Facility at UT Health San Antonio is supported by grant P30CA054174 from the National Cancer Institute.
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript, and the content is solely the responsibility of the authors.
REFERENCES
- 1.Calderone RA, Fonzi WA. 2001. Virulence factors of Candida albicans. Trends Microbiol 9:327–335. doi: 10.1016/S0966-842X(01)02094-7. [DOI] [PubMed] [Google Scholar]
- 2.Desai JV, Mitchell AP, Andes DR. 2014. Fungal biofilms, drug resistance, and recurrent infection. Cold Spring Harb Perspect Med 4:a019729. doi: 10.1101/cshperspect.a019729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gudlaugsson O, Gillespie S, Lee K, Vande Berg J, Hu J, Messer S, Herwaldt L, Pfaller M, Diekema D. 2003. Attributable mortality of nosocomial candidemia, revisited. Clin Infect Dis 37:1172–1177. doi: 10.1086/378745. [DOI] [PubMed] [Google Scholar]
- 4.Odds FC, Brown AJ, Gow NA. 2003. Antifungal agents: mechanisms of action. Trends Microbiol 11:272–279. doi: 10.1016/S0966-842X(03)00117-3. [DOI] [PubMed] [Google Scholar]
- 5.Arendrup MC, Perlin DS. 2014. Echinocandin resistance: an emerging clinical problem? Curr Opin Infect Dis 27:484–492. doi: 10.1097/QCO.0000000000000111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Perea S, Lopez-Ribot JL, Kirkpatrick WR, McAtee RK, Santillan RA, Martinez M, Calabrese D, Sanglard D, Patterson TF. 2001. Prevalence of molecular mechanisms of resistance to azole antifungal agents in Candida albicans strains displaying high-level fluconazole resistance isolated from human immunodeficiency virus-infected patients. Antimicrob Agents Chemother 45:2676–2684. doi: 10.1128/AAC.45.10.2676-2684.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ostrosky-Zeichner L, Casadevall A, Galgiani JN, Odds FC, Rex JH. 2010. An insight into the antifungal pipeline: selected new molecules and beyond. Nat Rev Drug Discov 9:719–727. doi: 10.1038/nrd3074. [DOI] [PubMed] [Google Scholar]
- 8.Perfect JR. 2017. The antifungal pipeline: a reality check. Nat Rev Drug Discov 16:603. doi: 10.1038/nrd.2017.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sudbery P, Gow N, Berman J. 2004. The distinct morphogenic states of Candida albicans. Trends Microbiol 12:317–324. doi: 10.1016/j.tim.2004.05.008. [DOI] [PubMed] [Google Scholar]
- 10.Uppuluri P, Pierce CG, Lopez-Ribot JL. 2009. Candida albicans biofilm formation and its clinical consequences. Future Microbiol 4:1235–1237. doi: 10.2217/fmb.09.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Whiteway M, Bachewich C. 2007. Morphogenesis in Candida albicans. Annu Rev Microbiol 61:529–553. doi: 10.1146/annurev.micro.61.080706.093341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu Y, Solis NV, Heilmann CJ, Phan QT, Mitchell AP, Klis FM, Filler SG. 2014. Role of retrograde trafficking in stress response, host cell interactions, and virulence of Candida albicans. Eukaryot Cell 13:279–287. doi: 10.1128/EC.00295-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bonhomme J, d’Enfert C. 2013. Candida albicans biofilms: building a heterogeneous, drug-tolerant environment. Curr Opin Microbiol 16:398–403. doi: 10.1016/j.mib.2013.03.007. [DOI] [PubMed] [Google Scholar]
- 14.Chandra J, Kuhn DM, Mukherjee PK, Hoyer LL, McCormick T, Ghannoum MA. 2001. Biofilm formation by the fungal pathogen Candida albicans: development, architecture, and drug resistance. J Bacteriol 183:5385–5394. doi: 10.1128/jb.183.18.5385-5394.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Desai JV, Mitchell AP. 2015. Candida albicans biofilm development and its genetic control. Microbiol Spectr 3:MB-0005-2014. doi: 10.1128/microbiolspec.MB-0005-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lopez D, Vlamakis H, Kolter R. 2010. Biofilms. Cold Spring Harb Perspect Biol 2:a000398. doi: 10.1101/cshperspect.a000398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lopez-Ribot JL. 2005. Candida albicans biofilms: more than filamentation. Curr Biol 15:R453–R455. doi: 10.1016/j.cub.2005.06.020. [DOI] [PubMed] [Google Scholar]
- 18.Mayer FL, Wilson D, Hube B. 2013. Candida albicans pathogenicity mechanisms. Virulence 4:119–128. doi: 10.4161/viru.22913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Almeida RS, Brunke S, Albrecht A, Thewes S, Laue M, Edwards JE, Filler SG, Hube B. 2008. The hyphal-associated adhesin and invasin Als3 of Candida albicans mediates iron acquisition from host ferritin. PLoS Pathog 4:e1000217. doi: 10.1371/journal.ppat.1000217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Phan QT, Myers CL, Fu Y, Sheppard DC, Yeaman MR, Welch WH, Ibrahim AS, Edwards JE Jr, Filler SG. 2007. Als3 is a Candida albicans invasin that binds to cadherins and induces endocytosis by host cells. PLoS Biol 5:e64. doi: 10.1371/journal.pbio.0050064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moyes DL, Wilson D, Richardson JP, Mogavero S, Tang SX, Wernecke J, Hofs S, Gratacap RL, Robbins J, Runglall M, Murciano C, Blagojevic M, Thavaraj S, Forster TM, Hebecker B, Kasper L, Vizcay G, Iancu SI, Kichik N, Hader A, Kurzai O, Luo T, Kruger T, Kniemeyer O, Cota E, Bader O, Wheeler RT, Gutsmann T, Hube B, Naglik JR. 2016. Candidalysin is a fungal peptide toxin critical for mucosal infection. Nature 532:64–68. doi: 10.1038/nature17625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kumamoto CA, Vinces MD. 2005. Contributions of hyphae and hypha-co-regulated genes to Candida albicans virulence. Cell Microbiol 7:1546–1554. doi: 10.1111/j.1462-5822.2005.00616.x. [DOI] [PubMed] [Google Scholar]
- 23.Aoki W, Kitahara N, Miura N, Morisaka H, Yamamoto Y, Kuroda K, Ueda M. 2011. Comprehensive characterization of secreted aspartic proteases encoded by a virulence gene family in Candida albicans. J Biochem 150:431–438. doi: 10.1093/jb/mvr073. [DOI] [PubMed] [Google Scholar]
- 24.Fallon K, Bausch K, Noonan J, Huguenel E, Tamburini P. 1997. Role of aspartic proteases in disseminated Candida albicans infection in mice. Infect Immun 65:551–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gropp K, Schild L, Schindler S, Hube B, Zipfel PF, Skerka C. 2009. The yeast Candida albicans evades human complement attack by secretion of aspartic proteases. Mol Immunol 47:465–475. doi: 10.1016/j.molimm.2009.08.019. [DOI] [PubMed] [Google Scholar]
- 26.Pichova I, Pavlickova L, Dostal J, Dolejsi E, Hruskova-Heidingsfeldova O, Weber J, Ruml T, Soucek M. 2001. Secreted aspartic proteases of Candida albicans, Candida tropicalis, Candida parapsilosis and Candida lusitaniae. Inhibition with peptidomimetic inhibitors. Eur J Biochem 268:2669–2677. doi: 10.1046/j.1432-1327.2001.02152.x. [DOI] [PubMed] [Google Scholar]
- 27.Pierce CG, Lopez-Ribot JL. 2013. Candidiasis drug discovery and development: new approaches targeting virulence for discovering and identifying new drugs. Expert Opin Drug Discov 8:1117–1126. doi: 10.1517/17460441.2013.807245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pierce CG, Srinivasan A, Ramasubramanian AK, Lopez-Ribot JL. 2015. From biology to drug development: new approaches to combat the threat of fungal biofilms. Microbiol Spectr 3:MB-0007-2014. doi: 10.1128/microbiolspec.MB-0007-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vila T, Romo JA, Pierce CG, McHardy SF, Saville SP, Lopez-Ribot JL. 2017. Targeting Candida albicans filamentation for antifungal drug development. Virulence 8:150–158. doi: 10.1080/21505594.2016.1197444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Romo JA, Pierce CG, Chaturvedi AK, Lazzell AL, McHardy SF, Saville SP, Lopez-Ribot JL. 2017. Development of anti-virulence approaches for candidiasis via a novel series of small-molecule inhibitors of Candida albicans filamentation. mBio 8:e01991-17. doi: 10.1128/mBio.01991-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Romo JA, Pierce CG, Esqueda M, Hung C-Y, Saville SP, Lopez-Ribot JL. 2018. In vitro characterization of a biaryl amide anti-virulence compound targeting Candida albicans filamentation and biofilm formation. Front Cell Infect Microbiol 8:227. doi: 10.3389/fcimb.2018.00227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Du H, Li X, Huang G, Kang Y, Zhu L. 2015. The zinc-finger transcription factor, Ofi1, regulates white-opaque switching and filamentation in the yeast Candida albicans. Acta Biochim Biophys Sin (Shanghai) 47:335–341. doi: 10.1093/abbs/gmv011. [DOI] [PubMed] [Google Scholar]
- 33.Nobile CJ, Mitchell AP. 2005. Regulation of cell-surface genes and biofilm formation by the C. albicans transcription factor Bcr1p. Curr Biol 15:1150–1155. doi: 10.1016/j.cub.2005.05.047. [DOI] [PubMed] [Google Scholar]
- 34.Chen C, Pande K, French SD, Tuch BB, Noble SM. 2011. An iron homeostasis regulatory circuit with reciprocal roles in Candida albicans commensalism and pathogenesis. Cell Host Microbe 10:118–135. doi: 10.1016/j.chom.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cheng X, Xu N, Yu Q, Ding X, Qian K, Zhao Q, Wang Y, Zhang B, Xing L, Li M. 2013. Novel insight into the expression and function of the multicopper oxidases in Candida albicans. Microbiology 159:1044–1055. doi: 10.1099/mic.0.065268-0. [DOI] [PubMed] [Google Scholar]
- 36.Noble SM. 2013. Candida albicans specializations for iron homeostasis: from commensalism to virulence. Curr Opin Microbiol 16:708–715. doi: 10.1016/j.mib.2013.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ramanan N, Wang Y. 2000. A high-affinity iron permease essential for Candida albicans virulence. Science 288:1062–1064. doi: 10.1126/science.288.5468.1062. [DOI] [PubMed] [Google Scholar]
- 38.Ding C, Festa RA, Sun TS, Wang ZY. 2014. Iron and copper as virulence modulators in human fungal pathogens. Mol Microbiol 93:10–23. doi: 10.1111/mmi.12653. [DOI] [PubMed] [Google Scholar]
- 39.Kuznets G, Vigonsky E, Weissman Z, Lalli D, Gildor T, Kauffman SJ, Turano P, Becker J, Lewinson O, Kornitzer D. 2014. A relay network of extracellular heme-binding proteins drives C. albicans iron acquisition from hemoglobin. PLoS Pathog 10:e1004407. doi: 10.1371/journal.ppat.1004407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weissman Z, Kornitzer D. 2004. A family of Candida cell surface haem-binding proteins involved in haemin and haemoglobin-iron utilization. Mol Microbiol 53:1209–1220. doi: 10.1111/j.1365-2958.2004.04199.x. [DOI] [PubMed] [Google Scholar]
- 41.Weissman Z, Shemer R, Conibear E, Kornitzer D. 2008. An endocytic mechanism for haemoglobin-iron acquisition in Candida albicans. Mol Microbiol 69:201–217. doi: 10.1111/j.1365-2958.2008.06277.x. [DOI] [PubMed] [Google Scholar]
- 42.Sascha T, Marianne K, Hyunsook P, Martin S, Filler SG, Bernhard H. 2007. In vivo and ex vivo comparative transcriptional profiling of invasive and non-invasive Candida albicans isolates identifies genes associated with tissue invasion. Mol Microbiol 63:1606–1628. doi: 10.1111/j.1365-2958.2007.05614.x. [DOI] [PubMed] [Google Scholar]
- 43.Liu NN, Flanagan PR, Zeng J, Jani NM, Cardenas ME, Moran GP, Kohler JR. 2017. Phosphate is the third nutrient monitored by TOR in Candida albicans and provides a target for fungal-specific indirect TOR inhibition. Proc Natl Acad Sci U S A 114:6346–6351. doi: 10.1073/pnas.1617799114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu N-N, Uppuluri P, Broggi A, Besold A, Ryman K, Kambara H, Solis N, Lorenz V, Qi W, Acosta-Zaldívar M, Emami SN, Bao B, An D, Bonilla FA, Sola-Visner M, Filler SG, Luo HR, Engström Y, Ljungdahl PO, Culotta VC, Zanoni I, Lopez-Ribot JL, Köhler JR. 2018. Intersection of phosphate transport, oxidative stress and TOR signalling in Candida albicans virulence. PLoS Pathog 14:e1007076. doi: 10.1371/journal.ppat.1007076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ikeh MA, Kastora SL, Day AM, Herrero-de-Dios CM, Tarrant E, Waldron KJ, Banks AP, Bain JM, Lydall D, Veal EA, MacCallum DM, Erwig LP, Brown AJ, Quinn J. 2016. Pho4 mediates phosphate acquisition in Candida albicans and is vital for stress resistance and metal homeostasis. Mol Biol Cell 27:2784–2801. doi: 10.1091/mbc.E16-05-0266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Martin R, Albrecht-Eckardt D, Brunke S, Hube B, Hünniger K, Kurzai O. 2013. A core filamentation response network in Candida albicans is restricted to eight genes. PLoS One 8:e58613. doi: 10.1371/journal.pone.0058613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sanglard D, Hube B, Monod M, Odds FC, Gow NA. 1997. A triple deletion of the secreted aspartyl proteinase genes SAP4, SAP5, and SAP6 of Candida albicans causes attenuated virulence. Infect Immun 65:3539–3546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Naglik JR, Challacombe SJ, Hube B. 2003. Candida albicans secreted aspartyl proteinases in virulence and pathogenesis. Microbiol Mol Biol Rev 67:400–428. doi: 10.1128/MMBR.67.3.400-428.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Naglik JR, Newport G, White TC, Fernandes-Naglik LL, Greenspan JS, Greenspan D, Sweet SP, Challacombe SJ, Agabian N. 1999. In vivo analysis of secreted aspartyl proteinase expression in human oral candidiasis. Infect Immun 67:2482–2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.White TC, Miyasaki SH, Agabian N. 1993. Three distinct secreted aspartyl proteinases in Candida albicans. J Bacteriol 175:6126–6133. doi: 10.1128/jb.175.19.6126-6133.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hoyer LL, Payne TL, Hecht JE. 1998. Identification of Candida albicans ALS2 and ALS4 and localization of als proteins to the fungal cell surface. J Bacteriol 180:5334–5343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.De Groot PW, Hellingwerf KJ, Klis FM. 2003. Genome-wide identification of fungal GPI proteins. Yeast 20:781–796. doi: 10.1002/yea.1007. [DOI] [PubMed] [Google Scholar]
- 53.Nantel A, Dignard D, Bachewich C, Harcus D, Marcil A, Bouin AP, Sensen CW, Hogues H, van Het Hoog M, Gordon P, Rigby T, Benoit F, Tessier DC, Thomas DY, Whiteway M. 2002. Transcription profiling of Candida albicans cells undergoing the yeast-to-hyphal transition. Mol Biol Cell 13:3452–3465. doi: 10.1091/mbc.e02-05-0272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lesiak-Markowicz I, Vogl G, Schwarzmüller T, Speth C, Lass-Flörl C, Dierich MP, Kuchler K, Würzner R. 2011. Candida albicans Hgt1p, a multifunctional evasion molecule: complement inhibitor, CR3 analogue, and human immunodeficiency virus-binding molecule. J Infect Dis 204:802–809. doi: 10.1093/infdis/jir455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Theiss S, Ishdorj G, Brenot A, Kretschmar M, Lan CY, Nichterlein T, Hacker J, Nigam S, Agabian N, Kohler GA. 2006. Inactivation of the phospholipase B gene PLB5 in wild-type Candida albicans reduces cell-associated phospholipase A2 activity and attenuates virulence. Int J Med Microbiol 296:405–420. doi: 10.1016/j.ijmm.2006.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bailey DA, Feldmann PJ, Bovey M, Gow NA, Brown AJ. 1996. The Candida albicans HYR1 gene, which is activated in response to hyphal development, belongs to a gene family encoding yeast cell wall proteins. J Bacteriol 178:5353–5360. doi: 10.1128/jb.178.18.5353-5360.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Staab JF, Bradway SD, Fidel PL, Sundstrom P. 1999. Adhesive and mammalian transglutaminase substrate properties of Candida albicans Hwp1. Science 283:1535–1538. doi: 10.1126/science.283.5407.1535. [DOI] [PubMed] [Google Scholar]
- 58.Ene IV, Bennett RJ. 2009. Hwp1 and related adhesins contribute to both mating and biofilm formation in Candida albicans. Eukaryot Cell 8:1909–1913. doi: 10.1128/EC.00245-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hope H, Bogliolo S, Arkowitz RA, Bassilana M. 2008. Activation of Rac1 by the guanine nucleotide exchange factor Dck1 is required for invasive filamentous growth in the pathogen Candida albicans. Mol Biol Cell 19:3638–3651. doi: 10.1091/mbc.e07-12-1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kadosh D, Johnson AD. 2005. Induction of the Candida albicans filamentous growth program by relief of transcriptional repression: a genome-wide analysis. Mol Biol Cell 16:2903–2912. doi: 10.1091/mbc.e05-01-0073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kadosh D, Lopez-Ribot JL. 2013. Candida albicans: adapting to succeed. Cell Host Microbe 14:483–485. doi: 10.1016/j.chom.2013.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Miramon P, Lorenz MC. 2017. A feast for Candida: metabolic plasticity confers an edge for virulence. PLoS Pathog 13:e1006144. doi: 10.1371/journal.ppat.1006144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Han T-L, Cannon RD, Villas-Bôas SG. 2012. The metabolic response of Candida albicans to farnesol under hyphae-inducing conditions. FEMS Yeast Res 12:879–889. doi: 10.1111/j.1567-1364.2012.00837.x. [DOI] [PubMed] [Google Scholar]
- 64.Han T-L, Cannon RD, Villas-Bôas SG. 2011. The metabolic basis of Candida albicans morphogenesis and quorum sensing. Fungal Genet Biol 48:747–763. doi: 10.1016/j.fgb.2011.04.002. [DOI] [PubMed] [Google Scholar]
- 65.Deveau A, Hogan DA. 2011. Linking quorum sensing regulation and biofilm formation by Candida albicans. Methods Mol Biol 692:219–233. doi: 10.1007/978-1-60761-971-0_16. [DOI] [PubMed] [Google Scholar]
- 66.Han T, Cannon RD, Villas-Bôas SG. 2012. Metabolome analysis during the morphological transition of Candida albicans. Metabolomics 8:1204. doi: 10.1007/s11306-012-0416-6. [DOI] [Google Scholar]
- 67.Rodaki A, Young T, Brown AJ. 2006. Effects of depleting the essential central metabolic enzyme fructose-1,6-bisphosphate aldolase on the growth and viability of Candida albicans: implications for antifungal drug target discovery. Eukaryot Cell 5:1371–1377. doi: 10.1128/EC.00115-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vylkova S, Carman AJ, Danhof HA, Collette JR, Zhou H, Lorenz MC. 2011. The fungal pathogen Candida albicans autoinduces hyphal morphogenesis by raising extracellular pH. mBio 2:e00055-11. doi: 10.1128/mBio.00055-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Priest SJ, Lorenz MC. 2015. Characterization of virulence-related phenotypes in Candida species of the CUG clade. Eukaryot Cell 14:931–940. doi: 10.1128/EC.00062-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Martinez P, Ljungdahl PO. 2004. An ER packaging chaperone determines the amino acid uptake capacity and virulence of Candida albicans. Mol Microbiol 51:371–384. doi: 10.1046/j.1365-2958.2003.03845.x. [DOI] [PubMed] [Google Scholar]
- 71.Martinez P, Ljungdahl PO. 2005. Divergence of Stp1 and Stp2 transcription factors in Candida albicans places virulence factors required for proper nutrient acquisition under amino acid control. Mol Cell Biol 25:9435–9446. doi: 10.1128/MCB.25.21.9435-9446.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zakikhany K, Naglik JR, Schmidt-Westhausen A, Holland G, Schaller M, Hube B. 2007. In vivo transcript profiling of Candida albicans identifies a gene essential for interepithelial dissemination. Cell Microbiol 9:2938–2954. doi: 10.1111/j.1462-5822.2007.01009.x. [DOI] [PubMed] [Google Scholar]
- 73.Lorenz MC, Bender JA, Fink GR. 2004. Transcriptional response of Candida albicans upon internalization by macrophages. Eukaryot Cell 3:1076–1087. doi: 10.1128/EC.3.5.1076-1087.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Brown AJ, Brown GD, Netea MG, Gow NA. 2014. Metabolism impacts upon Candida immunogenicity and pathogenicity at multiple levels. Trends Microbiol 22:614–622. doi: 10.1016/j.tim.2014.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bar-Yosef H, Vivanco Gonzalez N, Ben-Aroya S, Kron SJ, Kornitzer D. 2017. Chemical inhibitors of Candida albicans hyphal morphogenesis target endocytosis. Sci Rep 7:5692. doi: 10.1038/s41598-017-05741-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sudbery PE. 2011. Growth of Candida albicans hyphae. Nat Rev Microbiol 9:737–748. doi: 10.1038/nrmicro2636. [DOI] [PubMed] [Google Scholar]
- 77.Sudbery PE. 2001. The germ tubes of Candida albicans hyphae and pseudohyphae show different patterns of septin ring localization. Mol Microbiol 41:19–31. doi: 10.1046/j.1365-2958.2001.02459.x. [DOI] [PubMed] [Google Scholar]
- 78.Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. 1994. Current protocols in molecular biology. Wiley, New York, NY. [Google Scholar]
- 79.Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B 57:289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x. [DOI] [Google Scholar]
- 81.Mi H, Muruganujan A, Casagrande JT, Thomas PD. 2013. Large-scale gene function analysis with the PANTHER classification system. Nat Protoc 8:1551–1566. doi: 10.1038/nprot.2013.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K. 2017. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res 45:D353–D361. doi: 10.1093/nar/gkw1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Xie C, Mao X, Huang J, Ding Y, Wu J, Dong S, Kong L, Gao G, Li CY, Wei L. 2011. KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res 39:W316–W322. doi: 10.1093/nar/gkr483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork P, Jensen LJ, Mering CV. 2019. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 47:D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. 2003. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Doncheva NT, Morris JH, Gorodkin J, Jensen LJ. 2019. Cytoscape StringApp: network analysis and visualization of proteomics data. J Proteome Res 18:623–632. doi: 10.1021/acs.jproteome.8b00702. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sequenced reads obtained from the sample set from RNA sequencing experiments. Download Table S1, PDF file, 0.2 MB (252.1KB, pdf) .
Copyright © 2019 Romo et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Significantly overrepresented KEGG pathways downregulated in C. albicans cells during treatment with compound 9029936. Download Table S2, XLSX file, 0.01 MB (11.9KB, xlsx) .
Copyright © 2019 Romo et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Significantly overrepresented KEGG pathways upregulated in C. albicans cells during treatment with compound 9029936. Download Table S3, XLSX file, 0.01 MB (11.3KB, xlsx) .
Copyright © 2019 Romo et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Confirmatory qRT-PCR analysis of hypha-specific genes differentially expressed during treatment with compound 9029936. Levels of expression of PHO84, ALS3, and HWP1 were measured by qRT-PCR in order to confirm results from RNA-seq experiments. The primers used were TTTGTTGGGTTTGTTCGTCA (forward) and GCAATAATGGCACCGACTTT (reverse) for PHO84, CAACTTGGGTTATTGAAACAAAAACA (forward) and AGAAACAGAAACCCAAGAACAACCT (reverse) for ALS3, and TCAGCCTGATGACAATCCTC (forward) and GCTGGAGTTGTTGGCTTTTC (reverse) for HWP1. **, P < 0.0015; ***, P < 0.0008; ****, P < 0.0001. Download FIG S1, PDF file, 0.2 MB (253.3KB, pdf) .
Copyright © 2019 Romo et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data Availability Statement
The RNA sequencing data were deposited into the GEO database under accession number GSE136116.