We describe the complete genome assembly and sequence of a clinical Enterobacter isolate harboring both blaVIM-4 and mcr-9 recovered from a pediatric patient in the United States with a history of travel to Egypt. Moreover, to the best of our knowledge, this is the first report of an Enterobacter isolate harboring both blaVIM-4 and mcr-9 from the United States. The blaVIM-4 and mcr-9 genes are carried on the same IncH12 plasmid, pME-1a. The isolate tested susceptible to colistin, without observed induction of colistin resistance. The mcr-9 gene is located between two insertion elements, IS903 and IS1, but lacks the downstream regulatory genes (qseC and qseB) found in other isolates that harbor mcr-9.
KEYWORDS: carbapenem, carbapenemase, colistin, Enterobacter, mcr-9, VIM, VIM-4
ABSTRACT
An Enterobacter hormaechei isolate harboring blaVIM-4 and mcr-9 was recovered from a pediatric patient in a U.S. hospital. The blaVIM-4 and mcr-9 genes are carried on the same IncH12 plasmid, pME-1a. The isolate tested susceptible to colistin, without observed induction of colistin resistance. The mcr-9 gene is located between two insertion elements, IS903 and IS1, but lacks the downstream regulatory genes (qseC and qseB) found in other isolates that harbor mcr-9.
IMPORTANCE We describe the complete genome assembly and sequence of a clinical Enterobacter isolate harboring both blaVIM-4 and mcr-9 recovered from a pediatric patient in the United States with a history of travel to Egypt. Moreover, to the best of our knowledge, this is the first report of an Enterobacter isolate harboring both blaVIM-4 and mcr-9 from the United States. The blaVIM-4 and mcr-9 genes are carried on the same IncH12 plasmid, pME-1a. The isolate tested susceptible to colistin, without observed induction of colistin resistance. The mcr-9 gene is located between two insertion elements, IS903 and IS1, but lacks the downstream regulatory genes (qseC and qseB) found in other isolates that harbor mcr-9.
OBSERVATION
The rapid spread of carbapenemase-producing Enterobacteriaceae poses a significant global public health threat as therapeutic options are limited (1, 2). Carbapenemases are β-lactamases capable of hydrolyzing carbapenems and other β-lactams, leading to resistance to one of the most efficacious classes of antibiotics. These enzymes are grouped into molecular classes A, B, and D (3). Class B metallo-β-lactamases (MBLs) are capable of hydrolyzing penicillins, cephalosporins, and carbapenems but lack the ability to hydrolyze aztreonam (3). A member of this class, Verona integron-encoded MBL (VIM), has been reported in Pseudomonas aeruginosa, Acinetobacter baumannii, and members of the Enterobacteriaceae family (4). VIM-producing Enterobacteriaceae have been reported in several countries, especially in the Mediterranean region (4), including isolates from Kuwait (5) and the United Arab Emirates (4), but have rarely been described in the northeastern United States, where Klebsiella pneumoniae carbapenemase (KPC) is the predominate carbapenemase (1). As of 28 May 2019, 62 variants of blaVIM have been reported in the National Database of Antibiotic-Resistant Organisms (NDARO) (https://www.ncbi.nlm.nih.gov/pathogens/antimicrobial-resistance/). Here, we report the comprehensive characterization of a clinical Enterobacter hormaechei isolate harboring blaVIM-4 from a New York City (NYC) patient using both the Illumina and Oxford Nanopore DNA sequencing platforms. Surprisingly, the genomic analysis identified the presence of the mcr-9 gene on the same plasmid harboring blaVIM-4.
The E. hormaechei ME-1 isolate was recovered from a 3-year-old boy with the β-thalassemia trait who had a throat infection in Egypt 1 month prior to presentation at an NYC hospital in 2018. He was treated with an intramuscular injection of penicillin, but over the next 2 weeks he developed erythema and induration around the injection site. Two weeks later, he received 1 week of cephalexin, given the concern for cellulitis. He then presented to an emergency room in NYC with fever and a poorly healing wound with a central 1-cm eschar, and a surrounding rim of erythema was observed at the site of the prior intramuscular injection. He was found to have leukocytosis and an increasing percentage of bands on his blood count differential. A skin biopsy of the wound was performed. The Gram stain from the biopsy specimen demonstrated Gram-negative rods, and culture yielded an organism that was identified as a member of the Enterobacter cloacae complex by matrix-associated laser desorption ionization–time of flight mass spectrometry (MALDI Biotyper; Bruker Daltonics, Inc., Billerica, MA, USA), later identified as E. hormaechei by whole-genome sequencing (WGS). Using broth microdilution antibiotic susceptibility testing (6), the isolate tested resistant to ceftazidime, ceftriaxone, cefuroxime, doripenem, ertapenem, imipenem, meropenem, nitrofurantoin, piperacillin-tazobactam, ceftazidime-avibactam, ceftolozane-tazobactam, and trimethoprim-sulfamethoxazole; intermediate to tobramycin; and susceptible to amikacin, aztreonam, ciprofloxacin, doxycycline, levofloxacin, minocycline, tigecycline, tetracycline, and colistin. He was discharged prior to the availability of these culture results and was treated empirically with 2 weeks of oral clindamycin. The medical team was unable to reach his family after discharge to adjust his antimicrobial therapy. However, he was seen 3 months later, where his follow-up exam revealed that his rash had fully healed with a scar despite having never received treatment for E. hormaechei, suggesting that the organism was colonizing the wound but was not a pathogen.
The isolate was assayed on a research-use-only FilmArray antimicrobial resistance (AMR) panel (BioFire Diagnostics, LLC, Salt Lake City, UT, USA) utilizing the sample-to-answer BioFire system, and tested positive for both the blaCTX-M and blaVIM genes. To better understand the genetic structure of the blaVIM-harboring element and to confidently assemble all the plasmids contained in the strain, WGS was performed using a combination of the Illumina NextSeq (Illumina, San Diego, CA, USA) and MinION (Oxford Nanopore Technologies, Oxford, United Kingdom) platforms. Together, the sequence analysis generated high-quality assemblies and completely closed genomes of the isolate and all the plasmids harbored by the isolate.
Briefly, DNA was isolated from overnight cultures using a MasterPure Gram-positive DNA purification kit as recommended by the manufacturer (Epicentre, Madison, WI, USA). Libraries were prepared for sequencing using Illumina Nextera XT kits and sequenced on an Illumina NextSeq platform with paired 150-base sequence reads. In addition, a library for MinION sequencing was prepared using the Rapid Barcoding Sequencing kit (SQK-RBK004) and loaded onto an R9.4 flow cell. The run was performed on a MinION Mk1B device, and the sequencing reads were assembled using Unicycler (7).
The isolate contained a chromosome of 4,804,295 bp (Table 1) and 4 plasmids ranging in size from 2,317 bp to 276,502 bp. The average G+C content of the chromosome was 55.6%, and the chromosome harbored 5 prophages. The chromosome of ME-1 was compared to the genomes of the type strains of various Enterobacter species and found to belong to Enterobacter hormaechei subsp. steigerwaltii clade B Hoffmann VIII (8). In silico multilocus sequence typing (MLST) analysis assigned ME-1 to sequence type (ST) ST542 (allelic profile 178-4-4-6-37-4-6). In silico mining of antibiotic resistance genes revealed that the chromosome of ME-1 harbored blaACT-7, an intrinsic AmpC enzyme, and fosA, which confer resistance to some β-lactams and fosfomycin, respectively.
TABLE 1.
Key features of chromosome and plasmids harbored by ME-1
| Sample name | Size (bp) | MLST | Plasmid incompatibility type |
Antibiotic resistance gene(s) |
|---|---|---|---|---|
| Chromosome | 4,804,295 | ST542 | NAb | fosA, blaACT-7 |
| pME-1a | 276,520 | NA | IncHI2 |
aadA24, aadA2,a
sul1,a
dfrA1, dfrA16,a mcr-9, blaVIM-4, blaCTX-M-9, qnrA1 |
| pME-1b | 6,237 | NA | ColRNAI | NA |
| pME-1c | 2,496 | NA | ColRNAI | NA |
| pME-1d | 2,317 | NA | ColRNAI | NA |
Multiple copies on the plasmid.
NA, not applicable.
The isolate harbored a total of 4 plasmids (pME-1a, pME-1b, pME-1c, and pME-1d) belonging to incompatibility IncHI2 and ColE groups (Table 1). Analyses of acquired antibiotic resistance genes were performed using ResFinder 3.1 (9). Plasmid pME-1a harbored 12 antibiotic resistance genes conferring resistance to β-lactams (blaVIM-4 and blaCTX-M-9), aminoglycosides (aadA2 and aadA24), sulfonamide (sul1), trimethoprim (dfrA1 and dfrA16), and quinolones (qnrA1) (Fig. 1). Surprisingly, and with biological significance, a Blast analysis revealed that pME-1a also harbors the newly reported colistin resistance gene mcr-9, identical to the mcr-9 gene identified in a Salmonella isolate (10). The other plasmids (pME-1b, pME-1c, and pME-1d) were not found to harbor any antibiotic resistance genes.
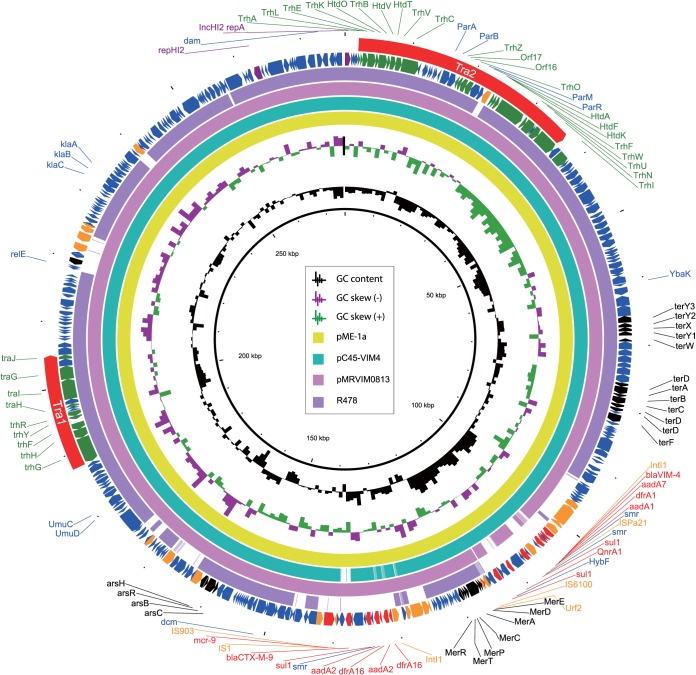
FIG 1.
Plasmid structure of blaVIM-4- and mcr-9-harboring plasmid pME-1a compared to other IncH12 plasmids. Open reading frames are indicated by arrows and are colored based on predicted gene function: purple arrows indicate plasmid replication-associated genes, blue arrows indicate plasmid scaffold regions, green arrows indicate genes associated with conjugative transfer, orange arrows indicate accessory genes, and black arrows indicate heavy metal resistance genes. The regions for conjugative transfer, Tra1 and Tra2, are indicated by red arrows on the outer circle.
Plasmid pME-1a is 276,520 bp in length with an average G+C content of 46.3% and encodes 356 predicted open reading frames (Fig. 1). Full plasmid sequence queries of pME-1a against the NCBI GenBank (https://www.ncbi.nlm.nih.gov/genbank/) showed that it has high similarity to other IncH12 plasmids, e.g., pMRVIM0813 (98% query coverage and 99.99% sequence identity; GenBank accession number KP975077), pC45-VIM4 (98% query coverage and 99.98% sequence identity; GenBank accession number LT991958), and IncHI2 prototype plasmid R478 (79% query coverage and 99.97% sequence identity; GenBank accession number BX664015) (11). Similarly to plasmid R478, the conjugative transfer genes in pME-1a are also present in two separate regions, Tra1 and Tra2, presumably facilitating the dissemination of this plasmid between various members of the Enterobacteriaceae (Fig. 1) (11). Like plasmid R478, the Tra2 region in pME-1a encodes the majority of the mating pair formation (Mpf) complex, required for intracellular DNA transfer during bacterial conjugation, but there is an insertion of the IS5 family transposase (IS1182) in the parR gene (11). The Tra1 region was similar to plasmid R478, harbored few Mpf complex genes, and encoded few OriT and relaxosome components (11).
The blaVIM-4 gene was identified on an In416 integron, with the cassette structure of blaVIM-4-aacA7-dfrA1-ΔssdA1-smr-ISPa21. The same integron has been found in an IncA/C plasmid containing blaVIM-4 in Klebsiella pneumoniae, Escherichia coli, and Enterobacter isolates from an outbreak in Kuwait (5). In416 appears to be a common integron associated with the spread of blaVIM (5). In addition, we found another novel integron with the cassette array of dfrA16-aadA2-dfrA16-aadA2-smr. Further examination of the plasmid sequences of pME-1a revealed the presence of a recently identified mobilized colistin resistance (mcr) gene, mcr-9 (10). The gene mcr-9 was initially recognized by in silico screening of sequenced Salmonella enterica subsp. enterica serotype Typhimurium (S. Typhimurium) genomes which revealed its sequence similarity to the mcr-3 gene (10). The S. Typhimurium isolate harboring mcr-9 had a colistin MIC value of 2 μg/ml, which is consistent with MICs observed for strains harboring other mcr alleles (12).
In contrast, using the broth microdilution method according to Clinical and Laboratory Standards Institute guidelines (6), the isolate described here had a much lower colistin MIC value (0.12 μg/ml) (13). In pME-1a, mcr-9 is located downstream of the blaCTX-M-9 gene (Fig. 1). Inspection of the regions surrounding pME-1 mcr-9 reveal that it is located between two insertion elements, IS903 (upstream) and IS1 (downstream) (Fig. 2). Sequence query of the region surrounding mcr-9 using NCBI GenBank showed similarity to other sequenced IncHI2 plasmids from Enterobacter: pCTXM9_020038 (GenBank accession number CP031724) and pMRVIM0813 (GenBank accession number KP975077) (Fig. 2). Analysis of the gene organization surrounding mcr-9 in the S. Typhimurium HUM_TYPH_WA_10_R9_3274 isolate (10) revealed a unique cupin fold metalloprotein, WbuC, downstream of mcr-9, and interestingly, the upstream flanking region showed sequence homology to the inverted repeat region right (IRR) associated with IS903 (GenBank accession number NZ_NAAN01000063) (Fig. 2). We are unable to determine whether a complete IS903 element is upstream because the mcr-9-harboring plasmid associated with the S. Typhimurium isolate was not completely sequenced and only a short mcr-9-bearing contig (2,661 bp) is available for comparison (GenBank accession number NZ_NAAN01000063) (Fig. 2). This limitation highlights the importance of using long-read nanopore sequencing coupled with hybrid assembly to identify and monitor the transfer and rapid evolution of antibiotic resistance genes among bacteria.
FIG 2.
Comparison of the mcr-9 region harbored by plasmids pCTXM9_020038 (GenBank accession number CP031724.1), pMRVIM0813 (GenBank accession number KP975077), and pME-1a (this study) and contigs from E. coli 68A (GenBank assembly GCA_900500325.1) and S. Typhimurium HUM_TYPH_WA_10_R9_3274 (NZ_NAAN01000063) (NCBI RefSeq accession number GCF_002091095.1). Blue shading denotes regions of shared homology among different plasmids and contigs. Colored arrows indicate open reading frames, with orange, yellow, and red arrows representing plasmid backbone genes, mobile elements, and antibiotic resistance genes, respectively. Purple arrowheads flanking IS903 indicate the location of IS903 inverted repeats.
A recent study from France reported a colistin-resistant isolate, E. coli 68A, harboring mcr-9 on an IncH12 plasmid (14). Unlike E. hormaechei ME-1, the sequenced plasmid contig did not appear to harbor other antibiotic resistance genes (14). Interestingly, the analysis of the region surrounding mcr-9 revealed the presence of insertion element IS903 upstream of mcr-9, similar to the organization of pME-1a, pCTXM9_020038, and pMRVIM0813 (Fig. 2). However, unlike pME-1a, the mcr-9-harboring plasmid from E. coli 68A has a different downstream region bearing wbuC, qseC, qseB, and an ATPase gene (Fig. 2). In this study, the investigators were able to achieve clinical levels of colistin resistance by inducing the two-component system encoded by qseB and qseC using subinhibitory concentrations of colistin (14). In contrast, plasmids pME-1a, pCTXM9_020038, and pMRVIM0813 lack the qseB and qseC regulatory genes (Fig. 2). To further evaluate whether this two-component system is necessary for mcr-9 induction in ME-1, we performed an induction assay and evaluated colistin resistance as described previously (14). Using subinhibitory concentrations of colistin at (0.03 and 0.06 μg/ml), the colistin MIC values of ME-1 did not change from 0.12 μg/ml, and gene expression of mcr-9 was not elevated compared to expression in the absence of colistin (data not shown). The chromosomally carried dnaJ gene was used to normalize gene expression. These results confirm the importance of the qseB/qseC two-component system in the induction of colistin resistance mediated by mcr-9.
Consistent with the ability of mcr-9 to confer colistin resistance, Carroll and colleagues placed mcr-9 under the control of an exogenous and inducible promoter and demonstrated colistin resistance in E. coli NEB5α (10). Together, this finding suggests that the low-level expression of mcr-9 in strain ME-1 is likely due to the absence of the qseB/qseC two-component regulators. However, we cannot rule out the possibility that the two insertion elements (IS903 and IS1) that flank the gene in the pME-1a plasmid influence the expression of this resistance gene.
In summary, we describe the complete genome assembly and sequence of a clinical Enterobacter isolate harboring both blaVIM-4 and mcr-9 recovered from a pediatric patient in the United States with a history of travel to Egypt. Moreover, to the best of our knowledge, this is the first report of an Enterobacter isolate harboring both blaVIM-4 and mcr-9 from the United States. Studies are under way to better understand the role of the mcr-9 gene and the contribution of the upstream IS903 insertion in strain ME-1.
Accession number(s).
The complete nucleotide sequences of the chromosome of ME-1, pME-1a, pME-1b, pME-1c, and pME-1d were deposited as GenBank accession numbers CP041733 to CP041737, respectively.
ACKNOWLEDGMENTS
This study was supported by grants from the National Institutes of Health (grants R01AI090155 and R21AI135250 to B.N.K., K23 AI114994 to M.J.S., and 1R01AI117035 to A.C.H.).
REFERENCES
- 1.Chen L, Mathema B, Chavda KD, DeLeo FR, Bonomo RA, Kreiswirth BN. 2014. Carbapenemase-producing Klebsiella pneumoniae: molecular and genetic decoding. Trends Microbiol 22:686–696. doi: 10.1016/j.tim.2014.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bonomo RA, Burd EM, Conly J, Limbago BM, Poirel L, Segre JA, Westblade LF. 2018. Carbapenemase-producing organisms: a global scourge. Clin Infect Dis 66:1290–1297. doi: 10.1093/cid/cix893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bush K. 2013. The ABCD’s of beta-lactamase nomenclature. J Infect Chemother 19:549–559. doi: 10.1007/s10156-013-0640-7. [DOI] [PubMed] [Google Scholar]
- 4.Sonnevend Á, Ghazawi A, Yahfoufi N, Al-Baloushi A, Hashmey R, Mathew M, Tariq WZ, Pál T. 2012. VIM-4 carbapenemase-producing Enterobacter cloacae in the United Arab Emirates. Clin Microbiol Infect 18:E494–E496. doi: 10.1111/1469-0691.12051. [DOI] [PubMed] [Google Scholar]
- 5.Sonnevend A, Yahfoufi N, Ghazawi A, Jamal W, Rotimi V, Pal T. 2017. Contribution of horizontal gene transfer to the emergence of VIM-4 carbapenemase producer Enterobacteriaceae in Kuwait. Infect Drug Resist 10:469–478. doi: 10.2147/IDR.S149321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.CLSI. 2015. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard, 10th ed CLSI document M07-A10 Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 7.Foong J, Girdea M, Stavropoulos J, Brudno M. 2015. Prioritizing clinically relevant copy number variation from genetic interactions and gene function data. PLoS One 10:e0139656. doi: 10.1371/journal.pone.0139656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sutton GG, Brinkac LM, Clarke TH, Fouts DE. 2018. Enterobacter hormaechei subsp. hoffmannii subsp. nov., Enterobacter hormaechei subsp. xiangfangensis comb. nov., Enterobacter roggenkampii sp. nov., and Enterobacter muelleri is a later heterotypic synonym of Enterobacter asburiae based on computational analysis of sequenced Enterobacter genomes. F1000Res 7:521. doi: 10.12688/f1000research.14566.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, Aarestrup FM, Larsen MV. 2012. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carroll LM, Gaballa A, Guldimann C, Sullivan G, Henderson LO, Wiedmann M. 2019. Identification of novel mobilized colistin resistance gene mcr-9 in a multidrug-resistant, colistin-susceptible Salmonella enterica serotype Typhimurium isolate. mBio 10:e00853-19. doi: 10.1128/mBio.00853-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gilmour MW, Thomson NR, Sanders M, Parkhill J, Taylor DE. 2004. The complete nucleotide sequence of the resistance plasmid R478: defining the backbone components of incompatibility group H conjugative plasmids through comparative genomics. Plasmid 52:182–202. doi: 10.1016/j.plasmid.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 12.Chew KL, La MV, Lin RTP, Teo J. 2017. Colistin and polymyxin B susceptibility testing for carbapenem-resistant and mcr-positive Enterobacteriaceae: comparison of Sensititre, MicroScan, Vitek 2, and Etest with broth microdilution. J Clin Microbiol 55:2609–2616. doi: 10.1128/JCM.00268-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chavda B, Lv J, Hou M, Chavda KD, Kreiswirth BN, Feng Y, Chen L, Yu F. 2018. Coidentification of mcr-4.3 and blaNDM-1 in a clinical Enterobacter cloacae isolate from China. Antimicrob Agents Chemother 62:e00649-18. doi: 10.1128/AAC.00649-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kieffer N, Royer G, Decousser JW, Bourrel AS, Palmieri M, Ortiz De La Rosa JM, Jacquier H, Denamur E, Nordmann P, Poirel L. 2019. mcr-9, an inducible gene encoding an acquired phosphoethanolamine transferase in Escherichia coli, and its origin. Antimicrob Agents Chemother 63:e00965-19. doi: 10.1128/AAC.00965-19. [DOI] [PMC free article] [PubMed] [Google Scholar]