Summary
Bone marrow stromal cells (BMSCs) that express high levels of stem cell factor (SCF) and CXC chemokine ligand 12 (CXCL12) are one crucial component of the hematopoietic stem cell (HSC) niche. While the secreted factors produced by BMSCs to support HSCs have been well described, little is known regarding the transcriptional regulators controlling the cell fate of BMSCs and thus indirectly maintaining HSCs. BMI1 is a polycomb group protein that regulates HSCs both cell intrinsically and extrinsically, but it is unknown in which cell type and how BMI1 functions to maintain HSCs extrinsically. Here we show that Bmi1 maintains HSCs by preventing adipogenic differentiation of BMSCs. Bmi1 is highly expressed in BMSCs but becomes downregulated upon adipogenic differentiation and during aging. Deleting Bmi1 from BMSCs increased marrow adipocytes, induced HSC quiescence and depletion, and impaired hematopoiesis. We found that BMI1 repressed multiple developmental programs in BMSCs by safeguarding the repressive epigenetic marks histone H2A ubiquitylation and H3 lysine 27 trimethylation. We identified a novel adipogenic program governed by Pax3, which BMI1 repressed in BMSCs. Our results establish Bmi1 as a critical regulator of BMSC cell fate that suppresses marrow adipogenesis to create a supportive niche for HSCs.
Keywords: BMI1, mesenchymal stromal cells, hematopoietic stem cells, niche, epigenetics, polycomb, PAX3, adipocytes
Graphical Abstract

Highlights
-
•
Bmi1 prevents adipogenesis and promotes osteogenesis by bone marrow stromal cells
-
•
Adipocyte expansion after Bmi1 loss in stroma depletes hematopoietic stem cells
-
•
Loss of Bmi1 reduces repressive histone marks on target genes Ink4a/Arf and Pax3
-
•
Derepressed Pax3 after Bmi1 ablation promotes adipocytic differentiation
Nakada and colleagues show that Bmi1 is highly expressed in bone marrow stromal cells, loss of which caused marrow adipocyte expansion and hematopoietic stem cell depletion. Bmi1 deletion diminished repressive histone marks on Pax3, leading to its expression and adipogenesis. Reduced Bmi1 expression with age may contribute to age-associated increase of marrow adipocytes.
Introduction
Bone marrow stromal cells (BMSCs) are multipotent progenitor cells that form osteoblasts and adipocytes in vivo, and have trilineage potential to form osteoblasts, adipocytes, and chondrocytes in vitro. Originally identified as cells forming fibroblast-like colonies (fibroblastic colony-forming units [CFU-F]), recent studies have identified multiple markers to prospectively identify BMSCs, including CD146 in human tissues (Sacchetti et al., 2007), platelet-derived growth factor receptor (PDGFR) (Morikawa et al., 2009), Cxcl12 (Ding and Morrison, 2013, Greenbaum et al., 2013, Omatsu et al., 2010, Sugiyama et al., 2006), Scf (Ding et al., 2012), Nestin-GFP (Kunisaki et al., 2013, Mendez-Ferrer et al., 2010), Mx1-Cre (Park et al., 2012), Leptin receptor (Lepr)-Cre (Ding et al., 2012, Omatsu et al., 2014, Zhou et al., 2014), and Prx1-Cre (Greenbaum et al., 2013, Zhou et al., 2014) in mice. These markers identify a largely overlapping population of BMSCs. For instance, stromal populations that express high levels of Cxcl12 (Cxcl12-abundant reticular [CAR] cells) (Sugiyama et al., 2006) largely overlap with cells that can be identified by Prx1-Cre labeling (Greenbaum et al., 2013), Lepr-Cre labeling (Ding et al., 2012), and Nestin-GFP expression (Zhou et al., 2014). Genetic manipulations with these tools are beginning to reveal important insights into the role of bone marrow BMSCs in bone maintenance and hematopoiesis.
BMSCs are a component of the perivascular HSC niche along with endothelial cells, and express high levels of HSC growth factors (Kfoury and Scadden, 2015, Morrison and Scadden, 2014). HSCs identified by highly selective markers were found in close proximity to endothelial cells and the surrounding BMSCs (Acar et al., 2015, Chen et al., 2016, Kiel et al., 2005, Mendez-Ferrer et al., 2010, Sugiyama et al., 2006). Depletion of BMSCs in Cxcl12-diphtheria toxin receptor (DTR) or in Nestin-Cre;inducible-DTR mice reduced the numbers of HSCs in the bone marrow, affirming the importance of BMSCs in HSC maintenance (Mendez-Ferrer et al., 2010, Omatsu et al., 2010). Moreover, deletion of HSC growth factors produced by BMSCs, such as stem cell factor (SCF) or CXCL12, depleted bone marrow HSCs, establishing that BMSCs support HSCs by providing these growth factors (Ding et al., 2012, Ding and Morrison, 2013, Greenbaum et al., 2013). In addition to producing HSC growth factors, the immature state of BMSCs is also important for HSC maintenance. Accumulation of marrow adipocyte is a hallmark of aging (Li et al., 2018), and this age-related expansion of adipocytes negatively affects HSCs (Ambrosi et al., 2017). Murine caudal vertebrae is enriched in adipocytes, making this location refractory for HSC expansion (Naveiras et al., 2009). Consistently, BMSC-specific deletion of Foxc1, a transcription factor highly expressed in BMSCs, results in the accumulation of adipocytes in the marrow and severely impairs hematopoiesis and HSC maintenance (Omatsu et al., 2014). On the other hand, stromal components committed to the adipocytic fate as defined by adiponectin expression play a positive role in hematopoietic recovery after myeloablation by producing SCF, possibly as an adaptive response to the damage (Zhou et al., 2017). Thus, not only the growth factor secretome but also the differentiation status of BMSCs is intimately linked to HSC maintenance. However, little is known regarding the mechanisms that regulate fate determination of BMSCs.
The polycomb group protein BMI1 is an epigenetic regulator involved in development and cell-fate determination (Schuettengruber et al., 2017). BMI1 is a component of the polycomb repressive complex 1 (PRC1) that catalyzes the repressive histone modification H2A ubiquitination (H2Aubi) (de Napoles et al., 2004, Wang et al., 2004a). The PRC1 was previously considered to depend on another polycomb complex, the PRC2, which catalyzes repressive histone H3 lysine 27 trimethylation (H3K27me3) to associate with chromatin (Wang et al., 2004b). However, recent studies have revealed a more complex interplay between PRC1 and PRC2, whereby PRC1-catalyzed H2Aubi recruits and stabilizes PRC2 association with chromatin (Blackledge et al., 2015, Schuettengruber et al., 2017). PRC1 and PRC2 govern developmental programs in pluripotent and tissue stem cells, as exemplified by the finding that H2Aubi and H3K27me3 decorates chromatin of gene loci that encodes developmental genes, preventing precocious expression of these genes in stem cells (Azuara et al., 2006, Bernstein et al., 2006, Boyer et al., 2006, Lee et al., 2006). Consistently, Bmi1-deficient mice exhibit multiple developmental defects including skeletal transformation, neuronal and hematopoietic defects, growth retardation, and postnatal lethality (Alkema et al., 1995, Lessard and Sauvageau, 2003, Molofsky et al., 2003, Park et al., 2003, van der Lugt et al., 1994). Bmi1 regulates self-renewal of HSCs both cell intrinsically in HSCs and cell extrinsically through its maintenance of the microenvironment (Oguro et al., 2006, Park et al., 2003). Since Bmi1-deficient mice exhibit myriad developmental defects in many organs, it remains unclear in which cell type Bmi1 functions to support HSCs cell extrinsically, in addition to its cell-intrinsic role in HSC self-renewal.
Here we show that Bmi1 is required in BMSCs to maintain HSCs, by suppressing the adipogenic differentiation of BMSCs. We found that BMSCs expressed high levels of Bmi1, and its expression level was downregulated upon adipocytic differentiation and during aging. Conditional deletion of Bmi1 from BMSCs increased the numbers of adipocytes in the bone marrow, depleting HSCs and hematopoietic cells. Bmi1 was required to epigenetically repress multiple developmental programs in BMSCs, including a novel adipogenic differentiation program mediated by Pax3. Our findings establish that Bmi1 maintains the HSC niche by suppressing adipogenesis of BMSCs.
Results
Bmi1 Is Expressed in Immature Bone Marrow BMSCs and Suppresses Senescence
To begin to study how Bmi1 in bone marrow stroma cells regulates HSCs cell extrinsically, we determined Bmi1 expression levels in BMSCs (CD140+CD45−Ter119−), non-BMSC stroma cells (CD140−CD45−Ter119−), and hematopoietic cells (CD45/Ter119+) by quantitative PCR (qPCR). CD140+CD45−Ter119− BMSCs expressed high levels of HSC niche factors Kitl (encoding SCF) and Cxcl12 (Ding et al., 2012, Ding and Morrison, 2013, Greenbaum et al., 2013), BMSC markers Lepr and Pdgfra (Ding et al., 2012, Morikawa et al., 2009), and transcription factors Foxc1 and Grem1 (Omatsu et al., 2014, Worthley et al., 2015) (Figures 1A and 1B). As shown in Figure 1C, Bmi1 mRNA was also highly expressed in BMSCs compared with non-BMSC stroma cells and hematopoietic cells. To examine whether expression of Bmi1 in BMSCs changes upon lineage commitment, we isolated BMSCs from wild-type bone marrow and cultured them with or without differentiation stimuli for adipocytes or osteoblasts, and measured Bmi1 expression levels by qPCR. Bmi1 expression was increased upon osteogenic stimuli, while it was decreased in cells that were induced to undergo adipocytic differentiation (Figure 1D). These results suggest that Bmi1 is highly expressed in undifferentiated BMSCs and that reduction in Bmi1 expression accompanies commitment to the adipocytic fate.
Figure 1.
Bmi1 Is Highly Expressed in BMSCs and Suppresses Senescence
(A) Schematic showing the gating strategy to isolate BMSCs (CD140a+CD45−Ter119−), non-BMSC stroma cells (CD140a−CD45−Ter119−), and hematopoietic cells (CD45+/Ter119+).
(B and C) qPCR showing the expression of HSC niche factors and regulators (B) and Bmi1 (C) in BMSCs, other stroma cells, and hematopoietic cells (Hem) (n = 3).
(D) Bmi1 expression 3 weeks after osteogenic stimulation (left) and adipocytic induction (right) (n = 7–9).
(E) qPCR of Bmi1 expression in purified BMSCs and HSCs from Prx1-Cre;Bmi1fl/fl mice (n = 3–4).
(F) The frequencies (left) and total numbers (right) of BMSCs in Bmi1fl/fl and Prx1-Cre;Bmi1fl/fl mice (n = 6).
(G) The numbers of CFU-F per 1 × 106 bone marrow cells following Bmi1 deletion (n = 6).
(H) Passaging of Bmi1fl/fl and Prx1-Cre;Bmi1fl/fl BMSCs (n = 5).
(I and J) The frequencies of SAβ-gal+ cells (n = 5) (I) and expression of Ink4a as determined by qPCR in Bmi1fl/fl and Prx1-Cre;Bmi1fl/fl BMSCs (normalized to 18S rRNA) (J) (n = 3).
In (B–E), expression values were normalized to β-actin. All data represent mean ± standard deviation (SD). ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001 by Student's t test. ns, not significant. See also Figure S1.
To study the role of Bmi1 in BMSC function, we conditionally deleted Bmi1 using Prx1-Cre. Prx1-Cre has been shown to be expressed in CAR cells (Sugiyama et al., 2006), Lepr+ stroma cells (Ding and Morrison, 2013, Greenbaum et al., 2013), Scf-expressing HSC niche cells (Ding et al., 2012), and Nestin+ BMSCs (Mendez-Ferrer et al., 2010). Prx1-Cre;Bmi1fl/fl mice were born normally and did not exhibit any changes in body weight, fat mass, or lean mass, although the amount of circulating adiponectin was lower than those in Bmi1fl/fl mice (referred to as control) (Figure S1A). qPCR to detect Bmi1 in purified BMSCs from control Bmi1fl/fl and Prx1-Cre;Bmi1fl/fl mice showed that Bmi1 expression was significantly depleted in the BMSCs from Prx1-Cre;Bmi1fl/fl mice, confirming efficient deletion of Bmi1 (Figure 1E). In young adult mice of 2–4 months of age, we did not detect any differences in the frequencies or total numbers of BMSCs in the long bones between Bmi1fl/fl and Prx1-Cre;Bmi1fl/fl mice (Figures 1F and S1B). Consistently, enumeration of BMSCs in a CFU-F assay showed that the numbers of CFU-F were not significantly affected by Bmi1 deletion (Figure 1G). We then serially passaged BMSCs in vitro to examine the proliferative capacity of Bmi1-deficient BMSCs. Whereas wild-type BMSCs expanded significantly over three passages, Bmi1-deficient BMSCs ceased proliferation upon passage 2 and failed to expand in later passages (Figure 1H). Since Bmi1 suppresses senescence by silencing the Ink4a/Arf locus (Jacobs et al., 1999), we examined whether Bmi1-deficient BMSCs undergo senescence. Bmi1-deficient BMSC cultures exhibited significantly more senescent-associated β-gal (SAβ-gal)+ cells and had increased expression of Ink4a, a marker of senescent cells (Figures 1I–1J and S1C). These results establish that Bmi1 is highly expressed in BMSCs and suppresses senescence in vitro.
Bmi1 Suppresses Adipogenic Differentiation of BMSCs
The finding that Bmi1 expression is reduced in BMSCs upon commitment to the adipocytic fate prompted us to test whether Bmi1 deletion promotes adipocyte differentiation of BMSCs. We cultured BMSCs from Prx1-Cre;Bmi1fl/fl and control Bmi1fl/fl mice and stimulated BMSCs to undergo adipocytic, osteoblastic, or chondrocytic differentiation in vitro. While wild-type BMSCs underwent osteoblastic differentiation after 2 weeks of induction, osteoblastic differentiation of Prx1-Cre;Bmi1fl/fl BMSCs was delayed, and fewer alizarin red-positive osteoblasts were observed after 4 and 6 weeks of induction (Figures 2A and S2A). Chondrogenesis occurred in Bmi1-deficient BMSCs similar to wild-type BMSCs (Figure S2B). In contrast, the accelerated adipocytic differentiation of Prx1-Cre;Bmi1fl/fl BMSCs compared with wild-type BMSCs was observed after only 1 week of induction (Figures 2B and S2C). These changes in differentiation potential were confirmed by gene expression changes. Consistent with the accelerated adipocytic differentiation of Bmi1-deficient BMSCs, genes involved in adipogenesis such as Pparg, Fabp4, and Adipoq were increased in Bmi1-deficient BMSCs 3 weeks after induction compared with control BMSCs (Figure 2C). Conversely, genes involved in osteogenesis such as Sp7 (Osterix) and Runx2 were reduced in Bmi1-deficient BMSCs upon osteoblastic differentiation compared with control BMSCs (Figure 2D). In vitro bromodeoxyuridine (BrdU) incorporation assays revealed that Bmi1 deletion does not affect proliferation of BMSCs or adipocyte progenitors (Figure S2D). These results suggest that Bmi1 deletion primes BMSCs to undergo adipocytic differentiation.
Figure 2.
Bmi1 Deletion from BMSCs Increased Adipogenesis In Vitro
(A and B) Representative images showing reduced osteogenesis (A) and increased adipogenesis (B) in Prx1-Cre;Bmi1fl/fl BMSCs upon in vitro induction (n = 3).
(C) Expression of adipogenic genes (Pparg, Fabp4, and Adipoq, normalized to 18S rRNA) in Bmi1fl/fl and Prx1-Cre;Bmi1fl/fl BMSCs (n = 5–7) 3 weeks after induction (n = 5), as determined by qPCR.
(D) Osteogenic gene (Osterix and Runx2, normalized to 18S rRNA) expression in Bmi1fl/fl and Prx1-Cre;Bmi1fl/fl BMSCs (n = 3).
All data represent mean ± SD. ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001 by Student's t test. See also Figure S2.
We then determined whether Bmi1 deletion from BMSCs affects the bone marrow environment by regulating osteogenesis or adipogenesis in vivo. We first performed microcomputed tomography (μCT) scanning of intact femurs from young adult mice to examine the bone parameters. Bmi1 deletion from BMSCs did not affect the thicknesses, densities, or the bone volume to total volume ratios of cortical and trabecular bones or the numbers of trabecular bones (Figures 3A, 3B, and S3A–S3G), suggesting that Bmi1 is largely dispensable for bone formation and maintenance. Since Bmi1 expression was attenuated upon adipogenic stimuli (Figure 1D), we tested whether Bmi1 deletion affects adipogenesis in the bone marrow. To quantitatively analyze the adipocyte volume in the marrow, we performed μCT scanning of osmium tetroxide-stained femurs (Figures 3C and 3D). Two- to 6-month-old wild-type mice exhibited some adipocytes in the epiphyses, whereas adipocytes expanded significantly in the epiphyses and extended to the diaphyses of femurs in Prx1-Cre;Bmi1fl/fl mice (Figures 3C, 3D, and S3H). Cross-sections of the femurs stained with osmium tetroxide confirmed this finding (Figure 3E). In contrast to the expansion of adipocytes in the long bones, we did not detect significant changes in adipocyte volume in the lumbar vertebrae (Figure S3I), consistent with the selective expression of Prx1-Cre in the limbs (Logan et al., 2002). These results establish that Bmi1 is required to suppress accumulation of bone marrow adipocytes.
Figure 3.
Bmi1 Deletion from BMSCs Increased Adipogenesis In Vivo
(A and B) μCT analyses of the cortical and trabecular bone thickness (A) and bone volume/total volume ratio (BV/TV) (B) (n = 6).
(C) Representative μCT image of femurs stained with osmium tetroxide.
(D) Quantification of the adipocyte volume to total volume ratio in 2-month-old Bmi1fl/fl and Prx1-Cre;Bmi1fl/fl mice (n = 3).
(E) Representative images of femur sections stained with osmium tetroxide (n = 4).
(F) Bmi1 expression (normalized to 18S rRNA) in BMSCs isolated from aged mice (18–24 months old, n = 4) compared with those isolated from young mice (2–4 months old, n = 5). Mice of both sexes were analyzed, with representative images from female mice shown.
All data represent mean ± SD. ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001 by Student's t test. ns, not significant. See also Figure S3.
Accumulation of marrow adipocytes is a hallmark of aging (Li et al., 2018). The finding that the bone marrow of young adult Prx1-Cre;Bmi1fl/fl mice become filled with adipocytes prompted us to examine Bmi1 expression in BMSCs isolated from mice with different ages. CD140+CD45−Ter119− BMSCs were isolated from 3- to 5-month-old and 18- to 24-month-old wild-type mice and Bmi1 expression was quantified by qPCR. We found that older mice had lower Bmi1 expression in BMSCs compared with young mice (Figure 3F). These results illustrate the links between aging, adipocyte expansion, and loss of Bmi1 expression; aging is accompanied by the expansion of marrow adipocytes and loss of Bmi1 expression in BMSCs, while Bmi1 deletion from BMSCs causes precocious accumulation of marrow adipocytes.
Deletion of Bmi1 from BMSCs Reduces HSCs and Attenuates Hematopoiesis
To determine the effect of Bmi1 deletion on the hematopoietic microenvironment, we analyzed hematopoiesis in the bone marrow of Prx1-Cre;Bmi1fl/fl mice. At 2 months of age, we observed a slight reduction in bone marrow cellularity, possibly due to the expansion of bone marrow adipocytes following Bmi1 deletion. In contrast to this minor reduction at early time points, a more severe phenotype was observed at 6 months of age (Figures 4A–4H and S4A–S4D). Bone marrow cellularity significantly increased in Bmi1fl/fl mice from 2 to 6 months of age, but this increase was not observed in Prx1-Cre;Bmi1fl/fl mice, which had significantly reduced bone marrow cellularity at 6 months compared with age-matched control mice (Figure 4A). Hematopoietic progenitor cells also expanded from 2 to 6 months of age in Bmi1fl/fl mice but not in Prx1-Cre;Bmi1fl/fl mice. Overall, Bmi1 deletion resulted in significant depletion of HSCs, common lymphoid progenitors (CLPs), common myeloid progenitors (CMPs), and granulocyte macrophage progenitors (GMPs) (Figures 4B–4H and S4A–S4D). Fractionating HSCs into long- and short-term (LT and ST) HSCs using CD34 and FLT3 revealed similar depletion of HSCs (Figures 4C and S4C). As expected, Prx1-Cre did not affect the expression of Bmi1 in HSCs (Figure 1E). Multipotent progenitors (MPPs) and megakaryocyte erythroid progenitors (MEPs) also showed trends toward becoming depleted following Bmi1 deletion (Figures 4D and 4G). Total numbers of hematopoietic colony-forming cells in hind limbs were significantly reduced in 6-month-old Prx1-Cre;Bmi1fl/fl mice (Figure 4I). Consistent with the depletion of hematopoietic progenitor cells, Prx1-Cre;Bmi1fl/fl mice had reduced white blood cells counts compared with Bmi1fl/fl mice (Figures 4J–4M). Despite the significant reduction in bone marrow hematopoiesis, we did not observe extramedullary hematopoiesis in Prx1-Cre;Bmi1fl/fl mice. The numbers of HSCs and other progenitors in the spleen and the liver were not different between Bmi1fl/fl and Prx1-Cre;Bmi1fl/fl mice (Figures S4E and S4F). CFU assays to detect hematopoietic progenitor activity in the spleen and the liver also failed to detect any differences between Bmi1fl/fl and Prx1-Cre;Bmi1fl/fl mice (Figure 4N). These results establish that deleting Bmi1 from BMSCs reduces the number of HSCs and other hematopoietic stem and progenitor cells (HSPCs) in the bone marrow, thereby impairing hematopoiesis in adult mice.
Figure 4.
Deletion of Bmi1 from BMSCs Impairs Steady-State Hematopoiesis
(A) Bone marrow (BM) cellularity in 2- and 6-month-old Bmi1fl/fl and Prx1-Cre;Bmi1fl/fl mice (n = 3–4).
(B–H) Absolute numbers of CD150+CD48−lineage−Sca-1+c-kit+ HSCs (B), LT-HSCs (CD34−Flt3−lineage−Sca-1+c-kit+ cells), and ST-HSCs (CD34+Flt3−lineage−Sca-1+c-kit+ cells) (C), MPPs (D), CMPs (E), GMPs (F), MEPs (G), and CLPs (H) in Bmi1fl/fl and Prx1-Cre;Bmi1fl/fl mice at 2 and 6 months (n = 4). Data in (C) are from 6-month-old mice.
(I–M) Total numbers of hematopoietic colonies (CFU-C) formed from bone marrow cells of hind limbs (n = 3) (I). White blood cell (WBC) (J), red blood cell (RBC) (K), and platelet (PLT) (L) counts, and lineage distribution (M) in the peripheral blood (PB) of Bmi1fl/fl (Ctrl) and Prx1-Cre;Bmi1fl/fl (KO) mice at 2 and 6 months (n = 4).
(N) Numbers of hematopoietic colonies formed per 200,000 cells isolated from the spleen and liver (n = 3). Mice of both sexes were used.
All data represent mean ± SD. ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001 by Student's t test. ns, not significant. See also Figure S4.
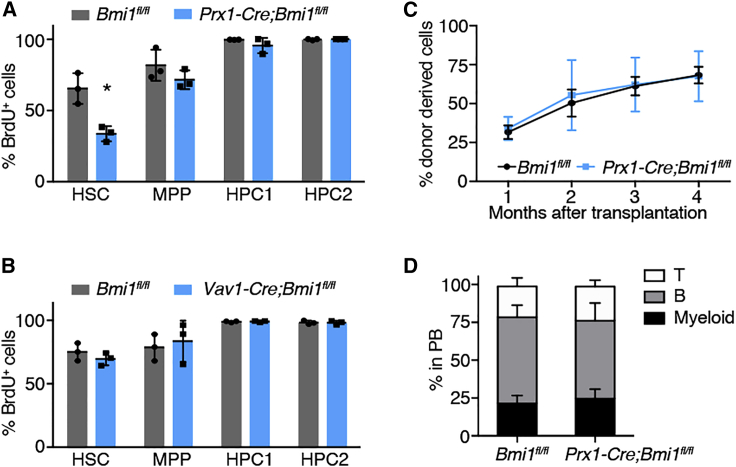
Previous reports have shown that adipocytes in the tail vertebrae promote quiescence and regenerative potential but reduce the numbers of HSCs (Naveiras et al., 2009). The expansion of adipocytes in Prx1-Cre;Bmi1fl/fl mice prompted us to examine the cell-cycle activity of HSCs in these mice by BrdU incorporation assays. Prx1-Cre;Bmi1fl/fl and control Bmi1fl/fl mice were treated with BrdU for 7 days and BrdU incorporation into HSCs was analyzed by flow cytometry. Interestingly, we found that BrdU incorporation specifically into HSCs was significantly reduced, whereas BrdU incorporation into other immature cells such as MPPs, HPC1, and HPC2 (Oguro et al., 2013) was not affected by deleting Bmi1 from BMSCs (Figures 5A and S4G). Importantly, deletion of Bmi1 from hematopoietic cells using Vav-Cre did not affect division of HSCs (Figure 5B), indicating that Bmi1 is required in the hematopoietic microenvironment, and not in HSCs themselves, to maintain HSC division. HSCs in Prx1-Cre;Bmi1fl/fl mice were capable of entering the cell cycle, since treating mice with cyclophosphamide and granulocyte-colony stimulating factor increased BrdU+ HSCs in Prx1-Cre;Bmi1fl/fl mice comparable with Bmi1fl/fl mice (Figure S4H). To examine the function of the residual HSCs, we isolated 50 HSCs from Prx1-Cre;Bmi1fl/fl and control Bmi1fl/fl mice and performed a competitive HSC transplantation assay. We found that HSCs from Prx1-Cre;Bmi1fl/fl mice do not exhibit any defects in long-term multilineage reconstitution compared with control HSCs (Figure 5C). We did not observe any differences in the lineage contribution of HSCs isolated from Prx1-Cre;Bmi1fl/fl or control Bmi1fl/fl mice (Figure 5D). Thus, deletion of Bmi1 from BMSCs causes quiescence and depletion of HSCs and reduces the numbers of many hematopoietic progenitor populations downstream of HSCs.
Figure 5.
Deletion of Bmi1 from BMSCs Causes Quiescence of HSCs
(A) Incorporation of BrdU during a 7-day pulse into HSC, MPP, HPC1, and HPC2 of Bmi1fl/fl and Prx1-Cre;Bmi1fl/fl mice (n = 3).
(B) BrdU incorporation into HSCs, MPPs, HPC1, and HPC2 in Bmi1fl/fl and Vav1-Cre;Bmi1fl/fl mice (n = 3).
(C and D) Long-term reconstitution assays with 50 HSCs isolated from Bmi1fl/fl or Prx1-Cre;Bmi1fl/fl mice (n = 3) (C). Lineage contribution from transplanted HSCs at 16 weeks after transplantation (n = 3) (D).
All data represent mean ± SD. ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001 by Student's t test.
Bmi1 Deletion Derepresses Developmental Programs in BMSCs
Since PRC1 is known to repress developmental gene expression in embryonic stem cells and tissue stem cells, we next analyzed the gene expression profiles of Bmi1fl/fl and Prx1-Cre;Bmi1fl/fl BMSCs. Freshly isolated BMSCs from Bmi1fl/fl and Prx1-Cre;Bmi1fl/fl mice were subjected to RNA sequencing (RNA-seq). Compared with whole bone marrow cells, many genes known to be expressed in BMSCs were highly enriched in our gene expression profiles, including Kitl, Cxcl12, Lepr, Pdgfra, Foxc1, and Grem1 (Figure 6A), attesting to the purity of the BMSCs we used for RNA-seq.
Figure 6.
Bmi1 Deletion Derepresses Developmental Programs in BMSCs
(A) Differentially expressed genes in BMSCs compared with whole bone marrow cells (n = 3).
(B and C) GSEA (B) and GO analysis (C) of Prx1-Cre;Bmi1fl/fl BMSCs compared with Bmi1fl/fl BMSCs (n = 3). NES, normalized enrichment score; FDR, false discovery rate.
(D) Differentially expressed genes in Prx1-Cre;Bmi1fl/fl BMSCs compared with Bmi1fl/fl BMSCs (n = 3).
(E and F) Pairwise GSEAs testing the enrichment of genes upregulated (E) or downregulated (F) in Prx1-Cre;Bmi1fl/fl BMSCs in CD45−CD31−Sca-1+CD24+ stem cell-like cells (MSCs), CD45−CD31−Sca-1−PDGFRα+ osteochondrogenic progenitor cells (OPCs), CD45−CD31−Sca-1+CD24− adipogenic progenitor cells (APCs), and CD45−CD31−Sca-1−ZFP423+ pre-adipocytes (PreAd). Left panels show the degree of enrichment in a pairwise comparison. Right panels show the cumulative enrichment scores (ES) representing the sum of enrichment scores for each cell type when compared across all other cell types (n = 3).
Data in (A) and (D) represent log2 fold change ± log2(standard error) and other data represent mean ± SD. ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001 by Student's t test.
To broadly examine the gene expression changes that occur in BMSCs after Bmi1 deletion, we performed a gene set enrichment analysis (GSEA) between wild-type and Bmi1-deficient BMSCs using the MSigDB collection of gene sets. Consistent with the role of BMI1 in the PRC1 complex, we found positive enrichment of gene sets that represent PRC2 target genes and genes with histone H3K27me3 modification in Bmi1-deficient BMSCs (Figure 6B and Table S1), indicating that polycomb target genes with H3K27me3 are being derepressed in the absence of Bmi1. Since the PRCs are involved in developmental programs in pluripotent stem cells, we assessed whether Bmi1 regulates developmental genes in BMSCs. GSEA revealed that gene ontology (GO) terms related to developmental processes, such as organ morphogenesis, pattern specification, and anterior/posterior patterning, were all enriched in Bmi1-deficient BMSCs (Figure 6C). We also identified gene sets representing fat cell differentiation and adiponectin-activated pathways as enriched in Bmi1-deficient BMSCs. Consistent with the dysregulation of developmental processes after deleting Bmi1, several transcription regulators involved in developmental processes, including posterior Hox genes (Hoxc10-Hoxc13), several Paired box transcription factors (Pax3 and Pax9), Engrailed 2 (En2), Gli1, and Sall1 were derepressed in Bmi1-deficient BMSCs (Figure 6D). These results suggest that Bmi1 is required to suppress developmental gene programs in BMSCs, possibly through its role in the PRC1 complex and in collaboration with the PRC2.
To determine whether Bmi1-deficient BMSCs exhibit transcriptional changes indicative of precocious adipogenic differentiation, we compared the expression profiles of Bmi1-deficient BMSCs with those from a recent study that identified four fractions of bone marrow stroma cells with osteogenic and/or adipogenic potential;CD45−CD31−Sca-1+CD24+ stem cell-like cells (MSCs), CD45−CD31−Sca-1−PDGFRα+ osteochondrogenic progenitor cells (OPCs), CD45−CD31−Sca-1+CD24− adipogenic progenitor cells, and CD45−CD31−Sca-1−ZFP423+ pre-adipocytes (PreAd) (Ambrosi et al., 2017). We created gene sets that are upregulated or downregulated in Bmi1-deficient BMSCs. We then compared MSCs, OPCs, APCs, and PreAd in a pairwise GSEA (Subramanian et al., 2005, Viny et al., 2015) and determined the enrichment scores of each population in regard to our custom gene sets. These analyses revealed that pre-adipocytes are enriched in genes upregulated in Bmi1-deficient BMSCs (Figure 6E), whereas osteochondrogenic progenitor cells were enriched in genes downregulated in Bmi1-deficient BMSCs (Figure 6F). These results suggest that Bmi1-deficient BMSCs share gene expression similarity with pre-adipocytes, consistent with the accelerated adipogenic and delayed osteogenic differentiation of these cells. Together, these results indicate that Bmi1 deletion initiates a precocious adipocytic differentiation program in BMSCs.
We then examined the role of Bmi1 in maintaining the epigenome of BMSCs. BMSCs were isolated from Bmi1fl/fl and Prx1-Cre;Bmi1fl/fl mice and chromatin immunoprecipitation sequencing (ChIP-seq) performed with antibodies against the PRC1 mark H2Aubi and PRC2 mark H3K27me3. The Hoxc10-Hoxc13 cluster, particularly on the centromere-proximal side with the Hoxc13 gene, was covered by the repressive H2Aubi and H3K27me3 modification in wild-type BMSCs (Figure 7A). In Bmi1-deficient BMSCs, the H2Aubi modification was reduced in this region, consistent with the notion that the Bmi1-containing PRC1 catalyzes H2A ubiquitylation (de Napoles et al., 2004, Wang et al., 2004a) (Figure 7A). Interestingly, H3K27me3 catalyzed by PRC2 was also reduced in this region in Bmi1-deficient BMSCs, suggesting that the BMI1-containing PRC1 is required to maintain the PRC2-catalyzed H3K27me3 modification on chromatin. Furthermore, we found that the Cdkn2a locus, which encodes the Ink4a/Arf tumor-suppressor genes that are known to be repressed by PRC1, was covered by H2Aubi and H3K27me2 in a Bmi1-dependent manner, consistent with the derepression of Ink4a and Arf (Figure 7B). Interestingly, among the genes that encode transcription factors involved in developmental processes, we found significant changes in both H2Aubi and H3K27me3 in the promoter region of Pax3 in Bmi1-deficient BMSCs compared with controls (Figure 7C). Bmi1 deletion decreased enrichment of H2Aubi and H3K27me3 in the Pax3 promoter region. Together with the finding that Pax3 expression is derepressed in the absence of Bmi1 (Figure 6D), this finding raised the possibility that Pax3 is a direct target of BMI1 in BMSCs.
Figure 7.
Derepressed Pax3 in the Absence of Bmi1 Promotes Adipogenic Differentiation of BMSCs
(A–C) Genomic snapshot at the Hoxc gene locus (A), Ink4a/Arf locus (B), and Pax3 locus (C) showing ChIP-seq profiles of H2Aubi and H3K27me3 in Bmi1fl/fl and Prx1-Cre;Bmi1fl/fl BMSCs (n = 3).
(D–F) Knockdown efficiency (D), changes in adipogenesis (E), and changes in osteogenesis (F) upon knockdown of Pax3 in Bmi1fl/fl and Prx1-Cre;Bmi1fl/fl mice (n = 3).
All data represent mean ± SD. ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001 by two-way ANOVA with Bonferroni post hoc tests. See also Figure S5.
Derepressed Pax3 in the Absence of Bmi1 Promotes Adipogenic Differentiation of BMSCs
PAX3 is a transcriptional regulator that integrates cell extrinsic signals to cell-fate determination, regulating multiple tissue stem/progenitor cells including neural crest stem cells, muscle satellite cells, and melanoblasts (Blake and Ziman, 2014, Lang et al., 2005, Young and Wagers, 2010). Pax3 was highly expressed in BMSCs compared with hematopoietic cells and non-BMSCs and its expression level did not change with age (Figure S5A). To determine the role of Pax3 in BMSCs, we knocked down Pax3 expression using short hairpin RNA (shRNA) and tested whether the derepressed Pax3 regulates adipogenesis in the absence of Bmi1. As controls, we also knocked down the expression of Pparg, a master regulator of fat cell fate, and Ink4a/Arf, a prototypical target of the PRC1/2 in Bmi1fl/fl and Prx1-Cre;Bmi1fl/fl mice. CD140+CD45−Ter119− BMSCs were isolated from Prx1-Cre;Bmi1fl/fl and control mice and infected with lentiviral shRNA constructs and stimulated to undergo adipogenic or osteoblastic differentiation. shRNA for these genes effectively knocked down the expression of Pax3, Pparg, and Ink4a/Arf in Bmi1-deficient BMSCs (Figures 7D, S5B, and S5C). Ink4a/Arf knockdown increased adipogenesis in both wild-type and Bmi1-deficient BMSCs (Figure S5D), suggesting that the derepressed Ink4a/Arf in Bmi1-deficient BMSCs counteracts rather than promotes adipogenesis. As expected, Pparg knockdown significantly attenuated the adipogenesis in both wild-type and Bmi1-deficient BMSCs (Figure S5E). Importantly, Pax3 knockdown also significantly blunted the increased adipogenesis observed in Bmi1-deficient BMSCs (Figures 7E and S5F), indicating that the derepressed Pax3 expression in Bmi1-deficient BMSCs is at least partly responsible for the enhanced adipogenesis. Pax3 knockdown also increased osteogenesis in both wild-type and Bmi1-deficient BMSCs (Figures 5F and S5G). Together, these results indicate that Pax3, which is repressed by BMI1 in immature BMSCs, promotes adipogenesis and suppresses osteogenesis in bone marrow BMSCs.
Discussion
Recent studies have established the role of bone marrow BMSCs as a crucial component of the HSC niche, responsible for the production of multiple secreted or membrane-bound factors to support HSCs, and elucidated their contribution toward osteogenic and adipogenic cells in the marrow (Ding et al., 2012, Ding and Morrison, 2013, Greenbaum et al., 2013, Himburg et al., 2018, Omatsu et al., 2010, Park et al., 2012, Sugiyama et al., 2006, Worthley et al., 2015, Zhou et al., 2014). However, relatively little is known of how the cell fate of BMSCs is regulated and the consequences of dysregulated BMSC cell fate on HSC maintenance. Here we show that Bmi1 is required in BMSCs to suppress adipogenic differentiation and thus maintain adult bone marrow HSCs. We found that Bmi1 is required for the repression of Pax3, which we identified as a novel adipogenic regulator of BMSCs.
Several studies have shown that Bmi1 expression is downregulated during aging in several tissues (Cordisco et al., 2010, Dhawan et al., 2009, Tschen et al., 2009). Consistent with these reports, we found that Bmi1 expression is reduced in BMSCs isolated from older mice. Furthermore, we found that Bmi1 expression was reduced upon exposing wild-type BMSCs to adipogenic stimuli, suggesting that reduced Bmi1 expression facilitates adipogenic differentiation. These studies corroborate the expansion of adipocytes in the marrow with age, known as the age-related conversion of red to yellow marrow (Li et al., 2018). As lineage-tracing studies indicate that bone marrow BMSCs are the major source of marrow adipocytes (Ambrosi et al., 2017, Zhou et al., 2014), our results are consistent with the model that Bmi1 expression is attenuated in bone marrow BMSCs with age, causing promiscuous differentiation of BMSCs to adipocytes.
Expansion of adipocytes after deleting Bmi1 from BMSCs impaired HSC niche function, reducing proliferation and the number of HSCs, suggesting that Bmi1-deficient BMSCs or increased adipocytes create a quiescent HSC niche. Prior studies have also demonstrated that adipocytes negatively regulate the numbers of murine HSCs (Ambrosi et al., 2017, Naveiras et al., 2009). The maintenance of human HSCs in vitro is also improved by co-culturing them with bone marrow adipocytes (Mattiucci et al., 2018). Interestingly, although the numbers of HSCs were depleted in Prx1-Cre;Bmi1fl/fl mice, residual HSCs were as functional as HSCs from control mice. As a consequence of HSC depletion, multiple downstream progenitor cells were also depleted and Prx1-Cre;Bmi1fl/fl mice had lower white blood cell counts compared with wild-type mice. The effects of Bmi1 deletion on peripheral blood cells were modest compared with the effects on bone marrow HSPCs, consistent with a study demonstrating that homeostatic blood production can be sustained with few HSPCs (Schoedel et al., 2016). The reduction in lymphocytes in the peripheral blood is consistent with aging; however, many changes that accompany HSC aging were not observed in Prx1-Cre;Bmi1fl/fl mice. For example, myeloid bias of HSCs after transplantation, a defining feature of HSC aging (Geiger et al., 2013), was not observed in HSCs isolated from Bmi1-deficient mice. Thus, while reduction in Bmi1 expression during aging may contribute to the age-related expansion of adipocytes in the HSC niche, it is insufficient to cause a major aging phenotype in HSCs. Additionally, whether Bmi1 reduction contributes to age-associated bone loss or BMSC loss remains unclear. Future investigation should shed light on additional age-associated changes in the niche, such as in the endothelial cells, as well as molecules produced by BMSCs that induce quiescence and aging of HSCs.
We found that deletion of Bmi1 from BMSCs reduces not only the H2Aubi mark catalyzed by the PRC1 but also the H3K27me3 mark decorated by the PRC2. These findings are consistent with the recently proposed model describing that the PRC1-catalyzed H2Aubi can stabilize PRC2 and the H3K27me3 modification on chromatin (Blackledge et al., 2015, Schuettengruber et al., 2017). Both H2Aubi and H3K27me3 were reduced on known polycomb target genes, such as the Hox and the Ink4a/Arf genes, causing derepression of these genes. We also identified Pax3, a master regulator of neural crest development (Blake and Ziman, 2014), as a potential target of BMI1/PRC1 whose promoter region is modified by H2Aubi and H3K27me2 in a Bmi1-dependent manner. Pax3 has also been found to distinguish brown adipocytes from white adipocytes (Hafner et al., 2016, Mohsen-Kanson et al., 2014), but whether Pax3 regulates adipogenesis of bone marrow BMSCs is unknown. Further work should delineate the role of Bmi1 and Pax3 in regulating the bone marrow niche for HSCs and age-related increase in adipogenesis.
Experimental Procedures
Mice
C57BL/Ka-Thy-1.2 (CD45.1) mice (8–12 weeks of age) were used for HSC transplantation assays. Bmi1fl/fl mice were described previously (Mich et al., 2014). Prx1-Cre (JAX: 005584) and Vav1-Cre (JAX: 008610) mice were obtained from The Jackson Laboratory. Age- and sex-matched pairs were analyzed in each experiment otherwise specified in figure legends. Mice were housed in AAALAC-accredited, specific-pathogen-free animal care facilities at Baylor College of Medicine (BCM). All procedures were approved by BCM Institutional Animal Care and Use Committees.
Statistics
Statistics were determined with a paired or unpaired Student's t test or ANOVA with Prism software (GraphPad). Group data represent mean ± standard deviation in all results. No randomization or blinding was used in any experiments. Experimental mice were not excluded from analysis in any experiments. Sample sizes were selected on the basis of previous experience with the degree of variance in each assay.
CFU-F Assays
One million enzymatically digested bone marrow cells were cultured per well of a 6-well plate in α-minimal essential medium (α-MEM) supplemented with 10% heat-inactivated fetal bovine serum (FBS) and 5% human platelet lysate (STEMCELL Technologies, Vancouver, Canada). After 10–14 days, the cultures were washed with PBS and stained with 1% crystal violet (C0775, Millipore Sigma, Burlington, MA) in methanol. Colonies with more than 50 cells were counted as a colony-forming unit. SAβ-gal staining was done with the Senescence β-Galactosidase Staining Kit (#9860, Cell Signaling Technologies, Danvers, MA).
Body Composition and Adiponectin Level Measurements
Body composition of 6-month-old mice was assessed by magnetic resonance imaging and circulating adiponectin levels were measured by a Mouse Adiponectin ELISA kit (EZMADP-60K, Millipore) according to the manufacturer's instructions, both by the BCM Mouse Metabolic and Phenotyping Core.
BMSC Differentiation Assays
To induce osteogenic differentiation, we cultured BMSCs in α-MEM supplemented with 20% heat-inactivated FBS, 10 nM dexamethasone (D1759), 100 mM ascorbic acid (49752), and 10 mM β-glycerophosphate (G9422, all from Millipore Sigma). For evaluation of osteogenesis, cells were fixed in 4% paraformaldehyde (PFA), washed twice with PBS, and stained with 2% Alizarin red S solution (pH 4.3; A5533, Millipore Sigma). Adipogenic differentiation was induced with Dulbecco’s modified Eagle’s medium (high glucose) supplemented with 10% heat-inactivated FBS, 0.5 mM indomethacin (I7378, Millipore Sigma), 1 μM dexamethasone, and 10 μg/mL insulin (91077C, Millipore Sigma). The differentiation medium was changed every 2–3 days. For evaluation of adipogenesis, cells were fixed in 4% PFA and stained with filtered oil red O solution for 15 min at 37°C. After washing with 60% isopropanol, cells were either imaged or oil red O extracted with 100% isopropanol and quantified by the absorption of 500 nm with a spectrophotometer. To induce chondrogenic differentiation, we cultured BMSCs in MSC chondrogenic differentiation medium (PT-3003, Lonza, Allendale, NJ) supplemented with 10 ng/mL TGFβ3 (PT-4124, Lonza) according to the manufacturer's instructions. For evaluation of chondrogenesis, cell aggregates were cryosectioned, fixed in 4% PFA, stained with toluidine blue O solution (pH 4.5), and imaged.
μCT Analysis
Femurs were fixed in 4% PFA for 48 h at room temperature, washed in PBS, and stored in 70% ethanol before scanning. To quantify bone marrow adipocytes in the bone marrow, we fixed femurs in 10% formalin at 4°C overnight and washed them in running water. Bones were decalcified in 14% EDTA for approximately 14 days. Decalcified bones were stained in 1% osmium tetroxide with 2.5% potassium dichromate for 48 h at room temperature before washing and scanning. μCT scanning was performed with the following parameters: energy of 55 kV, a diameter of 16,384 μm, an intensity of 145 μA, and a maximum isometric voxel size of 16 μm using a Scanco Medical Micro-CT-40 (Scanco Medical, Bruttisellen, Switzerland) and μCT V6.1 software. μCT scanning for osmium-stained samples was done with the voxel size of 8 μm.
Lentiviral shRNA Transduction
The pLKO shRNA plasmids were purchased from Sigma-Aldrich (St. Louis, MO). To generate lentiviral particles, we transiently transfected 293T cells with pLKO vectors with psPAX2 (12260) and pMD2.G (12259) from Addgene. Culture supernatant was collected after 48 h and used to transduce freshly isolated BMSCs for 48 h. After infection, medium was changed to α-MEM supplemented with 10% heat-inactivated FBS and 5% human platelet lysate.
HSC Transplantation
Fifty HSCs were sorted from mice and transplanted along with 200,000 competitor whole bone marrow cells from CD45.2 mice into lethally irradiated (two doses of 500 cGy) CD45.1 mice. Recipient mice were bled monthly and mononuclear cells were analyzed by flow cytometry.
Quantitative Real-Time PCR
BMSCs, stroma cells, and hematopoietic cells were sorted into TRIzol (Thermo Fisher Scientific, Waltham, MA) and RNA was isolated according to the manufacturer's instructions. cDNA was made with random primers and SuperScript IV VILO reverse transcriptase (Thermo Fisher Scientific). qPCR was performed using a ViiA7 Real-Time PCR System (Thermo Fisher Scientific). Primers are listed in Table S2.
Author Contributions
T.H., A.K., and V.L. performed most experiments. B.D. performed and analyzed CT scanning results under the supervision of B.H.L. K.A.H analyzed high-throughput sequencing data. T.H. and D.N. designed the experiments, analyzed the results, and wrote the manuscript.
Acknowledgments
This work was supported by the National Institutes of Health (NIH) (DK107413). MicroCT studies were supported by the Rolanette and Berdon Lawrence Bone Disease Program of Texas. Flow cytometry was partially supported by the NIH (NCRR grant S10RR024574, NIAID AI036211 and NCI P30CA125123) for the BCM Cytometry and Cell Sorting Core. Magnetic resonance imaging and adiponectin measurements were supported by the NIH (R01DK114356 and UM1HG006348) for the BCM Mouse Metabolic and Phenotyping Core. The authors declare no competing financial interests. We thank Catherine Gillespie for critical reading of the manuscript.
Published: June 27, 2019
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.stemcr.2019.05.027.
Accession Numbers
The RNA-seq and ChIP-seq are available at GEO with accession number GEO: GSE121288.
Supplemental Information
References
- Acar M., Kocherlakota K.S., Murphy M.M., Peyer J.G., Oguro H., Inra C.N., Jaiyeola C., Zhao Z., Luby-Phelps K., Morrison S.J. Deep imaging of bone marrow shows non-dividing stem cells are mainly perisinusoidal. Nature. 2015;526:126–130. doi: 10.1038/nature15250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alkema M.J., van der Lugt N.M., Bobeldijk R.C., Berns A., van Lohuizen M. Transformation of axial skeleton due to overexpression of bmi-1 in transgenic mice. Nature. 1995;374:724–727. doi: 10.1038/374724a0. [DOI] [PubMed] [Google Scholar]
- Ambrosi T.H., Scialdone A., Graja A., Gohlke S., Jank A.M., Bocian C., Woelk L., Fan H., Logan D.W., Schurmann A. Adipocyte accumulation in the bone marrow during obesity and aging impairs stem cell-based hematopoietic and bone regeneration. Cell Stem Cell. 2017;20:771–784.e6. doi: 10.1016/j.stem.2017.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azuara V., Perry P., Sauer S., Spivakov M., Jorgensen H.F., John R.M., Gouti M., Casanova M., Warnes G., Merkenschlager M. Chromatin signatures of pluripotent cell lines. Nat. Cell Biol. 2006;8:532–538. doi: 10.1038/ncb1403. [DOI] [PubMed] [Google Scholar]
- Bernstein B.E., Mikkelsen T.S., Xie X., Kamal M., Huebert D.J., Cuff J., Fry B., Meissner A., Wernig M., Plath K. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Blackledge N.P., Rose N.R., Klose R.J. Targeting Polycomb systems to regulate gene expression: modifications to a complex story. Nat. Rev. Mol. Cell Biol. 2015;16:643–649. doi: 10.1038/nrm4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blake J.A., Ziman M.R. Pax genes: regulators of lineage specification and progenitor cell maintenance. Development. 2014;141:737–751. doi: 10.1242/dev.091785. [DOI] [PubMed] [Google Scholar]
- Boyer L.A., Plath K., Zeitlinger J., Brambrink T., Medeiros L.A., Lee T.I., Levine S.S., Wernig M., Tajonar A., Ray M.K. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- Chen J.Y., Miyanishi M., Wang S.K., Yamazaki S., Sinha R., Kao K.S., Seita J., Sahoo D., Nakauchi H., Weissman I.L. Hoxb5 marks long-term haematopoietic stem cells and reveals a homogenous perivascular niche. Nature. 2016;530:223–227. doi: 10.1038/nature16943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordisco S., Maurelli R., Bondanza S., Stefanini M., Zambruno G., Guerra L., Dellambra E. Bmi-1 reduction plays a key role in physiological and premature aging of primary human keratinocytes. J. Invest. Dermatol. 2010;130:1048–1062. doi: 10.1038/jid.2009.355. [DOI] [PubMed] [Google Scholar]
- de Napoles M., Mermoud J.E., Wakao R., Tang Y.A., Endoh M., Appanah R., Nesterova T.B., Silva J., Otte A.P., Vidal M. Polycomb group proteins Ring1A/B link ubiquitylation of histone H2A to heritable gene silencing and X inactivation. Dev. Cell. 2004;7:663–676. doi: 10.1016/j.devcel.2004.10.005. [DOI] [PubMed] [Google Scholar]
- Dhawan S., Tschen S.I., Bhushan A. Bmi-1 regulates the Ink4a/Arf locus to control pancreatic beta-cell proliferation. Genes Dev. 2009;23:906–911. doi: 10.1101/gad.1742609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L., Morrison S.J. Haematopoietic stem cells and early lymphoid progenitors occupy distinct bone marrow niches. Nature. 2013;495:231–235. doi: 10.1038/nature11885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L., Saunders T.L., Enikolopov G., Morrison S.J. Endothelial and perivascular cells maintain haematopoietic stem cells. Nature. 2012;481:457–462. doi: 10.1038/nature10783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger H., de Haan G., Florian M.C. The ageing haematopoietic stem cell compartment. Nat. Rev. Immunol. 2013;13:376–389. doi: 10.1038/nri3433. [DOI] [PubMed] [Google Scholar]
- Greenbaum A., Hsu Y.M., Day R.B., Schuettpelz L.G., Christopher M.J., Borgerding J.N., Nagasawa T., Link D.C. CXCL12 in early mesenchymal progenitors is required for haematopoietic stem-cell maintenance. Nature. 2013;495:227–230. doi: 10.1038/nature11926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafner A.L., Contet J., Ravaud C., Yao X., Villageois P., Suknuntha K., Annab K., Peraldi P., Binetruy B., Slukvin I.I. Brown-like adipose progenitors derived from human induced pluripotent stem cells: identification of critical pathways governing their adipogenic capacity. Sci. Rep. 2016;6:32490. doi: 10.1038/srep32490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himburg H.A., Termini C.M., Schlussel L., Kan J., Li M., Zhao L., Fang T., Sasine J.P., Chang V.Y., Chute J.P. Distinct bone marrow sources of pleiotrophin control hematopoietic stem cell maintenance and regeneration. Cell Stem Cell. 2018;23:370–381.e5. doi: 10.1016/j.stem.2018.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs J.J., Kieboom K., Marino S., DePinho R.A., van Lohuizen M. The oncogene and Polycomb-group gene bmi-1 regulates cell proliferation and senescence through the ink4a locus. Nature. 1999;397:164–168. doi: 10.1038/16476. [DOI] [PubMed] [Google Scholar]
- Kfoury Y., Scadden D.T. Mesenchymal cell contributions to the stem cell niche. Cell Stem Cell. 2015;16:239–253. doi: 10.1016/j.stem.2015.02.019. [DOI] [PubMed] [Google Scholar]
- Kiel M.J., Yilmaz O.H., Iwashita T., Terhorst C., Morrison S.J. SLAM family receptors distinguish hematopoietic stem and progenitor cells and reveal endothelial Niches for stem cells. Cell. 2005;121:1109–1121. doi: 10.1016/j.cell.2005.05.026. [DOI] [PubMed] [Google Scholar]
- Kunisaki Y., Bruns I., Scheiermann C., Ahmed J., Pinho S., Zhang D., Mizoguchi T., Wei Q., Lucas D., Ito K. Arteriolar niches maintain haematopoietic stem cell quiescence. Nature. 2013;502:637–643. doi: 10.1038/nature12612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang D., Lu M.M., Huang L., Engleka K.A., Zhang M., Chu E.Y., Lipner S., Skoultchi A., Millar S.E., Epstein J.A. Pax3 functions at a nodal point in melanocyte stem cell differentiation. Nature. 2005;433:884–887. doi: 10.1038/nature03292. [DOI] [PubMed] [Google Scholar]
- Lee T.I., Jenner R.G., Boyer L.A., Guenther M.G., Levine S.S., Kumar R.M., Chevalier B., Johnstone S.E., Cole M.F., Isono K. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessard J., Sauvageau G. Bmi-1 determines the proliferative capacity of normal and leukaemic stem cells. Nature. 2003;423:255–260. doi: 10.1038/nature01572. [DOI] [PubMed] [Google Scholar]
- Li Z., Hardij J., Bagchi D.P., Scheller E.L., MacDougald O.A. Development, regulation, metabolism and function of bone marrow adipose tissues. Bone. 2018;110:134–140. doi: 10.1016/j.bone.2018.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan M., Martin J.F., Nagy A., Lobe C., Olson E.N., Tabin C.J. Expression of Cre Recombinase in the developing mouse limb bud driven by a Prxl enhancer. Genesis. 2002;33:77–80. doi: 10.1002/gene.10092. [DOI] [PubMed] [Google Scholar]
- Mattiucci D., Maurizi G., Izzi V., Cenci L., Ciarlantini M., Mancini S., Mensa E., Pascarella R., Vivarelli M., Olivieri A. Bone marrow adipocytes support hematopoietic stem cell survival. J. Cell. Physiol. 2018;233:1500–1511. doi: 10.1002/jcp.26037. [DOI] [PubMed] [Google Scholar]
- Mendez-Ferrer S., Michurina T.V., Ferraro F., Mazloom A.R., Macarthur B.D., Lira S.A., Scadden D.T., Ma'ayan A., Enikolopov G.N., Frenette P.S. Mesenchymal and haematopoietic stem cells form a unique bone marrow niche. Nature. 2010;466:829–834. doi: 10.1038/nature09262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mich J.K., Signer R.A., Nakada D., Pineda A., Burgess R.J., Vue T.Y., Johnson J.E., Morrison S.J. Prospective identification of functionally distinct stem cells and neurosphere-initiating cells in adult mouse forebrain. Elife. 2014;3:e02669. doi: 10.7554/eLife.02669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohsen-Kanson T., Hafner A.L., Wdziekonski B., Takashima Y., Villageois P., Carriere A., Svensson M., Bagnis C., Chignon-Sicard B., Svensson P.A. Differentiation of human induced pluripotent stem cells into brown and white adipocytes: role of Pax3. Stem Cells. 2014;32:1459–1467. doi: 10.1002/stem.1607. [DOI] [PubMed] [Google Scholar]
- Molofsky A.V., Pardal R., Iwashita T., Park I.K., Clarke M.F., Morrison S.J. Bmi-1 dependence distinguishes neural stem cell self-renewal from progenitor proliferation. Nature. 2003;425:962–967. doi: 10.1038/nature02060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morikawa S., Mabuchi Y., Kubota Y., Nagai Y., Niibe K., Hiratsu E., Suzuki S., Miyauchi-Hara C., Nagoshi N., Sunabori T. Prospective identification, isolation, and systemic transplantation of multipotent mesenchymal stem cells in murine bone marrow. J. Exp. Med. 2009;206:2483–2496. doi: 10.1084/jem.20091046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison S.J., Scadden D.T. The bone marrow niche for haematopoietic stem cells. Nature. 2014;505:327–334. doi: 10.1038/nature12984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naveiras O., Nardi V., Wenzel P.L., Hauschka P.V., Fahey F., Daley G.Q. Bone-marrow adipocytes as negative regulators of the haematopoietic microenvironment. Nature. 2009;460:259–263. doi: 10.1038/nature08099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oguro H., Ding L., Morrison S.J. SLAM family markers resolve functionally distinct subpopulations of hematopoietic stem cells and multipotent progenitors. Cell Stem Cell. 2013;13:102–116. doi: 10.1016/j.stem.2013.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oguro H., Iwama A., Morita Y., Kamijo T., van Lohuizen M., Nakauchi H. Differential impact of Ink4a and Arf on hematopoietic stem cells and their bone marrow microenvironment in Bmi1-deficient mice. J. Exp. Med. 2006;203:2247–2253. doi: 10.1084/jem.20052477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omatsu Y., Seike M., Sugiyama T., Kume T., Nagasawa T. Foxc1 is a critical regulator of haematopoietic stem/progenitor cell niche formation. Nature. 2014;508:536–540. doi: 10.1038/nature13071. [DOI] [PubMed] [Google Scholar]
- Omatsu Y., Sugiyama T., Kohara H., Kondoh G., Fujii N., Kohno K., Nagasawa T. The essential functions of adipo-osteogenic progenitors as the hematopoietic stem and progenitor cell niche. Immunity. 2010;33:387–399. doi: 10.1016/j.immuni.2010.08.017. [DOI] [PubMed] [Google Scholar]
- Park D., Spencer J.A., Koh B.I., Kobayashi T., Fujisaki J., Clemens T.L., Lin C.P., Kronenberg H.M., Scadden D.T. Endogenous bone marrow MSCs are dynamic, fate-restricted participants in bone maintenance and regeneration. Cell Stem Cell. 2012;10:259–272. doi: 10.1016/j.stem.2012.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park I.K., Qian D., Kiel M., Becker M.W., Pihalja M., Weissman I.L., Morrison S.J., Clarke M.F. Bmi-1 is required for maintenance of adult self-renewing haematopoietic stem cells. Nature. 2003;423:302–305. doi: 10.1038/nature01587. [DOI] [PubMed] [Google Scholar]
- Sacchetti B., Funari A., Michienzi S., Di Cesare S., Piersanti S., Saggio I., Tagliafico E., Ferrari S., Robey P.G., Riminucci M. Self-renewing osteoprogenitors in bone marrow sinusoids can organize a hematopoietic microenvironment. Cell. 2007;131:324–336. doi: 10.1016/j.cell.2007.08.025. [DOI] [PubMed] [Google Scholar]
- Schoedel K.B., Morcos M.N.F., Zerjatke T., Roeder I., Grinenko T., Voehringer D., Gothert J.R., Waskow C., Roers A., Gerbaulet A. The bulk of the hematopoietic stem cell population is dispensable for murine steady-state and stress hematopoiesis. Blood. 2016;128:2285–2296. doi: 10.1182/blood-2016-03-706010. [DOI] [PubMed] [Google Scholar]
- Schuettengruber B., Bourbon H.M., Di Croce L., Cavalli G. Genome regulation by polycomb and Trithorax: 70 years and counting. Cell. 2017;171:34–57. doi: 10.1016/j.cell.2017.08.002. [DOI] [PubMed] [Google Scholar]
- Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama T., Kohara H., Noda M., Nagasawa T. Maintenance of the hematopoietic stem cell pool by CXCL12-CXCR4 chemokine signaling in bone marrow stromal cell niches. Immunity. 2006;25:977–988. doi: 10.1016/j.immuni.2006.10.016. [DOI] [PubMed] [Google Scholar]
- Tschen S.I., Dhawan S., Gurlo T., Bhushan A. Age-dependent decline in beta-cell proliferation restricts the capacity of beta-cell regeneration in mice. Diabetes. 2009;58:1312–1320. doi: 10.2337/db08-1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Lugt N.M., Domen J., Linders K., van Roon M., Robanus-Maandag E., te Riele H., van der Valk M., Deschamps J., Sofroniew M., van Lohuizen M. Posterior transformation, neurological abnormalities, and severe hematopoietic defects in mice with a targeted deletion of the bmi-1 proto-oncogene. Genes Dev. 1994;8:757–769. doi: 10.1101/gad.8.7.757. [DOI] [PubMed] [Google Scholar]
- Viny A.D., Ott C.J., Spitzer B., Rivas M., Meydan C., Papalexi E., Yelin D., Shank K., Reyes J., Chiu A. Dose-dependent role of the cohesin complex in normal and malignant hematopoiesis. J. Exp. Med. 2015;212:1819–1832. doi: 10.1084/jem.20151317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Wang L., Erdjument-Bromage H., Vidal M., Tempst P., Jones R.S., Zhang Y. Role of histone H2A ubiquitination in Polycomb silencing. Nature. 2004;431:873–878. doi: 10.1038/nature02985. [DOI] [PubMed] [Google Scholar]
- Wang L., Brown J.L., Cao R., Zhang Y., Kassis J.A., Jones R.S. Hierarchical recruitment of polycomb group silencing complexes. Mol. Cell. 2004;14:637–646. doi: 10.1016/j.molcel.2004.05.009. [DOI] [PubMed] [Google Scholar]
- Worthley D.L., Churchill M., Compton J.T., Tailor Y., Rao M., Si Y., Levin D., Schwartz M.G., Uygur A., Hayakawa Y. Gremlin 1 identifies a skeletal stem cell with bone, cartilage, and reticular stromal potential. Cell. 2015;160:269–284. doi: 10.1016/j.cell.2014.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young A.P., Wagers A.J. Pax3 induces differentiation of juvenile skeletal muscle stem cells without transcriptional upregulation of canonical myogenic regulatory factors. J. Cell Sci. 2010;123:2632–2639. doi: 10.1242/jcs.061606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B.O., Yu H., Yue R., Zhao Z., Rios J.J., Naveiras O., Morrison S.J. Bone marrow adipocytes promote the regeneration of stem cells and haematopoiesis by secreting SCF. Nat. Cell Biol. 2017;19:891–903. doi: 10.1038/ncb3570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B.O., Yue R., Murphy M.M., Peyer J.G., Morrison S.J. Leptin-receptor-expressing mesenchymal stromal cells represent the main source of bone formed by adult bone marrow. Cell Stem Cell. 2014;15:154–168. doi: 10.1016/j.stem.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.