Summary
Induced pluripotent stem cells (iPSCs) provide a potential source for the derivation of smooth muscle cells (SMCs); however, current approaches are limited by the production of heterogeneous cell types and a paucity of tools or markers for tracking and purifying candidate SMCs. Here, we develop murine and human iPSC lines carrying fluorochrome reporters (Acta2hrGFP and ACTA2eGFP, respectively) that identify Acta2+/ACTA2+ cells as they emerge in vitro in real time during iPSC-directed differentiation. We find that Acta2hrGFP+ and ACTA2eGFP+ cells can be sorted to purity and are enriched in markers characteristic of an immature or synthetic SMC. We characterize the resulting GFP+ populations through global transcriptomic profiling and functional studies, including the capacity to form engineered cell sheets. We conclude that these reporter lines allow for generation of sortable, live iPSC-derived Acta2+/ACTA2+ cells highly enriched in smooth muscle lineages for basic developmental studies, tissue engineering, or future clinical regenerative applications.
Keywords: induced pluripotent stem cells, directed differentiation, smooth muscle cells, cell sheets, fluorochrome reporters
Graphical Abstract

Highlights
-
•
ACTA2 reporter iPSC lines allow isolation of iPSC-derived SMCs
-
•
iPSC-SMCs have transcriptomic profile reminiscent of immature or synthetic SMCs
-
•
Generated iPSC-SMC cell sheets with mechanical properties in physiological ranges
In this article, Wong, Kotton, and colleagues developed murine and human iPSC lines with fluorochrome reporters for ACTA2, allowing for isolation of putative iPSC-derived smooth muscle-like populations (iPSC-SMC) with transcriptomic profiles reminiscent of immature or synthetic SMCs. The iPSC-SMCs were used to generate cell sheets with mechanical properties within physiological ranges to serve as building blocks for multi-layered tissue constructs.
Introduction
Smooth muscle cells (SMCs) function to regulate the tone of blood vessels, airways, and intestines within the body, and SMC dysfunction is characteristic of prevalent diseases, including atherosclerosis (Bennett et al., 2016) and asthma (Pelaia et al., 2008). In either normal or diseased states, SMCs typically exist in defined layers, and emerging new engineering techniques present the possibility of generating layered tissue-like structures, such as vessel walls, including functional smooth muscle-like sheets to reconstitute normal function. Using autologous cells to engineer these sheets is a particularly compelling approach explored by some groups (Hibino et al., 2012, Kito et al., 2013, L’heureux et al., 1998, Syedain et al., 2011), but harvesting an expandable source of smooth muscle progenitors from patients has been limited by the tendency of adult SMCs to lose their phenotype during cell culture (Alexander and Owens, 2012).
Alternatively, induced pluripotent stem cells (iPSCs) (Takahashi and Yamanaka, 2006), which self-renew indefinitely in vitro, are poised to be an inexhaustible source of autologous smooth muscle cells (iPSC-SMCs) for these engineered tissue constructs. Their efficacy and functionality, however, depends on the ability to generate pure populations of SMCs from iPSCs. While many protocols exist for producing iPSC-SMCs, to date, their differentiation in vitro results in heterogeneous populations (Maguire et al., 2017), and there are no surface markers available for purification of live SMCs. Early differentiation protocols for producing SMCs from pluripotent stem cells (PSCs) use a single inductive medium recipe (Drab et al., 1997, Sinha et al., 2004). Other protocols rely on forced overexpression of regulators of SMC gene expression, such as myocardin (Raphel et al., 2012, Yoshida et al., 2004). Recognizing that SMCs are derived from multiple embryological lineages, such as neural crest and mesodermal lineages, several groups have more recently utilized developmental stage-specific approaches to pattern PSCs first toward neural crest, paraxial mesoderm, or lateral plate mesoderm before further specifying the cells toward an SMC lineage using serum-containing (Dash et al., 2016, Eoh et al., 2017, Ferreira et al., 2007, Lee et al., 2010, Marchand et al., 2014, Sinha et al., 2006, Syedain et al., 2011, Wang et al., 2012) or serum-free medium (Cheung et al., 2012, Yang et al., 2016). For example, Wang et al. (2012) used iPSC-derived neural crest cells to generate a population enriched in smooth muscle lineages through treatment with serum and transforming growth factor β (TGF-β). Cheung et al. (2012) described serum-free differentiation of PSCs to smooth muscle-like cells through neuroectoderm, lateral plate mesoderm, and paraxial mesoderm intermediates. Still other groups have differentiated cells through a kinase insert domain receptor (KDR)-expressing mesodermal intermediate (Gadue et al., 2006, Nostro et al., 2008, Prasain et al., 2014) or a similar mesodermal intermediate (Patsch et al., 2015) into smooth muscle-like cells. These effective stage-specific in vitro approaches recapitulate milestones of SMC development in vivo in order to derive putative iPSC-SMCs in vitro. Although these studies produce a high percentage yield of SMCs scored by intracellular staining of fixed specimens, no methods have yet been developed for the tracking and purification of live SMCs in directed differentiations.
Here, we have engineered mouse and human iPSC lines, carrying fluorochrome reporters for Acta2 expression, to accomplish the live cell tracking and purification of candidate SMCs. We utilize these purified derivatives to characterize their global molecular and functional phenotypes in comparison with primary cells, and produce the first known engineered cell sheets from iPSC-derived putative SMCs.
We have selected the mouse Acta2 (or human ACTA2) locus for reporter targeting for our developmental system: Acta2 is one of the earliest characteristic markers of SMCs broadly expressed during embryonic development (Owens et al., 2004). Our results indicate that developmental differentiation of iPSCs via mesoderm in serum-free defined conditions produces Acta2+/ACTA2+ cells expressing transcriptomic profiles reminiscent of immature or synthetic SMCs, with low levels of contractile SMC marker gene expression and functional capacity to form engineered cell sheets. In summary, we present a stem cell-based model system that recapitulates SMC development and produces highly characterized, purifiable SMC candidates for future regenerative medicine or tissue engineering applications.
Results
Generation of miPSC-Derived Acta2hrGFP+ Cells
To develop a method for derivation and purification of autologous candidate SMCs from iPSCs, we focused first on generating a murine iPSC line (miPSC) whose phenotype could be compared with in vivo SMCs of the same genetic background. A transgenic mouse expressing humanized Renilla reniformis green fluorescent protein (hrGFP) following a rat Acta2 promoter sequence (Acta2hrGFP) (Figure 1A) exhibits GFP fluorescence selectively in embryonic and adult SMCs (Ghosh et al., 2011). Hence, we reprogrammed tail tip fibroblasts from this mouse into iPSCs (Figures 1A and S1) using an excisable STEMCCA cassette we have previously published (Sommer et al., 2009). We employed the resulting miPSC line (hereafter Acta2hrGFP) to optimize a protocol (Figure 1B) for derivation of lateral plate mesodermal progenitors competent in forming candidate SMCs. In serum-free cultures, we tested varying doses and durations of bone morphogenetic protein 4 (BMP4), activin A, and Wnt3a (Figure S2), given prior reports establishing the role of activin A and Wnt3a in primitive streak formation and BMP signaling in promoting posterior primitive streak differentiation into KDR+ mesodermal progenitors (Gadue et al., 2006, Nostro et al., 2008). We found that 3 ng/mL Wnt3a, 3 ng/mL BMP4, and 2 ng/mL activin A induced KDR+ cells with an efficiency of 20% in our iPSC line, peaking on day 5 of differentiation (Figure S3A). The KDR+ population at this time point was enriched for Foxf1, a lateral plate mesoderm marker. Meanwhile, Pax2 (intermediate mesoderm), Tbx6 (paraxial mesoderm), and Foxa2 (endoderm) were only expressed at low levels in the KDR+ population (Figure S3B).
Figure 1.
Generation and Characterization of Mouse iPSC-Derived Acta2hrGFP+ Cells
(A) Schematic of the Acta2hrGFP reporter construct and Acta2hrGFP mouse fibroblast reprogramming to generate iPSCs, followed by differentiation into GFP-expressing cells.
(B) Schematic of mouse iPSC-directed differentiation protocol.
(C) Relative competence of day-5 KDR+ and KDR− populations to differentiate to an Acta2hrGFP+ population, scored by flow cytometry on day 13. Error bars represent average ± SD. ∗∗p ≤ 0.01 and ∗∗∗∗p ≤ 0.0001 by unpaired two-tailed Student's t test between KDR+ and KDR− populations. n = 3 replicates for PT condition and n = 6 replicates for serum condition from independent experiments.
(D) Flow-cytometry plot of Acta2hrGFP and KDR expression on day 13, with accompanying dot plot showing Acta2hrGFP+ efficiency on day 13.
(E) Gene expression of characteristic SMC markers as well as markers of endothelial cells, podocytes, and epithelial cells. qRT-PCR measurements of fold change (FC) of mRNA expression (2−ΔΔCt) is shown for presort, GFP+, and GFP− populations. Undifferentiated miPSC mRNA expression is defined as FC = 1. Error bars represent ±SD. ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001, ∗∗∗∗p ≤ 0.0001 by unpaired two-tailed Student's t test between GFP+ and GFP− populations. n = 3 independent experiments.
(F) Immunofluorescence images of hrGFP (green) and ACTA2 (red), with nuclear DAPI stain (blue). Scale bar, 50 μm.
See also Figures S1–S3.
Next, to understand the competence of mesodermal precursors to differentiate toward a smooth muscle-like cell, we sorted KDR+ and KDR− populations on day 5 and replated these cells in media containing previously published growth factors, platelet-derived growth factor BB (PDGF-BB), TGF-β, and fibroblast growth factor 2 (FGF2) (hereafter PT), known to promote generation of SMCs from iPSCs in vitro (Cheung et al., 2012). While little GFP (<1%) fluorescence was noted in KDR+ cells on day 5 (Figure 1C), GFP induction was easily detected in the outgrowth of KDR+ sorted cells within 3 days after exposure to these factors, increasing to 31.9% ± 1.1% by day 13. Acta2hrGFP+ cells were significantly enriched in the outgrowth of the KDR+ replated population as compared with the KDR− population (Figure 1C), and differentiation efficiency was higher in defined (PT) serum-free conditions compared with previously published conditions that employ serum (Figure 1C); thus, subsequent experiments were performed without sorting for KDR+ outgrowth. Compared with Acta2hrGFP− cells, day-13 Acta2hrGFP+ cells (26.4% ± 11.3% [Figure 1D]), purified by flow cytometry, showed enriched gene expression of characteristic SMC markers, such as Acta2, Tagln, Myh10, Col1a1, Eln, and Myh11. Synthetic SMC markers (Acta2, Col1a1, and Eln) were more highly expressed, while the contractile SMC marker (Myh11) was significantly enriched but lowly expressed. In contrast, the Acta2hrGFP+ population was depleted for markers of alternate mesodermal lineages, such as endothelial cells (VE-cad) or podocytes (Podxl), and depleted of epithelial cells (Cdh1) (Figure 1E). Immunofluorescence staining for ACTA2 in the iPSC differentiation revealed that all Acta2hrGFP+ cells expressed ACTA2 protein; however, there were ACTA2+ cells that were not marked by the hrGFP reporter (Figure 1F). Quantification of the immunofluorescent images revealed that approximately 98% of the hrGFP+ cells were ACTA2+ and approximately 66% of the ACTA2+ cells were hrGFP+, indicating that the hrGFP reporter is specific but not faithful.
Global Transcriptomic Analysis of Directed Differentiation of Mouse iPSCs
To characterize global gene expression kinetics of mouse iPSC-derived Acta2hrGFP+ cells during the directed differentiation process in comparison to primary aortic SMCs, we performed microarray expression analysis on three key stages of our directed differentiation: undifferentiated miPSCs (day 0), “lateral plate” KDR+ mesoderm (day 5), and Acta2hrGFP+ and Acta2hrGFP− smooth muscle-like cells (day 13). Primary aortic SMCs (Acta2hrGFP+) isolated from adult Acta2hrGFP mice were included as a positive control sample (Figure 2A).
Figure 2.
Global Transcriptomic Profiling of Primary Mouse Aortic SMCs and Key Stages of Acta2hrGFP miPSC-Directed Differentiation toward Smooth Muscle-like Cells
(A) Schematic of experiment. The following cell types were profiled using microarray expression analysis: undifferentiated miPSCs (D0), purified KDR+ mesodermal progenitors (D5 KDR+), purified Acta2hrGFP+ and Acta2hrGFP− cells (D13 GFP+ and D13 GFP−), and adult primary Acta2hrGFP+ aortic SMCs.
(B) Principal component (PC) analysis of all samples.
(C) Hierarchical clustering of gene expression using all 2,386 differentially expressed genes with FDR < 1 × 10−7 and absolute FC > 5 revealed 10 different gene clusters. Key genes from four of these clusters of interest (mature/contractile SMC, immature/synthetic SMC, mesoderm, and iPSC) are shown to the right.
(D) Dendrogram showing similarity between the samples through hierarchical clustering of all samples calculated across all genes.
Principal component analysis revealed changes in the global transcriptome of different stages of mouse smooth muscle differentiation as cells progress from undifferentiated miPSCs through KDR+ mesodermal intermediates to Acta2hrGFP+ cells (Figure 2B). Acta2hrGFP+ cells clustered closer to the primary aortic SMC population on the PC1 axis. Hierarchical clustering of all 15 samples by one-way ANOVA (2,386 top varying genes based on false discovery rate [FDR] adjusted p value, hereafter “FDR,” <1 × 10−7, and absolute fold change, hereafter “FC,” >5) revealed ten gene clusters. As expected, these clusters contained gene signatures indicative of undifferentiated iPSCs (cluster 8), lateral plate mesoderm (cluster 6), immature/synthetic smooth muscle (clusters 5 and 7), and mature/contractile smooth muscle (cluster 1) (Figure 2C and Table S1). For example, day-0 (or undifferentiated) mouse iPSCs expressed pluripotency markers such as Nanog, Zfp42, Klf5, Dppa4, and Dppa2. By day 5, iPSCs differentiated into KDR+ mesodermal intermediates, significantly downregulated these markers (Figure 2C), and upregulated mesodermal-associated transcripts, Kdr, T (primitive streak), Foxf1 (lateral plate mesoderm), and Pdgfrb. Following addition of PDGF and TGF-β ligands to further differentiate these cells until day 13, the Acta2hrGFP+ and Acta2hrGFP− populations both upregulated markers of immature/synthetic smooth muscle cells, including Acta2, Tagln, Col3a1, Col1a1, Pdgfrb, and Tpm1. The overall expression profiles of the Acta2hrGFP+ and Acta2hrGFP− cells were similar by clustering analysis (Figure 2D), potentially due to the presence of ACTA2+ cells that do not express GFP (Figure 1F); however, the Acta2hrGFP+ population expressed significantly higher levels of Tagln and Pdgfrb as well as higher levels of other synthetic smooth muscle markers such as Myl9, Itgb5, Actg2, and Aebp1 (Table S1; FDR < 0.05). While contractile smooth muscle markers (Tpm2, Itga8, Ecm1, Cnn1, Smtn, and Myh11) were only expressed at low levels compared with aorta controls, the expression level of Tpm2 was enriched in Acta2hrGFP+ compared with Acta2hrGFP− cells.
Next, we chose two approaches to compare gene expression programs between engineered (iPSC-derived Acta2hrGFP+) cells and primary aorta controls. First, we determined the number of differentially expressed genes (FDR < 0.05) between a common comparator (KDR+ day-5 progenitors) versus the engineered or versus the primary cells. We found that the majority of genes were commonly upregulated or downregulated (Figure 3A) in engineered and primary cells. Focusing only on transcription factors (TFs), we similarly found the majority of TFs were differentially expressed (FDR < 0.05) in common in either engineered or primary cells compared with KDR+ precursors (Figure 3B). Second, we sought to determine which signaling pathways were changing over time during the specification of our Acta2hrGFP+ cells from their precursors and whether these changes were similar in primary aortic smooth muscle cell controls. Gene set enrichment analysis (GSEA) revealed that the top five enriched gene sets were upregulated in both engineered cells and controls. These five gene sets were “epithelial mesenchymal transition,” “hypoxia,” “myogenesis,” “coagulation,” and “TNFA signaling via NFKB.” In addition, both comparisons showed enrichment of “angiogenesis” and “TGFbeta signaling” gene sets (Figure 3C). When looking at the top enriched genes within the “myogenesis” gene set in the Acta2hrGFP+ versus KDR+ comparison, a majority of the genes (11/16) were also highly expressed in aortic SMCs, such as Col3a1, Aebp1, Tagln, and Myom1 (Figure 3D, left). However, cardiomyocyte markers, Tnnt2 and Tnni1, were highly expressed in the in vitro Acta2hrGFP+ cells, but lowly expressed in primary aortic SMCs. Because Acta2 is also known to be expressed in developing cardiomyocytes (Owens et al., 2004), this raised the possibility of cardiomyocytes present in the Acta2hrGFP+ population. To quantify the presence of any contaminating cardiomyocytes, we used flow cytometry after staining for the cardiomyocyte marker, TNNT2, and found that 1.6% ± 0.6% of GFP+ cells were TNNT2+ (Figure S4). In contrast to our protocol, serum-containing protocols for SMC differentiation contained significantly higher numbers (4.6% ± 0.5% of GFP+ cells) of contaminating Acta2hrGFP+ cardiomyocytes (p < 0.008), and Wnt inhibition promoted differentiation of TNNT2+ cells, as expected based on prior reports (Kattman et al., 2011).
Figure 3.
Comparison of miPSC-Derived Populations with Primary Mouse Aortic SMCs
(A and B) Differentially expressed genes and transcription factors, respectively, with FDR < 0.05 when comparing Acta2hrGFP+ cells or aortic SMCs with KDR+ mesodermal progenitors. Numbers outside the circles represent number of genes differentially expressed between Acta2hrGFP+ versus KDR+ and Aorta versus KDR+ comparisons. Number in the intersection of circles represents the number of overlapping genes between the comparisons.
(C) Gene set enrichment analysis (GSEA) of Acta2hrGFP+ cells or aortic SMCs compared with KDR+ mesodermal progenitors.
(D) Heatmaps of top enriched genes in the “Myogenesis” gene set from the GSEA comparing Acta2hrGFP+ and KDR+ populations (left) or aortic SMC and KDR+ populations (right).
(E) GSEA showing enriched gene sets when comparing Acta2hrGFP+ cells and aortic SMCs.
Next, we sought to test for potential differences between in vitro engineered putative SMCs and primary in vivo aortic controls. In top-ranked genes within the “myogenesis” gene set from the aortic SMC versus KDR+ comparison using GSEA, we noted the expression of SMC contractile markers, such as Myh11 and Smtn, in aortic SMCs, but these markers were expressed at low levels in the iPSC-derived Acta2hrGFP+ population (Figure 3D, right). Comparing primary and engineered Acta2hrGFP+ cells head-to-head by GSEA, we found enrichment of cell-cycle and proliferation gene sets in the in vitro Acta2hrGFP+ population (Figure 3E) whereas myogenesis genes, such as Myh11 and Myocd, an SMC transcriptional co-activator, were both found in the top differentially expressed genes and TFs in primary aortic SMCs (GSEA based on 3,127 transcripts upregulated in aortic SMC and 4,444 downregulated in aortic SMC, FDR < 0.05).
Generation of Human iPSC-Derived ACTA2eGFP+ Cells
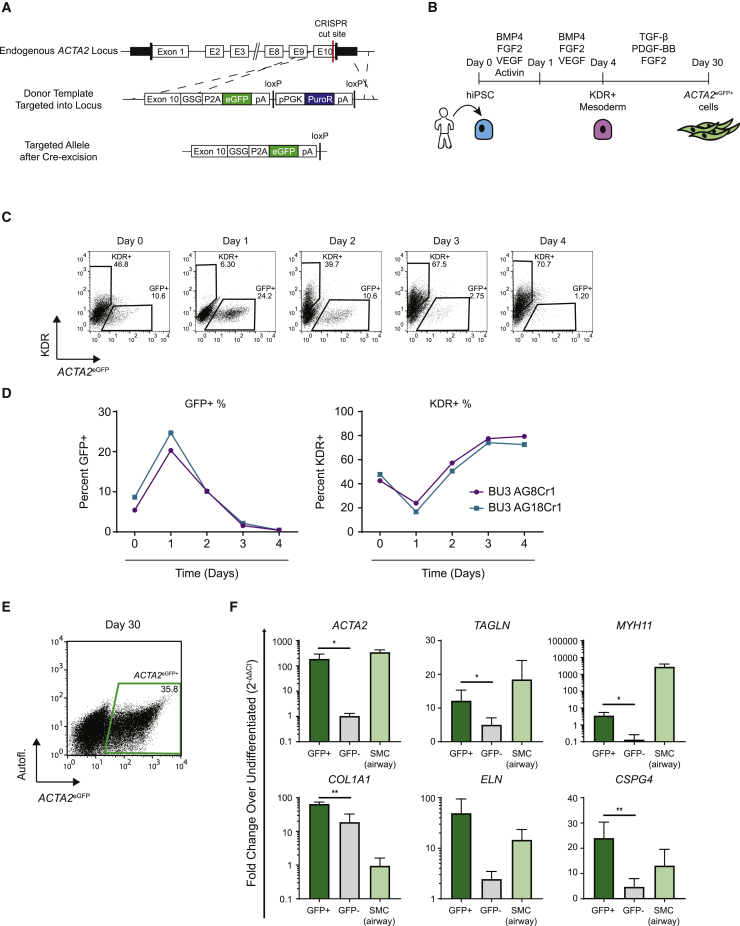
Next, we sought to engineer putative SMCs from human iPSCs (hiPSCs) in vitro. The reporter mouse iPSC system allowed us to isolate a population of ACTA2+ cells enriched in putative SMCs; however, the transgenic reporter did not mark all ACTA2+ cells. To address this issue, we used CRISPR/Cas9 gene editing (Ran et al., 2013) to target an enhanced GFP reporter (eGFP) to the endogenous human ACTA2 locus in normal human iPSCs (BU3 [Kurmann et al., 2015]; Figure 4A). Because smooth muscle α actin functions in cell movement and contraction of SMCs, we chose a targeting strategy designed to allow for translation of both ACTA2 and the eGFP reporter by introducing a P2A-eGFP cassette in place of the endogenous ACTA2 stop codon. Two clones were identified with successful monoallelic and biallelic targeting of the ACTA2 locus (BU3 AG8Cr1 and BU3 AG18Cr1, respectively; Figure S5).
Figure 4.
Human iPSC Reporter System for Generation of an ACTA2eGFP+ Smooth Muscle-like Cells
(A) Schematic showing targeting strategy to replace the endogenous stop codon of the ACTA2 locus with a 2A-eGFP cassette using the CRISPR/Cas9 system.
(B) Schematic of hiPSC differentiation toward smooth muscle-like cells.
(C and D) Flow-cytometry dot plots demonstrating the expression kinetics of KDR and eGFP over 4 days of directed differentiation into mesodermal progenitors using both monoallelically (BU3 AG8Cr1) and biallelically (BU3 AG18Cr1) targeted hiPSC lines. Line graphs in (D) represent n = 1 biological replicate for each hiPSC line.
(E) Flow-cytometry dot plot of ACTA2eGFP expression on day 30.
(F) Relative gene expression (qRT-PCR) of markers of SMCs in ACTA2eGFP+ and ACTA2eGFP− cells compared with freshly isolated uncultured primary human airway SMC controls. Error bars represent average ± SD. ∗p ≤ 0.05 and ∗∗p ≤ 0.01 by unpaired two-tailed Student's t test between GFP+ and GFP− populations. n = 3 independent experiments.
See also Figure S5.
To differentiate each iPSC clone, we followed similar developmental pathways and strategies as in our mouse system, focusing first on their differentiation into KDR+ mesoderm using a published serum-free medium (Prasain et al., 2014) supplemented with BMP4, vascular endothelial growth factor (VEGF), FGF2, and activin A for 24 h, followed by 72 h with BMP4, VEGF, and FGF2 (Figure 4B). In contrast to mouse iPSCs, we observed some expression of GFP in undifferentiated human iPSCs (Figure 4C). These ACTA2eGFP+/KDR− cells of unclear phenotype continued to be present from days 0 to 3, but this population became undetectable over 4 days of mesodermal differentiation as cells upregulated KDR with 70% efficiency, giving rise to an ACTA2eGFP−/KDR+ population of putative mesodermal cells, which were enriched for lateral plate mesoderm marker, FOXF1 (Figure 4C). As in our mouse model, hiPSCs further differentiated in response to PDGF-BB and TGF-β, giving rise to ACTA2eGFP+ cells (Figure 4D). By day 30, 36% of cells expressed the GFP reporter, and sorted GFP+ cells expressed the characteristic smooth muscle markers (ACTA2, TAGLN, COL1A1, ELN, and MYH11). ACTA2 and TAGLN were expressed at similar levels when compared with freshly isolated primary human airway SMC tissue controls. In addition, COL1A1 was expressed at higher levels in the ACTA2eGFP+ cells than in the primary SMCs, while MYH11 was expressed at significantly lower levels in the ACTA2eGFP+ cells. This suggests that the hiPSC-derived ACTA2eGFP+ smooth muscle-like population has an immature/synthetic phenotype, similar to the miPSC-derived Acta2hrGFP+ population.
To characterize the transcriptomic profile of the ACTA2eGFP+ and ACTA2eGFP− cells, we performed microarray analysis on both populations isolated on day 30 of directed differentiation (Figures 5A and 5B). The top differentially upregulated genes (FDR < 0.05) in the ACTA2eGFP+ cells included smooth muscle actins (ACTA2 and ACTG2) as well as collagens (COL8A1 and COL12A1) (Figure 5C). Furthermore, MYOCD, a transcription co-activator of the SMC lineage, was found in the top ten upregulated TFs (FDR < 0.05) in the ACTA2eGFP+ population (Figure 5D), and a variety of additional SMC markers were upregulated in the ACTA2eGFP+ cells (Figure 5E; FDR<0.05). GSEA also revealed enrichment in gene sets similar to the GSEA of the mouse microarray, including “myogenesis,” “TGFbeta signaling,” and “angiogenesis” (Figure 5F). Notably, top enriched genes in the “myogenesis” gene set included TAGLN, TPM2, MYH9, COL1A1, and COL3A1 (Figure 5G).
Figure 5.
Global Transcriptomic Profiling of Human iPSC-Derived ACTA2eGFP Populations
(A) Schematic of microarray experiment.
(B) Principal component (PC) analysis of ACTA2eGFP+ and ACTA2eGFP− populations.
(C and D) Heatmap of top 25 upregulated and top 25 downregulated expressed genes (C) and transcription factors (D) with FDR < 0.05 between ACTA2eGFP+ and ACTA2eGFP− populations.
(E) Heatmap of selected smooth muscle cell marker genes (FDR < 0.05).
(F) GSEA showing enriched gene sets in ACTA2eGFP+ cells compared with ACTA2eGFP− cells.
(G) Heatmap of top enriched genes in the “Myogenesis” gene set from the GSEA comparing ACTA2eGFP+ and ACTA2eGFP− populations.
Generation and Characterization of iPSC-SMC Cell Sheets
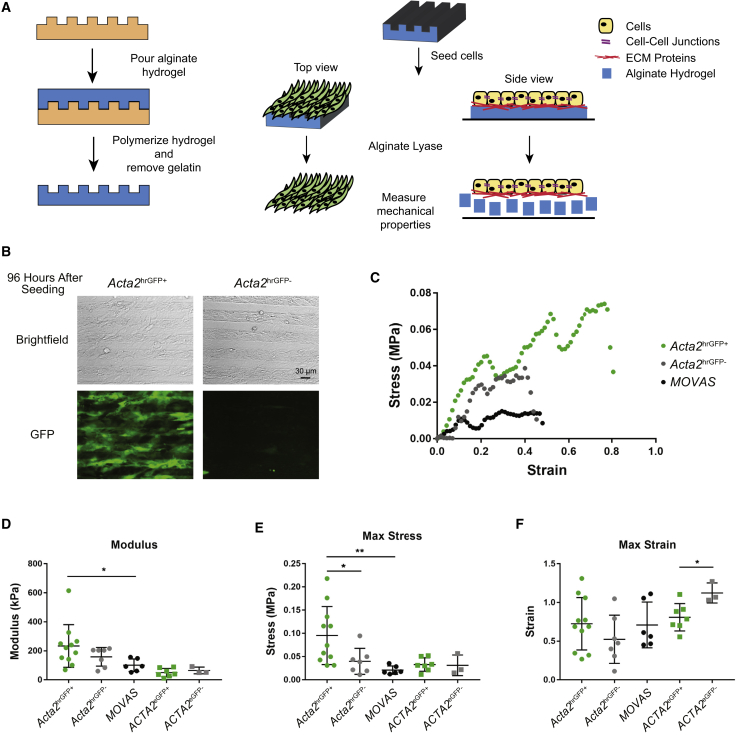
To determine whether our iPSC derivatives could serve as building blocks for engineered smooth muscle tissue constructs, we next utilized a micropatterned enzyme-degradable hydrogel system (Figure 6A) (Rim et al., 2018) to generate releasable aligned cell sheets that maintain cell-cell junctions and extracellular matrix (ECM) proteins. After seeding, sorted human or mouse iPSC-SMCs aligned in the direction of the micropatterns, consistent with our previously published work using multiple micropatterned substrate systems (Backman et al., 2017a, Rim et al., 2018). Notably, the Acta2hrGFP+ population retained expression of the GFP reporter following alignment on the alginate scaffold (Figure 6B). Additionally, expression of many characteristic SMC markers (Acta2, Tagln, Myh10, Myh11, Col3a1) remained unchanged, but expression of Cnn1 was upregulated and expression of Col1a1 and Eln was downregulated in Acta2hrGFP+ cells after cell sheet formation on alginate substrates (Figure S6). Due to effects of tissue stiffness on SMC phenotype (Owens et al., 2004) and detrimental effects of mechanical mismatch on engineered blood vessel patency (Salacinski et al., 2001), we were interested in characterizing mechanical properties of iPSC-derived cell sheets. Individual, single-layer cell sheets from mouse and human iPSC-derived cells as well as a control mouse aortic SMC line (MOVAS)—harvested after 4–6 weeks of culture on alginate substrates—underwent uniaxial tensile testing in the direction parallel to alignment to evaluate stress-strain responses (Figure 6C). As expected, cell sheets displayed non-linear mechanical behavior, although there was a minimal “toe region,” where the cell sheet undergoes strain while producing little stress. Furthermore, many samples underwent small breaks before complete failure, indicating multiple sites of failure, as shown by small decreases in stress around strains of 0.2 and 0.5. The stress-strain curves were analyzed to calculate elastic modulus, stress at failure (max stress), and strain at failure (max strain) (Figures 6D–6F). Mouse iPSC-derived Acta2hrGFP+ cell sheets had significantly higher elastic modulus compared with the MOVAS control cell sheets but were not significantly different from that of Acta2hrGFP− cell sheets. The moduli of the samples were in the 50- to 250-kPa range, which is in the range of elastic moduli of bovine vascular SMC sheets previously reported by our group (Backman et al., 2017a, Rim et al., 2018); however, this range of moduli is on the lower end of reported stiffnesses for blood vessels and engineered vascular grafts (0.1–50 MPa) (Guo and Kassab, 2003, Lashkarinia et al., 2018, Wagenseil and Mecham, 2009). The Acta2hrGFP+ cell sheets did, however, have significantly higher max stress at failure when compared with Acta2hrGFP− cell sheets and MOVAS cell sheets. Meanwhile, ACTA2eGFP+ and ACTA2eGFP− human cell sheets had comparable elastic moduli and max stress at failure, but higher max strains in the ACTA2eGFP− cell sheets at failure.
Figure 6.
Generation of iPSC-SMC Cell Sheets
(A) Schematic of alginate-tyramine hydrogel and cell sheet fabrication. Soft lithography techniques were used to fabricate micropatterned alginate-tyramine hydrogels to generate aligned cell sheets, and a hydrogel-specific enzyme was used to degrade the hydrogel and leave cell-cell junctions and ECM proteins intact.
(B) Images of Acta2hrGFP+ and Acta2hrGFP− cell sheets 96 h after seeding onto alginate-tyramine hydrogel substrates. Scale bar, 30 μm.
(C) Representative stress-strain curves for Acta2hrGFP+, Acta2hrGFP−, and immortalized mouse aortic vascular SMC (MOVAS) cell sheets undergoing uniaxial tensile tests.
(D–F) Elastic modulus (D), max stress (E), and max strain (F) of cell sheets derived from the following populations: Acta2hrGFP+, Acta2hrGFP−, MOVAS, ACTA2eGFP+, and ACTA2eGFP− cells. Error bars represent average ± SD. ∗p ≤ 0.05 and ∗∗p ≤ 0.05 by unpaired two-tailed Student's t test between GFP+ and GFP− cell sheets and between GFP+ and MOVAS cell sheets. n = 11, 7, 6, 7, and 3 for the Acta2hrGFP+, Acta2hrGFP−, MOVAS, ACTA2eGFP+, and ACTA2eGFP− cell sheets from independent experiments, respectively.
Discussion
In this study, we derived a smooth muscle-like population that expresses Acta2/ACTA2, an early but non-specific SMC marker. Generation of a transgenic mouse iPSC line with an hrGFP reporter for Acta2 allowed for enrichment of smooth muscle-like cells (~98% of hrGFP+ cells were ACTA2+; enrichment of Acta2 mRNA expression when comparing hrGFP+ and hrGFP− populations); however, not all ACTA2+ cells were marked by the transgenic reporter (~66% of ACTA2+ cells were hrGFP+), as is known to occur when promoter-reporter constructs are integrated outside their endogenous locus. To better understand purified SMC populations in a human system and to avoid issues related to transgenic reporters, we similarly generated a human iPSC line with a knockin eGFP reporter targeted to the endogenous ACTA2 locus, allowing purification of GFP+ cells that expressed ACTA2 and other SMC marker transcripts at levels equivalent to primary tissue controls. Our findings indicate that purified mouse and human iPSC-derived GFP+ populations had a transcriptomic profile reminiscent of an immature/synthetic SMC phenotype, with low-level expression of mature/contractile markers of SMCs. In addition, miPSC-derived Acta2hrGFP+ cells showed minimal change in surface area as a readout of contractility in response to agonists (carbachol and angiotensin II) (data not shown).
As the markers used to define maturity and synthetic/contractile phenotype of SMCs overlap, it is difficult to distinguish whether our in vitro derived cells are immature or synthetic compared with the mature and contractile primary SMC controls, but comparisons of the two populations suggests candidate pathways that might be harnessed to induce in vitro cells to become more like primary SMCs. Comparison of primary aortic SMCs and our in vitro Acta2hrGFP+ cells using GSEA revealed multiple signaling pathways that are enriched in the primary aortic SMCs, including Notch signaling, which plays a role in SMC development and phenotype switching (Fouillade et al., 2012). Modulation of these signaling pathways could enhance maturation or could induce phenotypic switching of the in vitro derived cells. Other studies have examined effects of biochemical and biomechanical cues on SMC phenotype switching and expression of contractile markers (Owens et al., 2004). Modulation of these cues can readily be explored to further enhance mature/contractile marker expression in our iPSC-derived cells. For example, a recent publication described a three-dimensional culture system for culturing and expanding iPSC-derived smooth muscle-like cells, which resulted in increased expression of contractile markers relative to the two-dimensional culture system (Lin et al., 2018b). Other studies utilized pulsatile flow/stretch bioreactors on engineered vascular grafts to modulate SMC phenotype (Niklason et al., 1999, Syedain et al., 2011), which could be adopted to enhance the contractile phenotype of our iPSC-SMC cell sheets. Although previous publications of iPSC-SMCs described expression of contractile markers either through staining or transcriptomic data, in contrast to our work, most prior reports have not included comparisons with uncultured primary SMC controls. Given the rapid loss of SMC programs known to occur in most primary SMC culture systems (Weissberg et al., 1995), it will be important in future studies to compare the contractile profile of cells with freshly isolated primary controls following modulation of these biochemical and biomechanical cues.
Even though the iPSC-SMCs in this study may be more immature or synthetic in phenotype, it is important to note that synthetic SMCs play a role in inflammation, atherosclerosis, and vascular remodeling (Lim and Park, 2014) through proliferation, and secretion of ECM proteins or inflammatory cytokines. Thus, iPSC-SMCs may be useful in studying and treating vascular disorders such as atherosclerosis. The synthetic SMC phenotype also plays a major role during development and vasculogenesis, whereby the cells are highly migratory and proliferative while also secreting ECM proteins including collagen, elastin, proteoglycans, cadherins, and integrins (Owens et al., 2004). This more developmental synthetic phenotype could be beneficial in our in vitro system to generate cell sheets as building blocks for multi-layered SMC tissue constructs.
Despite advancements facilitated by access to purified iPSC-derived putative SMCs, many questions still remain, including how to adequately establish SMC identity in engineered cell types and specifically how to determine which subtype of SMC is being derived. Acta2/ACTA2 is not specific to SMCs, as it is also found in myofibroblasts and cardiomyocytes. Based on our flow-cytometry quantification of the cardiomyocyte marker, TNNT2, as well as microarray analysis, we found a low but detectable presence of potential cardiac lineage cells within our Acta2hrGFP+ population. As Acta2 is not a specific marker, additional markers may need to be used to identify SMCs and completely exclude any potential cardiomyocyte presence. Recent work published by Sinha and colleagues had also begun to probe the effects of embryological origins on vascular SMC phenotype using an iPSC system (Cheung et al., 2012) in order to understand the role of those SMCs during disease. However, as with most prior reports, the potential presence of cardiomyocyte lineages, which can derive from some of these mesodermal intermediates, was not evaluated. There is also little literature investigating the differences between various SMC subtypes (e.g., vascular, airway), resulting in a paucity of markers to distinguish between those types. For example, to distinguish between vascular and airway SMCs, Notch3 (Ghosh et al., 2011) and Cspg4 (Paez-Cortez et al., 2013) have been previously reported as helpful markers of SMC subtypes, with the co-expression of CSPG4 and ACTA2 being restricted to arteries rather than veins (Murfee et al., 2005). In this study, we have compared our iPSC-SMCs with either primary aortic or airway SMCs with regard to characteristic SMC markers (Acta2, Tagln, Myh11, Col1a1, and Eln), which have not been shown to be different between SMC subtypes. However, it will be important to identify distinct markers of primary SMC populations to better characterize iPSC-SMC identity.
In this study, we utilized micropatterned enzyme-degradable hydrogels (Rim et al., 2018) to generate aligned cell sheets from iPSC-derived GFP+ cells, which can be used as building blocks to generate an engineered tissue construct composed of multiple aligned layers similar to the structure of blood vessels. Although many methods have now been described for generating cell sheets, including thermoresponsive, magnetic-based, and electroresponsive systems (Akintewe et al., 2017), this enzyme-degradable hydrogel system allows for modulation of substrate stiffness, which can be used as a biomechanical cue to alter SMC phenotype (synthetic versus contractile, immature versus mature) in future studies. In our studies, we saw an upregulation of a more contractile marker, Cnn1, with a downregulation of ECM proteins, Col1a1 and Eln, after cell sheet formation, suggestive of a possible shift toward a more mature or contractile phenotype. Compared with typical arterial stress-strain, our iPSC-derived SMC cell sheets also displayed non-linear mechanical behavior, with a much less pronounced “toe region,” where strain produces minimal stress. This “toe region” is due to the untangling of ECM proteins as well as the difference in mechanical properties of collagen and elastin (Roach and Burton, 1957). The small “toe region” could be due to a difference in ECM composition compared with that of the native blood vessel. The elastic moduli of the iPSC-derived cell sheets in this study were lower than in our previously reported work (Backman et al., 2017a, Rim et al., 2018), which may be due to the lack of treatment with ascorbic acid, an additive used in previous studies. The moduli of the iPSC-derived cell sheets was also lower than the modulus of blood vessels (Guo and Kassab, 2003, Lashkarinia et al., 2018, Wagenseil and Mecham, 2009); however, blood vessels have a complex architectural structure composed of multiple aligned SMC layers oriented in different directions, while only single-layer cell sheets were tested in this study. Previous work from our group (Rim et al., 2018) explored mechanical properties of multi-layered bovine vascular SMC sheets; future work will need to focus on characterizing mechanical properties of stacked iPSC-derived cell sheets. The blood vessel is also composed of multiple layers of ECM proteins such as collagen and elastin, so further characterization of ECM content and structure in iPSC-derived single- and multi-layered cell sheets is necessary. In addition, the max stress and max strain of the iPSC-derived cell sheets were also within physiologically relevant ranges, where blood vessels typically undergo ~20% strain (Fung and Liu, 1992) and wall stresses of 75–150 kPa (Chuong and Fung, 1986, Koullias et al., 2005). It is important to note that these studies described a distribution of stress and strains across the vessel wall, so it will be important to study the mechanical behavior of multi-layered iPSC-derived tissue constructs. A multi-layer cell sheet-based study characterized mechanical properties of fibroblast cell sheets rolled into multi-layered tubes with burst pressures of approximately 3,500 mmHg (Konig et al., 2009), which calculates to approximately 2–3 MPa. In addition to characterizing the mechanical behavior at baseline, it will be important to study the mechanical behavior of these tissue constructs in the presence of SMC agonists and antagonists, such as nitric oxide, prostacyclin, endothelin, and angiotensin, which we did not characterize due to the sensitivity of tensile testers, which are capable of measuring forces in the μN-to-N force range (Backman et al., 2017b). Cellular traction forces of SMCs have been reported in the 100-Pa range (Lin et al., 2018a), which is roughly in the nN force range when accounting for the area of a cell. Future studies of the iPSC-SMC constructs in the presence of agonists and antagonists will need to be performed using tools, such as microscopy and micropillars (Polacheck and Chen, 2016), for measuring cellular traction forces.
In summary, we have purified mouse and human iPSC-derived populations, which express characteristic SMC markers. Our transcriptomic profiling of these cells reveals a phenotype similar to immature or synthetic SMCs with low levels of contractile smooth muscle markers. In addition, the transcriptomic analysis highlights possible pathways and TFs enriched in primary SMCs as compared with iPSC-derived populations, providing hints as to biochemical cues that can be modulated to further mature iPSC-derived populations. Finally, we generated mouse and human iPSC-derived aligned cell sheets with elastic moduli, max stress, and max strains within physiological ranges to serve as building blocks for multi-layered smooth muscle tissue constructs.
Experimental Procedures
For detailed experimental procedures regarding microarray profiling of iPSC-derived populations, GSEA analysis, immunostainings, qRT-PCR, and statistical methods, please refer to Supplemental Experimental Procedures.
Generation of Acta2hrGFP Mouse iPSC and Knockin ACTA2eGFP Human iPSC Reporter Systems
All mouse studies including tissue harvests and iPSC derivation were approved by Boston University's Institutional Animal Care and Use Committee. An Acta2hrGFP reporter mouse iPSC line was generated by reprogramming tail tip fibroblasts isolated from adult female C57BL/6 Acta2hrGFP transgenic mice (Ghosh et al., 2011). The excisable lentiviral STEMCCA-loxP vector was employed as previously published (Sommer et al., 2009). In brief, following the completion of reprogramming, a single copy of the floxed STEMCCA-loxP vector was excised using transient expression of adenoviral Cre recombinase, and a normal female karyotype was documented by G-banding analysis (Cell Line Genetics, Madison, WI). Mouse iPSC lines were maintained on mouse embryonic fibroblasts in mouse embryonic stem cell medium for all experiments.
All human iPSCs were generated with informed consent and approval of Boston University's Institutional Review Board. ACTA2eGFP reporter lines were generated using the CRISPR/Cas9 system to target BU3 iPSCs, as detailed in Supplemental Experimental Procedures.
Directed Differentiation of Mouse iPSCs into Acta2hrGFP+ Cells
Mouse iPSCs were differentiated through a mesodermal progenitor expressing KDR and then further specified toward a smooth muscle-like lineage by modifying previously reported methods (Gadue et al., 2006, Nostro et al., 2008). In brief, 2 × 106 cells were plated in 10-cm non-adherent Petri dishes (Fisher), allowing embryoid bodies (EBs) to be formed during 48 h of leukemia inhibitory factor withdrawal in a complete serum-free differentiation medium (cSFDM) detailed in Supplemental Experimental Procedures. EBs were then treated with 2 ng/mL recombinant human (rh) activin A (R&D Systems, #338-AC), 3 ng/mL rhBMP4 (R&D Systems, #314-BP), and 3 ng/mL recombinant mouse (rm) Wnt3a (R&D Systems, #1324-WN-010) for 72 h to generate mesodermal progenitors and then plated without enzymatic digestion onto gelatin-coated plates (~500,000 cells in a well of a 6-well plate) in cSFDM supplemented with 10 ng/mL rhPDGF-BB (Life Technologies, #PHG0046), 10 ng/mL rmTGF-β (R&D Systems, #7666-MB-005), and 10 ng/mL rhFGF2 (R&D Systems, #233-FB). On day 13 of differentiation, cells were dissociated and sorted by gating on viable (calcein blue+) GFP-expressing cells. Where indicated in the text, day-5 cells were sorted on KDR after staining with APC rat anti-mouse KDR antibody (1:50 dilution, BD Biosciences, #560070).
Directed Differentiation of Human iPSCs into ACTA2eGFP+ Cells
Human iPSCs were differentiated into mesoderm and then further specified toward a smooth muscle-like lineage based on previously described literature (Prasain et al., 2014) with the following modifications. In brief, hiPSCs were dissociated into single-cell suspensions using Gentle Cell Dissociation Reagent, and 500,000 cells were plated onto a fresh Matrigel-coated well of a 6-well plate (Corning) in mTeSR supplemented with 10 μM Y-27632 (Fisher Scientific, #1254/50) for 48 h prior to directed differentiation. Human iPSCs were then treated with Stemline II Hematopoietic Stem Cell Expansion medium (Sigma-Aldrich, #S0192) supplemented with 10 ng/mL each of rhBMP4 (R&D Systems, #314-BP), rhActivin A (R&D Systems, #338-AC), rhVEGF (Fisher Scientific, #293-VE), and rhFGF2 (R&D System, #233-FB) for 24 h, followed by 72 h of treatment with 10 ng/mL each of rhBMP4, rhVEGF, and rhFGF2 to generate mesoderm. Cells were then dissociated with Gentle Cell Dissociation Reagent at 37°C for 3 min and passaged in small clumps onto fresh Matrigel-coated plates in cSFDM supplemented with 10 ng/mL rhPDGF-BB (Life Technologies, #PHG0046), 10 ng/mL rhTGF-β (Fisher Scientific, #240-B-010), and 10 ng/mL rhFGF2 (R&D Systems, #233-FB). Cells were dissociated and sorted based on viability (calcein blue+) and GFP expression for further analysis between days 20 and 30. Where indicated in the text, KDR expression was analyzed after staining with a PE mouse anti-human CD309 antibody (1:50 dilution, BD Biosciences, #560494).
Hydrogel Substrate and Cell Sheet Fabrication
Alginate-tyramine was synthesized as previously described (Rim et al., 2018), described in detail in Supplemental Experimental Procedures. Alginate hydrogels were generated in micropatterned 10% gelatin molds (30 μm wide × 30 μm spacing). Hydrogel substrates were coated using rat tail collagen type I before seeding with iPSC-SMCs or a control immortalized mouse aortic SMC line, MOVAS (ATCC, Manassas, VA). Cells were cultured using a common medium used for culturing primary SMCs, containing DMEM (Invitrogen, #11995-073), 10% fetal bovine serum (Fisher, #SH3007103), L-glutamine (final concentration 2 mM, Invitrogen, #25030-164), and primocin.
Mechanical Characterization of Cell Sheets
To characterize mechanical properties, we harvested cell sheets using 0.2 mg/mL alginate lyase (Sigma, A1603) and then utilized a custom-built uniaxial tensile tester, previously developed (Backman et al., 2017b) and described in further detail in Supplemental Experimental Procedures.
Author Contributions
G.K., J.Y.W., and D.N.K. designed the project, developed the experiments, analyzed the data, and wrote the paper. G.K., H.A.M., and Q.Y. performed differentiations and analyzed the experiments.
Acknowledgments
We are grateful to J.C. Jean for input regarding molecular cloning and gene-editing strategies. We thank Dr. Xingbin Ai (Brigham and Women's Hospital) for providing primary human SMC samples and for manuscript review. We also thank Nae Gyune Rim and Daniel Backman for help with cell sheet and tensile tester methodologies. We are grateful to Dr. Yanhang Zhang for helpful discussions regarding tissue mechanics. We thank Adam Gower and Eduard Drizik (Boston University Microarray and Sequencing Core, supported by NIH UL1TR001430) for microarray analysis. We are grateful to Brian Tilton and Patrick Autissier (Boston University Flow Cytometry Core, supported by NIH 1UL1TR001430) and Drs. Greg Miller and Marianne James (supported by NIH R24HL123828 and U01TR001810). This work was supported by NIH awards F31HL128085 and T32HL007969 (to G.K.), R01HL072900 (to J.Y.W.), and R01HL095993, R01HL122442, and R01HL128172 (to D.N.K.).
Published: August 15, 2019
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.stemcr.2019.07.014.
Contributor Information
Joyce Y. Wong, Email: jywong@bu.edu.
Darrell N. Kotton, Email: dkotton@bu.edu.
Accession Numbers
The datasets for the mouse and human microarray analyses have been deposited under accession numbers GEO: GSE126643 and GSE132454, respectively.
Supplemental Information
Gene expression data for hierarchical clustering (see Figure 2C) of all 15 samples by one-way ANOVA (2,386 top varying genes based on FDR adjusted p value < 1 × 10−7 and absolute fold change >5). Hierarchical clustering revealed ten different gene clusters. Clusters of interest: undifferentiated iPSCs (cluster 8), lateral plate mesoderm (cluster 6), immature/synthetic smooth muscle (clusters 5 and 7), and mature/contractile smooth muscle (cluster 1) populations.
References
- Akintewe O.O., Roberts E.G., Rim N.-G., Ferguson M.A.H., Wong J.Y. Design approaches to myocardial and vascular tissue engineering. Annu. Rev. Biomed. Eng. 2017;19:389–414. doi: 10.1146/annurev-bioeng-071516-044641. [DOI] [PubMed] [Google Scholar]
- Alexander M.R., Owens G.K. Epigenetic control of smooth muscle cell differentiation and phenotypic switching in vascular development and disease. Annu. Rev. Physiol. 2012;74:13–40. doi: 10.1146/annurev-physiol-012110-142315. [DOI] [PubMed] [Google Scholar]
- Backman D.E., LeSavage B.L., Shah S.B., Wong J.Y. A robust method to generate mechanically anisotropic vascular smooth muscle cell sheets for vascular tissue engineering. Macromol. Biosci. 2017;17:1600434. doi: 10.1002/mabi.201600434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backman D.E., LeSavage B.L., Wong J.Y. Versatile and inexpensive Hall-Effect force sensor for mechanical characterization of soft biological materials. J. Biomech. 2017;51:118–122. doi: 10.1016/j.jbiomech.2016.11.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett M.R., Sinha S., Owens G.K. Vascular smooth muscle cells in atherosclerosis. Circ. Res. 2016;118:692–702. doi: 10.1161/CIRCRESAHA.115.306361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung C., Bernardo A.S., Trotter M.W.B., Pedersen R.A., Sinha S. Generation of human vascular smooth muscle subtypes provides insight into embryological origin–dependent disease susceptibility. Nat. Biotechnol. 2012;30:165–173. doi: 10.1038/nbt.2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuong C.J., Fung Y.C. On residual stresses in arteries. J. Biomech. Eng. 1986;108:189. doi: 10.1115/1.3138600. [DOI] [PubMed] [Google Scholar]
- Dash B.C., Levi K., Schwan J., Luo J., Bartulos O., Wu H., Qiu C., Yi T., Ren Y., Campbell S. Tissue-engineered vascular rings from human iPSC-derived smooth muscle cells. Stem Cell Reports. 2016;7:19–28. doi: 10.1016/j.stemcr.2016.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drab M., Haller H., Bychkov R., Erdmann B., Lindschau C., Haase H., Morano I., Luft F.C., Wobus A.M. From totipotent embryonic stem cells to spontaneously contracting smooth muscle cells: a retinoic acid and db-cAMP in vitro differentiation model. FASEB J. 1997;11:905–915. doi: 10.1096/fasebj.11.11.9285489. [DOI] [PubMed] [Google Scholar]
- Eoh J.H., Shen N., Burke J.A., Hinderer S., Xia Z., Gerecht S. Enhanced elastin synthesis and maturation in human vascular smooth muscle tissue derived from induced-pluripotent stem cells. Acta Biomater. 2017;52:49–59. doi: 10.1016/j.actbio.2017.01.083. [DOI] [PubMed] [Google Scholar]
- Ferreira L.S., Gerecht S., Shieh H.F., Watson N., Rupnick M.A., Dallabrida S.M., Vunjak-Novakovic G., Langer R. Vascular progenitor cells isolated from human embryonic stem cells give rise to endothelial and smooth muscle–like cells and form vascular networks in vivo. Circ. Res. 2007;101:286–294. doi: 10.1161/CIRCRESAHA.107.150201. [DOI] [PubMed] [Google Scholar]
- Fouillade C., Monet-Lepretre M., Baron-Menguy C., Joutel A. Notch signalling in smooth muscle cells during development and disease. Cardiovasc. Res. 2012;95:138–146. doi: 10.1093/cvr/cvs019. [DOI] [PubMed] [Google Scholar]
- Fung Y.C., Liu S.Q. Strain distribution in small blood vessels with zero-stress state taken into consideration. Am. J. Physiol. 1992;262:H544–H552. doi: 10.1152/ajpheart.1992.262.2.H544. [DOI] [PubMed] [Google Scholar]
- Gadue P., Huber T.L., Paddison P.J., Keller G.M. Wnt and TGF-beta signaling are required for the induction of an in vitro model of primitive streak formation using embryonic stem cells. Proc. Natl. Acad. Sci. U S A. 2006;103:16806–16811. doi: 10.1073/pnas.0603916103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S., Paez-Cortez J.R., Boppidi K., Vasconcelos M., Roy M., Cardoso W., Ai X., Fine A. Activation dynamics and signaling properties of Notch3 receptor in the developing pulmonary artery. J. Biol. Chem. 2011;286:22678–22687. doi: 10.1074/jbc.M111.241224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X., Kassab G.S. Variation of mechanical properties along the length of the aorta in C57bl/6 mice. Am. J. Physiol. Circ. Physiol. 2003;285:H2614–H2622. doi: 10.1152/ajpheart.00567.2003. [DOI] [PubMed] [Google Scholar]
- Hibino N., Duncan D.R., Nalbandian A., Yi T., Qyang Y., Shinoka T., Breuer C.K. Evaluation of the use of an induced pluripotent stem cell sheet for the construction of tissue-engineered vascular grafts. J. Thorac. Cardiovasc. Surg. 2012;143:696–703. doi: 10.1016/j.jtcvs.2011.06.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kattman S.J., Witty A.D., Gagliardi M., Dubois N.C., Niapour M., Hotta A., Ellis J., Keller G. Stage-specific optimization of activin/nodal and BMP signaling promotes cardiac differentiation of mouse and human pluripotent stem cell lines. Cell Stem Cell. 2011;8:228–240. doi: 10.1016/j.stem.2010.12.008. [DOI] [PubMed] [Google Scholar]
- Kito T., Shibata R., Ishii M., Suzuki H., Himeno T., Kataoka Y., Yamamura Y., Yamamoto T., Nishio N., Ito S. iPS cell sheets created by a novel magnetite tissue engineering method for reparative angiogenesis. Sci. Rep. 2013;3:1418. doi: 10.1038/srep01418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konig G., McAllister T.N., Dusserre N., Garrido S.A., Iyican C., Marini A., Fiorillo A., Avila H., Wystrychowski W., Zagalski K. Mechanical properties of completely autologous human tissue engineered blood vessels compared to human saphenous vein and mammary artery. Biomaterials. 2009;30:1542–1550. doi: 10.1016/j.biomaterials.2008.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koullias G., Modak R., Tranquilli M., Korkolis D.P., Barash P., Elefteriades J.A. Mechanical deterioration underlies malignant behavior of aneurysmal human ascending aorta. J. Thorac. Cardiovasc. Surg. 2005;130:677.e1–677.e9. doi: 10.1016/j.jtcvs.2005.02.052. [DOI] [PubMed] [Google Scholar]
- Kurmann A.A., Serra M., Hawkins F., Zorn A.M., Hollenberg A.N., Kotton Correspondence D.N. Regeneration of thyroid function by transplantation of differentiated pluripotent stem cells. Cell Stem Cell. 2015;17:527–542. doi: 10.1016/j.stem.2015.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- L’heureux N., Pâquet S., Labbé R., Germain L., Auger F.A. A completely biological tissue-engineered human blood vessel. FASEB J. 1998;12:47–56. doi: 10.1096/fasebj.12.1.47. [DOI] [PubMed] [Google Scholar]
- Lashkarinia S.S., Piskin S., Bozkaya T.A., Salihoglu E., Yerebakan C., Pekkan K. Computational pre-surgical planning of arterial patch reconstruction: parametric limits and in vitro validation. Ann. Biomed. Eng. 2018;46:1292–1308. doi: 10.1007/s10439-018-2043-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T.-H., Song S.-H., Kim K.L., Yi J.-Y., Shin G.-H., Kim J.Y., Kim J., Han Y.-M., Lee S.H., Lee S.-H. Functional recapitulation of smooth muscle cells via induced pluripotent stem cells from human aortic smooth muscle cells. Circ. Res. 2010;106:120–128. doi: 10.1161/CIRCRESAHA.109.207902. [DOI] [PubMed] [Google Scholar]
- Lim S., Park S. Role of vascular smooth muscle cell in the inflammation of atherosclerosis. BMB Rep. 2014;47:1–7. doi: 10.5483/BMBRep.2014.47.1.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin F., Zhang H., Huang J., Xiong C. Contractility of airway smooth muscle cell in response to zinc oxide nanoparticles by traction force microscopy. Ann. Biomed. Eng. 2018;46:2000–2011. doi: 10.1007/s10439-018-2098-3. [DOI] [PubMed] [Google Scholar]
- Lin H., Qiu X., Du Q., Li Q., Wang O., Akert L., Wang Z., Anderson D., Liu K., Gu L. Engineered microenvironment for manufacturing human pluripotent stem cell-derived vascular smooth muscle cells. Stem Cell Reports. 2018;12:84–97. doi: 10.1016/j.stemcr.2018.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maguire E.M., Xiao Q., Xu Q. Differentiation and application of induced pluripotent stem cell–derived vascular smooth muscle cells. Arterioscler. Thromb. Vasc. Biol. 2017;37:2026–2037. doi: 10.1161/ATVBAHA.117.309196. [DOI] [PubMed] [Google Scholar]
- Marchand M., Anderson E.K., Phadnis S.M., Longaker M.T., Cooke J.P., Chen B., Reijo Pera R.A. Concurrent generation of functional smooth muscle and endothelial cells via a vascular progenitor. Stem Cells Transl. Med. 2014;3:91–97. doi: 10.5966/sctm.2013-0124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murfee W., Skalak T., Peirce S. Differential arterial/venous expression of ng2 proteoglycan in perivascular cells along microvessels: identifying a venule-specific phenotype. Microcirculation. 2005;12:151–160. doi: 10.1080/10739680590904955. [DOI] [PubMed] [Google Scholar]
- Niklason L.E., Gao J., Abbott W.M., Hirschi K.K., Houser S., Marini R., Langer R. Functional arteries grown in vitro. Science. 1999;284:489–493. doi: 10.1126/science.284.5413.489. [DOI] [PubMed] [Google Scholar]
- Nostro M.C., Cheng X., Keller G.M., Gadue P. Wnt, activin, and BMP signaling regulate distinct stages in the developmental pathway from embryonic stem cells to blood. Cell Stem Cell. 2008;2:60–71. doi: 10.1016/j.stem.2007.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owens G.K., Kumar M.S., Wamhoff B.R. Molecular regulation of vascular smooth muscle cell differentiation in development and disease. Physiol. Rev. 2004;84:767–801. doi: 10.1152/physrev.00041.2003. [DOI] [PubMed] [Google Scholar]
- Paez-Cortez J., Krishnan R., Arno A., Aven L., Ram-Mohan S., Patel K.R., Lu J., King O.D., Ai X., Fine A. A new approach for the study of lung smooth muscle phenotypes and its application in a murine model of allergic airway inflammation. PLoS One. 2013;8:e74469. doi: 10.1371/journal.pone.0074469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patsch C., Challet-Meylan L., Thoma E.C., Urich E., Heckel T., O’Sullivan J.F., Grainger S.J., Kapp F.G., Sun L., Christensen K. Generation of vascular endothelial and smooth muscle cells from human pluripotent stem cells. Nat. Cell Biol. 2015;17:994–1003. doi: 10.1038/ncb3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelaia G., Renda T., Gallelli L., Vatrella A., Busceti M.T., Agati S., Caputi M., Cazzola M., Maselli R., Marsico S.A. Molecular mechanisms underlying airway smooth muscle contraction and proliferation: implications for asthma. Respir. Med. 2008;102:1173–1181. doi: 10.1016/j.rmed.2008.02.020. [DOI] [PubMed] [Google Scholar]
- Polacheck W.J., Chen C.S. Measuring cell-generated forces: a guide to the available tools. Nat. Methods. 2016;13:415–423. doi: 10.1038/nmeth.3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasain N., Lee M.R., Vemula S., Meador J.L., Yoshimoto M., Ferkowicz M.J., Fett A., Gupta M., Rapp B.M., Saadatzadeh M.R. Differentiation of human pluripotent stem cells to cells similar to cord-blood endothelial colony-forming cells. Nat. Biotechnol. 2014;32:1151–1157. doi: 10.1038/nbt.3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raphel L., Talasila A., Cheung C., Sinha S. Myocardin overexpression is sufficient for promoting the development of a mature smooth muscle cell-like phenotype from human embryonic stem cells. PLoS One. 2012;7:e44052. doi: 10.1371/journal.pone.0044052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rim N.G., Yih A., Hsi P., Wang Y., Zhang Y., Wong J.Y. Micropatterned cell sheets as structural building blocks for biomimetic vascular patches. Biomaterials. 2018;181:126–139. doi: 10.1016/j.biomaterials.2018.07.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roach M.R., Burton A.C. The reason for the shape of the distensibility curves of arteries. Can. J. Biochem. Physiol. 1957;35:681–690. [PubMed] [Google Scholar]
- Salacinski H.J., Goldner S., Giudiceandrea A., Hamilton G., Seifalian A.M., Edwards A., Carson R.J. The mechanical behavior of vascular grafts: a review. J. Biomater. Appl. 2001;15:241–278. doi: 10.1106/NA5T-J57A-JTDD-FD04. [DOI] [PubMed] [Google Scholar]
- Sinha S., Hoofnagle M.H., Kingston P.A., McCanna M.E., Owens G.K. Transforming growth factor-β1 signaling contributes to development of smooth muscle cells from embryonic stem cells. Am. J. Physiol. Physiol. 2004;287:C1560–C1568. doi: 10.1152/ajpcell.00221.2004. [DOI] [PubMed] [Google Scholar]
- Sinha S., Wamhoff B.R., Hoofnagle M.H., Thomas J., Neppl R.L., Deering T., Helmke B.P., Bowles D.K., Somlyo A.V., Owens G.K. Assessment of contractility of purified smooth muscle cells derived from embryonic stem cells. Stem Cells. 2006;24:1678–1688. doi: 10.1634/stemcells.2006-0002. [DOI] [PubMed] [Google Scholar]
- Sommer C.A., Stadtfeld M., Murphy G.J., Hochedlinger K., Kotton D.N., Mostoslavsky G. Induced pluripotent stem cell generation using a single lentiviral stem cell cassette. Stem Cells. 2009;27:543–549. doi: 10.1634/stemcells.2008-1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syedain Z.H., Meier L.A., Bjork J.W., Lee A., Tranquillo R.T. Implantable arterial grafts from human fibroblasts and fibrin using a multi-graft pulsed flow-stretch bioreactor with noninvasive strength monitoring. Biomaterials. 2011;32:714–722. doi: 10.1016/j.biomaterials.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K., Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Wagenseil J.E., Mecham R.P. Vascular extracellular matrix and arterial mechanics. Physiol. Rev. 2009;89:957–989. doi: 10.1152/physrev.00041.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang A., Tang Z., Li X., Jiang Y., Tsou D.A., Li S. Derivation of smooth muscle cells with neural crest origin from human induced pluripotent stem cells. Cells Tissues Organs. 2012;195:5–14. doi: 10.1159/000331412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissberg P.L., Cary N.R., Shanahan C.M. Gene expression and vascular smooth muscle cell phenotype. Blood Press. Suppl. 1995;2:68–73. [PubMed] [Google Scholar]
- Yang L., Geng Z., Nickel T., Johnson C., Gao L., Dutton J., Hou C., Zhang J. Differentiation of human induced-pluripotent stem cells into smooth-muscle cells: two novel protocols. PLoS One. 2016;11:e0147155. doi: 10.1371/journal.pone.0147155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida T., Kawai-Kowase K., Owens G.K. Forced expression of myocardin is not sufficient for induction of smooth muscle differentiation in multipotential embryonic cells. Arterioscler. Thromb. Vasc. Biol. 2004;24:1596–1601. doi: 10.1161/01.ATV.0000137190.63214.c5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Gene expression data for hierarchical clustering (see Figure 2C) of all 15 samples by one-way ANOVA (2,386 top varying genes based on FDR adjusted p value < 1 × 10−7 and absolute fold change >5). Hierarchical clustering revealed ten different gene clusters. Clusters of interest: undifferentiated iPSCs (cluster 8), lateral plate mesoderm (cluster 6), immature/synthetic smooth muscle (clusters 5 and 7), and mature/contractile smooth muscle (cluster 1) populations.