Key Points
Question
In patients admitted to the hospital with serious infection, is there a significant association between PCSK9 genetic variation and risk of sepsis?
Findings
In this cohort study of 10 922 patients, the risk of sepsis was not significantly associated with PCSK9 functional variants, PCSK9 genetic risk score, or genetic estimation of PCSK9 expression levels.
Meaning
These findings suggest that PCSK9 genetic variations are not significantly associated with the risk of sepsis in patients hospitalized with infection.
This cohort study tests the associations between PCSK9 genetic variants, a PCSK9 genetic risk score, or genetically estimated PCSK9 expression levels and the risk of sepsis among patients admitted to a hospital with infection.
Abstract
Importance
Whether the PCSK9 gene is associated with the progress from infection to sepsis is unknown to date.
Objective
To test the associations between PCSK9 genetic variants, a PCSK9 genetic risk score (GRS), or genetically estimated PCSK9 expression levels and the risk of sepsis among patients admitted to a hospital with infection.
Design, Setting, and Participants
This retrospective cohort study used deidentified electronic health records to identify patients admitted to Vanderbilt University Medical Center, Nashville, Tennessee, with infection. Patients were white adults, had a code indicating infection from the International Classification of Diseases, Ninth Revision, Clinical Modification, or the International Statistical Classification of Diseases, Tenth Revision, Clinical Modification, and received an antibiotic within 1 day of hospital admission (N = 61 502). Data were collected from January 1, 1993, through December 31, 2017, and analyzed from April 1, 2018, to March 16, 2019.
Exposures
Four known PCSK9 functional variants, a GRS for PCSK9, and genetically estimated PCSK9 expression.
Main Outcomes and Measures
The primary outcome was sepsis; secondary outcomes included cardiovascular failure and in-hospital death.
Results
Of patients with infection, genotype information was available in 10 922 white patients for PCSK9 functional variants (5628 men [51.5%]; mean [SD] age, 60.1 [15.7] years), including 7624 patients with PCSK9 GRS and 6033 patients with estimated PCSK9 expression. Of these, 3391 developed sepsis, 835 developed cardiovascular failure, and 366 died during hospitalization. None of the 4 functional PCSK9 variants were significantly associated with sepsis, cardiovascular failure, or in-hospital death, with or without adjustment for (1) age and sex or (2) age, sex, and Charlson-Deyo comorbidities (in model adjusted for age, sex, and comorbidities, odds ratios for any loss-of function variant were 0.96 [95% CI, 0.88-1.04] for sepsis, 1.05 [95% CI, 0.90-1.22] for cardiovascular failure, and 0.89 [95% CI, 0.72-1.11] for death). Similarly, neither the PCSK9 GRS nor genetically estimated PCSK9 expression were significantly associated with sepsis, cardiovascular failure, or in-hospital death in any of the analysis models. For GRS, in the full model adjusted for age, sex, and comorbidities, the odds ratios were 1.01 for sepsis (95% CI, 0.96-1.06; P = .70), 1.03 for cardiovascular failure (95% CI, 0.95-1.12; P = .48), and 1.05 for in-hospital death (95% CI, 0.92-1.19; P = .50). For genetically estimated PCSK9 expression, in the full model adjusted for age, sex, and comorbidities, the odds ratios were 1.01 for sepsis (95% CI, 0.95-1.06; P = .86), 0.96 for cardiovascular failure (95% CI, 0.88-1.05; P = .41), and 0.99 for in-hospital death (95% CI, 0.87-1.14; P = .94).
Conclusions and Relevance
In this study, PCSK9 genetic variants were not significantly associated with risk of sepsis or the outcomes of sepsis in patients hospitalized with infection.
Introduction
Sepsis is a life-threatening organ dysfunction caused by a dysregulated host response to infection1 and is present in 35% of deaths that occur in US hospitals.2 There are no specific treatments for sepsis; management consists of cardiorespiratory resuscitation and treatment of infection.3 Studies suggest that drugs that inhibit the PCSK9 gene could have potential as a new treatment for sepsis.4,5,6,7
The PCSK9 gene increases low-density lipoprotein cholesterol (LDL-C) concentrations by targeting LDL receptors (LDLRs) for destruction, thereby decreasing transport of LDL into the liver and thus increasing circulating LDL-C concentrations.8,9 Therefore, as expected, gain-of-function (GOF) and loss-of-function (LOF) variants in the PCSK9 gene (OMIM 607786) are associated with increased and decreased LDL-C concentrations, respectively.10 Concordant with those observations, drugs that inhibit the PCSK9 gene lower LDL-C concentrations markedly.11,12,13 However, in addition to its role in the regulation of LDL-C concentrations, PCSK9 also appears to affect the outcomes of sepsis through LDLR-mediated effects on bacterial lipids.
Toxic lipids such as lipopolysaccharide (LPS) from gram-negative bacteria and lipoteichoic acid (LTA) from gram-positive bacteria trigger many of the manifestations of sepsis such as vasodilation, increased capillary permeability, and the production of inflammatory cytokines.14 Lipopolysaccharide binds to LDL-C and is taken up via LDLRs into hepatocytes, where it does not cause cell injury or cytokine production. The PCSK9 gene plays a role in detoxifying toxic bacterial lipids such as LPS15,16 because it controls the number of LDLRs available.14 Thus, PCSK9 could play a role in modulating responses to LPS and sepsis; several lines of evidence in animals and humans support this idea.4,5
In mice, deletion of Pcsk9 attenuated cytokine and physiological responses to LPS, and treatment with a PCSK9 inhibitor improved survival in a model of sepsis.4 Also, in patients with septic shock, the presence of PCSK9 LOF variants was associated with improved survival.4 Consequently, drugs that inhibit PCSK9 are under investigation as a therapy for sepsis.5 Moreover, because sepsis represents an extreme on a continuum of illness, the effects of PCSK9 inhibition could be more far-reaching and may not only treat sepsis but also prevent the progression from serious infection to sepsis.
We hypothesized that PCSK9 LOF variants protect patients with infection from developing sepsis, and, in those who did develop sepsis, that PCSK9 LOF variants would be associated with improved in-hospital survival. We addressed this hypothesis using 3 complementary approaches: (1) PCSK9 variants previously shown to be associated with survival in patients with sepsis4; (2) a PCSK9 genetic risk score (GRS)17; and (3) genetically determined PCSK9 expression levels imputed using GTEx (the Genotype-Tissue Expression project) expression quantitative traits loci data.18,19
Methods
Data Sources
Data were obtained from BioVU, a deidentified DNA biobank, and the Synthetic Derivative, a deidentified version of the electronic health record (EHR), for patients at Vanderbilt University Medical Center (VUMC). The Synthetic Derivative contains the deidentified records of approximately 2.9 million people, and their clinical information is updated every 1 to 3 months. Information available includes diagnostic and procedure codes, demographics, clinical notes, problem lists, laboratory values, and medications. BioVU is linked to the Synthetic Derivative and contains more than 285 000 DNA samples, of which approximately 87 000 have undergone genome-wide genotyping.20,21,22 BioVU accrues blood samples collected for clinical care that were otherwise scheduled to be discarded and applies a secure hash algorithm to deidentify the samples, which in part leads to a nonhuman subject designation with the institutional review board of VUMC. The program has received approval from the institutional review board of VUMC and was reviewed in detail by the federal Office for Human Research Protections, who agreed with the nonhuman subjects regulatory designation for the resource and subsequent research. From 2007 to 2014, BioVU operated under an opt-out accumulation model based on an opinion from the federal Office of Human Research Protection that discarded samples can be used for biomedical research without prospective consenting of each individual if the clinical data are deidentified. From 2015 to present, BioVU implemented a consented model, which adheres to the National Institutes of Health policy on genomic data sharing that was established in 2015. This study was approved by the VUMC institutional review board and followed the Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) reporting guideline.
Cohort Identification
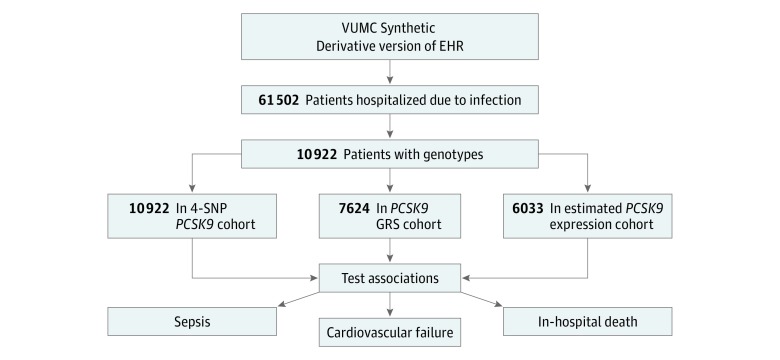
Data were collected from January 1, 1993, through December 31, 2017, and analyzed from April 1, 2018, to March 16, 2019. Using methods previously described in detail,23 we identified white patients 18 years and older who had been admitted to the hospital with an infection (Figure 1) and who had genome-wide genotyping available. The day of hospital admission was designated day 0. Infection was defined as (1) having a billing code indicating infection and (2) receiving an antibiotic within 1 day of hospital admission (ie, on day −1, 0, or +1). The International Classification of Disease, Ninth Revision, Clinical Modification (ICD-9-CM) codes were used for cohort construction and covariates, and ICD-9-CM and the International Statistical Classification of Diseases, Tenth Revision, Clinical Modification (ICD-10-CM) codes were used for outcomes. The ICD-9-CM codes for infection were based on the criteria of Angus and colleagues24 and Donnelly and colleagues,25 excluding nonbacterial infections.23 If more than 1 qualifying episode of infection was present in a patient’s EHR, only the first was included.
Figure 1. Study Design.
EHR indicates electronic health record; GRS, genetic risk score; SNP, single-nucleotide polymorphism; and VUMC, Vanderbilt University Medical Center.
Outcomes
The primary outcome was sepsis defined using the Third International Consensus Definitions for Sepsis of concurrent infection and organ dysfunction25 and identified with a validated algorithm2 (with minor modifications23) that uses billing codes and clinical variables to identify sepsis in the EHR (eFigure and eMethods in the Supplement). The algorithm is highly specific (98%) and moderately sensitive (70%).2 Most cases of sepsis are present on admission to the hospital2; therefore, to minimize the confounding effects of sepsis occurring secondary to procedures or events while in the hospital, we studied sepsis occurring within 1 day of hospital admission (days −1, 0, and +1). Individuals with infection met the definition of sepsis if they had septic shock or severe sepsis or met any organ dysfunction criterion.2,23 Septic shock and severe sepsis were defined by ICD-10-CM codes that are highly specific (99%).2 The following criteria were used to define organ dysfunction23: (1) cardiovascular failure as use of the vasopressor norepinephrine bitartrate (Levophed) or the vasopressors dobutamine hydrochloride or dopamine hydrochloride and a billing code for administration of a vasopressor; (2) respiratory failure as use of codes for ventilation and admission to the intensive care unit; (3) renal failure as doubling or greater increase of baseline creatinine concentration (defined as the lowest creatinine concentration from 1 year before admission to hospital discharge); (4) hepatic failure as total bilirubin level of at least 2.0 mg/dL (to convert to micromoles per liter, multiply by 17.104) and doubled from baseline (the baseline value was defined as the lowest total bilirubin level occurring from 1 year before admission to hospital discharge); and (5) hematologic failure as platelet count of less than 100 × 103/μL (to convert to ×109 per liter, multiply by 1.0) and at least 50% decline from a baseline that must have been at least 100 × 103/μL (the baseline value was defined as the highest platelet count occurring from 1 year before admission to hospital discharge). We also identified 2 secondary outcomes for exploratory purposes: cardiovascular failure as defined for the primary outcome (ie, requiring vasopressors) and in-hospital mortality. In-hospital death was extracted from discharge notes and death summaries. Event counts are summarized in eTable 1 and eTable 2 in the Supplement.
Covariates
Covariates, including age and sex, were extracted from the EHR. Age was ascertained at the time of the index hospital admission. We also established the presence or absence of the comorbidities that constitute the Charlson-Deyo Comorbidity Index.26,27,28 We also calculated 6 principal components for ancestry using SNPRelate, version 1.16.0 (Bioconductor). For each patient, relevant diagnostic comorbidity codes occurring in the year before the index hospital admission were grouped into PheCodes28 using the R PheWAS package.29 The PheCodes were further grouped into 16 Charlson-Deyo comorbidity categories, with the categories of diabetes and diabetes with complications combined into a single diabetes variable.23 We previously performed a manual review of the performance of the sepsis criteria in 50 random EHRs. All patients met the inclusion and exclusion criteria, and the algorithms performed well.23
Functional PCSK9 Gene Variants and the PCSK9 GRS
We used the following 3 approaches to examine the association between PCSK9 gene variants and sepsis: (1) 4 PCSK9 functional variants previously shown to affect survival in patients with sepsis4; (2) a PCSK9 GRS previously shown to detect an association with risk of cardiovascular events and diabetes30; and (3) GTEx data to estimate genetic expression of PCSK9.19 Four PCSK9 single-nucleotide polymorphisms (SNPs) were previously associated with sepsis outcomes4: rs505151 is a GOF variant, and the remainders, rs11591147, rs11583680, and rs562556, are reduced function (termed LOF) variants. Genotypes for these PCSK9 variants were extracted for 10 922 individuals who underwent genotyping on ExomeChip or genome-wide genotype platforms with imputation through the Michigan Imputation server31 with minimac3, using the Haplotype Reference Consortium reference panel, version r1.1.32,33
We constructed the PCSK9 GRS using the method established by Ference et al30 with a minor modification. Ference et al30 constructed a PCSK9 GRS with 7 SNPs (rs11206510, rs2479409, rs2149041, rs2479394, rs10888897, rs7552841, and rs562556); however, rs2479409 and rs2149041 were moderately correlated (R2 > 0.3) in our cohort. Therefore, as others who have constructed PCSK9 risk scores have also done,34 we removed rs2149041 and constructed a weighted 6-SNP GRS using the estimated effect size for each SNP as determined by the Global Lipids Genetics Consortium (eTable 4 in the Supplement).35,36
Expression of PCSK9 in liver was imputed for individuals based on their genotypes using the expression predictors previously trained on the GTEx V6p release of RNA sequencing data (eTable 5 in the Supplement).19 We tested the associations between median measured LDL-C levels and (1) 4 functional PCSK9 variants, (2) PCSK9 GRS, and (3) estimated PCSK9 expression (eTables 6-8 in the Supplement).
Statistical Analysis
We used logistic regression to test the associations between PCSK9 (4 functional variants, GRS, and genetically determined expression levels) and outcomes (sepsis, in-hospital death, and risk of cardiovascular failure). Concordant with analyses performed by others4 for the 4-SNP analysis, we also examined outcomes in the group of patients who carried any LOF variant. We used an additive model for single-variant association analysis and estimated odds ratios (ORs) and 95% CIs per 1-SD increase for the association between the PCSK9 GRS or PCSK9 expression levels and outcomes. All analyses were adjusted for (1) age and sex or (2) age, sex, and comorbidities. Models were adjusted for each comorbidity category individually because each category is likely to contribute to sepsis risk differently. To determine the association between comorbidity and outcomes, we constructed a single comorbidity score for each individual and tested the associations between this score and outcomes. Analyses were performed using PLINK, version 1.9,37 and R, version 3.4.4 (R Project for Statistical Computing). We conducted power calculation using principal component software (eMethods in the Supplement).38 The primary outcome was sepsis studied in 3 ways: (1) 4 functional PCSK9 variants, (2) PCSK9 GRS, and (3) genetically estimated PCSK9 expression; thus, 2-sided P < .0167 was considered statistically significant. Secondary outcomes and other analyses were regarded as exploratory.
Results
A total of 61 502 patients met the definition of infection, of whom 10 922 white adults (5294 women [48.5%] and 5628 men [51.5%]; mean [SD] age, 60.1 [15.7] years) had genotypes available for the PCSK9 functional variants (Table 1); of these, 3391 developed sepsis, 835 developed cardiovascular failure, and 366 died during hospitalization. Mean (SD) duration of hospitalization was 7.5 (9.1) days. The comorbidity score was associated with the outcomes of (1) sepsis (OR, 1.27; 95% CI, 1.26-1.29; P < 2 × 10−16); (2) cardiovascular failure (OR, 1.29; 95% CI, 1.26-1.31; P < 2 × 10−16); and (3) in-hospital death (OR, 1.39; 95% CI, 1.37-1.42; P < 2 × 10−16) (eTable 3 in the Supplement).
Table 1. Demographic Characteristics of Patients With Infection and Genotyping.
| Characteristic | Patient Data (n = 10 922) |
|---|---|
| Sex, No. (%) | |
| Female | 5294 (48.5) |
| Male | 5628 (51.5) |
| Age, mean (SD), y | 60.1 (15.7) |
| Comorbidities in the year preceding hospital admission, No. (%) | |
| Diabetes | 3274 (30.0) |
| Any malignant neoplasm, including lymphoma and leukemia, except malignant neoplasm of skin | 2831 (25.9) |
| Chronic pulmonary disease | 2694 (24.7) |
| Congestive heart failure | 2185 (20.0) |
| Cerebrovascular disease | 1741 (15.9) |
| Metastatic solid tumor | 1665 (15.2) |
| Myocardial infarction | 1464 (13.4) |
| Peripheral vascular disease | 952 (8.7) |
| Mild liver disease | 867 (7.9) |
| Renal disease | 820 (7.5) |
| Moderate or severe liver disease | 787 (7.2) |
| Rheumatic disease | 674 (6.2) |
| Peptic ulcer disease | 401 (3.7) |
| Hemiplegia or paraplegia | 301 (2.8) |
| Dementia | 268 (2.5) |
| AIDS/HIV | 133 (1.2) |
PCSK9 Functional Gene Variants and Outcomes
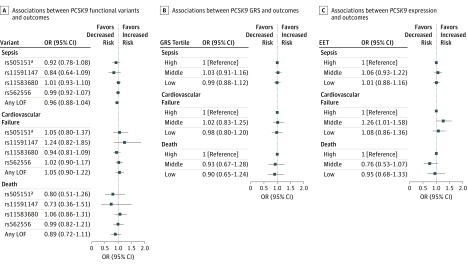
None of the 4 functional PCSK9 gene variants were significantly associated with sepsis, cardiovascular failure, or in-hospital death (Figure 2 and eTable 9 in the Supplement) with or without adjustment for (1) age and sex or (2) age, sex, and comorbidities (in model adjusted for age, sex, and comorbidities, odds ratios for any loss-of function variant were 0.96 [95% CI, 0.88-1.04] for sepsis, 1.05 [95% CI, 0.90-1.22] for cardiovascular failure, and 0.89 [95% CI, 0.72-1.11] for death) (eTable 9 in the Supplement). The group of patients who carried any LOF variant (n = 4965) did not have outcomes that differed significantly from noncarriers (Figure 2).
Figure 2. Associations Between PCSK9 Cohorts and Sepsis Outcomes.
Outcomes include sepsis, cardiovascular failure, and in-hospital death. A, Associations between PCSK9 functional gene variants and outcomes are adjusted for age, sex, and comorbidities. B and C, Associations between the PCSK9 genetic risk score (GRS) and estimated PCSK9 expression and outcomes are stratified by tertiles and adjusted for age, sex, and comorbidities. EET indicates estimated expression tertile; LOF loss-of-function variant; OR, odds ratio.
aIndicates a gain-of-function variant.
PCSK9 GRS and Outcomes
In white patients with infection in whom genotypes were available to construct the PCSK9 GRS (n = 7624), the GRS was not significantly associated with sepsis, in-hospital death, or cardiovascular failure in any of the models (Table 2). In the full model adjusted for age, sex, and comorbidities, the ORs were 1.01 for sepsis (95% CI, 0.96-1.06; P = .70), 1.03 for cardiovascular failure (95% CI, 0.95-1.12; P = .48), and 1.05 for in-hospital death (95% CI, 0.92-1.19; P = .50).
Table 2. Associations Between PCSK9 Genetic Risk Score and Sepsis-Related Adverse Outcomesa.
| Phenotype | Unadjusted | Adjusted for Age and Sex | Adjusted for Age, Sex, and Comorbidities | Adjusted for Age, Sex, Comorbidities, and 6 PCs | ||||
|---|---|---|---|---|---|---|---|---|
| OR (95% CI) | P Value | OR (95% CI) | P Value | OR (95% CI) | P Value | OR (95% CI) | P Value | |
| Sepsis | 1.01 (0.96-1.06) | .73 | 1.01 (0.96-1.06) | .79 | 1.01 (0.96-1.06) | .70 | 1.01 (0.96-1.06) | .70 |
| Cardiovascular failure | 1.03 (0.95-1.12) | .49 | 1.03 (0.95-1.11) | .55 | 1.03 (0.95-1.12) | .48 | 1.03 (0.95-1.12) | .46 |
| In-hospital death | 1.06 (0.93-1.20) | .41 | 1.05 (0.93-1.20) | .43 | 1.05 (0.92-1.19) | .50 | 1.05 (0.92-1.20) | .46 |
Abbreviations: OR, odds ratio; PCs, principal components.
Includes 7624 patients with a genetic risk score.
Genetically Estimated PCSK9 Expression Levels and Outcomes
In white patients with infection in whom PCSK9 expression levels could be calculated (n = 6033), expression was not significantly associated with sepsis, in-hospital death, or cardiovascular failure in any of the models (Table 3). In the full model adjusted for age, sex, and comorbidities, the ORs were 1.01 for sepsis (95% CI, 0.95-1.06; P = .86), 0.96 for cardiovascular failure (95% CI, 0.88-1.05; P = .41), and 0.99 for in-hospital death (95% CI, 0.87-1.14; P = .94).
Table 3. Associations Between Estimated PCSK9 Expression and Sepsis-Related Adverse Outcomesa.
| Phenotype | Unadjusted | Adjusted for Age and Sex | Adjusted for Age, Sex, and Comorbidities | Adjusted for Age, Sex, Comorbidities, and 6 PCs | ||||
|---|---|---|---|---|---|---|---|---|
| OR (95% CI) | P Value | OR (95% CI) | P Value | OR (95% CI) | P Value | OR (95% CI) | P Value | |
| Sepsis | 1.01 (0.95-1.06) | .83 | 1.01 (0.96-1.06) | .76 | 1.01 (0.95-1.06) | .86 | 1.01 (0.95-1.06) | .85 |
| Cardiovascular failure | 0.97 (0.89-1.06) | .47 | 0.97 (0.89-1.06) | .51 | 0.96 (0.88-1.05) | .41 | 0.96 (0.88-1.05) | .41 |
| In-hospital death | 1.01 (0.88-1.15) | .94 | 1.00 (0.87-1.15) | .96 | 0.99 (0.87-1.14) | .94 | 1.00 (0.87-1.15) | .99 |
Abbreviations: OR, odds ratio; PCs, principal components.
Includes 6033 patients with estimated PCSK9 expression.
For illustrative purposes, we show the risk of sepsis, in-hospital mortality, and risk of cardiovascular failure within different tertiles (low, middle, and high) of the PCSK9 GRS and genetically estimated PCSK9 expression levels. Compared with those in the highest tertile, patients in the low or middle tertiles of the PCSK9 GRS (Figure 2 and eTable 10 in the Supplement) and of genetically estimated PCSK9 expression levels (Figure 2 and eTable 11 in the Supplement) had a similar risk of sepsis, cardiovascular failure, and in-hospital mortality, except that the middle tertile of genetically estimated PCSK9 expression had a higher risk of cardiovascular failure at a nominal level of significance (OR, 1.26; 95% CI, 1.01-1.58; P = .04).
Discussion
The major finding of this study is that variants in the PCSK9 gene and estimated expression of PCSK9 were not associated with the risk of developing sepsis or the risk of poorer outcomes in patients admitted to hospital with infection. Low-density lipoprotein, through its ability to bind LPS and other toxic bacterial lipids, attenuated the severity of sepsis in animal studies.14 Low-density lipoprotein and its LPS cargo are internalized via LDLRs into hepatocytes, thus preventing the cellular and systemic toxic effects associated with LPS.14 Accordingly, because PCSK9 increases degradation of LDLRs, it decreases LPS uptake into the hepatocytes4,39; thus, strategies that target the PCSK9 gene would be expected to attenuate the toxic effects of LPS and sepsis. Indeed, 2 studies in animal models support this idea.4,40
Mice with a deletion of Pcsk9 had decreased production of interleukin-6 and other inflammatory cytokines compared with control animals after they received intraperitoneal LPS; they also had attenuated physiological responses to LPS with greater preservation of mean arterial pressure and ejection fraction.4 In another murine model of sepsis, Pcsk9 overexpression was associated with increased bacterial dissemination, inflammation, and histopathological damage compared with control animals, whereas mice with deletion of Pcsk9 were protected against these effects.40
Pharmacological inhibition of PCSK9 also protected against the effects of sepsis in mice; an antibody that blocked PCSK9 blunted inflammatory cytokines and improved survival in a polymicrobial peritonitis model.4 These beneficial effects were absent when the gene for the LDLR was deleted, indicating that PCSK9 inhibition altered responses to LPS through the LDLR.4 However, in a second study, neither Pcsk9 deletion nor PCSK9 antibodies protected mice against LPS-induced mortality.41
Less information is available to date about PCSK9 and its association with LPS clearance and sepsis in humans. Previous observations23,42 found that levels of LDL-C do not appear to be directly associated with the risk of sepsis in patients with infection. In healthy participants, peak and area-under-the-curve of interleukin 6 response to an intravenous injection of LPS were attenuated in those who carried a PCSK9 LOF allele compared with those who did not.4 In patients with sepsis, plasma PCSK9 concentrations were elevated compared with those without sepsis and were associated with decreased clearance of endotoxin.43 Moreover, in 2 cohorts of patients with septic shock, Walley and colleagues4,42 reported that the groups that carried any of 3 PCSK9 LOF variants had improved survival compared with the groups with no variants or a GOF variant. Conversely, groups with a GOF variant had worse survival than those with no variants or an LOF variant.4
Because sepsis is a continuum of severe illness after infection, and because studies in animals showed that PCSK9 was associated with not only survival after sepsis but also bacterial dissemination and physiological responses to LPS and infection,4,40 we hypothesized that PCSK9 decreased-function genetic variants would be associated with a lower risk of progression from infection to sepsis. We used 3 approaches. First, we studied the same 4 variants that were previously associated with altered survival in patients with septic shock.4 Second, we used a PCSK9 GRS (derived from the effect of variants on LDL-C levels) that previously detected associations between PCSK9 and risk of cardiovascular events and diabetes.30 Third, we estimated genetic PCSK9 expression levels using GTEx information. The 3 approaches yielded concordant results: PCSK9 variants were not significantly associated with risk of developing sepsis in patients with infection. Similarly, these variants were not significantly associated with the other outcomes.
Our study had several differences from that of Walley and colleagues4 that may account for the different conclusions. First, their patients had septic shock and were enrolled from 2 clinical trials; second, they included several ethnic groups with statistical adjustment for ancestry; and third, the outcome was 28-day mortality. The present study included not only patients with septic shock but also those with other manifestations of sepsis. Because we included all patients with sepsis at a large tertiary care hospital who had genotype information, the characteristics of these patients differ from those enrolled in clinical trials for septic shock. For example, to better establish the progressive association between infection and sepsis, we required that sepsis occur within 1 day of hospitalization for infection. Thus, patients admitted for a surgical procedure who subsequently developed sepsis in the hospital would not be included in our study. Also, we studied only white patients to minimize the effects of population stratification on any association between genotype and outcomes; however, this is unlikely to lead to a false-negative result because the sample size was large. In-hospital mortality was a secondary outcome because it is more clearly linked with the episode of sepsis, and we found no association between carriage of PCSK9 LOF or GOF variants and this outcome. Nevertheless, we cannot exclude the possibility that beneficial effects of PCSK9 variants on sepsis outcomes would be observed after patients are discharged from hospital.44
Strengths and Limitations
The study included patients who are representative of those admitted to a tertiary care hospital with infection, and the findings are therefore broadly generalizable to that population. However, we cannot exclude a potential effect of PCSK9 on outcomes in patients with sepsis associated with specific causes, or those with specific manifestations of sepsis. Second, information in the EHR linked to genotypes allowed us to perform the study efficiently; however, we did not perform a randomized clinical trial. One major advantage of randomization is to minimize unequal distribution of confounding variables among groups of interest. The genetic approaches used also have this advantage because constructing groups according to genotype should be minimally affected by unequal distribution of confounders. Third, we used 3 genetic approaches: the 4-SNP approach that previously found an association with sepsis outcomes,4 a PCSK9 GRS previously associated with cardiovascular outcomes,30 and genetically estimated gene expression levels.19 In BioVU, rs11591147 and rs505151 and the PCSK9 GRS were significantly associated with measured LDL-C; however, estimated PCSK9 expression was not (eTables 6-8 in the Supplement). Protein function could be more important than variability in gene expression in regulating LDL-C levels. Furthermore, although these approaches have previously been successful in other studies, the genetic signal may have been too small to affect the outcomes, and a clinical trial of a drug that inhibits PCSK9 in patients with sepsis could reach different conclusions. Fourth, we did not evaluate the association between statin use and risk of sepsis. The question is complex: higher LDL-C levels have been postulated to be protective in sepsis (although a previous study of LDL-C in sepsis23 did not support that idea), and statins, which lower LDL-C levels, have also been postulated to be beneficial in sepsis. The results have been inconclusive, and a systematic review and meta-analysis of randomized trials45 concluded that statin treatment was not associated with reduced mortality in sepsis. In addition, this study will require replication. The current algorithm included International Classification of Disease or Current Procedural Terminology codes, medications, laboratory test results, and knowledge of temporal associations; such information is not available in many databases. We believe that in the future, the algorithm can be modified and applied to other practice-based cohorts to test the generalizability of our observations.
Conclusions
In this study, genetic variants of PCSK9 were not significantly associated with the risk of sepsis or with the outcomes of sepsis in patients hospitalized with infection. We believe that future studies should modify the algorithm used and include practice-based cohorts.
eMethods. Cohort Identification and Power Calculations
eTable 1. Event Counts in Each Analysis
eTable 2. Event Counts by Genotypes
eTable 3. Associations Between Comorbidity Score and Sepsis and Related Outcomes
eTable 4. SNPs Included in PCSK9 Genetic Risk Score
eTable 5. SNPs Included in Estimated PCSK9 Expression
eTable 6. Associations Between Median Measured LDL-C Levels and 4 Functional PCSK9 Variants
eTable 7. Associations Between Median Measured LDL-C Levels and PCSK9 GRS
eTable 8. Associations Between Median Measured LDL-C Levels and PCSK9 Expression
eTable 9. Associations Between PCSK9 Candidate SNPs and Sepsis and Related Adverse Outcomes
eTable 10. Associations Between PCSK9 GRS and Sepsis and Related Adverse Outcomes
eTable 11. Associations Between Genetically Estimated PCSK9 Expression Tertiles and Sepsis and Related Adverse Outcomes
eFigure. Algorithm to Identify Sepsis Within Infection Cohort
eReferences.
References
- 1.Singer M, Deutschman CS, Seymour CW, et al. The Third International Consensus Definitions for Sepsis and Septic Shock (Sepsis-3). JAMA. 2016;315(8):-. doi: 10.1001/jama.2016.0287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rhee C, Dantes R, Epstein L, et al. ; CDC Prevention Epicenter Program . Incidence and trends of sepsis in US hospitals using clinical vs claims data, 2009-2014. JAMA. 2017;318(13):1241-1249. doi: 10.1001/jama.2017.13836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Angus DC, van der Poll T. Severe sepsis and septic shock. N Engl J Med. 2013;369(9):840-851. doi: 10.1056/NEJMra1208623 [DOI] [PubMed] [Google Scholar]
- 4.Walley KR, Thain KR, Russell JA, et al. PCSK9 is a critical regulator of the innate immune response and septic shock outcome. Sci Transl Med. 2014;6(258):258ra143. doi: 10.1126/scitranslmed.3008782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Momtazi AA, Banach M, Sahebkar A. PCSK9 inhibitors in sepsis: a new potential indication? Expert Opin Investig Drugs. 2017;26(2):137-139. doi: 10.1080/13543784.2017.1272570 [DOI] [PubMed] [Google Scholar]
- 6.Paciullo F, Fallarino F, Bianconi V, Mannarino MR, Sahebkar A, Pirro M. PCSK9 at the crossroad of cholesterol metabolism and immune function during infections. J Cell Physiol. 2017;232(9):2330-2338. doi: 10.1002/jcp.25767 [DOI] [PubMed] [Google Scholar]
- 7.Bray N. Sepsis: PCSK9 blockade helps clear pathogenic lipids. Nat Rev Drug Discov. 2014;13(12):886. doi: 10.1038/nrd4492 [DOI] [PubMed] [Google Scholar]
- 8.Urban D, Pöss J, Böhm M, Laufs U. Targeting the proprotein convertase subtilisin/kexin type 9 for the treatment of dyslipidemia and atherosclerosis. J Am Coll Cardiol. 2013;62(16):1401-1408. doi: 10.1016/j.jacc.2013.07.056 [DOI] [PubMed] [Google Scholar]
- 9.Peterson AS, Fong LG, Young SG. PCSK9 function and physiology. J Lipid Res. 2008;49(7):1595-1599. doi: 10.1194/jlr.CX00001-JLR200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cohen JC, Boerwinkle E, Mosley TH Jr, Hobbs HH. Sequence variations in PCSK9, low LDL, and protection against coronary heart disease. N Engl J Med. 2006;354(12):1264-1272. doi: 10.1056/NEJMoa054013 [DOI] [PubMed] [Google Scholar]
- 11.Robinson JG, Farnier M, Krempf M, et al. ; ODYSSEY LONG TERM Investigators . Efficacy and safety of alirocumab in reducing lipids and cardiovascular events. N Engl J Med. 2015;372(16):1489-1499. doi: 10.1056/NEJMoa1501031 [DOI] [PubMed] [Google Scholar]
- 12.Sabatine MS, Giugliano RP, Wiviott SD, et al. ; Open-Label Study of Long-Term Evaluation against LDL Cholesterol (OSLER) Investigators . Efficacy and safety of evolocumab in reducing lipids and cardiovascular events. N Engl J Med. 2015;372(16):1500-1509. doi: 10.1056/NEJMoa1500858 [DOI] [PubMed] [Google Scholar]
- 13.Giugliano RP, Pedersen TR, Park JG, et al. ; FOURIER Investigators . Clinical efficacy and safety of achieving very low LDL-cholesterol concentrations with the PCSK9 inhibitor evolocumab: a prespecified secondary analysis of the FOURIER trial. Lancet. 2017;390(10106):1962-1971. doi: 10.1016/S0140-6736(17)32290-0 [DOI] [PubMed] [Google Scholar]
- 14.Grin PM, Dwivedi DJ, Chathely KM, et al. Low-density lipoprotein (LDL)–dependent uptake of gram-positive lipoteichoic acid and gram-negative lipopolysaccharide occurs through LDL receptor. Sci Rep. 2018;8(1):10496. doi: 10.1038/s41598-018-28777-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Levels JHM, Abraham PR, van den Ende A, van Deventer SJH. Distribution and kinetics of lipoprotein-bound endotoxin. Infect Immun. 2001;69(5):2821-2828. doi: 10.1128/IAI.69.5.2821-2828.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Levels JHM, Abraham PR, van Barreveld EP, Meijers JCM, van Deventer SJH. Distribution and kinetics of lipoprotein-bound lipoteichoic acid. Infect Immun. 2003;71(6):3280-3284. doi: 10.1128/IAI.71.6.3280-3284.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ference BA, Majeed F, Penumetcha R, Flack JM, Brook RD. Effect of naturally random allocation to lower low-density lipoprotein cholesterol on the risk of coronary heart disease mediated by polymorphisms in NPC1L1, HMGCR, or both: a 2 × 2 factorial Mendelian randomization study. J Am Coll Cardiol. 2015;65(15):1552-1561. doi: 10.1016/j.jacc.2015.02.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ardlie KG, et al. ; GTEx Consortium . Human genomics: the Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348(6235):648-660. doi: 10.1126/science.1262110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gamazon ER, Wheeler HE, Shah KP, et al. ; GTEx Consortium . A gene-based association method for mapping traits using reference transcriptome data. Nat Genet. 2015;47(9):1091-1098. doi: 10.1038/ng.3367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wei W-Q, Denny JC. Extracting research-quality phenotypes from electronic health records to support precision medicine. Genome Med. 2015;7(1):41. doi: 10.1186/s13073-015-0166-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu H, Stenner SP, Doan S, Johnson KB, Waitman LR, Denny JC. MedEx: a medication information extraction system for clinical narratives. J Am Med Inform Assoc. 2010;17(1):19-24. doi: 10.1197/jamia.M3378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Denny JC, Bastarache L, Ritchie MD, et al. Systematic comparison of phenome-wide association study of electronic medical record data and genome-wide association study data. Nat Biotechnol. 2013;31(12):1102-1110. doi: 10.1038/nbt.2749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Feng Q, Wei WQ, Chaugai S, et al. Association between low-density lipoprotein cholesterol levels and risk for sepsis among patients admitted to the hospital with infection. JAMA Netw Open. 2019;2(1):e187223. doi: 10.1001/jamanetworkopen.2018.7223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Angus DC, Linde-Zwirble WT, Lidicker J, Clermont G, Carcillo J, Pinsky MR. Epidemiology of severe sepsis in the United States: analysis of incidence, outcome, and associated costs of care. Crit Care Med. 2001;29(7):1303-1310. doi: 10.1097/00003246-200107000-00002 [DOI] [PubMed] [Google Scholar]
- 25.Donnelly JP, Safford MM, Shapiro NI, Baddley JW, Wang HE. Application of the Third International Consensus Definitions for Sepsis (Sepsis-3) classification: a retrospective population-based cohort study. Lancet Infect Dis. 2017;17(6):661-670. doi: 10.1016/S1473-3099(17)30117-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deyo RA, Cherkin DC, Ciol MA. Adapting a clinical comorbidity index for use with ICD-9-CM administrative databases. J Clin Epidemiol. 1992;45(6):613-619. doi: 10.1016/0895-4356(92)90133-8 [DOI] [PubMed] [Google Scholar]
- 27.Charlson ME, Pompei P, Ales KL, MacKenzie CR. A new method of classifying prognostic comorbidity in longitudinal studies: development and validation. J Chronic Dis. 1987;40(5):373-383. doi: 10.1016/0021-9681(87)90171-8 [DOI] [PubMed] [Google Scholar]
- 28.Quan H, Sundararajan V, Halfon P, et al. Coding algorithms for defining comorbidities in ICD-9-CM and ICD-10 administrative data. Med Care. 2005;43(11):1130-1139. doi: 10.1097/01.mlr.0000182534.19832.83 [DOI] [PubMed] [Google Scholar]
- 29.Carroll RJ, Bastarache L, Denny JCR. R PheWAS: data analysis and plotting tools for phenome-wide association studies in the R environment. Bioinformatics. 2014;30(16):2375-2376. doi: 10.1093/bioinformatics/btu197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ference BA, Robinson JG, Brook RD, et al. Variation in PCSK9 and HMGCR and risk of cardiovascular disease and diabetes. N Engl J Med. 2016;375(22):2144-2153. doi: 10.1056/NEJMoa1604304 [DOI] [PubMed] [Google Scholar]
- 31.Das S, Forer L, Schönherr S, et al. Next-generation genotype imputation service and methods. Nat Genet. 2016;48(10):1284-1287. doi: 10.1038/ng.3656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McCarthy S, Das S, Kretzschmar W, et al. ; Haplotype Reference Consortium . A reference panel of 64,976 haplotypes for genotype imputation. Nat Genet. 2016;48(10):1279-1283. doi: 10.1038/ng.3643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Do R, Willer CJ, Schmidt EM, et al. Common variants associated with plasma triglycerides and risk for coronary artery disease. Nat Genet. 2013;45(11):1345-1352. doi: 10.1038/ng.2795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lyall D, Ward J, Banach M, et al. PCSK9 genetic variants, life-long lowering of LDL-cholesterol and cognition: a large-scale Mendelian randomization study. BioRxiv. Preprint posted May 31, 2018.
- 35.Willer CJ, Schmidt EM, Sengupta S, et al. ; Global Lipids Genetics Consortium . Discovery and refinement of loci associated with lipid levels. Nat Genet. 2013;45(11):1274-1283. doi: 10.1038/ng.2797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Teslovich TM, Musunuru K, Smith AV, et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466(7307):707-713. doi: 10.1038/nature09270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559-575. doi: 10.1086/519795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dupont WD, Plummer WD Jr. Power and sample size calculations: a review and computer program. Control Clin Trials. 1990;11(2):116-128. doi: 10.1016/0197-2456(90)90005-M [DOI] [PubMed] [Google Scholar]
- 39.Topchiy E, Cirstea M, Kong HJ, et al. Lipopolysaccharide is cleared from the circulation by hepatocytes via the low density lipoprotein receptor. PLoS One. 2016;11(5):e0155030. doi: 10.1371/journal.pone.0155030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dwivedi DJ, Grin PM, Khan M, et al. Differential expression of PCSK9 modulates infection, inflammation, and coagulation in a murine model of sepsis. Shock. 2016;46(6):672-680. doi: 10.1097/SHK.0000000000000682 [DOI] [PubMed] [Google Scholar]
- 41.Berger J-M, Loza Valdes A, Gromada J, Anderson N, Horton JD. Inhibition of PCSK9 does not improve lipopolysaccharide-induced mortality in mice. J Lipid Res. 2017;58(8):1661-1669. doi: 10.1194/jlr.M076844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Walley KR, Boyd JH, Kong HJ, Russell JA. Low low-density lipoprotein levels are associated with, but do not causally contribute to, increased mortality in sepsis. Crit Care Med. 2019;47(3):463-466. doi: 10.1097/CCM.0000000000003551 [DOI] [PubMed] [Google Scholar]
- 43.Boyd JH, Fjell CD, Russell JA, Sirounis D, Cirstea MS, Walley KR. Increased plasma PCSK9 levels are associated with reduced endotoxin clearance and the development of acute organ failures during sepsis. J Innate Immun. 2016;8(2):211-220. doi: 10.1159/000442976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Genga KR, Lo C, Cirstea MS, et al. Impact of PCSK9 loss-of-function genotype on 1-year mortality and recurrent infection in sepsis survivors. EBioMedicine. 2018;38:257-264. doi: 10.1016/j.ebiom.2018.11.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Deshpande A, Pasupuleti V, Rothberg MB. Statin therapy and mortality from sepsis: a meta-analysis of randomized trials. Am J Med. 2015;128(4):410-417.e1. doi: 10.1016/j.amjmed.2014.10.057 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
eMethods. Cohort Identification and Power Calculations
eTable 1. Event Counts in Each Analysis
eTable 2. Event Counts by Genotypes
eTable 3. Associations Between Comorbidity Score and Sepsis and Related Outcomes
eTable 4. SNPs Included in PCSK9 Genetic Risk Score
eTable 5. SNPs Included in Estimated PCSK9 Expression
eTable 6. Associations Between Median Measured LDL-C Levels and 4 Functional PCSK9 Variants
eTable 7. Associations Between Median Measured LDL-C Levels and PCSK9 GRS
eTable 8. Associations Between Median Measured LDL-C Levels and PCSK9 Expression
eTable 9. Associations Between PCSK9 Candidate SNPs and Sepsis and Related Adverse Outcomes
eTable 10. Associations Between PCSK9 GRS and Sepsis and Related Adverse Outcomes
eTable 11. Associations Between Genetically Estimated PCSK9 Expression Tertiles and Sepsis and Related Adverse Outcomes
eFigure. Algorithm to Identify Sepsis Within Infection Cohort
eReferences.