Summary
Soon after fertilization, the few totipotent cells of mammalian embryos diverge to form a structure called the blastocyst (BC). Although numerous cell types, including germ cells and extended-pluripotency stem cells, have been developed from pluripotent stem cells (PSCs) in vitro, generating functional BCs only from PSCs remains elusive. Here, we describe induced self-organizing 3D BC-like cysts (iBLCs) generated from mouse PSC culture. Resembling natural BCs, iBLCs have a blastocoel-like cavity and were formed with outer cells expressing trophectoderm lineage markers and with inner cells expressing pluripotency markers. iBLCs transplanted to pseudopregnant mice uteruses implanted, induced decidualization, and exhibited growth and development before resorption, demonstrating that iBLCs are implantation competent. iBLC precursor intermediates required the transcription factor Prdm14 and concomitantly activated the totipotency-related cleavage-stage MERVL reporter and 2C genes. Thus, our system may contribute to the understanding of molecular mechanisms underpinning totipotency, embryogenesis, and implantation.
Keywords: pluripotent stem cells, early embryo, implantation, cell biology, totipotency, reproduction, cell plasticity, blastocyst, reprogramming
Highlights
-
•
Primed pluripotent cells self-organize blastocyst-like cysts in defined conditions
-
•
Structures have several extraembryonic and embryonic characteristics of blastocysts
-
•
Structures can implant in the uterus and grow before resorption
-
•
Features of totipotency are implicated concomitantly in intermediate cell clusters
In this article, Kime and Tomoda worked with collaborators to expand their previous studies increasing primed pluripotent stem cell potency with defined media cell reprogramming. They induced unique 2C-like cells with the potential to self-organize blastocyst-like cysts that can implant and develop in utero for several days until resorbed.
Introduction
During early mammalian development, a fertilized egg (zygote) completely intersects the animal life cycle upon zygotic genome activation (ZGA), the event whereby gamete totipotent genomes of the pronucleus are epigenetically activated and rapidly enter cleavage (Seydoux and Braun, 2006, Wu et al., 2017). The zygote cleaves and later polarizes as symmetry bifurcates to form the blastocyst (BC) with emerging trophoblasts and pluripotent cells of the inner cell mass (ICM) (Hirate et al., 2015, Nishioka et al., 2009, Stephenson et al., 2010, Yu et al., 2016). The ICM further differentiates to the embryonic epiblast and primitive endoderm (PrE) while preparing for implantation (Guo et al., 2010, Plusa et al., 2008, Saiz and Plusa, 2013). BC implantation is crucial to natural development and is tightly regulated at several molecular and cellular levels that must occur in a short developmental window: failed implantation is a major cause of early pregnancy loss (Cha et al., 2012, Norwitz et al., 2001). Defective embryos can also fail later and begin resorption (Cossée et al., 2000, Flores et al., 2014).
The zygote and cleavage stages exhibit true totipotency, isogenically preceding all extraembryonic (ExEm, vegetal) and embryonic (Em, animal) cell bidirectional development toward entire organisms. From plant tissue cultures, specific cytokine, vitamin, and plant hormone (auxins) ratios are adjusted to induce totipotent transient cells for propagating isogenic embryos (Steward et al., 1958). In mammals, isogenic 3D BCs from differentiated cells are both attractive and elusive, and experiments inducing implantation-competent isogenic BCs entirely from pluripotent stem cells (PSCs) are unprecedented.
In PSC reprogramming and conversion experiments with specific cytokines, nutrient, and lipid (Kime et al., 2016), we observed cell organization and hemispherical cysts with features of BCs. Thereafter we developed a system to induce 8–16 cell iBLC precursors (iBLC-PCs) that self-organized into BC-like cysts in vitro, termed iBLCs. iBLC-PCs were found to be Prdm14-dependent and concomitantly expressed the murine endogenous retrovirus (MERVL) live 2C-state reporter, which suggested ZGA mechanisms related to Zscan4 expression (Macfarlan et al., 2012, Wu et al., 2017). Analysis of YAP (Yes-associated protein) in iBLC-PCs and early iBLCs revealed a transition from a nonpolarized state to a polarized cyst, closely resembling early pre-implantation embryogenesis (Bedzhov et al., 2014, Nishioka et al., 2009, Stephenson et al., 2010).
iBLCs were implantation competent and induced focal decidualization in the uterus that recruited surrogate blood supply and expanded the embryonic cavity. Implanted iBLCs could grow and produce many cell types like implanted embryos, but eventually failed in embryonic resorption. We anticipate that this system may lead to simplified isogenic embryo production for research, medicine, and uncovering the intricacies of totipotency and implantation.
Results
Defined Conditions Generate Early Embryo-like Tissues
In vitro pluripotency is characterized in two distinct states: a pre-implantation BC ICM-like state (naive) and a post-implantation epiblast-like state (primed). Naive female PSCs have two active X chromosomes (Xa/Xa), and primed female PSCs have inactivated one of those X chromosomes (Xa/Xi) (Payer et al., 2011). We employed a primed female mouse epiblast stem cell (mEpiSC) line with a constitutive green fluorescent protein transgene on the Xi chromosome (XGFP). XGFP is silent in the mEpiSCs and is expressed upon Xi reactivation to Xa, a sophisticated epigenetic reprogramming hallmark of naive pluripotency, the ICM, and often of cleavage-stage totipotent cells (Bao et al., 2009, Kime et al., 2016, Monk and Harper, 1979, Okamoto et al., 2004).
Under our defined conditions we greatly enhanced cell reprogramming, and robust primed- to naive-state PSC conversion experiments (Kime et al., 2016) also produced BC-like hemispheres and structures resembling early embryonic material among rapid X chromosome reactivation, reported here. The hemispheres had BC-like organization with important cell-lineage markers for trophoblasts, embryonic, and PrE cells (Figures S1 and S2; Video S1); we previously reported the embryonic potency of the XGFP+ cells(Kime et al., 2016). We observed NANOG+XGFP+ ICM-like cells with no bright DNA-stain puncta, which may indicate the loss of heterochromatin usually found in a rare transient Zscan4+ 2C-like state (Akiyama et al., 2015, Wu et al., 2016) (Figure S1A). The outer cells and cells of the inner face of the ICM-like mass were negative for XGFP and positive for TROMA-I, an ExEm lineage marker. We examined important PrE markers and found GATA4 enriched cells that were XGFP-negative and platelet-derived growth factor receptor A (PDGFRa)-positive co-localized at the inner face to resemble the hypoblast of hatching BCs (Figures S1D and S1E) (Plusa et al., 2008). GATA6, a PrE gene regulated alongside GATA4 (Figure S1D) (Morgani and Brickman, 2015, Plusa et al., 2008, Saiz and Plusa, 2013), was expressed among a population positioned similar to the GATA4+/PDGFRa+ cells(Figures S1D–S1F) (Guo et al., 2010, Morgani and Brickman, 2015, Plusa et al., 2008). Taken together, the BC-like hemispheres reflected the intricate regulation of X chromosome activity, gene expression, and cell organization of BCs including PrE formation. As such, the possibility that all embryonic cell lineages were induced inspired us to consider that transient total potential might be installed in some converting cells.
A late BC-like hemisphere imaged across the z dimension, visualized as a composite 3D model and animated for viewing from several angles. XGFP+NANOG+ cells are restricted to a polar mass of the fluid-filled dome surrounded by large flat cells with large flat nuclei. XGFP (green), NANOG (red), and DNA (blue).
Such primed-to-naive-state conversions strongly induced Prdm and Id family genes that broadly regulate the genome and are curiously related to the cleavage stage, early embryo, and germline preparation (Figure S2B) (Burton et al., 2013, Hiller et al., 2010, Luna-Zurita and Bruneau, 2013, Yamaji et al., 2008, Yang et al., 2017). Within this reprogramming context, we tested the SMAD2/3 signaling ALK5 inhibitor SB431542 that inhibits primed state ActivinA/TGFβ (transforming growth factor β) signaling and is described for in vitro germ-cell differentiation (Chen et al., 2012). The cultures released floating small cell clusters and cysts that we speculated could have BC-like properties similar to the related BC-like hemispheres. We then optimized phase-1 and -2 treatments of defined conditions (Figure 1A), which produced 5–30 floating BC-like cysts by day 7 (Figure S3A and Table S1). The BC-like cysts stuck together as they grew to resemble hatched BCs on day 8, so for most experiments we qualitatively assessed and isolated them on day 7 based on morphological similarity to early BCs (e.g., appropriate size, neatly round, trophectoderm (TE)-like outer cells, a putative ICM; Figure 1B). DNA staining of the cysts revealed a compact ICM-like mass and large flat TE-like cells surrounding the possible blastocoel (Figure S3B).
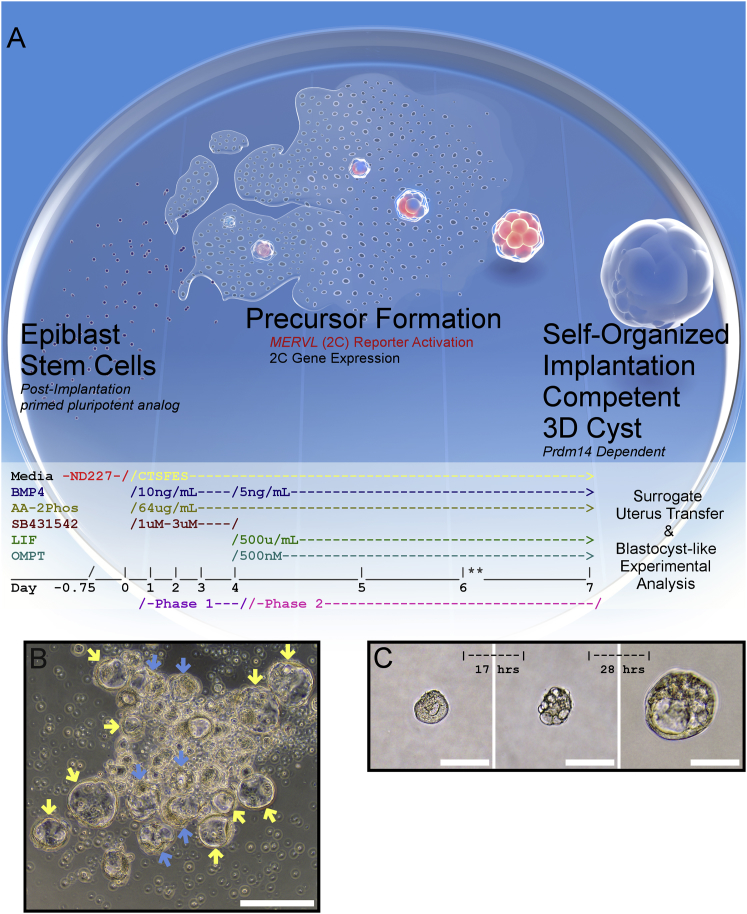
Figure 1.
Defined Conditions Induce Early Embryo-like Structures from Primed PSCs
(A) iBLC System Overview: Typically, ~40,000 primed mEpiSCs are plated and induced to MERVL::RFP+ cell cluster precursors (iBLC-PC) that differentiate into ~5–30 BC-like cysts (iBLCs). Two-phase iBLC induction media timing to induce mEpiSC to iBLCs. ∗∗Supernatant iBLC-PCs are collected to ultralow attachment (ULA) wells on day 6, and high-quality BC-like iBLCs are selected on day 7 by embryo pipette.
(B) iBLCs are qualified with BC-like characteristics (yellow arrows) or excluded (blue arrows) after pooling ULA plate for downstream experiments. Scale bar, 200 μm.
(C) Isolated iBLC-PC developing into iBLC over time. Scale bars, 100 μm.
To explore the origin of the BC-like cysts, we individually cultured the small clusters that appeared nonpolar on day 5.5 (Figure 1C, left panel). Some clusters (usually ~5%–50%) grew, changed morphology, and formed a blastocoel-like cavity as a cyst with BC-like morphology (Figure 1C). From these findings and the investigation described hereafter, we termed the day-7 floating structures “induced blastocyst-like cysts” (iBLCs) that grew, polarized, and self-organized from small cell clusters as iBLC precursors (iBLC-PCs).
iBLC-PCs Involve Early Embryonic Genes
BCs develop from a totipotent state. To gain molecular insights into iBLC induction, we prepared XGFP mEpiSC with the well-studied 2C MERVL live totipotency-related reporter (MERVL::RFP, Figure S3C) (Macfarlan et al., 2012, Wu et al., 2017), which was undetectable in all mEpiSCs as expected. On days 5–6 of iBLC induction, we observed RFP expression in some of the characteristic loci where iBLC-PCs emerge, and some RFP+ cells also expressed XGFP, perhaps consistent with Xa/Xa status of cleavage-stage embryos (Figure 2A) (Okamoto et al., 2004). Many iBLC-PCs were composed of several cells concurrently expressing MERVL::RFP (Figure 2B). In many cases, the RFP expression was weaker in the iBLC-PCs than in cells on the plate and XGFP was usually undetectable, suggesting that both reporters might be downregulated similarly to what is observed in compacting early embryos. Notably, RFP signals were further reduced in emergent iBLCs (Figure 2C) and, after iBLC-PC harvest, RFP+ cells with or without XGFP expression were variably maintained on iBLC generation plates for several days in phase-2 medium.
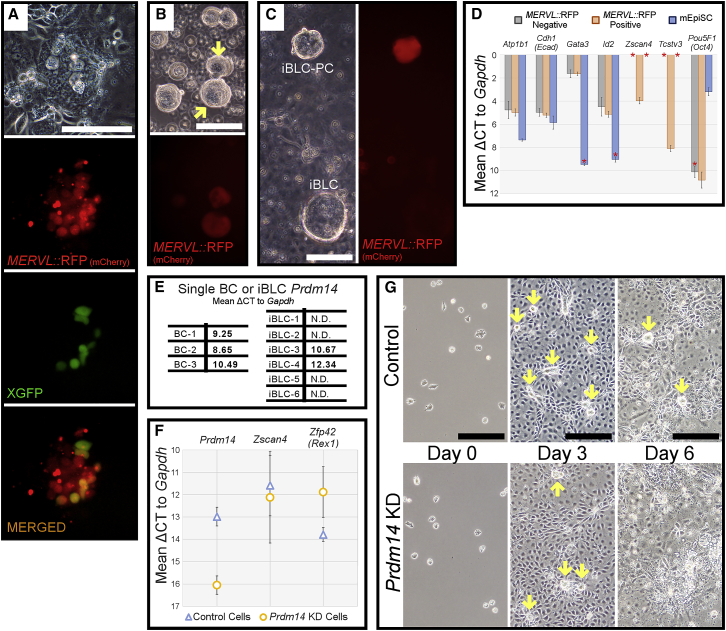
Figure 2.
iBLC Generation Activates 2C MERVL Reporter in iBLC-PCs and Requires Prdm14
(A) iBLC system day 6 co-localized expression of MERVL::RFP and XGFP reporters. Scale bar, 100 μm.
(B) MERVL::RFP+ iBLC-PCs (yellow arrows) in ULA plate on day 6. Scale bar, 100 μm.
(C) MERVL::RFP is expressed strongly in iBLC-PCs yet poorly in differentiating iBLC, observed on day 8. Scale bar, 100 μm.
(D) qRT-PCR of MERVL::RFP reporter-derived clusters on day 6, pooled by RFP− or RFP+ expression; shown as mean ΔCt to Gapdh. mEpiSC prepared for qRT-PCR by the same means were used as control. Data represent biological triplicate samples tested in technical triplicate, and error bars represent standard deviation. Asterisks denote sample detection notes: Gata3 detected in one mEpiSC sample, Id2 detected in two mEpiSC samples, Zscan4 not detected in RFP−/mEpiSC samples, Tcstv3 not detected in RFP−/mEpiSC samples, Pou5F1 (Oct4) detected in two RFP− samples.
(E) Single isolated BC and iBLC qRT-PCR for Prdm14.
(F) qRT-PCR of control and Prdm14 KD cell plate cDNA samples for Prdm14, Zscan4, and Zfp42 (Rex1) on day 6 of iBLC generation. Data represent biological triplicate samples tested in technical triplicate, and error bars represent standard deviation.
(G) Control and Prdm14 KD mEpiSC are plated for iBLC induction. Loci that originate iBLC-PCs are initiated in both experiments by day 3 (yellow arrows). Control cells maintain iBLC-PCs through day 6 (yellow arrows) and Prdm14 KD cells aborted iBLC-PCs. Scale bars, 200 μm.
To investigate the implications of the 2C MERVL::RFP reporter in our system, we pooled RFP+ or RFP− cell clusters resembling iBLC-PCs on day 6 and examined early embryonic gene expression. We found that only MERVL::RFP+ iBLC-PCs induced Zscan4 and Tsctv3, both critically important 2C markers (Falco et al., 2007, Macfarlan et al., 2011, Macfarlan et al., 2012), and with high relative expression (Figures 2D and S3D). These results validated that the 2C MERVL::RFP reporter represented a meaningful unique 2C-like gene expression in iBLC-PCs.
Next, we focused on Prdm14, a critical transcription factor shared in the germline and early embryo (Hackett et al., 2017, Nakaki and Saitou, 2014). Prdm14 was very low or undetectable in mEpiSCs yet could be induced in early iBLCs (Figure 2E). Consistent with these data, constitutive short hairpin RNA against Prdm14 (Prdm14 knockdown [KD]) did not have noticeable effects on mEpiSCs or in the first 5 days of iBLC generation (Figure 2G). However, on day 6 with efficient Prdm14 KD (Figure 2F), iBLC-PCs were nearly completely aborted and many peripheral cells of typical iBLC-PC loci degraded (Figure 2G), and supernatants did not generate iBLCs. We were curious as to whether the 2C marker Zscan4 would be lost from degrading cells, yet Prdm14 KD insignificantly but variably affected the induction of Zscan4 in the population while permitting consistently higher Zfp42 (Rex1) expression (Figure 2F). Collectively, these results suggest that a 2C-like state is induced in a population wherein iBLC-PC/iBLC survival may be Prdm14 dependent.
Induced Blastocyst-like Cysts Represent Pre-/Post-compacted Embryonic State
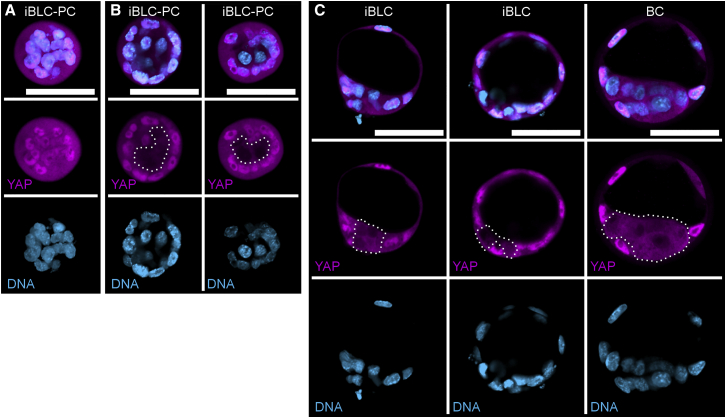
The transcription factor YAP governs positional information and polarization to bifurcate outer and inner cells during early embryonic development (Bedzhov et al., 2014, Nishioka et al., 2009). Thus, we examined cell positioning, along with YAP subcellular localization, in iBLC-PCs and iBLCs. Some iBLC-PCs resembled early embryos before compaction (Hirate et al., 2015, Nishioka et al., 2009, Yu et al., 2016) with morphologically homogeneous cells that equally expressed nuclear and cytosolic YAP (Figure 3A). Other iBLC-PCs implicated cell polarization with nuclear-enriched YAP among outer cells and nuclear-excluded YAP among inner cells (Figure 3B); emergent early iBLCs carried the same YAP pattern as early BCs (Figure 3C). Comprehensively, sample YAP expression resembled the developmental window where blastomeres polarize into compacting embryos and early BCs.
Figure 3.
iBLC-PCs and iBLCs May Follow Early Embryonic Polarization via YAP
(A and B) iBLC-PCs stained for YAP (magenta) and DNA (light blue). Scale bars, 50 μm. Nuclear-excluded YAP region is outlined with dotted white line in (B).
(C) Early iBLCs and early BCs stained for YAP (magenta) and DNA (light blue). Scale bars, 50 μm. Nuclear-excluded YAP region is outlined with dotted white line.
iBLCs Express Important Genes but Lack Full BC Potency
Early BCs have an outer layer of trophoblast cells and an ICM of pluripotent cells without distinct PrE. Thus, BCs express genes important for inducing and developing both lineages. Gene expression analyses revealed individual early BCs and particularly individual early iBLCs had variations in expression, suggesting there is a difference in the quality, developmental timing, or both, in each sample; iBLC-3 had notably higher expression of several genes closer to that of BCs. The genes we checked that are first activated in totipotent cleavage-stage cells (e.g., Atp1b1, Gata3, Id2, Zscan4, Bmp4) were strongly upregulated in iBLCs to match detection in BCs. Remarkably, Gata3 expression was high and even higher in iBLC-PC reprogramming samples (Figure 2D). Gata3 is expressed across early embryo development (Guo et al., 2010, Home et al., 2009) and was recently described as a master reprogramming factor that induced all three major in vitro cell culture equivalents of pre-implantation blastocysts (Benchetrit et al., 2019).
Genes involved in the outer cell-lineage development and/or function (e.g., Atp1b1, Cdx2, Gata3, and Krt8 [Troma-I]) were strongly induced in iBLCs (Figure 4A and S3E), although Cdx2 was usually much lower than in BCs. Critical pluripotent transcription factors Nanog and Pou5f1 (Oct4) were also low in iBLCs but were still expressed among other pre-implantation pluripotency genes (e.g., Tdgf1, Zfp42) (Figures 4A and S3E). Collectively, outer/inner cell fate-specifying genes (e.g., Cdx2, Eomes, Pou5F1 [Oct4]) were generally low in iBLCs (Figures 4A and S3E).
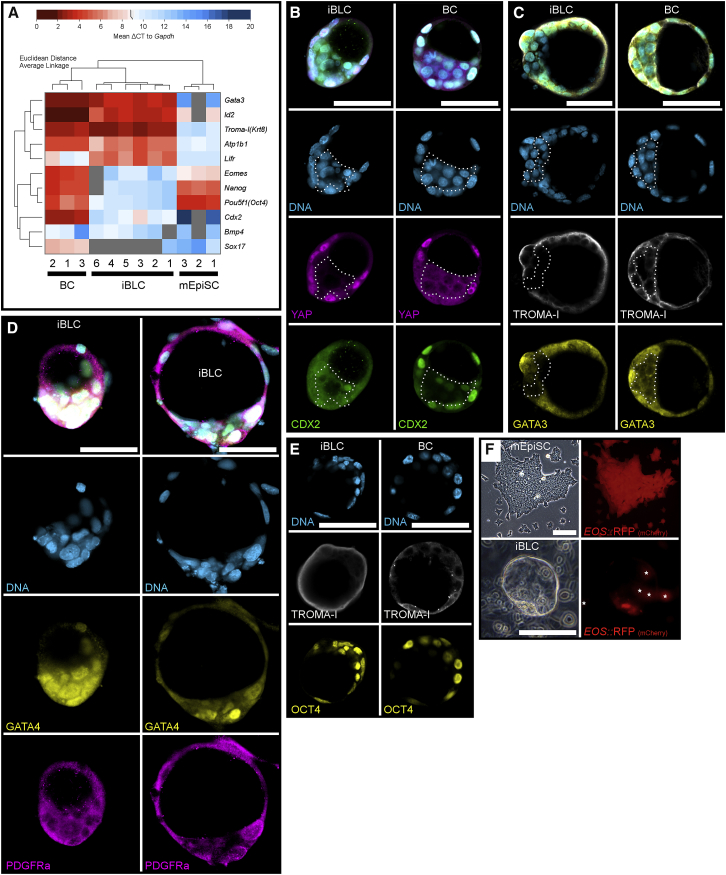
Figure 4.
iBLCs Share Many Characteristics with BCs
(A) qRT-PCR of individual early BC, early iBLC, and mEpiSC colony cDNA samples, with Euclidean distance and clustering by average linkage, represented as a heatmap of global ΔCt to Gapdh.
(B) Early iBLC and early BC stained for DNA (light blue), YAP (magenta), and CDX2 (green). Nuclear-excluded YAP region is outlined with dotted white line. Scale bars, 50 μm.
(C) Early iBLC and early BC stained for DNA (light blue), TROMA-I (white), and GATA3 (yellow). Downregulated and nuclear-excluded GATA3 region is outlined with dotted white line. Scale bars, 50 μm.
(D) Early iBLC (left) and late iBLC (right) stained for DNA (light blue), GATA4 (yellow), and PDGFRa (magenta). Scale bars, 50 μm.
(E) Early iBLC and early BC stained for DNA (light blue), TROMA-I (white), and OCT4 (yellow). Scale bars, 50 μm. Microscope gain settings from Table S2: Early iBLC DNA:398, TROMA-I:513, OCT4:731; Early BC DNA:363, TROMA-I:604, OCT4:601.
(F) EOS::RFP+ mEpiSCs and late iBLCs above culture with EOS::RFP expression in the putative ICM. White stars label out-of-focus EOS::RFP+ cells on the plate. Scale bars, 100 μm.
Low Cdx2 and Pou5f1 raised the question of whether iBLCs possess the TE-like outer and ICM-like mass. We also wondered whether iBLCs could regulate PrE genes as the cyst develops later. We examined detection and localization of CDX2, GATA3, OCT4, YAP, and the TE marker TROMA-I, in early iBLCs and early BCs by immunocytochemistry with well-characterized antibodies. For PrE regulation, we examined GATA4 and PDGFRa in early and late iBLCs to compare with early- and late-hatching BCs and previous reports (Guo et al., 2010, Plusa et al., 2008, Saiz and Plusa, 2013). The early iBLC inner cells downregulated CDX2, GATA3, and YAP like early BCs, although more extremely. GATA3 was found both cytosolic and nuclear in outer cells of both iBLCs and BCs while CDX2 in iBLC outer cells was mostly cytosolic and not enriched in many nuclei (Figures 4B and 4C), suggesting poor CDX2 phosphorylation in iBLCs (Rings et al., 2001). PrE genes GATA4 and PDGFRa are expressed in all cells of the early embryo (Guo et al., 2010), and GATA4 protein was still detectable in all cells of early BCs and early iBLCs (Figure 4D, left; Figure S1D, top). Late-hatching BCs begin to neatly regulate nuclear GATA4 and membrane PDGFRa to a subset of mostly polar mass cells that collect and form the PrE hypoblast at the inner face of the ICM (Guo et al., 2010, Plusa et al., 2008); such PrE-like cell regulation was confirmed in hatching BCs and reflected in the BC-like hemispheres (Figures S1D and S1E). Our day-8 late iBLCs could also regulate GATA4 and PDGFRa to subsets of cells mostly at the ICM-like mass, although less neatly (Figures 4D and S3F). GATA4 enriched cells in late iBLCs collected together, similar to PrE hypoblast formation yet oddly different from BCs and our BC-like hemispheres (Figures S1D and S1E), because the putative iBLC PrE-like cells collected to the outer face of the ICM-like mass and were frequently observed bulging away from the iBLC main body (Figure S3F). Still, early iBLC outer cells strongly expressed TROMA-I, and iBLC inner cells better enriched nuclear OCT4 like early BCs (Figure 4E) (Bulut-Karslioglu et al., 2016, Ralston and Rossant, 2008). We anticipated protein detection differences and designed a qualitative semi-quantitative microscopy experiment that compared detector gain settings for iBLC and BC samples imaged on common microscopes with comparable stain, laser, and confocal settings (Supplemental Experimental Procedures). As expected, early iBLC OCT4 protein may have been lower because detection required higher microscope gain settings than BCs. Furthermore, TROMA-I protein was better detected in iBLCs than in BCs, consistent with the finding that Krt8 (Troma-I) mRNAs were more highly expressed in iBLCs (Figures 4A–4E and S3E; Table S2).
To examine OCT4 and SOX2 functions we used the EOS-S(4+) live pluripotency reporter, which requires an OCT4/SOX2 heterodimer transcription activation. We cloned the reporter to drive red fluorescent proteins (EOS::RFP; Figure S3C and Experimental Procedures) and established EOS::RFP mEpiSCs that performed as expected (Hotta et al., 2009, Tomioka et al., 2002), with EOS::RFP+ PSCs that lost RFP when differentiated (Figures 4F, S2C, and S3G). iBLCs generated from EOS reporter cells often exhibited RFP signals (Figure S3H) that were stronger in the iBLC putative ICM, suggesting that these cells may have functional OCT4/SOX2-driven EOS::RFP expression (Figure 4F).
Lastly, mouse embryonic stem cells (ESCs) and trophoblast stem cells (TSCs) can be established from BCs under different conditions. In ESC derivation conditions (Czechanski et al., 2014), outgrowths from isolated iBLCs and iBLC-PCs on feeder cells proliferated and expressed both XGFP and EOS::D2nRFP (Figure S4A and Experimental Procedures). These cells could be thereafter cultured comparable with naive ESCs in terms of colony morphologies and pluripotency gene expressions (Figures 5A, 5B, and S4A). We also derived colonies of slow-growing TE-like cells expressing CDX2 without EOS expression (Figure 5C). Attempts to derive TSCs in defined conditions (Latos and Hemberger, 2016, Ohinata and Tsukiyama, 2014) failed in part, but developed binucleated TPBPA+ or PL-I+ trophoblast giant cell (TGC)-like cells (Figure S4B).
Figure 5.
iBLC System Outgrowths Generate Pluripotent and TE-Lineage Cells
(A) Mouse ESCs and iBLC/iBLC-PC-derived ESC-like cells stained for OCT4, NANOG, or YAP (magenta), and DNA (light blue). iBLC/iBLC-PC-derived ES-like cells demonstrate X chromosome reactivation (XGFP+) and express EOS::D2nRFP. Scale bars, 50 μm.
(B) qRT-PCR of mouse ESCs, iBLC/iBLC-PC-derived ES-like cells, and mEpiSC cDNA samples, shown as mean ΔCt to Gapdh. Data represent biological triplicate samples tested in technical triplicate, and error bars represent standard deviation. Asterisk indicates that Zscan4 was not detected in mEpiSC; double asterisk indicates that Zfp42 (Rex1) was detected in two mEpiSC samples.
(C) Left: live imaging of iBLC-derived TE-like cells. Right: TE-like cells stained for DNA (light blue) and CDX2 (green). Channels shown separately and merged. Scale bars, 100 μm.
Reproducibility of iBLC Generation
Our various XGFP mEpiSC reporter sublines performed similarly throughout iBLC induction. Two published mEpiSC lines reacted similarly throughout and produced iBLCs with lower yields (Figure S4C) (Ohtsuka et al., 2012, Tesar et al., 2007). Another mEpiSC line with obvious cell culture characteristic differences failed completely (Parchem et al., 2014). Therefore, iBLC generation should be possible with many but not all mEpiSC lines.
iBLCs Implant and Grow in Pseudopregnant Mice
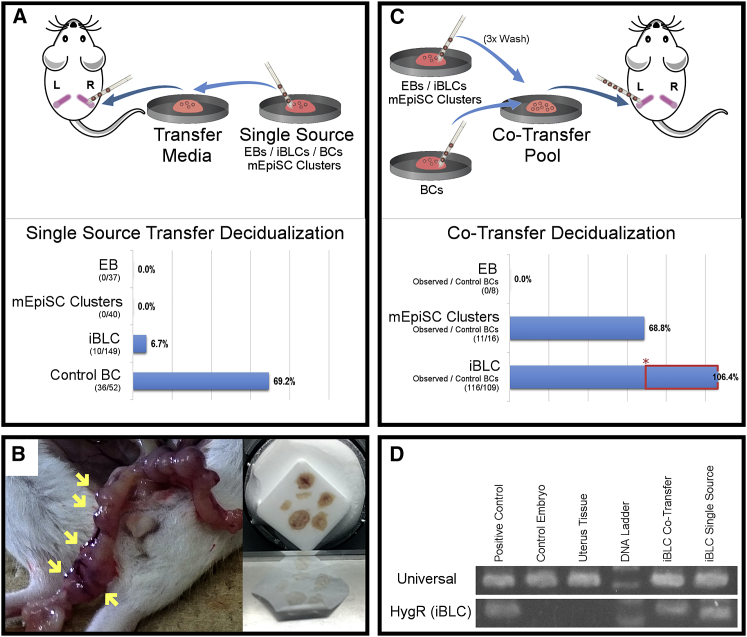
The similarities between iBLCs and BCs led us to examine iBLC implantation and developmental potency in utero. We transferred BCs, iBLCs, mEpiSC clusters, and embryoid bodies (EBs) into separate uterus horns of sterile-male bred pseudopregnant mice; only BCs and iBLCs implanted and induced deciduae thereafter (Figures 6A and 6B). Deciduae from iBLC transfer were similar in focal morphology but often smaller than deciduae from BCs. Importantly, many iBLC deciduae recruited large maternal blood vessels seen in the uterus, and sectioning showed red brown color in the decidua basalis region, like natural deciduae (Figure 6B). iBLCs and BCs induced deciduae at 6.7% (10/149) and 69.2% (36/52), respectively (Figure 6A). We did not observe any deciduae from the mEpiSC cluster and EB controls.
Figure 6.
iBLC Uterus Transfer Decidualization in Pseudopregnant Mice
(A) Diagram of single-source uterus transfer experiment. Observed deciduae in uterus horns with respect to EBs, mEpiSC clusters, iBLCs, or control BCs single-source uterus transfers.
(B) Uterus horn of mouse with iBLC-implanted deciduae (left, yellow arrows), prepared for cryosection (right).
(C) Diagram of co-transfer experiment. Observed deciduae from EB co-transfer, mEpiSC Cluster co-transfer, and iBLC co-transfer. Asterisk denotes control BC decidualization rate (69.2%, A); red box indicates decidualization gained from iBLCs.
(D) Laser capture microdissection (LCM) genomic DNA PCR test for mouse genomic DNA universal and iBLC-specific hygromycin resistance. LCM sample regions from H&E slides are shown in Figure S5A.
In (A) and (C), embryo pipettes indicate when a new pipette was used.
Co-transferring control embryos that easily implant increases implantation rates of difficult embryos in assisted reproductive settings (Mochida et al., 2014). Co-transferring iBLCs with BCs frequently yielded more focal deciduae than the total number of BCs transferred (Table S3), suggesting that iBLCs implanted more efficiently. Also, co-transfer yields often had more deciduae than the estimated sum based on BC-only and iBLC-only rates, suggesting that co-transfer increased iBLC or BC implantation, or both (Figure 6C and Table S3). Co-transferred mEpiSC clusters or EBs with BCs either inhibited or showed no improvement over estimated control BC implantation, highlighting the special ability of iBLCs (Figure 6C).
To confirm the origin of the deciduae from the iBLC experiments, we obtained genomic DNA from iBLC deciduae cryosections by laser capture microdissection (Figure S5A) and amplified a transgenic DNA region only in iBLCs (Figures S3C and 6D). This analysis confirmed that iBLC single-source and co-transfer experiments implanted to form deciduae with iBLC-derived tissue at the proper location for natural embryos. We therefore recognized that co-transferring BCs with iBLCs enhances the ability of iBLCs to implant and may prove more useful in later studies.
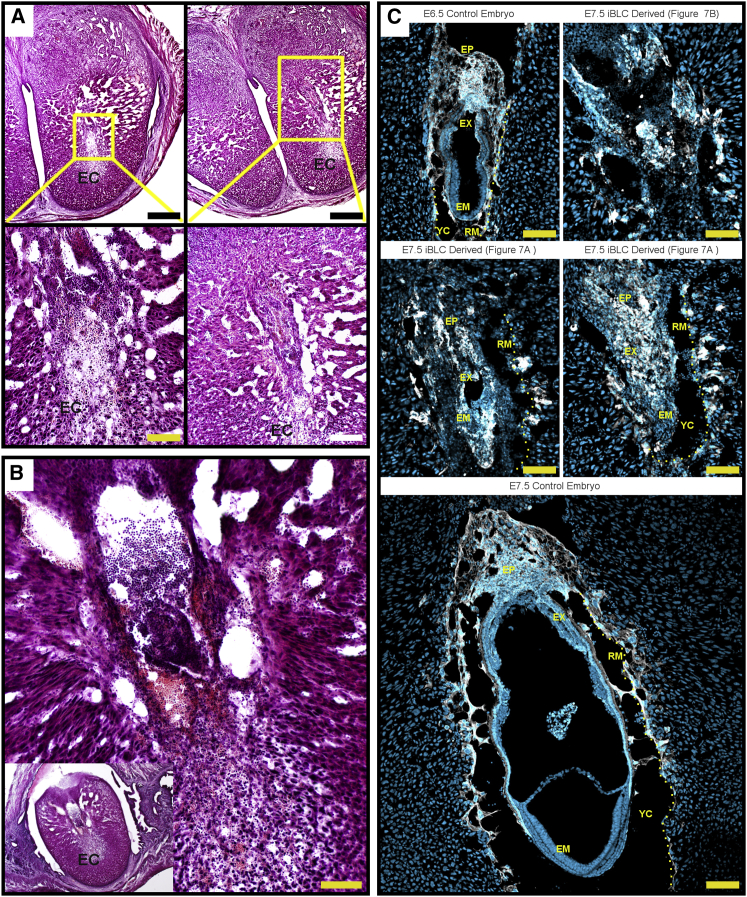
We performed hematoxylin and eosin (H&E) staining on cryosections of dissected deciduae from embryonic day 7.5 (E7.5) iBLC single-source transfer experiments (Figure 7). Like control deciduae (Figure S5B), iBLC-implanted deciduae were surrounded by uterine tissue and had distinct subregions; among which the decidua basalis showed vascular sinus foldings and red blood cells, confirming the maternal blood supply (Figures 7A, 7B, and S5B).
Figure 7.
iBLCs Implant and Partially Develop Before Resorption
(A) H&E-stained proximal cryosections of deciduae from iBLC single-source transfer. Higher magnification is indicated and shows iBLC-derived tissue resembling large cell masses of resorbing tissues. EC, embryonic cavity. Scale bars, 500 μm (top panels), 100 μm (bottom left), and 200 μm (bottom right).
(B) H&E-stained decidua from iBLC single-source transfer showed high presence of immune cells resorbing a mass of cells with ExEm-like and Em-like stain and morphology. Scale bar, 100 μm.
(C) Cryosection stain for TROMA-I (white) and DNA (light blue). E6.5 and E7.5 control embryos show healthy size and structure. E7.5 iBLC-derived tissues from cryosections proximal to (A) and (B) are labeled. EP, ectoplacental cone; EX, extraembryonic portion; EM, embryonic portion; YC, yolk sac cavity; RM and dotted line, Reichert's membrane labeled on one side for clarity, as previously described on resorbing embryos (Cossée et al., 2000). Scale bars, 100 μm.
While iBLC-derived E7.5 tissues were larger than E6.5 control embryo tissues, many cells appeared pycnotic (Gardner and Johnson, 1972) and lacked a healthy appearance, and were collectively smaller than an E7.5 control embryo (Figures 7 and S5B). We also observed many blood mononuclear cells around the disfigured pycnotic tissues, indicating embryo resorption (Cossée et al., 2000, Flores et al., 2014) (Figures 7B and S5C). Nevertheless, careful examination of resorbing iBLC-derived tissues showed markedly diverse cell morphology and localization strikingly similar to findings of a previous report of natural resorbing embryos (Figures 7A and 7B) (Cossée et al., 2000). The sections had distinct disfigured tissues in the presumptive embryonic region with surrounding ExEm-like cells and internal small dark stained cells resembling the embryonic portion (Figures 7A, 7B, S5B, and S5C). Consistently, immunostaining proximal cryosections of the same samples with the TROMA-I antibody showed positionally appropriate TROMA-I+ cells surrounding TROMA-I− cells that we speculated to be Em-portion cells from their location and H&E-stain characteristics (Figure 7C), and other TROMA-I+ cells had further invaded the deciduae with TGC-like morphology (Figure 7C).
To further examine the development of iBLC-derived ExEm tissues in the deciduae, we immunostained for TPBPA and PL-I. Both were expressed in the cells lining and surrounding the embryonic chamber of iBLC-implanted deciduae, similarly to those in BC-derived tissues (Figure S5D) (Chen et al., 2016, Peng et al., 2015), and PL-I was also detected on control embryo ExEm visceral endoderm as reported previously (Figure S5D) (Chen et al., 2016). Several peripheral TPBPA+ or PL-I+ cells had larger, more brightly stained nuclei and were scattered far from the cavity. These cells could be polyploid-scattering TGCs, which were also suggested by the invading TROMA-I+ cells (Figures 7C and S5D).
Discussion
We demonstrated here that PSCs can be reprogrammed to BC-like hemispheres with striking early embryonic implications, and we anticipate that cell conversions in such a context may be used to study early embryonic development in vitro. From this platform we modified the system to generate iBLCs with many similarities to BCs at morphological, developmental, molecular, and functional levels, although imperfect and perhaps less neatly regulated than the BC-like hemispheres (e.g., PrE regulation, Xi reactivation, pluripotency). Master transcription factors that establish, reprogram, and regulate cell fates in each lineage of BCs were expressed in iBLCs, and some (e.g., YAP, GATA3) were apparently correctly regulated. Notably, YAP is involved in both positional regulation and powerful gene regulation that can reprogram cell identity in numerous contexts (Panciera et al., 2016, Qin et al., 2016, Yu et al., 2016). iBLC system ICM-like masses had regulated nuclear-excluded YAP similar to natural BC ICM cells, which is a widely known characteristic difference from mouse ESCs in which YAP is nuclear enriched. iBLC implantation and growth in utero advances the prospect that important natural cues can be established in the system to later pass critically difficult natural barriers, even if dysregulation eventually caused embryonic resorption. The trend of the data suggests that developmental limitations in iBLCs may arise among heightened epigenetic plasticity that activates important genes weakly with poor regulation (e.g., OCT4, CDX2), perhaps causing latent or delayed lineage specification lacking distinct cell-identity resolution. The results point to diverging events for iBLCs and correcting the 2C-like establishment, and differentiation events could be the key to obtain fully functional iBLCs that develop further in utero. We provide comprehensive discussion of these data in Supplemental Discussion and a published preprint (Kime et al., 2018). Collectively, the current iBLC technology is a first step toward generating artificial isogenic BCs.
In previous work with mouse PSC-derived oogenesis, rare BC-like structures from >40-day long-term differentiation experiments, were briefly described (Hübner et al., 2003), although further characterization or competence in utero remains unknown. Two recently published systems for modeling early embryo development in vitro (Harrison et al., 2017, Rivron et al., 2018) expanded upon the reach of previously patented technology (Buhl et al., 2009). In such systems, ESCs and TSCs were precisely co-cultured to recapitulate embryo-like structures and highlight some of the increased developmental potential from lineage cooperation, as described from BC dissections decades prior (Gardner and Johnson, 1972). In great contrast, iBLCs self-organize and differentiate from 2C gene expressing iBLC-PCs, emerging only from primed PSCs in defined conditions.
Prdm14 is critical for germ-cell induction and fertility while important but dispensable in the embryonic PSC during development (Yamaji et al., 2008). On the contrary, Prdm14 may play a role in the earlier cleaving embryo where it is heterogeneously expressed, preceding discrete regulation to enrich the Em lineage during bifurcation (Burton et al., 2013). Given that the first phase of iBLC induction is akin to germ-cell induction from PSCs (Chen et al., 2012), we believe that the iBLC system failure from Prdm14 KD arises from compromising the germ-cell program. The implicit germ-like Prdm14 induction engaging iBLC-PC formation may be separated from the Prdm14 expression in iBLCs that may depend on the iBLC quality. qRT-PCR assay detection is valuable and Prdm14 detection in two out of six early iBLCs is a positive result, although not definitive.
The qRT-PCR assays used RNA extracted from single BCs and iBLCs. Unsurprisingly, iBLCs exhibited greater variations than BCs, yet one may appreciate that several important early embryo genes (Cripto, Zfp42, Zscan4, Cdx2, Sox17, and Gata3) were activated in some or many individual iBLCs. When several iBLCs activated these genes, there was a commonality in the expression level that allows us to distinguish the system from a true BC, to understand it better, and to consider how it is further distanced from the starting mEpiSCs.
iBLCs may advance research in isogenic increased potential from PSCs, and we speculate that partial germ-lineage induction may be a causal link to the unique 2C-like transient expression seen in iBLC-PCs. Therein, early embryonic mimicry is autonomously installed in potentiated cells by a subsequent treatment with defined small molecules: iBLC-PCs can differentiate to iBLCs in the traditional embryo medium KSOM (data not shown).
Defined conditions that induce primed PSCs toward synchronous expression of 2C/MERVL reporter in clusters (iBLC-PC/loci) of cells that subsequently self-organize BC-like cysts are unique in the field of totipotency/2C-like cell research (Wu et al., 2017). Zscan4 and Tcstv3 evidence herein further implicates a 2C totipotency-related program in our system. Until now, 2C-like gene expression was only transiently expressed in individual rare cells among naive ESC cultures, and although bidirectional contribution of the traditional 2C-like MERVL+ cells was shown in previous reports (Macfarlan et al., 2012), such cells remain unremarkably similar to ESCs in culture and require donor embryos to develop in utero: true isogenic totipotency from PSCs remains elusive. We anticipate that more precise control of cell reprogramming inputs may improve or stabilize the 2C-like program in iBLC-PCs to yield fully developed animals from PSCs alone and to maximize PSC reprogramming. Our methodology involves ~1 week of simplified defined media changes. Therefore, we envision that future iBLC technology may readily open avenues in several fields, such as embryology, 2C epigenetics, and implantation biology, in addition to its promise in early embryogenesis.
Experimental Procedures
Experimental Model Details
Please refer to Supplemental Information for exact reagent/resource ordering information.
Animal Use
Mouse handling and experiments were carried out with humane methods in compliance with animal ethical standards approved by RIKEN Kobe Safety Center. Sterile-male bred pseudopregnant (PP) surrogate CD-1 (ICR) female mice were prepared at PP2.5 and then control BCs, iBLCs, mEpiSC clusters, or EBs were transferred to the uterus using standard embryo in vitro fertilization (IVF) pipetting techniques. CD-1 (ICR) BCs and R26-H2B-EGFP BCs were used for control BC experiments (Abe et al., 2011).
Recombinant DNA Preparation
We prepared RFP as dsRed, mCherry, and also a modified mCherry under the EOS reporter by adding a mouse ornithine decarboxylase destabilization domain (D2) (Li et al., 1998) and nuclear localization tags to the RFP (EOS::D2nRFP). D2 drastically reduces the half-life of the D2nRFP, providing timely live RFP responsiveness to mRNA level changes: the D2nRFP signal more closely represents OCT4/SOX2 heterodimer transcriptional activity. We also cloned RFP under the 2C MERVL reporter promoter. All reporter systems were cloned in piggybac vector systems with 5′ and 3′ insulators.
mEpiSC Culture
mEpiSC culture medium (MCM) consisted of NDiff227 supplemented with 20 ng/mL ActivinA, 12 ng/mL basic fibroblast growth factor, and 1:100 penicillin/streptomycin. Media and supplements were stored separately at −20°C in aliquots, freshly thawed at least every 4 days, and stored at 4°C. mEpiSC were cultured on plates previously coated for 1 h at room temperature with 1:100 fibronectin/PBS. Medium was changed daily, and cells were passaged as small clumps every 2–3 days at ~1:10–20, never exceeding 30% confluence. Cell colonies remained less than 300–400 μm wide and largely resembled homogeneous mEpiSC colonies with few single cells. Cell passage was carried out, in brief, with PBS wash, fresh Accutase for 55 s, PBS wash, 2 mL of MCM, scraped, triturated 6–8 times in a conical vial, then dispersed ~1:10–20 in MCM. If cells exhibited signs of differentiation, the culture was discarded and replaced by a freshly thawed stock.
Method Details
CTSFES Medium Preparation for Working Medium
For preparation of ~1 L of CTSFES basal medium, 500 mL of DMEM:F12 + Glutamax, 500 mL of neurobasal medium, 10 mL of B27 supplement, 5 mL of N2 supplement, 5 mL of Glutamax supplement, and 670 μL of 7.5% BSA Frac V solution was filtered at 0.22 μm, aliquoted, and stored immediately at −20°C, thawed overnight at 4°C, and used for 1–8 days.
For CTSFES “working medium,” after thawing for experimental use, 1:100 penicillin/streptomycin, 1:1000 2-ME, and 64 μg/mL ascorbic acid 2-phosphate were added.
mEpiSC Preparation for Naive Conversion or iBLC Generation
Target wells of 6W plate were coated with 1.5 mL of 1:100 fibronectin/PBS substrate for 1 h at room temperature. Stock cultures of near-passage mEpiSC colonies were sourced for passage into the conversion experiment as follows. Cells were washed with PBS, then treated with freshly thawed room-temperature Accutase for 1 min which was gently aspirated, and then cells were washed again with equal volume of PBS while tapping the plate gently to release single cells, and then PBS was gently aspirated again and replaced with 37°C prewarmed fresh Accutase and incubated at 37°C for 5–7 min until cells floated and dispersed freely. PBS/MCM (5× volume, 1:1) was added and the volume triturated 10–20 times in 15-mL conical vial. Cells were centrifuged at 200 × g for 3 min, then the mEpiSC pellet was resuspended in 1–2 mL of MCM, and live cells were counted. Cells were diluted in MCM to yield ~20,000 cells/1.5 mL for naive conversions or 30–50,000 cells/1.5 mL for iBLC generation, mixed evenly. Fibronectin/PBS coating was aspirated from target plates and 1.5 mL of diluted cells in MCM were added per well. Cells were incubated at 37°C for 14–16 h before conversion medium was added; plates were often checked 2–3 h after plating to ensure cells plated as single evenly dispersed cells.
Naive Conversion Experiment
For naive conversion experiment medium (NCM) (8 days of changes), working medium + (10 ng/mL bone morphogenetic protein 4 [BMP4], 1,000 units/mL ESGRO leukemia inhibitory factor [LIF], 1 μM (1-oleoyl-2-methyl-sn-glycero-3-phosphothionate ammonium salt [OMPT]) were prepared fresh at least every 4 days. 6W wells plated with ~20,000 mEpiSC cells/well were fed 2 mL of NCM daily starting ~14–16 h after cells were plated with the aforementioned preparation.
iBLC Generation Experiment
For iBLC generation medium phase 1, day 0–3 medium (4 changes), working medium + (10 ng/mL BMP4, 1 μM SB43152) was prepared fresh on day 0, and SB431542 was increased to 3 μM for days 1–3.
For iBLC generation medium phase 2, day 4–6 medium (3 changes), working medium + (5 ng/mL BMP4, 500–1,000 units/mL ESGRO LIF, 0.5–1 μM OMPT) were prepared fresh on day 4.
6W wells plated with 30–50,000 mEpiSC cells/well were fed 2 mL of phase 1 medium daily at a similar time, starting 14–16 h after cells were plated using the aforementioned preparation method. From day 4, 2 mL of phase 2 medium was changed daily. On day 6 and day 7, iBLC-PCs and some emerging iBLCs were collected with ART P1000G wide-bore pipette tips. The iBLC generation plate was leaned at a 45° angle, and the upper 1 mL (primary) was harvested to one well of a 24-well ultralow attachment (ULA) plate; the lower 1 mL (secondary) was drawn up and cascaded over the plate once and then harvested to a separate well of a 24-well ULA plate. Two milliliters of phase 2 medium was replaced on the plate if the culture was observed or used later. Some iBLC experiments included 0.2 μM sodium pyruvate. In a few experiments, SB431542 was varied between 1 and 10 μM, and phase 1 and phase 2 media were mixed 1:1 on days 3, 4, or 5.
Primary and secondary harvests from one 6W well of iBLC generation were considered together, although secondary harvests contained more iBLC-PCs, iBLCs, and cell debris. Early on day 7, primary and secondary harvests were observed for brief periods, and the emergence of morula-like structures and early blastocyst-like structures from iBLC-PCs was noted on the 24W ULA plate lid. Working medium or phase 2 medium was placed in a Hydrocell 3.5-cm plate and incubated for ~1 h at 37°C. iBLCs were judged by morphology for blastocyst-like characteristics and isolated by embryo transfer pipette to the Hydrocell 3.5-cm plate and incubated for 1–3 h at 37°C. The Hydrocell 3.5-cm plate of near-completely purified iBLCs was then sourced for analysis or IVF transfer into PP2.5 sterile-male bred pseudopregnant mice. When iBLCs were transferred to pseudopregnant mice, they were washed three times by transfer into separate drops of standard embryo transfer medium. In such transfers, unique glass pipettes were used between each step to ensure sample-handling accuracy.
Author Contributions
Conceptualization, C.K.; Methodology, C.K. and K.T.; Cryosections, C.K. and E.K.; Validation, S.O.; Formal Analysis, C.K. and K.T.; Investigation, C.K., K.T., and H.K.; Resources, C.K., K.T., S.Y., and M.T.; Writing – Original Draft, C.K. and K.T.; Writing – Revision & Editing, C.K. and K.T.; Visualization, C.K.; Supervision, C.K. and K.T.; Project Administration, C.K.; Funding Acquisition, C.K., K.T., S.Y., M.A., and M.T.
Conflicts of Interest
C.K. and K.T. have patents related to this technology and extended works. S.Y. is a scientific advisor of iPS Academia Japan without salary.
Acknowledgments
We honor the help of Dr. Hitoshi Niwa for critical input and for providing the rabbit anti-mouse CDX2 antibody. We are grateful to Drs. Siqin Bao and Azim Surani for their female XGFP mEpiSC. We also thank Drs. Robert Blelloch and Paul Tesar for providing their mEpiSC for research. MERVL 2C and EOS-S(4+) reporter DNA was provided by Addgene (http://www.addgene.org) under MTA and subcloned into our systems. TROMA-I (Krt8) monoclonal antibody developed by Institut Pasteur was obtained from the Developmental Studies Hybridoma Bank (DSHB), created by the NICHD of the NIH and maintained at the Department of Biology, University of Iowa. We greatly thank the Yamanaka and Takahashi labs for the support and research environment that made this work possible. We also thank Michael Kime for producing the graphical abstract artwork in Figure 1. This work was supported by funding from RIKEN, Kyoto University, and the Gladstone Institutes; L.K. Whittier Foundation and the Roddenberry Foundation; National Heart, Lung, and Blood Institute/NIH grants U01-HL100406 and U01-HL098179; and from the California Institute for Regenerative Medicine (CIRM). The Gladstone Institutes received support from a National Center for Research Resources Grant RR18928-01. C.K. was supported in part by the RIKEN SPDR program. K.T. was supported in part by JSPS KAKENHI grant 17K07246 and the Suzuki Memorial Foundation. M.A was supported in part by an OMC Internal Research Grant.
Published: August 8, 2019
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.stemcr.2019.07.011.
Contributor Information
Cody Kime, Email: cody.kime@riken.jp.
Kiichiro Tomoda, Email: pha050@osaka-med.ac.jp.
Supplemental Information
References
- Abe T., Kiyonari H., Shioi G., Inoue K.-I., Nakao K., Aizawa S., Fujimori T. Establishment of conditional reporter mouse lines at ROSA26 locus for live cell imaging. Genesis. 2011;49:579–590. doi: 10.1002/dvg.20753. [DOI] [PubMed] [Google Scholar]
- Akiyama T., Xin L., Oda M., Sharov A.A., Amano M., Piao Y., Cadet J.S., Dudekula D.B., Qian Y., Wang W. Transient bursts of Zscan4 expression are accompanied by the rapid derepression of heterochromatin in mouse embryonic stem cells. DNA Res. 2015;22:307–318. doi: 10.1093/dnares/dsv013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao S., Tang F., Li X., Hayashi K., Gillich A., Lao K., Surani M.A. Epigenetic reversion of post-implantation epiblast to pluripotent embryonic stem cells. Nature. 2009;461:1292–1295. doi: 10.1038/nature08534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedzhov I., Graham S.J.L., Leung C.Y., Zernicka-Goetz M. Developmental plasticity, cell fate specification and morphogenesis in the early mouse embryo. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2014;369:20130538. doi: 10.1098/rstb.2013.0538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benchetrit H., Jaber M., Zayat V., Sebban S., Pushett A., Makedonski K., Zakheim Z., Radwan A., Maoz N., Lasry R. Direct induction of the three pre-implantation blastocyst cell types from fibroblasts. Cell Stem Cell. 2019;24:983–994. doi: 10.1016/j.stem.2019.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhl, S., Egert, A., Schorle, H., and Woynecki, T. (2009). Induced blastocyst-like structures, methods of production and uses of the same. European patent EP2088191A1, filed February 8, 2008.
- Bulut-Karslioglu A., Biechele S., Jin H., Macrae T.A., Hejna M., Gertsenstein M., Song J.S., Ramalho-Santos M. Inhibition of mTOR induces a paused pluripotent state. Nature. 2016;540:119–123. doi: 10.1038/nature20578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton A., Muller J., Tu S., Padilla-Longoria P., Guccione E., Torres-Padilla M.-E. Single-cell profiling of epigenetic modifiers identifies PRDM14 as an inducer of cell fate in the mammalian embryo. Cell Rep. 2013;5:687–701. doi: 10.1016/j.celrep.2013.09.044. [DOI] [PubMed] [Google Scholar]
- Cha J., Sun X., Dey S.K. Mechanisms of implantation: strategies for successful pregnancy. Nat. Med. 2012;18:1754–1767. doi: 10.1038/nm.3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W., Jia W., Wang K., Zhou Q., Leng Y., Duan T., Kang J. Retinoic acid regulates germ cell differentiation in mouse embryonic stem cells through a Smad-dependent pathway. Biochem. Biophys. Res. Commun. 2012;418:571–577. doi: 10.1016/j.bbrc.2012.01.078. [DOI] [PubMed] [Google Scholar]
- Chen C.-Y., Chan C.-H., Chen C.-M., Tsai Y.-S., Tsai T.-Y., Wu Lee Y.-H., You L.-R. Targeted inactivation of murine Ddx3x: essential roles of Ddx3x in placentation and embryogenesis. Hum. Mol. Genet. 2016;25:2905–2922. doi: 10.1093/hmg/ddw143. [DOI] [PubMed] [Google Scholar]
- Cossée M., Puccio H., Gansmuller A., Koutnikova H., Dierich A., LeMeur M., Fischbeck K., Dollé P., Kœnig M. Inactivation of the Friedreich ataxia mouse gene leads to early embryonic lethality without iron accumulation. Hum. Mol. Genet. 2000;9:1219–1226. doi: 10.1093/hmg/9.8.1219. [DOI] [PubMed] [Google Scholar]
- Czechanski A., Byers C., Greenstein I., Schrode N., Donahue L.R., Hadjantonakis A.-K., Reinholdt L.G. Derivation and characterization of mouse embryonic stem cells from permissive and nonpermissive strains. Nat. Protoc. 2014;9:559–574. doi: 10.1038/nprot.2014.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falco G., Lee S.-L., Stanghellini I., Bassey U.C., Hamatani T., Ko M.S.H. Zscan4: a novel gene expressed exclusively in late 2-cell embryos and embryonic stem cells. Dev. Biol. 2007;307:539–550. doi: 10.1016/j.ydbio.2007.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores L.E., Hildebrandt T.B., Kühl A.A., Drews B. Early detection and staging of spontaneous embryo resorption by ultrasound biomicroscopy in murine pregnancy. Reprod. Biol. Endocrinol. 2014;12:38. doi: 10.1186/1477-7827-12-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner R.L., Johnson M.H. An investigation of inner cell mass and trophoblast tissues following their isolation from the mouse blastocyst. Development. 1972;28:279–312. [PubMed] [Google Scholar]
- Guo G., Huss M., Tong G.Q., Wang C., Li Sun L., Clarke N.D., Robson P. Resolution of cell fate decisions revealed by single-cell gene expression analysis from zygote to blastocyst. Dev. Cell. 2010;18:675–685. doi: 10.1016/j.devcel.2010.02.012. [DOI] [PubMed] [Google Scholar]
- Hackett J.A., Kobayashi T., Dietmann S., Surani M.A. Activation of lineage regulators and transposable elements across a pluripotent spectrum. Stem Cell Reports. 2017;8:1645–1658. doi: 10.1016/j.stemcr.2017.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison S.E., Sozen B., Christodoulou N., Kyprianou C., Zernicka-Goetz M. Assembly of embryonic and extraembryonic stem cells to mimic embryogenesis in vitro. Science. 2017;356:eaal1810. doi: 10.1126/science.aal1810. [DOI] [PubMed] [Google Scholar]
- Hiller M., Liu C., Blumenthal P.D., Gearhart J.D., Kerr C.L. Bone morphogenetic protein 4 mediates human embryonic germ cell derivation. Stem Cells Dev. 2010;20:351–361. doi: 10.1089/scd.2010.0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirate Y., Hirahara S., Inoue K., Kiyonari H., Niwa H., Sasaki H. Par-aPKC-dependent and -independent mechanisms cooperatively control cell polarity, Hippo signaling, and cell positioning in 16-cell stage mouse embryos. Dev. Growth Differ. 2015;57:544–556. doi: 10.1111/dgd.12235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Home P., Ray S., Dutta D., Bronshteyn I., Larson M., Paul S. GATA3 is selectively expressed in the trophectoderm of peri-implantation embryo and directly regulates Cdx2 gene expression. J. Biol. Chem. 2009;284:28729–28737. doi: 10.1074/jbc.M109.016840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotta A., Cheung A.Y.L., Farra N., Vijayaragavan K., Séguin C.A., Draper J.S., Pasceri P., Maksakova I.A., Mager D.L., Rossant J. Isolation of human iPS cells using EOS lentiviral vectors to select for pluripotency. Nat. Methods. 2009;6:370–376. doi: 10.1038/nmeth.1325. [DOI] [PubMed] [Google Scholar]
- Hübner K., Fuhrmann G., Christenson L.K., Kehler J., Reinbold R., Fuente R.D.L., Wood J., Strauss J.F., Boiani M., Schöler H.R. Derivation of oocytes from mouse embryonic stem cells. Science. 2003;300:1251–1256. doi: 10.1126/science.1083452. [DOI] [PubMed] [Google Scholar]
- Kime C., Sakaki-Yumoto M., Goodrich L., Hayashi Y., Sami S., Derynck R., Asahi M., Panning B., Yamanaka S., Tomoda K. Autotaxin-mediated lipid signaling intersects with LIF and BMP signaling to promote the naive pluripotency transcription factor program. Proc. Natl. Acad. Sci. U S A. 2016;113:12478–12483. doi: 10.1073/pnas.1608564113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kime C., Kiyonari H., Ohtsuka S., Kohbayashi E., Asahi M., Yamanaka S., Takahashi M., Tomoda K. Implantation-competent blastocyst-like structures from mouse pluripotent stem cells. BioRxiv. 2018 doi: 10.1016/j.stemcr.2019.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latos P.A., Hemberger M. From the stem of the placental tree: trophoblast stem cells and their progeny. Development. 2016;143:3650–3660. doi: 10.1242/dev.133462. [DOI] [PubMed] [Google Scholar]
- Li X., Zhao X., Fang Y., Jiang X., Duong T., Fan C., Huang C.-C., Kain S.R. Generation of destabilized green fluorescent protein as a transcription reporter. J. Biol. Chem. 1998;273:34970–34975. doi: 10.1074/jbc.273.52.34970. [DOI] [PubMed] [Google Scholar]
- Luna-Zurita L., Bruneau B.G. Chromatin modulators as facilitating factors in cellular reprogramming. Curr. Opin. Genet. Dev. 2013;23:556–561. doi: 10.1016/j.gde.2013.07.002. [DOI] [PubMed] [Google Scholar]
- Macfarlan T.S., Gifford W.D., Agarwal S., Driscoll S., Lettieri K., Wang J., Andrews S.E., Franco L., Rosenfeld M.G., Ren B. Endogenous retroviruses and neighboring genes are coordinately repressed by LSD1/KDM1A. Genes Dev. 2011;25:594–607. doi: 10.1101/gad.2008511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macfarlan T.S., Gifford W.D., Driscoll S., Lettieri K., Rowe H.M., Bonanomi D., Firth A., Singer O., Trono D., Pfaff S.L. Embryonic stem cell potency fluctuates with endogenous retrovirus activity. Nature. 2012;487:57–63. doi: 10.1038/nature11244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochida K., Hasegawa A., Otaka N., Hama D., Furuya T., Yamaguchi M., Ichikawa E., Ijuin M., Taguma K., Hashimoto M. Devising assisted reproductive technologies for wild-derived strains of mice: 37 strains from five subspecies of Mus musculus. PLoS One. 2014;9:e114305. doi: 10.1371/journal.pone.0114305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monk M., Harper M.I. Sequential X chromosome inactivation coupled with cellular differentiation in early mouse embryos. Nature. 1979;281:311–313. doi: 10.1038/281311a0. [DOI] [PubMed] [Google Scholar]
- Morgani S.M., Brickman J.M. LIF supports primitive endoderm expansion during pre-implantation development. Development. 2015;142:3488–3499. doi: 10.1242/dev.125021. [DOI] [PubMed] [Google Scholar]
- Nakaki F., Saitou M. PRDM14: a unique regulator for pluripotency and epigenetic reprogramming. Trends Biochem. Sci. 2014;39:289–298. doi: 10.1016/j.tibs.2014.04.003. [DOI] [PubMed] [Google Scholar]
- Nishioka N., Inoue K., Adachi K., Kiyonari H., Ota M., Ralston A., Yabuta N., Hirahara S., Stephenson R.O., Ogonuki N. The hippo signaling pathway components lats and yap pattern tead4 activity to distinguish mouse trophectoderm from inner cell mass. Dev. Cell. 2009;16:398–410. doi: 10.1016/j.devcel.2009.02.003. [DOI] [PubMed] [Google Scholar]
- Norwitz E.R., Schust D.J., Fisher S.J. Implantation and the survival of early pregnancy. N. Engl. J. Med. 2001;345:1400–1408. doi: 10.1056/NEJMra000763. [DOI] [PubMed] [Google Scholar]
- Ohinata Y., Tsukiyama T. Establishment of trophoblast stem cells under defined culture conditions in mice. PLoS One. 2014;9:e107308. doi: 10.1371/journal.pone.0107308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsuka S., Nishikawa-Torikai S., Niwa H. E-Cadherin promotes incorporation of mouse epiblast stem cells into normal development. PLoS One. 2012;7:e45220. doi: 10.1371/journal.pone.0045220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto I., Otte A.P., Allis C.D., Reinberg D., Heard E. Epigenetic dynamics of imprinted X inactivation during early mouse development. Science. 2004;303:644–649. doi: 10.1126/science.1092727. [DOI] [PubMed] [Google Scholar]
- Panciera T., Azzolin L., Fujimura A., Di Biagio D., Frasson C., Bresolin S., Soligo S., Basso G., Bicciato S., Rosato A. Induction of expandable tissue-specific stem/progenitor cells through transient expression of YAP/TAZ. Cell Stem Cell. 2016;19:725–737. doi: 10.1016/j.stem.2016.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parchem R.J., Ye J., Judson R.L., LaRussa M.F., Krishnakumar R., Blelloch A., Oldham M.C., Blelloch R. Two miRNA clusters reveal alternative paths in late-stage reprogramming. Cell Stem Cell. 2014;14:617–631. doi: 10.1016/j.stem.2014.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payer B., Lee J.T., Namekawa S.H. X-inactivation and X-reactivation: epigenetic hallmarks of mammalian reproduction and pluripotent stem cells. Hum. Genet. 2011;130:265–280. doi: 10.1007/s00439-011-1024-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J., Fullerton P.T., Monsivais D., Clementi C., Su G.H., Matzuk M.M. Uterine activin-like kinase 4 regulates trophoblast development during mouse placentation. Mol. Endocrinol. 2015;29:1684–1693. doi: 10.1210/me.2015-1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plusa B., Piliszek A., Frankenberg S., Artus J., Hadjantonakis A.-K. Distinct sequential cell behaviours direct primitive endoderm formation in the mouse blastocyst. Development. 2008;135:3081–3091. doi: 10.1242/dev.021519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin H., Hejna M., Liu Y., Percharde M., Wossidlo M., Blouin L., Durruthy-Durruthy J., Wong P., Qi Z., Yu J. YAP induces human naive pluripotency. Cell Rep. 2016;14:2301–2312. doi: 10.1016/j.celrep.2016.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralston A., Rossant J. Cdx2 acts downstream of cell polarization to cell-autonomously promote trophectoderm fate in the early mouse embryo. Dev. Biol. 2008;313:614–629. doi: 10.1016/j.ydbio.2007.10.054. [DOI] [PubMed] [Google Scholar]
- Rings E.H.H.M., Boudreau F., Taylor J.K., Moffett J., Suh E.R., Traber P.G. Phosphorylation of the serine 60 residue within the Cdx2 activation domain mediates its transactivation capacity. Gastroenterology. 2001;121:1437–1450. doi: 10.1053/gast.2001.29618. [DOI] [PubMed] [Google Scholar]
- Rivron N.C., Frias-Aldeguer J., Vrij E.J., Boisset J.-C., Korving J., Vivié J., Truckenmüller R.K., van Oudenaarden A., van Blitterswijk C.A., Geijsen N. Blastocyst-like structures generated solely from stem cells. Nature. 2018;557:106–111. doi: 10.1038/s41586-018-0051-0. [DOI] [PubMed] [Google Scholar]
- Saiz N., Plusa B. Early cell fate decisions in the mouse embryo. Reproduction. 2013;145:R65–R80. doi: 10.1530/REP-12-0381. [DOI] [PubMed] [Google Scholar]
- Seydoux G., Braun R.E. Pathway to totipotency: lessons from germ cells. Cell. 2006;127:891–904. doi: 10.1016/j.cell.2006.11.016. [DOI] [PubMed] [Google Scholar]
- Stephenson R.O., Yamanaka Y., Rossant J. Disorganized epithelial polarity and excess trophectoderm cell fate in preimplantation embryos lacking E-cadherin. Development. 2010;137:3383–3391. doi: 10.1242/dev.050195. [DOI] [PubMed] [Google Scholar]
- Steward F.C., Mapes M.O., Mears K. Growth and organized development of cultured cells. II. Organization in cultures grown from freely suspended cells. Am. J. Bot. 1958;45:705–708. [Google Scholar]
- Tesar P.J., Chenoweth J.G., Brook F.A., Davies T.J., Evans E.P., Mack D.L., Gardner R.L., McKay R.D. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature. 2007;448:196–199. doi: 10.1038/nature05972. [DOI] [PubMed] [Google Scholar]
- Tomioka M., Nishimoto M., Miyagi S., Katayanagi T., Fukui N., Niwa H., Muramatsu M., Okuda A. Identification of Sox-2 regulatory region which is under the control of Oct-3/4–Sox-2 complex. Nucleic Acids Res. 2002;30:3202–3213. doi: 10.1093/nar/gkf435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Huang B., Chen H., Yin Q., Liu Y., Xiang Y., Zhang B., Liu B., Wang Q., Xia W. The landscape of accessible chromatin in mammalian preimplantation embryos. Nature. 2016;534:652–657. doi: 10.1038/nature18606. [DOI] [PubMed] [Google Scholar]
- Wu G., Lei L., Schöler H.R. Totipotency in the mouse. J. Mol. Med. 2017;95:1–8. doi: 10.1007/s00109-017-1509-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaji M., Seki Y., Kurimoto K., Yabuta Y., Yuasa M., Shigeta M., Yamanaka K., Ohinata Y., Saitou M. Critical function of Prdm14 for the establishment of the germ cell lineage in mice. Nat. Genet. 2008;40:1016–1022. doi: 10.1038/ng.186. [DOI] [PubMed] [Google Scholar]
- Yang S., Yuan Q., Niu M., Hou J., Zhu Z., Sun M., Li Z., He Z. BMP4 promotes mouse iPS cell differentiation to male germ cells via Smad1/5, Gata4, Id1 and Id2. Reproduction. 2017;153:211–220. doi: 10.1530/REP-16-0292. [DOI] [PubMed] [Google Scholar]
- Yu C., Ji S.-Y., Dang Y.-J., Sha Q.-Q., Yuan Y.-F., Zhou J.-J., Yan L.-Y., Qiao J., Tang F., Fan H.-Y. Oocyte-expressed yes-associated protein is a key activator of the early zygotic genome in mouse. Cell Res. 2016;26:275–287. doi: 10.1038/cr.2016.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A late BC-like hemisphere imaged across the z dimension, visualized as a composite 3D model and animated for viewing from several angles. XGFP+NANOG+ cells are restricted to a polar mass of the fluid-filled dome surrounded by large flat cells with large flat nuclei. XGFP (green), NANOG (red), and DNA (blue).