Abstract
Recently Bekker et al. [Bekker G‐J et al. Protein Sci. 2019;28:429–438.] described a computational strategy of applying molecular‐dynamics simulations to estimate the relative stabilities of single‐domain antibodies, and utilized their method to design changes with the aim of increasing the stability of a single‐domain antibody with a known crystal structure. The structure from which they generated potentially stabilizing mutations is an anti‐cholera toxin single domain antibody selected from a naïve library which has relatively low thermal stability, reflected by a melting point of 48°C. Their work was purely theoretical, so to examine their predictions, we prepared the parental and predicted stabilizing mutant single domain antibodies and examined their thermal stability, ability to refold and affinity. We found that the mutation that improved stability the most (~7°C) was one which changed an amino acid in CDR1 from an asparagine to an aspartic acid. This change unfortunately was also accompanied by a reduction in affinity. Thus, while their modeling did appear to successfully predict stabilizing mutations, introducing mutations in the binding regions is problematic. Of further interest, the mutations selected via their high temperature simulations, did improve refolding, suggesting that they were successful in stabilizing the structure at high temperatures and thereby decrease aggregation. Our result should permit them to reassess and refine their model and may one day lead to a usefulin silico approach to protein stabilization.
Keywords: molecular dynamics simulations, protein stabilization, single‐domain antibody, thermal stability
1. INTRODUCTION
One key attribute of single‐domain antibodies (sdAbs) is their ability to function following exposure to high temperatures. This is primarily due to their ability to refold following heat denaturation. Nonetheless, many efforts to provide sdAbs with enhanced thermal stability by raising their melting temperature have been undertaken.1 These efforts are not without merit, for in many instances sdAbs do not refold 100% and a fraction of the material aggregates irreversibly. Their failure to refold is a function of both intrinsic properties, such as the protein's isoelectric point, as well as the total number of charged amino acids on its surface and extrinsic properties such as protein concentration and solvent pH and ionic strength. Thus, efforts that succeed in raising the melting point of an sdAb, which retards unfolding until extreme temperatures, can prevent aggregation and loss of activity from occurring.
Amongst the methods examined to raise the melting point of sdAbs, CDR grafting onto know stable frameworks and making point mutations to residues identified to enhance stability from highly stable exemplars are two common methods. While perhaps the most straightforward method has been to insert a non‐canonical disulfide bond between framework regions.2, 3 This approach often yields an increase of 10–20°C in melting temperature. However, regardless of the approach the resulting mutant sdAb will need to have its affinity reevaluated, as all these methods can impact affinity and particularly in the case of an added disulfide bond, refolding often is negatively impacted.
In recent work, Bekker et al.4 took a different approach to sdAb thermal stabilization. They applied molecular dynamics (MD) simulations to estimate the relative stability for a set of 7 sdAbs for which both crystal structures and melting transition temperature (T ms) were available. Their approach was based on computing the fraction of native atomic contacts (denoted by Q) for the sdAbs at three different temperatures and drawing a correlation between Q and the experimental T m values. The best Q‐T m correlation from this strategy was observed at a simulation temperature of 400 K and yielded a Pearson correlation r = 0.79 from fitting to the set of sdAbs.
Applying this protocol to the PDB 4idl (designated as the A9 sdAb) structure at 400 K, several residues were identified as “unstable.” Two point mutations were predicted to stabilize A9 where the wild‐type (WT) form has a relatively low melting point. According to their model, both single mutations (N27D and R71I illustrated in Figure 1) slightly increased the stability with respect to the Q value over the WT and the double mutant even more. It was claimed that the stability of the double mutant would be comparable to sdAbs which have T m values of around 80°C. Unfortunately, the mutant sdAbs generated by in silico methods were not validated by producing the sdAbs and experimentally measuring their melting temperatures. As we had initially selected A9 from a naïve library, determined its crystal structure as well as measured its affinity and stability,5, 6 we elected to prepare A9 and the mutants predicted to stabilize its structure, to give Bekker et al.4 hard information from which they could reexamine their model to facilitate even better modeling approaches in the future.
Figure 1.
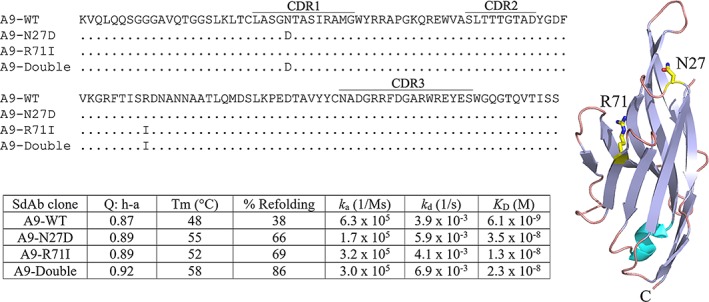
Sequences of A9 (PDB 4idl) and the mutants used in this study are shown. Changes in the mutants are indicated. The CDR identifications follow the usage in Bekker et al.4 The crystal structure of A9 is shown with the mutated amino acids rendered as a stick model. The table shows the melting point and affinity data reported in this study. “Q:h‐a” is the Q score for the set “hydrophilic ‐all” and is taken from Bekker et al.4
2. DISCUSSION
The DNA coding for A9 (PDB 4idl) and the mutants described by Bekker et al.4 was purchased from Eurofins Genomics. Soluble his‐tagged sdAb protein was produced in E. coli for each using the methods described previously.7 All four proteins produced well, giving yields that ranged from 29 to 39 mg/L. The circular dichroism (CD) data and surface plasmon resonance (SPR) data were obtained as described previously,7 and are reported in Figure 1.
It was clear from the good production that A9 and both single point mutants and the double mutant folded well. This was confirmed by the CD data, where A9 gave a T m of 48°C, within the error of what we had observed originally (47 ± 1.5°C),6 and the T ms of the mutants all showed improvement relative to A9 WT. The relative ordering of their T ms matched that predicted by Bekker et al.,4 in that T m of A9 (4idl) < R71I < N27D < double mutant. The double mutant yielded a T m increase of 10°C as determined by CD and was additive as predicted. For stabilizing mutations selected purely in silico these measured increases in thermal stability confirm the ability of their model to identify stabilizing changes. In addition to improved T ms, we found the refoldability of the mutant sdAbs also improved dramatically. The A9 WT (4idl) only refolded 39%. While we found the single point mutants improved to 66 and 69% refolding and again the improvement for the double mutant was “additive,” and gave excellent refolding at 86%. This finding may be just as important an observation as the improved T m, as the resistance to permanent denaturation while unfolded can also improve the ability of an sdAb to function in austere environments where transient exposure to high temperatures may occur.
What was not achieved by their model was the ability to obtain the level of thermal stability observed by VH‐B1a (3b9v)8 and VHH‐A3 (4tyu),9 which have T m values of around 80°C. Thus, while their current model successfully identified stabilizing mutations, the degree of stabilization achieved was not of the magnitude predicted. The other critical factor they failed to achieve was to improve stability while retaining wild type levels of affinity. The point mutant N27D with the change in CDR1 faired the worse, as they feared, increasing its dissociation constant (K D) by a factor >5, while the other point mutant R71I only increased slightly, at most a factor of 2, however this is near our ability to discriminate due to experimental error. In this case the double mutant did not appear additive, and measured as having a K D midpoint between N27D and R71I. This result is not surprising as it has already been demonstrated that stability and affinity cannot be independently modified without consideration of the effect on the other parameter.10
A method of applying Q values at a single temperature would appear to be ad hoc, a more rigorous simulation method would be to model the complete thermal unfolding profile as a function of the reaction coordinate Q. Nevertheless, this alternative approach is simply too demanding for a high throughput analysis. To help put the success by Bekker et al.4 in perspective, the application of computationally fast web‐based empirical methods described for modeling PDB 4tyu 11, 12 to evaluating N27D and R71I mutants showed inconsistent success. All of the fast empirical methods find N27D to be weakly destabilizing, while R71I improves stability. Notably, unlike most of the web‐based methods, the algorithm HoTMuSiC predicts direct changes in the T m from mutations.13 This server yields for N27D a ΔT m ~ −1 K change and for R71I, ΔT m of ~+2 K.
Despite the theoretical limitations, the in silico approach of Bekker et al.4 is a highly useful addition to the current stable of predictive stabilization methodologies, as it is able to identify mutations unlikely to be ascertained easily by any other means. Our results show that it is most beneficial for the modeling to direct the experimental, but that experimental results at the end of the day are required to refine the modeling.
CONFLICT OF INTEREST
The authors declare that they have no conflict of interest with the contents of this article.
ACKNOWLEDGMENT
This work was supported by US Naval Research Laboratory base funds. Authors are listed in reverse alphabetical order.
Zabetakis D, Shriver‐Lake LC, Olson MA, Goldman ER, Anderson GP. Experimental evaluation of single‐domain antibodies predicted by molecular dynamics simulations to have elevated thermal stability. Protein Science. 2019;28:1909–1912. 10.1002/pro.3692
REFERENCES
- 1. Goldman ER, Liu JL, Zabetakis D, Anderson GP. Enhancing stability of camelid and shark single domain antibodies: An overview. Front Immunol. 2017;8:865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Saerens D, Conrath K, Govaert J, Muyldermans S. Disulfide bond introduction for general stabilization of immunoglobulin heavy‐chain variable domains. J Mol Biol. 2008;377:478–488. [DOI] [PubMed] [Google Scholar]
- 3. Zabetakis D, Olson MA, Anderson GP, Legler PM, Goldman ER. Evaluation of disulfide bond position to enhance the thermal stability of a highly stable single domain antibody. PloS One. 2014;9:e115405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bekker G‐J, Ma B, Kamiya N. Thermal stability of single‐domain antibodies estimated by molecular dynamics simulations. Protein Sci. 2019;28:429–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Goldman ER, Anderson GP, Liu JL, Delehanty JB, Sherwood LJ, Osborn LE, Cummins LB, Hayhurst A. Facile generation of a heat‐stable antiviral and antitoxin single domain antibodies from a semisynthetic llama library. Anal Chem. 2006;78:8245–8255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Legler PM, Zabetakis D, Anderson GP, Lam A, Hol WGJ, Goldman ER. Structure of a low‐melting‐temperature anti‐cholera toxin: Llama VHH domain. Acta Crystallogr. 2013;F69:90–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Shriver‐Lake LC, Liu JL, Zabetakis D, Sugiharto VA, Lee CR, Defang GN, Wu SJ, Anderson GP, Goldman ER. Selection and characterization of anti‐dengue NS1 single domain antibodies. Sci Rep. 2018; 8:18086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Barthelemy PA, Raab H, Appleton BA, et al. Comprehensive analysis of the factors contributing to the stability and solubility of autonomous human VH domains. J Biol Chem. 2008;283:3639–3654. [DOI] [PubMed] [Google Scholar]
- 9. George J, Compton JR, Leary DH, Olson MA, Legler PM. Structural and mutational analysis of a monomeric and dimeric form of a single domain antibody with implications for protein misfolding. Proteins. 2014;82:3101–3116. [DOI] [PubMed] [Google Scholar]
- 10. Zabetakis D, Anderson GP, Bayya N, Goldman ER. Contributions of the complementarity determining regions to the thermal stability of a single‐domain antibody. PLoS One. 2013;8:e77678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Olson MA, Zabetakis D, Legler PM, Turner KB, Anderson GP, Goldman ER. Can template‐based protein models guide the design of sequence fitness for enhanced thermal stability of single‐domain antibodies? Protein Eng Des Sel. 2015;28:395–402. [DOI] [PubMed] [Google Scholar]
- 12. Olson MA, Legler PM, Zabetakis D, Turner TB, Anderson GP, Goldman ER. Sequence tolerance of a single‐domain antibody with a high thermal stability: Comparison of computational and experimental fitness profiles. ACS Omega. 2019;4:10444–10454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pucci F, Bourgeas R, Rooman M. Predicting protein thermal stability changes upon point mutations using statistical potentials: Introducing HoTMuSiC. Sci Rep. 2016;6:23257. [DOI] [PMC free article] [PubMed] [Google Scholar]


