Abstract
Invading cancer cells adapt their migration phenotype in response to mechanical and biochemical cues from the extracellular matrix. For instance, mesenchymal migration is associated with strong cell-matrix adhesions and an elongated morphology, while amoeboid migration is associated with minimal cell-matrix adhesions and a rounded morphology. However, it remains challenging to elucidate the role of matrix mechan-ics and biochemistry, since these are both dependent on ECM protein concentration. Here, we demonstrate a composite silk fibroin and collagen I hydrogel where stiffness and microstructure can be systematically tuned over a wide range. Using an overlay assay geometry, we show that the invasion of metastatic breast cancer cells exhibits a biphasic dependence on silk fibroin concentration at fixed collagen I concentration, first increasing as the hydrogel stiffness increases, then decreasing as the pore size of silk fibroin decreases. Indeed, mesenchymal morphology exhibits a similar biphasic depen-dence on silk fibroin concentration, while amoeboid morphologies were favored when cell-matrix adhesions were less effective. We used exogenous biochemical treatment to perturb cells towards increased contractility and a mesenchymal morphology, as well as to disrupt cytoskeletal function and promote an amoeboid morphology. Overall, we envision that this tunable biomaterial platform in a 96-well plate format will be widely applicable to screen cancer cell migration against combinations of designer biomaterials and targeted inhibitors.
Keywords: interpenetrating network, extracellular matrix, 3D culture, high content screening, overlay assay
1. Introduction
Cancer cell migration phenotype is regulated by mechanical and biochemical features of the extracellular matrix (ECM).1,2 Classically, mesenchymal migration has been associated with the actin-driven protrusion of the lamellipodium, followed by strong cell-matrix adhesion through integrins and subsequent retraction of the trailing end.3 In order to translocate through confined spaces, cancer cells apply large matrix deformations and secrete matrix metalloproteinases (MMPs) in order to remodel the ECM into a more accessible architec-ture.4 However, mesenchymal motility can be compromised by disruption of actin-driven protrusions,5–8 cell-matrix attachments,9–13 or MMPs.5,7,14–16 In response, cancer cells can switch to an amoeboid migration phenotype where they are propelled by actomyosin con-tractility in confined spaces, even with minimal cell-matrix adhesions.17 Biologically-derived ECM has enabled key insights into how cancer cells adapt their migration phenotype, but it remains challenging to systematically elucidate the role of ECM mechanics and biochemistry, since these properties are non-trivially coupled to ECM protein concentration.
Reconstituted collagen I hydrogels recapitulate many key features of ECM and have been widely utilized to investigate cell migration in a biomimetic microenvironment.18,19 Collagen I is a naturally-derived triple helical ECM protein that typically assembles into physically crosslinked fibrous networks. Cancer cells cultured in reconstituted 3D hydrogels typically exhibit increased motility from low to moderate collagen I concentration, which may be attributed to increasing ligand density or bulk stiffness.20–25 Nevertheless, cell migration is impeded at high collagen I concentrations, due to steric hindrance from the dense fiber net-works.15,26–30 These behaviors are qualitatively consistent with migratory behaviors observed in artificial hydrogel networks (e.g. PEG)31–33 but these biomaterials are nanoporous and lack the microstructural architectures observed in natural ECM. In order to independently tune mechanical stiffness and ligand density, composite interpenetrating networks have been demonstrated that consist of a cell-adhesive biopolymer (e.g. collagen I, reconstituted base-ment membrane) with a second, non-cell-adhesive biopolymer to tune mechanical stiffness (e.g. alginate, agarose).34–38 Nevertheless, one challenge is that these non-cell-adhesive poly-mers are not degradable by mammalian cells. Alternatively, silk fibroin is a naturally-derived block copolymer derived from silkworm cocoons, (Bombyx mori ) which fold into hydrophobic β-sheet forming blocks that crystallize together as effective crosslinks. Silk fibroin is promising for composite hydrogels since it is susceptible to enzymatic cleavage,39 mechanically tunable over a wide range, and non-cell-adhesive. 40 In particular, silk fibroin can be transiently dispersed by ultrasonication, which will then crosslink into a hydrogel by β-sheet stacking.41,42 It should be noted that this approach for silk fibroin hydrogels differs from the (salt-leached) porous silk fibroin scaffolds demonstrated by Kaplan and coworkers.43,44 Recent work has also utilized composite silk fibroin and collagen I hydrogels at constant ratios for encapsulating human mesenchymal stem cells. 45
In this article, we show that composite silk-fibroin and collagen I hydrogels can be used to elucidate breast cancer migration over a range of matrix stiffness and pore sizes. In particular, the collagen fiber architecture and silk fibroin porosity can be tuned by varying silk fibroin concentration at constant collagen I concentration. Breast cancer cell migration through these hydrogels can be biased from a contractile mesenchymal phenotype towards a propulsive amoeboid phenotype based on decreasing hydrogel pore size, forcing cells to “squeeze” through. These migration phenotypes can also be perturbed by biochemical treat-ment with growth factors or chemotherapeutic drugs. Overall, this biomimetic extracellular matrix can be used to elucidate the crosstalk of matrix and soluble cues on cancer cell invasion.
2. Methods
2.1. Materials and Reagents
Bombyx mori silkworm cocoons were purchased from Treenway Silks, Lakewood, CO. Rat-tail collagen I (354236), and Dulbecco’s modified Eagle’s medium containing L-glutamine, 4.5 g L-1 glucose and sodium pyruvate (DMEM, MT-10–013-CV) were purchased from Corning. ASTM Type II Deionized Distilled water (6442–88), sodium carbonate (S2127), lithium bromide (746479), Dulbecco’s Phosphate Buffered Saline (10X, Modified, without calcium chloride and magnesium chloride; D1408) were purchased from MilliporeSigma. Slide-A-Lyzer 3.5K MWCO G2 Dialysis Cassettes 30 mL dialysis cassettes (PI87725), sodium hydroxide (S320), acetic acid (A38), Fluorescent microparticles (0.5 μm diameter, red flu-orescent carboxyl-modified Fluospheres) (F8812), CellMask DeepRed (H32721), NucMask (H10325), and Hoechst pentahydrate (H3569) were purchased from ThermoFisher Scientific. DRAQ7 1 ml 0.3 mM (ab109202) was purchased from Abcam. Pepro Tech Inc Human Epidermal Growth Factor (EGF), Animal-Free Recombinant Protein (50–813-059), Triton X-100 (BP151–500) and Corning Penicillin-Streptomycin 100x solution (MT30002CI) were purchased from Fisher Scientific. Fetal Bovine Serum was purchased from GE Healthcare (SH30071.03HI). Paclitaxel (10461) was purchased from Cayman Chemicals. Optically clear sterile tissue culture treated 96 well CellCarrier imaging plates (6005550) were purchased from PerkinElmer. 6-well cell culture plates (25–105) were purchased from Genesee Scien-tific. 8 mm biopsy punches (504535) were purchased from World Precision Instruments. Dow Corning Sylgard 184 Silicone Encapsulant Clear 0.5 kg Kit (184 SIL ELAST KIT 0.5KG) was purchased from Ellsworth Adhesives.
2.2. Silk Fibroin Extraction
Silk fibroin solution was extracted and purified from silkworm (B. mori ) cocoons, as previ-ously described.46 Briefly, cocoons were cut into ≈1 cm2 pieces. To remove sericin, 5 g of silk cocoons were added to 2 L of boiling water containing 4.24 g sodium carbonate. Silk cocoons were boiled for 20 minutes and gently but continuously mixed using a metal spatula. After boiling, the silk cocoons were immediately placed into 2 L of purified water at room temperature for 20 min. The water was then regularly replaced with purified water, for a total of 3 water changes. The cocoons were then set to dry overnight, after which any clumps of silk fiber were manually pulled apart using tweezers. A 20% (w/v) solution was then made with silk fibers and 60°C LiBr, with a final volume of 15 mL. The silk and LiBr solution was left to sit at 60°C until all of the silk fibers had been solubilized. The solubilized silk solution was then dialyzed against 1 L of purified water for at least 48 hrs., changing the water a total of 6 times. Silk was then centrifuged to remove any large debris from cocoons. The purified silk solution was then stored at 4°C to limit β-sheet formation and crosslinking. The concentration of the purified silk solution was determined by pipetting 1 mL of the purified silk solution into a weigh boat and placing it in the 60°C oven. The dried silk film was then weighed to calculate the w/v concentration.
2.3. Preparation of Collagen I, Silk Fibroin, and Composite Hydrogels
All precursor solutions were prepared using culture media (DMEM with L-Glutamine, 4.5g/L Glucose and Sodium Pyruvate) supplemented with 10% fetal bovine serum, and 1% pen/strep).
Silk fibroin was typically stored as a stock solution of 50–80 mg/mL at 4 °C. For a 1 mL hydrogel, silk fibroin stock solution was added to cold cell culture media in order to reach final concentrations of 5 – 10 mg/mL. The solution was then adjusted to ensure the complete gel solution reached physiological pH (between 7.35 and 7.45) by adding 0–2 μL of 1 N acetic acid or 1 N sodium hydroxide, for consistency with collagen I preparation. Some batch-to-batch variability was observed for extracted silk fibroin, requiring some optimization of experimental conditions per batch. In particular, rapid silk fibroin β-sheet formation was initiated by sonication prior to gelation, and it was observed that longer sonication time typically resulted in faster gelation kinetics. Thus, an array of conditions was first tested for each batch in order to control gelation kinetics. A Fisher 100 Ultra Sonication Probe at Level 1 power was placed into contact with the precursor silk fibroin solution, avoiding contact with the tube walls. Silk fibroin solutions were prepared at varying concentrations (5.0, 7.5, and 10.0 mg/mL) and were typically sonicated from 5–15 seconds in order to form hydrogels after 30–40 min. For 1 mL total precursor solution, 20 μL fluorescent microparticles were sometimes added after sonication to visualize the hydrogel.
Composite silk fibroin-collagen I hydrogels were prepared by first resuspending silk fibroin stock solution into cold culture media and sonicating using a Fisher 100 Ultra Sonication Probe held at level 1 power. Next, collagen I stock solution was added to yield a final concentration of 1 mg/mL in a total volume of 1 mL. These two components were not sonicated together to avoid denaturing the collagen I monomers. To visualize the hydrogel, 20 μL of fluorescent microparticles were sometimes added at a final concentration of 2% (v/v) and lightly vortexed. This precursor solution was then adjusted to physiological pH (pH 7.35 – 7.45), by adding 0–3 μL of 1 N sodium hydroxide, then lightly vortexed.
Rat-tail collagen I was reconstituted and polymerized from 9–11 mg/mL stock solution through the following procedure: For a 1 mL hydrogel, 100 μL of 10x PBS was first added to 0.5 to 0.7 mL of cold cell culture media, calculated for a total volume of 1 mL at the desired collagen I concentration. Collagen I stock solution was added at varying volumes in order to reach final concentrations of 0.5 – 2.0 mg/mL collagen I at in 1 mL final volume. To visualize the hydrogel, 20 μL of fluorescent microparticles were sometimes added at a final concentration of 2% (v/v) and lightly vortexed. In order to initiate polymerization, this precursor solution was then adjusted to pH 7.4, by adding 0–3 μL of 1 N sodium hydroxide, then lightly vortexed.
The microstructure was further perturbed for hydrogels of comparable composition by incubating precursor solutions at 0°C, inspired by previous work by George and coworkers.47 Briefly, precursor solutions were prepared as detailed above, and split into two aliquots, with 500 μL each. One tube was placed on wet ice at 0°C for 30 minutes, while the other was immediately (aprox. 3–6 minutes) pipetted into a 96 well plate and placed into a 37°C incubator to polymerize. After 30 minutes, the well plate was removed from the incubator, and the 0°C precursor solution was vortexed lightly, then pipetted into a 96 well plate (Figure S8). The well plate was placed back into the incubator for 30 minutes (until gelation) and topped with media.
2.4. Bulk Rheology Measurements
Shear rheology of hydrogels was conducted using a TA Instruments AR-2000 rheometer equipped with a 40 mm diameter 2° acrylic cone geometry and a solvent trap to prevent hydrogel evaporation. Hydrogels were allowed to cure in situ on a temperature-controlled rheometer under conditions consistent with the cell invasion assays. Strain sweeps were performed to determine the linear viscoelastic regime.
For silk fibroin hydrogels, ≈600 μL of silk hydrogel precursor was pipetted onto the 25°C rheometer Peltier plate and the geometry was lowered into place. The solvent trap was put into place and the temperature held constant at 25°C for 10 min. The temperature was then ramped up to 37°C and held constant for 50 min, for a total of 60 min incubation time. A frequency sweep was then performed at 37°C from 0.01 – 1 Hz, at 1% strain. After the measurement, the Peltier plate temperature was returned to 25°C in preparation for the next sample.
For collagen I and composite silk fibroin-collagen I hydrogels, ≈600 μL of hydrogel precursor was pipetted onto the 25°C Peltier plate and the upper cone was lowered into place. The solvent trap was put into place and the temperature held constant at 25°C for 20 min. The temperature was then ramped up to 37°C and held constant for 20 min, for a total of 40 min incubation time. A frequency sweep was then performed at 37°C from 0.01 – 1 Hz, at 1% strain. After the measurement, the Peltier plate temperature was returned to 25°C in preparation for the next sample.
2.5. Unconfined Compression Stress Relaxation
Elastic modulus and stress relaxation half-life were measured using unconfined compression testing48 on an Instron 5944 MicroTester Precision Instrument with Instron Bluehill software, a 5 N load cell, and Screw Side Action Grips Rated to 500 N (Cat. 2710–004). Custom compression platens were 3D printed out of PLA using a Makerbot Replicator 3D Printer. To ensure a smooth compression surface a Fisherbrand Unbreakable 22 × 22mm Cover Slip was adhered to each platen surface using Scotch Removable Poster Tape (Cat. 109). Side-view images before and after compression were taken with a Logitech HD Pro Webcam C910. Compression data and images were analyzed in Matlab 2018.
Samples were crosslinked in 2 mL microcentrifuge tubes. After crosslinking, the bottom of the microcentrifuge tube was cut off using a scalpel, and the samples were removed from the tube. Each sample was cut to be 0.25 inch (6.35 mm) in height. The cross-sectional area was calculated from the side view images taken with the webcam. The unconfined compression stress relaxation protocol involved compressing the sample to 25% final compressive strain in increments of 5% strain at 8 mm / sec. Between compression steps the platens were held in place and the sample was allowed to relax for 10 minutes. Due to the sensitivity of the load cell and the lack of a vibration isolation table for the Instron, the load response data was smoothed in Matlab using a span of 10.
Elastic modulus was calculated by determining the slope of the stress-strain curve for each sample. This was determined using the compressive strain (mm/mm) and the residual stress at the end of each relaxation period. These 5 points were plotted and a linear regression was performed to determine the slope of this trendline. Stress relaxation half-life is defined as the point at which the stress had relaxed to 50% of the peak load. This was determined using the final compression step for each sample (from 20% to 25% compressive strain). The maximum stress was determined as the peak stress at the start of the relaxation period and the minimum stress was determined as the minimum stress at the end of the relaxation period. The half-maximum stress was determined from these values. Curve-fitting was then performed using a two-termed exponential model that showed good agreement with the relaxation data. Using this model, the stress relaxation half-life time point was calculated from the half-maximum stress.
2.6. Second Harmonic Generation Imaging and Analysis
For second harmonic generation (SHG) imaging in an upright microscope geometry, hydrogels were contained within open polydimethylsiloxane (PDMS) reservoirs bonded to a multiwell plate. Briefly, 5 mL of polydimethylsiloxane (PDMS) was cured in each well of a 6-well plate. PDMS was first mixed in a 10:1 ratio of elastomer to curing agent, degassed in a vacuum chamber for 15 minutes, and then cured in a 60°C oven for 1 hour. Smaller wells were then punched out of the PDMS using a 8 mm diameter biopsy punch, leaving a ≈5 mm deep reservoir. Roughly 200–250 μL of hydrogel was added into each small reservoir, which was then crosslinked in an incubator at 37°C for at least 1 hour.
SHG imaging was performed using a Olympus FV1000-MPE multiphoton laser scanning microscope (Brown University Leduc Bioimaging Facility) equipped with a Mai Tai HP tunable laser (690–1020 nm), four non-descanned detectors (2 PMTs and 2 GaAsP detectors), an encoded Prior Z deck with a scanning stage, a 405/40 SHG filter cube allowing selection of a stimulation wavelength between 780 and 840 nm, and an Olympus 25× 1.05 NA XLPlan Multiphoton water objective. In this upright geometry, images were acquired 150–450 μm from the top surface of the hydrogel. The laser was tuned to 800 nm and 3 W, the laser scanning speed was set to 10 μs / pixel, with a 4x line scan average, and a size of 2048 × 2048 pixels (507.9 μm x 507.9 μm). The photomultiplier (HV) was set between 525–675 Volts, Gain set to 2x, and Offset set to 9%.
CT-FIRE for Individual Fiber Extraction v2.0 software for Linux49 was used to extract collagen fiber angle, length, straightness, and width (Figure S4B). SHG tif images (n = 3 per condition) were first center cropped to an 800 × 800 pixel image to eliminate artifacts from non-uniform illumination. CT-FIRE parameters were optimized for the best segmen-tation of visible fibers and visually checked by overlaying detected fibers over a contrast optimized image. Parameters were kept consistent across images except for the following ranges, optimized per image manually: thresh_im2: 90–100, xlinkbox: 2–12. If CT-FIRE segmentation was satisfactory, CT-FIRE .mat files were read into CurveAlign, which output length, width, and density measurements.
The segmented fibers obtained from CT-FIRE and CurveAlign were further sub-divided in fiber segments and identified by a unique ID. The end-points of each fiber segment are connected together (by line segments) to visualize the the complete fiber. The probability density function of fiber length was plotted using CurveAlign frequency of fiber lengths.
The resulting distributions were compared using Kolmogorov-Smirnov tests and the p values corrected for multiple comparisons.
To quantify fiber density, the central ROI (800 px by 800 px) in each SHG image was further subdivided into square boxes and the number of distinct fibers within each box was computed by checking for the presence of at least one fiber segment endpoint within the box. The number of fibers per box was then calculated by counting the number of fibers with unique fiber ids per each box. For ease of visualization, the number of fibers per 50 × 50 pixel boxsize was converted into microns and divided by 2.53 scaling factor to yield an equivalent graph depicting the number of fibers per every 100 μm.
The mean number of fibers over all boxes was recorded at various box sizes (side lengths: 5, 10, 20, 40, 50, 80, 100, 160, 200, 400 and 800 pixels; 1 px = 0.318 μm). A relationship between box size (box area in μm2) and number of fibers was established by constructing a cubic interpolant. To estimate fiber density (and mesh area) in the image, the area corresponding to a single fiber was approximated using the cubic interpolation function. Pore diameter was then calculated by dividing by π, taking the square root and multiplying by 2.
Fiber widths were verified in NIS Elements (4.30.02). SHG representative images were imported from the Olympus FV1000-MPE multiphoton laser scanning microscope (Brown University Leduc Bioimaging Facility) into NIS Elements. Twenty representative fibers per replicate were measured using the length tool. Widths were measured as perpendicular to fiber longitudinal orientation. Consistent positive signal spanning the width of a fiber was required for the section to be considered one unit fiber width.
2.7. Scanning Electron Microscopy
Collagen and composite silk collagen gels were created as detailed above and pipetted into a 6 well plate and polymerized. Gels were a small volume in comparison to the surface area of the plate to enable a high solution to gel ratio. Gels were prepared for Scanning Electron Microscopy through chemical drying. First, samples were fixed using Karnovsky’s fixative overnight. Samples were then washed with sodium cacodylate buffer twice. Graded ethanol dilutions were then used to replace hydrogel water with ethanol. Increments of graded ethanol were 20, 40, 60, 80, 90, and 95 percent EtOH diluted with DI water. Samples were allowed to sit on rocker table for 30 minutes per wash. Samples were then moved into scintillation vials with aprox. 20 mLs of 100% EtOH. Samples were allowed to soak in 100% EtOH for aprox. 12 hours. This 12-hour soak was repeated after replacing with fresh 100% EtOH. An unopened bottle of high grade pure EtOH was opened for the final 100% EtOH steps. Samples infiltrated with 100% ethanol were then taken to the Leduc Bioimaging Facility for drying in the critical point dryer (Ladd Research Industries). Samples were sliced with an ultra-sharp blade such that the inside of the hydrogels was exposed. These sections were infiltrated with liquid CO2 in a pressure chamber at low temperature (<18 degrees) and vented multiple times to allow for liquid C02 to replace residual ethanol. Temperature and pressure were allowed to increase, driving the CO2 around its critical point. Instantaneous transformation of CO2 liquid into CO2 gas preserved gel ultrastructure. Samples were then mounted on SEM stubs and carbon tape, coated with Ag to increase sample conductance, and imaged or placed in a desiccator for future imaging. Images were acquired using Trinity Detectors on a Thermo Apreo VS SEM with no accessory. Samples were imaged under high vacuum. Image parameters were as follows. High Voltage (HV): 2.00 kV, current: 50 pA, WD: 9.7 mm, mag 1500 X, Horizontal Field Width (HFW): 138 μm, detector: T2, pressure: 10.00 e-4 Pa, dwell: 200.00.
Pore analysis was conducted on NIS Elements (version 4.30.02). Representative T2 images taken on the Thermo Apreo VS SEM were imported into NIS Elements and cropped to a focused region of 50 × 50 μm by calculating the ratio of pixels to microns for each image. The contrast LUTs was adjusted to better show pore depth and background contrast. Pores were then identified by stereopsis and traced with area tool in NIS Elements. 50 pores were traced per image, their area and equivalent spherical diameter recorded to be later imported into an csv file.
2.8. MDA-MB-231 Cell Culture
Highly metastatic breast adenocarcinoma cells (MDA-MB-231) stably transfected with green fluorescent protein fused to nuclear histones (H2B-GFP) were a gift from R.J. Giedt and R. Weissleder (Massachusetts General Hospital). Cells were routinely subcultured in culture media consisting of Corning DMEM with L-Glutamine, 4.5g/L Glucose and Sodium Pyru-vate) supplemented with 10% fetal bovine serum (GE Healthcare), and 1% pen/strep, and only used for passage numbers below 25.
2.9. Preparation and Cell Seeding in a 96-well plate for Overlay Assay
High content 96 well plates (PerkinElmer CellCarrier) were pre-coated with 5 μg/cm2 of rat-tail collagen I (Corning 354236) in order to improve cell adhesion. Briefly, the stock solution of 9–11 mg/mL collagen I was diluted to a final concentration of 0.028 mg/mL in 0.02 N acetic acid. Next, 60 μL of this collagen solution was pipetted into each well, corresponding to an areal density of 5 μg/cm2, ensuring each well bottom was completely covered in solution. The well plate was covered and left at room temperature for at least 45 minutes for coating. After at least 45 minutes, the dilute collagen solution was aspirated and the wells were washed three times with cold 1X PBS to remove any residual acetic acid. MDA-MB-231 cells stably transfected with green fluorescent protein labeling nuclear histones (GFP-H2B) were then trypsinized from a tissue culture flask, spun down then resuspended in 1 mL of media. The cell solution was then counted using a Nexcelom Cell Counter and a portion was used to make a final concentration of 105 cells/mL. Next, 60 μL of cell suspension was pipetted into each well to deposit about 6,000 cells per well; care was taken to ensure the solution evenly covered each well. The well plate was then placed into an incubator (37 °C, 5% CO2) to allow cells to adhere for 1–2 hours, during which precursor gel solutions could be prepared.
80 μL of each final precursor solution was pipetted into a 96 well plate on which cells have adhered (at 6,000 cells/ well). Precursor solutions with collagen were kept on ice to avoid premature gelation. Before pipetting any gel precursor solution, media was aspirated from each well. Careful consideration was taken to avoid disruption of the adhered cells. The completed plate was placed in a humidified incubator for 30–40 min (37 °C, 5% CO2) to enable gelation. After gelation, 100 μL of warmed media was pipetted gently onto the gels.
2.10. Confocal Microscopy for Live Cell Imaging
Cell migration was imaged for over 48 h using a Nikon Eclipse Ti fluorescence microscope with spinning-disk confocal head (Crest Optics X-light V2), with light-guide coupled solid state illumination system (Lumencor Spectra-X3), sCMOS camera (Andor Neo), 10x Plan Apo objective (NA 0.45), GFP/FITC Filter Set (Chroma 49002), TRITC/DSRed Filter Set (Chroma 49004), and maintained in a humidified environmental chamber at 37 °C, 5% CO2. NIS Elements was used for automated image acquisition with z-steps of 8 μm from the bottom of the well to a height of 100 μm under consistent exposure times, camera gain/gamma control, and aperture.
For nuclei detection, images were imported into Bitplane Imaris v8.2 and spot detection was applied with the following parameters: Enable Region of Interest = false, Enable Region Growing = false; Source Channel: Source Channel Index = 1, Estimated XY Diameter =14.0 um, Estimated Z Diameter = 18.0 um, Background Subtraction = true; Classify Spots: “Quality” above 10.0. Detected objects were manually verified to confirm that nuclei were correctly identified.
Within multiwell plates, the bottom surface of different wells varied slightly in vertical position relative to the objective. Moreover, the multiwell plate position could drift over time due to temperature changes and stage motion. These variations were automatically corrected over time by the autofocus mechanism of the microscope (Nikon PerfectFocus), so the absolute cell positions should not be affected by these spatial and temporal variations. The absolute invasion distance per well was zeroed against the average initial position of cells at the start of the experiment, when all cells were adherent to the bottom of the well.
2.11. Immunofluorescent Staining
After live cell imaging for 48 h, cells were fixed and immunofluorescently stained to visualize the cell morphology. Briefly, the excess media was first aspirated off. Cells were then fixed for 45 minutes in 100 uL each of 4% paraformaldehyde in phosphate buffered saline. The fixative was aspirated off and hydrogels were washed with 100 μl 1X Dulbecco’s Phosphate Buffer Solution (1X PBS) 3 times at 5 minute intervals. Hydrogels were then permeabilized using 0.25% TritonX-100 for 45 minutes, washed with PBS (3 times, five minute intervals) then placed overnight in 5% nonfat dry milk. The next day, hydrogels were washed with PBS (3 times, 10 minute intervals). Cells were then stained using HCS CellMask Deep Red Stain (1:2500 dilution in 1X PBS) and either NucMask (1:250 dilution) or Hoechst (1:200 dilution). Finally, hydrogels were washed twice with 1X PBS (2 times, 1 hour interval). Immunofluorescently stained cells were again imaged using a Nikon Eclipse Ti fluorescence microscope with spinning-disk confocal head, consistent with the previous description. How-ever, a 20x Plan Apo objective (NA 0.75) was used with the GFP/FITC Filter Set (Chroma 49002), Far Red/Cy5 Filter Set (Chroma49006), or DAPI Filter Set (Chroma 49000), and ranged from 2 to 10 um z steps across all replicates.
2.12. Perturbation with Epidermal Growth Factor and Taxol
Cell invasion behaviors were perturbed using 8.33 nM human epidermal growth factor (EGF) or 13 nM Taxol in cell culture medium. Both EGF and Taxol perturbations were pipetted onto respective polymerized hydrogels, at the beginning of the experiment. Gel volume was taken into account when calculating final drug concentrations for each perturbation. Stock solutions of Human EGF were reconstituted in sterile filtered deionized water as recommended by manufacturer to a stock concentration of 16 μM, then diluted to achieve a final concentration of 8.33 nM EGF50 in cell culture medium. Taxol was reconstituted in dimethyl sulfoxide as recommended by manufacturer to a concentration of 5.9 mM. The stock solution of Taxol was then diluted to achieve a final concentration of 13 nM Taxol51 and 0.05% DMSO in cell culture medium. Taxol treatment conditions had a paired control consisting of 0.05% DMSO in cell culture medium.
2.13. Live/Dead Quantification with DRAQ7
Live/dead cells were quantified using DRAQ7 dye per manufacturers instructions at a 1:100 (final concentration: 3 μM) dilution in media. DRAQ7 only enters the membrane of dead or permeabilized cells. Briefly, excess media (∼80 μl) was carefully removed from the hydrogels after 48 hours of imaging. DRAQ7 in media was pipetted onto each well. Dye was allowed to diffuse into the hydrogels and imaged for Far-Red signal (Far Red/Cy5 Filter Set, Chroma49006). Representative wells were cropped to a representative 500 × 500 pixel z-stack and the total number of cells per image was manually counted, indicated by GFP nuclei signal or a Far-Red DRAQ7 signal. The number of dead cells were also counted, indicated by total permeabilization of the nuclei by DRAQ7. The percentage of live cells was calculated by subtracting the number of dead cells from the total number of cells counted and dividing by the total number of cells counted.
2.14. Segmentation of Mesenchymal and Amoeboid Cell Morpholo-gies
Cell morphology was quantified as mesenchymal or amoeboid based on the following procedure. First, maximum intensity projections were created from confocal z-stack acquisitions (20X magnification, at a distance of 20 μm from the bottom of the hydrogel) and processed using a custom CellProfiler pipeline. A total of 672 cells (320 amoeboid and 352 mesenchymal cultured in media) were segmented manually by outlining the cell shape (using the IdentifyObjectsManually module) in the 2-D projection and their shape was characterized by computing non-dimensional shape factors (extent, solidity, compactness, etc.), area, perimeter, length of major and minor axes using CellProfiler 3.1.5. While performing manual segmentation each cell was labelled as amoeboid or mesenchymal, with up to 25 cells segmented per well. Care was taken when assigning labels based on morphology seen in the 2-D maximum intensity projection image by also simultaneously examining multiple z-slices (using NIS-Elements) to account for 3-D shape. Cell classification was performed by setting a threshold for binary classification on each shape factor and computing classification accuracy by comparing with manually assigned labels. Using compactness (the mean squared distance of the object’s pixels from the centroid divided by the area) and a threshold value of 1.25, amoeboid and mesenchymal cells were classified with 88% and 89% accuracy respectively.
Cells were also checked manually for mesenchymal or amoeboid morphological phenotype at the 48 h timepoint by first examining the topmost z slice (100 μm) and then checking successively downwards towards the bottom of the well. Amoeboid morphologies were defined as generally spherical morphology with little to no protrusions into the matrix, and minimal extension through successive z-slices. Mesenchymal morphologies were defined as an elongated, spindle-like morphology with at least one protrusion into the matrix as well as extension across successive z-slices. Some mesenchymal cells were elongated vertically in the z direction, which was checked by viewing the cell morphology at different heights. Care was taken to ensure that only cells which had invaded into 3D were counted by careful examination of confocal slices. Moreover, occasional cell debris not associated with a cell nucleus or connected to a cell body were not counted.
2.15. Scoring Metrics for Identification of Multinucleated Cells
Cells were also manually scored for aberrant nuclear morphologies, including multinucleated cells. Multinucleation was defined as one cell body containing multiple nuclei outlines that were disconnected from one another, or as one nucleus comprised of multiple rounded nuclear protrusions. The number of invaded and multinucleated cels were tallied beginning at the highest z slice for the hydrogel at about 100 μm and moving downward in z slices to 20 μm. Cytoplasm labeling was used to determine whether a multinucleated nucleus outline was contained within a single cell. Care was taken to ensure that only cells which had invaded into 3D were counted by careful examination of confocal slices. The percentage of multinucleated cells was determined by dividing the number of multinucleated cells by the total number of cells invaded per matrix composition.
2.16. Statistical Analysis
Statistical analysis was conducted using JMP(R) Pro 14.1.0 software (SAS Institute Incorporated). Full factorial logistic regression was used to examine the impact of collagen, silk, and collagen*silk on the odds of cancer cell invasion or cellular morphology. Due to significant interaction between the effect of collagen and silk on invasion, the impact of different silk concentration on the odds of cancer cell invasion by Chi-square analysis was examined independently by collagen concentration and drug perturbations. For the subpopulation of cells that invaded (invasion distance > 10 μm), the impact of silk concentration on invasion distance or cellular morphology were examined using Wilcoxon Krusal-Wallis test with non-parametric multiple comparisons using Steel-Dwass method or Contingency Analysis, respectively. The tests were conducted independently by collagen concentration and drug perturbations. Statistical significance was set at p = 0.05.
3. Results
3.1. Composite Hydrogels of Silk Fibroin and Collagen I
Composite hydrogels consisting of interpenetrating silk fibroin and collagen I were designed to independently tune mechanical stiffness and collagen concentration (Figure 1). Silk fibroin is a naturally-derived block copolymer with repeat units of [GAGAGS]n, which folds into hydrophobic β-sheet forming blocks linked by short hydrophilic segments (Figure 1A). The crystallization of these β-sheet domains permits physical crosslinking of silk fibroin into hydrogels. Briefly, silk fibroin was extracted from B. mori cocoons by boiling in sodium carbonate, rinsing in deionized water, dissolving in lithium bromide, dialyzing, then storing at 4°C until use.46 In order to form hydrogels, silk fibroin was sonicated for 5–15 seconds and dispensed into a multiwell plate.41,42 For this study, silk fibroin only hydrogels ranged in concentration from 5 – 10 mg/mL (Figure 1C).
Figure 1:
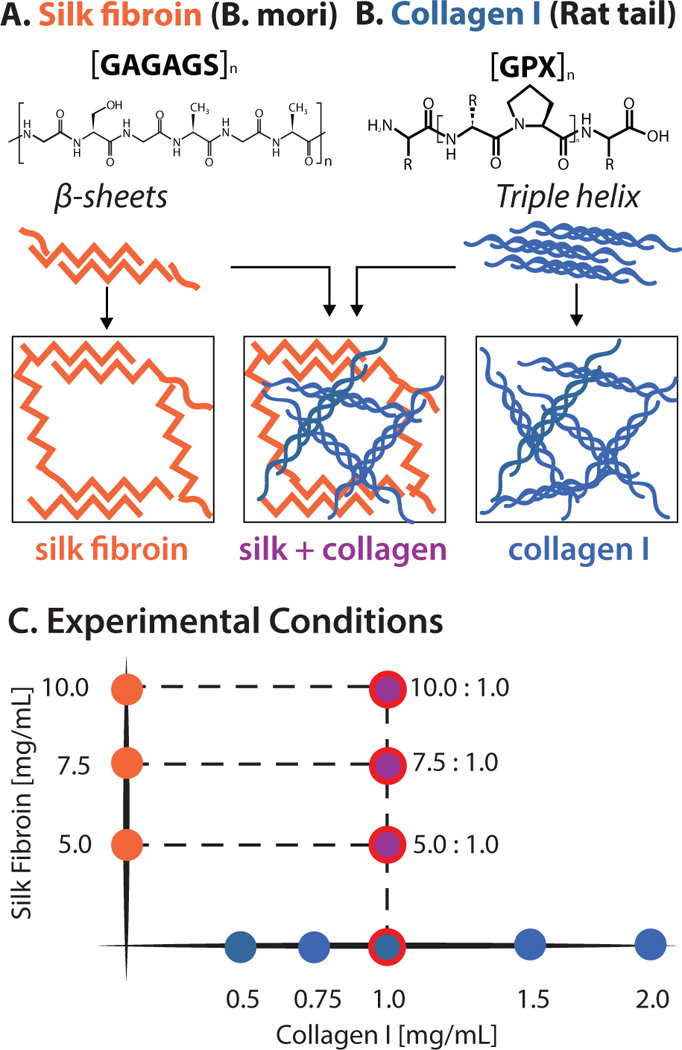
Chemical structures of (A) silk fibroin and (B) collagen I that were combined into a composite hydrogel with an interpenetrating network. (C) Hydrogel compositions for this study included silk fibroin only (5.0–10.0 mg/mL), and composite hydrogels of collagen I (1.0 mg/mL) and silk fibroin (5.0–10.0 mg/mL), and collagen I only (0.5 – 2.0 mg/mL). Red circles denote composite conditions of interest.
Collagen I has a typical chain composition of [GPX]n, where X is any amino acid other than glycine, proline, or hydroxyproline (Figure 1B).18,19 Reconstituted rat-tail collagen I typically polymerizes after adjusting pH to 7.4, particularly at increased temperature. Collagen I assembles into a triple helical structure, which can be hierarchically bundled and physically crosslinked into fibrous networks. For this study, collagen I only hydrogels ranged in concentration from 0.5 – 2.0 mg/mL (Figure 1C). It should be noted that 1.0 mg/mL collagen I hydrogels have been previously shown to be relatively soft with a larger mesh size that is permissive for cell migration.20,22,26
Finally, composite hydrogels were prepared with constant collagen I concentration (1 mg/mL) but varying silk fibroin concentrations (5–10 mg/mL) (Figure 1C). This approach allowed for hydrogels with constant ligand density from collagen I, while the hydrogel stiffness and microstructure could be tuned by silk fibroin concentration. In order to prepare interpenetrating networks, silk fibroin was first sonicated, then monomeric collagen I was added before dispensing into a multiwell plate.
3.2. Hydrogels Exhibit Greater Stiffness with Increasing Silk Fibroin
Hydrogels were first characterized using bulk rheology in order to elucidate how mechanical properties depended on silk fibroin and collagen I concentration. The shear storage modulus GI was characterized in the linear viscoelastic regime from 0.01 – 1 Hz at 0.1% strain using a 40 mm 2° acrylic cone and plate geometry (Figure S1). Silk fibroin hydrogels showed a slight frequency dependence for all conditions, with greater frequency dependence for 10 mg/mL (Figure 2A, S2A). For instance, at 5 mg/mL silk fibroin, GI(ω) increased from 45 Pa to 70 Pa, while for 7.5 mg/mL silk fibroin, GI(ω) increased by from 85 Pa to 130 Pa. Finally, 10 mg/mL silk fibroin hydrogels demonstrated strong frequency-dependent behavior with GI increasing from 90 Pa to 345 Pa.
Figure 2:
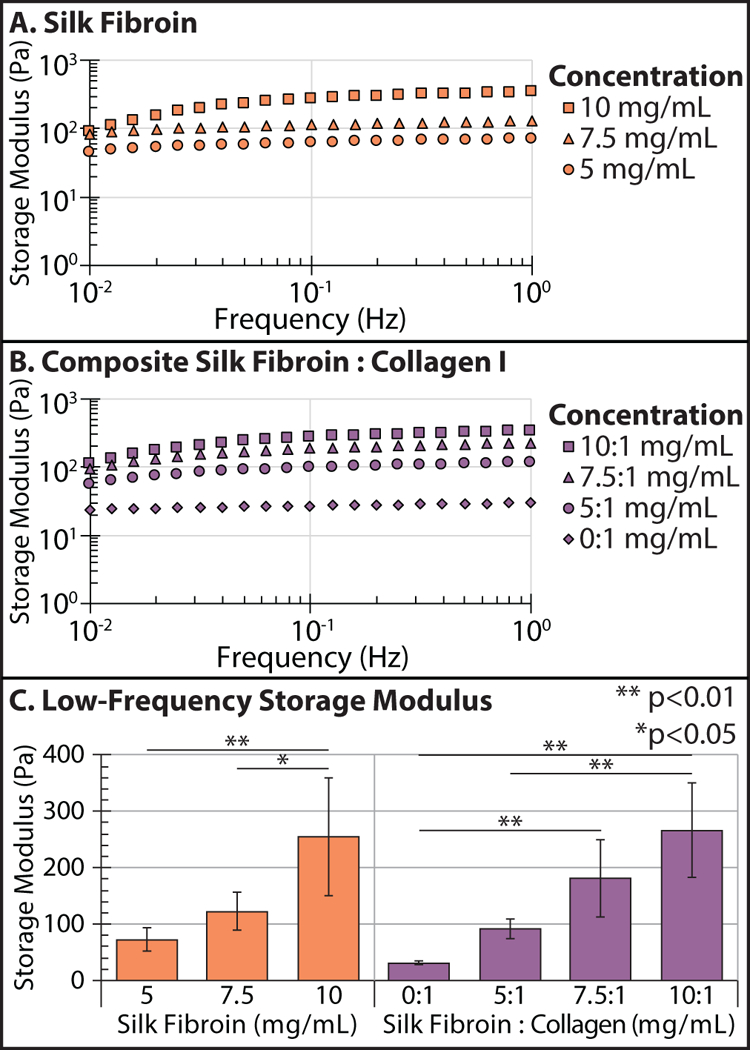
Frequency-dependent storage modulus Gt of silk fibroin (A), and composite silk fibroin-collagen I hydrogels (B) at varying concentration and composition. Comparison of low-frequency storage modulus Gt across conditions (C). Error bars = standard deviation. ** p < 0.01, * p < 0.05 (Student’s t-test)
Composite silk-collagen hydrogels exhibited more frequency-dependent behavior than silk fibroin only hydrogels (Figure 2B, S2B). For example, 5 mg/mL silk fibroin and 1 mg/mL collagen I hydrogels exhibited the least frequency dependence with GI(ω) increasing from 55 Pa to 115 Pa. For 7.5 mg/mL silk fibroin and 1 mg/mL collagen I hydrogels, the frequency dependence was slightly stronger with GI(ω) increasing from 90 Pa to 220 Pa. Finally, 10 mg/mL silk fibroin and 1 mg/mL collagen I hydrogels had the greatest frequency dependence with GI(ω) increasing from 110 Pa to 330 Pa. In comparison, collagen I only hydrogels exhibited slightly frequency-dependent behavior for all conditions (Figure 2C, S2C). For example, at 0.5 mg/mL collagen I, GI increased from 1.5 Pa to 2.5 Pa. For 1 mg/mL collagen I, GI(ω) increased from 20 Pa to 30 Pa. Finally, for 2 mg/mL collagen hydrogels GI(ω) increased from 110 Pa to 145 Pa (Figure S2A).
These results could be summarized by the low-frequency storage modulus GI. Silk fibroin hydrogels were stiffer over the concentrations tested, ranging from GI ≈ 70 Pa at 5 mg/mL up to GI ≈ 120 Pa at 7.5 mg/mL, then reaching GI ≈ 250 Pa for 10 mg/mL (Figure 2C). For collagen I hydrogels, GI ≈ 2 Pa at 0.5 mg/mL, increasing up to GI ≈ 30 Pa at 1 mg/mL, and up to GI ≈ 120 Pa at 2 mg/mL (Figure S2D). Finally, for composite silk-collagen hydrogels, GI was approximately the sum of GI for the collagen and silk-fibroin hydrogels at comparable concentrations. Thus, for composite hydrogels with 1 mg/mL collagen I, GI ≈ 90 Pa for 5 mg/mL silk fibroin, increasing to GI ≈ 190 Pa for 7.5 mg/mL silk fibroin, then reaching GI ≈ 270 Pa for 10 mg/mL silk fibroin (Figure 2D). Since silk fibroin was stiffer than collagen at these concentrations, it largely governed the stiffness of the composite hydrogel.
Finally, unconfined compression testing was used to measure elastic modulus and stress relaxation half life. 48 Elastic modulus E increased with increasing silk fibroin concentration (5–10 mg/mL) at constant collagen I concentration (1 mg/mL) (Figure S3A). Nevertheless, the stress relaxation half-life, defined as the characteristic timescale where the normalized load relaxed to 50% of its initial value, remained roughly consistent from 16–20 seconds as silk fibroin concentration was varied (Figure S3B,C), which is comparable to previous measurements of reconstituted collagen I hydrogels, silk fibroin, and soft tissues. 45,52,53
3.3. Collagen Fiber Architecture in Collagen I and Composite Hydrogels
Next, collagen I was imaged label-free using second harmonic generation (SHG) microscopy54 in order to determine how fiber architecture was affected by silk fibroin concentration in composite hydrogels. It should be noted that collagen I has a strong SHG signal since it is noncentrosymmetric and also highly crystalline,55 whereas the more amorphous silk fibroin microstructure tends to have minimal SHG signal. 56 Visually, the collagen I fiber density appeared to qualitatively increase with increasing silk fibroin concentration in composite hydrogels (Figure 3A).
Figure 3:
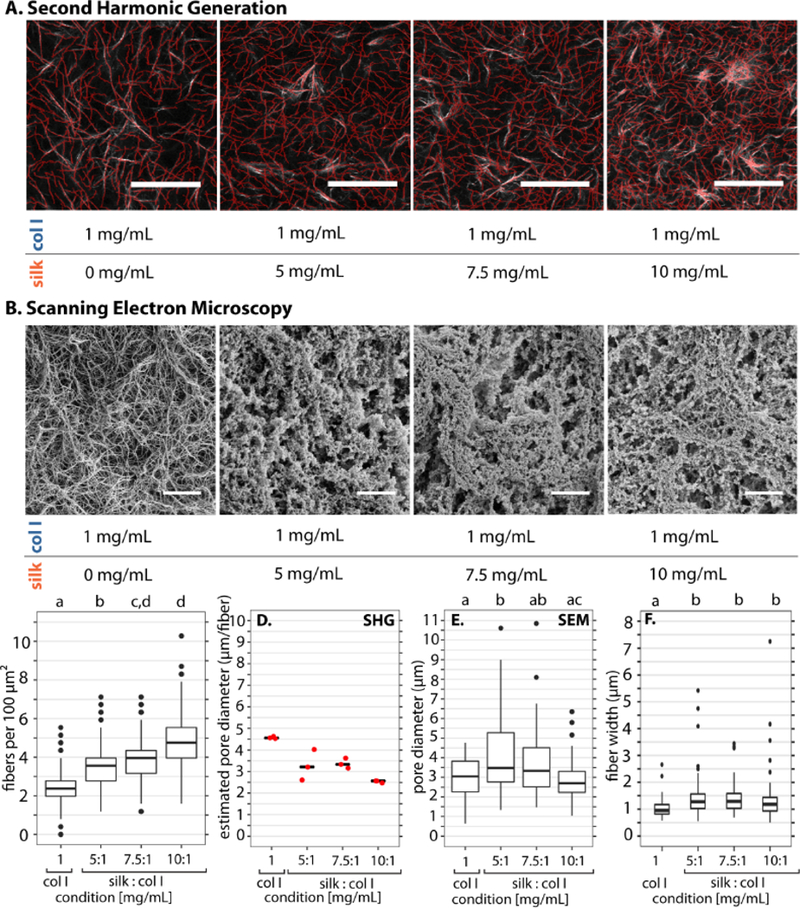
Representative images and analysis for collagen and composite silk collagen gels. (A) Represen-tative second harmonic generation (SHG) microscopy images of collagen I fibrillar architecture for 1 mg/mL collagen I with 0 – 10 mg/mL silk (white), with overlaid fiber detection (red). Scale bar: 50 μm (B) Scanning Electron Microscopy images of 1 mg/ml collagen gels and composite silk-collagen gels. (C) Number of detected collagen I fibers from second harmonic analysis using CurveAlign per 100 /mum2. Letters denote statistically different groups with p < 0.05, using Kolmogorov-Smirnov test corrected for multiple comparisons. (D) SHG pore diameter estimated interpolated for the average box that fits one fiber from CurveAlign density output. Pore diameter here is an estimate for collagen mesh alone in collagen and composite silk collagen gels. (E) SEM pore diameter from 50 pore segmentations from a 50 × 50 micron representative image. Statistic: Multiple comparison t-test corrected for multiple comparisons. (F) Analysis of SHG images for fiber width. Statistical test: Wilcoxen corrected for multiple comparisons. All boxplots show the median with lower (25%) and upper quartiles (75%). Whiskers further extend by 1.5X interquartile range from the lower and upper quartiles, with data points beyond this range plotted as outliers.
Scanning electron microscopy (SEM) was used to complement SHG by imaging the silk fibroin microstructure. Collagen I only hydrogels (1 mg/mL) exhibited a fibrillar architecture under SEM that was qualitatively consistent with SHG (Figure 3B). However, increasing silk fibroin concentration (5–10 mg/mL) at constant collagen I (1 mg/mL) resulted in decreasing porosity, with microstructures that were more rounded and less fibrous (Figure 3B).
In order to quantify the microstructure, SHG images were analyzed using the CT-FIRE software.49 Briefly, this analysis applied a binary threshold to detect fibers, performed a distance transform to detect nucleation points, extended candidate fiber branches outwards from these nucleation points, then linked adjacent fibers together and discarded short dangling ends (Figure S4A). Based on segmented fibers detected, CT-FIRE analysis quantified individual fiber parameters including length, width, angle, and curvature (Figure S4B). CT-FIRE parameters were optimized for detection of all visible collagen I fibers in each image, with all images set to comparable lookup table values. Subsequently, CurveAlign software was used to extract local fiber density.57
Quantitatively, the average collagen I fiber density increased with increasing silk fibroin concentration. Scaled for ease of comparison, there were ≈ 2.4 fibers within a 100 μm2 area or “box” for collagen I only (0.0 mg/mL silk fibroin) (Figure 3B). The addition of silk fibroin increased fiber density up to ≈ 3.6, 4.0 and 4.7 fibers per box for 5.0, 7.5 and 10.0 mg/mL silk fibroin, respectively (Figure 3C). Inverting this readout of fibers per box yielded an area per fiber, which can be mapped to a characteristic mesh size of the 2D fiber network. The pore area decreased from an average of 16 μm2 for 1 mg/mL collagen I only, down to 5 μm2 for 10 mg/mL silk fibroin with 1 mg/mL collagen I (Figure S5A). The square root of this value gives a characteristic pore diameter (mesh size), which decreased from 4.5 μm for 1 mg/mL collagen I only, down to 2.7 μm for 10 mg/mL silk fibroin with 1 mg/mL collagen I (Figure 3D). Since SHG does not resolve silk fibroin, the overall pore size was manually estimated from SEM images (Figure S6, S7). As silk fibroin concentration was increased at constant collagen I (1 mg/mL), the median of the pore diameter decreased from 3.47μm at 5 mg/mL silk fibroin down to 2.7 μm at 10 mg/mL (Figure 3E). It should be noted that SEM was performed on a dried hydrogel specimen, which is contracted relative to the hydrated state. Thus, SEM pore areas likely underestimate the actual pore areas, and cannot be directly compared with SHG. Finally, the fiber diameter remained roughly comparable at ~1 μm across all conditions (Figure 3E). This diameter corresponds to approximately 3 pixels for this objective (0.32 μm/pixel), so the differences in fiber diameter are within the uncertainty of the measurement. Nevertheless, an interesting possibility is to use this technique to modulate pore area at constant fiber diameter. Finally, the average fiber lengths were comparable at ≈16 μm across all hydrogels (Figure S5C). Nevertheless, the collagen I only hydrogel is visibly skewed towards longer fiber lengths, with a slight shift towards shorter fibers with increasing silk fibroin concentration (Figure S5C). This decrease in fiber length offsets the increased fiber density at constant collagen I concentration as silk fibroin concentration is varied.
An alternative method to control collagen I microstructure is to alter the gelation temperature, as shown by George and coworkers. Precursor solutions of unpolymerized collagen I and silk fibroin were gelled on ice at 0° for 30 min, then imaged using SHG and SEM (Figure S8). Qualitatively, SHG of collagen I only (1 mg/mL) hydrogels exhibited large pores and more variable fiber diameters (Figure S9A), consistent with previous observations.47 These effects were more pronounced with increasing silk fibroin with constant collagen I (1 mg/mL), due to the appearance of patchy regions of dense collagen I (Figure S9A). Moreover, SEM showed decreasing pore sizes with increasing silk fibroin concentration (Figure S9B). Overall, average fiber density across conditions remained comparable at 2 fibers per 100 μm2 (Figure S9C), while pore diameter decreased from 3.86 to 1.78 μm. (Figure S9D). Finally, SEM showed pore diameters for silk fibroin that were 2.5 μm or less (Figure S9E), which were comparable or smaller than the 10 mg/mL silk fibroin, 1 mg/mL collagen I prepared at 37° (Figure 3E).
Finally, collagen I hydrogels exhibited a relatively weak dependence of fiber density, pore area, and mesh size from 0.5 mg/mL to 1.5 mg/mL, with a more drastic decrease at 2 mg/mL (Figure S4C, S5A,B). These relatively similar values may result from a weak dependence of pore area and mesh size over this limited concentration range. It should be noted that gelation occurred in media with serum added, which may not be directly comparable to collagen I hydrogels prepared in media only.
3.4. Cancer Cell Invasion Exhibits Biphasic Dependence on Silk Fibroin Concentration in Composite Hydrogels
Cancer cell invasion was then characterized in hydrogels of varying silk fibroin and collagen I concentration using an overlay geometry. Briefly, metastatic breast adenocarcinoma cells (MDA-MB-231) stably transfected with green fluorescent protein fused to nuclear histones (H2B-GFP) were first plated in a a 96-well high content plate coated with a thin coating of collagen I (Figure 4A). After cells adhered for 1–2 h, media was aspirated away and the hydrogel precursor solution was overlaid in each well, which then polymerized and crosslinked at 37° C (Figure 4B). Finally, the multiwell plate was loaded into an environmentally controlled spinning disk confocal microscope and imaged at z-steps of 8 μm every hour over 48 h (Figure 4C). After the completion of time-lapse imaging, cells were fixed and immunofluorescently stained with a cytoplasmic dye (CellTracker Far Red) and then re-imaged using confocal microscopy (Figure 4D)
Figure 4:
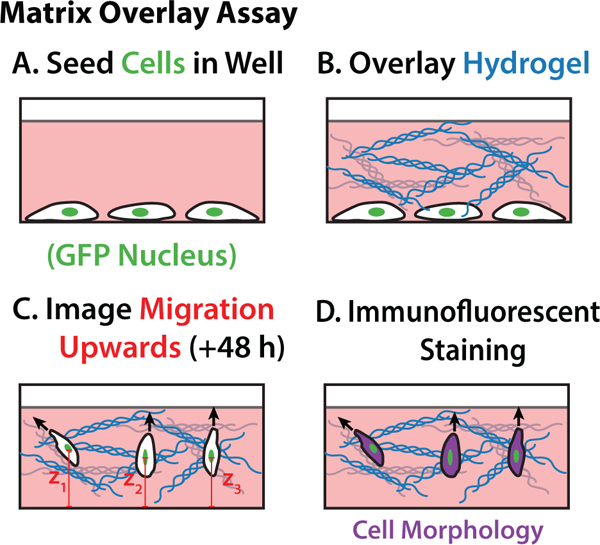
Overlay Assay Schematic. GFP nuclei MDA-MB-231 cells are seeded in a collagen coated 96 well plate and allowed to adhere for 1–2 hours (A.), media is aspirated and hydrogels are carefully placed on top of the cells (B.), upward migration observed over 48 hours (C.), wells are fixed and stained for cell body morphologies (D.)
For this assay, invasion was quantified using two metrics: the percentage of cells invaded (calculated based on the number of invasive cells relative to the total number of cells in the field of view) and the distance invaded by migratory cells. We defined “invasive” cells as those with nuclei detected at least 10 μm from the well bottom after 48 h. For ease of comparison, we plot these metrics for two experimental groups based on hydrogel composition: 1) silk fibroin only (5.0 mg/mL-10.0 mg/mL) and 2) composite collagen I (1 mg/mL) and silk fibroin (0.0 mg/mL-10.0 mg/mL).
For silk fibroin only hydrogels, the percentage of cells invaded was minimal at 5–6% for all concentrations (Figure 5A). For hydrogels that included collagen I, the percentage of cells invaded from 4% up to 38% then decreased down to 10% as silk concentration increased from 0.0 – 10.0 mg/mL, respectively (Figure 5A). Similarly, the distance invaded in silk fibroin only hydrogels was minimal at ≈15 μm (Figure 5B). In comparison, for composite silk-collagen hydrogels with 1 mg/mL collagen I, the median distance invaded increased from 24 μm to 65 μm and then decreased back to 34 μm and 18 μm for silk concentrations of 0.0, 5.0, 7.5 or 10.0 mg/mL, respectively (Figure 5B). Thus, both the percentage of cells invaded and distance invaded exhibited a biphasic dependence on silk fibroin concentration, with a maximum occurring for 5 mg/mL silk fibroin and 1 mg/mL collagen I. For comparison, the distance invaded in the collagen I only (1 mg/mL) hydrogel prepared at 0° increased relative to 37° (Figure S10), which was consistent with the increased collagen I pore size at 0° relative to 37° (Figure S9D, 3D). In comparison, the distance invaded was uniformly low for 5–10 mg/mL silk fibroin with 1 mg/mL collagen I at 0° relative to 37°, where the pore size was consistently small at at 0° relative to 37° (Figure S9E, 3E).
Figure 5:
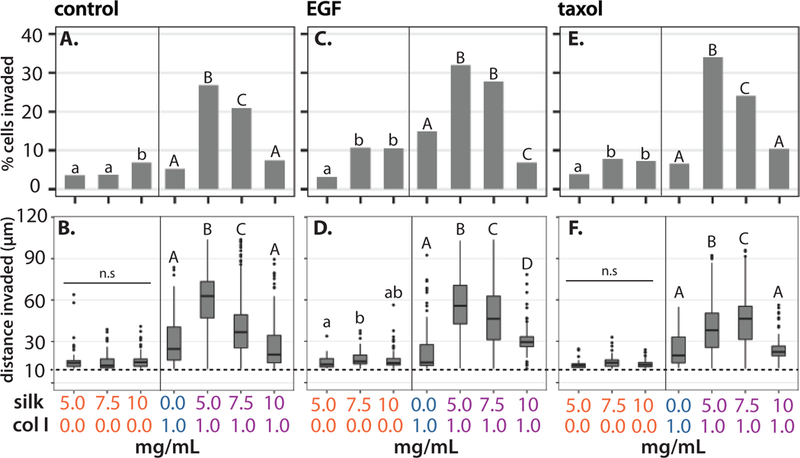
Percent of cells invaded in silk fibroin only hydrogels, or composite hydrogels with 1 mg/mL collagen I in control media or 0.05% DMSO (A.), with 8.33 nM EGF (C.), and 13 nM Taxol (E.). Box plot of cells invaded in silk fibroin only hydrogels or composite hydrogels with 1 mg/mL collagen I in control media or 0.05% DMSO (B.), with 8.33 nM EGF (D.), and 13 nM Taxol (F.). Boxplots show the median with lower (25%) and upper quartiles (75%). Whiskers further extend by 1.5X interquartile range from the lower and upper quartiles, with data points beyond this range plotted as outliers. Conditions with matching letter and style indicate non-significant differences with p < 0.05, based on Wilcoxon Krusal-Wallis test with non-parametric multiple comparisons.
For comparison, cell invasion was characterized after treatment with exogeneous epidermal growth factor (EGF), which is associated with increased motility and contractility in MDA-MB-231 cells. 50 After hydrogels were crosslinked, 8.33 nM EGF in media was added to fill the well. In silk fibroin only hydrogels, the percent of cells invaded and distance invaded remained minimal with EGF, at ≈10% and ≈15 μm, respectively (Figure 5C,D), remaining comparable to the untreated control. In comparison, for hydrogels that included collagen I, the percent of cells invaded increased from 15% for 0 mg/mL silk fibroin, up to 30% for 5 mg/mL silk fibroin, then back down to 7% for 10 mg/mL silk fibroin (Figure 5C). Similarly, the median invasion distance increased from 15 μm in 1 mg/ml collagen gels (0 mg/ml silk) to 50 μm with the addition of 5 mg/ml silk. Increasing silk concentration to 7.5 mg/ml decreased median invasion distance to 42 μm. Increasing silk protein further to 10 mg/ml silk decreased median invasion distance to 27 μm (Figure 5D). Thus, after EGF treatment, both the percentage of cells invaded and distance invaded again exhibited a biphasic dependence on silk fibroin concentration, with a maximum occurring for 5 mg/mL silk fibroin and 1 mg/mL collagen I.
Finally, cell invasion was characterized after sublethal treatment with the chemotherapeutic agent paclitaxel (Taxol), which stabilizes microtubules and disrupts cytoskeletal function.58 13 nM Taxol in media was again added over the crosslinked overlaid hydrogels. This treatment resulted in appreciable cell death, although at 70–90% of the population re-mained alive after 48 h based on live/dead staining with DRAQ7 (Figure S11, S12). In silk fibroin only hydrogels, the percent of cells invaded and distance invaded remained minimal with Taxol, at ≈7% and ≈15 μm, respectively (Figure 5E), remaining comparable to the untreated control. In comparison, for hydrogels that included collagen I, the percent of cells invaded increased from 5% for 0 mg/mL silk fibroin, up to 34% or 5 mg/mL silk fibroin, then back down to 10% for 10 mg/mL silk fibroin (Figure 5F). Thus, after Taxol treatment, both the percentage of cells invaded and distance invaded exhibited a biphasic dependence that was qualitatively consistent with the untreated and EGF treatments. In comparison, the percentage of cells invaded and distance invaded exhibited a relatively weak concentration dependence in collagen I only hydrogels, at least within a limited range between 0.5 – 2.0 mg/mL, for untreated, EGF, and Taxol treated cells (Figure S13).
3.5. Transitions from Amoeboid to Mesenchymal Morphology Occur with Collagen I as well as EGF
Single cell morphology was further characterized in hydrogels of varying silk fibroin and collagen I composition, using immunofluorescent staining of the cytoplasm at the completion of the experiment. In particular, some cells exhibited a compact and rounded morphology consistent with an amoeboid phenotype, propelled by actomyosin contractility (Figure 6A–C). Alternatively, other cells exhibited an elongated, spindle-like morphology consistent with a mesenchymal phenotype, based on actin-driven protrusions with strong cell-matrix adhesions (Figure 6A–C).
Figure 6:
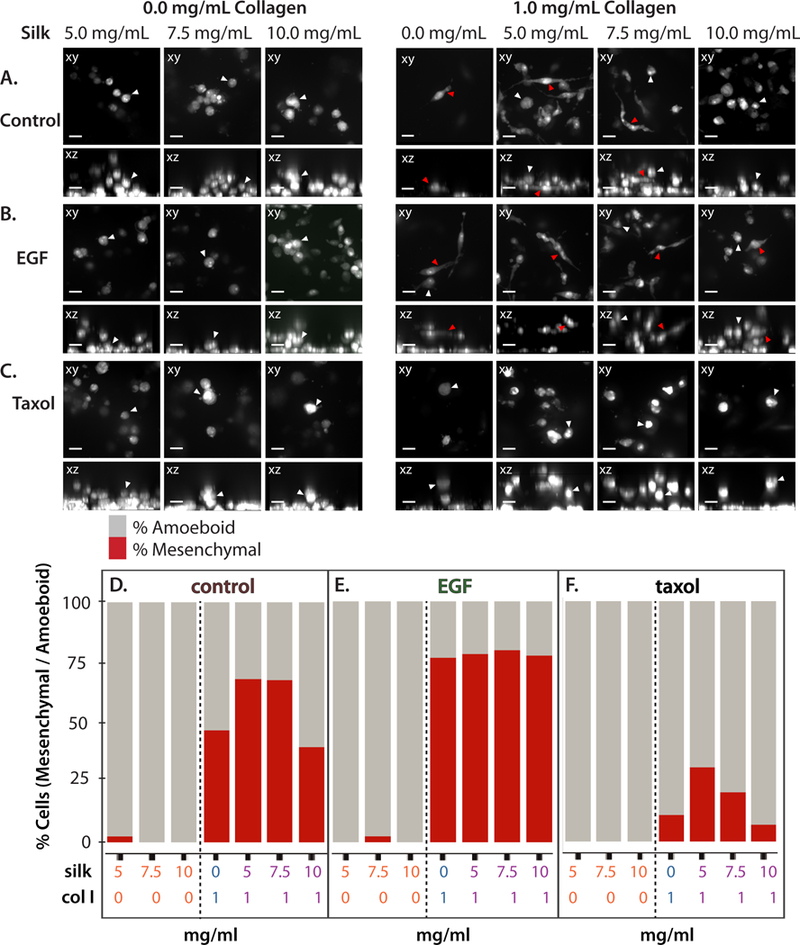
Representative xy and xz rendered mesenchymal (elongated) or amoeboid (compact) morphologies for cells in different silk fibroin concentrations only, and with 1 mg/mL collagen I in control media (A), with 8.33 nM EGF (B), and 13 nM Taxol (C). Amoeboid morphologies highlighted with white arrows, mesenchymal morphologies highlighted with red arrows. Percent cells with mesenchymal or amoeboid morphologies in different silk fibroin concentrations only, and composite hydrogels with 1 mg/mL collagen I in control media (D), with 8.33 nM EGF (E), and 13 nM Taxol (F).
Cells were classified as either amoeboid or mesenchymal phenotypes based on compactness (the mean squared distance of the object’s pixels from the centroid divided by the area), where circular morphologies have a compactness closer to 1 (Figure S14). relative morphological elongation. In silk fibroin only hydrogels, greater than 98% of all invaded cells exhibited rounded amoeboid morphologies, which was consistent for untreated controls, as well as EGF and Taxol treatments (Figure 6D,E,F). It should be noted that the absolute number of cells invaded in silk was typically quite small (5–15 cells per well, with 4+ technical replicates, for 20–60 cells total), so that the observed amoeboid cells represent an exceptional phenotype that was unusually effective at migrating with minimal cell-matrix adhesions.
For hydrogels that incorporated collagen I, cells typically exhibited a mixture of mesenchymal and amoeboid morphology (Figure 6A). In particular, for 1 mg/mL collagen I only, the cells were roughly half mesenchymal and half amoeboid (Figure 6D). As silk fibroin increased to 5.0 and 7.5 mg/mL in the composite hydrogels, the percentage of mesenchymal morphologies increased to ≈70%, decreasing back to about half for 10.0 mg/mL silk fibroin in the composite hydrogel (Figure 6D). Overall, this biphasic concentration dependence of percentage mesenchymal morphology on silk concentration (in composite hydrogels) in untreated controls is highly reminiscent of the biphasic concentration dependence of percent cells invaded and invasion distance observed previously.
In comparison, treatment with exogeneous EGF biased towards mesenchymal morphologies of ≈75% for all hydrogels that incorporated collagen I (Figure 6B,E; S15). Instead, treatment with Taxol biased cell morphology towards amoeboid morphologies, decreasing the mesenchymal morphology to ≈10–25% (Figure 6C,F; S15). It was also observed that Taxol treatment further resulted in aberrant nuclear morphologies, particularly multinucleation associated with mitotic defects (Figure S16A,B,C). The percentage of multinucleated cells ranged from 25–75% after Taxol treatment, but there were no obvious trends for multinu-cleation based on hydrogel composition (Figure S16D). Finally, the cells in collagen I only hydrogels exhibited a mixture of mesenchymal and amoeboid morphologies with a relatively weak concentration dependence (Figure S17). Nevertheless, EGF treatment in collagen I only hydrogels also biased towards a greater percentage of cells with mesenchymal morphologies, while Taxol treatment consistently biased towards cells with a greater percentage of amoeboid morphologies (Figure S15).
4. Discussion
Bulk rheology of these composite silk fibroin and collagen I hydrogels showed that the low-frequency storage modulus GI increased with silk concentration (for small strains), consistent with silk fibroin only hydrogels. These measurements were conducted with cell-free hydrogels, which represent the initial state of the overlaid hydrogel. Nevertheless, it is likely that migratory cells locally remodel the ECM over the course of the 48 h experiment through strong deformations, secretion of matrix metalloproteases, or deposition of their own ECM. Since cells were plated at relatively low densities and the experiments are relatively short, we anticipate that these structural changes will not significantly affect the bulk rheology. Nevertheless, local microstructural alterations on cellular length scales can greatly affect cell migration. An important question for future work is to understand how cancer cell mechanotransduction occurs in the context of spatiotemporal changes in ECM architecture and microrheology.
Both the percentage of cells invaded as well as the median distance invaded exhibited a biphasic dependence on silk concentration in composite hydrogels with fixed collagen I concentration. It was observed that the largest percentage and median distance increased from 0 up to 5 mg/mL silk and decreased thereafter from 7.5 to 10 mg/mL silk (with 1 mg/mL collagen I). The corresponding low-frequency storage modulus GI increased from 30 Pa to 90 Pa for 0 to 5 mg/mL silk, and further increased to 185 Pa and 265 Pa for 7.5 to 10 mg/mL silk (with 1 mg/mL collagen). The maximum observed invasion thus corresponds to an intermediate stiffness of GI ≈ 90 Pa. Interestingly, Shenoy and coworkers observed a similar biphasic dependence on concentration for melanoma cells in collagen I hydrogels of varying concentration.30 Their observed optimum stiffness occurred at GI ≈153 Pa (E ≈ 430 Pa, assuming ν = 0.4) with diminished invasion at higher (GI ≈ 267 Pa) stiffness. This trend was theoretically justified based on the decrease in mesh size, which would sterically hinder cell invasion.15,26,28,29,31–33 These hydrogels consist of an interpenetrating network of collagen I and silk fibroin, where collagen I polymerization may be sterically impeded by the presence of silk fibroin. As a consequence, collagen I fibers are slightly shorter with increasing silk fibroin concentration, which contributes to a smaller pore size (Figure 3). It should be noted that the densest composite hydrogel (10 mg/mL silk fibroin, 1 mg/mL collagen I) exhibited an average mesh size of 5 μm2, which is below the 7 μm2 threshold reported as a physical limit for migration,15 and consistent with the diminished invasion measured here. These observations were corroborated by gelation at lower temperature with SEM, which resulted in smaller silk fibroin pore sizes that further impeded cell migration (Figure S9, S10). Thus, our observations of a biphasic dependence of migration on silk fibroin concentration is analogous to previous reports that report a biphasic dependence on collagen I concentration.
Transitions from mesenchymal to amoeboid migration phenotype have been associated with microenvironmental conditions that impede cell-matrix adhesions, ECM remodeling, or cytoskeletal protrusion formation. For example, cells migrating in silk fibroin gels almost exclusively exhibited rounded, amoeboid phenotypes. In comparison, composite silk fibroin-collagen I hydrogels biased towards mesenchymal morphology for the lower silk fibroin concentrations (5.0–7.5 mg/mL), but towards amoeboid morphology at the highest silk fibroin concentration (10.0 mg/mL). These results suggest that amoeboid migration is favored when cell-matrix adhesions are less effective, particularly with substantial concentrations of silk fibroin, which lacks cell-adhesive ligands. Exogenous EGF treatment increased cytoskeletal contractility and cell-matrix adhesions, strongly biasing towards mesenchymal morphology (~ 80%) in the composite and collagen I only hydrogels. Instead, Taxol treatment to suppress microtubule dynamics and disrupt cytoskeletal function biased towards an amoeboid phenotype in the composite and collagen I only hydrogels. Nevertheless, in both EGF and Taxol treatment conditions, cells always exhibited an amoeboid morphology in silk fibroin hydrogels, likely since effective cell-matrix adhesions could not occur. These trends could also be explained by increased resistance to MMP remodeling with increasing silk fibroin concentration. These silk fibroin hydrogels exhibit some susceptibility to MMP degradation (MMP2, MMP3), particularly at the “crosslinking” β-sheet crystalization sites,39 but it is likely that MMPs are less effective at cleaving (non-mammalian) silk fibroin relative to collagen I. These differences in MMP-susceptibility should become less pronounced over longer durations as secreted MMPs have more time to degrade the matrix.
Malignant carcinomas arise from tightly connected epithelial tissues, and can exhibit a diverse spectrum of individual and collective migration phenotypes.59 An epithelial-mesenchymal transition (EMT) has been associated with weakened cell-cell junctions and enhanced motility, enabling cancer cell detachment and dissemination.60 For these experiments, the highly metastatic breast adenocarcinoma cells (MDA-MB-231) represents an extreme mesenchymal phenotype with minimal cell-cell junctions, which rarely reverts to a more epithelial phenotype. Nevertheless, other epithelial cell lines that form cell-cell junctions could exhibit unexpected behaviors in these hydrogels. For instance, epithelial cells adhered as multi-cellular clusters in non-adherent confinement in vitro do not exhibit amoeboid migration, suggesting that EMT is a prerequisite for cells of epithelial origin to further transition to an amoeboid migration phenotype.13 This crosstalk between cell-cell and cell-matrix signaling in tumor invasion also represents an important question for future studies.
Finally, this overlay geometry represents a facile assay for 3D invasion that is implemented using standardized multiwell plates. These experiments have been analyzed at endpoint measurements with a primary readout of upward invasion distance, and can be supplemented with high-content measurements of cell morphology or biomarker expression. In principle, this approach could be conducted with automatic robotic dispensing as a high-throughput screen, going up to 384 well plates. Alternatively, with appropriate fluorescent labeling, this experimental system is compatible with longitudinal tracking through live-cell imaging and 3D traction force microscopy. This geometry was designed for ease of imaging using inverted confocal microscopy, since cells remain relatively localized within ~100 μm of the bottom of the plate. Nevertheless, one issue that may arise at longer times is that cells may become difficult to image if they migrate too far upwards into the hydrogel where optical adsorption and scattering dominate. Moreover, the multiwell plate represents a very rigid substrate relative to the hydrogel, which may mechanically bias cells to remain adherent on the surface. Future work could reduce this mechanical mismatch by coating the well with a soft, optically transparent substrate (e.g. polyacrylamide), which would then be overlaid with hydrogel (e.g. collagen I), as demonstrated elsewhere. 61
5. Conclusion
In this article, we demonstrate a mechanically-tunable composite silk fibroin and collagen I hydrogel that can be used to elucidate phenotypic plasticity in breast cancer migration. We show that the mechanical stiffness can be systematically varied with silk concentration at constant collagen concentration from GI = 30 Pa up to 300 Pa. We show that cancer cell invasion in an overlay assay geometry exhibits a biphasic dependence on silk concentration, with maximum invasion observed at intermediate stiffness with GI ≈ 100 Pa. Invasion is reduced at higher silk fibroin concentrations, due to steric hindrance with smaller mesh sizes. We further show that that cells can be biased towards mesenchymal migration by treatment with exogeneous EGF, which increases cell contractility and enhances cell-matrix adhesions. Instead, cells can be biased towards amoeboid migration by treatment with Taxol, which disrupts cytoskeletal function by stabilizing microtubules. Overall, we envision that this platform will enable higher-throughput preclinical screens of cancer cell invasion against panels of targeted inhibitors and engineered 3D matrix.
Supplementary Material
Acknowledgement
We thank A.M. Hruska for careful reading of the manuscript and A. Tripathi for access to the rheometer. We acknowledge funding from the National Institutes of Health (R21CA212932, T32ES007272, P30GM110759), the U.S. Department of Education (P200A150037), and Brown University (Start-Up Funds, Karen T Romer Undergraduate Research and Teaching Award).
Footnotes
6 Supporting Information
Strain and frequency sweeps from shear rheology, elastic modulus and stress relaxation from unconfined compression testing, collagen I fiber detection workflow, collagen I mesh size estimates, scanning electron micrographs of silk-collagen hydrogels, representative images and analysis of silk-collagen hydrogels prepared at 0°, distance invaded into silk-collagen hydrogels prepared at 0°, live/dead staining and cell viability, percent and distance invaded in collagen I only hydrogels, quantification of amoeboid and mesenchymal morphology, analysis of nuclear morphology, morphological analysis in collagen I only hydrogels.
References
- (1).Even-Ram S; Yamada KM Cell migration in 3D matrix. Curr Opin Cell Biol 2005, 17, 524–532, DOI: 10.1016/j.ceb.2005.08.015. [DOI] [PubMed] [Google Scholar]
- (2).Leggett SE; Khoo AS; Wong IY Multicellular tumor invasion and plasticity in biomimetic materials. Biomater. Sci. 2017, 5, 1460–1479, DOI: 10.1039/C7BM00272F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Parsons JT; Horwitz AR; Schwartz MA Cell adhesion: integrating cytoskeletal dynamics and cellular tension. Nat Rev Mol Cell Biol 2010, 11, 633–643, DOI: 10.1038/nrm2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Wolf K; Friedl P Extracellular matrix determinants of proteolytic and non-proteolytic cell migration. Trends Cell Biol 2011, 21, 736–744, DOI: 10.1016/j.tcb.2011.09.006. [DOI] [PubMed] [Google Scholar]
- (5).Sahai E; Marshall CJ Differing modes of tumour cell invasion have distinct requirements for Rho/ROCK signalling and extracellular proteolysis. Nat Cell Biol 2003, 5, 711–719, DOI: 10.1038/ncb1019. [DOI] [PubMed] [Google Scholar]
- (6).Vial E; Sahai E; Marshall CJ ERK-MAPK signaling coordinately regulates activity of Rac1 and RhoA for tumor cell motility. Cancer Cell 2003, 4, 67–79, DOI: 10.1016/S1535-6108(03)00162-4. [DOI] [PubMed] [Google Scholar]
- (7).Wyckoff JB; Pinner SE; Gschmeissner S; Condeelis JS; Sahai E ROCK-and Myosin-Dependent Matrix Deformation Enables Protease-Independent Tumor-Cell Invasion In Vivo. Curr Biol 2006, 16, 1515–1523, DOI: 10.1016/j.cub.2006.05.065. [DOI] [PubMed] [Google Scholar]
- (8).Sanz-Moreno V; Gadea G; Ahn J; Paterson H; Marra P; Pinner S; Sahai E; Marshall CJ Rac activation and inactivation control plasticity of tumor cell movement. Cell 2008, 135, 510–523, DOI: 10.1016/j.cell.2008.09.043. [DOI] [PubMed] [Google Scholar]
- (9).Lämmermann T; Bader BL; Monkley SJ; Worbs T; Wedlich-Söldner R; Hirsch K; Keller M; Förster R; Critchley DR; Fässler R; Sixt M Rapid leukocyte migration by integrin-independent flowing and squeezing. Nature 2008, 453, 51–55, DOI: 10.1038/nature06887. [DOI] [PubMed] [Google Scholar]
- (10).Renkawitz J; Schumann K; Weber M; Lämmermann T; Pflicke H; Piel M; Polleux J; Spatz JP; Sixt M Adaptive force transmission in amoeboid cell migration. Nat Cell Biol 2009, 11, 1438–1443, DOI: 10.1038/ncb1992. [DOI] [PubMed] [Google Scholar]
- (11).Toyjanova J; Flores-Cortez E; Reichner JS; Franck C Matrix confinement plays a pivotal role in regulating neutrophil-generated tractions, speed, and integrin utilization. J Biol Chem 2015, 290, 3752–3763, DOI: 10.1074/jbc.M114.619643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Ruprecht V; Wieser S; Callan-Jones A; Smutny M; Morita H; Sako K; Barone V; Ritsch-Marte M; Sixt M; Voituriez R; Heisenberg C-P Cortical contractility triggers a stochastic switch to fast amoeboid cell motility. Cell 2015, 160, 673–685, DOI: 10.1016/j.cell.2015.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Liu Y-J; Le Berre M; Lautenschlaeger F; Maiuri P; Callan-Jones A; Heuzé M; Takaki T; Voituriez R; Piel M Confinement and low adhesion induce fast amoeboid migration of slow mesenchymal cells. Cell 2015, 160, 659–672, DOI: 10.1016/j.cell.2015.01.007. [DOI] [PubMed] [Google Scholar]
- (14).Wolf K; Mazo I; Leung H; Engelke K; von Andrian UH; Deryugina EI; Strongin AY; Bröcker E-B; Friedl P Compensation mechanism in tumor cell migration: mesenchymal-amoeboid transition after blocking of pericellular proteolysis. J Cell Biol 2003, 160, 267–277, DOI: 10.1083/jcb.200209006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Wolf K; Te Lindert M; Krause M; Alexander S; Te Riet J; Willis AL; Hoff-man RM; Figdor CG; Weiss SJ; Friedl P Physical limits of cell migration: control by ECM space and nuclear deformation and tuning by proteolysis and traction force. J Cell Biol 2013, 201, 1069–1084, DOI: 10.1083/jcb.201210152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Suh J; Kim D-H; Surh Y-J Resveratrol suppresses migration, invasion and stemness of human breast cancer cells by interfering with tumor-stromal cross-talk. Arch. Biochem. Biophys. 2018, 643, 62–71, DOI: 10.1016/j.abb.2018.02.011. [DOI] [PubMed] [Google Scholar]
- (17).Paluch EK; Aspalter IM; Sixt M Focal Adhesion–Independent Cell Migration. Annu Rev Cell Dev Biol 2016, 32, 469–490, DOI: 10.1146/annurev-cellbio-111315-125341. [DOI] [PubMed] [Google Scholar]
- (18).Hapach LA; VanderBurgh JA; Miller JP; Reinhart-King CA Manipulation of in vitro collagen matrix architecture for scaffolds of improved physiological relevance. Phys. Biol. 2015, 12, 1–13, DOI: 10.1088/1478-3975/12/6/061002. [DOI] [PubMed] [Google Scholar]
- (19).Sapudom J; Pompe T Biomimetic tumor microenvironments based on collagen matrices. Biomater. Sci. 2018, 6, 2009–2024, DOI: 10.1039/c8bm00303c. [DOI] [PubMed] [Google Scholar]
- (20).Paszek MJ; Zahir N; Johnson KR; Lakins JN; Rozenberg GI; Gefen A; Reinhart-King CA; Margulies SS; Dembo M; Boettiger D; Hammer DA; Weaver VM Tensional homeostasis and the malignant phenotype. Cancer Cell 2005, 8, 241–254, DOI: 10.1016/j.ccr.2005.08.010. [DOI] [PubMed] [Google Scholar]
- (21).Provenzano PP; Inman DR; Eliceiri KW; Keely PJ Matrix density-induced mechanoregulation of breast cell phenotype, signaling and gene expression through a FAK-ERK linkage. Oncogene 2009, 28, 4326–4343, DOI: 10.1038/onc.2009.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Carey SP; Martin KE; Reinhart-King CA Three-dimensional collagen matrix induces a mechanosensitive invasive epithelial phenotype. Sci Rep 2017, 7, 42088, DOI: 10.1038/srep42088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Plou J; Juste-Lanas Y; Olivares V; Del Amo C; Borau C; García-Aznar JM From individual to collective 3D cancer dissemination: roles of collagen concentration and TGF-β. Sci Rep 2018, 8, 12723, DOI: 10.1038/s41598-018-30683-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Casson J; Davies OG; Smith C-A; Dalby MJ; Berry CC Mesenchymal stem cell-derived extracellular vesicles may promote breast cancer cell dormancy. J Tissue Eng 2018, 9, 2041731418810093, DOI: 10.1177/2041731418810093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Suh YJ; Hall MS; Huang YL; Moon SY; Song W; Ma M; Bonassar LJ; Segall JE; Wu M Glycation of collagen matrices promotes breast tumor cell invasion. Integr Biol 2019, DOI: 10.1093/intbio/zyz011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Yang Y.-l.; Motte S; Kaufman LJ Pore size variable type I collagen gels and their interaction with glioma cells. Biomaterials 2010, 31, 5678–5688, DOI: 10.1016/j.biomaterials.2010.03.039. [DOI] [PubMed] [Google Scholar]
- (27).Carey SP; Kraning-Rush CM; Williams RM; Reinhart-King CA Biophysical control of invasive tumor cell behavior by extracellular matrix microarchitecture. Biomaterials 2012, 33, 4157–4165, DOI: 10.1016/j.biomaterials.2012.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Guzman A; Ziperstein MJ; Kaufman LJ The effect of fibrillar matrix architecture on tumor cell invasion of physically challenging environments. Biomaterials 2014, 35, 6954–6963, DOI: 10.1016/j.biomaterials.2014.04.086. [DOI] [PubMed] [Google Scholar]
- (29).Fraley SI; Wu P-H; He L; Feng Y; Krisnamurthy R; Longmore GD; Wirtz D Three-dimensional matrix fiber alignment modulates cell migration and MT1-MMP utility by spatially and temporally directing protrusions. Sci Rep 2015, 5, 14580, DOI: 10.1038/srep14580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Ahmadzadeh H; Webster MR; Behera R; Jimenez Valencia AM; Wirtz D; Weeraratna AT; Shenoy VB Modeling the two-way feedback between contractility and matrix realignment reveals a nonlinear mode of cancer cell invasion. Proc. Natl. Acad. Sci. U.S.A. 2017, 114, E1617–E1626, DOI: 10.1073/pnas.1617037114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Zaman MH; Trapani LM; Sieminski AL; Siemeski A; Mackellar D; Gong H; Kamm RD; Wells A; Lauffenburger DA; Matsudaira P Migration of tumor cells in 3D matrices is governed by matrix stiffness along with cell-matrix adhesion and proteolysis. Proc. Natl. Acad. Sci. U.S.A. 2006, 103, 10889–10894, DOI: 10.1073/pnas.0604460103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Gill BJ; Gibbons DL; Roudsari LC; Saik JE; Rizvi ZH; Roy-bal JD; Kurie JM; West JL A Synthetic Matrix with Independently Tunable Biochemistry and Mechanical Properties to Study Epithelial Morphogenesis and EMT in a Lung Adenocarcinoma Model. Cancer Res 2012, 72, 6013–6023, DOI: 10.1158/0008-5472.CAN-12-0895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Ehrbar M; Sala A; Lienemann P; Ranga A; Mosiewicz K; Bittermann A; Rizzi SC; Weber FE; Lutolf MP Elucidating the role of matrix stiffness in 3D cell migration and remodeling. Biophys J 2011, 100, 284–293, DOI: 10.1016/j.bpj.2010.11.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Suri S; Schmidt CE Photopatterned collagen–hyaluronic acid interpene-trating polymer network hydrogels. Acta Biomater 2009, 5, 2385–2397, DOI: 10.1016/j.actbio.2009.05.004. [DOI] [PubMed] [Google Scholar]
- (35).Ulrich TA; Jain A; Tanner K; MacKay JL; Kumar S Probing cellular mechanobiology in three-dimensional culture with collagen–agarose matrices. Biomaterials 2010, 31, 1875–1884, DOI: 10.1016/j.biomaterials.2009.10.047. [DOI] [PubMed] [Google Scholar]
- (36).Sun J; Xiao W; Tang Y; Li K; Fan H Biomimetic interpenetrating polymer network hydrogels based on methacrylated alginate and collagen for 3D preosteoblast spreading and osteogenic differentiation. Soft Matter 2012, 8, 2398–7, DOI: 10.1039/c2sm06869a. [DOI] [Google Scholar]
- (37).Chaudhuri O; Koshy ST; Branco da Cunha C; Shin J-W; Verbeke CS; Alli-son KH; Mooney DJ Extracellular matrix stiffness and composition jointly regulate the induction of malignant phenotypes in mammary epithelium. Nature Mat 2014, 13, 970–978, DOI: 10.1038/nmat4009. [DOI] [PubMed] [Google Scholar]
- (38).Branco da Cunha C; Klumpers DD; Li WA; Koshy ST; Weaver JC; Chaud-huri O; Granja PL; Mooney DJ Influence of the stiffness of three-dimensional alginate/collagen-I interpenetrating networks on fibroblast biology. Biomaterials 2014, 35, 8927–8936, DOI: 10.1016/j.biomaterials.2014.06.047. [DOI] [PubMed] [Google Scholar]
- (39).Brown J; Lu C-L; Coburn J; Kaplan DL Impact of silk biomaterial structure on proteolysis. Acta Biomater 2015, 11, 212–221, DOI: 10.1016/j.actbio.2014.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Hu X; Cebe P; Weiss AS; Omenetto F; Kaplan DL Protein-based composite materials. Mater Today 2012, 15, 208–215, DOI: 10.1016/S1369-7021(12)70091-3. [DOI] [Google Scholar]
- (41).Wang X; Kluge JA; Leisk GG; Kaplan DL Sonication-induced gela-tion of silk fibroin for cell encapsulation. Biomaterials 2008, 29, 1054–1064, DOI: 10.1016/j.biomaterials.2007.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Hu X; Lu Q; Sun L; Cebe P; Wang X; Zhang X; Kaplan DL Biomaterials from ultrasonication-induced silk fibroinhyaluronic acid hydrogels. Biomacromolecules 2010, 11, 3178–3188, DOI: 10.1021/bm1010504. [DOI] [PubMed] [Google Scholar]
- (43).Wang X; Zhang X; Sun L; Subramanian B; Maffini MV; Soto A; Sonnen-schein C; Kaplan DL Preadipocytes Stimulate Ductal Morphogenesis and Func-tional Differentiation of Human Mammary Epithelial Cells on 3D Silk Scaffolds. Tissue Eng Part A 2009, 15, 3087–3098, DOI: 10.1089/ten.tea.2008.0670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Wang X; Sun L; Maffini MV; Soto A; Sonnenschein C; Kaplan DL A complex 3D human tissue culture system based on mammary stromal cells and silk scaffolds for modeling breast morphogenesis and function. Biomaterials 2010, 31, 3920–3929, DOI: 10.1016/j.biomaterials.2010.01.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Buitrago JO; Patel KD; El-Fiqi A; Lee J-H; Kundu B; Lee H-H; Kim H-W Silk fibroin/collagen protein hybrid cell-encapsulating hydrogels with tunable gelation and improved physical and biological properties. Acta biomater 2018, 69, 218–233, DOI: 10.1016/j.actbio.2017.12.026. [DOI] [PubMed] [Google Scholar]
- (46).Rockwood DN; Preda RC; Yücel T; Wang X; Lovett ML; Kaplan DL Materials fabrication from Bombyx mori silk fibroin. Nat Protoc 2011, 6, 1612, DOI: 10.1038/nprot.2011.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Raub CB; Suresh V; Krasieva T; Lyubovitsky J; Mih JD; Putnam AJ; Tromberg BJ; George SC Noninvasive assessment of collagen gel microstructure and mechanics using multiphoton microscopy. Biophys J 2007, 92, 2212–2222, DOI: 10.1529/biophysj.106.097998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Zhao X; Huebsch N; Mooney DJ; Suo Z Stress-relaxation behavior in gels with ionic and covalent crosslinks. J. Appl. Phys. 2010, 107, 63509, DOI: 10.1063/1.3343265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Bredfeldt JS; Liu Y; Pehlke CA; Conklin MW; Szulczewski JM; In-man DR; Keely PJ; Nowak RD; Mackie TR; Eliceiri KW Computational segmentation of collagen fibers from second-harmonic generation images of breast cancer. J Biomed Opt 2014, 19, 016007, DOI: 10.1117/1.JBO.19.1.016007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Geum DT; Kim BJ; Chang AE; Hall MS; Wu M Epidermal growth factor promotes a mesenchymal over an amoeboid motility of MDA-MB-231 cells embedded within a 3D collagen matrix. Eur. Phys. J. Plus 2016, 131, 8–10, DOI: 10.1140/epjp/i2016-16008-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Yu D; Liu B; Jing T; Sun D; Price JE; Singletary SE; Ibrahim N; Hortobagyi GN; Hung M-C Overexpression of both p185 c-erbB2 and p170 mdr-1 renders breast cancer cells highly resistant to Taxol. Oncogene 1998, 16, 2087, DOI: 10.1038/sj.onc.1201729. [DOI] [PubMed] [Google Scholar]
- (52).Chaudhuri O; Gu L; Klumpers D; Darnell M; Bencherif SA; Weaver JC; Huebsch N; Lee H.-p.; Lippens E; Duda GN; Mooney DJ Hydrogels with tunable stress relaxation regulate stem cell fate and activity. Nature Mat 2016, 15, 326–334, DOI: 10.1038/nmat4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Nam S; Hu KH; Butte MJ; Chaudhuri O Strain-enhanced stress relaxation impacts nonlinear elasticity in collagen gels. Proc Natl Acad Sci USA 2016, 201523906– 6, DOI: 10.1073/pnas.1523906113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Williams RM; Zipfel WR; Webb WW Interpreting second-harmonic generation images of collagen I fibrils. Biophys J 2005, 88, 1377–1386, DOI: 10.1529/biophysj.104.047308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Chen X; Nadiarynkh O; Plotnikov S; Campagnola PJ Second harmonic generation microscopy for quantitative analysis of collagen fibrillar structure. Nat Protoc 2012, 7, 654–669, DOI: 10.1038/nprot.2012.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Rice WL; Firdous S; Gupta S; Hunter M; Foo CW; Wang Y; Kim HJ; Ka-plan DL; Georgakoudi I Non-invasive characterization of structure and morphology of silk fibroin biomaterials using non-linear microscopy. Biomaterials 2008, 29, 2015–2024, DOI: 10.1016/j.biomaterials.2007.12.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Liu Y; Keikhosravi A; Mehta GS; Drifka CR; Eliceiri KW In Fibrosis: Methods and Protocols; Rittié L, Ed.; Springer; New York: New York, NY, 2017; pp 429–451, DOI: 10.1007/978-1-4939-7113-8_28. [DOI] [Google Scholar]
- (58).Horwitz SB Taxol (paclitaxel): mechanisms of action. Ann Oncol 1994, 5, S3–6. [PubMed] [Google Scholar]
- (59).Friedl P; Locker J; Sahai E; Segall JE Classifying collective cancer cell invasion. Nat Cell Biol 2012, 14, 777–783, DOI: 10.1038/ncb2548. [DOI] [PubMed] [Google Scholar]
- (60).Wong IY; Javaid S; Wong EA; Perk S; Haber DA; Toner M; Irimia D Collective and individual migration following the epithelial-mesenchymal transition. Nat Mater 2014, 13, 1063–1071, DOI: 10.1038/nmat4062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Arevalo RC; Kumar P; Urbach JS; Blair DL Stress Heterogeneities in Sheared Type-I Collagen Networks Revealed by Boundary Stress Microscopy. PLOS ONE 2015, 10, e0118021–12, DOI: 10.1371/journal.pone.0118021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


