Abstract
Aim:
Long noncoding RNAs (lncRNAs) have been reported to influence multiple gene regulatory processes. Technological advances in RNA-seq platforms allow detection of low-abundance RNA species such as lncRNAs. This study examined the relationship between expression of lncRNAs and their putative partner mRNAs.
Methods:
We analyzed total RNA-seq data from mouse macrophages under various inflammatory and intervention conditions.
Results:
The macrophage expression of lncRNAs is strongly regulated by an inflammatory stimulus. Moreover, the expression of a majority of lncRNAs was correlated or anti-correlated with the partner mRNA(s), across the different treatment conditions. This relationship was maintained even in cells from a distinct genotype.
Conclusion:
These results suggest a previously unappreciated tight coupling of lncRNA and mRNA expression during macrophage responses to various microenvironmental perturbations.
Keywords: : bioinformatics, computational biology, computational epigenomics, functional genomics, gene regulation
Long noncoding RNAs (lncRNAs) have been implicated in diverse aspects of the gene regulatory hierarchy, including transcription, epigenetic modification, splicing, mRNA stability and translation [1]. Although progress has been made in our understanding of these mechanisms, the functional significance and prevalence of the reported mechanisms are unclear, because the data are largely based on a relatively small number of specific RNAs. While focused studies are necessary to understand the mechanisms of lncRNA function, genome-scale analyses will help uncover the relative prevalence and overall impact of individual modes of lncRNA action.
Immune cells are often the context for the investigation of global gene regulation, partly because the cellular constituents of the mammalian immune system must reprogram their transcriptome rapidly in response to danger signals such as invading pathogens or tissue injury. Macrophages are early effector cells of the innate immune system. They help maintain tissue homeostasis by clearing endogenous debris and preventing their toxic accumulation. When macrophages encounter pathogens, they recognize the shared molecular patterns in microbial products and launch immediate inflammatory responses. The rapid inflammatory responses are mediated by global chromatin and transcriptional changes induced by pathogen-derived stimuli [2,3].
Here we used recent total RNA-seq data [4] on bone marrow-derived macrophages (BMDMs) stimulated with lipopolysaccharide (LPS) with or without dexamethasone (Dex) to analyze the expression patterns of lncRNAs in relation to their putative partner mRNAs. This study was motivated by increasingly accurate annotation of lncRNA databases and consequently expanded repertoire of lncRNAs and inquiring whether the standard total RNA-seq may be used to reveal useful ‘lncRNA-seq’ data without custom enrichment for lncRNAs. Our analysis revealed an unexpectedly strong correlation or anti-correlation between their expression levels of lncRNAs and putative mRNA targets across perturbation conditions.
Methods
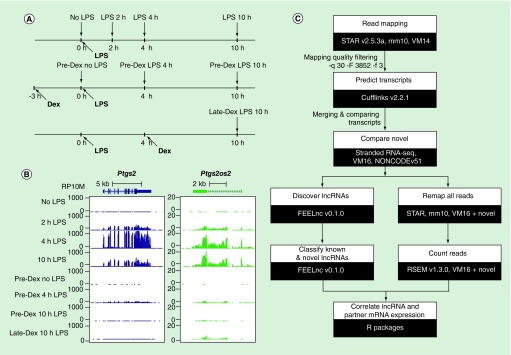
The full experimental design of the samples is available in our original study [4]. Experimental procedures with mice were carried out according to NIH guidelines and were approved by the NIAID Animal Care and Use Committee (NIH, MD, USA). Briefly, mouse BMDMs were stimulated with LPS for 2, 4 or 10 h or left unstimulated. Additional samples were pre- or late-treated with Dex, a synthetic ligand of the glucocorticoid receptor (Figure 1A & B, see [4] for details). All samples were in duplicates (16 samples in total). RNA sequencing reads were downloaded from the GEO database (GSE93739) and mapped to the mouse reference mm10 with the STAR v2.5.3a [5]. The full analysis pipeline is presented in Figure 1C. New transcripts were identified with the Cufflinks v2.2.1 software [6–8]. We applied stringent filtering criteria for the new lncRNA transcripts to exclude potential artifacts: length >200 bp; >1 exon; intron chain confirmed by another LPS-stimulated BMDM-derived stranded RNA-seq dataset (Oh KS et al., unpublished data). We also filtered out transcripts identified on unplaced scaffolds. The novel transcripts were then compared with the newest Gencode gene annotation available (VM16) [9,10] and a noncoding RNA gene database, NONCODEv5 [11] to identify previously annotated transcripts. The reads were then remapped using combined gene annotations (VM16, NONCODEv5 confirmed by our data, and newly identified lncRNA transcripts). Gene expression was calculated with the RSEM v1.3.0 tool [12]. Genes with the length >200 bp and with expression ≥0.5 FPKM in at least two samples were subjected to a differential gene expression analysis with the edgeR v3.18.1 package [13] and R v3.4.2 [14]. We used contrasts to compare stimulated with unstimulated cells and Dex-treated with nontreated cells from matching LPS time points. A gene was considered as differentially expressed if the false discovery rate (FDR) <0.05 and |log2 fold-change| ≥1.
Figure 1. . Study design and analysis workflow.
(A) The experimental design of the cell culture used in the original study. The vertical black arrows indicate time points for RNA extractions followed by RNA sequencing. (B) A track view of an example of correlated mRNA and lncRNA genes from the University of California, Santa Cruz Genome Browser (http://genome.ucsc.edu).
(C) The analysis flow chart used for this study.
Dex: Dexamethasone; LPS: Lipopolysaccharide.
Novel, NONCODEv5 and other noncoding transcripts from VM16 were also subjected to an analysis with the FEELnc v0.1.0 software [15] to extract lncRNAs. For this analysis, the following gene types were considered as lncRNAs: 3′ overlapping ncRNA, processed transcript, sense intronic, sense overlapping, antisense RNA, lincRNA, macro lncRNA and bidirectional promoter lncRNA. From these, only 4246 transcripts with an annotation support level of 1 were used as training set (known as lncRNAs) and remaining 9841 were merged with the novel candidate lncRNAs. As potential partner mRNA genes, we only considered 28,885 protein coding transcripts with the annotation support level of 1. We used the default setting for FEELnc_filter.pl and FEELnc_codpot.pl modules. The coding potential score cutoff was measured by the random forest and was set to 0.4988. Candidate lncRNAs were then classified based on their position to neighboring partner mRNA genes (100 kb window) to genic or intergenic, and sense or antisense (see [15] for details). The lncRNA:mRNA pairs identified on a transcript level were then merged to obtain interactions on a gene level. All pairs found were subjected to further correlation analysis (the FEELnc ‘isBest’ score 0 and 1).
Pearson’s correlation between lncRNA:mRNA pairs was calculated with corAndPvalue function of the WGCNA v1.6.3 R package [16,17] using log2 (FPKM + 0.1) gene expression values of all 16 samples. Significance cutoff was set to p-value <0.05. Gene set enrichment analysis was performed using Panther v13.1 tool [18,19] and the ClusterProfiler v3.6.0 R v3.4.3 package [20] using all mouse genes dataset (Gencode VM16) as a reference set.
To determine predictive values of the lncRNA expression, linear models were built using the 797 lncRNA:mRNA pairs based on the expression of the 16 samples (GSE93739) [4]. The models were then used to predict the expression of mRNAs based on its partner lncRNA expression level in a genetically distinct dataset of eight samples (GSE93602) [2]. Pearson’s correlation was used to assess the similarity between predicted and observed values in the second dataset.
Results
Identification & classification of lncRNAs
To classify any novel and known lncRNAs expressed in BMDMs, we first assembled transcripts for each of the 16 RNA-seq samples, from a transcriptional profiling study of BMDMs treated with LPS for 2, 4 or 10 h, with or without Dex pre- or late-treatment (Figure 1A, see the ‘Methods’ section for details). In total, we detected 5433 potential transcripts (coding and noncoding) and performed a series of filtering with stringent criteria to include any novel lncRNAs in our analysis (see the ‘Methods’ section; Figure 1 and Supplementary Figure 1A). We identified 24 novel lncRNA transcripts which were not previously annotated in either the NONCODE v5 [11] or Gencode (VM16) [9,10] databases. From the merged set of novel and known lncRNAs (total 9841 transcripts), 7190 passed the FEELnc filtering step; 6689 were considered as candidate lncRNAs after coding potential estimate and merged with the 4246 known lncRNAs for the subsequent pairing with mRNAs. For 2755 transcripts, FEELnc reported no putative partner mRNAs. We then merged the 16,568 putative pairs identified on the transcript level to obtain 7977 pairs on a gene level. The majority of the pairs were classified as intergenic with 2178 sense and 2755 antisense pairs. Among the genic pairs, 255 were sense and 2789 were antisense. From these, only 1421 lncRNA:mRNA gene pairs (1010 unique lncRNA and 1297 unique mRNA genes) were considered for subsequent analyses after filtering based on expression levels (see the ‘Methods’ section). Overall, the lncRNA gene expression was highly reproducible with replicate correlation coefficients ranging from 0.90 to 0.95 (Supplementary Figure 1B).
lncRNA expression is regulated by a pro-inflammatory stimulus in macrophages
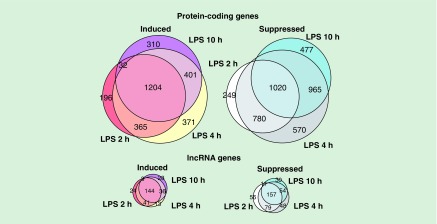
We determined protein-coding and lncRNA genes differentially expressed between LPS-stimulated and unstimulated BMDMs. Although lncRNAs are generally expressed at lower levels than protein-coding genes (mean log2 (FPKM + 0.1) of -0.37 in lncRNAs versus 2.66 in mRNAs in our data, Wilcoxon’s test p-value = 2.2e-16, Supplementary Figure 2), the lncRNAs showed similar patterns as protein-coding genes in terms of the number of LPS-regulated genes with a large overlap of LPS-regulated genes among different time points (Figure 2). For example, the proportions of genes upregulated at 4 h (68%) and at 10 h (81%) among all LPS-induced genes were not significantly different (p-value >0.05) between protein-coding and lncRNA genes (73% and 81%, respectively). Among the LPS-suppressed genes, the proportions of genes downregulated at 10 h after LPS stimulation were similar between protein-coding and lncRNAs (61% vs 66%). This result suggests that lncRNAs exhibit expression kinetic patterns resembling those of protein-coding genes, and may have an important role in a functional response to LPS.
Figure 2. . lncRNAs expression is tightly regulated by lipopolysaccharide stimulation in primary macrophages.
The Venn diagrams represent the numbers of significantly induced or suppressed protein-coding and lncRNA genes upon stimulation with LPS for 2, 4 or 10 h. The area of each circle is proportional to the number of constituents in the subset.
LPS: Lipopolysaccharide.
Expression of lncRNAs is correlated or anti-correlated with their partner mRNAs
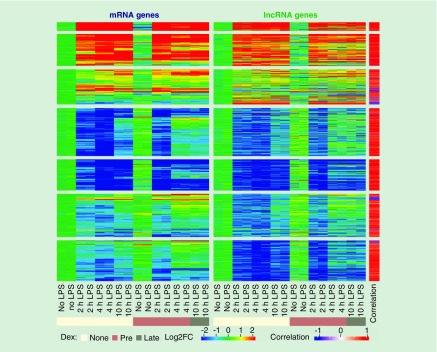
We next classified lncRNAs with FEELnc classifier (see the ‘Methods’ section for details) and identified 1421 lncRNA:mRNA pairs, from which 951 (67%) had significantly correlated expression levels (p-value <0.05) between the paired genes. Reasoning that longer stimulations can induce secondary effects, we focused our analysis on the early responsive 797 pairs that included at least one gene differentially expressed upon stimulation with LPS for 2 or 4 h (no Dex treatment) and were significantly correlated (Figure 3 & Supplementary Table 1). Among these 797, 226 were also responsive to Dex treatments. The majority of these (n = 750, 95%) were positively correlated and only 47 pairs showed an anti-correlation; the proportion of anti-correlations was slightly higher in intergenic lncRNAs (Supplementary Table 1 and Supplementary Figure 3). To assess whether the extent of such correlated behaviors from lncRNAs and putative partner mRNAs is distinct from that for proximally located mRNAs only, we calculated correlations of mRNA:mRNA pairs within 100 kb windows, as was applied for lncRNA:mRNA pairing (see the ‘Methods’ section). The absolute values of lncRNA:mRNA correlations were similar to or even slightly higher than those of mRNA:mRNA pairs (Supplementary Figure 4A & B). The proportion of significantly correlated pairs among the lncRNA:mRNA putative pairs was also slightly higher than for mRNA:mRNA pairs (67% vs. 64%, chi-squared test p = 0.017). Interestingly, the strength of correlation decayed rapidly for the mRNA pairs as a function of the genomic distance, while the correlation strength was rather independent of the distance for lncRNA:mRNA pairs (Supplementary Figure 4C). We verified that the correlation decay of mRNA:mRNA pairs remained the same after subsampling of the pairs to match the smaller numbers of lncRNA:mRNA pairs (data not shown). These results suggest that the correlated expression of lncRNA:mRNA cannot be explained solely by the proximity of the two loci.
Figure 3. . The heatmap showing the expression profiles of 797 correlated mRNA and lncRNA gene pairs.
The color in the main heatmap represents the log2 fold change relative to the mean of the two control samples from unstimulated cells, calculated based on (FPKM + 0.1) gene expression values. An additional bar on the right side of the heatmap shows the Pearson’s correlation coefficients between the expression of lncRNA:mRNA pairs.
LPS: Lipopolysaccharide.
Among the lncRNAs showing a strong correlation with its partner mRNA was a previously reported Ptgs2os2 (also known as lincRNA-Cox2). The mRNA partner of this gene, Ptsg2 is highly induced upon stimulation with LPS (Figures 1B & 4B). Ptgs2os2 has been also reported as a regulator of immune responses [21,22]. Moreover, our correlated pairs included 12 of 24 previously reported lncRNA:mRNA correlations in mouse BMDMs based on expression microarray analysis [23] (Supplementary Figure 5).
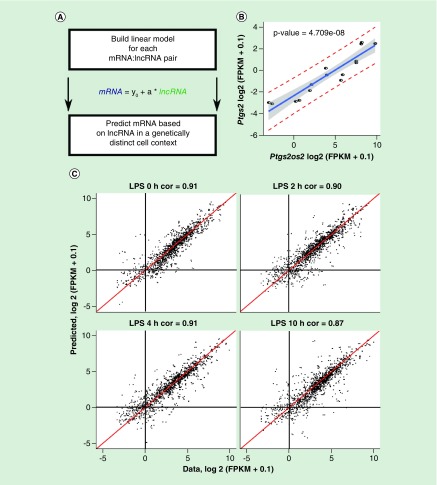
Figure 4. . The expression of lncRNAs is highly predictive of the partner mRNAs in a genetically distinct cell context across different conditions.
(A) Schematic overview of the workflow. (B) An example of lncRNA:mRNA expression modeling based on the initial dataset. (C) The correlation of predicted and observed mRNA expression values in the genetically distinct dataset (four time points in duplicates). Each dot represents a lncRNA:mRNA pair in each of the two biological replicates.
LPS: Lipopolysaccharide.
Correlated or anti-correlated lncRNA:mRNA pairs were significantly enriched for Gene Ontology (GO) Biological Process terms related to catabolic process and apoptosis regulation (Supplementary Figure 6), and apoptosis signaling Panther Pathway (Fisher's exact test corrected for FDR = 2.34e-4). The lack of enrichment in immune-related pathways among all mRNAs from significant pairs may reflect that these genes are involved in more systemic processes. For example, some genes under the GO categories ‘apoptotic signaling’ or ‘catabolic processes’ may still be relevant for certain immune mechanisms. Indeed, among the GO categories were immune-related genes such as Tnf, Bcl2a1s, Bcl2a1b and Relb.
lncRNA expression allows prediction of the partner mRNA abundance
The observed quantitative relationship between the lncRNAs and partner mRNAs was tight enough to be captured with linear models for all the 797 correlated pairs (Figure 4A). Next, we asked whether the individual pair-specific linear model can be used to predict the abundance of mRNA based on the expression of its partner lncRNA in a new dataset. To address this, the linear models with the estimated parameters were used to predict the expression of mRNAs based on their partner lncRNA expression levels in LPS-stimulated BMDMs from a genetically distinct context, in other words, Ikaros knock-out mice (GSE93602) [2] (Figure 4B). The Ikaros knock-out macrophages showed globally altered transcriptional response to LPS, with substantial proportions of LPS-responsive genes affected either positively or negatively. Despite such a massive difference in the transcriptional responses between the original and knock-out macrophages, the lncRNA expression was predictive of the partner mRNA expression in the knock-out RNA-seq data, based on the wild-type (WT)-derived linear models. The predicted and observed mRNA expression values were highly correlated at each of the LPS stimulation time point, indicating a high predictive value of lncRNA expression (Figure 4C).
Discussion
We took advantage of a total RNA-seq dataset generated from a number of treatment conditions and the latest annotation information about lncRNAs, and found a tight relationship between the expression of lncRNAs and their putative partner mRNAs. This relationship could be represented by a log linear model. Although there are previous reports relating lncRNA:mRNA expression, genome-wide analysis in the context of immune responses has been so far only based on microarray data [23], which does not capture novel lncRNA genes. The pairwise relationship shown here was validated in a new cellular context where the transcriptome is dramatically altered due to the lack of a transcriptional regulator.
The majority of the lncRNA:mRNA pairs showed correlated expression patterns, with only a small number of lncRNAs, preferentially antisense than sense, whose expression were anti-correlated to their putative partner mRNAs. Our study describes the genome-wide relationship between lncRNAs and partner mRNAs and suggests potential modes of lncRNA action, e.g. enhancement versus inhibition, in terms of their orientation or location with respect to each other.
Conclusion
The lncRNA expression is tightly coupled with their partner mRNA expression during macrophage responses to various microenvironmental perturbations. This observation was supported in a genetically different cell context and was not affected by the genomic distance between the genes paired.
Future perspective
These findings raise questions for future investigations: it has to be determined for each lncRNA:mRNA pair, if the two RNA species are invariantly co-expressed across different tissues, whether the lncRNA regulates the expression of its partner mRNA or if both are co-regulated by a third factor. Our list of lncRNA:mRNA relationships represents a useful resource for functional characterization of specific mRNA:lncRNA pairs which may fine-tune macrophage inflammatory responses. The mechanisms that govern lncRNA:mRNA relationships will likely vary for individual pairs in a given cell type. With improvements in methods to perturb lncRNA expression, future studies will be able to uncover more insights about functional significance of lncRNAs.
Summary points.
Long noncoding RNA (lncRNA) genes, similar to protein-coding genes, show transcriptional reprogramming during inflammatory responses in innate immune effector cells.
The expression of lncRNAs and their neighboring partner mRNAs are correlated.
lncRNA:mRNA correlation seems independent of the distance between the two loci.
lncRNA:mRNA relationship is preserved in cells with dramatically altered transcriptome.
lncRNA expression can be used to predict the expression of their partner mRNA.
Supplementary Material
Acknowledgments
This work utilized the computational resources of the NIH high-performance computing including the Biowulf cluster (http://hpc.nih.gov). The authors thank K-S Oh, E Martin and members of the Sung laboratory and the Myriam Gorospe Laboratory for helpful discussions and critical reading of the manuscript.
Footnotes
Authors’ contributions
MH Sung and A Pacholewska conceived the study. A Pacholewska implemented the computational methods. MH Sung supervised the study. A Pacholewska and MH Sung wrote the paper.
Financial & competing interests disclosure
This work was supported by the Intramural Research Program of the NIH at National Institute on Aging. NIH Grant number: AG000390. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Ethical conduct of research
The authors state that they have obtained appropriate institutional review board approval or have followed the principles outlined in the Declaration of Helsinki for all animal experimental investigations.
References
Papers of special note have been highlighted as: • of interest
- 1.Elling R, Chan J, Fitzgerald KA. Emerging role of long noncoding RNAs as regulators of innate immune cell development and inflammatory gene expression. Eur. J. Immunol. 46(3), 504–512 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Oh K-S, Gottschalk RA, Lounsbury NW. et al. Dual roles for ikaros in regulation of macrophage chromatin state and inflammatory gene expression. J. Immunol. 201(2), 757–771 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]; • Reports data used here for assessing lncRNAs as predictors of their partner mRNAs.
- 3.Tong AJ, Liu X, Thomas BJ. et al. A stringent systems approach uncovers gene-specific mechanisms regulating inflammation. Cell 165(1), 165–179 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oh KS, Patel H, Gottschalk RA. et al. Anti-inflammatory chromatinscape suggests alternative mechanisms of glucocorticoid receptor action. Immunity 47(2), 298–309.e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]; • Original study that describes RNA-seq samples used in this study.
- 5.Dobin A, Davis CA, Schlesinger F. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29(1), 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Trapnell C, Roberts A, Goff L. et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7(3), 562–78 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roberts A, Pimentel H, Trapnell C, Pachter L. Identification of novel transcripts in annotated genomes using RNA-Seq. Bioinformatics 27(17), 2325–2329 (2011). [DOI] [PubMed] [Google Scholar]
- 8.Trapnell C, Williams B, Pertea G. et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28(5), 511–515 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Derrien T, Johnson R, Bussotti G. et al. The GENCODE v7 catalogue of human long non-coding RNAs: analysis of their structure, evolution and expression. Genome Res. 22, 1775–1789 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mudge JM, Harrow J. Creating reference gene annotation for the mouse C57BL6/J genome assembly. Mamm. Genome 26(9–10), 366–378 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fang S, Zhang L, Guo J. et al. NONCODEV5: a comprehensive annotation database for long non-coding RNAs. Nucleic Acids Res. 46(D1), D308–D314 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12(1), 1–16 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26(1), 139–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.R Core Team. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: (2017). http://www.r-project.org/ [Google Scholar]
- 15.Wucher V, Legeai F, Hédan B. et al. FEELnc: a tool for long non-coding RNA annotation and its application to the dog transcriptome. Nucleic Acids Res. 45(8), e57 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]; • Describes the bioinformatics tool used for identifying lncRNA:mRNA pairs.
- 16.Langfelder P, Horvath S. Fast R functions for robust correlations and hierarchical clustering. J. Stat. Softw. 46(11), pii: i11 (2012). [PMC free article] [PubMed] [Google Scholar]
- 17.Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9, 559 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mi H, Muruganujan A, Casagrande JT, Thomas PD. Large-scale gene function analysis with the PANTHER classification system. Nat. Protoc. 8(8), 1551–66 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mi H, Poudel S, Muruganujan A, Casagrande JT, Thomas PD. PANTHER version 10: expanded protein families and functions, and analysis tools. Nucleic Acids Res. 44(D1), D336–D342 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu G, Wang L-G, Han Y, He Q-Y. clusterProfiler: an R package for comparing biological themes among gene clusters. Omi. A J. Integr. Biol. 16(5), 284–7 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guttman M, Amit I, Garber M. et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 458(7235), 223–227 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carpenter S, Aiello D, Atianand MK. et al. A long noncoding RNA mediates both activation and repression of immune response genes. Science 341(6147), 789–792 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mao A-P, Shen J, Zuo Z. Expression and regulation of long noncoding RNAs in TLR4 signaling in mouse macrophages. BMC Genomics 16(1), 45 (2015). http://www.biomedcentral.com/1471-2164/16/45 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.