Abstract
Radial glia cells both generate neurons and physically guide nascent neurons to their target destination in the cortex, and as such they are essential for CNS development. It has been proposed that in the developing cerebellum, neuronal contact induces radial glia formation, however, the mechanisms involved in this process are not well understood. Here we demonstrate that neuronal induction of radial glia formation is the result of sequential signaling through Notch1 and erbB receptors. First, Notch1 activation by neuronal contact induces the glial expression of the brain lipid binding protein (BLBP) and erbB2 genes. Interestingly, two different signaling pathways mediate these effects of Notch1 on transcription, BLBP expression being dependent on Su(H), whereas erbB2 is regulated by a yet unidentified Notch1 pathway. The subsequent increase in erbB2 receptor expression makes the glia more responsive to neuronal NRG, which then induces the morphological transformation into radial glia. Thus, these results unveil some of the mechanisms underlying radial glia formation.
Keywords: Notch1, Su(H), erbB, cerebellum, glia, BLBP
Introduction
Cell migration is a major step in the development of the vertebrate CNS. Newborn neurons, which are generated in the germinal layers of the neural tube, must move to their final destination in the cortex and other areas of the CNS, where they establish appropriate synaptic connections. These cell movements are critical for normal brain development and function. Radial glia constitute a population of specialized cells that play critical roles in this neuronal migration. Radial glia are present from early stages of neural tube formation, and they possess thin processes that, in most cases, span the entire thickness of the developing cerebral wall. Early studies indicate that radial glia fibers act as scaffolds to guide migrating newborn neurons from their birthplaces to their final destinations in the developing cortex (Rakic and Sidman, 1970; Hatten and Mason, 1990). More recently, studies have suggested that radial glia in the cerebral cortex also function as neuronal precursors (Alvarez-Buylla et al., 1990; Malatesta et al., 2000; Noctor et al., 2001). Thus, radial glia contribute to the major processes in brain formation. After neurogenesis and neuronal migration are completed, most radial glia cells disappear, either by differentiating into astrocytes or by undergoing cell death (for review, see Chanas-Sacre et al., 2000). The mechanisms underlying radial glia formation, function, and final disappearance remain poorly understood.
Studies on cerebellar development indicate that contact between neurons and glia is a critical event for the formation of radial glia. Using cocultures of cerebellar granule cells and astroglia, it has been shown that these neurons induce astrocytes to undergo a morphological, functional, and molecular differentiation into radial glia. For example, granule cell contact induces cerebellar astrocytes to extend a process along which neurons can migrate (Hatten, 1985). Moreover, neuronal contact also induces expression of a radial glia gene, brain lipid binding protein (BLBP) (Feng and Heintz, 1995). These results suggested that cell–cell contact might be a key event in the initial formation of radial glia in the developing brain and that this in vitro system can help in the identification of the factors regulating genesis of radial glia.
Using cerebellar cells, we previously demonstrated that neuron–glia contact induces the morphological differentiation of radial glia via neuregulin (NRG)–erbB receptor signaling, and that erbB receptor signaling in radial glia is necessary for neuronal migration (Rio et al., 1997). Similar results were obtained with cells from the cerebral cortex (Anton et al., 1997). However, the observation that radial glia are present in mice lacking erbB2 receptors, but their morphology is defective (Anton et al., 1997) also suggested that this pathway is necessary for late events in radial glia differentiation, and that other signaling pathways may act at earlier stages of radial glia formation.
The Notch receptors and their ligands, molecules best known for influencing cell fate decisions through direct cell–cell contact (Nye and Kopan, 1995; Weinmaster, 1997), are strong candidates to regulate early events in radial glia formation. For example, Gaiano et al. (2000) reported that activation of Notch1 signaling in cells of the embryonic neuroepithelium induces radial glia formation. However, the mechanisms by which Notch1 activation may regulate radial glia formation remain unclear.
The most widely accepted model for Notch signaling proposes that after ligand binding to Notch, the receptor undergoes proteolytic cleavages within its extracellular and intramembrane sequences, releasing the Notch intracellular domain (NICD) from the membrane. A group of proteins collectively known as the CSL proteins, which include Suppressor of Hairless [Su(H)] in Drosophila and its homologous genes in Xenopus [X-Su(H)], mammals (RBP-J/CBF1/KBF2) and Caenorhabditis elegans (LAG-1) are important mediators of Notch signaling. These proteins bind DNA and act as transcriptional repressors, but through direct interaction with the cytoplasmic domain of Notch they are converted into transcriptional activators. Accordingly, Notch is a clear example of a transmembrane receptor that functions not only in ligand binding but also as a signal transducer that directly activates gene transcription.
To investigate the roles of Notch1 signaling in the induction of radial glia formation, we used the well characterized in vitro system of cerebellar astrocytes and granule neuron cocultures. Here we report that contact with cerebellar granule neurons activates Notch1 signaling in cerebellar astroglia and that activation of Notch1 is sufficient to induce the morphological differentiation of radial glia and the expression of the radial glia gene BLBP. Analysis of the interactions between Notch1 and erbB receptor signaling showed that Notch1 signaling is upstream of erbB signaling and that Notch1-induced radial glia formation is mediated by its effects on erbB receptor expression and function. Our results show that radial glia differentiation requires sequential activation of Notch1 and erbB receptor signaling and that the Notch1 effects on radial glia differentiation require signaling through two intracellular pathways, one that requires CSL and one that does not.
Materials and Methods
Histology and immunostaining. Long–Evans postnatal day 6 (P6) rat pups (Charles River Laboratories, Wilmington, MA) were fixed by intracardial perfusion with 4% paraformaldehyde in PBS, and the cerebellum was dissected, cryoprotected, and cut in 20 μm parasagittal sections. Tissue sections were blocked with 3% BSA and 0.1% Triton X-100 in PBS for 1 hr at room temperature, followed by incubation with mouse anti-glial fibrillary acidic protein (anti-GFAP; Boehringer Mannheim, Indianapolis, IN) and an affinity-purified rabbit anti-Notch1 polyclonal antibody (Ab 93–4, 1:100) (Shawber et al., 1996b) diluted in blocking solution at 4°C overnight. Sections were washed with PBS, and detection was performed using the appropriate fluorescent secondary antibodies (Jackson ImmunoResearch), for 1 hr at room temperature.
Cells in culture were fixed in 4% paraformaldehyde for 10 min, rinsed with PBS, blocked as above, and then labeled with a rabbit anti-GFAP (Dako, Carpinteria, CA) and/or mouse monoclonal anti-erbB4 antibody (NeoMarkers, Fremont, CA). Nuclei were stained with Hoechst 33342 (Molecular Probes, Eugene, OR) or SYBR Green (Molecular Probes).
Cell culture. Primary astroglial and neuronal cells were purified from Long–Evans P6 rat cerebella (Charles River) by selective preplating. Cerebella were excised, and the meninges were removed in Dulbecco's PBS (Invitrogen, Gaithersburg, MD), under a dissecting microscope. The tissue was cut into small pieces and incubated in 1% trypsin (Sigma, St. Louis, MO) and 0.02% deoxyribonuclease (DNase; Worthington, Free-hold, NJ) in PBS for 10 min at 37°C. After centrifugation, the pellet was triturated using successively decreasing bore size fire-polished glass Pasteur pipettes in astrocyte media (DMEM with high glucose), 10% fetal bovine serum (FBS), and 2 mm l-glutamine (Invitrogen) with 0.02% DNase. Cell were pelleted, resuspended in fresh medium, plated onto two 100 mm tissue culture dishes, and incubated at 37°C for 20 min to allow fibroblasts to attach to the plate. Then, the unattached cells (neurons and glia) were removed from the dishes and plated onto one poly-d-lysine (PDL; 0.5 μg/ml; BD Biosciences) -coated dish. After 1 hr incubation at 37°C, unattached cells (primarily neurons) were removed, and the dish was washed gently with PBS to remove any loosely attached cells. The remaining attached cells, primarily glia, were incubated in astrocyte media. The supernatant from the first PDL-coated dish was plated onto a second PDL-coated dish, and the steps described above were repeated to generate another dish of glial cells. The washout of the second PDL dish was used as the source of purified granule cells. The neurons were centrifuged and resuspended in astrocyte media with 1% N2 supplement (Invitrogen). Glia were passaged 3–4 d later, a step that removed any remaining neurons, producing a glial culture >95% pure. All cells were incubated at 37°C with 5% CO2. Parental fibroblasts (SRα) and Jagged1-expressing (SN3T) (Lindsell et al., 1995) cells were grown in astrocyte media on PDL-coated dishes.
Plasmids. FCDN1 and OCDN1 cDNAs were cloned into mammalian expression vector pEF1a-BOS (Mizushima and Nagata, 1990), as described in Shawber et al. (1996b). The 1.7 kb BLBP promoter (Feng and Heintz, 1995), the 4.5 kb erbB2 promoter (White and Hung, 1992), and the CBF1-dependent reporter (Hsieh et al., 1996) were subcloned into a pGL3basic Luciferase reporter vector (Promega, Madison, WI). DN-erbB4 was cloned into pcDNA3, as described in Rio et al. (1997). Xenopus DN-Su(H) was cloned into pCS2+, as described in Wettstein et al. (1997).
Recombinant NRG. Recombinant epidermal growth factor (EGF)-like domain of rat NDFβ1 (NDFβ1[177–246]) was used in all experiments (Wen et al., 1994).
Transfections. Primary astrocytes at 70% confluence were transfected with Fugene 6 reagent (Boehringer Mannheim), according to the recommended protocol. To detect the transfected cells, cells were cotransfected with a plasmid encoding green fluorescent protein (GFP) at a molar ratio of 3:1 (plasmid:GFP plasmid). The level of coexpression of the genes of interest and GFP was assessed by immunostaining with antibodies against erbB4 (NeoMarkers) or Notch1 (Ab 93–4, 1:100) (Shawber et al., 1996b). In these cases we found that between 96 ± 2% and 98 ± 2% of GFP-positive cells also expressed the genes of interest (FCDN1 and erbB4, respectively). These results indicate that our protocol resulted in very high levels of coexpression.
Luciferase assay. Luciferase assays were performed 2 d after transfection using the Dual Assay Luciferase kit (Promega). Cotransfected TK–renilla luciferase was used to normalize samples for transfection efficiency and sample handling. Cells were lysed, and Luciferase activity was measured following the recommended protocol.
Astroglial morphology assay. Glia were plated at 10,000 cells/200 mm 2, and freshly dissociated neurons were added at a ratio of 10:1. The radial glia morphology was determined as described in Rio et al. (1997) with minor modifications. If morphology was to be determined after transfection or treatment with NRG or neurons, glial morphology was assayed 24 hr after treatment. If the experiment required neuron–glia cocultures of transfected cells, neurons were added to the glia 24 hr after transfection, and morphology was analyzed 24 hr later. Cells were fixed with 4% paraformaldehyde and labeled with a rabbit anti-GFAP antibody. Nuclei were stained with Hoechst 33342 (Molecular Probes) or SYBR Green (Molecular Probes). An erbB4 monoclonal antibody was used when cells were transfected with DN–erbB4. Cells were then incubated with appropriate secondary antibodies, washed, and mounted. The morphology of cells in random fields was assessed looking only at the GFAP staining. Only then, the transfected cells were identified based on GFP or DN–erbB4 expression. Glia were considered to be radial when they had at least one thin process longer than 50 μm. This criterion was based on previous studies (Hatten, 1985; Rio et al., 1997). Each assay was repeated two to five times, and 125–225 cells were examined for each assay.
For glia–fibroblast coculture experiments, P6 cerebellar astroglia were plated at 30,000 cells/200 mm 2 in astrocyte media. Twenty-four hours later, control (SRα) or Jagged1-expressing (SN3T) fibroblasts were added at a ratio of 1.6:1. The cocultures were then incubated at 37°C for 24 or 48 hr. Cells were fixed with 4% paraformaldehyde and labeled with a rabbit anti-GFAP antibody. Nuclei were stained with Hoechst 33342 (Molecular Probes). Only glia in contact with fibroblasts were analyzed for radial morphology, as described above. Assays were repeated two or three times, and at least 120 cells were examined for each assay.
Quantitative Western blot analysis. Primary astrocytes grown to 70% confluency were transfected with FCDN1 or the control plasmid (PBOS). Two days later cells were lysed in radioimmunoprecipitation assay buffer, and samples (6 μg of protein of each) were separated by SDS-PAGE on 4–20% gels (Invitrogen), transferred to nitrocellulose membranes (Bio-Rad, Hercules, CA), and probed with a rabbit polyclonal anti-erbB2 C terminus antibody (Santa Cruz Biotechnology, Santa Cruz, CA; sc-284) or a rabbit polyclonal anti-BLBP antibody (a gift of N. Heintz) followed by an HRP-conjugated anti-rabbit antibody (Jackson ImmunoResearch). Blots were developed using Western Lightning chemiluminescence reagent (PerkinElmer Life Sciences, Emeryville, CA), and images were captured with the Fujifilm (Fuji, Tokyo, Japan) Intelligent Dark Box II LAS-1000 plus. Quantitation of band intensities was performed using IP Lab Gel H software. After the erbB2 and BLBP images were obtained, the membranes were reprobed with anti-actin (Oncogene Sciences, Uniondale, NY) or anti-GAPDH antibodies (Chemicon, Temecula, CA). The intensity of the actin or GAPDH bands was used for normalization of the erbB2 and BLBP signals. Both normalizers gave identical results.
Statistical analysis. Statistical significance for the astroglial morphological assays was determined by χ 2 using the Statview program. Statistical significance for the glia–fibroblast morphological assays was determined by Student's t test. Statistical significance for the Luciferase assays was determined by Wilcoxon Signed Rank test. Statistical significance for the quantitative Westerns was determined by paired t test.
Results
Notch1 and Jagged1 expression in the developing cerebellum
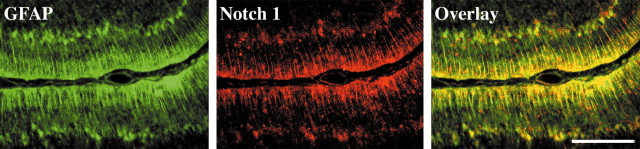
To determine the possible roles of Notch1 in postnatal cerebellar development and the migration of cerebellar granule cells along Bergmann radial glia, we studied the pattern of expression of Notch1 and its ligand, Jagged1. At P6, a time of maximal granule cell migration, Notch1 immunoreactivity was concentrated in Bergmann glia fibers, colocalizing with immunostaining for GFAP (Fig. 1). This is in agreement with in situ hybridization studies showing that Notch1 mRNA appears to be localized to Bergmann glia (Irvin et al., 2001). Northern blot analysis of granule cells and astrocytes purified from P6 cerebellum confirmed that Notch1 mRNA is most strongly expressed by cerebellar glia (data not shown) and that Jagged1 mRNA is primarily expressed by cerebellar granule cells, as shown by Solecki et al. (2001). Because granule cells and Bergmann glia are in contact during migration, these results suggested that Jagged1-expressing migrating granule cells might activate Notch1 receptors within glia.
Figure 1.
Notch1 is expressed by Bergmann glia fibers in the developing cerebellum. Parasagittal sections through the P6 rat cerebellum stained with antibodies against GFAP (green) and Notch1 (red). The overlay image demonstrates colocalization of GFAP and Notch1 staining in Bergmann radial glia. Scale bar, 250 μm.
Granule neurons activate Notch signaling in cerebellar glia
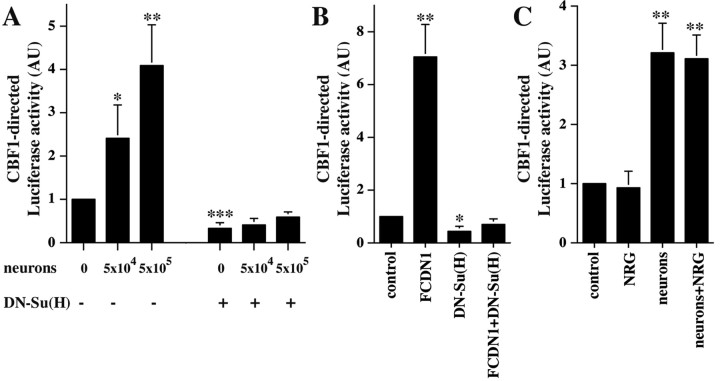
To determine if neuronal contact activates Notch1 signaling in cerebellar astrocytes, we measured the effects of neuron–glia contact on the Notch-dependent transcriptional activation of the CSL reporter construct. We also tested if these effects are blocked by a dominant-negative Suppressor of Hairless [DN-Su(H)], a mutant Su(H) that interacts with Notch but cannot bind DNA (Wettstein et al.,1997). Cerebellar astroglia were transfected with a plasmid encoding 4 tandem arrayed CBF1 binding sites upstream of the Firefly Luciferase gene (Hsieh et al., 1996) alone or together with a DN-Su(H) expression plasmid. Twenty-four hours later, granule cells were isolated, dissociated, added to the cultures, and incubation was continued for an additional 24 hr, after which Luciferase activity was measured (Fig. 2A). The level of Luciferase activity was dramatically increased by neuronal contact in a dose-dependent manner, suggesting that neuronal contact leads to activation of Notch1 receptors in glia. These effects were completely blocked by expression of DN-Su(H) in the astroglia (Fig. 2A). Interestingly, DN-Su(H) expression by itself reduced the basal level of CBF1 transcription, suggesting that a basal level of Notch signaling occurs in astroglia in culture. To further asses the effectiveness of DN-Su(H) in glial cells, we tested its ability to block activation the CBF1 reporter through a constitutively activated form of Notch1 (FCDN1) (Nofziger et al., 1999). FCDN1 expression in astrocytes significantly induced CBF1-mediated transcription, and this was completely abolished by DN-Su(H) (Fig. 2B).
Figure 2.
Contact with granule cells activates Notch signaling in cerebellar glia. A, Purified P6 cerebellar glia were cotransfected with plasmids encoding the CBF1-Luciferase reporter and with either DN-Su(H) or a control plasmid. Twenty-four hours later, purified P6 cerebellar granule cells (5 × 104 or 5 × 105) were added to the culture. The next day cells were lysed, and Luciferase activity was measured. The results represent the mean ± SEM of three independent experiments. Asterisks mark which conditions are significantly different from the control. *p < 0.05; **p < 0.005; ***p < 0.0005. AU, Arbitrary units. B, Purified P6 cerebellar glia were cotransfected with plasmids encoding the CBF1-Luciferase reporter and FCDN1, DN-Su(H), or a control plasmid, or both FCDN1 and DN-Su(H) together. Two days later, cells were lysed, and Luciferase activity was measured. The results represent the mean ± SD of two independent experiments. Asterisks mark which conditions are significantly different from the control. *p < 0.003; **p < 0.0005. C, Purified P6 cerebellar glia were transfected with a plasmid encoding the CBF1-Luciferase reporter. Twenty-four hours later, purified P6 cerebellar granule cells or NRG (1 nm) were added to the culture. The next day cells were lysed, and Luciferase activity was measured. The results represent the mean ± SD of two independent experiments. Asterisks mark which conditions are significantly different from the control, p < 0.005.
To study further the specificity of the CBF1 reporter activation, we tested if NRG-erbB receptor signaling, a signaling pathway shown to mediate granule cell–glia interactions (Rio et al., 1997), could affect CBF1 reporter activity. Treatment of transfected glia with NRG did not result in activation of the CBF1 reporter construct, and when tested in neuron–glia cocultures it did not alter the neuronal-induced Notch1 signaling in astroglial cells (Fig. 2C). These results show that neuronal contact activates Notch signaling in glia and that this leads to transcriptional activation.
Notch signaling induces radial glia formation
It has been well documented that contact with granule cells induces cerebellar astroglia in culture to adopt radial glia morphology (Hatten, 1985). The finding that Notch signaling in glia is activated by neuronal contact suggested that this signaling pathway might participate in the neuronal induction of radial glia formation. Therefore, we tested if activation of Notch1 signaling in glia is sufficient to induce a radial glia phenotype. Expression of FCDN1 in cerebellar astroglia resulted in a significant increase in the percentage of radial glia cells, but this effect was not as robust as that induced by neuronal contact (Fig. 3A). Importantly, the morphological change induced by FCDN1 expression was indistinguishable from that elicited by either neuronal contact or NRG treatment (Fig. 3B).
Figure 3.
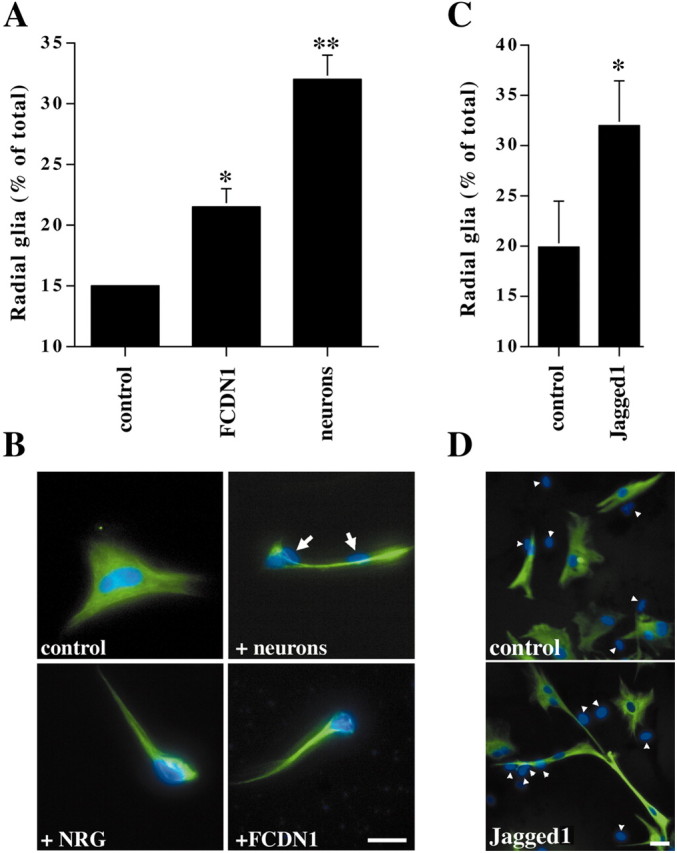
Activated Notch1 and Jagged1 induce radial formation. A, Purified P6 cerebellar glia were transfected with a plasmid encoding either FCDN1 or a control plasmid. Twenty-four hours later, purified P6 cerebellar granule cells (1 × 105) were added, and the next day glial morphology was examined. Radial glia were identified as GFAP(+) cells with at least one thin process longer than 50 μm. The results represent the mean ± SEM of four independent experiments. Asterisks mark which conditions are significantly different from the control. *p < 0.005; **p < 0.0001. B, Images show representative glia under control conditions, with neurons (white arrows indicate neuronal nuclei), with NRG (1 nm), or transfected with a plasmid encoding FCDN1. Glia were stained with antibodies against GFAP (green), and nuclei were stained with Hoechst (blue). The morphology of the radial glia is similar under all stimuli. Scale bar, 20 μm. C, Jagged1-expressing or parental fibroblasts were added to cultures of purified P6 cerebellar glia. Twenty-four hours later, radial glia morphology was examined. The results represent the mean ± SEM of three independent experiments; p = 0.008. D, Images show representative glia cocultured with parental fibroblasts or Jagged1-expressing fibroblasts (white arrows indicate fibroblast nuclei). Scale bar, 20 μm. Glia were stained with antibodies against GFAP (green), and nuclei were stained with Hoechst (blue).
Because Northern blot analysis indicated that cerebellar granule cells express Jagged1, we tested if this ligand by itself could induce radial glia formation. Cerebellar astroglia were cocultured either with Jagged1-expressing or parental fibroblasts, and their morphology was quantified. Contact with Jagged1-expressing cells induced morphological changes consistent with radial glia, suggesting that Jagged1-induced Notch signaling is sufficient to induce radial glia formation (Fig. 3C,D). There were no significant differences between the levels of radial glia morphology resulting from expression of FCDN1 and contact with Jagged1-expressing cells (p = 0.466).
Both Notch and erbB receptor signaling are necessary for neuronal-induced radial glia formation
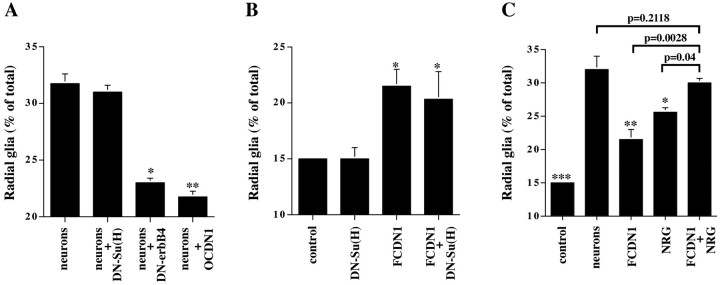
To determine if Notch1 signaling is necessary for the neuronal induction of radial glia morphology, we used two published Notch1 signaling antagonists; dominant-negative forms of Notch1 (OCDN1) and Suppressor of Hairless (DN-Su(H)) (Wettstein et al., 1997; Nofziger et al., 1999; Redmond et al., 2000). We tested the ability of these reagents to block neuronal induction of radial glia formation, and we compared their effects to that of a dominant-negative erbB receptor (DN-erbB4), which inhibits NRG activation of the erbB2, erbB3, and erbB4 receptors (Prevot et al., 2003) and blocks neuronal induction of radial glia formation (Rio et al., 1997). Cerebellar astroglia were transfected with plasmids encoding one of the dominant-negative molecules or a control plasmid. After 24 hr, purified granule neurons were added to the transfected glia, which were examined the next day for morphological changes indicative of radial glial differentiation (Fig. 4A). Both OCDN1 and DN-erbB4 blocked to a similar extent the ability of neurons to induce radial glia formation, while not affecting the morphology of the unstimulated glia (Fig. 5). In contrast, DN-Su(H) did not block neuron-induced radial glia formation. In a similar way, expression of DN-Su(H) did not block the induction of radial glia morphology by FCDN1 expression (Fig. 4B) or contact with Jagged1-expressing cells (data not shown). A number of studies have suggested that not all of the biological effects of Notch signaling require Su(H)/CBF1 activation (Shawber et al., 1996a,b; Nofziger et al., 1999; Yamamoto et al., 2001; Endo et al., 2002). Therefore, these results indicate that induction of radial glia morphology by Notch1 occurs through an undefined pathway that functions independently of CSL activation.
Figure 4.
Notch and erbB receptor signaling are both necessary for neuronal induction of radial glia. A, Purified P6 cerebellar glia were transfected with plasmids encoding DN-Su(H), DN-erbB4, OCDN1, or a control plasmid. Twenty-four hours later, purified P6 granule neurons were added, and morphology was examined the next day. The results represent the mean ± SEM of four independent experiments. Asterisks mark which conditions are significantly different from the neuronal treatment. *p < 0.004; **p < 0.0007. B, Purified P6 cerebellar glia were transfected with plasmids encoding DN-Su(H), FCDN1, or a control plasmid. Two days later morphology was examined. The results represent the mean ± SEM of three independent experiments. Asterisks mark which conditions are significantly different from the control. *p < 0.004. C, Purified P6 cerebellar glia were transfected with plasmids encoding either FCDN1 or a control plasmid. Twenty-four hours later, purified P6 granule neurons or NRG (1 nm) was added, and morphology was examined the next day. The results represent the mean ± SEM of three independent experiments. Asterisks mark which conditions are significantly different from the neuronal treatment. *p < 0.02; **p < 0.0005; ***p < 0.0001.
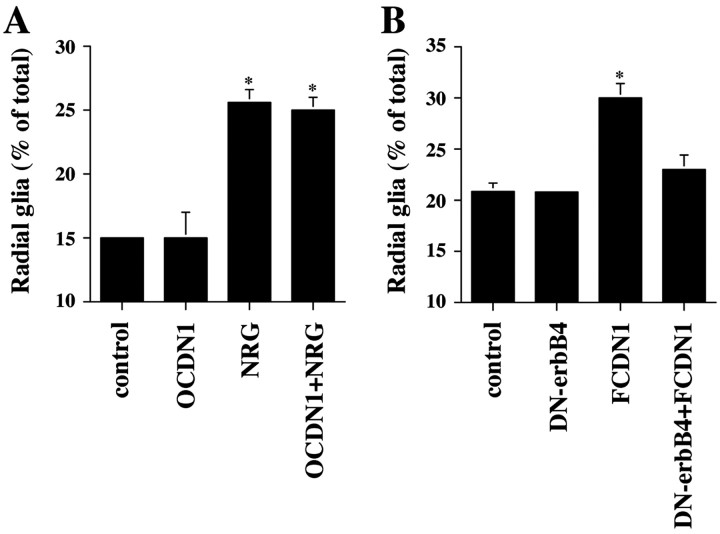
Figure 5.
Notch1 and erbB act sequentially to induce radial glia formation. A, Purified P6 cerebellar glia were transfected with either a plasmid encoding OCDN1 or a control plasmid. Twenty-four hours later, NRG (1 nm) was added to the culture, and morphology was examined the next day. The results represent the mean ± SEM of three independent experiments. Asterisks mark which conditions are significantly different from the control; p < 0.0003. B, Purified P6 cerebellar glia were transfected with plasmids encoding DN-erbB4, or FCDN1, or both DN-erbB4 and FCDN1 together, or a control plasmid. Twenty-four hours later, glial morphology was examined. The results represent the mean ± SEM of three independent experiments. The asterisk marks which condition is significantly different from the control; p < 0.008.
Independent activation of erbB or Notch1 signaling is capable of inducing radial glia formation, however to a lesser extent than that elicited by neuronal contact. Because neurons express ligands for both of these receptors, we tested if simultaneous activation of both pathways is similar to the effects of neurons. Cerebellar astroglia were transfected with a control plasmid and 24 hr later treated with neurons or NRG or transfected with the plasmid encoding FCDN1 and either left untreated or treated with NRG. While independent activation of the each of these pathways, Notch1 or erbB, induced radial glia formation to a lesser extent than that induced by neuronal contact, simultaneous activation of both pathways resulted in a robust response that was indistinguishable from the effects of neurons (Fig. 4C). In similar manner, stimulation with soluble NRG and coculture with Jagged1-expressing cells mimicked the effects of neurons (data not shown).
Notch and erbB receptors act sequentially to induce radial glia formation
The results presented above indicated that both Notch1 and erbB receptor signaling pathways are necessary for the induction of radial glia morphology by neuronal contact. Moreover, coactivation of both pathways simultaneously led to a greater level of radial glia formation than independent activation of each pathway. A likely explanation for these observations is that Notch and erbB function in the same cascade of events that occur subsequent to neuron–glia contact. For example, one pathway could be necessary for the effective activation of the second one, to direct radial glia formation. To test this idea and to define the order in which these signaling molecules are activated to direct glial differentiation, we determined if blockade of one pathway prevents induction of radial glia through the other. When cerebellar glia were transfected with the plasmid encoding OCDN1, the ability of NRG to induce radial glia formation was unaffected (Fig. 5A). In contrast, coexpression of DN-erbB4 blocked the ability of FCDN1 (Fig. 5B) and Jagged1 (data not shown) to induce radial glia formation. These results are consistent with the idea that the Notch1 signaling pathway is upstream of erbB receptor signaling during neuronal induction of radial glia formation.
Neurons activate erbB2 expression in cerebellar glia through Notch1 signaling
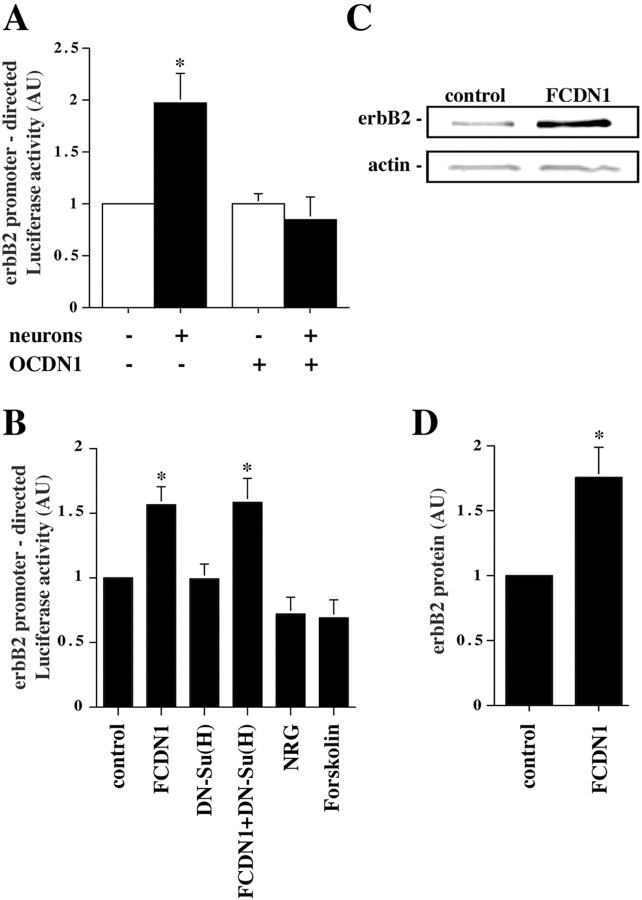
The epistasis observed between Notch1 and erbB signaling suggested that Notch1 signaling might regulate some aspects of erbB signaling. For example, it is possible that Notch1 signaling induces erbB receptor expression in the glia, which in turn makes the astroglia more responsive to NRG. To test this possibility, we determined whether granule cells induce the expression of erbB2 in the glia, and if the induction is dependent on Notch1 signaling. A plasmid encoding the 4.5 kb erbB2 upstream sequences (White and Hung, 1992), the only characterized erbB receptor promoter, was cloned upstream of the Luciferase gene and transfected into cerebellar astrocytes. The levels of Luciferase activity in glia alone or in glia after the addition of neurons indicated that neuronal contact induced a twofold increase in erbB2 promoter activity (Fig. 6A). Importantly, the effect of neurons on erbB2 promoter activity was completely abolished by cotransfection with OCDN1, indicating that the neuronal effects were mediated through ligand-induced Notch signaling. Similar to the induction of radial glia morphology, expression of FCDN1 was sufficient to induce erbB2 promoter activity, and this was not affected by expression of DN-Su(H) (Fig. 6B). These results indicate that Notch1 induces erbB2 expression through a pathway independent of Su(H). Moreover, these effects are specific because neither NRG nor forskolin have effects on erbB2 promoter activity (Fig. 6B).
Figure 6.
Notch1 signaling mediates the neuronal induction of erbB2 expression in glia. A, Purified P6 cerebellar glia were cotransfected with plasmids encoding the erbB2-Luciferase reporter and either OCDN1 or a control plasmid. Twenty-four hours later, purified P6 granule neurons were added to the culture, and the next day cells were lysed and Luciferase activity measured. The results represent the mean ± SEM of five independent experiments. The asterisk marks which condition is significantly different from the control; p < 0.028. B, Purified P6 cerebellar glia were cotransfected with plasmids encoding the erbB2-Luciferase reporter and FCDN1, DN-Su(H), or a control plasmid. Twenty-four hours later, NRG (1 nm) or forskolin (5 μm) was added to the culture. The next day cells were lysed, and Luciferase activity was measured. The results represent the mean ± SEM of at least three independent experiments. Asterisks mark which conditions are significantly different from the control; p < 0.04. C, Purified P6 cerebellar glia were transfected with either FCDN1 or a control plasmid, lysed 48 hr later, and equal amounts of protein were subjected to quantitative Western blot analysis with rabbit polyclonal anti-erbB2 and mouse monoclonal anti-actin antibodies for normalization. D, Intensities of the erbB2 immunoreactivity normalized to the actin control. The data represent the mean ± SEM of four independent experiments; p < 0.007.
To further characterize the effects of Notch1 activation on erbB2 expression, we tested if this signaling pathway leads to increases of endogenous glial erbB2 protein. Glia were transfected with FCDN1 or a control plasmid and incubated for 2 d. Then, cells were lysed and erbB2 protein levels examined by quantitative Western blot analysis. FCDN1 expression induced a 1.75 ± 0.23-fold (p < 0.007) increase of erbB2 protein in glia (Fig. 6C,D). Because the transfection efficiency for glia in these experiments was 30%, this most likely reflects a 3.5-fold increase of erbB2 protein in the transfected cells. In these cell extracts we also tested for the possibility that expression of FCDN1 leads to increases in erbB activation, using phospho-erbB2 and phospho-tyrosine antibodies. These experiments did not detect Notch1-induced changes in erbB2 phosphorylation (data not shown). However, because ligand induced-erbB receptor phosphorylation is transient, lasting <1 hr, it was expected that chronic Notch1 signaling would not result in significant long-term erbB2 phosphorylation.
Neuronal-induction of BLBP expression in glia is mediated by Notch activation
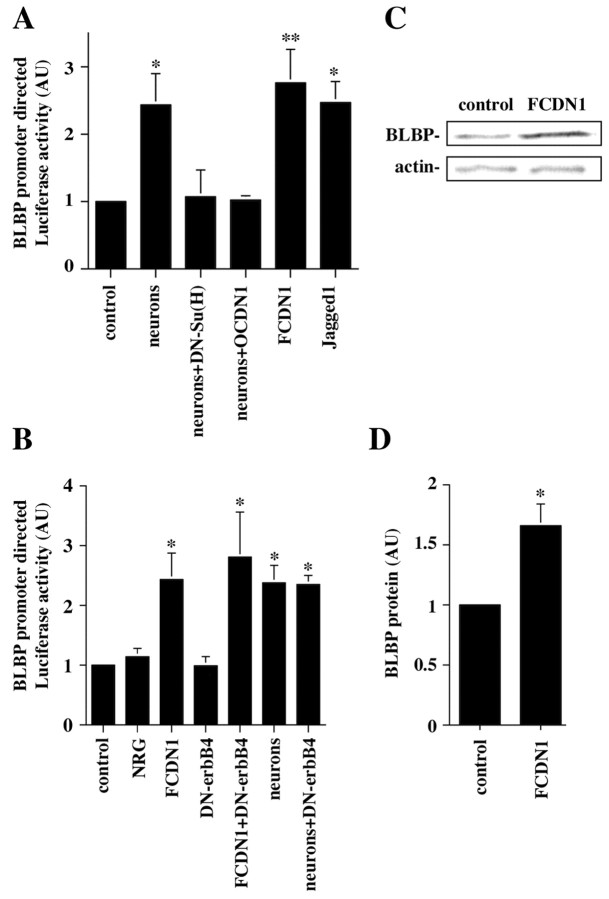
Previous studies have shown that neuronal contact upregulates the expression of BLBP, a gene expressed by radial glia that has been proposed to be important for neuronal migration (Feng et al., 1994; Feng and Heintz, 1995). We therefore examined roles of Notch1 signaling in the neuronal induction of BLBP. Cerebellar astroglia were cotransfected with a plasmid containing the 1.7 kb BLBP promoter (Feng and Heintz, 1995) upstream of the Luciferase gene and plasmids encoding either OCDN1, DN-Su(H), or a control plasmid. After 24 hr, granule neurons were added, and Luciferase activity was determined the following day. As previously shown, neurons induced BLBP promoter activity (Fig. 7A). Remarkably, blockade of Notch1 signaling, either by DN-Su(H) or OCDN1 completely abolished the ability of neurons to induce BLBP expression, indicating that Notch1 signaling through CSL activation is necessary for this. To determine if Notch1 activation is sufficient to induce BLBP transcription, glia were transfected with the FCDN1 expression plasmid or cocultured with Jagged1-expressing cells. Both treatments induced BLBP expression to the same extent as neuronal contact (Fig. 7A), showing that Notch1 signaling is sufficient for the induction of BLBP expression.
Figure 7.
Notch1 signaling mediates the neuronal induction of BLBP promoter transcription in glia. A, Purified P6 cerebellar glia were cotransfected with plasmids encoding the BLBP-Luciferase reporter and FCDN1, OCDN1, DN-Su(H), or a control plasmid. Twenty-four hours later, purified P6 granule neurons or Jagged1-expressing fibroblasts were added to the culture, and the next day cells were lysed and Luciferase activity was measured. The results represent the mean ± SEM of at least three independent experiments. Asterisks mark which conditions are significantly different from the control. *p < 0.028; **p < 0.018. B, Purified P6 cerebellar glia were cotransfected with plasmids encoding the BLBP-Luciferase reporter and FCDN1, DN-erbB4, or a control plasmid. Twenty-four hours later, purified P6 granule neurons or NRG (1 nm) was added to the culture. The next day cells were lysed, and Luciferase activity was measured. The results represent the mean ± SEM of three independent experiments. Asterisks mark which conditions are significantly different from the control. *p<0.04. C, Purified P6 cerebellar glia were transfected with either FCDN1 or a control plasmid, lysed 48 hr later, and equal amounts of protein were subjected to quantitative Western blot analysis with rabbit polyclonal anti-BLBP and mouse monoclonal anti-actin antibodies for normalization. D, Intensities of the BLBP immunoreactivity normalized to the actin control. The data represent the mean ± SEM of three independent experiments; p = 0.046.
Studies by Anton et al. (1997) showed that NRG increased the levels of BLBP immunostaining in cortical astrocytes in culture. We therefore tested the effects of NRG on BLBP expression in cerebellar astroglia. Surprisingly, NRG did not affect the activity of the BLBP reporter by itself (Fig. 7B). Moreover, blockade of erbB signaling by expression of DN-erbB4 did not affect the ability of FCDN1 or neurons to induce BLBP expression (Fig. 7B). These results indicate that Notch1 signaling, but not erbB signaling, mediates the neuronal induction of BLBP transcription.
To further characterize the effects of Notch1 activation on BLBP expression we tested if this signaling pathway leads to increases in endogenous glial BLBP protein. Glia were transfected with FCDN1 or a control plasmid and incubated for 2 d. Then, cells were lysed, and BLBP protein levels were examined by quantitative Western blot analysis. FCDN1 expression induced a 1.66 ± 0.18-fold (p = 0.046) increase of BLBP protein in glia (Fig. 7C,D). Because the transfection efficiency for glia in these experiments was 30%, this most likely reflects a 3.1-fold increase of BLBP protein in the transfected cells.
Discussion
Radial glia play critical roles in brain development, acting as neuronal precursors (Malatesta et al., 2000; Noctor et al., 2001), and guiding newborn neurons to their final destination in the cortex (Rakic and Sidman, 1970; Hatten and Mason, 1990). Radial glia are born early during embryonic development and, in many cases, transform into astrocytes at later stages (Culican et al., 1990). Because there is clear evidence that mature astrocytes can be induced to revert to the embryonic radial glia phenotype in vitro (Hatten, 1985, 1999; Hunter and Hatten, 1995) and in vivo (Leavitt et al., 1999), understanding the mechanisms that mediate this reversion may provide insights into the mechanisms of radial glia formation during development.
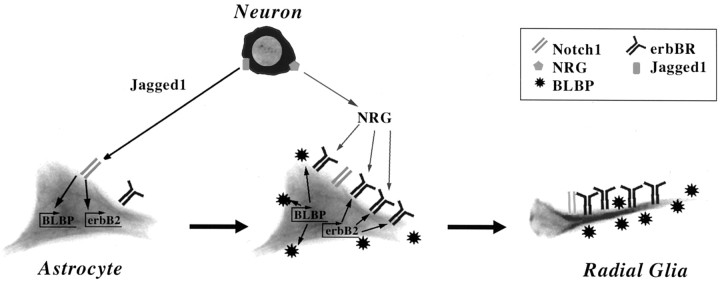
Our results show that cerebellar granule cells induce astrocytes to become radial glia by sequentially activating the Notch and the erbB receptor signaling pathways. Together, these signaling events promote the molecular, morphological, and functional differentiation of these cells (Fig. 8). Our data suggest a role for Notch1 signaling in radial glia formation that involves the transcriptional activation of radial glia-specific genes as well as erbB receptors. In contrast, the role of erbB receptor signaling appears to be the induction of morphological transformation. However, the possibility that erbB receptor signaling has effects on the expression of radial glia proteins, either by transcriptional or post-transcriptional mechanisms, remains open. Although our conclusions are based on results obtained with cerebellar cells, other observations (Anton et al., 1997; Gaiano et al., 2000) suggest that the same mechanism could be involved in the differentiation of cortical radial glia.
Figure 8.
A model for neuronal induction of radial glia by sequential signaling through Notch and erbB pathways. Initial contact by a Jagged1-expressing neuron activates astrocytic Notch receptors. Notch signaling then induces expression of BLBP and erbB2 in the glia. The increase in erbB receptor expression makes the astrocyte more responsive to neuron-derived NRG, which subsequently induces the glia to adopt a radial morphology and to support neuronal migration.
DN-erbB4 has also been demonstrated to act very specifically, blocking signaling through erbB2, erbB3, and erbB4 but not through the EGF receptor (erbB1) (Prevot et al., 2003). Therefore, the ability of DN-erbB4 to abolish the effects of Notch1 activation on glial morphology suggests that Notch1-induction of radial glia formation depends on erbB signaling. Moreover, the inability of DN-erbB4 to affect the Notch1-induced expression of BLBP shows that this step in radial glia differentiation does not require sequential signaling, that NRG-erbB signaling is not implicated in the regulation of BLBP expression, and indicates that increasing BLBP expression is not sufficient to induce radial glia morphology.
The finding that Notch1 activation is sufficient to induce radial glia formation, but that this requires erbB receptor signaling, suggests that Notch1 signaling, in some manner, regulates or induces erbB receptor signaling. This could be the result of a Notch1-induced increase in astrocyte expression of either erbB receptors, erbB receptor ligands, or both. Our results suggest that a Notch1-mediated induction of erbB receptor expression is part of this process. Whereas we only analyzed the effects of Notch1 signaling on erbB2 expression, it is possible that Notch1 signaling is involved in the regulation of erbB3 or erbB4 expression as well. Like erbB2, these erbB receptors are expressed by cerebellar and cortical radial glia (Anton et al., 1997; Rio et al., 1997) (our unpublished results). It has been shown that overexpression of erbB receptors can lead to ligand-independent receptor activation. Thus, Notch1-induced increases in erbB receptor expression could lead to levels of signaling sufficient to induce radial glia formation. Nevertheless, the mechanisms by which Notch1 regulates erbB signaling in glia require further studies. For example, experiments with inducible or cell-specific Notch1 mutant mice could add to the understanding of the relationship between Notch1 and erbB receptor signaling.
These previously unknown interactions between Notch1 and erbB receptor signaling may also be significant for other cell types in which both Notch1 and erbB receptors have important biological effects, including muscle, oligodendrocytes, and Schwann cells. Notch and erbB signaling influence in similar manner the differentiation of neural crest cells into Schwann cells (Shah et al., 1994; Morrison et al., 2000), and both have been reported to inhibit the differentiation of oligodendrocyte precursors (Canoll et al., 1996; Wang et al., 1998). Thus, as found for radial glia formation, sequential activation of these pathways could be involved in these processes.
Several studies showed that Notch and erbB receptors as well as their ligands are expressed in the postnatal cerebellum. For example, Bergmann glia express mRNA for all Notch receptors (Irvin et al., 2001; Tanaka et al., 1999), whereas granule neurons express Notch1 and Notch 2 (Irvin et al., 2001; Solecki et al., 2001). Jagged1 mRNA has been reported to be expressed by cerebellar granule cells (Solecki et al., 2001). Rio et al. (1997) showed that during the period of granule cell migration NRG1 is expressed by the granule cells, and erbB receptors by the Bergmann glia. After the granule cells reach their destination, NRG1 expression is downregulated, and these neurons begin to express erbB receptors at high levels (Rieff et al., 1999). Further analysis of the pattern of expression of these molecules, particularly at embryonic stages, could provide important insights into their roles in cerebellar radial glia formation.
Until now, the only radial glial gene whose expression had been shown to be induced by neuronal contact was BLBP (Feng and Heintz, 1995). Here we show that erbB2 expression is also regulated by neuronal contact and that these two events depend on Notch1 signaling. However, an important difference between these effects of Notch1 signaling is that the Notch1-induced BLBP transcription requires the Su(H)/CBF1 pathway, whereas Notch1-induced erbB2 expression, and radial glia formation, appear to be independent of this signaling pathway. This suggests that Notch1 signaling acts through two pathways to induce radial glia formation. This is similar to the Notch1-mediated inhibition of myoblast differentiation, where Notch1 signaling has been shown to act through both Su(H)/CBF1-dependent and -independent pathways (Shawber et al., 1996b; Nofziger et al., 1999). Neural crest formation in chick also relies on Notch signaling independent of Su(H) activation (Endo et al., 2002). Yamamoto et al. (2001) suggested that Notch inhibits differentiation of neural progenitors through a downstream transcriptional regulator, Deltex-1, which acts independently of Su(H)/CBF1. Thus, there is increasing evidence that Notch may act through other pathways, separate from Su(H)/CBF1, to regulate cell differentiation.
BLBP is not only a useful marker for radial glia but it is also an important protein for granule cell migration, given the observation that anti-BLBP antibodies block the migration of granule cells along radial glia fibers (Feng et al., 1994). Thus, our results suggest that Notch1 signaling in glia is an important step in preparing radial glia for neuronal migration. In a previous study we showed that erbB receptor signaling in radial glia is also necessary for neuronal movement along radial glia fibers (Rio et al., 1997). Because we now show that NRG does not induce BLBP expression in these cells, it is possible that the erbB receptor-mediated regulation of neuron movement results from increases in glial BLBP levels by post-transcriptional mechanisms or from the induction of other yet unidentified radial glia genes.
A significant difference between Bergmann glia and cortical radial glia is that although the latter disappear after migration is completed, Bergmann glia persist in the adult cerebellum, maintaining their radial morphology. However, Bergmann glia cease to express BLBP after migration is finished. Irvin et al. (2001) showed that Notch1 mRNA is present in adult Bergmann glia. Our results indicate that the effects of Notch1 on radial glia morphology and BLBP expression are mediated by different pathways. Thus, a downregulation in the adult of the Su(H) pathway, but preservation of the yet unidentified pathways that mediates Notch1 effects on glial morphology, may be the reason for the loss of BLBP expression without a loss in radial morphology.
It is clear that during the development of the vertebrate nervous system several extracellular signaling molecules contribute to particular cellular events, such as glial or neuronal differentiation. However, in most cases, the hierarchies of these signals, or the order in which they need to act, are often difficult to establish. Our results show that sequential signaling through Notch1 and then erbB receptors mediates the induction of radial glia formation by neuronal contact. Similar mechanisms may underlie the functions of Notch1 and erbB receptors in other developmental events, and sequential signaling of other extracellular molecules may serve a variety of processes such as axonal and dendritic outgrowth and pathfinding or synapse formation.
Footnotes
This work was supported in part by National Institute of Neurological Disorders and Stroke Grant R01 NS35884 (G.C.), The EJLB Foundation (G.C.), National Alliance for Research on Schizophrenia and Depression (G.C.), Mental Retardation Research Center Grant NIH P30-HD 18655 (G.C.), ARSEP (J.M.P.), and predoctoral National Institutes of Health training Grant T-32 AG00222 (B.P.). We thank Sally Temple for valuable discussions, Nathaniel Heintz for the BLBP promoter and antibody, and Phillip Leder for the erbB2 promoter.
*B.A.P. and J.M.P. contributed equally to this work.
Correspondence should be addressed to Dr. Gabriel Corfas, Division of Neuroscience, Children's Hospital, 300 Longwood Avenue, Boston, MA 02115. E-mail: gabriel.corfas@tch.harvard.edu.
J. M. Peyrin's present address: Laboratoire d'Immunologie des Tumeurs, Faculté de Pharmacie (Université Paris XI), 5 rue Jean Baptiste Clément, 92296 Chatenay Malabry Cedex, France.
Copyright © 2003 Society for Neuroscience 0270-6474/03/236132-09$15.00/0
References
- Alvarez-Buylla A, Theelen M, Nottebohm F ( 1990) Proliferation “hot spots” in adult avian ventricular zone reveal radial cell division. Neuron 5: 101–109. [DOI] [PubMed] [Google Scholar]
- Anton ES, Marchionni MA, Lee KF, Rakic P ( 1997) Role of GGF/neuregulin signaling in interactions between migrating neurons and radial glia in the developing cerebral cortex. Development 124: 3501–3510. s [DOI] [PubMed] [Google Scholar]
- Canoll PD, Musacchio JM, Hardy R, Reynolds R, Marchionni MA, Salzer JL ( 1996) GGF/neuregulin is a neuronal signal that promotes the proliferation and survival and inhibits the differentiation of oligodendrocyte progenitors. Neuron 17: 229–243. [DOI] [PubMed] [Google Scholar]
- Chanas-Sacre G, Rogister B, Moonen G, Leprince P ( 2000) Radial glia phenotype: origin, regulation, and transdifferentiation. J Neurosci Res 61: 357–363. [DOI] [PubMed] [Google Scholar]
- Culican SM, Baumrind NL, Yamamoto M, Pearlman AL ( 1990) Cortical radial glia: identification in tissue culture and evidence for their transformation to astrocytes. J Neurosci 10: 684–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo Y, Osumi N, Wakamatsu Y ( 2002) Biomodal functions of Notch-mediated signaling are involved in neural crest formation during avian ectoderm development. Development 129: 863–873. [DOI] [PubMed] [Google Scholar]
- Feng L, Heintz N ( 1995) Differentiating neurons activate transcription of the brain lipid-binding protein gene in radial glia through a novel regulatory element. Development 121: 1719–1730. [DOI] [PubMed] [Google Scholar]
- Feng L, Hatten ME, Heintz N ( 1994) Brain lipid-binding protein (BLBP): a novel signaling system in the developing mammalian CNS. Neuron 12: 895–908. [DOI] [PubMed] [Google Scholar]
- Gaiano N, Nye JS, Fishell G ( 2000) Radial glial identity is promoted by Notch1 signaling in the murine forebrain. Neuron 26: 395–404. [DOI] [PubMed] [Google Scholar]
- Hatten ME ( 1985) Neuronal regulation of astroglial morphology and proliferation in vitro. J Cell Biol 100: 384–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatten ME ( 1999) Central nervous system neuronal migration. Annu Rev Neurosci 22: 511–539. [DOI] [PubMed] [Google Scholar]
- Hatten ME, Mason CA ( 1990) Mechanisms of glial-guided neuronal migration in vitro and in vivo. Experientia 46: 907–916. [DOI] [PubMed] [Google Scholar]
- Hsieh JJ, Henkel T, Salmon P, Robey E, Peterson MG, Hayward SD ( 1996) Truncated mammalian Notch1 activates CBF1/RBPJk-repressed genes by a mechanism resembling that of Epstein-Barr virus EBNA2. Mol Cell Biol 16: 952–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter KE, Hatten ME ( 1995) Radial glial cell transformation to astrocytes is bidirectional: regulation by a diffusible factor in embryonic forebrain. Proc Natl Acad Sci USA 92: 2061–2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irvin DK, Zurcher SD, Nguyen T, Weinmaster G, Kornblum HI ( 2001) Expression patterns of Notch1, Notch2, and Notch3 suggest multiple functional roles for the Notch-DSL signaling system during brain development. J Comp Neurol 436: 167–181. [PubMed] [Google Scholar]
- Leavitt BR, Hernit-Grant CS, Macklis JD ( 1999) Mature astrocytes transform into transitional radial glia within adult mouse neocortex that supports directed migration of transplanted immature neurons. Exp Neurol 157: 43–57. [DOI] [PubMed] [Google Scholar]
- Lindsell CE, Shawber CJ, Boulter J, Weinmaster G ( 1995) Jagged: a mammalian ligand that activates Notch1. Cell 80: 909–917. [DOI] [PubMed] [Google Scholar]
- Malatesta P, Hartfuss E, Gotz M ( 2000) Isolation of radial glial cells by fluorescent-activated cell sorting reveals a neuronal lineage. Development 127: 5253–5263. [DOI] [PubMed] [Google Scholar]
- Mizushima S, Nagata S ( 1990) pEF-BOS, a powerful mammalian expression vector. Nucleic Acids Res 18: 5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison SJ, Perez SE, Qiao Z, Verdi JM, Hicks C, Weinmaster G, Anderson DJ ( 2000) Transient Notch activation initiates an irreversible switch from neurogenesis to gliogenesis by neural crest stem cells. Cell 101: 499–510. [DOI] [PubMed] [Google Scholar]
- Noctor SC, Flint AC, Weissman TA, Dammerman RS, Kriegstein AR ( 2001) Neurons derived from radial glial cells establish radial units in neocortex. Nature 409: 714–720. [DOI] [PubMed] [Google Scholar]
- Nofziger D, Miyamoto A, Lyons KM, Weinmaster G ( 1999) Notch signaling imposes two distinct blocks in the differentiation of C2C12 myoblasts. Development 126: 1689–1702. [DOI] [PubMed] [Google Scholar]
- Nye JS, Kopan R ( 1995) Developmental signaling. Vertebrate ligands for Notch. Curr Biol 5: 966–969. [DOI] [PubMed] [Google Scholar]
- Prevot V, Rio C, Cho GJ, Ma YJ, Neville C, Rosenthal N, Heger S, Ojeda SR, Corfas G ( 2003) Normal female sexual development requires NRG-erbB receptor signaling in hypothalamic astrocytes. J Neurosci 23: 230–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakic P, Sidman RL ( 1970) Histogenesis of cortical layers in human cerebellum, particularly the lamina dissecans. J Comp Neurol 139: 473–500. [DOI] [PubMed] [Google Scholar]
- Redmond L, Oh SR, Hicks C, Weinmaster G, Ghosh A ( 2000) Nuclear Notch1 signaling and the regulation of dendritic development. Nat Neurosci 3: 30–40. [DOI] [PubMed] [Google Scholar]
- Rieff HI, Raetzman LT, Sapp DW, Yeh HH, Siegel RE, Corfas G ( 1999) Neuregulin induces GABA(A) receptor subunit expression and neurite outgrowth in cerebellar granule cells. J Neurosci 19: 10757–10766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rio C, Rieff HI, Qi P, Khurana TS, Corfas G ( 1997) Neuregulin and erbB receptors play a critical role in neuronal migration. Neuron 19: 39–50. [DOI] [PubMed] [Google Scholar]
- Shah NM, Marchionni MA, Isaacs I, Stroobant P, Anderson DJ ( 1994) Glial growth factor restricts mammalian neural crest stem cells to a glial fate. Cell 77: 349–360. [DOI] [PubMed] [Google Scholar]
- Shawber C, Boulter J, Lindsell CE, Weinmaster G ( 1996a) Jagged2: a serrate-like gene expressed during rat embryogenesis. Dev Biol 180: 370–376. [DOI] [PubMed] [Google Scholar]
- Shawber C, Nofziger D, Hsieh JJ, Lindsell C, Bogler O, Hayward D, Weinmaster G ( 1996b) Notch signaling inhibits muscle cell differentiation through a CBF1-independent pathway. Development 122: 3765–3773. [DOI] [PubMed] [Google Scholar]
- Solecki DJ, Liu X, Tomoda T, Fang Y, Hatten ME ( 2001) Activated Notch2 signaling inhibits differentiation of cerebellar granule neuron precursors by maintaining proliferation. Neuron 31: 557–568. [DOI] [PubMed] [Google Scholar]
- Tanaka M, Kadokawa Y, Hamada Y, Marunouchi T ( 1999) Notch2 expression negatively correlates with glia differentiation in the postnatal mouse brain. J Neurobiol 41: 524–539. [PubMed] [Google Scholar]
- Wang S, Sdrulla AD, diSibioG, Bush G, Nofziger D, Hicks C, Weinmaster G, Barres BA ( 1998) Notch receptor activation inhibits oligodendrocyte differentiation. Neuron 21: 63–75. [DOI] [PubMed] [Google Scholar]
- Weinmaster G ( 1997) The ins and outs of notch signaling. Mol Cell Neurosci 9: 91–102. [DOI] [PubMed] [Google Scholar]
- Wen D, Suggs SV, Karunagaran D, Liu N, Cupples RL, Luo Y, Janssen AM, Ben-Baruch N, Trollinger DB, Jacobsen VL ( 1994) Structural and functional aspects of the multiplicity of Neu differentiation factors. Mol Cell Biol 14: 1909–1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wettstein DA, Turner DL, Kintner C ( 1997) The Xenopus homolog of Drosophila Suppressor of Hairless mediates Notch signaling during primary neurogenesis. Development 124: 693–702. [DOI] [PubMed] [Google Scholar]
- White MR, Hung MC ( 1992) Cloning and characterization of the mouse Neu promoter. Oncogene 7: 677–683. [PubMed] [Google Scholar]
- Yamamoto N, Yamamoto S, Inagaki F, Kawaichi M, Fukamizu A, Kishi N, Matsuno K, Nakamura K, Weinmaster G, Okano H, Nakafuku M ( 2001) Role of Deltex-1 as a transcriptional regulator downstream of the Notch receptor. J Biol Chem 276: 45031–45040. [DOI] [PubMed] [Google Scholar]