Abstract
The suprachiasmatic nucleus (SCN) is the neuroanatomical locus of the mammalian circadian pacemaker. Here we demonstrate that an abrupt shift in the light/dark (LD) cycle disrupts the synchronous oscillation of circadian components in the rat SCN. The phases of the RNA cycles of the period genes Per1 and Per2 and the cryptochrome gene Cry1 shifted rapidly in the ventrolateral, photoreceptive region of the SCN, but were relatively slow to shift in the dorsomedial region. During the period of desynchrony, the animals displayed increased nighttime rest, the timing of which was inversely correlated with the expression of Per1 mRNA in the dorsomedial SCN. Molecular resynchrony required ∼6 d after a 10 hr delay and 9∼13 d after a 6 hr advance of the LD cycle and was accompanied by the reemergence of normal rest–activity patterns. This dissociation and slow resynchronization of endogenous oscillators within the SCN after an LD cycle shift suggests a mechanism for the physiological symptoms that constitute jet lag.
Keywords: suprachiasmatic nucleus, jet lag, Per1, Per2, Cry1, desynchrony, resynchronization
Introduction
Circadian rhythms are generated by endogenous timekeeping mechanisms that are conserved across evolution. In mammals, the principal circadian oscillator exists in the hypothalamic suprachiasmatic nucleus (SCN) (Klein et al., 1991). Within the SCN, individual neurons are equipped with autonomous oscillators that generate circadian firing rhythms (Welsh et al., 1995; Liu et al., 1997; Herzog et al., 1998).
Recent genetic dissection of circadian rhythms in several organisms has provided insights into the nature of these intracellular oscillators. The basic clock mechanism consists of a feedback loop in which the expression of clock genes is suppressed periodically by their protein products (Dunlap et al., 1999; Panda et al., 2002). The major negative elements are homologs of the Drosophila period genes (Per1 and Per2) and cryptochrome genes (Cry1 and Cry2), whereas positive elements are CLOCK and BMAL1. All these genes are abundantly expressed in the SCN. Per1, Per2, Cry1, and Bmal1 transcripts show dynamic endogenous oscillations, whereas Clock and Cry2 are constitutively expressed in the SCN (Reppert and Weaver, 2001). Mutant mice lacking these genes show disruption of circadian rhythms (King et al., 1997; van der Horst et al., 1999; Vitaterna et al., 1999; Bunger et al., 2000; Bae et al., 2001; Zheng et al., 2001), indicating that these genes are essential components of the circadian pacemaking system.
Circadian oscillations in the neuronal firing rate can be recorded directly from populations of SCN neurons, either through multiple-unit recordings (Inouye and Kawamura, 1979; Bouskila and Dudek, 1993) or single-unit recordings (Green and Gillette, 1982; Groos and Hendriks, 1982; Shibata et al., 1982). Using such methods, it is clear that a population of SCN neurons shows a single stable circadian rhythm with a robust periodicity of ∼24 hr. This rhythm is reflected in daily fluctuations of behavioral and physiological events such as locomotor activity, temperature, wake/sleep cycles, and hormonal rhythms. However, studies using primary dispersed cell cultures indicate that individual SCN neurons are rhythmic, but they show different intrinsic periodicities (Welsh et al., 1995; Honma et al., 2000). The robust oscillations observed in vivo or in SCN slice preparations must result from interneural communication that synchronizes neurons in the SCN.
Although synchronized, the SCN can be divided into two distinct subdivisions, ventrolateral (VLSCN) and dorsomedial (DMSCN), based on cell size, neuropil volume, afferent input and efferent output, and transmitter content (van den Pol, 1980). The ventrolateral photoreceptive region corresponds to the terminal field of the retinal ganglion cells and expresses high levels of vasoactive intestinal peptide and gastrin-releasing peptide (Van Esseveldt et al., 2000). The clock genes Per1 and Per2 are induced in the ventrolateral SCN in response to light pulses delivered during the subjective night (Shigeyoshi et al., 1997; Yan et al., 1999). Photic resetting in mammals possibly occurs through this induction of Per1 and Per2 genes (Shigeyoshi et al., 1997; Akiyama et al., 1999; Albrecht et al., 2001). The spatial restriction of Per1 and Per2 gene induction in the VLSCN led us to propose that there are two types of oscillators in the SCN: ventrolateral light-responsive and dorsomedial light-unresponsive (Shigeyoshi et al., 1997). Given that only one of these is light-responsive, it is possible that shifts in the light/dark cycle cause transient dissociation of the two oscillators. To determine whether this is the case, we shifted rats from one light/dark cycle to a second that was 10 hr delayed or 6 hr advanced with respect to the first. This treatment, which basically simulated the changes associated with travel to a different time zone, resulted in the desynchrony of clock gene expression in the two oscillators. At the same time, the rest–activity behavior was disrupted such that there was increased rest during nighttime hours. Resynchronization of the two oscillators was accompanied by resumption of the normal pattern of rest and activity.
Materials and Methods
Animals and light conditions. Male Wistar rats (JACJO, Osaka, Japan) were purchased 5 weeks after birth. In the initial phase of the experiment, rats were on a 12 hr light (400 lux)/dark (LD) cycle with lights on at 7 A.M. and lights off at 7 P.M. After 2 weeks of entrainment under this lighting condition, rats underwent a phase delay of 10 hr or a phase advance of 6 hr. The delaying phase shift was accomplished by extending the night so that lights on changed from 7 A.M. to 5 P.M. The advancing shift was accomplished by ending the dark period 6 hr early, resulting in lights on from 1 P.M. to 1 A.M.
Behavior analysis. For assessment of locomotor activity, rats were housed individually; their locomotor activity rhythm was measured by area sensors (FA-05 F5B; Omron, Tokyo, Japan) with a thermal radiation detector system; and data were stored on a personal computer. Additional rats were kept under the same conditions, and two or three were killed at each time point for in situ hybridization and immunohistochemical analysis.
In situ hybridization analysis. Rats were deeply anesthetized with ether and were intracardially perfused with 50 ml of saline and 100 ml of a fixative containing 4% paraformaldehyde in 0.1 m phosphate buffer, pH 7.4. We hybridized all frontal sections containing the SCN of rats killed between days 0 and 4 and on day 7. Rat Per1 (rPer1, nucleotides 736∼1720 of rPer1), rPer2 (nucleotides 1390–2915 of rPer2), rCry1 (a fragment corresponding to the nucleotides 530–1688 of mouse Cry1), and rc-fos (nucleotides 500–958 of rat c-fos) cDNA fragments were obtained by PCR and cloned into PGEM-T easy vector. The rPer1, rPer2, rCry1, and rc-fos cRNA probes were labeled with digoxigenin-UTP through in vitro transcription using T7 or SP6 RNA polymerase. The in situ hybridization method using digoxigenin-labeled probes was described previously (Yan et al., 1999). We successfully used rPer1 cRNA probes for in situ hybridization in our former study (Yan et al., 1999). For rPer2 and rCry1 cRNA probes, we performed control studies. No significant signals were detected by using the sense probe of rPer2 and rCry1. RNase treatment of sections gave out no signals. The localization and circadian oscillations of rPer2 and rCry1 mRNA we observed by using antisense cRNA probes were consistent with previous in situ hybridization studies (Thresher et al., 1998; Kume et al., 1999; Yan et al., 1999).
Stored tissues were cut to a thickness of 30 μm on the coronal plane by a cryostat and processed using the free-floating in situ hybridization method as described previously (Ban et al., 1997). Briefly, tissue sections were sequentially transferred through 2× SSC for 10 min, proteinase K (1 μg/ml in 0.1 m Tris buffer, pH 8.0, including 50 mm EDTA) for 5 min at 37°C, and 4% paraformaldehyde in 0.1 m phosphate buffer (PB) for 10 min. Then sections were treated with 0.25% acetic anhydride in 0.1 m triethanolamine for 10 min and 2× SSC for 10 min. Sections were then incubated in the hybridization buffer (60% formamide, 10% dextran sulfate, 20 mm Tris-HCl, pH 8.0, 5 mm EDTA, pH 8.0, 0.3 m NaCl, 10 mm Na-PB, pH 8.0, and 10 mm dithiothreitol) containing rPer1, rPer2, or rCry1 (∼20 ng/100 ml) for 12 hr at 60°C. After two rinses in 2× SSC and 50% formamide at 60°C, sections were treated with a solution containing 20 μg/ml RNase A, 10 mm Tris-HCl, pH 8.0, 1 mm EDTA, and 0.5 m NaCl for 30 min at 37°C. The sections were further washed twice in 2× SSC and 50% formamide and then twice in 0.4× SSC at 60°C (15 min for each wash). They were then transferred into buffer 1 (100 mm Tris-HCl, pH 7.5 and 150 mm NaCl, room temperature, 5 min) and buffer 2 (buffer 1 containing 1.5% blocking reagent) for 1 hr. Sections were then incubated with alkaline phosphatase-labeled anti-digoxigenin serum (Roche Diagnostics, 1:1000 diluted in buffer 1) for 16 hr. The sections were washed first in buffer 1 and then in buffer 3 (100 mm Tris-HCl containing 50 mm NaCl, pH 9.5) for 3 min each. Finally, the hybridization was visualized as a blue signal by treating with nitroblue tetrazolium salt (0.34 mg/ml) and 5-bromo-4-chloro-3-indolyl phosphate toluidinium salt (0.18 mg/ml) in buffer 3 at room temperature for 10 hr. The sections were analyzed under a bright-field microscope.
The number of rPer1 mRNA-positive cells was counted twice, separately in DMSCN and VLSCN, in slices with clear separation of the subregions (three or four sections per animal). An observer blind to experimental treatment counted twice the number of positive cells in the SCN. Cells were counted only when the cytoplasm was clearly labeled against the background. As a result, there was good agreement in the numbers between the two rounds of counting. The average numbers of positive cells per single nucleus at each time point were plotted. The DMSCN was distinguished from the VLSCN by using morphological criteria described previously (van den Pol, 1980).
Immunohistochemistry. Adult Wistar rats were anesthetized by diethyl ether and intracardially perfused with 50 ml of ice-cold saline followed by 100 ml of 0.1 m PB, pH 7.4, containing 4% paraformaldehyde. Brains were removed, immersed in the same fixative for 24 hr at 4°C, and transferred to 20% sucrose in PB for 48 hr at 4°C. Brains were frozen in dry ice and coronally sectioned with a cryostat at a thickness of 20 μm. Free-floating sections were rinsed several times with 0.02 m PBS, pH 7.4, and incubated for 1 hr at 4°C in PBS containing 1% normal goat serum and 0.3% Triton X-100. They were then bleached for 30 min with 50% methanol containing 1.5% H2O2 and rinsed three times with PBS for 15 min each time. The sections were incubated for 4 d at 4°C in the primary anti-Cry1 (Alpha Diagnostic International) diluted 1:500 in PBS containing 0.3% Triton X-100. After rinsing with PBS, sections were incubated for 24 hr at 4°C in biotinylated anti-rabbit goat IgG (Nichirei) diluted 1:75 in PBS. They were then washed three times for 20 min each with PBS and incubated in peroxidase-conjugated streptavidin (Nichirei) diluted 1:75 with PBS for 24 hr at 4°C. Sections were washed three times with 0.05 m Tris-HCl buffer, pH 7.4, for 10 min each. They were then treated with 0.035% diaminobenzidine (DAB) and 0.05 m Tris-HCl buffer in the presence of 0.003% hydrogen peroxide for 10–15 min at room temperature. After the DAB reaction, they were rinsed with 0.05 m Tris-HCl buffer three times (for 10 min each). Free-floating sections were mounted onto 1% gelatin-coated glass slides. After being air-dried, they were dehydrated with a graded series of ethanol rinses, immersed in xylene, and embedded in Entellan (Merck, Darmstadt, Germany).
The number of positive cells was counted in the same way as for in situ hybridization, described above. Positive cell numbers were counted separately in DMSCN and VLSCN in slices with clear separation of the subregions. The average number of positive cells per single nucleus at each time point was plotted. DMSCN was distinguished from VLSCN by using morphological criteria described previously (van den Pol, 1980).
Results
A shift in photoperiod results in desynchrony between the VLSCN and DMSCN
To determine the effect of a large shift in the photoperiod on the phase relationship of the VLSCN and the DMSCN, we evaluated rPer1 expression in rat brains. We used rPer1 as a marker of internal clock phase because of its clear and consistent cycling in the SCN. Expression of rPer1 mRNA is high during the daytime and low during the night (Sun et al., 1997; Tei et al., 1997).
We first needed to establish the phase relationship of the VLSCN and the DMSCN under entrained conditions. To accomplish this, rPer1 mRNA expression was evaluated in the brains of rats housed under a 12 hr LD cycle (lights on at 7 A.M. and lights off at 7 P.M.). Under these conditions, the VLSCN and the DMSCN showed similar profiles of rPer1 expression (Figs. 1A–F,M, 2B). At 11 A.M. [Zeitgeber time 4 (ZT4), 4 hr after lights on], rPer1 was strongly expressed throughout both subregions of the SCN. Modest expression was seen in both subregions at ZT8 (3 P.M.). By lights off (ZT12, 7 P.M.), rPer1 expression was sparse in the entire SCN and remained low throughout the rest of the night. The VLSCN and the DMSCN exhibited similar time courses of staining, indicating that these two subregions are synchronized in the 12 hr LD cycle.
Figure 1.
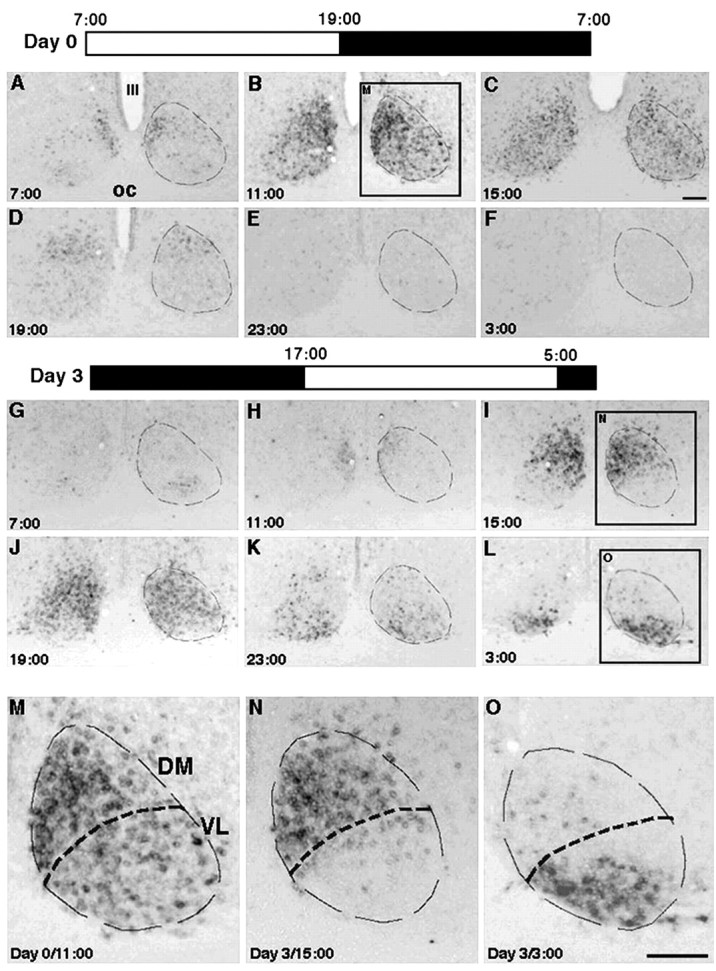
rPer1 expression in the SCN before and after a shift in the light/dark cycle. Coronal sections of the rat bilateral SCN were processed for in situ hybridization using a rPer1 probe. Dashed lines indicate the SCN boundaries and denote DM and VL subdivisions within the SCN. Scale bar, 200 μm. A–F, M, Brains from rats maintained on the light/dark cycle shown as Day 0. Rat brains were collected every 4 hr over a 24 hr period. Clock time is denoted in each panel. Before the photoperiod shift, the subdivisions of the SCN are synchronized with respect to their rPer1 expression. This is particularly evident when viewed at a higher magnification (M), at which the DM and VL SCN are easily delineated. G–L, N, O, Brains taken every 4 hr from rats on the third day after a 10 hr shift in the light/dark cycle. The new photoperiod is shown in Day 3. The abrupt shift in the lighting cycle causes the SCN subregions to become out of phase with each other. This is especially clear at higher magnification (N, O). OC, Optic chiasm; III, the third ventricle.
Figure 2.
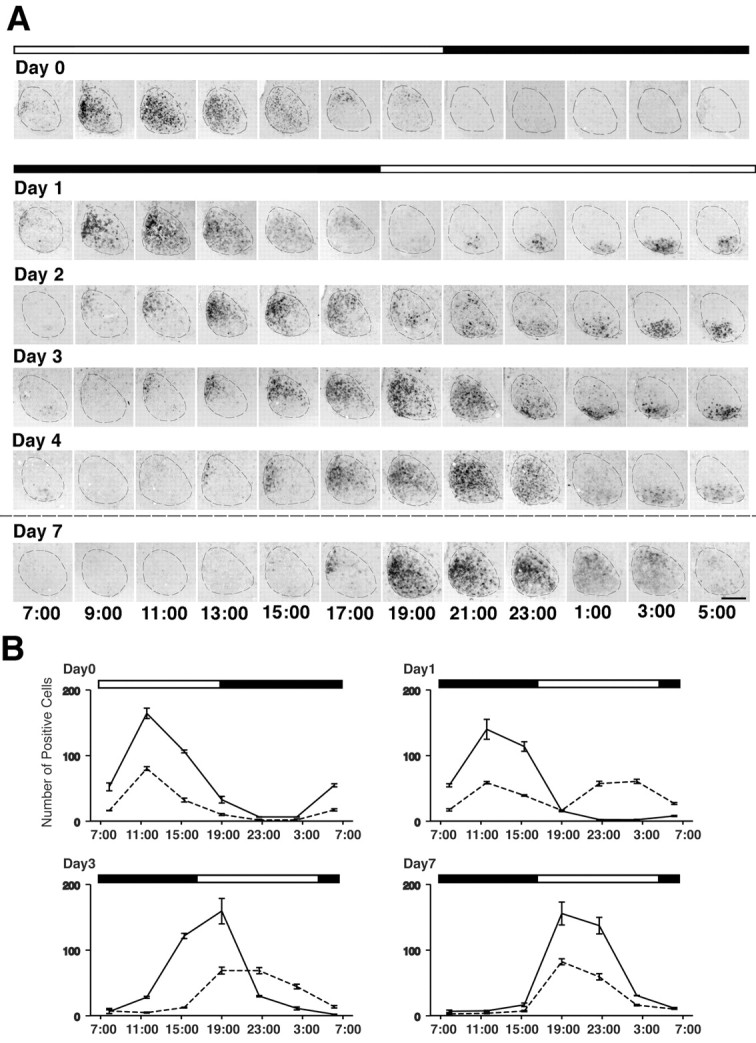
Detailed time course of rPer1 expression in the SCN after a delaying shift in the light/dark cycle. A, Representative coronal brain sections of the SCN processed for rPer1 in situ hybridization. The top black-and-white bar indicates the original LD cycle, and the bottom bar shows the new light cycle. The dashed line indicates the SCN. Clock time is displayed below each column of photomicrographs. On the day before the photoperiod shift (Day 0), the VLSCN and DMSCN are synchronized with respect to their rPer1 expression. On the days after the shift, the two subregions are initially desynchronized but eventually become completely resynchronized by Day 7. Labeled cells in the VLSCN are reset to the new photoperiod by Day 1, whereas this process takes several days for the cells in the DMSCN. Scale bar, 200 μm. B, Graphs showing the number of SCN cells expressing rPer1 mRNA in the DMSCN (solid line) or the VLSCN (dashed line) under entrained conditions (Day 0) or in the days after a 10 hr photoperiod shift (Day 1, Day 3, Day 7). Each time point indicates the mean ± SEM of three rats.
To determine the effects of a large shift in the photoperiod on the phase relationship between the VLSCN and the DMSCN, a second group of entrained rats was transferred to an LD cycle that was delayed 10 hr with respect to the previous cycle (lights on at 5 P.M. and lights off at 5 A.M.). On the third day of the new photoperiod, the VLSCN and the DMSCN showed dramatically different profiles of rPer1 mRNA expression (Fig. 1G–L,N,O). Expression of rPer1 was initially seen in the medial portion of the DMSCN at ZT18 (11 A.M.) and was high at ZT22 (3 P.M.), at which time the dense labeling had spread throughout the entire subregion. In stark contrast, the VLSCN was devoid of label at this time point. At ZT10 (3 A.M.), 12 hr later, rPer1 mRNA was high in the VLSCN, but the DMSCN was essentially devoid of labeling. These findings suggest that 3 d after a shift to a new LD cycle, the DMSCN and the VLSCN are out of phase with each other by ∼12 hr. The rPer1 mRNA expression in the VLSCN is synchronized to the new photoperiod, peaking during the new subjective day. In contrast, rPer1 expression in the DMSCN is desynchronized with respect to the LD cycle, its peak occurring during the night of the new photoperiod. The 10 hr shift in the LD cycle may cause this dissociation by causing a rapid shift of the oscillators in the VLSCN and a much slower shift in the DMSCN.
Time course of resynchronization between the DMSCN and the VLSCN
To determine the time course of the resynchronization process, we monitored the expression profile of rPer1 mRNA in the SCN every 2 hr from day 1 to day 4 and on day 7 of the new photoperiod (Fig.). There was an immediate change in the expression profile in the VLSCN after the 10 hr shift in the LD cycle. During the first day of the new cycle, the rPer1 oscillation in the VLSCN shifted by ∼12 hr such that it was expressed between ZT4 and ZT12 (9 P.M. and 5 A.M.) with a peak in the late day at approximately ZT10 (3 A.M.). A similar pattern of expression was observed on day 2. By day 4 of the new photoperiod, the cycling of rPer1 in the VLSCN stabilized to its normal oscillation profile with a peak at approximately ZT2–ZT4 (7–9 P.M.). Thus, although the rPer1 oscillation in the VLSCN shifted rapidly, the signal intensity of the rPer1 mRNA in the VLSCN was high during the late daytime for the first 3 d of the new photoperiod, indicating that the expression initially overshoots the phase predicted by the light/dark cycle.
In contrast, the expression of rPer1 mRNA in the DMSCN was still predominantly restricted to the subjective nighttime hours on day 2 of the new photoperiod. On this day, a cluster of positive neurons was located in the medial portion of the DMSCN at ZT16 (9 A.M.), and the expression peaked between ZT20 and ZT0 (1 and 5 P.M.). Eight hours later at ZT6 (11 P.M.), the DMSCN was practically devoid of labeled neurons. The timing of the rPer1 peak in the DMSCN was delayed ∼2 hr each day during days 1–4 after the shift in the LD cycle. On day 7 of the new photoperiod, peak expression in the DMSCN was between ZT2 and ZT4 (7 and 9 P.M.). The timing of this peak on day 7 was comparable with that observed under initial entraining conditions and was similar in both subregions, indicating the end of the resynchronization process. These data suggest that light exposure causes a rapid phase shift in the VLSCN, resulting in internal desynchrony of the circadian oscillators in the VLSCN and DMSCN. Light is much less effective in shifting the oscillators of the DMSCN, which required 5–7 d under the new photoperiod to shift its molecular cycle and to reach a stable phase relationship with the VLSCN.
One-way ANOVA applied to the data of days 0, 1, 3, and 7 (n = 3; Fig. 2B) revealed significant differences over time (p < 0.001) in both subregions. Furthermore, we analyzed the differences over days. In the DMSCN, two-way ANOVA revealed a significant overall effect of days (p < 0.001) except between days 0 and 1 (p = 0.13). This is consistent with the lack of an acute effect of light on the expression profile of rPer1 in the DMSCN. In the VLSCN also, there are overall effects of day. However, the F value of day 3 compared with day 7 was much smaller (F = 8.4) than the F values of all the other days (0, 1, and 3) compared with each other and also of each of the other days (0 and 1) compared with day 7 (F > 68), supporting our conclusion that the resetting of the VLSCN was mostly accomplished by day 3.
Oscillations of rat Per2 mRNA, rat Cry1 mRNA, and rat CRY1 protein expression show similar desynchrony in the VLSCN and the DMSCN in response to an abrupt change in photoperiod
To further verify the disparate phases of the circadian oscillators in the VLSCN and DMSCN and, consequently, the internal desynchronization within the SCN, produced by a 10 hr photoperiod shift, we examined the expression profiles of rPer2, rCry1, and rCry1 protein. If a change in photoperiod results in the desynchronization of the VLSCN and DMSCN, we would expect to see similar asynchronous expression for these clock molecules as we demonstrate for rPer1.
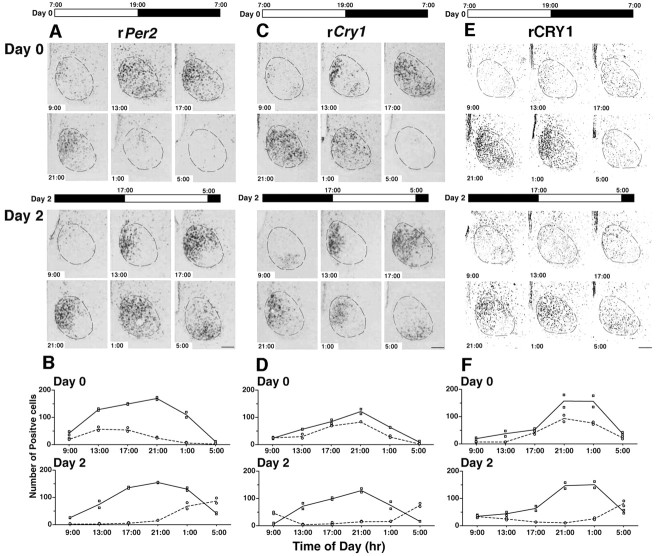
Under stable LD conditions, the distribution and circadian profile of rPer2 mRNA were similar but somewhat delayed with respect to the expression pattern of rPer1 (Fig. 3A,B, Day 0). rPer2 expression was initially detected at ZT2 (9 A.M.) in the medial portion of the DMSCN and peaked between ZT6 and ZT10 (1–5 P.M.), at which point labeled cells extended throughout the entire SCN. By ZT18 (1 A.M.), most of the SCN was devoid of rPer2-labeled cells. The circadian profile of rPer2 expression was identical between the DMSCN and VLSCN; however, cells were more intensely labeled in the DMSCN, indicating that the amplitude of the oscillation is greater in this region.
Figure 3.
Expression of additional clock genes and CRY1 protein before and after a delaying shift in the LD cycle. A, C, E, Photomicrographs of coronal sections through a unilateral SCN processed for in situ hybridization (A, rPer2; C, rCry1) or immunohistochemistry (E, rCRY1). The light/dark cycles used before (Day 0) and after (Day 2) a 10 hr photoperiod shift are depicted above each set of photomicrographs. Clock times are displayed in each micrograph. On Day 0, the two SCN subdivisions are synchronized with respect to the expression of rPer2 and rCry1 transcripts and CRY1 protein. However, on Day 2, the peak of RNA and protein expression in the VLSCN has undergone a dramatic phase shift, whereas the peak expression in the DMSCN has not changed phase compared with Day 0. Scale bar, 200 μm. B, D, F, Graphs showing the number of SCN cells expressing rPer2 mRNA (B), rCry1 mRNA (D), or CRY1 protein (F) at 4 hr intervals on Day 0 and Day 2. The line is drawn through the mean of two independent animals per time point. Individual data points for the DMSCN (square) or VLSCN (circle) are shown.
Similar to what was observed for rPer1 mRNA, a 10 hr shift in the photoperiod resulted in a circadian profile of rPer2 in the DMSCN that was dissimilar to that in the VLSCN (Fig. 3A,B, Day 2). On the second day of the new photoperiod (day 2), rPer2 was expressed in a small number of cells in the medial portion of the DMSCN at ZT16 (9 A.M.). By ZT20 (1 P.M.), the number of positive cells had increased substantially, and the peak occurred at approximately ZT0–ZT4 (5–9 P.M.). The VLSCN was devoid of rPer2 signal at ZT0 (5 P.M.). Labeled cells began to appear at approximately ZT4 (9 P.M.), and by ZT8 (1 A.M.), modest expression was seen in both regions of the SCN. Four hours later at ZT12 (5 A.M.), rPer2 expression reached its peak in the VLSCN. Labeled cells in the DMSCN were rare at this time point. This pattern of dissociation is comparable with that observed for rPer1 mRNA after the shift in the LD cycle, providing further support for a desynchronization of the two SCN regions.
rCry1 mRNA expression also oscillates robustly in the SCN under entrained conditions (Thresher et al., 1998; Kume et al., 1999) (Fig. 3C,D, Day 0). However, the timing of rCry1 expression is slightly different from what we observed with rPer1 and rPer2. rCry1 mRNA-containing neurons were rarely observed at ZT2 (9 A.M.) when the other RNAs had already reached detectable levels of expression. At ZT6 (1 P.M.), rCry1 was expressed in the medial DMSCN and lateral portion of the VLSCN. The area expressing rCry1 increased, and cells throughout the entire SCN expressed modest amounts of label between ZT10 and ZT14 (5 and 9 P.M.). At ZT18 (1 A.M.), the intensity of the signal in the SCN began to decrease and was almost gone by ZT22 (5 A.M.).
As a result of a 10 hr shift in the photoperiod, there was dissociation between the rCry1 expression pattern in the DMSCN versus the VLSCN (Fig. 3C,D, Day 2). On the second day of the new LD cycle, rCry1 mRNA was detected in the VLSCN at ZT16 (9 A.M.), but the DMSCN did not display any detectable signal until ZT20 (1 P.M.). The signal in the DMSCN peaked between ZT0 and ZT4 (5 and 9 P.M.) and was still present at ZT8 (1 A.M.). During the peak of rCry1 in the DMSCN, there was little or no detectable signal in the VLSCN. rCry1 started to rise again in the VLSCN when the DMSCN was devoid of any mRNA staining at ZT12 (5 A.M.). The rCRY1 protein also showed a robust oscillation in the SCN (Kume et al., 1999) (Fig. 3E,F, Day 0). Under stable LD conditions, the circadian profile of the rCRY1 protein is delayed with respect to that of rPer1 and rPer2 but similar to that of rCry1 mRNA. In experiments in which rCRY1 expression was monitored every 4 hr from ZT2 (9 A.M.), rCRY1 protein appeared at ZT6 (1 P.M.) in the medial portion of the DMSCN. Four hours later at ZT10 (5 P.M.), rCRY1-positive cells appeared in the VLSCN. The peak expression for both subregions was ZT14 (9 P.M.). At ZT18 (1 A.M.), the number of immunoreactive neurons in the VLSCN decreased substantially, although there was no apparent change in the DMSCN. At ZT22 (5 A.M.), rCRY1-immunoreactive neurons were sparse in both regions.
A 10 hr shift in the photoperiod caused dissociation of rCRY1 expression between the DMSCN and VLSCN (Fig. 3E,F, Day 2). The rCRY1 expression profile in the DMSCN on day 2 did not show much change from that on day 0 despite the change in the LD cycle. rCRY1-immunoreactive neurons were first detected in the DMSCN at ZT20 (1 P.M.). The staining density increased to peak at ZT4–ZT8 (9 P.M.–1 A.M.). By ZT12 (5 A.M.), only a few labeled cells could be localized to this region. The VLSCN demonstrated rCRY1 immunoreactivity with a significantly different time course. At ZT16 (9 A.M.), a few neurons in the VLSCN showed moderate staining. At ZT4 (9 P.M.), a time of high rCRY1 expression in the VLSCN on day 0, rCRY1-immunopositive neurons were not detected. Labeled neurons were sparse at ZT8 (1 A.M.), but numerous intensely stained neurons were observed in the VLSCN at ZT12 (5 A.M.).
rc-fos mRNA is transiently induced in the VLSCN after an abrupt shift of the LD cycle
One major factor that affects Per1 mRNA levels is exposure to light (Albrecht et al., 1997; Shigeyoshi et al., 1997; Zylka et al., 1998). When animals are exposed to light during the dark period, a rapid induction of Per1 mRNA occurs exclusively in the retinorecipient ventrolateral SCN region (Moore and Lenn, 1972; Shigeyoshi et al., 1997; Yan et al., 1999). It has been suggested that the induction of Per1 mRNA is an initial event of the photic phase-shifting mechanism (Shigeyoshi et al., 1997; Akiyama et al., 1999). We infer that the dissociation reported here is primarily attributable to the rapid response of the VLSCN Per1 to light and the subsequent rapid shift in this oscillator. Because daytime of the new photoperiod represents subjective night in the previous LD schedule, it is difficult to tell whether the expression we observe after the LD shift is attributable to the light induction of Per1 in a clock that is still running with the previous phase or a true shift in the circadian oscillators. To distinguish the light induction of rPer1 mRNA from the autonomous oscillation, we examined the expression of rc-fos mRNA in the SCN. Similar to the light induction of rPer1, the induction of rc-fos mRNA occurs only during the subjective night and correlates with a shift in the locomotor activity rhythm (Kornhauser et al., 1990). However, unlike rPer1, rc-fos is not a component of the clock and therefore does not display robust free-running oscillations.
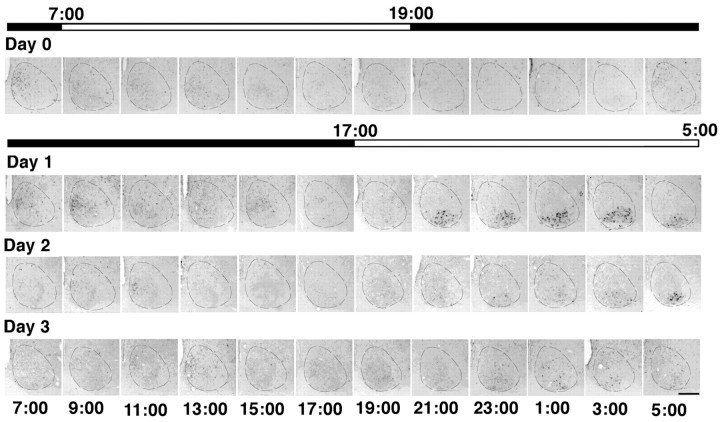
During the initial entraining conditions (day 0), rc-fos mRNA was weakly expressed in the DMSCN at ZT0 (7 A.M.) and not at all in the VLSCN (Fig. 4). The day after an abrupt shift in the light cycle, there was a robust signal in the VLSCN from ZT4 to ZT12 (9 P.M.–5 A.M.), a time that corresponds to the dark period in the previous LD cycle. This is in contrast to postshift day 2, on which rc-fos expression was sparse in the VLSCN except for the staining of a small number of cells at ZT12 (5 A.M.). Cells expressing rc-fos were rarely observed for the remainder of the experiment. These data indicate that light-induced changes in the VLSCN do not persist beyond day 2, so the cyclic expression of rPer1 on subsequent days reflected the endogenous oscillation.
Figure 4.
rc-fos expression occurs primarily on Day 1 after a 10 hr shift in the light/dark cycle. Representative coronal brain sections of the SCN processed for rc-fos in situ hybridization are shown. The top black-and-white bar indicates the original LD cycle, and the bottom bar shows the new light cycle. The dashed line indicates the SCN. Clock time is displayed below each column of photomicrographs. Only during the light phase on Day 1 after the photoperiod shift is there robust rc-fos expression in the VLSCN. Modest rc-fos expression is seen late in the day on Day 2 but is sparse at all other time points throughout the experiment. Scale bar, 200 μm.
An advance of the LD cycle is also associated with the desynchronization and subsequent resynchronization of the VLSCN and the DMSCN
We subjected entrained rats to a 6 hr advance of the LD cycle and then monitored the expression profile of rPer1 mRNA in the SCN every 2 hr on days 1–6, 9, and 13 of the new photoperiod (lights on at 1 A.M. and lights off at 1 P.M.; Fig. 5). An advance of the LD cycle resulted in the dissociation and subsequent resynchronization of the DMSCN and the VLSCN similar to what was observed after a delaying phase shift. However, with only a 6 hr advance, the resynchronization process took longer than it did with a 10 hr delay.
Figure 5.
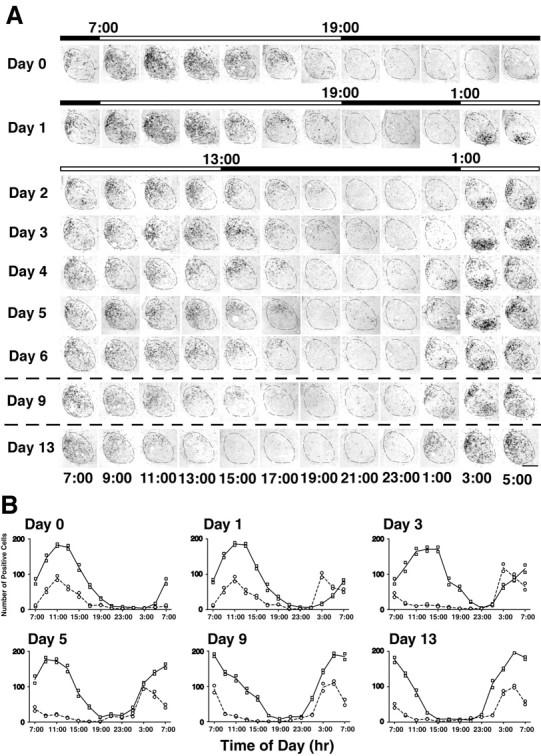
Time course of rPer1 expression in the SCN after a 6 hr advance of the light/dark cycle. A, Representative coronal brain sections of the SCN processed for rPer1 in situ hybridization. The top black-and-white bar indicates the original LD cycle, and the bottom bar shows the new light cycle. The dashed line indicates the SCN. Clock time is displayed below each column of photomicrographs. On the day before the photoperiod shift (Day 0), the VLSCN and DMSCN are synchronized with respect to their rPer1 expression. On the days after the shift, the two subregions are initially desynchronized, but eventually become completely resynchronized by Day 13 of the experiment. Labeled cells in the VLSCN are reset to the new photoperiod by Day 1, whereas this process takes 9–13 d for the cells in the DMSCN. Scale bar, 200 μm. B, Graphs showing the number of SCN cells expressing rPer1 mRNA at 2 hr intervals on Day 0, Day 1, Day 3, Day 5, Day 9, and Day 13. The line is drawn through the mean of two independent animals per time point. Individual data points for the DMSCN (square) or VLSCN (circle) are shown.
In response to the new photoperiod, the rPer1 oscillation in the VLSCN showed a rapid change such that during the first day of the new cycle (day 2), it had already advanced by ∼8 hr with its expression restricted to early daytime (ZT2–ZT6, 3–7 A.M.). From day 2 to day 9, the peak of rPer1 expression in the VLSCN was maintained at ZT2 (3 A.M.), with dense labeling throughout the region. On all days subsequent to the photoperiod shift, the VLSCN was practically devoid of positive neurons from ZT8 (9 A.M.) to ZT22 (11 P.M.). Beginning on day 5, rPer1 expression in the VLSCN was first seen slightly earlier, at ZT0 (1 A.M.), just before “lights on,” although the peak appeared to remain the same. On day 13, when the oscillation had stably entrained to the new regime, rPer1 expression was initiated again at ZT2 (3 A.M.) in the VLSCN and peaked at ZT4 (5 A.M.).
In contrast, the expression of rPer1 mRNA in the DMSCN shifted slowly. From day 0 to day 4, rPer1 expression in the DMSCN showed less than a 2 hr shift in the expression profile. Although expression was first detected at ZT2 (3 A.M.), it did not peak until much later, at approximately ZT8–ZT10 (9–11 A.M.), for the first 3 d after the shift. On day 5, a cluster of positive neurons was detected in the medial portion of the DMSCN at ZT0 (1 A.M.), which suggests a shift of only ∼4 hr by this time. However, on day 6 there were still labeled cells detectable in the DMSCN at ZT16 (5 P.M.), indicating that the shift was still not complete. By day 9, the DMSCN appeared to have shifted several more hours because the peak was now at approximately ZT4 (5 A.M.), and no labeled cells were found between ZT16 and ZT22 (5 and 11 P.M.). On day 13, rPer1 was expressed from ZT 22 to ZT10 (11 P.M.–11 A.M.) with a peak at ZT2–ZT4 (3–5 A.M.). The expression pattern of rPer1 in the VLSCN and DMSCN on day 13 was similar to the expression seen on day 0, indicating the end of the resynchronization process.
rPer1 expression and locomotor activity are inversely correlated
Rats are nocturnal species concentrating their locomotor activity during the dark portion of their day. The activity phase of their circadian cycle on average lasts ∼12 hr when rats are maintained in a 12 hr LD cycle. As a result of a 10 hr phase delay in the LD cycle, there was a considerable shortening of their active phase during the night (Fig. 6A). The shortening of the nighttime active phase was associated with an increase of activity during the late daytime, both of which are typical of transients of the rodent locomotor activity rhythm after LD cycle shifts (Aschoff et al., 1975). During the first day after the shift, activity was restricted to the beginning of the dark phase, and there was an increase in daytime activity. The length of the nocturnal active phase gradually increased ∼2 hr/d until day 6, when the active phase extended through the entire dark portion of the cycle.
Figure 6.
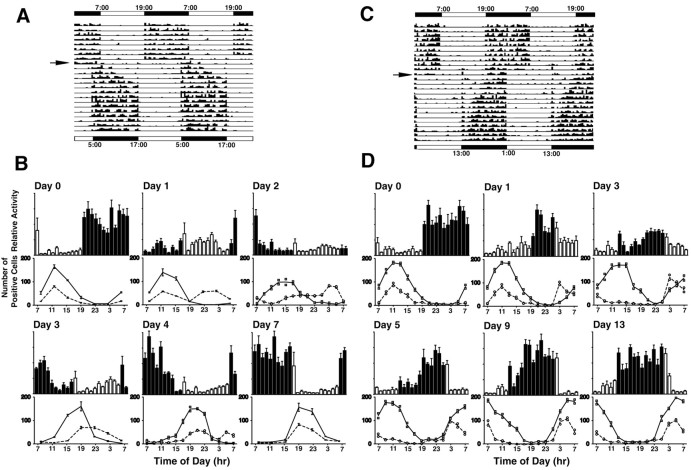
Relationship between locomotor activity and expression of rPer1 in the DMSCN. A, C, Representative locomotor activity records from a Wistar rat after a large phase delay (A) or phase advance (C) of the light/dark cycle. Representative activity of rats was double-plotted. The original LD cycle (indicated by the bar at the top) was shifted, as indicated by the arrow (new light cycle indicated by black-and-white bars at the bottom). After the photoperiod shift, the nighttime activity is compressed and restricted to the early part (after delay) or latter part (after advance) of the night for several days. The daytime activity is also increased until the animal finishes reentraining to the new lighting cycle. B,D, Top, Histograms indicating the mean locomotor activity ± SEM (n = 8). Black bars indicate nighttime points, and white bars indicate day time points. Bottom, Numbers of rPer1-positive cells in the DMSCN (squares, solid lines) and the VLSCN (circles, dashed lines). In B, for Day 0, Day 1, Day 3, and Day 7, the data from Figure 2 B were used to indicate the relationship of locomotor activity to the number of rPer1-labeled cells. Day 2 and Day 4 were evaluated in a separate experiment (n = 2). In D, the numbers of positive cells were from two animals. The data show an inverse correlation between the activity level and the number of labeled cells in the DMSCN throughout the experiment.
Under entrained conditions, we noted that the phase of rPer1 mRNA cycling was opposite that of the locomotor activity cycle. Furthermore, after a 10 hr phase delay, the rPer1 oscillation in the DMSCN appeared to follow a similar time course for stabilization to the new LD cycle, as did locomotor activity, with the peak consistently occurring at the opposite phase. This suggested an inverse relationship between the phase of peak locomotor activity and that of rPer1 mRNA expression in the DMSCN. To determine whether these parameters were truly correlated, we calculated the number of activity counts per hour and compared that with the number of rPer1-labeled cells in the DMSCN in rats under stable entrainment conditions and up to 7 d after a 10 hr phase delay (Fig. 6B). We found that over the course of the experiment, the average number of hourly running wheel revolutions was inversely correlated with the average number of labeled cells (all days, r = -0.60; p < 0.0001). On days when both running wheel activity and the molecular oscillators are completely entrained (days 0 and 7), these two parameters are highly correlated (day 0, r = -0.83; p = 0.001; day 7, r = -0.84; p = 0.001). On days 3 and 4, when both the locomotor activity and the rPer1 expression in the DMSCN are in the process of shifting, the correlation between these two parameters remains robust (day 3, r = -0.66; p = 0.02; day 4, r = -0.68; p = 0.015). On the 2 d immediately after the shift, the correlation is not significant, probably because of the masking effects of light on daytime activity, which, in the presence of limited nighttime activity, results in low overall activity levels. Despite this, the daytime activity does increase on day 1, when the peak of rPer1 mRNA occurs during the subjective night, compared with day 0. Furthermore, on days 1 and 2, there is clearly an absence of locomotor activity during the latter part of the night when rPer1 expression is at its highest.
A 6 hr phase advance in the LD cycle also resulted in a shortening of the active phase during the night (Fig. 6C). Similar to the reentrainment of the mPer1 expression in the DMSCN, the recovery of steady locomotor activity also took longer in an advanced cycle than it did in a delayed cycle. From day 2 to day 5, animals were inactive through the early half of the dark period. In addition, there was an increase in daytime activity. By day 7, there was an extension of the nocturnal active period, but it did not commence until several hours after the onset of dark. The reentrainment process for the locomotor activity took ∼12–14 d. Consistent with what was seen after a delaying shift, there was a significant inverse correlation between the number of positive cells in the DMSCN and the locomotor activity counts (Fig. 6D). These parameters were highly correlated when the animals were under the original entraining conditions (day 0, r = -0.87; p < 0.003), as well as during the days after the shift when reentrainment to the new LD cycle was occurring (day 1, r = -0.72; p = 0.008; day 3, r = -0.72; p = 0.026; day 5, r = -0.85; p < 0.0005; day 9, r = -0.94; p < 0.00001). After the reentrainment process was complete, the correlation between the two parameters was also highly significant (day 13, r = 0.93; p = 0.00002).
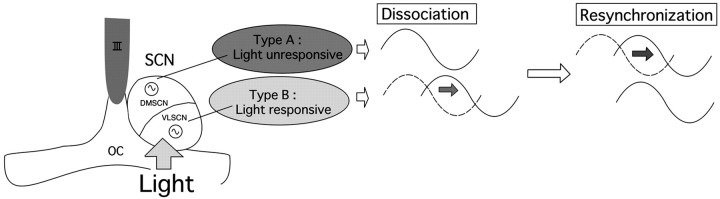
Taken together with our previous results, these data suggest that locomotor activity is an output regulated by the clock cells in the DMSCN, and that the failure of these cells to entrain as rapidly as those in the VLSCN may result in the desynchrony between overt behavior and the environmental light cycle seen in conditions such as jet lag (Fig. 7).
Figure 7.
Schematic representation of the dissociation and slow resynchronization of endogenous oscillators within the rat SCN after an LD cycle shift. The light-responsive and -unresponsive clocks in the SCN are dissociated by light exposure in the subjective night. The phase of the light-responsive clock in the ventrolateral region of the SCN shifts rapidly, but the phase of the light-unresponsive one in the dorsomedial region is relatively slow to shift. During the period of desynchrony, the animals display increased nighttime rest. OC, Optic chiasm; III, the third ventricle.
Discussion
In the present study, we found that either an advancing or a delaying shift in the LD cycle caused an immediate dissociation of synchronous clock gene oscillations within subregions of the SCN. In response to the change in the light cycle, the retinorecipient VLSCN underwent a rapid large shift, whereas the DMSCN shifted slowly but steadily each day, taking several days to resynchronize to the new light cycle. After a 10 hr delaying shift, it took 5–7 d for the molecular oscillators of the DMSCN and the VLSCN to resynchronize with each other. The internal dissociation of these SCN subregions was demonstrated by the desynchrony of rPer1, rPer2, and rCry1 mRNA and rCRY1 protein expression in the DMSCN and the VLSCN. These findings support the hypothesis that there are two spatially distinct oscillators in the SCN: a light-responsive one and a light-unresponsive one. This idea was initially based on the finding that light-induced rPer1 mRNA in the SCN is spatially limited to the VLSCN (Shigeyoshi et al., 1997; Yan et al., 1999). Unlike the VLSCN, the DMSCN cells do not receive direct retinal innervation. The entrainment of these clock cells is likely to be accomplished via afferent connections originating in the retinal recipient VLSCN (Watanabe et al., 2000).
The robust rPer1 gene expression in the VLSCN after the LD shift may, in part, be attributable to its response to light. On day 1 after a delaying shift, rPer1 was expressed in the VLSCN during both day and night, which suggests that the light exposure during subjective night induced rPer1 RNA expression. However, from day 2 onward, the expression was limited to daytime, indicating that the clocks in the VLSCN had adjusted to the new LD regimen by this time. Note that the oscillation in the VLSCN shifted dramatically before adopting a stable phase, presumably also because of this acute light response. rc-fos was expressed in the VLSCN primarily on the first day, with only limited expression on days 2 and 3. This demonstrates that the effects of light are limited primarily to the first day, and oscillations observed thereafter are clock-controlled. In support of the idea that the rPer1 expression in the VLSCN represents an endogenous oscillation, rPer1 was detected just before lights on from day 4 to day 9 in the 6 hr-advanced animal (Fig. 5A). This finding suggests that rPer1 expression in the VLSCN is spontaneous, driven by the endogenous oscillator. However, another finding indicates that some neurons in the VLSCN were photo-responsive for several days after the LD cycle shift; a few cells in the VLSCN showed a very strong rPer1 signal during the day that was comparable with light-induced expression on day 1. Thus, the expression of rPer1 in the VLSCN during the resynchronization process may represent a combination of light-induced and spontaneous expression.
The extended rest after the shift in the LD cycle in our experiments could be caused by at least two factors. During the day, locomotor activity is suppressed by light, a phenomenon called masking (Aschoff, 1999; Mrosovsky, 1999). In addition, after the shift, nighttime activity is suppressed. We propose that the SCN suppresses nighttime activity in the shifted animals by transmitting “subjective day” signals based on the expression of rPer1. This is supported by the inverse correlation we found between rPer1 expression in the DMSCN and locomotor activity not only during entrained conditions but also during the transient period. Normally, rPer1 is expressed during the day when it may function similarly to suppress activity or to promote rest in nocturnal animals. The finding that electrical activity in the rat SCN is high during the day (Gillette, 1991; Miller and Fuller, 1992), despite the nocturnal behavior of this animal, supports the active control of rest. This role of the SCN in suppressing locomotor activity was also demonstrated by transplants of SCN tissues in hamsters (Vogelbaum and Menaker, 1992).
Previously, Yamazaki et al. (2000) reported a transgenic rat line in which luciferase is expressed under the control of the mouse Per1 promoter. SCN slices as well as peripheral body tissues showed rhythmic expression of luciferase, and a shift in the LD cycle resulted in an immediate shift of the luciferase oscillation in the SCN. However, luciferase oscillations were slower to shift in peripheral oscillators, and this desynchrony between the SCN and oscillators in the body was proposed as a mechanism for jet lag. Although these results appear to pose a discrepancy with the present data, we believe the differences can be explained. One possible explanation is that the mouse Per1 promoter in the perluc construct lacks some critical elements involved in the regulation of Per1. Tracking the expression of the luciferase gene and the endogenous rPer1 genes in transgenic rats might reveal differences between the two that could account for the differential response to shifts in the LD cycle. Another possibility is that, because of methodological limitations, any dichotomy within the SCN may have been overlooked. At the same time, it should be recognized that jet lag consists of a wide range of symptoms, and the desynchrony between the SCN and peripheral tissues may also contribute to these symptoms. On the other hand, Reddy et al. (2002) attributed jet lag in the mouse to a rapid shift of Per1 and Per2 oscillations and a relatively slow shift of Cry1 after an acute LD cycle shift. They did not report any dichotomy in the SCN. The difference in the anatomy of the SCN between rats and mice may account for this difference. In the mouse, the separation within the VLSCN and DMSCN subdivisions is not obvious compared with the rat; the retinohypothalamic tract projects more diffusely throughout the ventrodorsal extent of the mouse SCN (Castel et al., 1993). Therefore, the dichotomy of light-responsive and -unresponsive components in the mouse SCN may not have been detected.
The shift of rPer1 expression in the VLSCN was associated with the shift of rPer2 and rCry1 oscillatory expression. Previous studies (Miyamoto and Sancar, 1999; Okamura et al., 1999) demonstrated that acute light pulses have barely any effect on rCry1 expression in the SCN, which is seemingly contradictory to our observation of rCry1 expression in the VLSCN on day 1. It is likely that a large LD cycle shift, which is different from pulsatile light exposure during the night, causes a chronic change of the rCry1 circadian oscillation. However, the mechanisms that would translate apparent Per1 or Per2 induction by light to the shift of Cry1 oscillatory expression are not known. The expression pattern of rCry1 in the VLSCN on day 2 was similar to that in steady LD conditions; the VLSCN started to express rCry1 during the late daytime and ceased to express it during the early daytime, suggesting that the change in Cry1 expression did not result from acute induction but resulted from a shift of the circadian feedback loop.
The transient activity pattern, elongated rest and increased daytime activity, during the period of resynchronization can be compared with jet lag in humans. Jet lag is caused by transient dissociation between environmental and internal times after an abrupt shift of the LD cycle (Redfern et al., 1994; Waterhouse et al., 1997). In other words, a certain population of internal clocks fails to catch up immediately with the new environmental light/dark cycle. The comparison of the time course of rPer1 mRNA expression in the SCN with the rest during dark periods after the LD cycle shift suggests that the dissociation of internal clocks in the SCN is closely related to a specific behavioral pattern in rats.
The present study shows that two distinct oscillators are evident in the SCN after an abrupt change in the light environment. Two-oscillator models have been used to describe peculiar phenomena of circadian rhythmicity, such as the splitting of free-running activity rhythms into two distinct components (Daan and Berde, 1978; Kawato and Suzuki, 1980). Some two-oscillator models hypothesize the existence of light-responsive and -unresponsive oscillators. Such models have been used to account for the desynchrony of the sleep/wake cycle and the core body temperature rhythms of humans (Kronauer et al., 1982), photoperiodic time measurement (Pittendrigh, 1972), and jet lag (Gander et al., 1985). Gander et al. (1985) used the two-oscillator model to explain the dissociation between the circadian oscillation in body temperature and the sleep/wake cycle in humans. Our work indicates that this dissociation between oscillators can take place even within a single SCN and can have profound physiological consequences.
Footnotes
This work was supported by research grants from the Japanese Ministry of Education, Science, Sports and Culture, the Novartis Foundation (Japan) for the Promotion of Science, and the Inamori foundation. We thank Dr. J. Dunlap and Dr. H. Okamura for useful discussions and comments on this manuscript.
Correspondence should be addressed to Dr. Yasufumi Shigeyoshi, Department of Anatomy and Neurobiology, Kinki University School of Medicine, 377-2 Ohno-Higashi, Osakasayama City, Osaka 589-8511, Japan. E-mail: shigey@med.kindai.ac.jp.
Copyright © 2003 Society for Neuroscience 0270-6474/03/236141-11$15.00/0
References
- Akiyama M, Kouzu Y, Takahashi S, Wakamatsu H, Moriya T, Maetani M, Watanabe S, Tei H, Sakaki Y, Shibata S ( 1999) Inhibition of light- or glutamate-induced mPer1 expression represses the phase shifts into the mouse circadian locomotor and suprachiasmatic firing rhythms. J Neurosci 19: 1115–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albrecht U, Sun ZS, Eichele G, Lee CC ( 1997) A differential response of two putative mammalian circadian regulators, mPer1 and mPer2, to light. Cell 91: 1055–1064. [DOI] [PubMed] [Google Scholar]
- Albrecht U, Zheng B, Larkin D, Sun ZS, Lee CC ( 2001) Mper1 and mper2 are essential for normal resetting of the circadian clock. J Biol Rhythms 16: 100–104. [DOI] [PubMed] [Google Scholar]
- Aschoff J ( 1999) Masking and parametric effects of high-frequency light-dark cycle. Jpn J Physiol 49: 11–18. [DOI] [PubMed] [Google Scholar]
- Aschoff J, Hoffmann K, Pohl H, Wever R ( 1975) Re-entrainment of circadian rhythms after phase-shifts of the Zeitgeber. Chronobiologia 2: 23–78. [PubMed] [Google Scholar]
- Bae K, Jin X, Maywood ES, Hastings MH, Reppert SM, Weaver DR ( 2001) Differential functions of mPer1, mPer2, and mPer3 in the SCN circadian clock. Neuron 30: 525–536. [DOI] [PubMed] [Google Scholar]
- Ban Y, Shigeyoshi Y, Okamura H ( 1997) Development of circadian VIP rhythm in the rat suprachiasmatic nucleus. J Neurosci 17: 3920–3931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouskila Y, Dudek FE ( 1993) Neuronal synchronization without calcium-dependent synaptic transmission in the hypothalamus. Proc Natl Acad Sci USA 90: 3207–3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunger MK, Wilsbacher LD, Moran SM, Clendenin C, Radcliffe LA, Hogenesch JB, Simon MC, Takahashi JS, Bradfield CA ( 2000) Mop3 is an essential component of the master circadian pacemaker in mammals. Cell 103: 1009–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castel M, Belenky M, Cohen S, Ottersen OP, Storm-Mathisen J ( 1993) Glutamate-like immunoreactivity in retinal terminals of the mouse suprachiasmatic nucleus. Eur J Neurosci 5: 368–381. [DOI] [PubMed] [Google Scholar]
- Daan S, Berde C ( 1978) Two coupled oscillators: simulations of the circadian pacemaker in mammalian activity rhythms. J Theor Biol 70: 297–313. [DOI] [PubMed] [Google Scholar]
- Dunlap JC, Loros JJ, Crosthwaite SK, Liu Y ( 1999) Eukaryotic circadian systems: cycles in common. Genes Cells 4: 1–10. [DOI] [PubMed] [Google Scholar]
- Gander PH, Kronauer RE, Graerer RC ( 1985) Phase shifting two coupled circadian pacemakers: implications for jet lag. Am J Physiol 249: R704–R719. [DOI] [PubMed] [Google Scholar]
- Gillette MU ( 1991) SCN electrophysiology in vitro: rhythmic activity and endogenous clock properties. In: Suprachiasmatic nucleus: the mind's clock (Klein DC, Moore RY, Reppert SM, eds.), pp 125–143. New York: Oxford UP.
- Green DJ, Gillette R ( 1982) Circadian rhythm of firing rate recorded from single units in the rat suprachiasmatic brain slice. Brain Res 245: 198–200. [DOI] [PubMed] [Google Scholar]
- Groos GA, Hendriks J ( 1982) Circadian rhythms in electrical discharge of rat suprachiasmatic neurons recorded in vitro. Neurosci Lett 34: 283–288. [DOI] [PubMed] [Google Scholar]
- Kawato M, Suzuki R ( 1980) Two coupled neural oscillators as a model of the circadian pacemaker. J Theor Biol 86: 547–575. [DOI] [PubMed] [Google Scholar]
- Kronauer RE, Czeisler CA, Pilato SF, Moore-Ede MC, Weitzmann ED ( 1982) Mathematical model of the human circadian system with two interacting oscillators. Am J Physiol 242: R3–R17. [DOI] [PubMed] [Google Scholar]
- Herzog ED, Takahashi JS, Block GD ( 1998) Clock controls circadian period in isolated suprachiasmatic nucleus. Nat Neurosci 1: 708–713. [DOI] [PubMed] [Google Scholar]
- Honma S, Shirakawa T, Nakamura W, Honma K ( 2000) Synaptic communication of cellular oscillations in the rat suprachiasmatic neurons. Neurosci Lett 17: 113–116. [DOI] [PubMed] [Google Scholar]
- Inouye S-IT, Kawamura H ( 1979) Persistence of circadian rhythmicity in a hypothalamic “island” containing the suprachiasmatic nucleus. Proc Natl Acad Sci USA 76: 5962–5966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King DP, Zhao Y, Sangoram AM, Wilsbacher LD, Tanaka M, Antoch MP, Steeves TD, Vitaterna MH, Kornhauser JM, Lowrey PL, Turek FW, Takahashi JS ( 1997) Positional cloning of the mouse circadian clock gene. Cell 89: 641–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein DC, Moore RY, Reppert SM, eds ( 1991) Suprachiasmatic nucleus: the mind's clock. New York: Oxford UP.
- Kornhauser JM, Nelson DE, Mayo KE, Takahashi JS ( 1990) Photic and circadian regulation of c-fos gene expression in the hamster suprachiasmatic nucleus. Neuron 5: 127–134. [DOI] [PubMed] [Google Scholar]
- Kume K, Zylka MJ, Sriram S, Shearman LP, Weaver DR, Jin X, Maywood ES, Hastings MH, Reppert SM ( 1999) mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell 98: 193–205. [DOI] [PubMed] [Google Scholar]
- Liu C, Weaver DR, Strogatz SH, Reppert SM ( 1997) Cellular construction of a circadian clock: period determination in the suprachiasmatic nucleus. Cell 91: 855–860. [DOI] [PubMed] [Google Scholar]
- Miller JD, Fuller CA ( 1992) Isoperiodic neuronal activity in suprachiasmatic nucleus of the rat. Am J Physiol 263: R51–R58. [DOI] [PubMed] [Google Scholar]
- Miyamoto Y, Sancar A ( 1999) Circadian regulation of cryptochrome gene in the mouse. Brain Res Mol Brain Res 71: 238–243. [DOI] [PubMed] [Google Scholar]
- Moore RY, Lenn NJ ( 1972) A retinohypothalamic projection in the rat. J Comp Neurol 146: 1–14. [DOI] [PubMed] [Google Scholar]
- Mrosovsky N ( 1999) Masking: history, definitions, and measurement. Chronobiol Int 16: 415–429. [DOI] [PubMed] [Google Scholar]
- Okamura H, Miyake S, Sumi Y, Yamaguchi S, Yasui A, Muijtjens M, van der Hoeijmakers JH ( 1999) Photic induction of mPer1 and mPer2 in Cry-deficient mice lacking a biological clock. Science 286: 2531–2534. [DOI] [PubMed] [Google Scholar]
- Panda S, Hogenesch JB, Kay SA ( 2002) Circadian rhythms from flies to human. Nature 417: 329–335. [DOI] [PubMed] [Google Scholar]
- Pittendrigh CS ( 1972) Circadian surfaces and the diversity of possible roles of circadian organization in photoperiodic induction. Proc Natl Acad Sci USA 69: 2734–2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy AB, Field MD, Maywood ES, Hastings MH ( 2002) Differential resynchronization of circadian clock gene expression within the suprachiasmatic nuclei of mice subjected to experimental jet lag. J Neurosci 22: 7326–7330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redfern P, Minors D, Waterhouse J ( 1994) Circadian rhythms, jet lag, and chronobiotics: an overview. Chronobiol Int 11: 253–265. [DOI] [PubMed] [Google Scholar]
- Reppert SM, Weaver DR ( 2001) Molecular analysis of mammalian circadian rhythms. Annu Rev Physiol 63: 647–676. [DOI] [PubMed] [Google Scholar]
- Shibata S, Oomura Y, Kita H, Hattori K ( 1982) Circadian Rhythmic changes in neuronal activity in the suprachiasmatic nucleus of the rat hypothalamic slice. Brain Res 247: 154–158. [DOI] [PubMed] [Google Scholar]
- Shigeyoshi Y, Taguchi K, Yamamoto S, Takekida S, Yan L, Tei H, Moriya T, Shibata S, Loros JJ, Dunlap JC, Okamura H ( 1997) Light-induced resetting of a mammalian circadian clock is associated with rapid induction of the mPer1 transcript. Cell 91: 1043–1053. [DOI] [PubMed] [Google Scholar]
- Sun ZS, Albrecht U, Zhuchenko O, Bailey J, Eichele G, Lee CC ( 1997) RIGUI, a putative mammalian ortholog of the Drosophila period gene. Cell 90: 1003–1011. [DOI] [PubMed] [Google Scholar]
- Tei H, Okamura H, Shigeyoshi Y, Fukuhara C, Ozawa R, Hirose M, Sakaki Y ( 1997) Circadian oscillation of a mammalian homologue of the Drosophila period gene. Nature 389: 512–516. [DOI] [PubMed] [Google Scholar]
- Thresher RJ, Vitaterna MH, Miyamoto Y, Kazantsev A, Hsu DS, Petit C, Selby CP, Dawut L, Smithies O, Takahashi JS, Sancar A ( 1998) Role of mouse cryptochrome blue-light photoreceptor in circadian photoresponses. Science 282: 1490–1494. [DOI] [PubMed] [Google Scholar]
- van den Pol AN ( 1980) The hypothalamic suprachiasmatic nucleus of rat: intrinsic anatomy. J Comp Neurol 191: 661–702. [DOI] [PubMed] [Google Scholar]
- van der Horst GT, Muijtjens M, Kobayashi K, Takano R, Kanno S, Takao M, de Wit J, Verkerk A, Eker AP, van Leenen D, Buijs R, Bootsma D, Hoeijmakers JH, Yasui A ( 1999) Mammalian Cry1 and Cry2 are essential for maintenance of circadian rhythms. Nature 398: 557–558. [DOI] [PubMed] [Google Scholar]
- Van Esseveldt LKE, Lehman MN, Boer GJ ( 2000) The suprachiasmatic nucleus and the circadian time-keeping system revisited. Brain Res Rev 33: 34–77. [DOI] [PubMed] [Google Scholar]
- Vitaterna MH, Selby CP, Todo T, Niwa H, Thompson C, Fruechte EM, Hitomi K, Thresher RJ, Ishikawa T, Miyazaki J, Takahashi JS, Sancar A ( 1999) Differential regulation of mammalian period genes and circadian rhythmicity by cryptochromes 1 and 2. Proc Natl Acad Sci USA 96: 12114–12119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelbaum MA, Menaker M ( 1992) Temporal chimeras produced by hypothalamic transplants. J Neurosci 12: 3619–3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe K, Vanecek J, Yamaoka S ( 2000) In vitro entrainment of the circadian rhythm of vasopressin-releasing cells in suprachiasmatic nucleus by vasoactive intestinal polypeptide. Brain Res 877: 361–366. [DOI] [PubMed] [Google Scholar]
- Waterhouse J, Reilly T, Atkinson G ( 1997) Jet-lag. Lancet 350: 1611–1616. [DOI] [PubMed] [Google Scholar]
- Welsh DK, Logothetis DE, Meister M, Reppert SM ( 1995) Individual neurons dissociated from rat suprachiasmatic nucleus express independently phased circadian firing rhythms. Neuron 14: 697–706. [DOI] [PubMed] [Google Scholar]
- Yamazaki S, Numano R, Abe M, Hida A, Takahashi R, Ueda M, Block GD, Sakaki Y, Menaker M, Tei H ( 2000) Resetting central and peripheral circadian oscillators in transgenic rats. Science 288: 682–685. [DOI] [PubMed] [Google Scholar]
- Yan L, Takekida S, Shigeyoshi Y, Okamura H ( 1999) Per1 and Per2 gene expression in the rat suprachiasmatic nucleus: circadian profile and the compartment-specific response to light. Neuroscience 94: 141–150. [DOI] [PubMed] [Google Scholar]
- Zheng B, Albrecht U, Kaasik K, Sage M, Lu W, Vaishnav S, Li Q, Sun ZS, Eichele G, Bradley A, Lee CC ( 2001) Nonredundant roles of the mPer1 and mPer2 genes in the mammalian circadian clock. Cell 105: 683–694. [DOI] [PubMed] [Google Scholar]
- Zylka MJ, Shearman LP, Weaver DR, Reppert SM ( 1998) Three period homolog in mammals: differential light responses in the suprachiasmatic circadian clock and oscillating transcripts outside of brain. Neuron 20: 1103–1110. [DOI] [PubMed] [Google Scholar]