Abstract
Altered regulation of 5-HT1A receptors is implicated in mood disorders such as anxiety and major depression. To provide insight into its transcriptional regulation, we previously identified a novel DNA element [14 bp 5′-repressor element (FRE)] of the 5-HT1A receptor gene that mediates repression in neuronal and non-neuronal cells (Ou et al., 2000). We have now cloned a novel DNA binding protein [five′ repressor element under dual repression binding protein-1 (Freud-1)] that binds to FRE to mediate repression of the 5-HT1A receptor or heterologous promoters. Freud-1 is evolutionarily conserved and contains two DM-14 basic repeats, a predicted helix-loop-helix DNA binding domain, and a protein kinase C conserved region 2 (C2)/calcium-dependent lipid binding (CalB) calcium/phospholipid binding domain. An intact CalB domain was required for Freud-1-mediated repression. In serotonergic raphe cells, overexpression of Freud-1 repressed the 5-HT1A promoter and decreased 5-HT1A receptor protein levels, whereas transfection of antisense to Freud-1 derepressed the 5-HT1A gene and increased 5-HT1A receptor protein expression. Calcium-dependent signaling blocked Freud-1-FRE binding and derepressed the 5-HT1A promoter. Treatment with inhibitors of calmodulin or CAM-dependent protein kinase reversed calcium-mediated inhibition of Freud-1. Freud-1 RNA and protein were present in raphe nuclei, hippocampus, cortex, and hypothalamus, and Freud-1 protein was colocalized with 5-HT1A receptors, suggesting its importance in regulating 5-HT1A receptors in vivo. Thus, Freud-1 represents a novel calcium-regulated repressor that negatively regulates basal 5-HT1A receptor expression in neurons and may play a role in the altered regulation of 5-HT1A receptors associated with anxiety or major depression.
Keywords: serotonin, transcription, silencer, raphe, autoreceptor, calcium
Introduction
The 5-HT1A receptor is widely expressed at postsynaptic sites including cortex, hippocampus, limbic system, and hypothalamus (Albert et al., 1990; Törk, 1990; Pompeiano et al., 1992) and is implicated in major depression, anxiety, and suicide (Mann, 1999; Pineyro and Blier, 1999; Veenstra-VanderWeele et al., 2000). Importantly the 5-HT1A receptor functions as a somatodendritic autoreceptor (Riad et al., 2001) to inhibit the firing of serotonergic raphe neurons (Pineyro and Blier, 1999). 5-HT1A receptor knock-out mice display enhanced serotonergic neurotransmission attributable to the loss of presynaptic 5-HT1A receptors (Ase et al., 2000) and have increased anxiety-related behaviors (Heisler et al., 1998; Parks et al., 1998; Ramboz et al., 1998). Downregulation of 5-HT1A autoreceptors is implicated in the clinical efficacy of chronic antidepressant treatments, which enhance serotonergic neurotransmission (Albert et al., 1996; Pineyro and Blier, 1999). Acute desensitization by receptor uncoupling (seconds), internalization (seconds to minutes), or degradation (hours) occurs too rapidly to explain the 2-3 week time course for 5-HT1A autoreceptor downregulation in vivo (Albert et al., 1996; Riad et al., 2001), suggesting a role for transcriptional silencing. Conversely, 5-HT1A autoreceptor levels are increased in the midbrain of suicide victims with major depression compared with control subjects (Stockmeier et al., 1998), suggesting that the level of basal expression of this receptor may predispose subjects to depression or suicide. Therefore, we have addressed the transcriptional mechanisms that regulate the basal expression of the 5-HT1A receptor gene in raphe cells.
Transcriptional regulation is an important determinant of the basal neuron-specific expression of 5-HT1A receptors (Parks and Shenk, 1996; Hendricks et al., 1999; Storring et al., 1999; Ou et al., 2000). We have used rat RN46A cells as a model of raphe neurons to examine the transcriptional regulation of the rat 5-HT1A receptor gene (Storring et al., 1999; Ou et al., 2000). RN46A cells are embryonic day 13 (e13) rat raphe neurons that are transformed reversibly and retain essential characteristics of raphe neurons including expression of tryptophan hydroxylase, 5-HT transporters, and 5-HT1A receptors (Eaton et al., 1995; Storring et al., 1999). Analysis using 5-HT1A promoter-luciferase reporter constructs revealed that rat, mouse, and human 5-HT1A receptor genes contain a conserved dual repressor element (DRE) (31 bp DRE, -1555/-1524 bp) that conferred strong basal repression in non-neuronal and neuronal cells (Ou et al., 2000). Deletion or inactivation of the DRE resulted in 10-fold induction of basal 5-HT1A transcriptional activity. The 5-HT1A DRE is composed of two elements: a 14 bp 5′-repressor element (FRE) and an adjacent 12 bp 3′-repressor element (TRE). The FRE is active in neuronal cells that normally express the 5-HT1A receptor such as raphe RN46A or septal SN-48 cells, as well as in non-neuronal cells. The TRE is active only in 5-HT1A receptor-negative cells such as rat L6 myoblasts. Here we identify a novel FRE-binding protein, five′ repressor element under dual repression binding protein-1 (Freud-1), as the first transcriptional repressor for neuronal regulation of a serotonin system gene, the 5-HT1A receptor.
Materials and Methods
Yeast one-hybrid screening. The Matchmaker One-Hybrid System (Clontech, Palo Alto, CA) was used to screen for DRE-binding cDNA clones. A concatenated probe with three copies of 31 bp 5-HT1A DRE (-1555/-1524 bp) was subcloned into EcoRI-XmaI-digested pHISi and pLacZi vectors (Clontech). The constructs were linearized and stably integrated into the genome of YM4271 and plated onto synthetic dropout (SD)His-Ura plates to establish the dual-construct yeast strain YM4271-LZ3-HIS3. Subsequent transformation with a mouse brain Matchmaker cDNA library (>1 × 10 6 clones) fused to GAL4-AD allowed for selection of resistant clones, which were tested by the β-galactosidase plate assay, retransformed for verification, and sequenced. 5′ rapid amplification of cDNA ends (RACE) was used to isolate full-length Freud-1 using the primer 5′-GCACGAATGGCGTCT-TGGTATTGCTTGACA-3′. 5′ RACE was performed using the Mouse brain Marathon-Ready cDNA and the Advantage 2 PCR Kit (Clontech). PCR products were gel-purified, subcloned into the pGEM-T Easy Vector (Promega, Madison, WI), and sequenced using the ABI/PRISM automated system.
Plasmids. 5-HT1A reporter constructs were derived from a 2.7 kb fragment of the rat 5-HT1A receptor promoter (-2719-luciferase), which has been described previously (Storring et al., 1999; Ou et al., 2000). Freud-1 expression plasmids were subcloned by digestion of pACT2 clones obtained from yeast one-hybrid screening with BglII and subsequent ligation with BamHI-digested pcDNA3 (Invitrogen, Burlington, Ontario, Canada). The Freud-1 coding sequence was obtained by PCR and verified by DNA sequence analysis. Subsequently, the PCR product was subcloned into the pGEM-T Easy vector (Promega) before subcloning in the EcoR1 site of pET30A (Invitrogen) for protein expression and purification. A Freud-1 deletion mutant (Freud-1 del), lacking amino acids 341-348 within the CalB portion of the C2 domain, was made by site-specific mutagenesis of the Freud-1 sequence in pcDNA3, Flag-pcDNA3, or pET30A plasmids using QuikChange XL Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA). The Flag epitope (DYKDDDK) was incorporated at the N terminus of Freud-1 or Freud-1 del and subcloned into the EcoRI site of pcDNA3.
Freud-1 protein expression and purification. Escherichia coli BL21 (DE3) cells were transformed with Freud-1 expression plasmid pET30A-Freud-1, grown overnight, and induced with 1 mm isopropyl-β-d-thiogalactopyranoside at 37°C for 3-4 hr. Cells were harvested and purified by TALON metal affinity resin (Clontech). Fractions containing Freud-1 were dialyzed against a buffer containing 20 mm HEPES, pH 7.9, 150 mm KCl, 0.2 mm EDTA, 0.5 mm dithiothreitol, 20% glycerol, and stored frozen at -80°C.
Cell culture. Rat L6 myoblast and rat raphe RN46A cells were maintained and transfected as described previously (Ou et al., 2000). For Western blots of 5-HT1A receptor, 75% confluent RN46A cells were differentiated at 39°C, and the medium was changed to 1:1 DMEM/Ham's F12 medium containing 1% (v/v) fetal bovine serum, 1 μg/ml insulin, 1 μg/ml transferrin, and 30 ng/ml nerve growth factor for 3 d (White et al., 1994). Differentiated RN46A cells were transiently transfected using Superfect (Qiagen, Mississauga, Ontario, Canada) for 72 hr, and cell extracts were harvested. Transfection efficiency was determined in separate cultures by counting X-gal-stained cells after transfection of pCMV-βGal as described (Albert et al., 1999). Rat hippocampal and cortical cells were dissected from Sprague Dawley fetuses at 18 d of gestation (Banker and Cowan, 1977). Cells were dispersed, resuspended in Neurobasal media containing B27 supplement (Invitrogen), 0.5 mm l-glutamine, 1% penicillin/streptomycin, and 25 μm glutamate (Brewer et al., 1993), plated on poly-d-lysine (MW 30,000 -70,000)-coated vessels at 37°C in 5% CO2 for 13 d, and then fixed in 4% paraformaldehyde. Less than 0.2% GFAP-positive cells were present, suggesting that the cultures were 99.8% neuronal (Brewer et al., 1993).
Electrophoretic mobility shift assay. Nuclear proteins were extracted, and electrophoretic mobility shift assay (EMSA) was performed as described previously (Ou et al., 2000). Nuclear extracts (20 μg per sample) or purified Freud-1 (4 μg per sample) were preincubated with or without competitor 5-HT1A DNA oligonucleotides, antibody, peptide, CaCl2 (100 μm), and/or ATP (150 μm) or different protein kinase inhibitors (50 μm) in a 30 μl reaction containing DNA binding buffer (20 mm HEPES, 0.2 mm EDTA, 0.2 mm EGTA, 100 mm KCl, 5% glycerol, and 2 mm DTT, pH 7.9), 2 mg of poly (dI-dC) at room temperature for 25 min. 32P-labeled DRE or FRE probe (60,000 cpm per sample) was added and incubated for an additional 20 min. The locations, sizes, and forward sequences of rat 5-HT1A receptor gene primers were as follows: DRE (-1555/-1524 bp, 31 bp): 5′-CGGCATAAGCAAGCCCTTATTGCACAGAGCT-3′; FRE: (-1554/-1540 bp, 14 bp): 5′-GGCATAAGCAAGCC-3′; TRE (-1543/-1532 bp, 12 bp): 5′-GCCCTTATTGCA-3′ (Ou et al., 2000). The mutation in the 2300m1 and DREmut/SV40 constructs changes the FRE sequence to 5′-GG ACATAG AGCACC-3′.
Luciferase and β-galactosidase assays. Cells were extracted with 150 μl Reporter Lysis Buffer (Promega) 48 hr after transfection with promoter-luciferase, pCMV-βGal, and expression plasmids. The extracts were centrifuged, collected, and assayed for luciferase and β-galactosidase activities as described previously (Ou et al., 2000). The ratio of luciferase/β-galactosidase activity was determined in triplicate samples and normalized to vector-transfected (pGL3-basic) extracts. All data are presented as the mean ± SEM of at least three independent experiments.
Northern and Western blot analyses. For Northern blot analysis, tissues were dissected from Sprague Dawley rats (Gispen et al., 1972) and RNA isolated using RNA Isolation Reagent (Ambion, Austin, TX), electrophoresed, blotted, and probed using a full-length Freud-1 cDNA probe (Albert et al., 1990). For production of antibodies, the peptide KEALYRRNLVESELQR (Freud-1 amino acids 577-592) was synthesized as a multiple-antigenic peptide to immunize rabbits (Cedarlane, Hornby, Ontario, Canada). Freud-1 antiserum was purified by Affi-Gel blue 50-100 mesh (Bio-Rad Laboratories, Hercules, CA) for albumin removal, followed by purification on peptide-conjugated Affi-Gel 15 (Bio-Rad). For Western blot, nuclear proteins (20 μg) or purified Freud-1 (1 μg) were separated by SDS-PAGE on a 10% polyacrylamide gel. Immunoblotting was performed as described previously (Ghahremani et al., 1999). Freud-1 antibody was used at a dilution of 1:1000 followed by a 1:2000 dilution of the secondary horseradish peroxidase (HRP)-linked rabbit antibody (Bio/Can Scientific, Mississauga, ON). Anti-Flag M2 mouse monoclonal antibody (Sigma, Oakville, ON) was used at 1:1000, and HRP-conjugated anti-mouse (Amresco, Santa Cruz, CA) was used as the secondary antibody at a 1:8000 dilution. For detection of 5-HT1A receptor, whole-cell lysates (60 μg) were electrophoresed on a 12% Tris glycine polyacrylamide gel (Invitrogen/Novex Electrophoresis, Carlsbad, CA), transferred onto a nitrocellulose membrane, and incubated with Tris-buffered saline (TBS) and 5% milk for 1 hr and overnight at 4°C with primary rabbit polyclonal anti-5-HT1A antibody (Santa Cruz Biotechnology, Santa Cruz, CA; catalog number sc 10801) at a 1:300 dilution. The blots were then washed three times and incubated with HRP-conjugated donkey anti-rabbit secondary antibody (Amersham, Arlington Heights, IL) at 1:3000. The reactive bands were visualized by chemiluminescence using the ECL-kit (Amersham) after exposure to Hyperfilm (Amersham). An antiserum to β-actin (Santa Cruz Biotechnology) was also tested on blots and served as a control for equal loading of samples.
Immunofluorescence. Adult male (250-300 gm) Sprague Dawley rats were anesthetized (Somnatol, 100 mg/kg, i.p.), perfused with saline, 4% paraformaldehyde, and decapitated. Brains were removed and postfixed overnight at 4°C, transferred to 10% sucrose/0.02% sodium azide, and frozen. Tissues were sectioned (12 μm), mounted on slides, and kept at -80°C or stored as floating sections at 4°C in PBS containing 0.02% sodium azide. Cultured cells were fixed in 4% paraformaldehyde for 1 hr at 37°C. Brain sections or cells were permeabilized and blocked in 1% BSA, 5% goat serum, and 0.05% Triton X-100 and incubated for 48 hr at 4°C with rabbit anti-Freud-1 primary antibody in combination with guinea pig anti-5-HT1A (Chemicon International, Temecula, CA), guinea pig anti-5-HT (Dr. A. A. Verhogstad, Nijmegen, Sweden), or mouse anti-tyrosine hydroxylase (TH) (Chemicon) (1:100, 1:500, and 1:100 dilutions in blocking buffer, respectively). Immunostaining was detected using corresponding secondary antibodies, Texas Red-coupled anti-rabbit (Calbiochem, San Diego, CA), anti-guinea pig FITC (Chemicon), and anti-mouse FITC (Sigma) (1:100, 1:500, and 1:100 dilutions, respectively, in 1% BSA, 1% goat serum, 0.05% Triton X-100 for 1 hr at room temperature). RN46A cells transfected with DsRed (pDsRed2-C1, Clontech) were fixed and stained for 5-HT1A receptor (as above), and the red fluorescent protein (DsRed) was detected under the red channel by fluorescence microscopy. Immunofluorescence was visualized under a Zeiss Axioscop 2 microscope, and images were captured using a Sony Hyper Had color camera and Northern Eclipse software (Emphix imaging).
In situ hybridization. Sense (5′-CAGCGGAAGGCACGCATGCATGAACGAATTGTCAAGCAAT-3′) or antisense (5′-ATTGCTTGACAATTCGTTCATGCATGCGTGCCTTCCGCTG-3′) Freud-1 oligonucleotides (1 μg per reaction) were labeled using 30 U terminal transferase, 5 nmol digoxigenein-11-dUTP in tailing buffer/CoCl2 (Roche, Laval, Quebec, Canada) in 50 μl for 90 min at 37°C, and stored at -20°C. Slides were heated (45°C, 10 min), permeabilized in 5 μg/ml proteinase K/0.1 m Tris-HCl, pH 7.5 (37°C, 4 min), rinsed, and hybridized overnight at room temperature with 75 μl of hybridization buffer per slide (500 pg/μl of sense or antisense oligonucleotide in 50% deionized formamide, 6× SSC, 50 mm Tris-HCl, pH 7.0, 2× Denhardt's solution, 0.2% SDS). Sections were washed in 2× and 0.5× SSC (two times for 10 min, room temperature), and incubated with anti-digoxygenin-AP (Roche) (1:500, 2 hr, room temperature). The alkaline phosphatase reaction was developed in nitroblue tetrazolium/5-bromo-4-chloro-3 indoyl phosphate (NBT/BCIP) (10 mg/5 mg; Boehringer-Ingelheim, Laval, Quebec, Canada) and 8 mg levamisole (15 hr, room temperature) in 30 ml of 100 mm Tris-HCl (pH 9.5)/NaCl/MgCl2. After washing, sections were dehydrated in a series of ethanol washes and mounted, and images were captured as described above.
Statistical analysis. The significance of values compared with control was set at p < 0.05 as determined by t test using Prism 3.0 (GraphPad Software, San Diego, CA).
Results
Molecular cloning of Freud-1
To identify regulatory proteins that target the 5-HT1A DRE, three copies of the DRE sequence were placed upstream of LacZ or various selectable markers to screen a mouse brain cDNA library for DRE-binding proteins by the yeast one-hybrid approach. Three different positive clones were identified, two of which encoded a novel 595 aa protein (NP 666082; predicted molecular weight 66.5 kDa) that we named Freud-1. By GenBank search, human (BAA91194), Drosophila melanogaster (NP 609488), Anopheles gambieae (EEA09979), and Caenorhabditis elegans (NP 493412) homologs were identified with 85, 33, 31, or 28% amino acid identity to the murine Freud-1, respectively. As shown in Figure 1A, Freud-1 contains a number of known protein domains, including two DM-14 domains (boxed) and a protein kinase C (PKC) conserved region 2 (C2 domain) (dashed box). The DM-14 domain is a conserved 60 aa repeat sequence of unknown function that is specific to Freud-1-species homologs (Fig. 1B). The C2 domain (via its central 43 aa CalB domain) mediates Ca2+-dependent lipid binding of PKC, phospholipases, and synaptotagmin and protein-protein interactions (Clark et al., 1991). The C2 domain is present in all Freud-1 species homologs, suggesting a conserved calcium-dependent regulation of Freud-1. Freud-1 also contains a putative helix-loop-helix (HLH) DNA binding domain (HELIXTURNHELIX; http://xblast.tamu.edu/pise/helixturnhelix-simple.html) that could mediate its DNA binding function (Fig. 1A). In addition, a consensus PKA/calmodulin-dependent protein kinase (CAMK) phosphorylation site and nine putative PKC phosphorylation sites were identified (ProSearch; http://bioweb.pasteur.fr/seqanal/interfaces/prosearchsimple.html) (Fig. 1A). Although a consensus nuclear localization signal was not identified, Freud-1 localization is predicted to be nuclear (ProtComp program; http://www.softberry.com/berry.phtml?topic=proteinloc&prg=ProtCompA), consistent with a transcriptional regulatory function.
Figure 1.
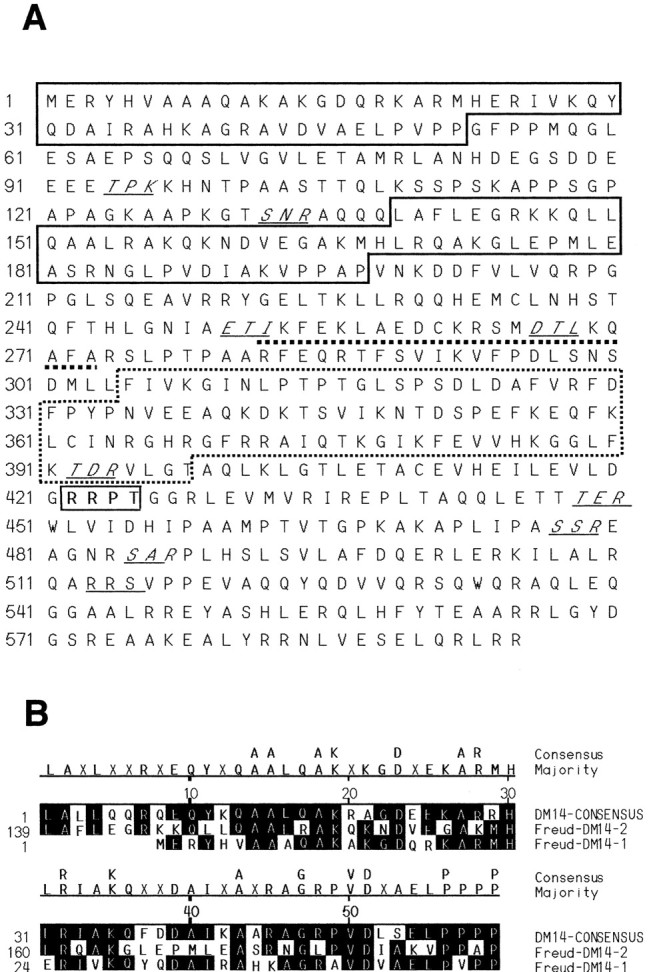
Protein structure of Freud-1. A, Predicted structural motifs in Freud-1 are shown. Indicated are consensus sequences for DM-14 (boxed) and C2 domains (dashed box), helix-loop-helix DNA binding domain (underlined), cAMP phosphorylation site (bold and outlined), and PKC phosphorylation sites (italic and underlined). B, Alignment of Freud-1 DM-14 sequence with consensus sequence.
Freud-1 binding to 5-HT1A FRE
We examined the DNA binding specificity of purified recombinant His-tagged Freud-1 protein to double-stranded primers of 5-HT1A repressor elements by EMSA (Fig. 2). A single specific species (band 1) was observed that was competed by unlabeled 5-HT1A-FRE or -DRE oligonucleotides (Fig. 2A, lanes 3, 5), indicating that Freud-1 binds specifically to FRE. Incubation of purified Freud-1 with an antibody against its N terminal super-shifted the protein-DRE complex, band 2 (Fig. 2A, lane 4), and this supershifted complex was partially disrupted by Freud-1 peptide antigen leading to reappearance of the original complex, band 1 (Fig. 2A, lane 6). Purified Freud-1 also bound as a single complex to labeled FRE that was competed by unlabeled FRE oligonucleotides but not by TRE primers (Fig. 2B). To address the role of the C2 domain in Freud-1, a deletion mutant (Freud-1 del) lacking amino acids 341-348 within the CalB domain was examined. Mutation of the C2 domain attenuated but did not abolish Freud-1 DNA binding activity, suggesting a role for this domain in Freud-1-DNA interaction (Fig. 2C). Thus recombinant Freud-1 binds specifically to the FRE of the 5-HT1A receptor gene.
Figure 2.

Specific binding of Freud-1 to 5-HT1A-FRE requires an intact CalB motif. The sequences of oligonucleotides for DRE (31 bp), FRE (14 bp), and TRE (12 bp) are indicated. A, Recombinant Freud-1-DRE binding was assessed by EMSA using 5-HT1A-DRE (31 bp) as a probe incubated with 4 μg of purified Freud-1 (lanes 1-6) or without (lane 7) as indicated. A single specific species (band 1) was retarded in the presence of 10 μg of anti-Freud-1 antibody (band 2, lane 4). The retarded complex was blocked by coincubation with 4 μg of peptide antigen (lane 6). Double-stranded unlabeled competitor oligonucleotides (at 100-fold molar excess) were included as indicated. FRE was sufficient to compete for Freud-1-DRE complex. B, Recombinant Freud-1-FRE binding: 4 μg of purified Freud-1 interacted specifically with FRE (band 1, lane 1). Freud-1-FRE interaction was abolished by preincubation with 100-fold unlabeled FRE (lane 2) but not by TRE (lane 3). C, Recombinant mutated Freud-1-DRE binding: EMSA using DRE as a probe incubated with 8 μg of purified Freud-1 with a disrupted CalB domain (Freud-1 del) alone (lane 1) or with unlabeled 14 bp FRE (lane 2) as indicated. A disrupted CalB motif reduced affinity of Freud-1 for the DRE. D, Presence of Freud-1 in nuclear extracts of L6 and RN46A cells. EMSA using the 31 bp DRE as a probe incubated without (lane 1) or with nuclear extracts from L6 (lanes 2, 3) or RN46A cells (lanes 4, 5) as indicated. The higher mobility complex (band 1) was displaced rather than retarded in the presence of anti-Freud-1 antibody in extracts from both cell lines (lanes 3 and 5, respectively). The second protein/DRE complex present in L6 cells (band 2) was not affected by Freud-1 antibody (lane 3) and represents an additional unknown repressor.
To determine whether Freud-1 is present in cells and can bind to the 5-HT1A DRE, EMSA was done using nuclear extracts (Fig. 2D). As we reported previously (Ou et al., 2000), in 5-HT1A-expressing raphe RN46A nuclear extracts only one protein-DRE complex was observed (band 1, lane 4), whereas in 5-HT1A receptor-negative L6 myoblasts, two complexes were present (lane 2). Nuclear extracts were preincubated with anti-Freud-1 antibody before EMSA to assess the presence of Freud-1 in protein-DRE complexes. Addition of anti-Freud-1 specifically displaced the lower FRE-specific complex (band 1) in both RN46A and L6 cells but did not affect the upper protein-TRE complex (band 2) present in L6 cells (Fig. 2D, lane 3). Previous studies showed that in 5-HT1A-negative cells such as L6 myoblasts, this second protein-DRE complex maintains repression of the 5-HT1A receptor gene after mutation of the FRE (Ou et al., 2000). Antibody alone did not bind the DRE probe (Fig. 2D, lane 6). In contrast to the supershift of recombinant Freud-1 by anti-Freud-1 (Fig. 2A), incubation with anti-Freud-1 inhibited DRE binding by the protein complex from nuclear extracts (Fig. 2D, lanes 3, 5), suggesting that anti-Freud-1 destabilizes the Freud-1 complex present in cell nuclei. These results indicate that Freud-1 is present in nuclear extracts and exhibits sequence-specific DNA binding to the 31 bp DRE.
Freud-1 repression of raphe 5-HT1A receptor expression
To test whether Freud-1 represses transcriptional activity of the rat 5-HT1A receptor gene, the Freud-1 expression plasmid was cotransfected with the DRE-containing -2300 or -1590 5-HT1A promoter-luciferase reporter constructs in RN46A and L6 cells (Fig. 3). In both cell types, Freud-1 decreased the transcriptional activity of the -2300 5-HT1A construct, but the effect was greater in the 5-HT1A-expressing raphe RN46A cells (Fig. 3A). Transrepression of the 5-HT1A promoter by Freud-1 was abolished by disruption of the CalB domain in the Freud-1 del mutant, although this mutant could still bind weakly to the DRE (Fig. 2C). Importantly, both Freud-1 and Freud-1 del were expressed at similar levels in L6 and RN46A cells as detected by Western blot (Fig. 3A, blot). Thus Freud-1 represses the 5-HT1A receptor gene, especially in raphe RN46A cells, and the CalB domain is required for this repression.
Figure 3.
Freud-1 represses basal 5-HT1A receptor expression in raphe RN46A cells via FRE. Luciferase activity was normalized to that of β-galactosidase and is expressed as relative light units and normalized to control. *p < 0.05 compared with control by t test. A, CalB-dependent Freud-1 repression. The FRE-containing -2300 5-HT1A-luciferase was transiently cotransfected in L6 myoblast or raphe RN46A cells with vector (pcDNA3, Control), Freud-1, or Freud-1 del mutant of the C2 (CalB) domain. Above, A Western blot of 30 μg of nuclear extracts was probed with anti-Flag to show equal expression of Flag-Freud-1 or Flag-Freud-1 del in these experiments. B, FRE dependence of Freud-1 repression. Transfections as above were done with FRE-inactivated mutant 2300m1, which was insensitive to Freud-1. C, FRE-dependent repression of SV40 promoter by Freud-1. The 31 bp DRE (DRE/SV40) or the FRE-mutant DRE (DREmut/SV40) was placed upstream of the SV40 promoter and cotransfected with vector (pcDNA3) or Freud-1 expression construct. D, E, Freud-1 inhibits transcriptional activity of the 5-HT1A receptor gene. L6 myoblast or raphe RN46A cells were transiently cotransfected with the DRE-containing (-1590luc) 5-HT1A-luciferase reporter or vector (pcDNA3, Control), and sense (D) or antisense (E) Freud-1 expression vectors. Freud-1 protein expression in each cell line after transfection was detected by Western blot using specific anti-Freud antibody (shown above). β-Actin immunoreactivity was tested to confirm equal loading. F, G, Freud-1 protein inhibits 5-HT1A receptor expression in raphe RN46A cells. F, Proteins prepared from RN46A cells transfected with Freud-1 sense or antisense expression vectors and differentiated for 72 hr were subjected to anti-5-HT1A receptor immunoblotting. β-Actin immunoreactivity was tested to confirm equal loading. Shown is a representative blot of three independent experiments. The relative intensity was quantified by an Automated Digitizing System (UN-SCAN-IT, Silk Scientific Inc.). Data represent the mean ± SE of three independent experiments. G, RN46A cells cotransfected with 1 μg of Red Fluorescent Protein (DsRed) and 5 μg of Freud-1 sense or antisense expression plasmids as indicated were stained for 5-HT1A immunoreactivity (see Materials and Methods). Arrows indicate representative DsRed-positive cells that express either high or low 5-HT1A immunoreactivity (for antisense or sense Freud-1, respectively) compared with cells not transfected with DsRed.
The importance of an intact FRE for Freud-1-induced repression was examined using the -2300m1 and DREmut/SV40 constructs containing mutations that specifically inactivate the FRE, while the downstream TRE remains intact (Ou et al., 2000). In contrast to the -2300 construct, transfection of Freud-1 did not affect the luciferase activity of the -2300m1 construct in either L6 or RN46A cells (Fig. 3B). The DRE/SV40 construct contains the 5-HT1A DRE upstream of a heterologous promoter (SV40) and mediates repression in RN46A cells (Ou et al., 2000). Cotransfection of Freud-1 induced a further 50% repression of this construct (Fig. 3C, DRE/SV40), but Freud-1 had no effect when the FRE was inactivated (DREmut/SV40). These results demonstrate that Freud-1 requires an intact FRE to mediate repression of the 5-HT1A or heterologous promoters.
To further study its importance in transcriptional regulation of the 5-HT1A receptor gene, Freud-1 protein expression was modulated using sense and antisense Freud-1 cDNA constructs (Fig. 3D-G). On cotransfection with sense Freud-1, transcriptional activity of the DRE-containing -1590 5-HT1A promoter-luciferase construct was significantly decreased compared with control in both L6 and RN46A cells (by 25 or 50%, respectively), as observed for the larger -2300 5-HT1A construct (Fig. 3D). The DRE-deficient -1519-luciferase construct lacked Freud-1-induced repression (data not shown). Transfection of Freud-1 plasmid increased Freud-1 protein content by 1.8- and 2.2-fold in L6 and RN46A cells, respectively (Fig. 3D, blot). The greater repression by Freud-1 in RN46A cells may reflect the importance of the single Freud-1-DRE complex in these cells compared with two protein-DRE complexes in L6 (Fig. 2D) and other nonneuronal cells (Ou et al., 2000). Cotransfection of an antisense Freud-1 cDNA construct depleted endogenous Freud-1 protein levels in L6 and RN46A by approximately the same extent (50 vs 55%) (Fig. 3E, blot). Cotransfection of antisense Freud-1 derepressed the 5-HT1A promoter construct only in RN46A cells, indicating that endogenous Freud-1 plays a role in basal regulation of the 5-HT1A receptor gene in these neuronal cells.
To address the role of Freud-1 in regulating receptor expression, we measured 5-HT1A receptor levels in raphe RN46A cells transiently transfected with sense or antisense Freud-1 cDNA constructs (Fig. 3F). The level of endogenous 5-HT1A receptor protein in differentiated RN46A cells was decreased significantly after transient transfection with sense Freud-1, but increased with antisense to Freud-1, parallel to the changes observed in 5-HT1A gene transcription. Because the transfection efficiency in these experiments was 30% (see Materials and Methods), alterations in Freud-1 and 5-HT1A receptor expression would occur in a maximum of only 30% of cells. To examine changes specifically in transfected cells, cotransfection of sense or antisense Freud-1 constructs was done with a DsRed construct as a fluorescent marker for transfected cells (Fig. 3G). DsRed fluorescence filled the transfected cells consistent with its cytosolic localization, whereas 5-HT1A staining was strongest in perinuclear regions, perhaps reflecting internalized receptors. On transfection of sense Freud-1, decreased 5-HT1A immunostaining was observed in DsRed-positive cells relative to DsRed-negative cells, whereas anti-sense to Freud-1 led to increased 5-HT1A receptor staining in DsRed-labeled cells. Although dramatic effects were seen in a majority of DsRed-positive cells, immunofluorescence represents a qualitative method that complements the quantitative findings observed using Western blot (Fig. 3F). These experiments indicate that Freud-1 represses 5-HT1A gene transcription to decrease 5-HT1A protein expression in raphe RN46A cells.
Inactivation of Freud-1 by calcium/ATP
Because the CalB domain is thought confer Ca2+-dependent lipid binding, we examined the action of calcium and ATP on protein-DRE binding activity in nuclear extracts from L6 and RN46A cells by EMSA. On addition of Ca2+ and ATP, the specific interaction of the Freud-1-containing nuclear protein-DRE complex (band 1) was decreased in RN46A or abolished in L6 extracts (Fig. 4A). By contrast, the protein-TRE complex from L6 cells (band 2) was sensitive to ATP alone. Thus the combination of calcium and ATP specifically interferes with Freud-1 binding to DNA. The role of calcium-dependent phosphorylation in Ca2+/ATP-mediated inhibition of the Freud-1-DRE interaction was assessed by addition of protein kinase inhibitors (Fig. 4B,C). The inhibitors of CAMK (KN93) and CAM [calmidazolium (CMZ)] blocked the inhibitory action of Ca2+/ATP on Freud-1-DRE binding, leading to a recovery of the Freud-1-DRE complex (Fig. 4B, lanes 4, 7). In L6 cells, these inhibitors specifically rescued the Freud-1-DRE complex (Fig. 4C, band 1), whereas the second protein-TRE complex (band 2) was not rescued. However, the inactive analog of KN93 (KN92), PKC inhibitor Calphostin C (CalC), or PKA inhibitor H-8 failed to influence Freud 1-DNA binding. These results suggest that CAMKIV (the predominant nuclear CAMK), but not PKC or PKA, mediated the inhibitory action of calcium/ATP on Freud-1 interaction with FRE.
Figure 4.

CAM kinase attenuates Freud-1-mediated repression. A, Ca2+ and ATP in combination interfere with Freud-1/DRE interaction. EMSA using the 31 bp DRE as a probe incubated with nuclear extracts from RN46A (lanes 1-4) or L6 cells (lanes 5-8) as indicated. CaCl2 (lanes 2, 6), ATP (lanes 3, 7), or both (lanes 4, 8) were added into incubation buffer for 20 min before incubation with DRE probe. B, C, CAM kinase activation in nuclear extracts interferes with Freud-1-DRE binding. EMSA using DRE as a probe and nuclear extracts from RN46A cells (B) and L6 cells (C) are shown. Both CaCl2 and ATP were added into the incubation before adding probe (lanes 2-7). Treatment was with 10 μm (except 50 nm CalC) KN92 (negative control for KN93; lane 3), KN93 (CAM kinase inhibitor; lane 4), CalC (PKC inhibitor; lane 5), H-8 (PKA inhibitor, lane 6), and CMZ (CAM inhibitor; lane 7), as indicated. In L6 cells (C), CAM or CAMK inhibitors rescued only the Freud-1-containing species (band 1). D, Calcium signaling enhances 5-HT1A promoter activity in a Freud-1-FRE-dependent manner. 5-HT1A promoter-luciferase reporter constructs containing FRE (-1590 or -2300) or lacking FRE (-1519, -2300m1) were transfected into RN46A cells. Twenty-four hours after transfection, the cells were treated with 40 mm KCl or with 40 mm KCl and 1 μm ionomycin without or with 10 μm KN92, KN93, or CMZ in the medium for 16 hr. Luciferase activity is expressed as relative light units normalized to control (untreated) samples. *p < 0.05 in comparison with control. Note that calcium mobilizing agents had no effect in FRE-lacking reporter constructs (-1519 or -2300m1) and were blocked by CAM or CAMK inhibitors.
We tested whether calcium-mediated inhibition of Freud-1 binding to FRE can activate 5-HT1A receptor gene transcription. RN46A cells were treated with agents to increase [Ca2+]i, and the activity of FRE-containing (-1590- and -2300-luciferase), FRE-deleted (-1519-luciferase), or FRE-inactivated (-2300m1) 5-HT1A reporter constructs was examined (Fig. 4D). After incubation with 40 mm KCl to induce calcium influx, the luciferase activity of -1590 and -2300, but not the -1519 or -2300m1 constructs, was increased significantly. Co-addition of ionomycin (1 μm), a calcium ionophore that induces calcium influx and releases calcium stores (Albert and Tashjian, 1986; Szonyi et al., 2001), further enhanced the luciferase activity of the -1590 and -2300 5-HT1A promoter constructs. Thus calcium signaling enhances 5-HT1A receptor transcription in raphe cells via a Freud-1-FRE-dependent mechanism. Inhibitors KN93 and CMZ, but not the inactive analog KN92, blocked calcium-mediated induction of the 5-HT1A promoter constructs, indicating a specific role for CAMK and CAM. Thus, calcium/CAM-dependent activation of CAMK inhibits Freud-1 function, resulting in enhanced transcriptional activity of the 5-HT1A receptor gene.
Freud-1 RNA and protein expression in brain
The role of Freud-1 in vivo was addressed initially by examining the expression profile of Freud-1 mRNA by Northern blot analysis of rat tissues and brain regions (Fig. 5). A single 3.8 kb mRNA species was strongly expressed in several rat brain areas, including frontal cortex, cortex, mesencephalon, hypothalamus, hippocampus, and midbrain. Freud-1 RNA was also detected in peripheral tissues such as testis and at low levels in pituitary, liver, and kidney. Freud-1 RNA and protein expression in rat brain were analyzed further by in situ hybridization and immunohistochemistry (Fig. 6A-C). Hybridization of antisense (a′-c′) versus sense (a-c) Freud-1 oligonucleotide probes demonstrated specific Freud-1 mRNA expression that corresponded with regions of strong Freud-1 immunoreactivity (a″-c″). The highest levels of Freud-1 RNA were detected in hippocampal pyramidal cells (Fig. 6B) and raphe nuclei [including dorsal, medial (Fig. 6A), and magnus (data not shown)]. Discrete subsets of cells were labeled intensely in the hypothalamus and cortex, with weaker hybridization in the thalamus. In pyramidal cells of hippocampal CA1, CA2, CA3, and dentate gyrus regions, Freud-1 RNA was present in the cell body and proximal dendrites, whereas Freud-1-like immunoreactivity was predominant in the nucleus (Fig. 6B). A few pyramidal cells of the prefrontal cortex also displayed intense Freud-1 RNA staining and immunoreactivity (Fig. 6C). Freud-1 was also strongly expressed in cells of the dorsal and intermediate nuclei of the lateral septum, medial septum, and hypothalamus (data not shown). Each of these brain areas that express Freud-1 RNA and protein also express 5-HT1A receptor RNA (Pompeiano et al., 1992), consistent with a role for Freud-1 to regulate the 5-HT1A gene in vivo.
Figure 5.
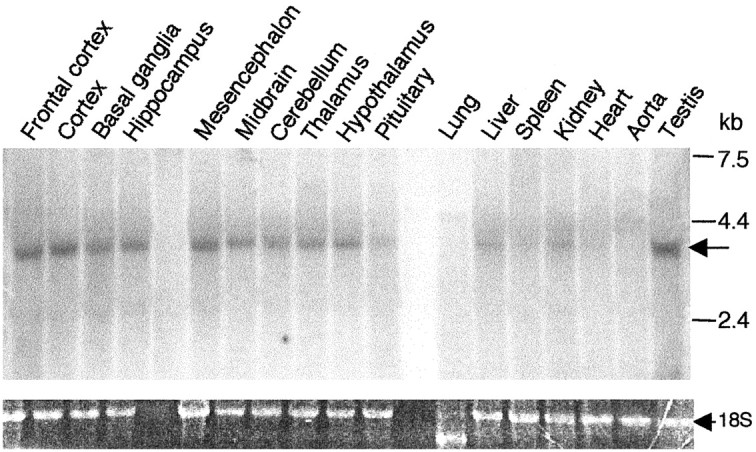
Tissue distribution of Freud-1 RNA expression. Freud-1 mRNA expression in rat tissues. RNA prepared from 17 different rat tissues was used for Northern blot analysis. The position of molecular mass markers is shown on the right. An arrow indicates hybridization to Freud-1 probe. Ethidium bromide-stained 18S RNA was used as a loading control.
Figure 6.
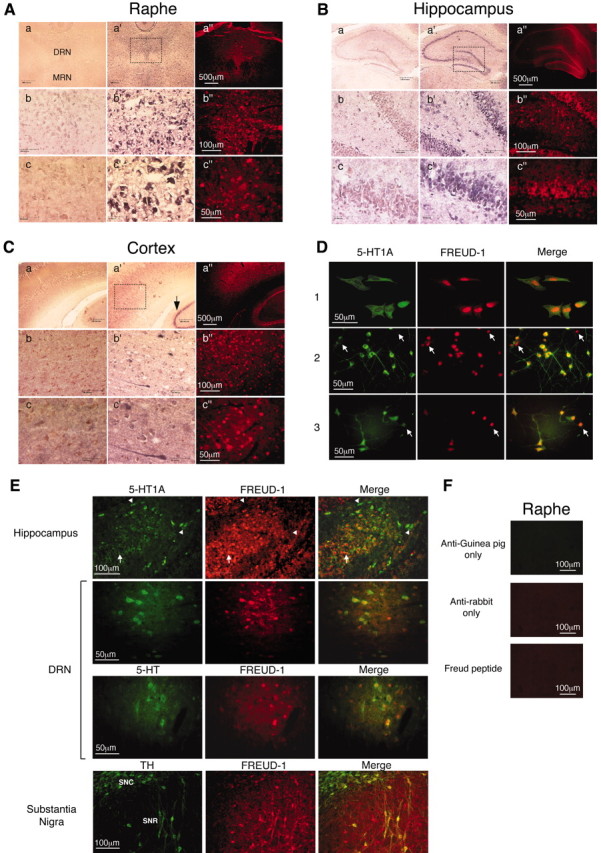
Colocalization of Freud-1 with 5-HT1A receptor. A-C, Coronal brain sections were hybridized with sense (a-c) or antisense (a′-c′) digoxigenin-labeled Freud-1 oligonucleotides and stained as described in Materials and Methods or were assayed for Freud-1 immunoreactivity (a″-c″). Boxed regions are displayed at successively higher magnification in a-c, a′-c′, and a″-c″. Scale bars: a, a′, a″, 500 μm; b, b′, b″, 100 μm; c, c′, c″, 50 μm. Freud-1 staining was observed in the following regions. A, Raphe nuclei. Staining was most intense in cells of the dorsal raphe nucleus (DRN) but also present in the medial raphe nucleus (MRN) and raphe magnus (data not shown). The dorsal raphe nucleus is magnified showing cytoplasmic Freud-1 RNA and nuclear Freud-1 protein. B, Hippocampus. Staining is prominent in pyramidal cells of CA1, CA3, and the dentate gyrus. Dentate gyrus cells are magnified to show pyramidal cell morphology. C, Primary sensory cortex. The arrow indicates hippocampus for comparison. D, Nuclear localization of Freud-1. Colocalization of Freud-1 (red) and 5-HT1A receptors (green) in raphe RN46A cells (1), and primary cultures of embryonic cortical (2) and hippocampal (3) cells. Arrows indicate cells expressing high levels of Freud-1 and low levels of 5-HT1A receptor. E, Colocalization studies. Dual immunofluorescence was used to detect colocalization of Freud-1 with various markers. Freud-1 was colocalized with 5-HT1A receptor staining in sections from hippocampus and dorsal raphe nucleus (DRN), as indicated. In DRN, Freud-1 was also colocalized with staining for 5-HT, a marker of serotonergic neurons. In the substantia nigra pars compacta (SNC) or especially pars reticulata (SNR), Freud-1 was colocalized with TH, a marker for dopaminergic neurons. Arrowheads indicate cells expressing low Freud-1 and high 5-HT1A receptor levels. F, Specificity of 5-HT1A and Freud-1 staining. Raphe tissue sections that were incubated with anti-guinea pig or anti-rabbit secondary antibodies alone or in the presence of 100 μg of Freud-1 peptide antigen displayed only background immunoreactivity.
To address directly whether Freud-1 protein is expressed in 5-HT1A receptor-positive cells, dual immunofluorescent staining of cells in culture and brain sections was done. In RN46A cells as well as primary hippocampal and cortical cultures (Fig. 6D), Freud-1 protein was present in the nucleus of cells expressing 5-HT1A receptors in the cell body and processes. Especially in the cortical cultures, some cells expressed Freud-1 but not 5-HT1A receptors (arrows); however, the majority of 5-HT1A-expressing cells also expressed Freud-1. In tissue sections (Fig. 6E), Freud-1 staining was predominantly nuclear and was colocalized by dual immunofluorescence with extra-nuclear staining for 5-HT1A receptors [in hippocampus and dorsal raphe nucleus (DRN)], 5-HT (DRN), or TH (substantia nigra). In the raphe, Freud-1 staining was weaker than in the hippocampus and was colocalized primarily with 5-HT1A receptors and 5-HT staining, suggesting its presence in 5-HT1A-positive serotonergic neurons. Interestingly, some cells that stained weakly for Freud-1 displayed strong 5-HT1A staining (Fig. 6D,E, arrowheads), and vice-versa (arrows), suggesting that Freud-1 protein inhibits 5-HT1A receptor expression in vivo, as we observed after transfection of sense or antisense Freud-1 in RN46A cells (Fig. 3G). Staining for Freud-1 in substantia nigra was strongest in TH-positive cells of the pars reticulata, suggesting a role in a subset of dopaminergic neurons. In control studies, fixed sections and cells that were incubated with primary or secondary antibody alone or preincubated with Freud-1 peptide antigen displayed only background immunoreactivity, demonstrating the specificity of Freud-1 staining (Fig. 6F) (data not shown). Colocalization of Freud-1 and 5-HT1A receptors in presynaptic and postsynaptic neurons of the serotonin system is consistent with a role for Freud-1 as a repressor to negatively regulate 5-HT1A gene expression in vivo.
Discussion
Freud-1: a novel repressor of the 5-HT1A receptor gene expressed in brain
Previous studies have suggested the importance of regulation of 5-HT1A receptor expression in mental illnesses such as anxiety and major depression and in suicide (Albert et al., 1996; Mann, 1999; Pineyro and Blier, 1999; Artigas et al., 2001). Antidepressant compounds target the serotonin system, and chronic down-regulation of the somatodendritic 5-HT1A autoreceptor on raphe neurons has been proposed as an obligatory step to their clinical efficacy. Conversely, genetic oblation of the 5-HT1A receptor results in increased anxiety behaviors in three different strains of mice (Saudou et al., 1994; Heisler et al., 1998; Parks et al., 1998; Ramboz et al., 1998). Importantly, in postmortem brains from depressed suicides, the basal level of 5-HT1A receptor expression was increased specifically in the raphe nuclei, suggesting that altered regulation of the 5-HT1A autoreceptor may predispose subjects to depression or suicide (Stockmeier et al., 1998). Thus, we have studied the transcriptional mechanisms of 5-HT1A receptor regulation in raphe cells and identified the neuronal repressor element FRE (Ou et al., 2000). We have now used the yeast one-hybrid approach to identify a novel protein, Freud-1, that binds to the FRE site of the 5-HT1A promoter to repress 5-HT1A receptor expression.
Several pieces of evidence indicate that Freud-1 acts as a neuronal repressor of the 5-HT1A gene by binding to the FRE site. First, the two independent Freud-1 clones were isolated on the basis of the trans activation of the DRE in the yeast system. Second, recombinant purified His-tagged Freud-1 bound specifically to the 5′ FRE 14 bp segment of the DRE, and this complex was retarded by a specific antibody directed against Freud-1. Additionally, in RN46A or L6 nuclear extracts, the formation of the FRE-protein complex was inhibited by this antibody, indicating the presence of Freud-1 in the complex. Third, Freud-1 repressed various 5-HT1A or heterologous promoter-luciferase constructs in a FRE-dependent manner in raphe RN46A cells to a greater extent than in non-neuronal L6 cells, suggesting a greater role for Freud-1 in raphe cells. In combination with other repressors such as the TRE-specific protein complex, Freud-1 mediates silencing of the gene in non-neuronal cells (Ou et al., 2000). Depletion of Freud-1 protein using an antisense construct derepressed the 5-HT1A receptor gene in RN46A cells but not L6 cells, suggesting that Freud-1 is a crucial regulator of this gene in raphe cells. Fourth, Freud-1 expression decreased 5-HT1A receptor protein levels whereas antisense to Freud-1 increased receptor expression in RN46A cells, indicating that Freud-1 is a key repressor of the 5-HT1A receptor in raphe cells. Thus Freud-1 mediates neuronal repression of the 5-HT1A receptor gene by binding to the FRE.
The subcellular and brain regional distribution of Freud-1 further supports its role in neuronal regulation of the 5-HT1A receptor gene. The colocalization of Freud-1 with both 5-HT and the 5-HT1A receptor in raphe nuclei strongly suggest that Freud-1 is involved in regulation of the expression of somatodendritic 5-HT1A autoreceptor on serotonergic neurons. Interestingly, Freud-1 was also expressed in TH-positive cells of the substantia nigra, not known to express 5-HT1A receptors, suggesting other roles for Freud-1 in addition to regulation of the 5-HT1A receptor. The substantia nigra is known to express the dopamine-D2 receptor, and an FRE-like sequence is present in the second intron of the human D2 receptor gene, suggesting that D2 receptors could be an additional target for regulation by Freud-like proteins.
The presence of a human Freud-1 homolog suggests a role in regulation of the human 5-HT1A receptor. Within a strong repressor region of the human 5-HT1A gene located between -1624/-1570 bp we have identified two adjacent functional DREs with 80% identity to the rat DRE (S. Lemonde and P. R. Albert, unpublished observations). The human DRE binds to Freud-1 and mediates repression of the human 5-HT1A gene, suggesting a role for Freud-1 in humans. For example, altered regulation of Freud-1 in the raphe nuclei could mediate the increase in 5-HT1A autoreceptors observed in depressed suicide victims (Stockmeier et al., 1998), perhaps implicating Freud-1 in predisposition to suicide or depression. Expression of Freud-1 in serotonergic and 5-HT1A-positive neurons of the raphe nuclei suggests that Freud-1 could play a role in regulating human 5-HT1A autoreceptor expression in vivo.
Structural domains and calcium-dependent regulation of Freud-1
Freud-1 represents a novel DNA binding protein with no significant homology to other families of known DNA binding proteins. All Freud-1 species homologs contain multiple repeats of the basic DM-14 domain, a predicted HLH domain, and a conserved C2 domain. By analogy with known basic HLH transcription factors, the HLH domain of Freud-1 may mediate DNA binding in combination with the basic DM-14 repeats. The nuclear localization of Freud-1 could also be mediated in part by the basic DM-14 domains, given the lack of a consensus nuclear localization sequence. Deletion of the first nine amino acids of the C2 domain reduced DNA binding of Freud-1 and abolished its repressor activity, suggesting its role in DNA binding but especially in repressor function of Freud-1. Interestingly, the calcium-independent C2 domain of phosphatase and tensin homolog (PTEN) mediates protein-protein interaction with the tumor suppressor p53 (Freeman et al., 2003). By analogy the C2 domain of Freud-1 may also interact with nuclear proteins to localize Freud-1 to the nucleus or to recruit co-repressors. The Freud-1 C2 domain may be calcium insensitive because calcium alone did not alter Freud-1 function, and the Freud-1 C2 domain lacks several acidic residues that mediate calcium binding of the PKC C2 domain. In addition, the Freud-1 C2 domain contains a poly-basic insert (five of eight basic residues between residues 365 and 372) that is not present in calcium-dependent C2 domains and may function as a nuclear localization signal as reported for the double-C2 γ protein (Fukuda et al., 2001). Further experiments will be required to identify the precise function of the Freud-1 C2 domain.
We found that although not directly regulated by calcium, Ca2+-ATP inactivated Freud-1 binding to DNA in nuclear extracts. Pharmacological analysis of calcium-mediated inhibition of Freud-1 implicated CAMKII or more likely CAMKIV, which is located predominantly in the nucleus and known to mediate transcriptional activation associated with long-term memory (Soderling, 2000; Kasahara et al., 2001; Impey et al., 2002). It remains unclear whether CAMK directly phosphorylates Freud-1 or phosphorylates an associated protein to inactivate Freud-1 function. Unlike repressors that are inhibited directly by calcium binding such as DREAM (Carrion et al., 1999; Mellstrom and Naranjo, 2001; Cheng et al., 2002), Freud-1 provides a new example of a repressor that is inactivated by CAMK-mediated phosphorylation. Consistent with this, calcium mobilization by high K+ depolarization and calcium ionophore increased 5-HT1A transcriptional activity in RN46A cells. Calcium signaling in raphe neurons can be initiated via glutamatergic input, which activates NMDA receptors to enhance calcium-dependent 5-HT release (Celada et al., 2001; Lee et al., 2003). Conversely, activation of 5-HT1A autoreceptors decreases [Ca2+]i (Chen and Penington, 1996; Bayliss et al., 1997a,b), which would activate Freud-1 to repress the 5-HT1A receptor gene. Activation of 5-HT1A autoreceptors by antidepressant treatment could use calcium- and Freud-1-dependent mechanisms to desensitize the 5-HT1A autoreceptor. Thus, Freud-1 can regulate 5-HT1A receptor transcription in two ways: the level of Freud-1 protein negatively regulates basal 5-HT1A expression, whereas CAMK-mediated signaling enhances 5-HT1A transcription by inactivation of Freud-1. Thus, either Freud-1 protein levels or calcium-mediated signaling determines Freud-1 activity to regulate 5-HT1A receptor expression.
It should be emphasized that Freud-1 is one of several transcription factors that regulate 5-HT1A receptor expression, including enhancers such as Sp1/MAZ (Parks and Shenk, 1996), PET-1 (Hendricks et al., 1999), and nuclear factor κB (NFκB) (Abdouh et al., 2001), which positively regulate the gene. Other negative regulators include the putative TRE-binding protein (Ou et al., 2000), the neural restriction silencing factor REST/ NRSF (Lemonde and Albert, unpublished observations), and a novel protein complex that recognizes a palindrome DNA sequence in the repressor region of the 5-HT1A gene (Lemonde and Albert, unpublished observations). In addition, a negative mineralocorticoid response element confers inhibitory regulation of 5-HT1A transcription by glucocorticoids (Ou et al., 2001). However, Freud-1 appears to play an important role in neuronal regulation of the 5-HT1A receptor, whereas factors like REST or TRE-binding proteins appear more important to silence the gene in non-neuronal tissues. The recent finding that PET-1 regulates the differentiation of precursor raphe cells to a serotonergic phenotype (Hendricks et al., 2003) suggests that in raphe cells Freud-1 and PET-1 may coordinately regulate 5-HT1A receptor expression, whereas in non-serotonergic neurons, Freud-1 may coordinate with other as yet unknown factors. Thus, the transcription factors that underlie the tissue-specific and regulated expression of 5-HT1A receptors may include, but are not limited to, Freud-1.
In summary, Freud-1 is a novel transcription factor that negatively regulates the basal expression of the 5-HT1A receptor gene in neuronal cells. The importance of Freud-1 in repressing basal 5-HT1A transcription and receptor levels in raphe cells suggests a potential role in altered regulation of 5-HT1A receptors in disorders such as anxiety or major depression.
Footnotes
This research was supported by Canadian Institutes of Health Research (CIHR). X.O. was supported by a postdoctoral fellowship from the Ontario Mental Health Foundation and is a Young Investigator Awardee of the National Alliance for Research on Schizophrenia and Depression. S.L. is supported by a CIHR Studentship; P.R.A. holds the Novartis/CIHR Michael Smith Chair in Neurosciences. We thank Dr. William Staines for assistance with immunohistochemistry.
Correspondence should be addressed to Paul R. Albert, Ottawa Health Research Institute, Neuroscience, University of Ottawa, 451 Smyth Road, Ottawa, Ontario, Canada K1H-8M5. E-mail: palbert@uottawa.ca.
X.-M. Ou's current address: Department of Molecular Pharmacology and Toxicology, University of Southern California, Los Angeles, CA 90089-9121.
H. Jafar-Nejad's current address: Baylor College of Medicine, One Baylor Plaza, Houston, TX 77030.
Copyright © 2003 Society for Neuroscience 0270-6474/03/237415-11$15.00/0
X.O. and S.L. contributed equally to this work.
References
- Abdouh M, Storring JM, Riad M, Paquette Y, Albert PR, Drobetsky E, Kouassi E ( 2001) Transcriptional mechanisms for induction of 5-HT1A receptor mRNA and protein in activated B and T lymphocytes. J Biol Chem 276: 4382-4388. [DOI] [PubMed] [Google Scholar]
- Albert PR, Tashjian Jr AH ( 1986) Ionomycin acts as an ionophore to release TRH-regulated Ca 2+ stores from GH4C1 cells. Am J Physiol 251: C887-891. [DOI] [PubMed] [Google Scholar]
- Albert PR, Zhou QY, Van Tol HH, Bunzow JR, Civelli O ( 1990) Cloning, functional expression, and mRNA tissue distribution of the rat 5-hydroxy-tryptamine1A receptor gene. J Biol Chem 265: 5825-5832. [PubMed] [Google Scholar]
- Albert PR, Lembo P, Storring JM, Charest A, Saucier C ( 1996) The 5-HT1A receptor: signaling, desensitization, and gene transcription. Neuropsychopharmacology 14: 19-25. [DOI] [PubMed] [Google Scholar]
- Albert PR, Sajedi N, Lemonde S, Ghahremani MH ( 1999) Constitutive G(i2)-dependent activation of adenylyl cyclase type II by the 5-HT1A receptor. Inhibition by anxiolytic partial agonists. J Biol Chem 274: 35469-35474. [DOI] [PubMed] [Google Scholar]
- Artigas F, Celada P, Laruelle M, Adell A ( 2001) How does pindolol improve antidepressant action? Trends Pharmacol Sci 22: 224-228. [DOI] [PubMed] [Google Scholar]
- Ase AR, Reader TA, Hen R, Riad M, Descarries L ( 2000) Altered serotonin and dopamine metabolism in the CNS of serotonin 5-HT(1A) or 5-HT(1B) receptor knockout mice. J Neurochem 75: 2415-2426. [DOI] [PubMed] [Google Scholar]
- Banker GA, Cowan WM ( 1977) Rat hippocampal neurons in dispersed cell culture. Brain Res 126: 397-442. [DOI] [PubMed] [Google Scholar]
- Bayliss DA, Li YW, Talley EM ( 1997b) Effects of serotonin on caudal raphe neurons: activation of an inwardly rectifying potassium conductance. J Neurophysiol 77: 1349-1361. [DOI] [PubMed] [Google Scholar]
- Bayliss DA, Li YW, Talley EM ( 1997a) Effects of serotonin on caudal raphe neurons: inhibition of N- and P/Q-type calcium channels and the after-hyperpolarization. J Neurophysiol 77: 1362-1374. [DOI] [PubMed] [Google Scholar]
- Brewer GJ, Torricelli JR, Evege EK, Price PJ ( 1993) Optimized survival of hippocampal neurons in B27-supplemented neurobasal, a new serum-free medium combination. J Neurosci Res 35: 567-576. [DOI] [PubMed] [Google Scholar]
- Carrion AM, Link WA, Ledo F, Mellstrom B, Naranjo JR ( 1999) DREAM is a Ca 2+-regulated transcriptional repressor. Nature 398: 80-84. [DOI] [PubMed] [Google Scholar]
- Celada P, Puig MV, Casanovas JM, Guillazo G, Artigas F ( 2001) Control of dorsal raphe serotonergic neurons by the medial prefrontal cortex: involvement of serotonin-1A, GABA(A), and glutamate receptors. J Neurosci 21: 9917-9929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Penington NJ ( 1996) Differential effects of protein kinase C activation on 5-HT1A receptor coupling to Ca 2+ and K+ currents in rat serotonergic neurones. J Physiol (Lond) 496: 129-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng HY, Pitcher GM, Laviolette SR, Whishaw IQ, Tong KI, Kockeritz LK, Wada T, Joza NA, Crackower M, Goncalves J, Sarosi I, Woodgett JR, Oliveira-dos-Santos AJ, Ikura M, van der Kooy D, Salter MW, Penninger JM ( 2002) DREAM is a critical transcriptional repressor for pain modulation. Cell 108: 31-43. [DOI] [PubMed] [Google Scholar]
- Clark JD, Lin LL, Kriz RW, Ramesha CS, Sultzman LA, Lin AY, Milona N, Knopf JL ( 1991) A novel arachidonic acid-selective cytosolic PLA2 contains a Ca(2+)-dependent translocation domain with homology to PKC and GAP. Cell 65: 1043-1051. [DOI] [PubMed] [Google Scholar]
- Eaton MJ, Staley JK, Globus MY, Whittemore SR ( 1995) Developmental regulation of early serotonergic neuronal differentiation: the role of brain-derived neurotrophic factor and membrane depolarization. Dev Biol 170: 169-182. [DOI] [PubMed] [Google Scholar]
- Freeman DJ, Li AG, Wei G, Li HH, Kertesz N, Lesche R, Whale AD, Martinez-Diaz H, Rozengurt N, Cardiff RD, Liu X, Wu H ( 2003) PTEN tumor suppressor regulates p53 protein levels and activity through phosphatase-dependent and -independent mechanisms. Cancer Cell 3: 117-130. [DOI] [PubMed] [Google Scholar]
- Fukuda M, Saegusa C, Kanno E, Mikoshiba K ( 2001) The C2A domain of double C2 protein gamma contains a functional nuclear localization signal. J Biol Chem 276: 24441-24444. [DOI] [PubMed] [Google Scholar]
- Ghahremani MH, Cheng P, Lembo PM, Albert PR ( 1999) Distinct roles for Galphai2, Galphai3, and Gbeta gamma in modulation of forskolin- or Gs-mediated cAMP accumulation and calcium mobilization by dopamine D2S receptors. J Biol Chem 274: 9238-9245. [DOI] [PubMed] [Google Scholar]
- Gispen WH, Schotman P, de Kloet ER ( 1972) Brain RNA and hypophysectomy: a topographical study. Neuroendocrinology 9: 285-296. [DOI] [PubMed] [Google Scholar]
- Heisler LK, Chu HM, Brennan TJ, Danao JA, Bajwa P, Parsons LH, Tecott LH ( 1998) Elevated anxiety and antidepressant-like responses in serotonin 5-HT1A. Proc Natl Acad Sci USA 95: 15049-15054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendricks T, Francis N, Fyodorov D, Deneris ES ( 1999) The ETS domain factor Pet-1 is an early and precise marker of central serotonin neurons and interacts with a conserved element in serotonergic genes. J Neurosci 19: 10348-10356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendricks TJ, Fyodorov DV, Wegman LJ, Lelutiu NB, Pehek EA, Yamamoto B, Silver J, Weeber EJ, Sweatt JD, Deneris ES ( 2003) Pet-1 ETS gene plays a critical role in 5-HT neuron development and is required for normal anxiety-like and aggressive behavior. Neuron 37: 233-247. [DOI] [PubMed] [Google Scholar]
- Impey S, Fong AL, Wang Y, Cardinaux JR, Fass DM, Obrietan K, Wayman GA, Storm DR, Soderling TR, Goodman RH ( 2002) Phosphorylation of CBP mediates transcriptional activation by neural activity and CaM kinase IV. Neuron 34: 235-244. [DOI] [PubMed] [Google Scholar]
- Kasahara J, Fukunaga K, Miyamoto E ( 2001) Activation of calcium/ calmodulin-dependent protein kinase IV in long term potentiation in the rat hippocampal CA1 region. J Biol Chem 276: 24044-24050. [DOI] [PubMed] [Google Scholar]
- Lee HS, Kim MA, Valentino RJ, Waterhouse BD ( 2003) Glutamatergic afferent projections to the dorsal raphe nucleus of the rat. Brain Res 963: 57-71. [DOI] [PubMed] [Google Scholar]
- Mann JJ ( 1999) Role of the serotonergic system in the pathogenesis of major depression and suicidal behavior. Neuropsychopharmacology 21: 99S-105S. [DOI] [PubMed] [Google Scholar]
- Mellstrom B, Naranjo JR ( 2001) Mechanisms of Ca(2+)-dependent transcription. Curr Opin Neurobiol 11: 312-319. [DOI] [PubMed] [Google Scholar]
- Ou XM, Jafar-Nejad H, Storring JM, Meng JH, Lemonde S, Albert PR ( 2000) Novel dual repressor elements for neuronal cell-specific transcription of the rat 5-HT1A receptor gene. J Biol Chem 275: 8161-8168. [DOI] [PubMed] [Google Scholar]
- Ou XM, Storring JM, Kushwaha N, Albert PR ( 2001) Heterodimerization of mineralocorticoid and glucocorticoid receptors at a novel negative response element of the 5-HT1A receptor gene. J Biol Chem 276: 14299-14307. [DOI] [PubMed] [Google Scholar]
- Parks CL, Shenk T ( 1996) The serotonin 1a receptor gene contains a TATA-less promoter that responds to MAZ and Sp1. J Biol Chem 271: 4417-4430. [DOI] [PubMed] [Google Scholar]
- Parks CL, Robinson PS, Sibille E, Shenk T, Toth M ( 1998) Increased anxiety of mice lacking the serotonin1A receptor. Proc Natl Acad Sci USA 95: 10734-10739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pineyro G, Blier P ( 1999) Autoregulation of serotonin neurons: role in antidepressant drug action. Pharmacol Rev 51: 533-591. [PubMed] [Google Scholar]
- Pompeiano M, Palacios JM, Mengod G ( 1992) Distribution and cellular localization of mRNA coding for 5-HT1A receptor in the rat brain: correlation with receptor binding. J Neurosci 12: 440-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramboz S, Oosting R, Amara DA, Kung HF, Blier P, Mendelsohn M, Mann JJ, Brunner D, Hen R ( 1998) Serotonin receptor 1A knockout: an animal model of anxiety-related disorder. Proc Natl Acad Sci USA 95: 14476-14481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riad M, Watkins KC, Doucet E, Hamon M, Descarries L ( 2001) Agonist-induced internalization of serotonin-1a receptors in the dorsal raphe nucleus (autoreceptors) but not hippocampus (heteroreceptors). J Neurosci 21: 8378-8386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saudou F, Amara DA, Dierich A, LeMeur M, Ramboz S, Segu L, Buhot MC, Hen R ( 1994) Enhanced aggressive behavior in mice lacking 5-HT1B receptor. Science 265: 1875-1878. [DOI] [PubMed] [Google Scholar]
- Soderling TR ( 2000) CaM-kinases: modulators of synaptic plasticity. Curr Opin Neurobiol 10: 375-380. [DOI] [PubMed] [Google Scholar]
- Stockmeier CA, Shapiro LA, Dilley GE, Kolli TN, Friedman L, Rajkowska G ( 1998) Increase in serotonin-1A autoreceptors in the midbrain of suicide victims with major depression: postmortem evidence for decreased serotonin activity. J Neurosci 18: 7394-7401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storring JM, Charest A, Cheng P, Albert PR ( 1999) TATA-driven transcriptional initiation and regulation of the rat serotonin 5-HT1A receptor gene. J Neurochem 72: 2238-2247. [DOI] [PubMed] [Google Scholar]
- Szonyi M, He DZ, Ribari O, Sziklai I, Dallos P ( 2001) Intracellular calcium and outer hair cell electromotility. Brain Res 922: 65-70. [DOI] [PubMed] [Google Scholar]
- Törk I ( 1990) Anatomy of the serotonergic system. Ann NY Acad Sci 600: 9-34. [DOI] [PubMed] [Google Scholar]
- Veenstra-VanderWeele J, Anderson GM, Cook EH Jr ( 2000) Pharmacogenetics and the serotonin system: initial studies and future directions. Eur J Pharmacol 410: 165-181. [DOI] [PubMed] [Google Scholar]
- White LA, Eaton MJ, Castro MC, Klose KJ, Globus MY, Shaw G, Whittemore SR ( 1994) Distinct regulatory pathways control neurofilament expression and neurotransmitter synthesis in immortalized serotonergic neurons. J Neurosci 14: 6744-6753. [DOI] [PMC free article] [PubMed] [Google Scholar]



