Abstract
The circadian clock nucleus of the mammalian brain is composed of thousands of oscillator neurons, each driven by the cell-autonomous action of a defined set of circadian clock genes. A critical question is how these individual oscillators are organized into an internal clock that times behavior and physiology. We examined the neural organization of the suprachiasmatic nucleus (SCN) through time-lapse imaging of a short-half-life green fluorescent protein (GFP) reporter of the circadian clock gene Period 1 (Per1). Using brain slice preparations, Per1 promoter rhythms were resolved at the level of the SCN, and in individual neurons within the SCN, to determine the temporal patterns of rhythmicity resulting from exposure of mice to light/dark cycle (LD) and constant darkness (DD) conditions. Quantitative imaging and patch-clamp electrophysiology were used to define the relationship of Per1 gene expression to neurophysiological output on an individual neuron basis. We found that in both LD and DD, the overall rhythm of the clock nucleus is composed of individual cellular rhythms that peak in distinct phase groups at 3-4 hr intervals. However, the phase relationships of Per1 oscillations to locomotor activity and the phase relationships among individual neuronal oscillators within the SCN are different in LD and DD. There was a positive, linear correlation of Per1 transcription with neuronal spike frequency output, thus Per1::GFP rhythms are representative of physiological rhythmicity. Our results reveal multiple phase groupings of SCN oscillators and suggest that light regulation of oscillator interactions within the SCN underlies entrainment to the photoperiod.
Keywords: suprachiasmatic nucleus, circadian rhythms, GFP, transgenic mice, confocal microscopy, electrophysiology, time-lapse imaging, gene expression, Period 1, entrainment
Introduction
In mammals, the principal circadian pacemakers driving daily behavioral rhythms are the bilateral suprachiasmatic nuclei (SCNs) of the hypothalamus. Each nucleus contains ∼10,000 neurons, many of which are individually competent circadian oscillators (Welsh et al., 1995). A defined set of genes, including the Period 1 gene (Per1), constitutes the fundamental circadian oscillation mechanism (Albrecht, 2002). It is clear that these “clock genes” produce ∼24 hr rhythms in individual cells through interlocked transcription-translation feedback loops (Albrecht, 2002). However, the organization of these individual molecular oscillators into a “biological clock” controlling behavior and physiology remains to be fully elucidated.
A prevailing view of SCN organization is that individual SCN neurons oscillate in synchrony, and the resulting physiological output is a monophasic waveform (Liu et al., 1997). In such models, the SCN pacemaker is suggested to produce a single, coherent output representing the mean period of its many individual oscillators (Liu et al., 1997; Herzog et al., 1998; Low Zeddies and Takahashi, 2001). An alternative view is that oscillators of the SCN are organized in a multiphasic manner, and that phase relationships among oscillators can be modulated by environmental input or experience (Pittendrigh and Daan, 1976; Jagota et al., 2000; Mrugala et al., 2000). To experimentally distinguish these two models of SCN organization, we have monitored Per1 gene rhythmicity of individual neurons in acute brain slices of the SCN in vitro using a dynamic green fluorescent protein (GFP) reporter (Kuhlman et al., 2000) and have tested the effects of cyclic light input on the temporal organization of the clock nucleus.
Materials and Methods
Gene expression imaging of SCN. Coronal hypothalamic slices (300 μm) containing the SCN were harvested from mice hemizygous for the mouse Per1 (mPer1)::d2EGFP transgene (Kuhlman et al., 2000) and were imaged according to previously published methods (LeSauter et al., 2003). For SCN from light/dark cycles (LD), animals 1-3 weeks old were maintained on a 14/10 hr LD. Zeitgeber time (ZT) 12 was defined as the time of lights off, whereas ZT22 was the time of lights on. SCNs from constant darkness (DD) animals were taken from adult mice individually monitored for wheel-running activity. They were first entrained to a 12 hr LD cycle and then released into constant darkness for at least 4 d before slices were harvested. Wheel-running activity was monitored using Chronobiology Kit software (Stanford Scientific, Santa Cruz, CA). Slices were harvested using infrared goggles. Circadian Time (CT) was predicted for the cycle during which the SCN was harvested by extrapolating a regression line through the activity onsets for the previous 4 d. CT12 was defined as the time of predicted locomotor activity onset. The time scales for in vitro recordings of LD slices are shown in ZT to indicate their reference to the previous light/dark cycle, whereas the time scales for DD slices are indicated in CT to indicate their reference to the previous wheel-running activity. For comparison of phases across LD and DD conditions, the time scales were aligned at the time of lights off/wheel running activity onset (ZT12/CT12). The timing of locomotor activity onsets on the first cycle in DD from animals entrained to 14/10 hr LD did not exhibit significant masking effects, suggesting that this time scale alignment reliably indicates pacemaker phase in the two conditions.
Gene expression imaging of individual neurons. For imaging of individual cells, animals 16-21 d old were used because their SCNs were found to be highly suitable for confocal imaging. At this age, the photic input to the mouse SCN is fully developed (Llamosas et al., 2000), whereas, in the absence of a light cycle, the activity rhythms of the pups are tightly grouped around the phase of the mother (Viswanathan, 1999). For LD, mothers and pups were maintained in 14/10 hr LD. For DD, mothers and pups were placed in constant darkness for at least 6 d before slice harvesting from pups, and the wheel-running activity of the mother was monitored to predict CT12 for the pups. SCNs were imaged using a laser-scanning confocal microscope (Leica, Bannockburn, IL). The 488 nm laser line was used for excitation; emission was detected between 500 and 525 nm. Every 15-30 min, six to eight optical z-plane sections were acquired through 50-70 μm. For analysis, individual cells were delimited by a region of interest tool, and their fluorescence intensity was tracked over time with either NIH Image or IP Lab software (Scanalytics, Fairfax, VA). The time of peak was defined as the time of maximum fluorescence, where the values for the preceding and following time points were at least 95% of the maximum. Cells that exhibited a 1.4-fold difference in intensity between peak and nadir, the empirically determined threshold for reliable rhythm detection from our conventional imaging experiments, were considered rhythmic. Cells in which the fluorescence intensity was sufficiently elevated above background so that we could reliably detect and accurately quantify the Per1::GFP signal (i.e., at least 1.2-fold above the lowest cell nadir in the same slice) but that exhibited time-dependent changes that were <1.4-fold in amplitude were classified as nonrhythmic. Those cells in which the fluorescence intensity was not sufficient for accurate quantification over time (i.e., <1.2-fold above the lowest cell nadir) were not included in the data set. Phase analysis of individual cell rhythms was performed in the first circadian cycle in vitro.
To test the extent to which cells losing their viability contributed to the GFP signals from the SCN, propidium iodide (PI; 3-5 μm), a stain for cells with compromised membrane integrity, was added at different intervals in vitro. We then assayed for overlap between the GFP and PI fluorescence signals. At the start of recording, 3 of 64 GFP + cells were also PI +; after 10 hr, 2 of 75 GFP + were PI +; and after 24 hr, 1 of 12 GFP + cells were PI + (n = 3 slices). Thus the proportion of GFP +/PI + cells remained at <10% across 24 hr, and the preponderance of GFP signals we recorded were from healthy, viable neurons.
Electrophysiology. Fluorescence intensity and spike frequency were determined for individual SCN neurons as described by Kuhlman et al. (2003). Electrophysiological methods for N2A cells were similar to those of Wagner et al. (1998). For transfection of N2A cells, 5 μg of the P1PG plasmid carrying the Per1::GFP construct used to generate the reporter mice (Kuhlman et al., 2000) was transfected into cultures by the calcium phosphate method. Experiments were performed 24-72 hr after transfection and reproduced on two separate rounds of transfection.
Results
Per1-driven GFP rhythms in the in vitro SCN
The Per1-driven GFP transgene faithfully reports the endogenous circadian rhythm of mPer1 promoter activity in the SCN in vivo and in vitro (Kuhlman et al., 2000, LeSauter et al., 2003). Coronal brain slices containing the SCN, removed from animals kept on 14/10 hr LD cycles, showed robust rhythms of Per1-driven GFP fluorescence (Fig. 1A,B). SCN fluorescence peaked 2 hr before the time of lights off on the preceding LD cycle (ZT, 10.0 ± 2.5 hr) on the first day in vitro, regardless of whether the slices were prepared early in the day (ZT, 22 ± 2 hr; n = 5) or late in the day (ZT, 3 ± 7 hr; n = 4), indicating that slice preparation itself did not perturb the endogenous clock. The interval between first and second cycles was 23.55 ± 2.18 hr (n = 9), which is identical to the free-running period of behavioral activity rhythms in this transgenic line (Kuhlman et al., 2000). The peak of fluorescence intensity averaged 2.1-and 1.5-fold higher than the nadir on the first and second days, respectively. Slices from animals maintained in DD (Fig. 1C) also exhibited Per1-driven GFP cycles (Fig. 1D), with the mean time of peak fluorescence synchronous with predicted locomotor activity onset (CT, 11.8 ± 1.1 hr; n = 4; slices made between CT1.5 and CT3.5). The interval between first and second cycle peaks for SCN from DD animals was 23.2 ± 0.5 hr (n = 4). Non-SCN hypothalamic areas from transgenic mice and the SCN of nontransgenic mice did not show rhythms in fluorescence intensity (data not shown).
Figure 1.
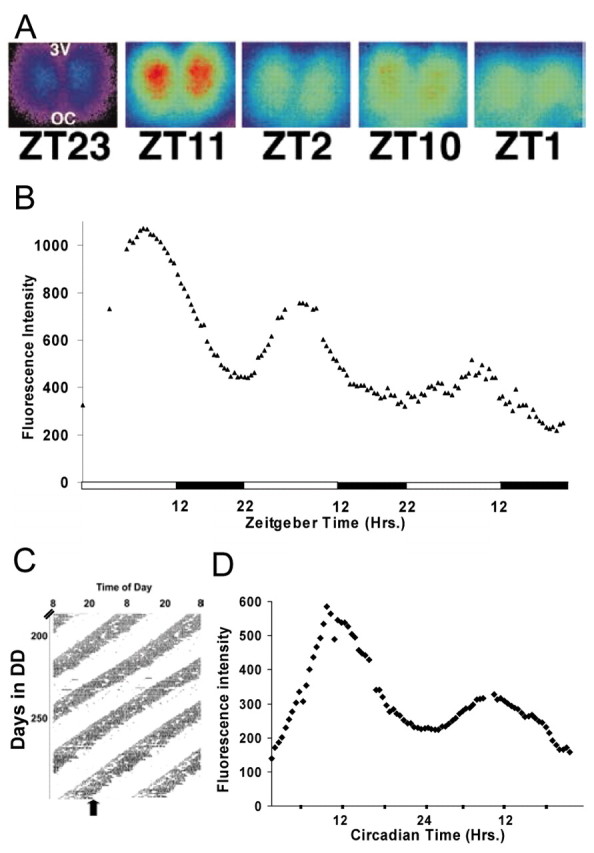
Time-lapse imaging of SCN Per1-driven GFP rhythms. A, Individual pseudocolored fluorescence images from a time-lapse recording of Per1-driven GFP fluorescence rhythms from an SCN brain slice in vitro. B, Fluorescence rhythms in the in vitro SCN from an animal housed in a 14/10 hr LD cycle. Images were captured every 30 min. The open and filled bars indicate the previous light/dark cycle for the animal. C, Wheel-running activity, double-plotted, from an animal housed in constant darkness. Vertical marks show times when the animal was active. The filled arrow marks the time the SCN slice was prepared for imaging. D, GFP fluorescence recording of the SCN from the animal in C. Images were captured every 30 min for 2 d.
Spike activity correlates with Per1 promoter activity
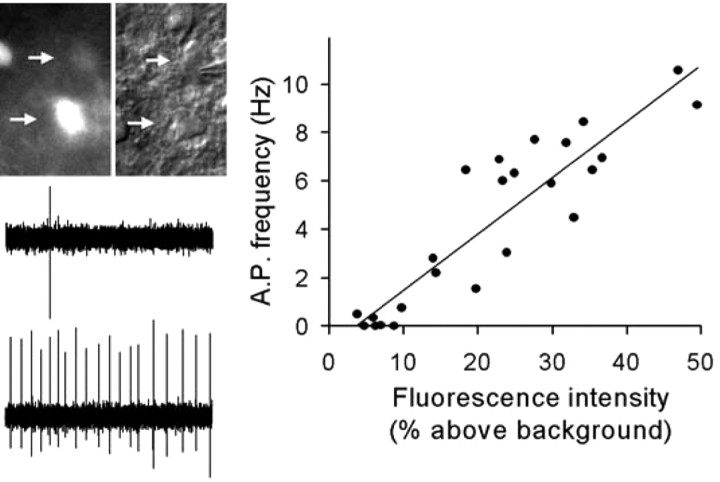
To assess the relationship between Per1 promoter activity and neurophysiological activity, we performed loose patch electro-physiology while quantifying the Per1-driven GFP fluorescence of individual neurons. When examined in the midday (ZT4-7), a 13-fold range in neuronal fluorescence intensities was highly correlated with a 10 Hz range in spike frequency (r = 0.91; p < 0.001; Fig. 2). There was a positive, linear correlation of Per1 promoter activity with the spike frequency output of circadian clock neurons. Neurons with high levels of Per1-driven gene expression were more active than those with low levels; thus, the gene expression activity we observed in our imaging experiments is also reflected in neural activity during the day phase.
Figure 2.
Per1-driven fluorescence intensity correlates with spike frequency in SCN neurons. Right, Fluorescence intensity, measured as percent above background, versus action potential (AP) frequency plotted for SCN neurons (n = 24, 3 slices). r = 0.91; p < 0.001. Left, Example of spike output from neighboring neurons; top arrow, low fluorescence intensity (6.9% above background, 0.03 Hz); bottom arrow, high fluorescence intensity (49.5% above background, 9.14 Hz).
The expression of GFP has been previously demonstrated to have no significant effects on neuronal properties (Smith et al., 1997). However, as a direct control for the possibility that GFP levels per se could alter electrophysiological parameters, we transfected mouse N2A neuroblastoma cells with the GFP construct and then performed combined patch-clamp and quantitative imaging experiments similar to those we had performed on SCN neurons. Low-GFP-expressing cells (dim) and high-GFP-expressing cells (bright) within the same transfections were selected, and current-voltage relationships from -120 through +75 mV were determined (see Materials and Methods). On average, the high-expressing cells exhibited sevenfold higher fluorescence intensity, yet the current-voltage curves for the two populations were identical (supplemental Fig. 1, available at www.jneurosci.org), with both peak inward and outward currents being indistinguishable in amplitude (low-intensity cells, -44.0 ± 7.3 and 397.6 ± 57.0 pA; high-intensity cells, -49.6 ± 7.0 and 364.9 ± 27.4 pA). These results indicate that cellular GFP concentrations per se do not influence electrophysiological properties, and, thus, the correlation of Per1::GFP expression with spike rate in SCN neurons is likely attributable to the action of this reporter gene as a faithful indicator of underlying circadian rhythms in mPer1 gene expression.
Individual neuronal rhythms in SCN in vitro
Using time-lapse confocal microscopy, we recorded the gene expression rhythms of individual SCN neurons within SCN slices from animals housed in either LD or DD conditions. Imaging was directed to one SCN from each animal to assess intra-SCN organization. Figure 3A shows single-cell gene expression rhythms from two cells in a DD SCN in which the Per1 promoter activity was tracked as GFP fluorescence intensity. Individual SCN neurons in both LD and DD SCN exhibited circadian cycles of Per1-driven gene expression that averaged approximately threefold in amplitude and 21.4 ± 0.5 hr in period (n = 27). We did not identify any oscillations in individual cells having multiple peaks during a circadian cycle. Eleven percent of imaged neurons in LD SCN and 26% of neurons in DD SCN exhibited nonrhythmic Per1::GFP expression (see Materials and Methods).
Figure 3.
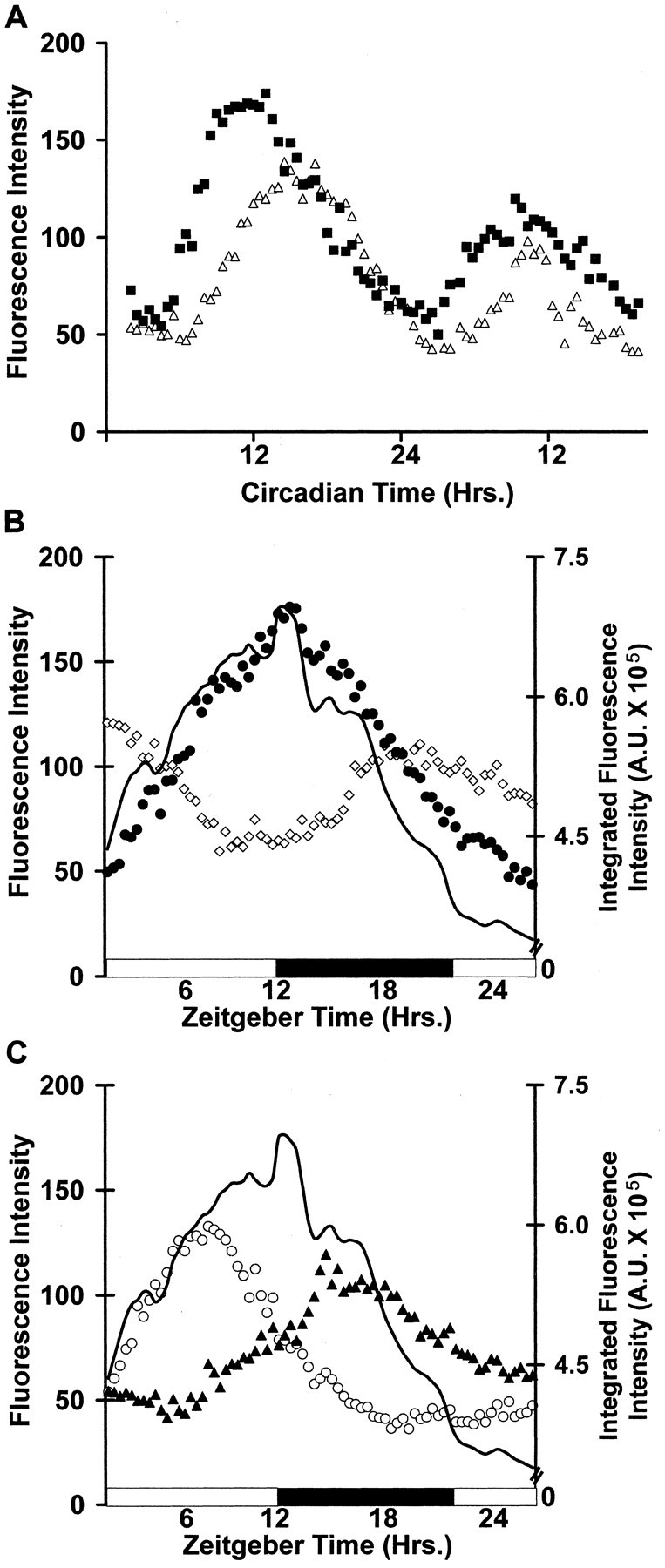
Individual neuron gene expression rhythms in the SCN. A, Per1-driven GFP rhythms from two individual SCN neurons in an in vitro DD SCN obtained by confocal time-lapse imaging. B, C, Example gene expression rhythms from four individual neurons in an in vitro SCN from a LD animal. Each set of symbols represents the measured fluorescence for an individual cell during the circadian cycle. The black solid line indicates the integrated overall fluorescence rhythms of the SCN as a whole.
The individual neuronal rhythms within each SCN exhibited a wide range of peak times (Fig. 3B,C; also see time-lapse video in supplemental Fig. 2). In SCN from both LD and DD animals, approximately half of the individual cell rhythms peaked in synchrony, forming a principal phase group that gave rise to the overall peak in Per1-driven GFP fluorescence from the nucleus as a whole (Fig. 3B,C, black line). These rhythms were typified by the cell plotted in in Figure 3B, filled circles (52% of rhythmic cells at ZT8 of LD and 63% of rhythmic cells at CT12 of DD; n = 126 cells for LD; n = 131 cells for DD; n = 5 SCNs each). The peak times of the remaining 48 or 37% of rhythmic cells in LD and DD SCN were not aligned with the principal phase group (Fig. 3C, open circles, filled triangles) and included neurons that cycled in antiphase to the principal phase peak (Fig. 3B, open diamonds).
Many neurons outside the main phase group also clustered their peak times at distinct circadian phases. In both individual and group data (Fig. 4A-C) peak time histograms exhibited clear multiple-phase peaks. Peak times could not be described by a single normal distribution (Kolmogorov-Smirnov test, p > 0.05) but were, in fact, best fit by three-peak Gaussian functions. For SCNs from animals experiencing an LD cycle, groupings of cell rhythms consistently occurred before and after the main phase group, at ZT5 and ZT11 (13% each; Fig. 4B). In the absence of a light cycle, groupings of cell rhythms consistently occurred after the main peak at CT15 and CT18 (19 and 10%, respectively; Fig. 4C). Thus, in LD SCN, 78% of neuronal peak times were contained in three distinct phase groups at ZT5, ZT8, and ZT11, whereas in DD SCN, 92% of neuronal peak times were contained in phase groups at CT12, CT15, and CT18. Twenty-two percent of rhythmic SCN neurons in LD slices and 8% of rhythmic neurons in DD slices peaked at times outside the three organized phase clusters, their peak times being primarily distributed throughout the late night and into the early day (Fig. 4B,C, points not fit by smooth curves).
Figure 4.
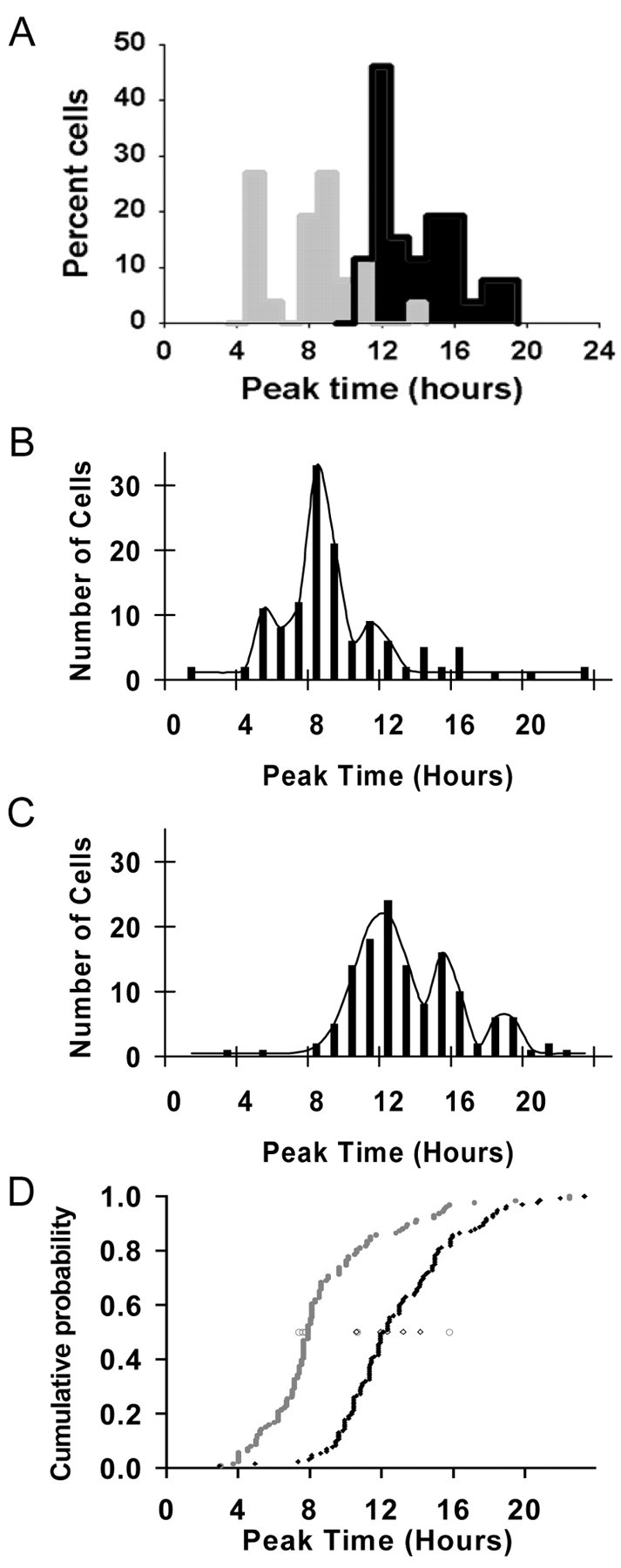
Neuronal phase relationships are dependent on light history. A, Histograms of individual neuron fluorescence peak times in an SCN slice from an LD animal (gray bars) and an SCN slice from a DD animal (black bars). Cell peak times were binned at 1 hr intervals; n = 26 neurons for the LD slice; n = 37 neurons for the DD slice. B, Histogram of individual neuron fluorescence peak times summed from SCN slices from five LD animals. Peak times are in 1 hr bins. The black line indicates the best fit curve showing phase group peak times of ZT5, ZT8, and ZT11. C, Histogram of individual neuron fluorescence peak times summed from SCN slices from five DD animals. The black line indicates the best fit curve showing phase group peak times of CT12, CT15, and CT18. D, Cumulative probability plot of peak times from animals housed in the LD cycle (filled circle, gray line) and peak times from DD (filled diamond, black line). The median time of cellular peaks for each animal is plotted as an open symbol (circle, LD; diamond, DD).
Although the temporal gene expression patterns of SCN networks from LD and DD animals were both characterized by three phase groups at 3-4 hr intervals, they were distinct in terms of the phase relationship of Per1 rhythms to locomotor activity and the phase relationships among individual SCN oscillator neurons. The cumulative probability distribution of cell peak times in vitro, referenced to the time of lights off/activity onset of the slice donor animal on previous circadian cycles, were significantly earlier in the LD SCN compared with the DD SCN (Fig. 4D; Kolmogorov-Smirnov test, p < 0.001). In addition, as described above, in LD SCN, one accessory phase group preceded the main peak and the other followed it, whereas in DD SCN, both accessory phase groups followed the main phase group.
In addition to the different temporal patterns of cellular gene expression rhythms, LD and DD SCN slices also exhibited distinct anatomical distributions of rhythmic cells. When the position of rhythmic cells (Fig. 5A) relative to the lateral-medial axis of the SCN was examined, LD slices exhibited nearly equal proportions of lateral and medial rhythmic cells (54% lateral vs 46% medial), whereas DD slices had twice as many rhythmic cells in the medial half of the SCN (66% medial vs 34% lateral). In the dorsal-ventral axis, LD slices had approximately twice as many rhythmic cells in the ventral region (64% ventral vs 36% dorsal), whereas DD slices had more dorsal rhythmic cells, resulting in a more equal distribution (57% dorsal vs 43% ventral). With regard to the spatial distribution of cells in the various phase groups, there was a lateral-to-medial gradient that correlated with the peak time of phase groups (Fig. 5B). The earliest phase group, ZT5 of LD slices, had the highest proportion of laterally located cells, whereas the latest phase group, CT18 of DD slices, had the highest proportion of medial cells. Intermediate phase groups had intermediate proportions that followed a general trend of increasing medial and decreasing lateral cells with later peak times. The small numbers of “antiphase” rhythmic neurons and nonrhythmic neurons precluded meaningful analysis of their spatial distributions.
Figure 5.
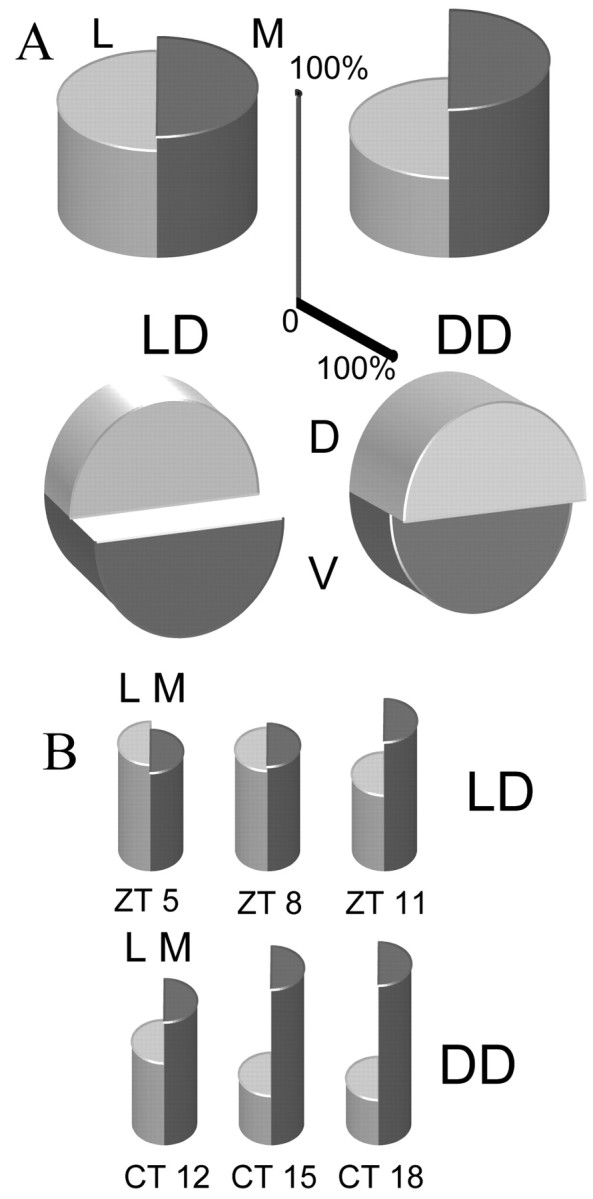
Rhythmic neurons by SCN region. Split cylinders shown in a three-dimensional perspective rendering represent the percentage of rhythmic neurons in the SCN mapped by SCN region. The height of each half-cylinder indicates the percentage of rhythmic SCN neurons in that region. Scale bars indicate the percentage scale for the lateral-medial axis (vertical) and the dorsal medial axis (horizontal). Total cells are n = 126 cells for LD slices and 131 cells for DD slices. A, Overall distributions of rhythmic neurons. Top row, Percentage of rhythmic SCN neurons mapped in the lateral-medial axis for LD (left) and DD (right) slices. Bottom row, Percentage of rhythmic SCN neurons mapped in the dorsal-medial axis for LD (left) and DD (right) slices. B, Distributions of rhythmic neurons by phase group. Top row, Percentage of rhythmic SCN neurons from the three LD phase groups mapped in the lateral-medial axis. Bottom row, Percentage of rhythmic SCN neurons from the three DD phase groups mapped in the lateral-medial axis.
Discussion
A prevailing model of SCN tissue level organization is a “monophasic” view in which SCN neurons form a single coherent phase population (Liu et al., 1997; Herzog et al., 1998). Recent findings of light cycle-dependent biphasic SCN electrical output suggested a more multiphasic SCN organization but could not distinguish between light-induced changes in the waveform of coherent cellular oscillators vs changes in the relative phase of individual oscillators (Jagota et al., 2000; Mrugala et al., 2000). Our time-lapse imaging studies of Per1 gene dynamics directly demonstrate that the individual neuronal molecular rhythms comprising the SCN clock waveform are organized as multiple-phase groups of neurons with distinct peak times. Simultaneous patch recording and quantitative imaging revealed that heterogeneity in the phase of molecular oscillations is reflected in the neurophysiological output of Per1-expressing neurons. In addition, we demonstrated that entrainment of animals to a light cycle redefines the phase relationships among oscillator ensembles within the SCN. These data suggest that the SCN is organized as multiphasic oscillator ensembles, and that light-driven changes in the phase relationships between constituent oscillators underlie pacemaker entrainment to the photoperiod.
The SCN clock comprises discrete phase groups
The SCN comprises multiple individual cellular oscillators, with a bare majority of neurons (50-60%) forming a main phase group. A consistent finding of our studies was that, in addition to this main activity peak, the remaining neuronal rhythms in an SCN were clustered into two additional well defined phase groups, each of which contained 10-20% of the rhythmic neurons. The three phase groups peaked at 3-4 hr intervals. Interestingly, previous in vivo studies have documented similar accessory peaks in SCN multiunit spike frequency at 3-4 hr intervals (Meijer et al., 1997). Together, the evidence strongly suggests that discrete phase groups, now experimentally defined by both molecular and electrical activity, are characteristics of the mammalian circadian pacemaker.
In addition to the robust accessory peaks, a less numerous group of neurons cycled with peak times primarily in the night phase. The presence of cells cycling in approximate antiphase is not entirely unexpected because PER1 immunocytochemistry has demonstrated a subpopulation of SCN neurons that peak in antiphase to most of the nucleus (Bae et al., 2001; King et al., 2003). Furthermore, spike frequency rhythms of individual units within organotypic cultures of SCN also exhibit antiphase rhythmicity (Herzog et al., 1997; Nakamura et al., 2001).
There are three principal mechanisms by which the phase heterogeneity in SCN neuronal rhythms could arise: differences in the periods of neuronal rhythms, differences in the amplitude neuronal rhythms, and differences in the coupling interactions among neurons. Variations in the period and amplitude of SCN neuron rhythms have indeed been documented (Liu et al., 1997; Herzog et al., 1998; Honma et al., 1998; this paper). Although more detailed studies are warranted, the data, at present, suggest that period and amplitude vary in continuous distributions; thus, although they could engender monophasic distributions in neuronal phase, they are not likely to cause the multiphasic clustering we observed. Therefore, we suggest that differential coupling interactions between SCN neurons, perhaps expressed as differential responsiveness to intra-SCN coupling signals, such as neuropeptides, or GABA, likely play key roles in establishing the multiphasic nature of the SCN clock. The organized phase groups we have defined could represent specific outputs of the SCN or SCN subnetworks tuned to specific inputs. Additional studies are needed to determine the precise relationship between phase diversity and cell type heterogeneity within the SCN.
The patterns of phase heterogeneity among SCN neurons could also be stable or dynamic. We have used acute SCN slice preparations and have primarily analyzed neuronal rhythms on the initial circadian cycle in vitro to assess the effects of animal light experience during pacemaker organization. Our data set of neuronal rhythms from the second cycle in vitro is more limited but shows that the general patterns of phase relationships among neurons are maintained, and that individual neurons are not rigidly phase-locked to each other (Fig. 3A). Although we have demonstrated that the pattern of neuronal gene rhythms on the initial cycle reflects the light history of the donor animal, the patterns on subsequent cycles also likely reflect reorganization of the SCN slice attributable to deafferentation and maintenance in in vitro conditions. Thus, our experiments provide a unique “snapshot” of experience-dependent SCN organization but are less well suited for determining the stability of individual neuron phase relationships. Although this issue should ultimately be addressed with in vivo experiments, electrophysiological neuronal rhythms in long-term organotypic cultures of SCN show significant cycle-to-cycle variations in relative phase (Nakamura et al., 2001), suggesting that dynamic phase relationships among SCN neurons may be the norm.
Light-driven changes in oscillator ensembles
Two defining hallmarks of circadian pacemakers are that they generate rhythms endogenously and that they are synchronized to the external environment. We tested directly the hypothesis that entrainment of the SCN pacemaker occurs through light-driven changes in the relative phase of individual oscillators. By examining the effects of the animal's previous light history on SCN network dynamics, resolved at the level of individual neurons, we determined that the presence of a light cycle induced a reorganization of individual neuron peak times. Importantly, light changed the phases but not waveforms of individual neuronal rhythms. This is a direct demonstration that light input to the SCN indeed influences the relative phase relationships of its constituent oscillators, as previously suggested by behavioral observations (Pittendrigh and Daan, 1976) and in vitro electrical recordings (Jagota et al., 2000; Mrugala et al., 2000).
We found that light input has two distinct effects on SCN neuronal rhythms: (1) altering the absolute phase relationship between SCN rhythmic activity and locomotor activity onset (Fig. 4D) and (2) reorganizing the relative phases of constituent oscillators within the SCN (Fig. 4B,C). Comparison of the absolute phase relationships of SCN rhythms and activity rests, in part, on the assumption that in DD pups, activity rhythms are synchronized to their mother's. Previous results from mice, rats, and hamsters indicate the widespread nature of maternal entrainment of neonatal rodents, both for locomotor behavior (Takahashi and Deguchi, 1983; Davis and Gorski, 1985; Viswanathan, 1999) and for SCN rhythms (Reppert and Schwartz, 1983). The fact that in our experiments the patterns of phase peaks observed in an individual SCN (Fig. 4A) are preserved in summed data from multiple SCNs (Fig. 4B,C) indicates that the pups were indeed synchronized to a common Zeitgeber time. However, we cannot rigorously exclude the possibility that in our mouse line, there could be a systematic phase difference between mother and pup that could contribute to the overall phase differences between LD and DD slices. Thus, we must be cautious in interpreting the apparent effect of light input on the absolute phase relationship between SCN rhythms and locomotor activity, but this does not affect our measurements of phase reorganization within the SCN.
Cyclic light input to the SCN could induce phase reorganization of SCN oscillators by directly resetting the phase of SCN neurons, by increasing the amplitude of neuronal oscillations (and thus affecting coupling angles), by driving SCN neuron populations that are not self-sustained pacemakers, or by a combination thereof. There is considerable heterogeneity in SCN neuron responsiveness to light, including a specific subpopulation enriched for vascoactive intestinal polypeptide-containing neurons that exhibits persistent increases in neurophysiological activity after Per1 induction (Kuhlman et al., 2003) and a subpopulation of hamster SCN neurons that is arhythmic in Per1 expression in the absence of cyclic light input (Hamada et al., 2002). We found that the proportion of nonrhythmic Per1-expressing neurons was higher in DD SCN than in LD SCN, suggesting the presence of a light-driven subpopulation of neurons in the mouse SCN that can continue to oscillate on the initial cycles in vitro. In addition, rhythmic neurons were more prevalent in the ventral-lateral region of LD SCN than in DD SCN, similar to the regionalization of rhythmic electrical activity in the presence and absence of a light cycle in the rat SCN (Shibata et al., 1984). Thus, light-driven neurons are a general feature of mammalian SCN organization and may play critical roles in entrainment.
Summary
On the basis of our findings, we propose a “multiphasic” view of SCN organization in which the individual neurons participate in oscillator ensembles that adopt varying phase angles to each other through coupling processes influenced by light input. Reconciling variations in the characteristics of individual oscillators with a multiphasic overall pacemaker organization of phase clusters, rather than with strict monophasic synchronization, provides a framework for understanding critical aspects of SCN function. This multiphasic view better accounts for many properties of circadian organization, including the ability of the SCN to produce multiple-phase peak rhythms in SCN electrophysiology in vitro and in vivo (Meijer et al., 1997; Yamazaki et al., 1998; Jagota et al., 2000; Mrugala et al., 2000) to time various physiological output rhythms, and to respond to seasonal variations in the photoperiod by altering the coupling of its constituent oscillators (Pittendrigh and Daan, 1976).
Footnotes
This work was supported by National Institutes of Health Grant MH063341 (D.G.M.). We thank Dr. Carl Johnson, Dr.TerryPage, Dr.RaeSilver, and Dr.Shin Yamazaki for comments on previous versions of this manuscript; Dr.Dao-Qi Zhang for performing the N2A recordings; and Jeff Stone for excellent technical assistance.
Correspondence should be addressed to Dr. Douglas G. McMahon, Department of Biological Sciences, Vanderbilt University, 1210 MRBIII, VU Station B, Box 35-1634, Nashville, TN 37235-1634. E-mail:douglas.g.mcmahon@vanderbilt.edu.
J. E. Quintero's present address: Center for Sensor Technology, University of Kentucky Medical Center, Lexington, KY 40536-0098.
S. J. Kuhlman's present address: Beckman Neuroscience, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY 11724.
Copyright © 2003 Society for Neuroscience 0270-6474/03/238070-07$15.00/0
J.E.Q. and S.J.K. contributed equally to this work.
References
- Albrecht U ( 2002) Functional genomics of sleep and circadian rhythm: invited review. Regulation of mammalian circadian clock genes. J Appl Physiol 92: 1348-1355. [DOI] [PubMed] [Google Scholar]
- Bae K, Jin X, Maywood ES, Hastings MH, Reppert SM, Weaver DR ( 2001)Differential functions of mPer1, mPer2, and mPer3 in the SCN circadian clock. Neuron 30: 525-536. [DOI] [PubMed] [Google Scholar]
- Davis FC, Gorski RA ( 1985) Development of hamster circadian rhythms. I. Within-litter synchrony of mother and pup activity rhythms at weaning. Biol Reprod 33: 353-362. [DOI] [PubMed] [Google Scholar]
- Hamada T, LeSauter J, Venuti JM, Silver R ( 2002) Expression of Period genes: rhythmic and non-rhythmic compartments of the suprachiasmatic nucleus pacemaker. J Neurosci 21: 7742-7750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzog ED, Geusz ME, Khalsa SB, Straume M, Block GD ( 1997) Circadian rhythms in mouse suprachiasmatic nucleus explants on multimicroelectrode plates. Brain Res 757: 285-290. [DOI] [PubMed] [Google Scholar]
- Herzog ED, Takahashi JS, Block GD ( 1998) Clock controls circadian period in isolated suprachiasmatic nucleus neurons. Nat Neurosci 1: 708-713. [DOI] [PubMed] [Google Scholar]
- Honma S, Shirakawa T, Katsuno Y, Namihira M, Honma K ( 1998) Circadian periods of single suprachiasmatic neurons in rats. Neurosci Lett 250: 157-160. [DOI] [PubMed] [Google Scholar]
- Jagota A, de la Iglesia HO, Schwartz WJ ( 2000) Morning and evening circadian oscillations in the suprachiasmatic nucleus in vitro. Nat Neurosci 3: 372-376. [DOI] [PubMed] [Google Scholar]
- King VM, Chahad-Ehlers S, Shen S, Harmar AJ, Maywood ES, Hastings MH ( 2003) A hVIPR transgene as a novel tool for the analysis of circadian function in the mouse suprachiasmatic nucleus. Eur J Neurosci 17: 822-832. [DOI] [PubMed] [Google Scholar]
- Kuhlman SJ, Quintero JE, McMahon DG ( 2000) GFP fluorescence reports Period 1 circadian gene regulation in the mammalian biological clock. NeuroReport 11: 1479-1482. [PubMed] [Google Scholar]
- Kuhlman SJ, Silver R, Le Sauter J, Bult-Ito A, McMahon DG ( 2003) Phase resetting light pulses induce Per1 and persistent spike activity in a subpopulation of biological clock neurons. J Neurosci 23: 1441-1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeSauter J, Yan L, Vishnubhotla B, Quintero J, Kuhlman S, McMahon D, Silver R ( 2003) A short half-life GFP mouse model for analysis of SCN organization. Brain Res 964: 279-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Weaver DR, Strogatz SH, Reppert SM ( 1997) Cellular construction of a circadian clock: period determination in the suprachiasmatic nuclei. Cell 91: 855-860. [DOI] [PubMed] [Google Scholar]
- Llamosas MM, Heurta JJ, Cernuda-Cernuda R, Garcia-Fernandez JM ( 2000) Ontogeny of a photic response in the retina and suprachiasmatic nucleus in the mouse. Dev Brain Res 120: 1-6. [DOI] [PubMed] [Google Scholar]
- Low Zeddies SS, Takahashi JS ( 2001) Chimera analysis of the Clock mutation in mice shows that complex cellular integration determines circadian behavior. Cell 105: 25-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijer JH, Schaap J, Watanabe K, Albus H ( 1997) Multiunit activity recordings in the suprachiasmatic nuclei: in vivo versus in vitro models. Brain Res 753: 322-327. [DOI] [PubMed] [Google Scholar]
- Mrugala M, Zlomanczuk P, Jagota A, Schwartz WJ ( 2000) Rhythmic multiunit neural activity in slices of hamster suprachiasmatic nucleus reflect prior photoperiod. Am J Physiol 278: R987-R994. [DOI] [PubMed] [Google Scholar]
- Nakamura W, Honma S, Shirakawa T, Honma K ( 2001) Regional pacemakers composed of multiple oscillator neurons in the rat suprachiasmatic nucleus. Eur J Neurosci 14: 666-674. [DOI] [PubMed] [Google Scholar]
- Pittendrigh CS, Daan S ( 1976) A functional analysis of circadian pacemakers in nocturnal rodents. V. Pacemaker structure: a clock for all seasons. J Comp Physiol [A] 106: 333-355. [Google Scholar]
- Reppert SM, Schwartz WJ ( 1983) Maternal coordination of the fetal biological clock in utero. Science 220: 969-971. [DOI] [PubMed] [Google Scholar]
- Shibata S, Liou S, Ueki S, Oomura Y ( 1984) Influence of environmental light-dark cycle and enucleation on activity of suprachiasmatic neurons in slice preparations. Brain Res 302: 75-81. [DOI] [PubMed] [Google Scholar]
- Smith GM, Berry RL, Yang J, Tanelian D ( 1997) Electrophysiological analysis of dorsal root ganglion neurons pre- and post-coexpression of green fluorescent protein and functional 5-HT3 receptor. J Neurophysiol 77: 3115-3121. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Deguchi T ( 1983) Entrainment of the circadian rhythms of blinded infant rats by nursing mothers. Physiol Behav 31: 373-378. [DOI] [PubMed] [Google Scholar]
- Viswanathan N ( 1999) Maternal entrainment in the circadian activity rhythm of laboratory mouse (C57BL/6J). Physiol Behav 68: 157-162. [DOI] [PubMed] [Google Scholar]
- Wagner TL, Beyer EC, McMahon DG ( 1998) Cloning and functional expression of a novel gap junction channel from the retina of Danio aquipinnatus. Vis Neurosci 15: 1137-1144. [DOI] [PubMed] [Google Scholar]
- Welsh DK, Logothetis DE, Meister M, Reppert SM ( 1995) Individual neurons dissociated from rat suprachiasmatic nucleus express independently phased circadian firing rhythms. Neuron 14: 697-706. [DOI] [PubMed] [Google Scholar]
- Yamazaki S, Kerbeshian MC, Hocker CG, Block GD, Menaker M ( 1998) Rhythmic properties of the hamster suprachiasmatic nucleus in vivo J Neurosci 18: 10709-10723. [DOI] [PMC free article] [PubMed] [Google Scholar]