Abstract
Protein ubiquitination has been implicated recently in neural development, plasticity, and degeneration. We previously identified ZNRF1/nin283, a protein with a unique, evolutionarily conserved C-terminal domain containing a juxtaposed zinc finger/RING finger combination. Here we describe the identification of a closely related protein, ZNRF2, thus defining a novel family of ZNRF E3 ubiquitin ligases. Both ZNRF1 and ZNRF2 have E3 ubiquitin ligase activity and are highly expressed in the nervous system, particularly during development. In neurons, ZNRF proteins are located in different compartments within the presynaptic terminal: ZNRF1 is associated with synaptic vesicle membranes, whereas ZNRF2 is present in presynaptic plasma membranes. Mutant ZNRF proteins with a disrupted RING finger, a domain necessary for their E3 function, can each inhibit Ca2+-dependent exocytosis in PC12 cells. These data suggest that ZNRF proteins play a role in the establishment and maintenance of neuronal transmission and plasticity via their ubiquitin ligase activity.
Keywords: RING finger, synaptic transmission, endocytosis, endosome/lysosome, proteosome, spermatogenesis
Introduction
The conjugation of ubiquitin to proteins at lysine residues (ubiquitination), thereby targeting them to the proteosome, is essential for the degradation of many proteins in eukaryotic cells. Ubiquitination is controlled by a multi-enzyme cascade that involves E1 (ubiquitin-activating enzyme), E2 (ubiquitin-conjugating enzyme), and E3 (ubiquitin-ligating enzyme) activities (Pickart, 2001). E3 proteins recruit substrates to the ubiquitination machinery and play a crucial role in specifying which proteins are selected for ubiquitination. There are several amino acid sequence motifs associated with E3 ligases, including the RING finger, HECT (homologous to the E6-AP carboxyl terminus), F box, and U box domain (von Arnim, 2001). The RING finger is a zinc-binding protein domain that was initially characterized as a protein interaction domain (Borden, 1998). More recently, it has become clear that a large number of proteins containing RING finger motifs function as E3 ligases, with the RING finger motif itself serving to recruit specific E2s (Pickart, 2001).
The classical view of ubiquitination is to target proteins for degradation by a multi-subunit, ATP-dependent protease termed the proteosome (Pickart, 2001). However, there is increasing evidence that the addition of a single ubiquitin molecule, mono-ubiquitination, is an important post-translational mechanism of modifying protein function (Hicke, 2001). For instance, mono-ubiquitination of transmembrane proteins, such as receptors for trophic factors and ligand-gated ion channels, often serves as an internalization signal and thereby modulating the activity of signaling pathways (Hicke, 2001). Mono-ubiquitination is sufficient to trigger internalization of plasma membrane proteins into primary endocytic vesicles; however, few E3 ubiquitin ligases that target membrane proteins (e.g., Cbl) have been identified (Sanjay et al., 2001). In the nervous system, in which cell surface molecules such as receptors for neurotrophic factors and neurotransmitters play important roles, it has been suggested that ubiquitin-mediated protein metabolism may regulate a variety of processes, including long-term memory formation (Lopez-Salon et al., 2001), synapse formation (DiAntonio et al., 2001), and the establishment of neuropathic pain (Moss et al., 2002). Furthermore, some RING finger-containing proteins bind presynaptic terminal proteins necessary for exocytosis, such as syntaxin 1, and play a role in regulating synaptic signal transmission (Chin et al., 2002).
During our efforts to identify molecules whose expression is induced in Schwann cells after peripheral nerve injury, we identified a novel RING finger protein, which we initially termed nin283 (Araki et al., 2001) (nin283 has been renamed ZNRF1 to conform to the standardized nomenclature used in National Center for Biotechnology Information LocusLink). ZNRF1 contains a conserved C-terminal domain that is composed of a unique zinc finger-RING finger combination. Proteins homologous to ZNRF1 are present in a wide range of species including Caenorhabditis elegans and Drosophila. Here we designate ZNRF proteins as a family of molecules by the identification of a second mammalian protein, ZNRF2, which is highly similar to ZNRF1 in the zinc finger-RING finger region. The widespread expression of ZNRF proteins in the nervous system along with their involvement in exocytosis in presynaptic terminals that we demonstrate here suggests that they may participate in the regulation of proteins involved in presynaptic exocytosis and/or synaptic vesicle recycling.
Materials and Methods
Isolation and sequence analysis of ZNRF2. The full-length cDNA sequence of human and mouse ZNRF2 was determined by analyzing expressed sequence tag (EST) clones (wh57e11, wi35c02, and xs53e14 for human; ux04e08, my52g11, and mu26f10 for mouse) obtained from the Washington University-Merck EST project. These clones were sequenced completely, and the sequence information was combined with those present in GenBank (Unigene clusters Hs.127294, Hs.209125, Hs.288088, Hs.300734, and Hs.351657 for human and Mm.423 and Mm.158534 for mouse) to create contiguous full-length sequences. Sequence editing, mapping, alignment, and contig production was performed using the DNASTAR (Madison, WI) and Vector NTI Suite 6 (InforMax, Frederick, MD) software packages. Human genome sequences for ZNRF1 and ZNRF2 were obtained from the database (accession number AC099508 for ZNRF1; accession number AC006978 for ZNRF2) and compared with cDNA sequences of ZNRF1 and ZNRF2 using BLAST2 (basic local alignment search tool).
Construction of expression plasmids and mutagenesis. Plasmids encoding E2 ubiquitin-conjugating enzymes (pT7-7-Ubc2A, -Ubc2B, -Ubc3, -Ubc4, -UbcH5C, -UbcH6, -UbcH7, and -UbcH8) were kindly provided by Dr. K. Nakayama (Kyushu University, Fukuoka, Japan). The fusion proteins consisting of ZNRF1 or ZNRF2 and enhanced green fluorescent protein (EGFP) were generated by amplifying the coding region of the human ZNRF1 or ZNRF2 cDNA using Herculase (Stratagene, La Jolla, CA) and ligating the product upstream and in-frame with EGFP in pEPECsNeo (a gift from J. Wu, Washington University, St. Louis, MO) at the BamHI site. Deletion mutants were generated by amplifying ZNRF1 nucleotides corresponding to residues 1-141, 11-142, and 142-227 or ZNRF2 residues 1-156 by PCR (a methionine residue was added at the N terminus for the last two ZNRF1 mutants) and fused to EGFP as above. ZNRF1 mutants (C145A and C184A) and a ZNRF2 mutant (C199A) were generated by PCR-mediated site-directed mutagenesis (Ausubel, 2001). A plasmid containing +3 to +2150 of the human growth hormone (hGH) gene [pBSdBam/colon/hGH#78 (Saam and Gordon, 1999)] was obtained from Dr. J. Gordon (Washington University, St. Louis, MO). The hGH gene was subcloned into pEPECsNeo (pCMV-hGH). The integrity of all the constructs was confirmed by sequencing.
Real-time quantitative reverse transcription-PCR analysis. Human total RNAs from pooled tissue sources were purchased from Clontech (Cambridge, UK). First-strand cDNA templates were prepared from 1 μg of RNA using standard methods. Two independent cDNA syntheses were performed for each RNA sample. Quantitative reverse transcription (RT)-PCR was performed by monitoring in real-time the increase in fluorescence of the SYBR-GREEN dye on a TaqMan 7700 Sequence Detection System (Applied Biosystems, Foster City, CA) as described previously (Araki et al., 2001). Normalization of the data and statistical analysis were done as described previously (Araki et al., 2001).
Animals and surgical procedures. Surgical procedures to obtain rat sciatic nerve samples were performed according to National Institutes of Health guidelines for care and use of laboratory animals at Washington University. Sciatic nerves were transected and recovered as described previously (Araki and Milbrandt, 1996).
Generation of anti-ZNRF2 antibodies. Synthetic peptides containing an N-terminal cysteine and ZNRF2 residues 1-15 or 16-30 were separately conjugated to keyhole limpet hemocyanine using glutaraldehyde and m-maleimidobenzoyl-n-hydroxysuccinimide ester and were mixed together. The conjugated proteins were used to immunize rabbits following standard procedures (Animal Pharm Services). Anti-ZNRF2 antibodies were purified by chromatography over an affinity column in which the immunizing peptides was linked to SulfoLink Gel (Pierce, Rockford, IL) per instructions of the manufacturer. The specificity of the affinity-purified ZNRF2 antibodies was confirmed by immunoblotting using lysates of naive NIH3T3 cells and those expressing ZNRF1 or ZNRF2 (data not shown).
Immunohistochemistry and in situ hybridization analysis. Sense and antisense digoxygenin-labeled RNA probes were transcribed from a fragment of the ZNRF1 or ZNRF2 cDNAs. The probes were hybridized to fresh frozen tissue samples as described previously, and signal was visualized using alkaline phosphatase substrate nitroblue-tetrazolium-chloride and 5-bromo-4-chlor-indolyl-phosphate (Roche Products, Hertforshire, UK). Immunohistochemical analysis of rat and mouse tissues was performed on 10 μm sections of fresh frozen tissues using standard methods. Purified anti-ZNRF1 and anti-ZNRF2 antibodies were both used at 1:1000 dilution. Tissue sections were incubated with primary antibodies or with FITC-conjugated α-bungarotoxin (Molecular Probes, Eugene, OR) in some experiments at 4°C overnight and were subsequently incubated with biotinylated anti-mouse IgG and Cy3-conjugated Streptavidin (Jackson ImmunoResearch, West Grove, PA).
In vitro ubiquitination assay. For in vitro ubiquitination assays, E2 enzymes fused to hexahistidine were generated in Escherichia coli as described previously (Hatakeyama et al., 1997). The production of these E2 enzymes was confirmed by immunoblot analysis using anti-pentahistidine antibody (Qiagen, Hilden, Germany).
Wild-type and mutant forms of ZNRF1 and ZNRF2 used in the in vitro ubiquitination assays were generated by in vitro transcription-translation using the TNT-coupled reticulocyte lysate system (Promega, Madison, WI) and [35S]methionine. All proteins were generated as fusion proteins with EGFP-hexahistidine and purified using Ni-NTA agarose (Qiagen) per the protocol of the manufacturer. A fraction of the in vitro translated proteins were separated by SDS-PAGE and subjected to autoradiography for confirmation of protein production. In vitro ubiquitination assays were performed as described previously (Hatakeyama et al., 2001).
Cell culture and hGH release assays. NIH3T3 cells were grown in DMEM supplemented with 10% fetal calf serum. The cells were grown on six-well plates and transfected with ZNRF-EGFP fusion protein expression constructs using Superfect reagent (Qiagen) per the protocol of the manufacturer. To determine the subcellular localization of the fusion proteins, transfected cells were incubated 24 hr later with Lysotracker, Mitotracker, or ER-Tracker (1 nm; Molecular Probes) for 30 min, washed once in culture medium, and examined under a fluorescent microscope.
Primary hippocampal neuron cultures were performed as described previously with minor modifications (Kim et al., 2002). Mouse hippocampal neurons were cultured from embryonic day 16 CD1 fetuses in serum-free Neurobasal medium (Invitrogen, San Diego, CA) plus B27 supplement (Invitrogen) and 25 μm glutamate. At 3 d in vitro, cytosine arabinoside (Sigma, St. Louis, MO) was added to the medium to inhibit proliferation of non-neuronal cells. Cells were subjected to either immunocytochemistry or transfection by Lipofectamine2000 (Invitrogen) between 14 and 21 d in vitro. For presynaptic staining, cells were loaded for 1 min with FM 4-64 (15 μm; Molecular Probes) in HBSS with high K+ and washed in culture media for 5 min. For synaptic vesicle localization, cells were immunostained with an antibody to SV2 (Developmental Studies Hybridoma Bank, University of Iowa, Iowa City, IA) and Alexa488-conjugated anti-mouse IgG (Molecular Probes) by standard procedures.
PC12 cell transfection and secretion assays were performed essentially as described previously (Sugita et al., 1999). In brief, PC12 cells were grown in DMEM supplemented with 10% fetal calf serum and 5% horse serum. Transfection was performed in 12-well plates by Lipofectamine2000 per the protocol of the manufacturer. Two days after transfection, cells from each transfected well were divided into two cultures. One day after replating, secretion experiments were performed. The secretion of hGH was induced in one population by incubation for 20 min with high K+ saline solution containing the following (in mm): 95 NaCl, 56 KCl, 2.2 CaCl2, 0.5 MgCl2, 5.6 glucose, and 15 HEPES, pH 7.4. Cells from the other well were used as control by treating with physiological saline solution containing 145 mm NaCl and 5.6 mm KCl. In some experiments, hGH secretion was induced by permeabilization with 8 μm digitonin for 5 min, followed by incubation with KGEP (in mm: 139 potassium glutamate, 5 EGTA, and 20 PIPES, pH 6.6) containing 2 mm MgATP and 4.76 mm CaCl2. Under these conditions, the calculated final free Ca2+ is 26.6 μm (Goldstein, 1979). After recovery of the medium, cells were incubated with TBS containing 1% Triton X-100 at 4°C for 10 min to determine the amount of hGH remaining inside the cell (i.e., not secreted). The hGH released into the medium, as well as that retained inside the cells, was determined using an ELISA kit (Roche Products). All secretion experiments were performed in duplicate, and the average percentage of hGH release was calculated. Numbers shown are mean ± SD of a duplicated experiment (performed four times independently). Statistical analysis was performed by Student's t test.
Myristoylation assays. NIH3T3 cells transfected with the indicated expression constructs were grown in 100-mm-diameter dishes and starved for 1 hr in DMEM supplemented with 1 mml-glutamine and 10 μg/ml bovine serum albumin (Sigma), [3H-9, 10(n)]myristic acid (100 μCi per plate; PerkinElmer Life Sciences, Emeryville, CA) was added, and the cells were incubated for an additional 4 hr. Cells were lysed, and ZNRF1-EGFP fusion proteins were immunoprecipitated using anti-EGFP antibody (Abcam, Cambridge, UK) and protein A Sepharose (Invitrogen). The immunoprecipitated proteins were separated by SDS-PAGE, and the [3H]-myristoylated proteins were detected by autoradiography.
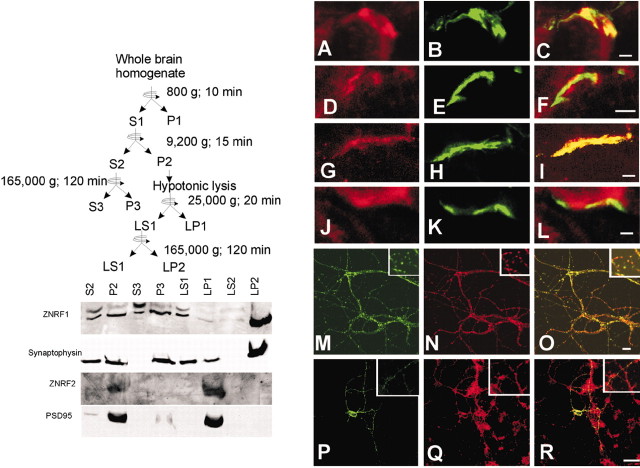
Subcellular fractionation. Fractionation of adult mouse brain into synaptic membrane and vesicle fractions was performed by sequential centrifugation as described previously (Huttner et al., 1983) (also shown in Fig. 5). Protein samples were subjected to SDS-PAGE and immunoblotting using antibodies against ZNRF1, ZNRF2, synaptophysin (monoclonal; BD Biosciences, San Jose, CA), and PSD95 (postsynaptic density 95) (monoclonal; Affinity BioReagents, Golden, CO).
Figure 5.
ZNRF proteins are located in the presynaptic region in neurons in vivo. Top left, Adult mouse brain homogenate was sequentially centrifuged as schematically represented. Bottom, Immunoblot analysis to examine localization of ZNRF1, ZNRF2, as well as synaptophysin and PSD95. An equivalent amount of protein from each fractionation step was subjected to SDS-PAGE and immunoblot analysis using antibodies to the indicated proteins. Note that the profiles of ZNRF1 and ZNRF2 resemble those of synaptophysin and PSD95, respectively. ZNRF1 is present in LP2 (enriched for synaptic vesicles), whereas ZNRF2 is found in LP1 (synaptic heavy membrane). A-L, Top right, Both ZNRF1 and ZNRF2 are located in presynaptic terminals at the neuromuscular junction. Sections from adult mouse anterior tibial muscle tissue were immunostained using antibodies to ZNRF1 (A, G) and ZNRF2 (D, J) (visualized by Cy3) and compared with localization of SV2 immunoreactivity (B, H; a presynaptic marker, visualized by Alexa488) and binding site of FITC-conjugated α-bungarotixin (E,K; a postsynaptic marker). C,F,I, and L are merged images. Note that immunoreactivity for both ZNRF1 and ZNRF2 is colocalized with SV2 but not with α-bungarotixin-binding site. Scale bars, 5 μm. M-R, Bottom right, ZNRF1 and ZNRF2 are located in synaptic vesicle and on presynaptic terminal, respectively, in primary cultured hippocampal neurons. For synaptic vesicle localization, primary cultured hippocampal neurons were stained by antibodies against ZNRF1 (M) and synaptophysin (N; a synaptic vesicle marker). For presynaptic terminal localization, neurons transiently transfected with a ZNRF2-EGFP expression plasmid (shown in P) were loaded with FM 4-64 dye (Q; a presynaptic terminal marker). O and R are merged images. Scale bars, 20 μm.
Results
Identification and sequence analysis of ZNFR2
To identify additional mammalian proteins containing the unique zinc finger-RING finger motif present in ZNRF1, we used the BLAST to search dbEST and genomic DNA sequence databases using the full-length human ZNRF1 cDNA sequence as a query. We identified multiple human and murine EST clones (as well as genomic clones) that corresponded to a single transcript with high homology to ZNRF1. The full-length cDNA sequence of this transcript was determined by sequencing multiple EST clones identified by the search and merging them with EST sequences available in the database. The predicted open reading frame encodes 242 and 238 residues in human and mouse, respectively (Fig. 1). The sequence similarity between ZNRF1 and this novel protein, termed ZNRF2, was observed primarily in the C-terminal region, which contains a unique combination zinc finger-RING finger motif. Human ZNRF1 and ZNRF2 proteins share 80% identity in the region containing the zinc finger-RING finger motif, whereas the overall identity between them is 43%. We previously identified a single protein in both C. elegans and Drosophila that contains a similar zinc finger-RING finger domain, indicating that this domain is evolutionarily conserved and suggesting that it is functionally significant (Araki et al., 2001). The identification of a second protein (ZNRF2) containing this domain in human and mouse suggested that there may be a family of mammalian proteins with this domain, but numerous attempts to identify additional members and splice variants of this family in the databases were unsuccessful, indicating that there are only two ZNFR proteins in the human genome.
Figure 1.
ZNRF2 contains a zinc finger-RING finger motif homologous to that of ZNRF1. A, Alignment of human ZNRF1 and ZNRF2 amino acid sequences. Residues conserved between ZNRF1 and ZNRF2 are shaded. Cys and His residues of the zinc finger (boxed; residues 145-166 in ZNRF1 and 160-179 in ZNRF2) and RING finger (boxed; residues 184-224 in ZNRF1 and 199-239 in ZNRF2) are in bold. Arrows indicate sites of introns within the coding regions of ZNRF proteins. The ZNRF2 sequence data are available from GenBank/European Molecular Biology Laboratory/DNA Data Bank of Japan under accession numbers AF513707 and AF513708. B, Conserved exon-intron organization in the ZNRF1 and ZNRF2 genes. Genome sequences of ZNRF1 and ZNRF2 at the three intron-exon junctions within the coding region are shown. Nucleotides of the exon are shown in uppercase. The corresponding amino acids are numbered and shown above the nucleotide sequence.
To further explore their homology at the genomic level, we determined the exon-intron organization of both ZNRF1 and ZNRF2 within the coding region by analyzing the corresponding human genomic clones (Fig. 1B). This analysis revealed an identical structure for the human ZNRF1 and ZNRF2 genes, with all three exon-intron junctions of the coding region located within the conserved zinc finger-RING finger motif. A BLAST search against the genome sequence database revealed the location of human ZNRF2 on chromosome 7, region p15.1. A search of the OMIM (On-line Mendelian Inheritance in Men) database showed that human diseases of unknown pathogenesis have been linked to this region, including spinal muscular atrophy, cystoid macular dystrophy, autosomal dominant deafness 5, and retinitis pigmentosa 9.
ZNRF2 is highly expressed in the nervous system
ZNRF1 is expressed predominantly in the nervous system during development and in adulthood. The nearly ubiquitous neuronal expression in both the CNS and PNS suggested that ZNRF1 may be central to basic neuronal functions. To obtain similar expression information for ZNRF2, we analyzed ZNRF2 mRNA levels in a panel of human tissues using quantitative RT-PCR (qRT-PCR). We found that ZNRF2 is highly expressed in the brain, with higher expression during development than in adult, similar to that observed for ZNRF1 (Fig. 2A). Additionally, many organs outside of the nervous system express ZNRF2, including mammary gland, testis, colon, and kidney. To determine the cell-type-specific expression of ZNRF2 and compare it with ZNRF1 expression, we performed in situ hybridization on postnatal day 0 (P0) mouse pups and immunohistochemistry on adult mouse tissues (Fig. 2F,K). At P0, expression of both ZNRF1 and ZNRF2 was most prominent in the nervous system. In the CNS, ZNRF2 was expressed in the entire brain and the spinal cord, with the most intense signal detected in the cortical plate in which differentiated neurons are located, a pattern that is strikingly similar to that of ZNRF1. In the adult, immunohistochemical analysis revealed that ZNRF2 is expressed in the entire CNS. However, the expression of ZNRF2 was more intense in the granular cell layer of hippocampus, Purkinje cell layer of the cerebellum, and the granular cell layer of the olfactory bulb, whereas ZNRF1 showed a uniform staining pattern throughout the brain (Fig. 2B,C).
Figure 2.
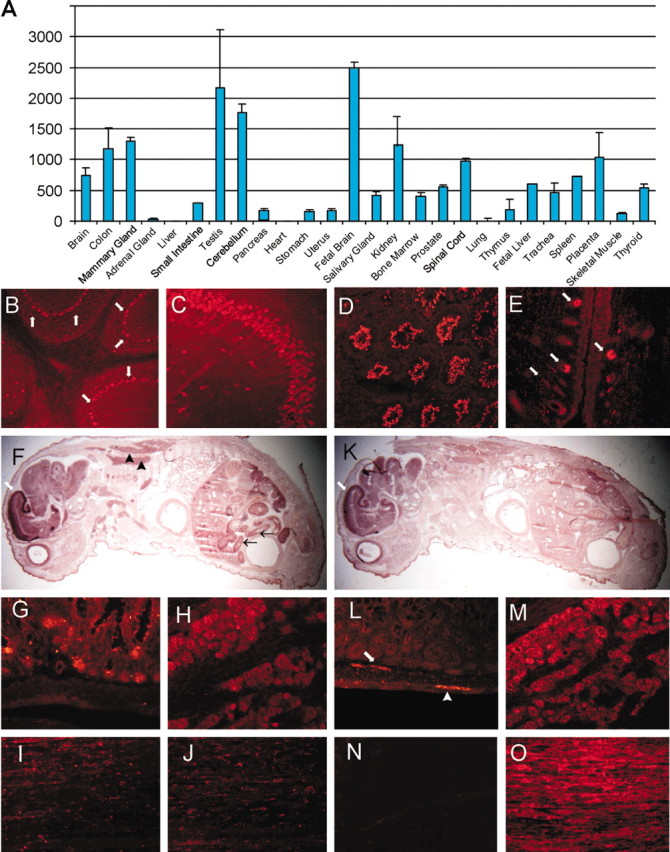
ZNRF2 is highly expressed in the nervous system. A, ZNRF2 mRNA levels in human tissues were determined by qRT-PCR. The expression level was normalized to glyceraldehyde-3-phosphate dehydrogenase expression in each sample and is indicated relative to the expression level in liver. All qRT-PCR reactions were performed in duplicate, and the SD is indicated. B-E, G-O, Immunohistochemical analysis of ZNRF2 (B-E, G-J) and ZNRF1 (L-O) in adult mouse (B-M) and rat (I-O) tissues. Signals in cerebellum (B), hippocampus (C), testis (D), hair follicles (E), gut (G, L), trigeminal ganglion (H, M), normal sciatic nerve (I, N), and sciatic nerve (J, O) 7 d after transection are shown. Arrows denote signals in Purkinje cell layer in B, hair matrix (basal portion of hair follicles) in E, and submucosal enteric ganglia in L. Arrowhead in L denotes myenteric ganglia. F, K, Expression of ZNRF2 (F) and ZNRF1 (K) in P0 mouse was examined using in situ hybridization. White arrows denote intense expression of ZNRF2 (F) and ZNRF1 (K) in the cortical plate. Black arrows and arrowheads denote ZNRF2 expression in the gut and adipose tissue, respectively. Note that the hybridization signal is observed predominantly in brain at P0 for both ZNRF1 and ZNRF2 and that the distribution patterns inthe brain are very similar.
Because ZNRF1 expression is induced in Schwann cells after nerve injury and it is also expressed in most neurons of the PNS, we examined the expression of ZNRF2 in PNS ganglia and peripheral nerves. In P0 and adult animals, ZNRF2 was detected in sensory neurons but was not expressed in sympathetic or enteric neurons (Fig. 2G-M). In peripheral nerve, ZNRF2 was weakly expressed in myelinating Schwann cells, and its expression did not change after peripheral nerve injury, in contrast to the induction of ZNRF1 expression in Schwann cells after nerve injury (Fig. 2I-O). These results indicate that, although ZNRF1 and ZNRF2 have overlapping patterns of expression in the CNS, the expression of these two proteins is distinct in the PNS. As observed in the qRT-PCR survey, ZNRF2 is expressed in many tissues outside of the nervous system, including the testis. Immunohistochemical analysis of this tissue showed that mature sperm, but not developing spermatogonia, strongly express ZNRF2. Other regions that strongly express ZNRF2 include adipose tissue, columnar epithelial cells of the gut, and hair follicles (Fig. 2D-G). These results are in contrast to those obtained with ZNRF1, which is almost exclusively expressed in the nervous system.
ZNRF proteins are E3 ubiquitin ligases
Many proteins containing RING finger(s) function as E3 ubiquitin ligases; we therefore tested whether ZNRF proteins, which contain a zinc finger-RING finger combination motif, had ubiquitin ligase activity. We performed in vitro ubiquitination assays to determine whether ZNRF1 and ZNRF2 could transfer ubiquitin to other proteins in the presence of the proper E2 ubiquitin-conjugating enzyme (Fig. 3A). ZNRF1 and ZNRF2 proteins were generated by in vitro transcription-translation-coupled reaction as molecules fused to EGFP-hexahistidine and purified by Ni-affinity chromatography. Purified ZNRF1 or ZNRF2 was incubated along with ubiquitin, ATP, recombinant E2 proteins produced in E. coli (which does not express components of ubiquitin conjugating system), and purified E1 ubiquitin-activating enzyme. Among the eight different E2 ubiquitin-conjugating enzymes tested, both ZNRF1 and ZNRF2 exhibited E3 activity with only Ubc4 and UbcH5C. To ensure the veracity of the observed ubiquitination, additional experiments were performed to confirm that all of enzymatic components [i.e., E1, E2 (UbcH5C), ubiquitin, and ZNRF protein] were required for ubiquitination (Fig. 3B). Together, these data demonstrate that both ZNRF1 and ZNRF2 have E3 ubiquitin ligase activity.
Figure 3.
ZNRF1 and ZNRF2 are E3 ubiquitin ligases. A, In vitro ubiquitination assays were performed with ZNRF1 (top) or ZNRF2 (bottom) in the absence (Mock) or presence of bacterial lysates containing the indicated E2 proteins. Polyubiquitinated proteins generated by in vitro ubiquitination were detected by immunoblot analysis using anti-ubiquitin (Anti-Ub) antibodies. B, In vitro ubiquitination assays were performed with ZNRF1, E1, bacterial lysate containing E2 (UbcH5C), and ubiquitin or in reactions lacking the indicated component (denoted by Δ). Assay mixtures were incubated for 2 hr at 30°C and separated by SDS-PAGE, and polyubiquitinated proteins were detected by immunoblot analysis using anti-ubiquitin antibodies.
To determine whether the ubiquitin ligase activity of ZNRF proteins requires the RING finger domain as has been found in previously characterized RING finger-containing E3 ubiquitin ligases, we tested a number of ZNRF1 mutants for their ability to ubiquitinate substrates (Fig. 4). We found that deletion of the entire zinc finger-RING finger domain [mutant ZNRF1(1-141)] abolished E3 activity. Conversely, a mutant containing this domain alone, ZNRF1(142-227), was fully capable of E3 activity. Furthermore, mutation of a cysteine residue that disrupts the RING finger structure [ZNRF1(C184A)] resulted in the loss of E3 activity, whereas a mutation disrupting the zinc finger structure [ZNRF1(C145A)] did not alter E3 activity. Collectively, these data indicate that both ZNRF1 and ZNRF2 function as E3 ubiquitin ligases and that only the RING finger of the zinc finger-RING finger motif is required for this activity.
Figure 4.

ZNRF protein ubiquitin ligase activity requires the RING finger motif. A, Schematic representation of wild-type and mutant ZNRF proteins. B, In vitro ubiquitination assays were performed using the E3 proteins indicated in A. Assay mixtures were separated by SDS-PAGE, and polyubiquitinated proteins were detected by immunoblot analysis using anti-ubiquitin (Anti-Ub) antibodies (top). The ZNRF proteins were produced using in vitro transcription-translation-coupling reactions using [35S]methionine-cysteine mixture. Aliquots of the reaction mixtures were separated by SDS-PAGE and subjected to autoradiography to confirm production of the indicated proteins (bottom).
ZNRF proteins are differentially located in presynaptic terminals of differentiated neurons
The subcellular location of a newly discovered protein can provide valuable insights into its potential function. This is particularly true for E3 ubiquitin ligases because they are necessarily colocalized with their target protein(s). Because ZNRF1 and ZNRF2 are highly expressed in neurons, their function in these cells is of particular interest. To determine the subcellular localization of ZNRF proteins in neurons, we first used sequential centrifugation to fractionate extracts of adult mouse brain. Initial separation experiments of brain homogenate indicated that both ZNRF proteins are located in the crude synaptosomal fraction (data not shown). We therefore performed a standard synaptosomal fractionation (Huttner et al., 1983) as schematically shown in Figure 5 to further examine their subcellular localization. Immunoblot analysis of separated fractions showed that ZNRF1 is present in the LP2 fraction, which is enriched for synaptic vesicle membranes. Copurification of ZNRF1 with synaptophysin, a marker for synaptic vesicles, further supported the localization of ZNRF1 to synaptic vesicle membranes. On the other hand, ZNRF2 is predominantly detected in the LP1 fraction, which is enriched for synaptic plasma membranes. This fraction also contains PSD95, a protein found in the postsynaptic density, a structure important for signal transduction in the postsynaptic membrane. Despite the overall homology of ZNRF proteins, these fractionation studies using brain homogenates demonstrate that ZNRF1 and ZNRF2 are differentially localized within the synaptic region.
The synaptosomal fractions obtained by sequential centrifugation contain both presynaptic and postsynaptic components. Therefore, to clearly determine the synaptic localization of the two ZNRF proteins, we examined two well characterized synapses: the neuromuscular junction, a distinct structure with well characterized presynaptic and postsynaptic molecular components, and primary cultured hippocampal neurons, in which subcellular localization of synaptic components has been well studied. For analysis of the neuromuscular junction, cross-sections of mouse anterior tibial muscle was costained using antibodies against ZNRF proteins and either SV2 (a presynaptic marker) or α-bungarotoxin (a postsynaptic marker) (Fig. 5). We found that both ZNRF1 and ZNRF2 colocalize with SV2 but not with α-bungarotoxin binding, indicating that they are present in a presynaptic compartment. For analysis of synaptic connections in hippocampal neuron cultures, we performed immunohistochemistry using antibodies against ZNRF proteins and synaptophysin (a synaptic vesicle marker). We found that ZNRF1, but not ZNRF2, colocalizes with synaptophysin (Fig. 5 and data not shown). We also examined whether ZNRF proteins are present in the presynaptic terminal. The hippocampal neurons were transfected with a construct expressing either ZNRF1-EGFP or ZNRF2-EGFP fusion protein. Neurons overexpressing these reporter proteins were then stained with FM 4-64 to identify the presynaptic region. We found that the ZNRF2-EGFP, but not ZNRF1-EGFP, overlapped with the FM 4-64 presynaptic dye. We also attempted to examine the synaptic terminal localization of ZNRF proteins by electron microscopy using this system, but the anti-ZNRF antibodies were not suitable for this type of analysis. Together, the body of evidence obtained from the immunohistochemical results as well as the fractionation results collectively indicates that both ZNRF1 and ZNRF2 are located in the presynaptic region, with ZNRF1 located on synaptic vesicles and ZNRF2 on presynaptic plasma membranes.
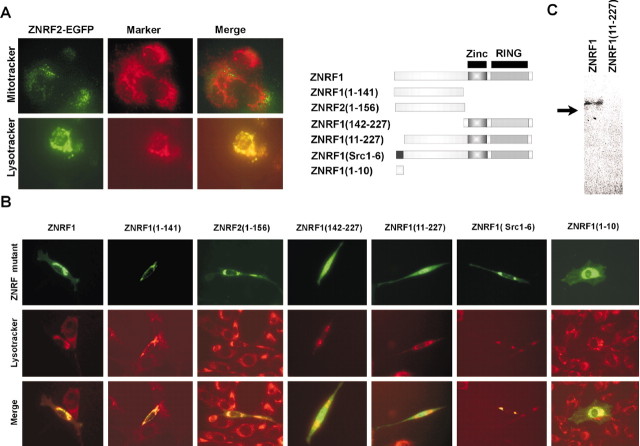
Both ZNRF1 and ZNRF2 are N-myrisotoylated and located in the endosome-lysosome compartment in fibroblasts
In addition to their abundant expression in neurons, both ZNRF proteins are also expressed in non-neuronal tissues. The synaptic membrane fractions of brain homogenates also include endocytic vesicles or endosomes located at synaptic terminals. Indeed, a number of synaptic vesicle molecules are located in endosomes in non-neuronal cells (Feany et al., 1993; Stenius et al., 1995; Wheeler et al., 2002). We transfected ZNRF expression constructs into fibroblasts to examine their subcellular localization and to determine the sequence motifs involved in their compartmentalization. For these experiments, we used ZNRF-EGFP fusion proteins, composed of either wild-type or mutant ZNRF1 or ZNRF2. These expression vectors were transiently transfected into NIH3T3 cells and stained with organelle-specific markers. The cells were then examined under a fluorescent microscope 24-48 hr later to determine the subcellular localization of EGFP reporters (Fig. 6). We found that ZNRF2-EGFP colocalized with Lysotracker, an endosome-lysosome-specific dye, but not with mitochondrial or endoplasmic reticulum markers (Fig. 6 and data not shown). These results indicate that ZNRF2, like ZNRF1 (Araki et al., 2001), is targeted to the endosome-lysosome compartment in fibroblasts.
Figure 6.
ZNRF proteins are located in the endosome-lysosome compartment by N-myristoylation. A, NIH3T3 cells expressing the ZNRF2-EGFP fusion protein were stained with the indicated organelle-specific markers. Note that the EGFP signal colocalized with Lysotracker, a marker for endosome-lysosomes (bottom). Mitotracker, a mitochondrial marker, did not colocalize with the EGFP signal (top). Merge signifies the merged ZNRF2-EGFP and organelle-specific signals. B, NIH3T3 cells were transfected with wild-type or mutant ZNRF1-EGFP fusion protein expression constructs (schematically represented). Twenty-four hours after transfection, the cells were stained with Lysotracker, an endosome-lysosome-specific dye. Note that wild-type ZNRF1, ZNRF1(1-141), ZNRF2(1-156), and ZNRF1(Src1-6) showed speckled patterns and colocalized well with Lysotracker, but ZNRF1(142-227) and ZNRF1(11-227) were located throughout the entire cell. ZNRF1(1-10) associated with membrane structures within the cell but was not located exclusively in the endosome-lysosome compartment. C, NIH3T3 cells transfected with either ZNRF1 or ZNRF1(11-227)-EGFP expression construct (see schematic figure in B) were metabolically labeled with [3H]myristic acid for 4 hr. The cells were lysed, and ZNRF-EGFP fusion proteins were immunoprecipitated using anti-EGFP antibodies. The immunoprecipitated proteins were separated by SDS-PAGE, and myristoylated proteins were detected by autoradiography. Myristoylated ZNRF1 protein is indicated by an arrow.
The targeting of both ZNRF proteins to the endosome-lysosome compartment in fibroblasts suggests that they share a sequence motif that directs them to this subcellular location. However, the shared zinc finger-RING finger domain does not contain sequences, such as the FYVE motif, which directs localization to the endosome-lysosome (Raiborg et al., 2002). To identify regions important for the subcellular localization of ZNRF proteins, we constructed a series of mutants containing various regions of ZNRF1 fused to EGFP. These mutants were transfected into NIH3T3 cells, and their subcellular location was determined using fluorescence microscopy. We initially tested ZNRF1(1-141)-EGFP, which contained the N-terminal 142 residues (non-zinc finger-RING finger region) and ZNRF1(142-227)-EGFP, which contained the C-terminal 85 residues (zinc finger-RING finger region) (Fig. 6B). We found that the N-terminal 142 residue fragment was capable of supporting localization to the endosome-lysosome, whereas the C-terminal zinc finger-RING finger domain EGFP fusion protein was distributed throughout the entire cell in a pattern similar to EGFP itself. We also generated a ZNRF2 deletion mutant that removed the zinc finger-RING finger domain [ZNRF2(1-156)-EGFP] and found that this molecule was also localized to the endosome-lysosome compartment. These results indicated that the N-terminal domains of both ZNRF proteins contain sequence motif(s) that determine their subcellular distribution.
We further scrutinized the amino acid sequences of the ZNRF1 and ZNRF2 N-terminal regions and found a region of weak homology (residues 1-10). Because this region contains an N-myristoylation signal (Maurer-Stroh et al., 2002a,b), we designed mutants to distinguish whether N-myristoylation itself or the conserved sequence around the N-myristoylation consensus is important for endosomal localization. We generated ZNFR(11-227), which lacks the entire N-terminal conserved region, and ZNRF1(Src1-6), in which the putative N-myristoylation signal of ZNRF1 (residues 1-6) was exchanged with that of human c-Src, a well characterized N-myristoylated protein with no sequence homology to ZNRF proteins. We found that the deletion of the first 10 residues in ZNRF1 had a profound effect on subcellular localization because the ZNRF1(11-227)EGFP protein was observed throughout the entire cell, including the nucleus. Conversely, the signal from ZNRF1(Src1-6)-EGFP overlapped with Lysotracker and appeared similar to that of full-length ZNRF1, indicating that a myristoylation signal is necessary for proper localization. To determine whether the N terminus of ZNRF proteins is myristoylated, we metabolically labeled NIH3T3 cells transfected with either ZNRF1-EGFP or ZNRF1(11-227)-EGFP with [3H]myristic acid. The ZNRF-EGFP fusion proteins were immunoprecipitated using anti-EGFP antibody, and myristoylated proteins were detected by autoradiography (Fig. 6C). [3H]-Labeled protein was found in lysates of cells transfected with ZNRF1-EGFP but not with ZNRF1(11-227)-EGFP. These results indicate that residues near the N terminus of ZNRF proteins are required for myristoylation and subsequent localization to endosome-lysosomes.
Although N-myristoylation supports attachment to membranous structures within the cell, it is possible that additional signals are also necessary for appropriate endosomal localization. To further examine whether residues near the N terminus are sufficient for endosome-lysosome localization, we fused the first 10 residues of ZNRF1 directly to EGFP to create mutant ZNRF1(1-10)-EGFP (Fig. 6B). The subcellular location of this mutant was found to be associated with various membranous structures in the cytoplasm, including the endosome-lysosome compartment, as demonstrated by partial colocalization with Lysotracker. These results indicate that the N-myristoylation signal is necessary but not sufficient for the restricted endosome-lysosome localization of ZNRF proteins and that additional residues within the non-conserved N-terminal domain are also involved.
ZNRF proteins play a role in regulating Ca2+-dependent exocytosis
The presynaptic localization of ZNRF proteins suggests that they play a role in the regulation of molecules related to secretion from and/or recycling of synaptic vesicles. Molecular aspects of exocytosis have been extensively studied by monitoring secretion of hGH from transfected PC12 cells in response to depolarization (Wick et al., 1993). Using this paradigm, alterations in exocytosis have been observed consequent to overexpression of important synaptic vesicle molecules (e.g., synaptophysin and synaptogyrin), indicating that proper levels of these proteins are crucial for exocytotic function. To examine whether perturbation of ZNRF-regulated protein metabolism results in abnormal exocytosis, plasmids encoding either wild-type ZNRF1 or ZNRF2 or their dominant-negative mutant counterparts, ZNRF1(C184A) and ZNRF2(C199A) that contain point mutations in a consensus cysteine residue of the RING finger, were transiently transfected into PC12 cells along with an hGH reporter plasmid (Fig. 7). We found that overexpression of either ZNRF1 or ZNRF2 mutants dramatically inhibits hGH secretion, whereas the wild-type ZNRF proteins had no effect. The inhibitory effects on secretion by the dominant-negative ZNRF mutants occurred in a dose-dependent manner, indicating the direct relationship between ZNRF activity and exocytosis. The inhibitory effect by mutant ZNRF proteins was similarly observed when hGH secretion was induced by incubating the transfected PC12 cells with high potassium saline or in medium containing 4.76 mm calcium under permeabilized conditions, indicating that the inhibitory effect was mediated directly via calcium-dependent exocytosis and was not dependent on voltage-gated potassium channels. The effects of ZNRF overexpression on secretion were not attributable to defects in the loading of hGH into exocytotic vesicles, because immunocytochemistry on NGF-stimulated transfected PC12 cells showed that hGH was similarly compartmentalized into subcellular vesicular structures in the presence or absence of mutant or wild-type ZNRF proteins (data not shown). Overexpression of either wild-type or dominant-negative ZNRF proteins did not appear to affect the general cellular metabolism of PC12 cells because there was no change in their growth rate or in NGF-mediated differentiation (e.g., neurite outgrowth) (data not shown).
Figure 7.
ZNRF proteins play a role in exocytosis. A, PC12 cells were transfected with either hGH expression plasmid alone (indicated by hGH only) or together with expression plasmids for native and mutated ZNRF proteins [WT, wild type; ZNRF1 Mut, ZNRF1(C184A); ZNRF2 Mut, ZNRF2(C199A)]. Transfected cells were incubated at 37°C for 20 min with physiological saline (indicated by PBS), high K+ solution (56 mm) (indicated by High K), or Ca2+ (4.76 mm) under a permeablized condition (indicated by Ca). The amounts of hGH secreted into the medium versus retained in the cells was measured by ELISA, and hGH release was calculated from four independent experiments performed in duplicate as a percentage of total hGH synthesized. **p < 0.01, indicates significant decrease of secreted hGH. B, PC12 cells were cotransfected with a constant amount of hGH plasmid and a varying amount of the indicated ZNRF expression plasmids. Secretion and measurement of hGH and calculation of secreted portion of hGH were performed as described in A. *p < 0.05 and **p < 0.01, indicate significant decrease of secreted hGH.
Finally, to examine the specificity of the effects of mutant ZNRF expression on exocytosis, we performed analogous experiments in which wild-type ZNRF1 or ZNRF2 was cotransfected with mutant ZNRF1 or mutant ZNRF2. We found that inhibition of secretion by mutant ZNRF1 could be reversed by expression of wild-type ZNRF1 but not wild-type ZNRF2. Similarly, mutant ZNRF2 inhibition could only be reversed by wild-type ZNRF2. These results indicate that both ZNRF1 and ZNRF2 play a role in regulation of exocytosis-related molecules and that they appear to have independent target molecules.
Discussion
The ubiquitination pathway plays a crucial role in the degradation of proteins involved in a variety of cellular processes, including differentiation, proliferation, and apoptosis (Pickart, 2001). In the present study, we defined a novel family of E3 ubiquitin ligases, the ZNRF proteins, through the identification of ZNRF2, a protein homologous to the previously identified nin283/ZNRF1. The ZNRF proteins share a novel functional domain containing a combination of a zinc finger and a RING finger, which we determined is required for their role as E3 ubiquitin ligases. ZNRF proteins are also similar in other aspects because they are both highly expressed in CNS and PNS neurons during development and in adulthood. In the brain and PNS ganglia, ZNRF proteins are uniformly and ubiquitously expressed without large regional variation or cell-type expression specificity. This pattern of expression, along with the presynaptic localization of ZNRF proteins and their effects on exocytosis, suggest that they are involved in general presynaptic mechanisms shared by both CNS and PNS neurons.
A number of recently identified molecules involved in Ca2+-dependent vesicular fusion are conserved among a wide range of species, indicating their central role in cell biology. These include Sec18/NSF (N-ethylmaleimide-sensitive fusion protein) homologues, SNAPs (soluble NSF attachment proteins), SNARE (SNAP receptor) complex [SNAP25 (synaptosome-associated protein 25 kDa], syntaxin, and synaptobrevin), Sec1/Munc18-like (SM) proteins, small GTPases of Rab family, and other synaptic vesicle-associated molecules, such as synaptotagmin. In some cases, such as synaptogyrin and synaptophysin, overexpression of these proteins strongly inhibits exocytosis, indicating that the proper stoichiometry of these proteins is important in maintaining exocytotic function. The regulation of many proteins involved in presynaptic exocytosis (e.g., syntaxin1 and SNAP25) occurs via targeted proteolysis, becuase they are ubiquitinated by specific E3 ubiquitin ligases and degraded in the proteasome. The regulated degradation of these proteins is important in establishing proper synaptic signal transmission and maintaining neuronal integrity, because a naturally occurring mutation in the USP14 de-ubiquitinating enzyme in mice results in ataxia secondary to defects in synaptic transmission (Wilson et al., 2002). Experiments in Drosophila also support the importance of ubiquitination of presynaptic proteins, because neuronal overexpression of the de-ubiquitinating enzyme fat facets as well as loss-of-function of highwire, a putative synaptic E3 ubiquitin ligase, lead to disruption of synaptic growth control (Hegde and DiAntonio, 2002). The expression of ZNRF ubiquitin ligases during neuronal development, their presence at presynaptic terminals, and their role in exocytosis all lead to the suggestion that they are involved in establishing and/or maintaining synaptic transmission or plasticity. In accord, impaired ZNRF activity may contribute to neurological disorders. In this regard, it is interesting that degenerative diseases of unknown etiology, including spinal muscular atrophy, cystoid macular dystrophy, and retinitis pigmentosa 9, are linked to the ZNRF2 locus.
Whereas ubiquitination has been linked to many crucial processes in the CNS, the roles of ubiquitin-mediated protein metabolism in the PNS are poorly documented. ZNRF1 and ZNRF2 are both expressed in PNS neurons, and ZNRF1 is highly induced in Schwann cells surrounding damaged axons after peripheral nerve injury (Araki et al., 2001). These cells go from a quiescent state with a stable myelin structure to a proliferative and trophic state that creates an environment conducive to nerve regeneration after the damaged axons have degenerated. In addition to ZNRF1, we identified additional proteins induced after nerve injury that are also involved in ubiquitination, such as Praja1 and E2 ubiquitin-conjugating enzymes E2N (Araki et al., 2001). These data, along with experiments linking synaptogenesis with ubiquitination, suggest that ubiquitination plays an important role in the PNS, particularly during nerve degeneration and regeneration. Indeed, a mutation in the ubiquitination factor 4B has been identified in Wld mice (Conforti et al., 2000), a naturally occurring mouse mutant with markedly slower Wallerian degeneration of injured axon. Thus, it appears that ubiquitin-mediated processes, such as those potentially mediated by ZNRF proteins, are important for proper development and regeneration after injury in the PNS.
Although ZNRF proteins are located in the presynaptic terminal of differentiated neurons, they are also found in the endosome-lysosome compartment of non-neuronal cells. Among these tissues, ZNRF1 and ZNRF2 levels are highest in the testis. We observed that ZNRF1 is expressed in developing spermatogonia (Araki et al., 2001), whereas ZNRF2 is expressed in mature spermatozoa. Ubiquitination is crucial for proper spermatogenesis, as evidenced by the infertility phenotype of mice lacking molecules associated with ubiquitination, such as Siah1a (an E3 ubiquitin ligase) (Dickins et al., 2002) or Uch-L1 (a de-ubiquitinating enzyme) (Kwon et al., 2003). Although a role for histone ubiquitination during meiosis is clear (Sutovsky, 2003), recent reports suggest that alterations in ubiquitination can also modify signal transduction and produce changes in the microtubule network (Escalier et al., 2003). In addition, ubiquitination of proteins on the surface of spermatozoa in the epididymis may play a role in the recognition and elimination of defective spermatozoa (Sutovsky et al., 2001). Interestingly, the E2 ligases used by ZNRF proteins, UBC4 and UBC5, are both induced during spermatogenesis (Wing et al., 1996). Additional studies to determine ZNRF expression in the different stages of spermatogenesis as well as to examine their subcellular localization in these cells will be necessary to determine the roles of ZNRF proteins in sperm development.
The discovery that ZNRF proteins are localized to the presynaptic region and endosomes suggest that they are involved in regulating molecules important for exocytosis and cell signaling. Functional deficits in these proteins may result in abnormal accumulations of these target proteins and lead to abnormal signaling and disease.
Footnotes
This work was supported by National Institute of Neurological Disorders and Stroke Grant NS40745 and a grant from the Alzheimer's Disease Research Center at Washington University. We thank Drs. Kei-Ichi Nakayama and J. Gordon for plasmids encoding E2 enzymes and hGH, respectively, and Nam Le for comments on this manuscript.
Correspondence should be addressed to Jeffrey Milbrandt, Department of Pathology and Immunology, Washington University School of Medicine, 660 South Euclid Avenue, Campus Box 8118, St. Louis, MO 63110. E-mail: jeff@pathbox.wustl.edu.
Copyright © 2003 Society for Neuroscience 0270-6474/03/239385-10$15.00/0
References
- Araki T, Milbrandt J ( 1996) Ninjurin, a novel adhesion molecule, is induced by nerve injury and promotes axonal growth. Neuron 17: 353-361. [DOI] [PubMed] [Google Scholar]
- Araki T, Nagarajan R, Milbrandt J ( 2001) Identification of genes induced in peripheral nerve after injury. Expression profiling and novel gene discovery. J Biol Chem 276: 34131-34141. [DOI] [PubMed] [Google Scholar]
- Ausubel FM ( 2001) Current protocols in molecular biology, Vol 1, Chap 8. New York: Wiley. [Google Scholar]
- Borden KL ( 1998) RING fingers and B-boxes: zinc-binding protein-protein interaction domains. Biochem Cell Biol 76: 351-358. [DOI] [PubMed] [Google Scholar]
- Chin LS, Vavalle JP, Li L ( 2002) Staring, a novel E3 ubiquitin-protein ligase that targets syntaxin 1 for degradation. J Biol Chem 277: 35071-35079. [DOI] [PubMed] [Google Scholar]
- Conforti L, Tarlton A, Mack TG, Mi W, Buckmaster EA, Wagner D, Perry VH, Coleman MP ( 2000) A Ufd2/D4Cole1e chimeric protein and overexpression of Rbp7 in the slow Wallerian degeneration (WldS) mouse. Proc Natl Acad Sci USA 97: 11377-11382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiAntonio A, Haghighi AP, Portman SL, Lee JD, Amaranto AM, Goodman CS ( 2001) Ubiquitination-dependent mechanisms regulate synaptic growth and function. Nature 412: 449-452. [DOI] [PubMed] [Google Scholar]
- Dickins RA, Frew IJ, House CM, O'Bryan MK, Holloway AJ, Haviv I, Traficante N, de Kretser DM, Bowtell DD ( 2002) The ubiquitin ligase component Siah1a is required for completion of meiosis I in male mice. Mol Cell Biol 22: 2294-2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escalier D, Bai XY, Silvius D, Xu PX, Xu X ( 2003) Spermatid nuclear and sperm periaxonemal anomalies in the mouse Ube2b null mutant. Mol Reprod Dev 65: 298-308. [DOI] [PubMed] [Google Scholar]
- Feany MB, Yee AG, Delvy ML, Buckley KM ( 1993) The synaptic vesicle proteins SV2, synaptotagmin and synaptophysin are sorted to separate cellular compartments in CHO fibroblasts. J Cell Biol 123: 575-584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein DA ( 1979) Calculation of the concentrations of free cations and cation-ligand complexes in solutions containing multiple divalent cations and ligands. Biophys J 26: 235-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatakeyama S, Jensen JP, Weissman AM ( 1997) Subcellular localization and ubiquitin-conjugating enzyme (E2) interactions of mammalian HECT family ubiquitin protein ligases. J Biol Chem 272: 15085-10592. [DOI] [PubMed] [Google Scholar]
- Hatakeyama S, Yada M, Matsumoto M, Ishida N, Nakayama KI ( 2001) U box proteins as a new family of ubiquitin-protein ligases. J Biol Chem 276: 33111-33120. [DOI] [PubMed] [Google Scholar]
- Hegde AN, DiAntonio A ( 2002) Ubiquitin and the synapse. Nat Rev Neurosci 3: 854-861. [DOI] [PubMed] [Google Scholar]
- Hicke L ( 2001) Protein regulation by monoubiquitin. Nat Rev Mol Cell Biol 2: 195-201. [DOI] [PubMed] [Google Scholar]
- Huttner WB, Schiebler W, Greengard P, De Camilli P ( 1983) Synapsin I (protein I), a nerve terminal-specific phosphoprotein. III. Its association with synaptic vesicles studied in a highly purified synaptic vesicle preparation. J Cell Biol 96: 1374-1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim AH, Yano H, Cho H, Meyer D, Monks B, Margolis B, Birnbaum MJ, Chao MV ( 2002) Akt1 regulates a JNK scaffold during excitotoxic apoptosis. Neuron 35: 697-709. [DOI] [PubMed] [Google Scholar]
- Kwon J, Kikuchi T, Setsuie R, Ishii Y, Kyuwa S, Yoshikawa Y ( 2003) Characterization of the testis in congenitally ubiquitin carboxy-terminal hydrolase-1 (Uch-L1) defective (gad) mice. Exp Anim 52: 1-9. [DOI] [PubMed] [Google Scholar]
- Lopez-Salon M, Alonso M, Vianna MR, Viola H, Mello e Souza T, Izquierdo I, Pasquini JM, Medina JH ( 2001) The ubiquitin-proteasome cascade is required for mammalian long-term memory formation. Eur J Neurosci 14: 1820-1826. [DOI] [PubMed] [Google Scholar]
- Maurer-Stroh S, Eisenhaber B, Eisenhaber F ( 2002a) N-terminal N-myristoylation of proteins: refinement of the sequence motif and its taxon-specific differences. J Mol Biol 317: 523-540. [DOI] [PubMed] [Google Scholar]
- Maurer-Stroh S, Eisenhaber B, Eisenhaber F ( 2002b) N-terminal N-myristoylation of proteins: prediction of substrate proteins from amino acid sequence. J Mol Biol 317: 541-557. [DOI] [PubMed] [Google Scholar]
- Moss A, Blackburn-Munro G, Garry EM, Blakemore JA, Dickinson T, Rosie R, Mitchell R, Fleetwood-Walker SM ( 2002) A role of the ubiquitin-proteasome system in neuropathic pain. J Neurosci 22: 1363-1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickart CM ( 2001) Mechanisms underlying ubiquitination. Annu Rev Biochem 70: 503-533. [DOI] [PubMed] [Google Scholar]
- Raiborg C, Bache KG, Gillooly DJ, Madshus IH, Stang E, Stenmark H ( 2002) Hrs sorts ubiquitinated proteins into clathrin-coated microdomains of early endosomes. Nat Cell Biol 4: 394-398. [DOI] [PubMed] [Google Scholar]
- Saam JR, Gordon JI ( 1999) Inducible gene knockouts in the small intestinal and colonic epithelium. J Biol Chem 274: 38071-38082. [DOI] [PubMed] [Google Scholar]
- Sanjay A, Horne WC, Baron R (2001) The Cbl family: ubiquitin ligases regulating signaling by tyrosine kinases. Science's STKE (http://stke.sciencemag.org/cgi/content/full/sigtrans;2001/110/pe40). [DOI] [PubMed]
- Stenius K, Janz R, Sudhof TC, Jahn R ( 1995) Structure of synaptogyrin (p29) defines novel synaptic vesicle protein. J Cell Biol 131: 1801-1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugita S, Janz R, Sudhof TC ( 1999) Synaptogyrins regulate Ca2+-dependent exocytosis in PC12 cells. J Biol Chem 274: 18893-18901. [DOI] [PubMed] [Google Scholar]
- Sutovsky P ( 2003) Ubiquitin-dependent proteolysis in mammalian spermatogenesis, fertilization, and sperm quality control: killing three birds with one stone. Microsc Res Tech 61: 88-102. [DOI] [PubMed] [Google Scholar]
- Sutovsky P, Moreno R, Ramalho-Santos J, Dominko T, Thompson WE, Schatten G ( 2001) A putative, ubiquitin-dependent mechanism for the recognition and elimination of defective spermatozoa in the mammalian epididymis. J Cell Sci 114: 1665-1675. [DOI] [PubMed] [Google Scholar]
- von Arnim AG ( 2001) A hitchhiker's guide to the proteosome. Sci STKE 97: PE2. [DOI] [PubMed] [Google Scholar]
- Wheeler TC, Chin LS, Li Y, Roudabush FL, Li L ( 2002) Regulation of synaptophysin degradation by mammalian homologues of seven in absentia. J Biol Chem 277: 10273-10282. [DOI] [PubMed] [Google Scholar]
- Wick PF, Senter RA, Parsels LA, Uhler MD, Holz RW ( 1993) Transient transfection studies of secretion in bovine chromaffin cells and PC12 cells. Generation of kainate-sensitive chromaffin cells. J Biol Chem 268: 10983-10989. [PubMed] [Google Scholar]
- Wilson SM, Bhattacharyya B, Rachel RA, Coppola V, Tessarollo L, Householder DB, Fletcher CF, Miller RJ, Copeland NG, Jenkins NA ( 2002) Synaptic defects in ataxia mice result from a mutation in Usp14, encoding a ubiquitin-specific protease. Nat Genet 32: 420-425. [DOI] [PubMed] [Google Scholar]
- Wing SS, Bedard N, Morales C, Hingamp P, Trasler J ( 1996) A novel rat homolog of the Saccharomyces cerevisiae ubiquitin-conjugating enzymes UBC4 and UBC5 with distinct biochemical features is induced during spermatogenesis. Mol Cell Biol 16: 4064-4072. [DOI] [PMC free article] [PubMed] [Google Scholar]