Abstract
Previous studies of oxytocin (OT) and vasopressin (VP) cell-specific gene expression in the hypothalamus using transgenic mouse and rat models focused attention on the intergenic region (IGR) as the site of critical enhancer elements. In this study, we used organotypic slice-explant cultures of rat hypothalamus as in vitro models, and particle-mediated gene transfer (biolistics) transfection methods to identify critical DNA sequences in the IGR between the OT and VP genes responsible for hypothalamic-specific gene expression. Reducing the 5′ flanking region in the mouse VP gene from 3.5 kbp to 288 bp did not alter the efficacy of its expression in hypothalamic slices. All subsequent VP constructs were based on this 288 bp VP gene construct with changes made only to the IGR. These studies, which used various constructs with OT and VP promoters driving enhanced green fluorescent protein reporter gene expression, demonstrated that the IGR is necessary for OT and VP gene expression in hypothalamic slices in vitro. The DNA sequences in the IGR responsible for both OT and VP gene expression were located in a 178 bp domain immediately downstream of exon 3 of the VP gene. In addition, another domain in the IGR, 430 bp immediately downstream of exon 3 of the OT gene, contained a positive regulatory element for OT gene expression in the hypothalamus. Alignment of the DNA sequences in the 178 and 430 bp domains reveals four common sequences (motifs) that may be candidates for the putative enhancers in the IGR that regulate OT and VP gene hypothalamic-specific expression.
Keywords: hypothalamus, organotypic culture, biolistics, gene expression, vasopressin, oxytocin
Introduction
The neuropeptides oxytocin (OT) and vasopressin (VP) were first identified as neurohormones in the hypothalamo-neurohypophysial system (HNS) (Bargmann and Scharrer, 1951; Du Vigneaud, 1954). These peptides are now known to act in other regions of the nervous system and to control complex behaviors (Reijmers et al., 1998; Wang et al., 1998; Insel and Young, 2001). Although OT and VP fibers can be found throughout the CNS, expression of the OT and VP genes occurs almost exclusively in the hypothalamus. The principal sources of these peptides in the hypothalamus are the OT and VP magnocellular neurons (MCNs) of the HNS; the parvocellular corticotropin-releasing hormone (CRH)-synthesizing neurons in the paraventricular nucleus (PVN), which coexpress VP that is secreted into the portal circulation to stimulate release of ACTH from anterior pituitary corticotropes (Gillies et al., 1982; Rivier et al., 1984; Antoni, 1993); neurons in the suprachiasmatic nucleus, the central circadian clock in mammals (Klein et al., 1991), where VP is a major output controlling diurnal endocrine rhythms (Kalsbeek et al., 1996; Reppert and Weaver, 2001); as well as other parvocellular neurons in the PVN that regulate autonomic functions via projections to the brainstem and spinal cord (Swanson and Sawchenko, 1980, 1983; Sofroniew, 1985).
The OT and VP genes each contain three exons and two introns and are found on the same chromosome in opposite transcriptional orientations (Burbach et al., 2001). The domain separating the OT and VP genes has been called the intergenic region (IGR), and ranges from ∼ 3.6 kbp in the mouse (Sausville et al., 1985; Hara et al., 1990; Ratty et al., 1996) to ∼ 11 kbp in the rat and human (Mohr et al., 1988; Gainer et al., 2001). Cell-specific OT and VP gene expression can only be obtained in the hypothalamus of transgenic animals when constructs containing some of the IGR sequence downstream of exon 3 in the rodent vasopressin gene are used (Gainer, 1998; Waller et al., 1998; for review, see Burbach et al., 2001; Gainer and Young, 2001; Murphy and Wells, 2003). Consequently, the IGR has been postulated to contain important cis-elements involved in the cell-specific expression of these genes, and this view has been referred to as the IGR hypothesis (Gainer, 1998; Gainer and Young, 2001). Deletion analyses to further dissect the sequences in the IGR that regulate the cell-specific expression have been hampered by the absence of homologous cell lines, which could be useful as models for the OT- and VP-expressing hypothalamic neurons.
To further elucidate the putative regulatory elements within the IGR for OT and VP gene hypothalamus-specific expression, we used an alternative in vitro approach involving the combination of organotypic cultures and biolistics, in which we transfected primary neurons with constructs of the OT and VP genes linked to an enhanced green fluorescent protein (EGFP) reporter and containing varying segments of the IGR. This in vitro approach allowed us to explicitly test the IGR hypothesis and identified novel domains in the IGR that participate in hypothalamic-specific gene expression.
Materials and Methods
Organotypic slice explants
Organotypic cultures were prepared from postnatal day 6-8 Sprague Dawley rat pups, as described previously (House et al., 1998). Rats were decapitated, and their brains were quickly removed in accordance with National Institutes of Health guidelines for the care and use of animals study protocol approved by the National Institute of Neurological Disorders and Stroke Animal Care and Use Committee. Briefly, hypothalamic, hippocampal, and brainstem areas were isolated and sectioned into 350 μm slices and placed on top of Millicell-CM filter inserts (pore size, 0.4 μm; diameter, 30 mm; Millipore, Bedford, MA). Each filter insert contained three to five slices of a specific brain region from a single animal, and these were placed in a Petri dish (35 mm) containing 1.2 ml of culture medium (50% Eagle's basal medium with Earle's salts, 25% heat-inactivated horse serum, 25% HBSS, 0.5% glucose, and 25 U/ml penicillin-streptomycin). The osmotic pressure of the standard medium was 314 mOsm/l. Incubation of the cultures was stationary in 5% CO2-enriched air at 35°C, and the media was changed every 3 d. In some experiments, CNTF was added to the culture medium to increase the survival of magnocellular neurons in the slices, as described previously (Rusnak et al., 2002). Nine to 11 d after transfection, cultures were fixed using 10% formalin in PBS and assayed for expression of EGFP, VP, or OT by immunohistochemistry (IHC). Figure 1 shows examples of organotypic rat hypothalamic cultures immunohistochemically stained with a monoclonal antibody, PS 45 (Ben-Barak et al., 1985; Whitnall et al., 1985), that cross-reacts with both OT and VP neurophysins (NPs).
Figure 1.
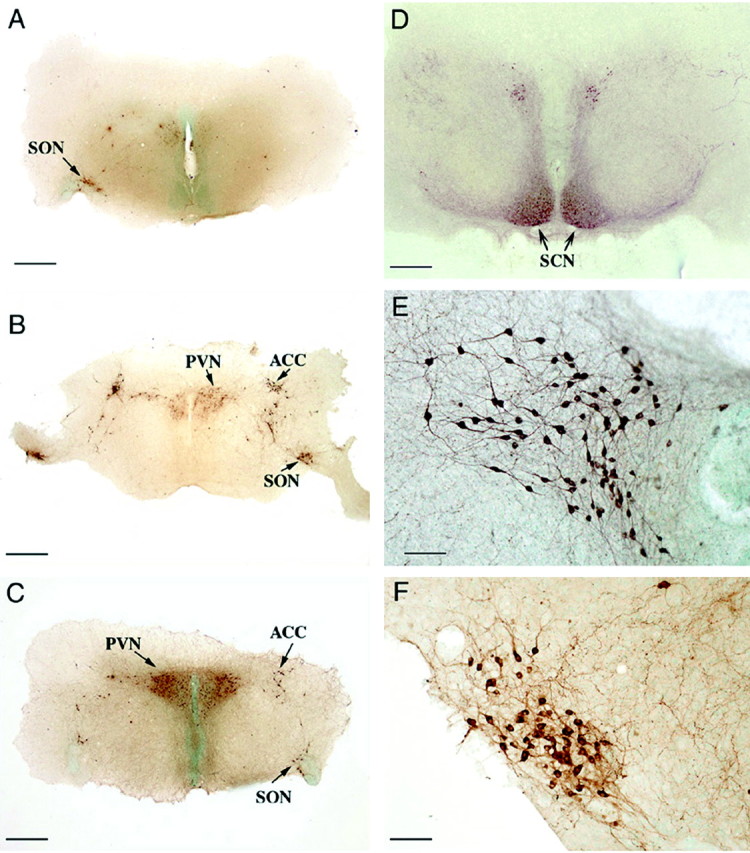
Illustration of a typical organotypic rat hypothalamic culture after 2 weeks in vitro and immunostained with a monoclonal antibody (PS 45) that cross-reacts with both OT and VP neurophysin. A-D illustrate the hypothalamic nuclei that express OT and VP. E and F show higher magnifications of the PVN and SON, respectively, and show the immunoreactive magnocellular neurons. Scale bars: A-D, 800 μm; E, F, 100 μm. ACC, Accessory hypothalamic nuclei; SCN, suprachiasmatic nucleus.
Biolistic transfections
Two days after preparation of the slices and their initial culturing, the cultures were transfected by biolistics using the Helios Gene Gun (Bio-Rad, Hercules, CA) using methods described previously (Wellmann et al., 1999; McAllister, 2000; Gainer et al., 2002). Each culture was “shot” twice to cover all of the slices on the filter with 1 μm gold particles containing 2 μg of plasmid DNA per milligram of gold (vector sequences are not shown in the gene constructs section) using 180 psi at a distance of 17 mm. The slice cultures were typically fixed for immunohistochemical assay (see below) 9 -11 d after the transfections.
Gene constructs
Neuronal-specific (positive control) construct
An α-tubulin promoter linked to the EGFP reporter construct was made from the pTA1:nlacZ plasmid obtained from Dr. F. D. Miller (McGill University, Montreal, Canada), which exhibited neuron-specific expression in transgenic mice (Gloster et al., 1994). The nlacZ in the construct was replaced by EGFP according to the methods of Wang et al. (1996).
Vasopressin gene constructs
All VP constructs were derived from VPIII.CAT.IGR2.1 (Jeong et al., 2001).
3.5VPIII.EGFP.IGR2.1. The 748 bp Bsp120I-NotI band containing the EGFP gene from pEGFP-N2 (Clontech, Palo Alto, CA) was ligated into VPIII.CAT.IGR2.1, which had been digested with NotI to remove the chloramphenicol acetyl transferase (CAT) gene (see Fig. 3A).
Figure 3.
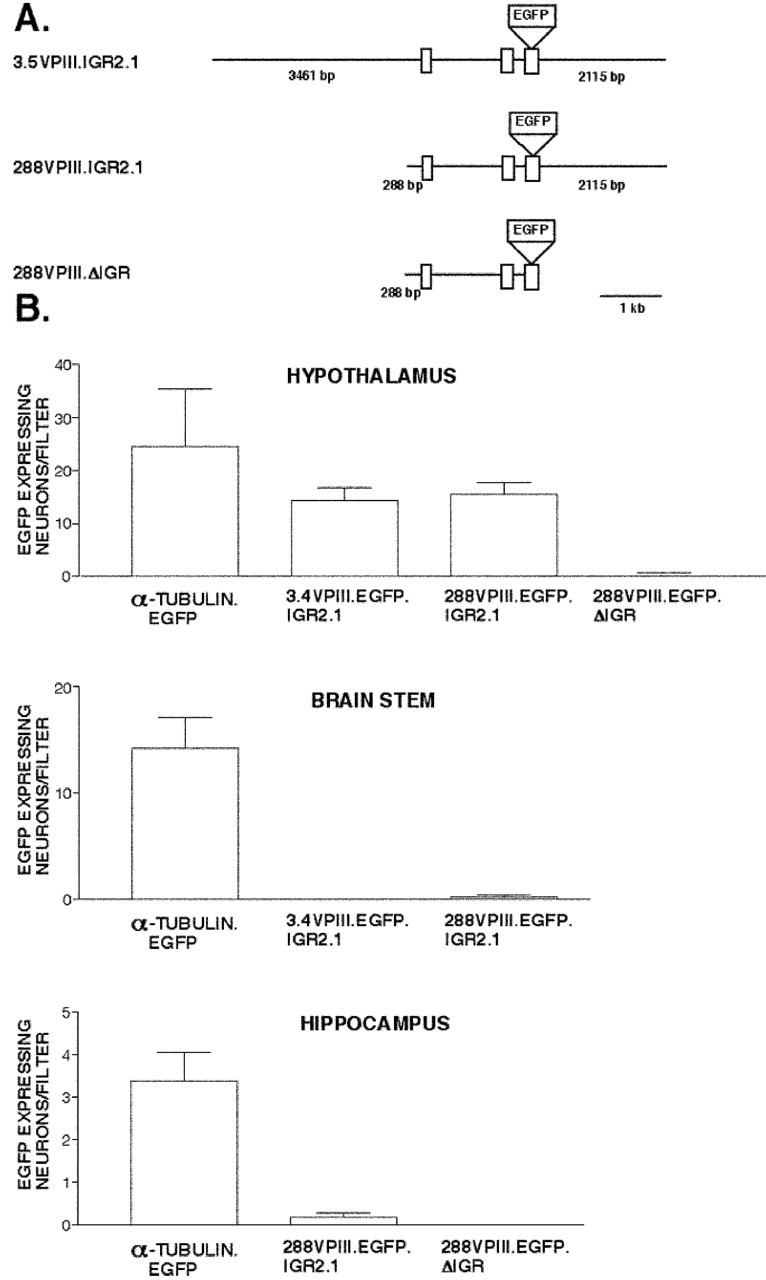
Efficacy and specificity of the vasopressin gene constructs used in the biolistic transfections. A, The 3.5VPIII.EGFP.IGR2.1 contains 3461 bp of sequence upstream of exon 1 followed by exon 1-3 with the EGFP reporter gene inserted into exon 3 at the end of the neurophysin coding sequence, followed by the first 2115 bp of sequence downstream of exon 3. The 288VPIII.EGFP.IGR2.1 construct was made by reducing the 5′ flanking sequence of 3.5VPIII.EGFP.IGR2.1 to 288 bp. The 288VPIII.EGFP.ΔIGR construct was made by removing the IGR sequence from the 288VPIII.EGFP.IGR2.1 construct (see Materials and Methods). B, Expression of the VP gene constructs shown in A, as well as plasmid constructs containing the α-tubulin promoter linked to the EGFP gene (positive control), were biolistically transfected into different regions of the rat brain. Cultures were assayed for EGFP by IHC fluorescent microscopy, and the EGFP expressing neurons on each filter were counted. Graphs show data for average numbers of EGFP-expressing neurons per filter ± SEMs in the hypothalamus, brainstem, and hippocampus cultures. Each filter contained tissues from a single neonatal rat. Note that although the α-tubulin promoter EGFP construct was expressed in all three brain regions, the two VP constructs containing the IGR were only expressed in the hypothalamus, with no difference in efficacy between the constructs containing either the 3.5 kb or the 288 bp 5′ upstream flanking region. Note also that the VP construct lacking the IGR (288VPIII.EGFP.ΔIGR) was not expressed in the hypothalamus.
288VPIII.EGFP.IGR2.1. The VPIII.EGFP.IGR2.1 was digested with BstEII to remove 86 bp of the pSE280 vector and 3173 bp of the 5′ flanking region of the VP gene (see Fig. 3A).
288VPIII.EGFP.ΔIGR. The VPIII.EGFP.IGR2.1 construct was digested with NotI and SpeI, and the 2178 bp band containing the mouse IGR along with 58 bp of the noncoding region of exon 3 was removed. The N-S linker oligos (forward strand, 5′-ATAAGAATGCGGCCGCAACTACTGAGCCATCGCCCCCACGCCTCGCCCCTACAGCATGGAAAATAAACTTTTAAAAACCGCGGCCAGTCACTAGTACACGCT-3′) were digested by NotI and SpeI and then ligated into the digested construct above to restore the exon 3 sequence and provide restriction sites for the insertion of a linker oligo. This gave the intermediate construct 288VPIII.EGFP.IGRN-S. The 288VPIII.EGFP.IGRN-S was digested with SacII and SpeI, and the VPIII linker oligos (forward strand, 5′-GTTCCCGCGGATGCGATCGCATTGCATTCGATATCATGCATCGAGTTAACTCGACTAGTGACC-3′) were digested by SacII and SpeI and then ligated into the digested 288VPIII.EGFP.IGRN-S to yield 288VPIII.EGFP.ΔIGR (see Fig. 3A).
288VPIII.EGFP.IGR376, 288VPIII.EGFP.IGR298, 288VPIII.EGF-P.IGR446, and 288VPIII.EGFP.IGR834. The 2178 bp NotI-SpeI band described above, which contained 2018 bp of the mouse IGR, was ligated into the NotI-SpeI sites of pSE280 (Invitrogen Carlsbad, CA) to give pSEIGR. Aliquots of pSEIGR were digested with PvuI and BsmI, BsmI and AvaIII, AvaIII and HpaI, and HpaI and SpeI to produce the 374, 298, 446, and 834 bp bands, which were isolated (described as bands B, C, D, and E in Fig. 6 A) and ligated into aliquots of 288VPIII.EGFP.ΔIGR that were digested by the above enzymes to yield the constructs 288VPIII.EG-FP.IGR374, 288VPIII.EGFP.IGR298, 288VPIII.EGFP.IGR446, and 288VPIII.EGFP.IGR834, respectively (see Fig. 6 A).
Figure 6.
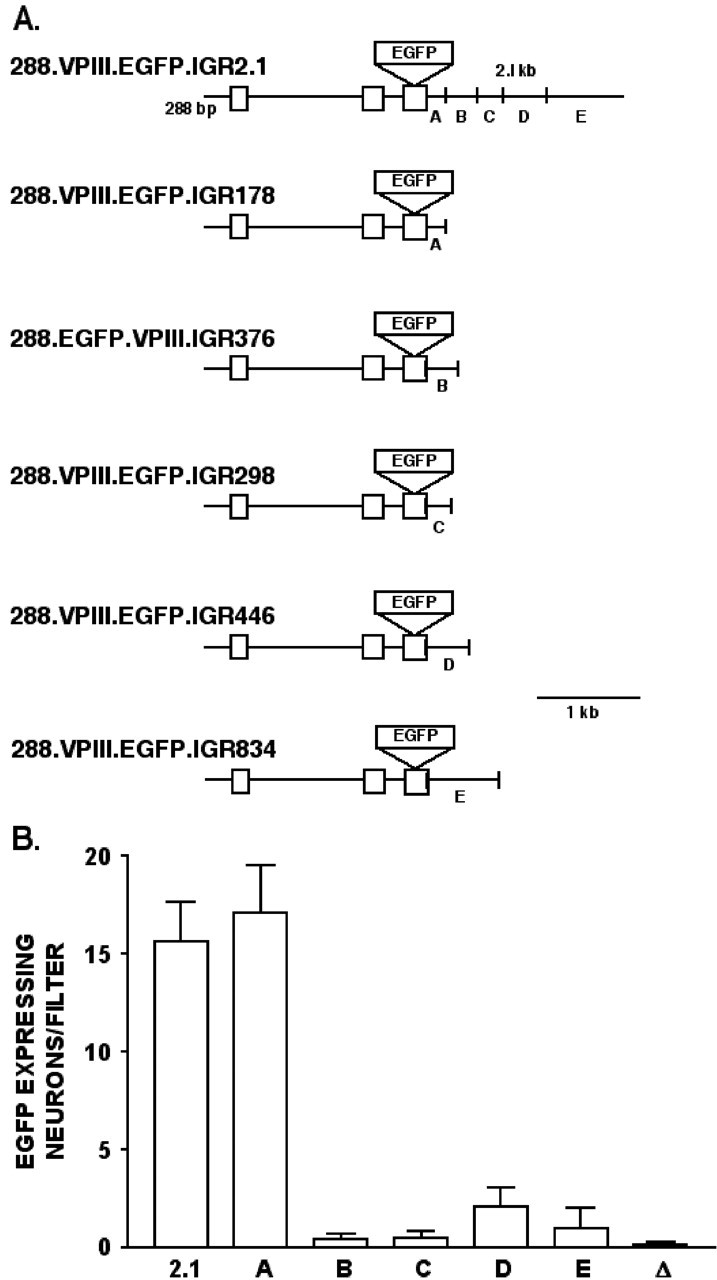
Analysis of regulatory elements in the IGR responsible for hypothalamic VP gene expression. A, The VP constructs used in this IGR analysis were based on the 288VPIII.EGFP.IGR2.1 construct in which the 2.1 kb IGR was divided into five segments (A-E). Each segment was linked to 288VPIII.EGFP. ΔIGR construct contained no IGR sequence (Fig. 5), thereby producing the other constructs with IGR inserts (A-E) of length 178, 376, 298, 446, or 834 bp, respectively (see Materials and Methods). B, Results from the biolistic transfection of rat hypothalamic cultures using the VP constructs shown in A. Cultures were assayed by IHC of EGFP using fluorescent microscopy, and EGFP-expressing neurons were counted per filter. Data are expressed as averages ± SEMs. Note that the enhancer activity in the 2.1 kb IGR appears to be primarily located in the 178 bp segment (A). 2.1, 288VPIII.EGFP.IGR2.1; A, 288VPIII.EGFP.IGR178; B, 288VPIII.EGFP.IGR376; C, 288VPIII.EGFP.IGR298; D, 288VPIII.EGFP.IGR446; E, 288VPIII.EGFP.IGR834.
288VPIII.EGFP.IGR178. The 288-VPIII.EGFP.IGR2.1 construct was digested with SpeI and then partially digested with PvuI. The 6752 band was isolated, which removed all but the first 178 bp of the IGR downstream of exon 3 of VP. The digested restriction sites were filled in with Klenow enzyme (Invitrogen), and the plasmid was ligated back together (see Fig. 6 A).
Oxytocin gene constructs
All OT constructs were derived from OTIII.CAT.IGR3.6 (Jeong et al., 2001).
OTIII.EGFP.IGR3.6. The EGFP gene was removed from pEGFP-N1 (Clontech) by Bsp120I-NotI digestion and was ligated into OTIII.CAT.IGR3.6 after removal of the CAT gene with NotI (see Fig. 4 A).
Figure 4.
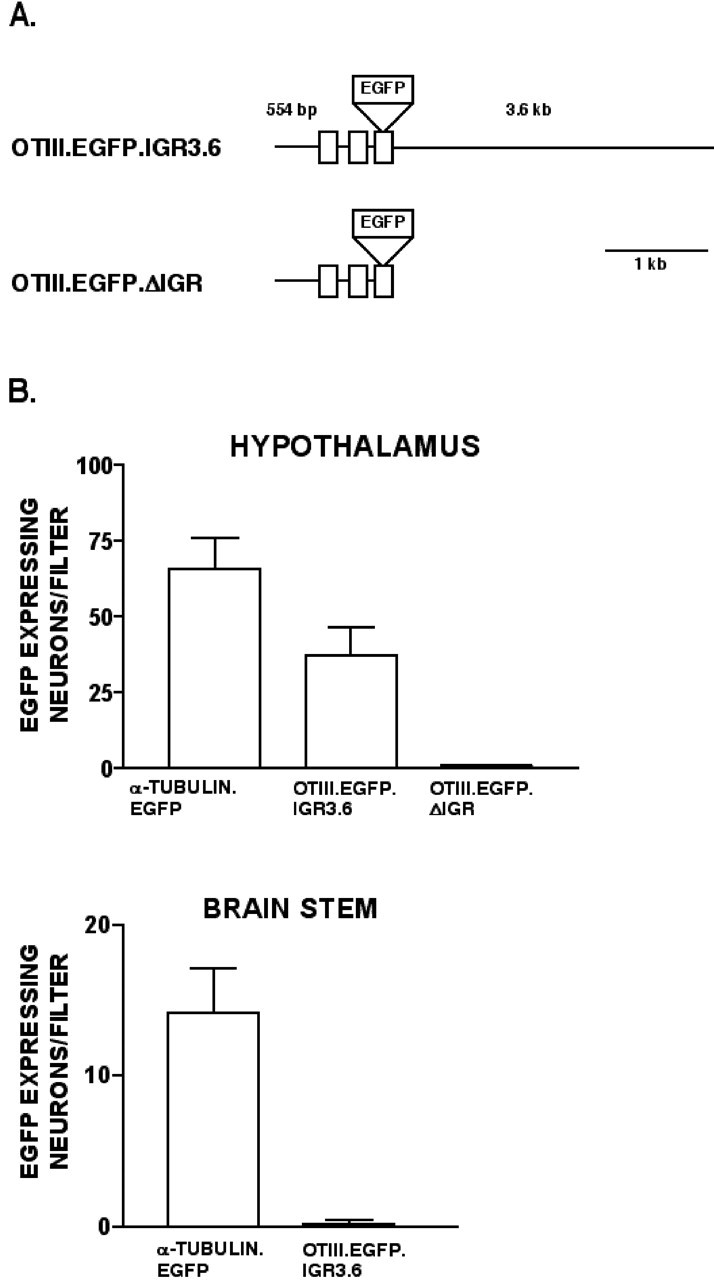
Efficacy and specificity of oxytocin gene constructs used in biolistic transfections. A, The OTIII.EGFP.IGR3.6 contains 554 bp of sequence upstream of exon 1 followed by exons 1-3 of the OT gene with the EGFP reporter gene inserted into exon 3 at the end of the neurophysin coding sequence, followed by the entire 3.6 kb of the mouse IGR sequence. The OTIII.EGFP.ΔIGR construct was made by removing the IGR sequence from the OTIII.EGFP.IGR3.6 construct (see Materials and Methods). B, The OT gene constructs shown in A were biolistically transfected into rat hypothalamus and brainstem slices in the cultures. Plasmid constructs containing the α-tubulin promoter linked to the EGFP gene were also used as a positive control. Cultures were assayed for EGFP expression by IHC using fluorescent microscopy, and EGFP-expressing neurons on each filter were counted. Graphs show data for the average number of EGFP-expressing neurons per filter ± SEMs in the hypothalamic and brainstem cultures, in which each filter contained the brain tissues from a single neonatal rat. Note that although the α-tubulin promoter EGFP construct was expressed in both tissue types, the OTIII.EGFP.IGR3.6 construct was only expressed in the hypothalamus, and the OT construct lacking the IGR was not expressed in the hypothalamus.
OTIII.EGFP.IGR430. The OTIII.EGFP.IGR3.6 construct was digested with EcoRI, and the 3235 bp band containing the entire mouse IGR, except for 430 bp downstream from OT exon 3, was removed (see Fig. 7A).
Figure 7.
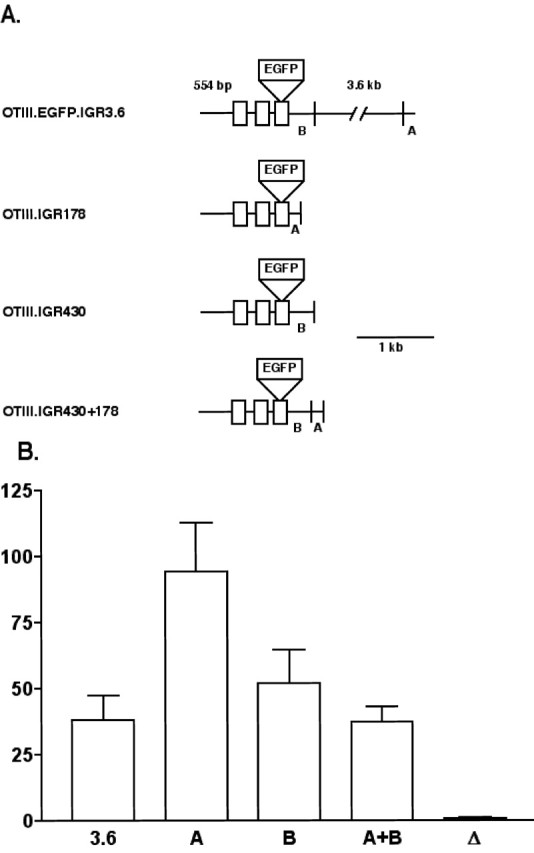
Analysis of the regulatory domains in the IGR responsible for hypothalamic OT gene expression. A, The OT constructs used in this analysis were based on the OTIII.EGFP.IGR3.6 construct. Two domains in the 3.6 kb mouse IGR (A, B) were excised and linked to the OTIII.EGFP.Δ-IGR construct (Fig. 4), which contained no IGR sequence (see Materials and Methods). B, Results from the biolistic transfection of rat hypothalamic cultures using the OT constructs shown in A. Cultures were assayed by IHC using fluorescent microscopy, and EGFP-expressing neurons per filter were counted. Data are expressed as averages ± SEMs. Note that a robust enhancer activity is located in the 178 bp segment A domain, and that significant enhancer activity is also present in the 430 bp B domain (see Results). 3.6, OTIII.EGFP.IGR2.1; A, OTIII.EGFP.IGR178; B, OTIII.EGFP.IGR430; A+B, OTIII.EGFP.IGR430 plus OTIII.EGFP.IGR178; Δ, OTIII.EGFP.ΔIGR.
OTIII.EGFP.ΔIGR. The OTIII.EGFP.IGR430 construct was partially digested with BssHII, and the 4953 bp band was isolated. This removed the 614 bp band containing 88 bp of OT exon 3, 430 bp of the IGR, and 106 bp of the vector. The OTIII.EGFP.ΔIGR oligos (forward strand, 5′-ATTGGCGCGCTTCCTTCGTTCCCCATGGCCACTGCCAGAAAAAAAAAAAAAAAAGAAAAGAAAAGAAAAGAAAAGAAAAATAAAGTAGATTTCGAATTCGCGCGCCCAT-3′), which contained an EcoRI site near the 3′ end, were digested by BssHII and ligated into the 4953 bp band to give OTIII.EGFP.ΔIGR (see Fig. 4 A).
OTIII.EGFP.IGR178. The 178-5′ primer (5′-CCGGAATTCCTGCACCCTGGTGTCTGTCTCTATTT-3′) and 178-3′ primer (5′-CCGGAATTCGATCGCTTCCTTTATTCTATAAGACTTACAGG-3′) were used to amplify the 178 bp IGR band (178 bp immediately downstream from exon 3 of VP) by PCR from OTIII.EGFP.IGR3.6. Amplified product was then digested with EcoRI and ligated into the EcoRI site of OTIII.EGFP.ΔIGR to yield the OTIII.EGFP.IGR178 construct (see Fig. 7A).
OTIII.EGFP.IGR430 and OTIII.EGFP.IGR178. The 178 bp EcoRI-digested PCR product used to construct OTIII.EGFP.IGR178 was ligated into the EcoRI site of OTIII.EGFP.IGR430 to add the 178 bp band to OTIII.EGFP.IGR430 (see Fig. 7A).
Immunohistochemical assays
After the biolistic transfection and 9 -11 additional days of culture, the slice explants on the filters were fixed in 4% formaldehyde in PBS for 1 hr, rinsed three times for 10 min each in PBS, and then placed into cryoprotectant medium (Watson et al., 1986), in which they were stored at 4°C until they were used for immunohistochemistry. For immunostaining, the filters containing the fixed slices were excised from the inserts using a scalpel and then placed in Netwell carriers (Costar, Cambridge, MA). Filters were then thoroughly rinsed in PBS and blocked in 10% normal goat serum and 0.3% Triton X-100 for 2 hr at room temperature to prevent nonspecific binding. Double immunofluorescence staining was performed sequentially. The first immunostaining was against the EGFP that was expressed in the neurons and used a rabbit polyclonal antibody Ab290 (Abcam, Cambridge, UK) at a dilution of 1:1000 overnight at 4°C, which was subsequently visualized by Alexa 488 conjugated goat anti-rabbit (Molecular Probes, Eugene, OR) second antibody at 1:1000 dilution. For the second immunostaining, to identify VP neurons, a monoclonal antibody against VP-NP, PS 41, was used at a dilution of 1:10 (of supernatant) overnight at 4°C, and was visualized by Alexa 594-conjugated goat anti-mouse (Molecular Probes) second antibody at 1:500 dilution. Alternatively, to label OT neurons, a mouse monoclonal antibody against OT-neurophysin (OT-NP), PS 38, was used at a dilution of 1:25 and followed by Alexa 594-conjugated goat anti-mouse second antibody (Molecular Probes) staining at 1:1000 dilution. The specificities of these monoclonal antibodies for their respective neurophysins have been described previously (Ben-Barak et al., 1985; Whitnall et al., 1985). An important aspect of their specificity is that the C-terminal epitopes in the neurophysins, which are recognized by PS 38 and PS 41, are not recognized when the C terminals of the neurophysins are linked to the EGFP reporter in the OT and VP gene constructs. For the DAB staining of OT and VP neurons in the hypothalamic cultures, a mouse monoclonal antibody that cross-reacts with both the OT and VP neurophysins, PS 45, was used at a dilution of 1:25. The secondary antibodies were removed by washing three times for 10 min each in PBS, followed by incubation in avidin-biotinylated horseradish peroxidase (Vectastain Elite ABC kit; Vector Laboratories, Burlingame, CA), and then subsequently visualized by DAB.
EGFP-labeled and OT- and VP-identified fluorescent neurons were viewed using an epifluorescence microscope (Nikon Eclipse 400 Labophot; Nikon, Melville, NY). Counts of the EGFP-expressing neurons are reported as the average number of EGFP-expressing neurons per filter. Each filter contained approximately equivalent amounts of hypothalamic, brainstem, or hippocampal tissue derived from a single animal.
Statistical analysis
Quantitative data are expressed as means ± SEM from at least three independent experiments. Multiple comparisons against a single control group were made by one-way ANOVA (non-parametric) followed by Newman-Keuls multiple comparison tests using the Prism program (version 3.0; Graph Pad, San Diego, CA). The data were also analyzed by the nonparametric, Mann-Whitney U test, also using the Prism software. In all cases, p < 0.05 was considered to be a statistically significant difference.
Results
The in vitro paradigm
Figure 1A-D shows low-magnification views of OT-NP and VP-NP immunoreactive neurons in regions of a typical rat hypothalamic organotypic culture after 13 d in vitro. These neurons are located in identifiable hypothalamic nuclei that normally express these peptides. This indicates that these organotypic cultures contain many of the key neuronal phenotypes that are found in the hypothalamus that express OT or VP. Higher magnifications of the immunoreactive neurons found in the PVN (Fig. 1E) and supraoptic nucleus (SON) (Fig. 1F) show that in these areas in the cultures, they closely resemble the differentiated OT and VP neurons that are found in vivo.
Figure 2 illustrates the types of neurons expressing EGFP at low (Fig. 2A,C,E) and high (Fig. 2B,D,F) magnification after their biolistic transfection with various constructs containing an EGFP reporter in the hypothalamic organotypic cultures. These included an α-tubulin promoter linked to the EGFP gene (Fig. 2A,B), which was expressed nonspecifically in most of the neurons in the cultures and served as a positive control in all of the experiments. The neurons that expressed EGFP from this promoter showed varied morphology and were often multipolar with dendritic spines (Fig. 2B). In contrast, the neurons that expressed EGFP from 3.5VPIII.EGFP.IGR2.1 (Fig. 2C,D) and OTIII.EGFP.IGR3.6 (Fig. 2E,F) tended to be located around the third ventricle (Fig. 2C,E), with simpler bipolar or monopolar dendritic morphology and no dendritic spines. The VP-EGFP and OT-EGFP constructs used for the transfections in Figure 2 are similar to the VPIII.CAT.IGR2.1 and OTIII.CAT.IGR3.6 constructs, which exhibited cell-specific expression in the hypothalamus in our previous transgenic studies (Jeong et al., 2001), with the only change being the replacement of the CAT reporter by the EGFP reporter. Because these OT and VP constructs containing an EGFP reporter produced hypothalamic expression comparable with that found in the transgenic studies, we used these constructs as the starting point for our subsequent analysis of tissue-specific OT and VP gene expression in the organotypic cultures and for experiments directed at identifying the putative cis-acting elements contained within the IGR.
Figure 2.
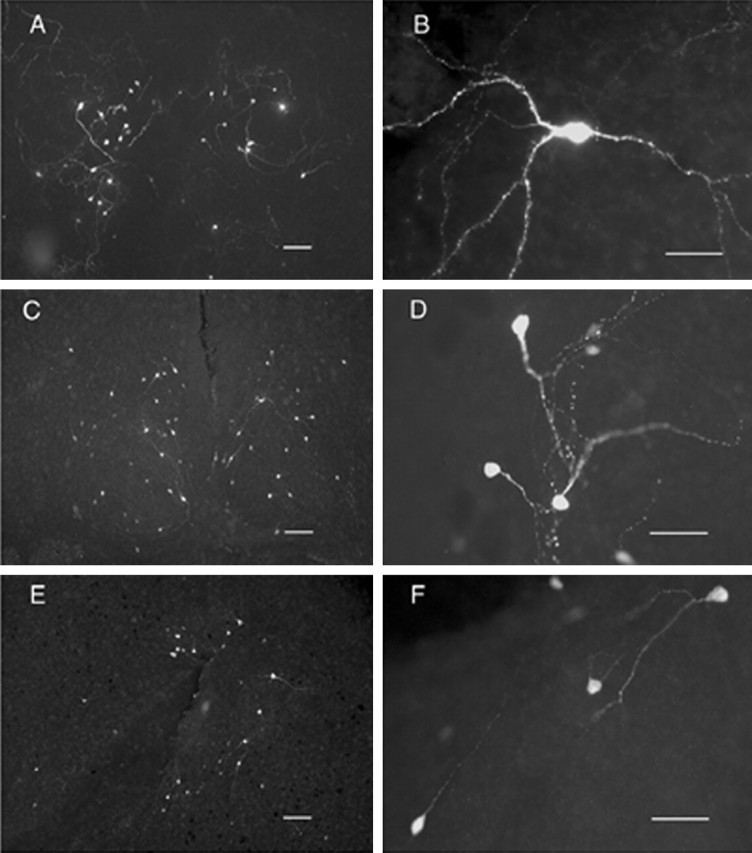
EGFP expression in hypothalamic organotypic cultures after biolistic transfection with α-tubulin. EGFP (A, B), 3.5VPIII.EGFP.IGR2.1 (C, D), or OTIII.EGFP.IGR3.6 (E, F) constructs. See Materials and Methods for description of the EGFP constructs. Scale bars: A, C, E, 100μm; B, D, F, 50 μm.
Efficacy and hypothalamic-specificity of OT and VP gene expression
To determine whether the expression of the OT and VP promoter-driven gene constructs was restricted to the hypothalamus, the only region of the brain in which these peptide genes are appreciably expressed, cultures derived from the hypothalamus as well as other brain areas (e.g., brainstem and hippocampus) were prepared and transfected with the α-tubulin, VP, and OT promoter-driven constructs and evaluated for EGFP expression. In addition to evaluating OT and VP constructs that contained IGR sequences suggested as being important for hypothalamic-specific gene expression in vivo (Jeong et al., 2001), we also transfected similar constructs in which the IGR was removed (Figs. 3, 4). Use of the latter constructs provided a novel test of the IGR hypothesis.
Figure 3 shows the results of transfecting various promoter-driven EGFP constructs into the hypothalamus, hippocampus, and brainstem slice explant cultures. Figure 3A shows the structures of the VP constructs that were used to test for tissue-specific expression. These included the starting 3.5VPIII.EGFP.IGR2.1 construct, which contains the first 2.1 kbp of the mouse IGR downstream of exon 3 of the VP gene, the 288VPIII.EGFP.IGR2.1 construct, in which the 5′ flanking region of the VP gene is drastically reduced from 3.5 kbp to 288 bp, and the 288VPIII.EGFP.ΔIGR construct, in which the entire IGR sequence is removed. Separate transfections using the α-tubulin-EGFP (positive control) plasmid were also done with each brain region. Figure 3B shows the results of transfecting all of these constructs in the hypothalamus, hippocampus, and brainstem cultures, expressed as the average number of neurons expressing EGFP per filter (each filter representing tissues from an individual rat pup) for each construct. Although the α-tubulin-EGFP was expressed in all three tissues, the 3.5VPIII.EGFP.IGR2.1 and the 288VPIII.EGFP.IGR2.1 constructs were only expressed in the hypothalamus and were not at all expressed either in the brainstem or hippocampal cultures. In addition, there was no significant difference in efficacy of EGFP expression between the VP constructs containing either the 3.5 kbp or the 288 bp 5′ flanking region. These data indicate that these VP constructs are specifically expressed in the hypothalamus. In this context, it is important to note that the removal of the IGR in the 288VPIII.EGFP.ΔIGR construct eliminated its ability to express the EGFP reporter in the hypothalamus, thereby providing support for the IGR hypothesis.
A similar set of experiments was done using the two OT constructs shown in Figure 4A. Both OT constructs had the same 5′ flanking region consisting of 554 bp. Figure 4B compares the results of transfecting these OT constructs into the hypothalamus and brainstem, expressed as the average number of neurons expressing EGFP per filter (each filter representing and individual rat pup) for each construct. The OTIII.EGFP.IGR3.6 construct contains the entire mouse IGR and produced EGFP expression in the hypothalamus but not the brainstem, indicating a hypothalamic specificity for this construct. In contrast, the α-tubulin-EGFP (positive control) construct was expressed in both tissue types. The identical construct, OTIII.EGFP.ΔIGR, although lacking any IGR sequence, was also transfected in the hypothalamic slices but was not significantly expressed, consistent with predictions from the IGR hypothesis.
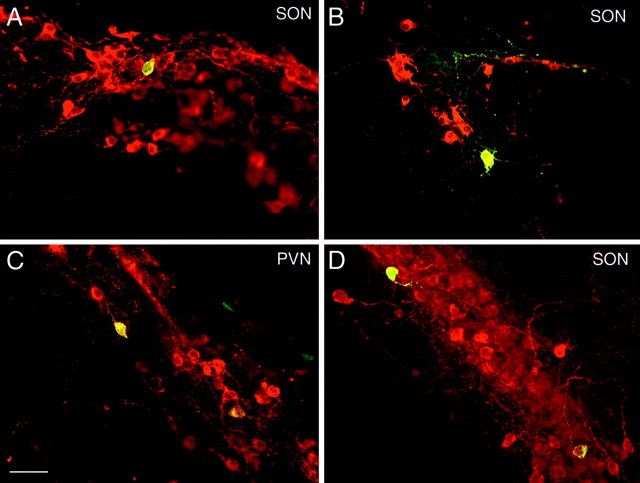
Although the data in Figures 3 and 4 indicate hypothalamic-specific expression of the OT and VP constructs and a dependency of this expression on the IGR, these data do not speak to the cell specificity of the gene expression. In the transgenic mouse studies using similar constructs containing a CAT reporter, we found that the CAT expression from the OT and VP promoter-driven constructs was specifically expressed in the immunohistochemically identified OT and VP MCNs, respectively, but also in some ectopic sites (Jeong et al., 2001). We also evaluated cell specificity in the MCNs in these in vitro experiments by doing double-label immunofluorescence analysis. Figure 5 shows hypothalamic cultures biolistically transfected with the 3.5VPII-I.EGFP.IGR2.1 (A, SON; C, PVN), OTIII.EGFP.IGR3.6 (B, SON), and 288VPIII.EGFP.IGR2.1 (D, SON) constructs. The slices were immunocytochemically stained for EGFP (green fluorescence) and for VP-NP or OT-NP (red fluorescence). Double labeling of cells can be seen in Figure 5 for all three constructs, indicating that these OT and VP constructs are expressed in the appropriate MCN phenotypes. We never observed the reverse labeling (i.e., evidence of VP-EGFP gene expression in OT MCNs or OT-EGFP gene expression in VP MCNs). However, we did find considerable EGFP expression from the OT and VP promoter-driven constructs containing the IGR in neurons that did not contain either OT-NP or VP-NP immunoreactivity. We quantified the percentages of identified OT or VP neurons that had colocalized EGFP compared with all the neurons in the slices that expressed EGFP after transfection with the different VP-EGFP and OT-EGFP gene constructs illustrated in Figs. 6 and 7, respectively. Of the 1193 hypothalamic neurons that we found expressing EGFP after transfection of the various OT-EGFP constructs shown in Figure 7, we found that 315 (26%) of these neurons also contained OT immunoreactivity. In addition, there were no significant differences in this percentage of coexpression between the various OT-EGFP constructs (Fig. 7) that were used. In a similar analysis of 304 hypothalamic neurons that we found expressing EGFP after transfection of the 3.5VPIII.EGFP.IGR2.1 or 288VPIII.EGFP.IGR2.1 constructs (Fig. 3) or the 288VPII-I.EGFP.IGR178 construct (Fig. 6), we found that 77 of these neurons (25%) also contained VP immunoreactivity. Here, too, there were no significant differences in this percentage of coexpression between the VP-EGFP constructs that were used. These data demonstrate that the OT and VP constructs produced substantial expression of EGFP in OT- and VP-expressing hypothalamic neurons, respectively, but that there also were large numbers of unidentified neurons in the hypothalamic slices that expressed the reporter. The expression of the EGFP in the latter could reflect expression in parvocellular neurons that normally express these peptides at very low levels but which are not usually detectable in vivo or in vitro unless axonal transport in the neuron is blocked by colchicine (Vutskits et al., 1998). These parvocellular neurons are abundant in these cultures (Bertini et al., 1993; Arima et al., 2001), and the detection of the EGFP reporter in these neurons in our in vitro experiments in the absence of colchicine could be a result of the high sensitivity of the biolistic method (see Discussion). We were unable to perform these studies in the presence of colchicine, because in preliminary experiments we found that extensive cell death was produced in the hypothalamic cultures as a result of this treatment.
Figure 5.
Hypothalamic cultures biolistically transfected with 3.5VPIII.EGFP.IGR2.1 (A, C), OTIII.EGFP.IGR3.6 (B), and 288VPIII.EGFP.IGR2.1 (D) constructs. Cells in the supraoptic nucleus (A, B, D) and in the paraventricular nucleus were immunohistochemically stained for VP-NP (A, C, D) or OT-NP (B), seen as red fluorescence, and for EGFP. The double-labeled neurons are seen as yellow in this merged view using a combined red and green filter. Scale bar, 40 μm.
Evaluation of IGR sequences involved in VP and OT gene expression
Previous studies on transgenic mice have suggested that the key sites in the IGR responsible for the cell- and tissue-specific expression of the OT and VP genes resided within the first 2.1 kbp downstream of exon 3 of the VP gene (Gainer and Young, 2001; Jeong et al., 2001). Figures 6 and 7 illustrate in vitro experiments directed at the additional elucidation of the specific enhancer sites within the IGR.
Figure 6A shows five specific segments of the 2.1 kbp IGR (labeled A-E), which were selected on the basis of comparative genomic considerations (Gainer et al., 2001) to be tested for their ability to mimic the efficacy of the 2.1 kbp IGR to affect VP promoter expression in hypothalamus. The structures of each of these constructs are shown in Figure 6A, with sequences ranging in length from 178 bp in segment A to 834 bp in segment E. The expression data in Figure 6B clearly show that only the 178 bp (segment A) contains the enhancer activity for VP gene expression, comparable with the entire 2.1 kbp IGR segment. None of the other segments (B-E) produced a level of expression that was significantly greater (p < 0.05) than that observed with the 288.VPIII.EGFP.ΔIGR construct (symbolized by Δ in Fig. 6; full structure of the construct is shown in Fig. 3) that lacked an IGR. Thus, a hypothalamic-specific VP enhancer appears to be located 178 bp immediately downstream of exon 3 of the vasopressin gene.
A similar type of experiment was performed using the OT gene constructs shown in Figure 7A. Here, two specific segments were chosen for evaluation based on predictions from previous transgenic data (Gainer and Young, 2001) and comparative genomic analyses (Gainer et al., 2001). These were the same 178 bp (A) segment used in the studies shown in Figure 6 and a 430 bp IGR domain downstream of exon 3 of the OT gene (B), which was found to have highly conserved sequences between the mouse and the human IGR (Gainer et al., 2001). The data in Figure 7B show that the 178 bp (A) segment had enhancer activity that exceeded the total 3.6 IGR (p < 0.05), whereas the 430 bp (B) segment also had OT gene enhancer activity but only equivalent to the entire 3.6 kbp IGR domain (p > 0.05). Interestingly, when both the A and B (430 plus 178 bp) segments were present together in the OT gene construct, the enhancer effect was at the same level as when the 430 bp segment was used alone. All four constructs showed significantly greater expression (p < 0.05) than the OTIII.EGFP.ΔIGR construct, which lacked the IGR (symbolized by Δ in Fig. 7; the structure of the construct is shown in Fig. 4). The data in Figure 7B suggest that significant enhancer activity for OT gene expression is located at both ends of the IGR, in both the 178 bp (A) and 430 bp (B) segments, but that the 430 bp segment may also contain a suppressor-like element that is operative in the complete 3.6 kbp IGR.
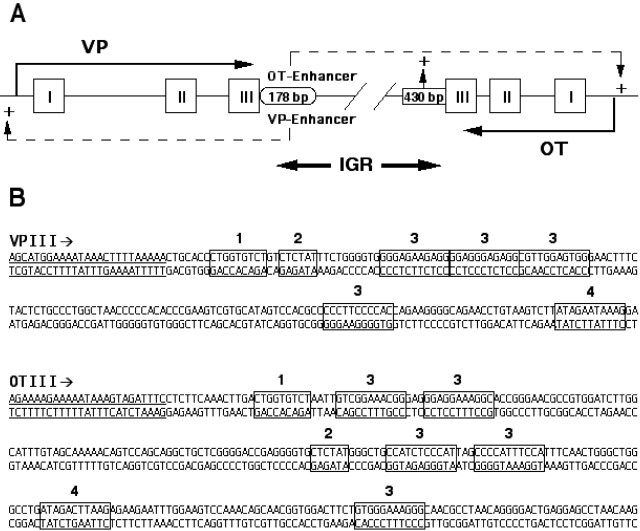
Given the above observation that the OT gene enhancer activity is present in both the 178 bp domain of the IGR downstream of the VP gene and in the 430 bp domain downstream of the OT gene (Fig. 8A), we then examined whether there were any common DNA sequences present in these two domains. Using a motif alignment and search tool (MAST) (Bailey and Gribskov, 1998), available as MAST software online at http://meme.sdsc.edu, we found four distinct common sequences that were present in both domains. These are illustrated in Figure 8B downstream of exon 3 (underlined) of the OT and VP genes, and were CTGGTGTGT (shown in box 1), CTCTAT (shown in box 2), GTGGGAAAGGGG (shown in box 3), and ATAGACTTAAG (shown in box 4).
Figure 8.
Regulatory domains in the mouse IGR. A, Diagram illustrating that the putative enhancer domains in the mouse IGR that are involved in hypothalamic-specific gene expression of the OT and VP genes reside in a 178 bp segment immediately downstream of exon 3 of the VP gene. A second putative OT enhancer domain also appears to be located within the 430 bp segment immediately downstream of exon 3 of the OT gene (see Results). B, Distribution of common DNA sequences found in the 180 and 430 bp domains in the mouse IGR, as determined by sequence alignment analysis. Exon 3 of the OT and VP genes is shown underlined, and the four common sequences found in the downstream sequences of both are indicated by blocks of bases numbered from 1 to 4 (see Results).
Using the same MAST software, we also searched for specific DNA sequences in the IGR regions shown in Fig. 8B that might be conserved between animal species. By comparing mouse and human OT and VP genes with the related isotocin and vasotocin genes, respectively, in pufferfish (Venkatesh and Brenner, 1995; Venkatesh et al., 1997; Murphy et al., 1998), we found that one consensus sequence, TCTGTCTCTATCTCT, which corresponded to boxes 1 and 2 in the 178 bp VP sequence in Figure 8B, was conserved in all three species. Another consensus sequence that we found to be conserved between all three species, TGCTGTTTTGACTTCCACATC, was located between boxes 3 and 4 in the 430 bp region downstream of exon 2 of the OT gene.
Discussion
Experiments in transgenic mice and rats have led to the proposal that critical enhancers for cell-specific OT and VP gene expression in the hypothalamus are present in the IGR within the first 2.1 kb downstream of the VP gene (for review, see Burbach et al., 2001; Gainer and Young, 2001; Murphy and Wells, 2003) [referred to as the IGR hypothesis (Gainer, 1998; Gainer and Young, 2001)]. To further test this hypothesis and to dissect the IGR into the minimal DNA sequences that are responsible for the hypothalamic-specific expression of the OT and VP genes, we used a simpler and less lengthy method than transgenic analysis. Because there are no homologous cell lines available that adequately exhibit the properties of the OT- and VP-synthesizing neurons that are present in the mature hypothalamus, we turned to organotypic slice-explant cultures in which all these neuronal phenotypes are maintained in an easily identifiable form even after many weeks in culture (House et al., 1998; Rusnak et al., 2002) (Fig. 1). To transfect the neurons in these slice cultures with exogenous gene constructs, we used particle-mediated gene transfer (biolistics). Such an approach has been successfully used with cerebellar slices to uncover a calcium-responsive element in the promoters of the calbindin D28 and calmodulin II genes, which are expressed in Purkinje cells (Arnold et al., 1994; Arnold and Heintz, 1997), in the analysis of cis-regulatory elements of genes expressed in the cornea (Wang et al., 2002), and in Langerhans (dendritic) cells in the lymphoid immune system (Morita et al., 2001).
Using the above in vitro strategy, we were able to perform novel tests of the IGR hypothesis as well as to evaluate the 5′ and 3′ flanking regions of the VP gene for DNA sequences necessary for hypothalamic gene expression. We first evaluated the efficacy of the biolistics technique by transfecting hypothalamic neurons in organotypic cultures with constructs that were based on OT and VP gene constructs that had been shown previously to produce successful cell-specific gene expression in vivo in transgenic mice (Jeong et al., 2001). These positive control constructs were the 3.5VPIII.EG-FP.IGR2.1 and OTIII.EGFP.IGR3.6 constructs described in Materials and Methods. The results of these experiments showed that after biolistic transfection, the EGFP was selectively expressed only in neurons that closely resembled the parvocellular and magnocellular cells that endogenously expressed VP in these organotypic hypothalamic cultures, and not the predominately multipolar neurons that were visualized when the α-tubulin promoter was used to drive reporter gene expression (Fig. 2B). The specificity of the expression is also indicated by the fact that biolistic transfection of the control constructs in brainstem and hippocampal slices, which have no OT- or VP-expressing neurons, yielded no neurons that expressed the EGFP reporter after transfection (Figs. 3, 4). Furthermore, although glia are the predominant cells in the slices being penetrated by the gold particles (Gainer et al., 2002), no glial expression of EGFP was found using the OT and VP promoter-driven EGFP constructs. We also found that the 3461 and 288 bp upstream regions in the VP-EGFP construct were equivalent in their transfection efficiencies in the hypothalamus (Fig. 3). An important finding in this group of experiments was the demonstration that the IGR was necessary for the hypothalamic-specific gene expression of the OT and VP gene constructs (Figs. 3, 4), providing, for the first time, a direct experimental test of the IGR hypothesis. However, it should be noted that the IGR regions that were deleted in these experiments also could have removed the so-called G- and GU-rich sequences that are usually located 20-40 bases downstream of the poly(A) signal sequence, AAUAAA, which are believed to have a role in pre-mRNA 3′ end [poly(A)] formation (Colgan and Manley, 1997; Barabino and Keller, 1999; Proudfoot et al., 2002).
The expression of the OT and VP constructs was hypothalamic-specific (Figs. 3, 4) and was also in identified endogenous OT- and VP-containing neurons (Fig. 5). However, as noted in Results, many unidentified cells in the hypothalamus, particularly near the third ventricle, also expressed the EGFP reporter. We believe that many of these are cryptic OT- and VP-expressing cells, such as the CRH cells in the PVN that are known to coexpress VP but require colchicine inhibition of axonal transport to permit detection of the neuropeptides by immunohistochemistry (Vutskits et al., 1998). We did not use colchicine in these experiments because of its known proapoptotic effects (Kristensen et al., 2003) and because of the special vulnerability of the MCNs in culture to apoptosis (Vutskits et al., 1998; Rusnak et al., 2002). Indeed, in preliminary experiments, we found that addition of as little as 0.1 μm colchicine to the media caused extensive cell death in the hypothalamic slice cultures. In transgenic studies, the absence of parvocellular neuronal expression in vivo is usually interpreted as being attributable to the absence of relevant enhancer elements in the transgenic constructs (Waller et al., 1998; Murphy and Wells, 2003). However, another possible interpretation is that the efficiency of the expression of the transgenic constructs is typically very low compared with endogenous genes, and in the parvocellular neurons, in which the endogenous expression is already quite low, any expression of the transgene would therefore be even more difficult to detect. Our ability to detect expression of the exogenous constructs used here in vitro could be related to the fact that in biolistics, each gold particle transfecting the cells carries 100 -200 copies of the plasmid (Gainer et al., 2002), thereby providing a very sensitive assay.
Given the demonstration that the IGR was essential for OT and VP gene expression in these in vitro assays (Figs. 3, 4), we next examined which parts of the IGR sequence were responsible. The data in Figures 6 and 7 show unequivocally that the first 178 bp downstream of the third exon of the VP gene (domain A) in the IGR contained regulatory elements that were essential for the VP and OT expression of the gene constructs in the hypothalamic cultures. Consistent with this finding is the recent report (Davies et al., 2003) that vasopressin constructs containing only 200 bp of 3′ flanking region downstream of exon 3 of the VP gene were able to produce cell-specific gene expression in VP MCNs in transgenic rats. What was interesting in our experiments was that although the 178 bp domain was equivalent to the 2.1 IGR region in producing VP gene expression (Fig. 6B), the same 178 bp domain was even more effective than the full 3.6 kbp IGR region in producing OT gene expression (Fig. 7B), suggesting that there was a suppressor element for OT expression in the full 3.6 kbp mouse IGR.
We also examined the IGR region 430 bp immediately downstream of the OT gene, because our comparative genomics analysis of the IGR (Gainer et al., 2001) identified this area as having evolutionary conserved sequences, and because this sequence was present in all of the OT constructs that had been successfully expressed in transgenic mouse experiments to date (Gainer and Young, 2001; Jeong et al., 2001). Moreover, no construct equivalent to the OTIII.EGFP.IGR430 construct used here was ever tested in transgenic mice, although many efforts were made to do so, but no mouse was shown to integrate this construct and survive to term (Young et al., 1990; Gainer and Young, 2001; Murphy and Wells, 2003). In our in vitro studies, the OTIII.EGFP.IGR430 construct was found to be as effective as the full 3.6 kbp IGR-containing construct in evoking EGFP expression but significantly less than the 178 bp-containing construct (Fig. 7B). Interestingly, combining the 178 bp (A) and 430 bp (B) domains in the same OT construct produced OT gene expression, but only at the level of the 430 bp domain (Fig. 7B), suggesting that the aforementioned putative suppressor element in the 3.6 IGR might be present in the 430 bp domain.
The presence of putative enhancer elements for the OT gene in the 3′ flanking regions of both the OT and VP genes is consistent with current views about the evolution of these mammalian genes (i.e., that they were formed by gene duplication and inversion) (Urano et al., 1992; Burbach et al., 2001). If the 3′ flanking region of the primordial gene was also duplicated and inverted, then one might expect to find duplicated, common sequences in these two domains. Figure 8A shows the location of the 178 and 430 bp domains in the IGR, and Figure 8B shows four common sequences in the 3′ flanking regions of the OT and VP genes containing these domains. It remains to be determined in future experiments which of these sequences, if any, represent the DNA sequences that act as enhancer elements for the two genes. One interesting question is whether the 430 bp domain, if attached to the 288VPIII.EGFP.ΔIGR construct, would give robust hypothalamic expression in vitro. It will also be important to determine whether the putative elements identified in vitro will function similarly as in in vivo models such as transgenic mice.
Footnotes
We thank Dr. Milan Rusnak for his help in the preparation of rat brainstem organotypic cultures.
Correspondence should be addressed to Dr. Harold Gainer, National Institute of Neurological Disorders and Stroke, National Institutes of Health, Building 36, Room 4D-04, Bethesda, MD 20892. E-mail: gainerh@ninds.nih.gov.
Copyright © 2003 Society for Neuroscience 0270-6474/03/237801-09$15.00/0
References
- Antoni FA ( 1993) Vasopressinergic control of pituitary adrenocorticotropin secretion comes of age. Front Neuroendocrinol 14: 76-122. [DOI] [PubMed] [Google Scholar]
- Arima H, House SB, Gainer H, Aguilera G ( 2001) Direct stimulation of arginine vasopressin gene transcription by cyclic adenosine monophosphate in parvocellular neurons of paraventricular nucleus in organotypic cultures. Endocrinology 142: 5027-5030. [DOI] [PubMed] [Google Scholar]
- Arnold D, Feng L, Kim J, Heintz N ( 1994) A strategy for the analysis of gene expression during neural development. Proc Natl Acad Sci USA 91: 9970-9974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold DB, Heintz N ( 1997) A calcium responsive element that regulates expression of two calcium binding proteins in Purkinje cells. Proc Natl Acad Sci USA 94: 8842-8847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Gribskov M ( 1998) Combining evidence using p-values: application to sequence homology searches. Bioinformatics 14: 48-54. [DOI] [PubMed] [Google Scholar]
- Barabino SML, Keller W ( 1999) Last but not least: regulated poly (A) tail formation. Cell 99: 9-11. [DOI] [PubMed] [Google Scholar]
- Bargmann W, Scharrer E ( 1951) The site of origin of the hormones of the posterior pituitary. Am Sci 39: 255-259. [PubMed] [Google Scholar]
- Ben-Barak Y, Russell JT, Whitnall MH, Ozato K, Gainer H ( 1985) Neurophysin in the hypothalamo-neurohypophysial system. I. Production and characterization of monoclonal antibodies. J Neurosci 5: 81-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertini LT, Kursner C, Gaillard RC, Corder R, Kiss JZ ( 1993) A tissue culture model of the hypophysiotropic CRF producing neuronal system. Neuroendocrinology 57: 716-728. [DOI] [PubMed] [Google Scholar]
- Burbach JP, Luckman SM, Murphy D, Gainer H ( 2001) Gene regulation in the magnocellular hypothalamo-neurohypophysial system. Physiol Rev 81: 1197-1267. [DOI] [PubMed] [Google Scholar]
- Colgan DF, Manley JL ( 1997) Mechanism and regulation of mRNA polyadenylation. Genes Dev 11: 2755-2766. [DOI] [PubMed] [Google Scholar]
- Davies J, Waller S, Zeng Q, Wells S, Murphy D ( 2003) Further delineation of the sequences required for the expression and physiological regulation of the vasopressin gene in transgenic rat hypothalamic magnocellular neurones. J Neuroendocrinol 15: 42-50. [DOI] [PubMed] [Google Scholar]
- Du Vigneaud V ( 1954) Hormones of the posterior pituitary gland: oxytocin and vasopressin. Harvey Lect 1954 50: 1-26. [PubMed] [Google Scholar]
- Gainer H ( 1998) Cell-specific gene expression in oxytocin and vasopressin magnocellular neurons. Adv Exp Med Biol 449: 15-27. [DOI] [PubMed] [Google Scholar]
- Gainer H, Young III WS ( 2001) Transgenic models for studies of oxytocin and vasopressin. In: Transgenic models in endocrinology, Chap 2 (Castro MG, ed), pp 25-46. Boston/Dordrecht/London: Kluwer-Academic.
- Gainer H, Fields RL, House S ( 2001) Vasopressin gene expression: experimental models and strategies. Exp Neurol 171: 190-199. [DOI] [PubMed] [Google Scholar]
- Gainer H, Fields RL, House SB ( 2002) Neuronal transfection using particle-mediated gene transfer. In: Cellular and molecular methods in neuroscience (Merighi A, Carmignoto G, eds), pp 67-84. New York: Springer.
- Gillies G, Linton EA, Lowry PF ( 1982) Corticotropin releasing activity of the new CRF is potentiated several times by vasopressin. Nature 299: 355-357. [DOI] [PubMed] [Google Scholar]
- Gloster A, Wu W, Speelman A, Weiss S, Causing C, Pozniak C, Reynolds B, Chang E, Toma JG, Miller FD ( 1994) The Tα-1 α-1 tubulin promoter specifies gene expression as a function of neuronal growth and regeneration in transgenic mice. J Neurosci 14: 7319-7330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara Y, Battey J, Gainer H ( 1990) Structure of mouse vasopressin and oxytocin genes. Mol Brain Res 8: 319-324. [DOI] [PubMed] [Google Scholar]
- House SB, Thomas A, Kusano K, Gainer H ( 1998) Stationary organotypic cultures of oxytocin and vasopressin magnocellular neurons from rat and mouse hypothalamus. J Neuroendocrinol 10: 849-861. [DOI] [PubMed] [Google Scholar]
- Insel TR, Young LJ ( 2001) The neurobiology of attachment. Nat Rev Neurosci 2: 129-136. [DOI] [PubMed] [Google Scholar]
- Jeong SW, Castel M, Zhang BJ, Fields RL, Paras P, Arnheiter H, Chin H, Gainer H ( 2001) Cell-specific expression and subcellular localization of neurophysin-CAT-fusion proteins expressed from oxytocin and vasopressin gene promoter-driven constructs in transgenic mice. Exp Neurol 171: 255-271. [DOI] [PubMed] [Google Scholar]
- Kalsbeek A, van Heerikhuize JJ, Wortel J, Buijs RM ( 1996) A diurnal rhythm of stimulatory input to the hypothalamo-pituitary-adrenal system as revealed by timed intrahypothalamic administration of vasopressin V1 antagonist. J Neurosci 16: 5555-5565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein DC, Moore RY, Reppert SM ( 1991) Suprachiasmatic nucleus. In: The mind's clock, pp 1-467. New York: Oxford UP.
- Kristensen BW, Noer H, Gramsbergen JB, Zimmer J, Noraberg J ( 2003) Colchicine induces apoptosis in organotypic hippocampal slice cultures. Brain Res 964: 264-278. [DOI] [PubMed] [Google Scholar]
- McAllister AK (2000) Biolistic transfection of neurons. www.stke./org/cgi/content/full/oc_sigtrans;2000/51/pl1. [DOI] [PubMed]
- Mohr E, Schmitz E, Richter D ( 1988) A single rat genomic DNA fragment encodes both the oxytocin and vasopressin genes separated by 11 kilobases and oriented in opposite transcriptional directions. Biochimie 70: 649-654. [DOI] [PubMed] [Google Scholar]
- Morita A, Ariizumi K, Ritter III R, Jester JV, Kumamoto T, Johnston SA, Takashima A ( 2001) Development of a Langerhans cell-targeted gene therapy format using a dendritic cell-specific promoter. Gene Ther 8: 1729-1737. [DOI] [PubMed] [Google Scholar]
- Murphy D, Wells S ( 2003) In vivo gene transfer studies on the regulation and function of the vasopressin and oxytocin genes. J Neuroendocrinol 15: 109-125. [DOI] [PubMed] [Google Scholar]
- Murphy D, Si-Hoe SL, Brenner S, Venkatesh B ( 1998) Something fishy in the rat brain: molecular genetics of the hypothalamo-neurohypophysial system. BioEssays 20: 741-749. [DOI] [PubMed] [Google Scholar]
- Proudfoot NJ, Furger A, Dye MJ ( 2002) Integrating mRNA processing with transcription. Cell 108: 501-512. [DOI] [PubMed] [Google Scholar]
- Ratty A, Jeong SW, Nagle JW, Chin H, Gainer H, Murphy D, Venkatesh B ( 1996) A systematic survey of the intergenic region between the murine oxytocin and vasopressin encoding genes. Gene 174: 71-78. [DOI] [PubMed] [Google Scholar]
- Reijmers LG, van Ree JM, Spruijt BM, Burbach JP, De Wied D ( 1998) Vasopressin metabolites: a link between vasopressin and memory? Prog Brain Res 119: 523-535. [DOI] [PubMed] [Google Scholar]
- Reppert SM, Weaver DR ( 2001) Molecular analysis of mammalian circadian rhythms. Annu Rev Physiol 63: 647-676. [DOI] [PubMed] [Google Scholar]
- Rivier C, Rivier J, Mormede P, Vale WW ( 1984) Studies on the nature of the interaction between vasopressin and corticotropin releasing factor on adrenocorticotropin release in the rat. Endocrinology 115: 882-886. [DOI] [PubMed] [Google Scholar]
- Rusnak M, House SB, Arima H, Gainer H ( 2002) Ciliary neurotrophic factor increases the survival of magnocellular vasopressin and oxytocin neurons in rat supraoptic nucleus in organotypic cultures. Microsc Res Tech 56: 101-112. [DOI] [PubMed] [Google Scholar]
- Sausville ED, Carney D, Battey J ( 1985) The human vasopressin gene is linked to the oxytocin gene and is selectively expressed in a cultured lung cancer cell line. J Biol Chem 260: 10236-10241. [PubMed] [Google Scholar]
- Sofroniew MV ( 1985) Vasopressin, oxytocin and their related neurophysins. In: Handbook of chemical neuroanatomy, Vol 4, Pt I (Bjorklund A, Hökfelt T, eds), pp 93-165. Amsterdam: Elsevier Science. [Google Scholar]
- Swanson LW, Sawchenko PE ( 1980) Paraventricular nucleus: a site for the integration of neuroendocrine and autonomic mechanisms. Neuroendocrinology 31: 410-417. [DOI] [PubMed] [Google Scholar]
- Swanson LW, Sawchenko PE ( 1983) Hypothalamic integration: organization of the paraventricular and supraoptic nuclei. Annu Rev Neurosci 6: 269-324. [DOI] [PubMed] [Google Scholar]
- Urano A, Hyodo S, Suzuki M ( 1992) Molecular evolution of neurohypophysial hormone precursors. Prog Brain Res 92: 39-46. [DOI] [PubMed] [Google Scholar]
- Venkatesh B, Brenner S ( 1995) Structure and organization of the isotocin and vasotocin genes from teleosts. Adv Exp Med Biol 395: 629-638. [PubMed] [Google Scholar]
- Venkatesh B, Si-Ho S-L, Murphy D, Brenner S ( 1997) Transgenic rats reveal remarkable functional conservation of regulatory controls between the fish isotocin and rat oxytocin genes. Proc Natl Acad Sci USA 94: 12462-12466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vutskits L, Bartanusz V, Schulz MF, Kiss JZ ( 1998) Magnocellular vasopressinergic neurons in explant cultures are rescued from cell death by ciliary neurotrophic factor and leukemia inhibiting factor. Neuroscience 87: 571-582. [DOI] [PubMed] [Google Scholar]
- Waller SJ, Ratty A, Burbach JP, Murphy D ( 1998) Transgenic and transcriptional studies on neurosecretory cell gene expression. Cell Mol Neurobiol 18: 149-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang IJ, Carlson EC, Liu CY, Kao CW, Hu FR, Kao WW ( 2002) Cis-regulatory elements of the mouse Krt. 1.12 gene. Mol Vision 8: 94-101. [PubMed] [Google Scholar]
- Wang, S, Wu H, Jiang J, Delohery TM, Isdell F, Goldman SA ( 1998) Isolation of neuronal precursors by sorting embryonic forebrain transfected with GFP regulated by the Tα1 tubulin promoter. Nat Biotechnol 16: 198-201. [DOI] [PubMed] [Google Scholar]
- Wang Z, Young LJ, De Vries GJ, Insel TR ( 1998) Voles and vasopressin: a review of molecular, cellular, and behavioral studies of pair bonding and paternal behaviors. Prog Brain Res 119: 483-499. [DOI] [PubMed] [Google Scholar]
- Watson Jr RE, Wiegand SJ, Clough RW, Hoffman GE ( 1986) Use of cryoprotectant to maintain long-term peptide immunoreactivity and tissue morphology. Peptides 7: 155-159. [DOI] [PubMed] [Google Scholar]
- Wellmann H, Kaltschmidt B, Kaltschmidt C ( 1999) Optimized protocol for biolistic transfection of brain slices and dissociated cultured neurons with a hand-held gene gun. J Neurosci Methods 92: 55-64. [DOI] [PubMed] [Google Scholar]
- Whitnall MH, Key S, Ben-Barak Y, Ozato K, Gainer H ( 1985) Neurophysin in the hypothalamo-neurohypophysial system. II. Immunocytochemical studies of the ontogeny of oxytocinergic and vasopressinergic neurons. J Neurosci 5: 98-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young III WS, Reynolds K, Shepard EA, Gainer H, Castel M ( 1990) Cell specific expression of the rat oxytocin gene in transgenic mice. J Neuroendocrinol 2: 917-925. [DOI] [PubMed] [Google Scholar]