Abstract
The transcription factor Engrailed (En) controls axon pathfinding and synaptic target choice in an identified neuron (6m) of the cockroach cercal sensory system. Knock-out of En using double-stranded RNA interference (dsRNAi) transforms 6m so that it resembles a neighboring neuron that normally does not express the en gene, has a different arbor anatomy, and makes different connections. Like many animals, the cockroach has two En paralogs, Pa-En1 and Pa-En2. In this study we tested the hypothesis that the paralogs have different effects on axon guidance and synaptic target recognition, using RNAi to knock out each one individually. Using dye injections into 6m and intracellular recordings from target interneurons, we obtained evidence that both Pa-En1 and Pa-En2 determine the axonal arborization, but only Pa-En1 controls synaptic connections. However, because immunocytochemical quantification of En protein in 6m after RNAi showed that Pa-En1 represents 65% of the total En activity and Pa-En2 only 35%, our results could be caused by dosage effects. We measured the effects of diluting the mixture of both dsRNAs on the amounts of En protein. From this dose-response curve, we calculated the appropriate dilutions of the dsRNA mixture that would titrate total En protein to levels equivalent to knock-out of either paralog. RNAi using these dilutions showed that Pa-En1 and Pa-En2 both contribute toward the control of axonal guidance and confirmed that Pa-En1 has the paralog-specific function of controlling synaptic target recognition.
Keywords: Engrailed, RNA interference, homeodomain, synaptic specificity, axon guidance, evolution
Introduction
Engrailed (En) is an ubiquitous transcriptional regulator, probably present in all bilaterian metazoa (Webster and Mansour, 1992; Gibert, 2002). In arthropods En plays a crucial part in the patterning of body segments and limbs (Morata and Lawrence, 1975; Kornberg, 1981; Lawrence and Struhl, 1982); it also has a role in vertebrate limb development (Wurst et al., 1994). However, the most conserved role of En appears to be in neuronal development. En has a role in cerebellar patterning (Baader et al., 1999), and the gradient of En in the tectum activates ephrin expression and guides retinal axons (Friedman and O'Leary, 1996; Itasaki and Nakamura, 1996; Logan et al., 1996; Shigetani et al., 1997). En regulates axonal pathfinding in spinal cord interneurons (Saueressig et al., 1999) and may influence their synaptic connections to motor neurons (Wenner et al., 2000). In addition, it affects the survival of dopaminergic midbrain neurons through its effects on α-synuclein (Simon et al., 2001). In the insect nervous system, En controls neuronal identity, axon pathfinding, and synaptic target recognition (Condron et al., 1994; Lundell et al., 1996; Siegler and Jia, 1999; Marie et al., 2000; Siegler et al., 2001; Jia and Siegler, 2002; Marie et al., 2002).
Many animals have more than one copy of the engrailed gene. Mammals have two paralogs, en-1 and en-2, as do many insects such as Drosophila (engrailed and invected). Spatially or temporally separate patterns of expression may account for the different phenotypes of en-1 and en-2 mutations because, at least on a gross anatomical level, they appear to have redundancy of biochemical function (Hanks et al., 1995). In Drosophila, En and Invected (Inv) are largely coexpressed, but there is evidence that they have separate functions in wing patterning (Simmonds et al., 1995). Some CNS neurons express only Inv, whereas others express both, also indicating that they may have different roles (Siegler and Jia, 1999). It is possible therefore that En paralogs may also have subtly different roles in the control of axon guidance and synaptic target selection.
The second larval stage of the cockroach, Periplaneta americana, has a well defined array of wind-sensitive sensory neurons on each cercus. These neurons form synapses with subsets of giant interneurons (Thompson et al., 1992), mediating the animal's escape response. Persistent expression of En by medial neurons is necessary to determine their characteristic axonal arborization and choice of synaptic targets (Marie et al., 2000, 2002), summarized in Figure 1. This insect also has two en genes, Pa-en1 and Pa-en2 (Marie and Bacon, 2000). The paralogs are coexpressed in embryos, and there is evidence for coexpression in cercal sensory neurons (Marie et al., 2000). Here we use an identified neuron to test the hypothesis that the En paralogs have different effects on axonal pathfinding and synaptic target selectivity. Our results show that Pa-En1 and Pa-En2 act together to control axon guidance but that only Pa-En1 controls synaptic target recognition.
Figure 1.
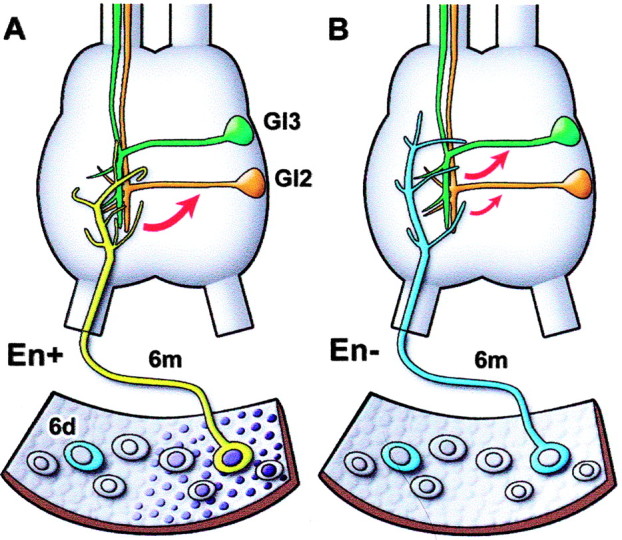
A diagram representing the results of a previous study (Marie et al., 2000), illustrating a portion of the cercal epidermis and its array of second instar neurons. A, Medial sensory neurons express En (dark nuclei). The identified neuron studied, 6m, has a characteristic axonal arbor in the terminal abdominal ganglion. 6m normally forms synaptic connections (red arrow) with certain target interneurons such as GI2 (orange) and not with others such as GI3 (green). B, After En knock-out with dsRNA injection shortly after hatching, the arbor type of 6m is altered so that it resembles that of a neighboring, laterally positioned neuron 6d (blue) that does not normally express En. The pattern of synaptic connections made by 6m is also altered, reducing the strength of its normal connections and forming new synapses with inappropriate targets.
Materials and Methods
Double-stranded RNA interference. Double-stranded RNAs (dsRNAs) from nonconserved regions of Pa-en1 and Pa-en2 were synthesized as described previously (Marie et al., 2000). Briefly, a 609-bp-long Pa-en1 PCR fragment and a 546 bp Pa-en2 PCR fragment were subcloned in both orientations into the pCR 2.1 vector (TA cloning kit; Invitrogen, Gaithersburg, MD). In vitro transcription using T7 RNA polymerase was performed as specified by the manufacturer (Maxiscript kit; Ambion, Austin, TX) to produce both sense and antisense RNA strands for each of the fragments. The quantity of synthesized RNA was determined by optical density, and equimolar amounts (30-60 pmol) of complementary strands were mixed. RNA mixes were denatured and annealed as previously described (Kennerdell and Carthew, 1998). The dsRNAs were resuspended in injection buffer (20 mm Tris, pH 7.8; 150 mm NaCl). When used singly, the dsRNAs were diluted to a concentration of 3 μm; when mixed, each was at 1.5 μm to give the same total dsRNA concentration. Newly hatched first instar cockroaches (Periplaneta americana) were immobilized on ice and injected twice (injection volume is 150 nl) through the broken tips of the cerci, using wide-bore glass micropipettes. Double-stranded RNA and other tracers introduced by this method into the animal circulate freely throughout the hemolymph but do not penetrate the CNS blood-brain barrier (Marie et al., 2000). The specificity of double-stranded RNA interference (dsRNAi) in this system was demonstrated previously (Marie et al., 2000) by showing that Pa-en dsRNA does not affect the expression of other proteins, by application of Pa-en1 and Pa-en2 dsRNAs separately, and by application of injection buffer only.
Dissection. Second instar animals were placed in a Sylgard-lined Petri dish containing saline of the following composition (in mm): 150 NaCl, 3.1 KCl, 5.4 CaCl2, 2 MgCl2, 5 MOPS buffer, 50 sucrose, and 5 glucose, pH 7.4. The CNS and cerci were isolated and then transferred to a Sylgard-walled chamber constructed on a glass microscope slide. The connective tissue sheath surrounding the terminal abdominal ganglion was carefully removed using finely sharpened forceps. Distal segments 1-5 were removed from the right cercus, then all of the filiform hairs, except that of 6m, and all the major mechanosensory bristles, were plucked from the remaining segments with the forceps; the remaining sockets were coated with petroleum jelly to prevent residual movements. The left cercal nerve was crushed to remove all inputs. The slide was then transferred to the fixed stage of an upright microscope equipped with differential interference contrast optics. All experiments were performed at room temperature (21-23°C).
Intracellular recording and filiform hair stimulation. Electrodes for intracellular recording from giant interneuron (GI) cell bodies were filled with 1 m KCl and had a resistance of 60-120 MΩ. Cell bodies of GIs were identified by using the standard criteria of size, appearance, and position in relation to ganglionic landmarks (Blagburn, 1989; Blagburn and Thompson, 1990). The GI was designated as ipsilateral or contralateral according to the position of its cell body, not of its axon. Recordings were rejected if the resting membrane potential stabilized at values more positive than -60 mV; most GIs had membrane potentials of -65 to -80 mV. The sensory neuron was stimulated by pushing its hair in the excitatory direction briefly, using a micropipette with a petroleum jelly-covered tip mounted on a loudspeaker connected to a pulse generator. The resulting burst of action potentials invariably evokes a corresponding burst of monosynaptic EPSPs with a short (4-6 msec), constant latency in the reference GI (Thompson et al., 1992). Removal of the other hairs during dissection ensures that no other sensory neurons are stimulated. Data were recorded with a computer data acquisition system (Axoscope; Axon Instruments, Foster City, CA) with which the amplitude of the first unitary EPSP in ∼20 such bursts was measured.
Lucifer Yellow injections. After electrophysiological experiments, the 6m sensory neuron was identified with Nomarski optics and impaled with a glass microelectrode backfilled with 4% Lucifer Yellow (Molecular Probes, Eugene, OR). Hyperpolarizing current (-2 nA) was applied through the microelectrode for 8-10 min, then the preparation was fixed in 4% paraformaldehyde in 0.075 m PBS for 1 hr. After thorough washing in PBS, preparations were preincubated in normal goat serum, and then incubated in 1:2000 anti-Lucifer Yellow antibody (Molecular Probes) for 15-20 hr at 4°C. After washing, the preparations were incubated in 1:200 Cy3-conjugated anti-rat antibody (Jackson ImmunoResearch, West Grove, PA) for 2 hr at room temperature. After washing, the ganglia were cleared and mounted in Vectashield (Vector Laboratories, Burlingame, CA) and observed with a Zeiss Axioskop equipped with epifluorescence. A through-focus series of images was made of each arborization with a Zeiss Axiocam CCD camera, then in-focus regions were combined using the layer mask option of Adobe Photoshop (Adobe Systems, San Jose, CA).
En immunocytochemistry. First-instar animals were placed in saline, and the dorsal tergites and gut were removed. The distal two segments of the cerci were cut off, and fixative was added to the dissection chamber. The dorsal side of each cercus was then cut off, exposing the interior. Tracheae, the cercal nerves, connective tissue, and as many fat body cells as possible were removed with fine forceps to improve access and visibility of the sensory neurons. Fat body cells in particular tend to show strong granular avidin-binding in the cytoplasm. The CNS and cerci were fixed for 1 hr in 4% paraformaldehyde in 0.075 m PBS, before washing thoroughly in PBS. After a preincubation in normal horse serum in PBS plus 0.3% Triton X-100 (PBST) for 30 min, monoclonal 4D9 anti-En antibody, obtained from the Developmental Studies Hybridoma Bank (University of Illinois) or as a gift from Dr. Corey Goodman (University of California, Berkeley), was applied at a dilution of 1:20 in PBST for 15-20 hr at 4°C. After 3 × 10 min washes, biotinylated horse anti-mouse antibody (Vector Laboratories) was applied at a dilution of 1:200 for 1 hr at room temperature, and the tissue was again washed three times. After incubation in avidin-peroxidase complex (Vector Laboratories) for 1 hr at room temperature, the tissue was washed, a solution of 0.1% DAB with 0.03% H2O2 was added to the wells, and the reaction was allowed to proceed for 20 min. The specimens were washed in PBS then cleared and mounted in 70% glycerol. Images were captured with the CCD camera. Percentage grayscale levels of the sample areas (nuclei of sensory neurons or the medial epidermis of segments 5-7) and the corresponding background areas (nucleus of lateral neurons in the same segment or the lateral epidermis of segments 5-7) were measured using Adobe Photoshop. Background percent gray levels were subtracted from the sample values.
Results
More Pa-En1 is present in sensory neurons than Pa-En2
In our previous studies of this system (Marie et al., 2000, 2002) we eliminated expression of both en genes using dsRNAi (Fire et al., 1998). Here we use RNAi to prevent expression of each paralog separately, using dsRNAs synthesized from nonconserved regions of Pa-en1 and Pa-en2. There are no antibodies that label specifically either Pa-En1 or Pa-En2, and there are technical obstacles that prevent in situ hybridization to the cerci; we therefore used the monoclonal antibody 4D9 (Patel et al., 1989), which recognizes a conserved epitope in the homeodomain of both Pa-En1 and Pa-En2, to measure the total amounts of En protein. The density of antibody staining in the nuclei of cercal sensory neurons, and in the medial cercal epidermis, was measured and expressed as a percentage of the maximum staining intensity, allowing the standardization of data from different batches of cerci. As reported previously (Blagburn et al., 1995; Marie et al., 2000), control cerci showed a stereotyped pattern of staining intensities, with neurons located near the midline of the cercus (neurons 4h, 5h, 6h) showing low levels of immunoreactivity, and those near the medial edge (neurons 4m, 5m, 6m, 6p) exhibiting the most intense staining (Fig. 2A,F).
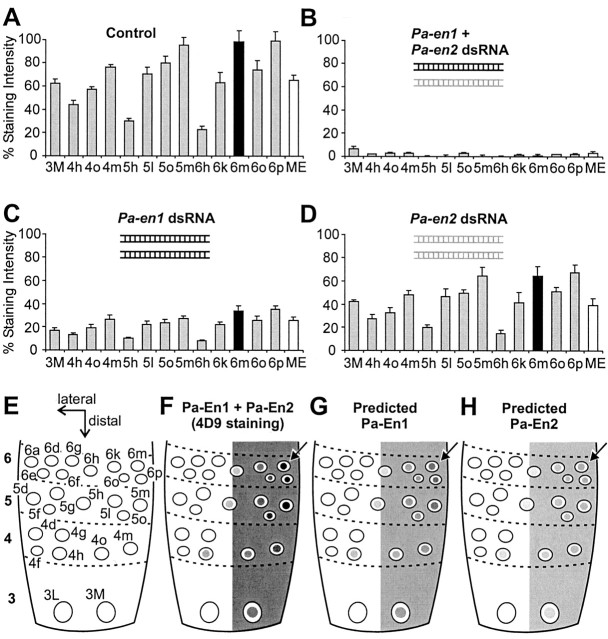
Figure 2.
Pa-En1 and Pa-En2 are colocalized in the cercal neuroepithelium. A-D, Mean percentage staining intensities of En antibody in identified neurons of cercal segments 3-6 (identified in E) and of the medial epidermis (ME). Neuron 6m is indicated with a filled bar. A, Control cerci (n = 16). B, An equimolar mixture of Pa-en1 (black icon) and Pa-en2 (gray icon) dsRNA (1.5 μm each) abolishes En immunoreactivity (n = 13). C, Pa-en1 dsRNA alone (3 μm) reduces total En staining intensity in neurons to ∼30-36% of control values (n = 17). The staining intensity of 6m (filled bar) is reduced to 35% of control. D, Pa-en2 dsRNA alone (3 μm) reduces total En staining intensity in neurons to ∼60-68% of control values (n = 12). The staining intensity of 6m (filled bar) is reduced to 65% of control. E, Identified cercal sensory neurons in incipient segments 3-6. Lateral and distal axes are indicated by arrows. F, Diagrammatic representation of the actual distribution of En protein (Pa-En1 + Pa-En2), as detected with the 4D9 antibody. Percentage gray levels represent the mean percentage staining intensities shown in A above. The position of neuron 6m is indicated by an arrow. G, The predicted distribution of the Pa-En1 paralog alone, based on the knock-out experiments described above. H, Predicted distribution of the Pa-En2 paralog.
Because our preparations were processed together using the same antibody, it allowed us to compare quantitatively the levels of 4D9 staining in control cerci to those in double and single Pa-En1 and Pa-En2 knock-outs. It should be noted that, because we used the same total amount of dsRNA in single as in double knock-outs, an animal injected with a single en dsRNA received twice the number of copies of this particular dsRNA than an animal injected with both. Injection of a mixture of Pa-en1 and Pa-en2 dsRNAs abolished all staining in the sensory neurons and medial epidermis (Fig. 2B). However, injection of Paen1 dsRNA alone, at the same total dsRNA concentration, did not completely abolish staining in neurons and the epidermis but substantially reduced it to 30-36% of the control levels (Fig. 2C). Injection of Pa-en2 dsRNA alone had even less effect, reducing staining significantly to 60-68% of the control intensities (Fig. 2D). In neuron 6m, the identified neuron used previously to quantify the effects of En on neuronal phenotype, Pa-en1 dsRNA reduced 4D9 staining to 35% of control, and Pa-en2 dsRNA reduced it to 65%. The fact that we see only partial knock-outs with excess quantities of Paen1 or Pa-en2 dsRNA rules out the possibility that any significant cross-reaction is taking place between dsRNA from one paralog and mRNA of the other.
The peroxidase staining method is highly sensitive but it is commonly thought to saturate with high levels of antigen, although quantitative tests have shown the reaction is linear (Nibbering et al., 1986). In our case, midline neurons in the terminal ganglion stain with intensities that are approximately twice what we see in cercal neurons (data not shown), so we can be reasonably confident that the reaction is not saturated and that the relative staining intensities in the cercal neurons are indicative of the relative amounts of En protein. Taken together, the only parsimonious explanation of these results is that all sensory neurons coexpress both Engrailed paralogs, with, in neuron 6m, Pa-En1 protein representing 65% of the total Engrailed activity (Fig. 2G), and Pa-En2 representing only 35% (Fig. 2H).
Knock-out of Pa-En1 or Pa-En2 has different effects on axonal anatomy
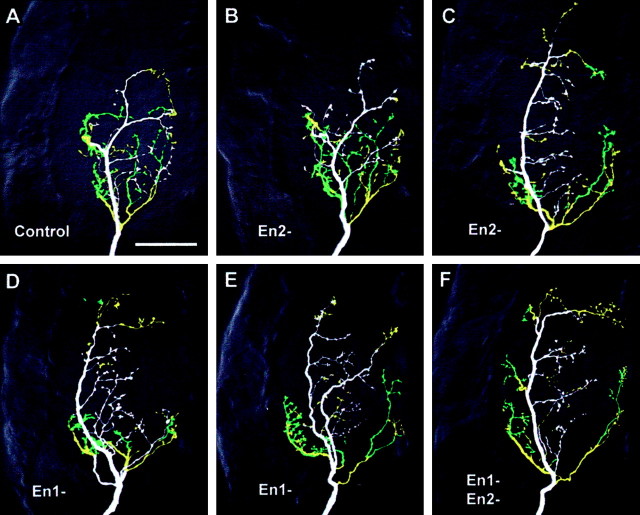
Are there paralog-specific effects on axonal anatomy? Knock-out of Pa-En2 alone had variable effects on the arbor of 6m, as revealed with Lucifer Yellow injections followed by immunocytochemical intensification. In 60% of the cases there was no obvious qualitative effect, with 6m retaining an M-type axon trajectory (Fig. 3B) similar to that of controls (Fig. 3A). In the remaining 40% of cases, the arbor appeared to be transformed to L-type (Fig. 3C), similar to that seen in double knock-outs (Fig. 3F). RNAi of Pa-En1 was more effective, with apparent transformation to L-type arbors in 90% of the cases (Fig. 3D). In one interesting preparation, the arbor appeared to have an intermediate morphology, with the axon following both L and M pathways (Fig. 3E).
Figure 3.
Effects of En paralog knock-out on the axonal arbor of neuron 6m. The panels show ventral views of whole-mount terminal ganglia containing antibody-intensified Lucifer Yellow fills of 6m, colored according to branch depth within the neuropil (white represents most ventral, green, most dorsal). The anterior of the ganglion is toward the top and the midline to the right of each panel. A, Typical control medial-type arborization. B, C, Arborizations after Pa-en2 dsRNA injection (En2-). The arbor in B appears similar to control, whereas that in C is lateral-type. D, E, Arborizations of neurons treated with Pa-en1 dsRNA (En1-). D illustrates the lateral-type appearance of most of the arbors, whereas E shows an unusual arbor with mixed characteristics and axon branches in both lateral and medial tracts. F, Typical 6m lateral-type arbor after application of both en dsRNAs.
These sensory neuron arborizations have several characteristic, quantifiable features, allowing us to measure subtle differences in the effects of the two Engrailed proteins. The first of these is the angle of the medial bend (Fig. 4A), formed when the axonal growth cone chooses to follow the pre-existing tract of M afferents rather than the L afferents (Marie et al., 2002). L-type axons do not make this abrupt change in pathway and therefore exhibit a smaller axon angle. It should be noted that intermediate pathway choices are not available, so the range of possible axon angles is bimodally distributed. A second characteristic is that, partly because of this pathway choice, M-type axons do not innervate the anterior-lateral region of the cercal glomerulus (Fig. 4B), whereas L-type axons do. The third characteristic feature is the quantity of axonal branches in the dorsal region of neuropil (Fig. 4C). M-type axons tend to form many such branches, whereas L-type axons do not.
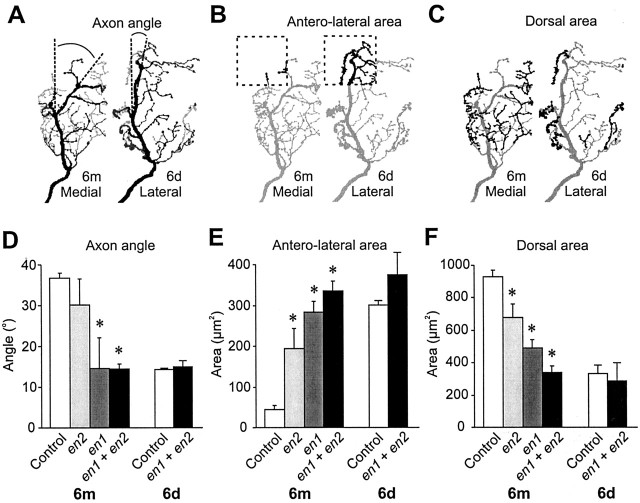
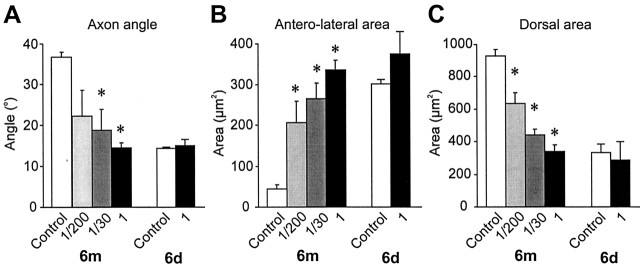
Figure 4.
Quantification of the effects of En paralog knock-out on the axonal arbor of neuron 6m. A-C, Diagrams illustrating the three parameters of axon arbor that were measured pointing out the differences between a medial-type Engrailed-expressing neuron (6m) and a lateral-type Engrailed-negative neuron (6d). A, Angle of the axon bend, measured between defined branch points. The ventral branches are dark gray, and the most dorsal are light gray. B, Area of branches (black) in the anterolateral corner of the cercal glomerulus (dashed box). C, Area of branches in the dorsal third of the neuropil (black). D-F, Histograms of each characteristic, comparing arbors from control 6m (n = 10) and 6d (n = 5) to those from neurons treated with Pa-en1 (n = 5) or Pa-en2 dsRNA alone (n = 3), and from neurons treated with a mixture of both (n = 9). Asterisks indicate significant differences from control values. D, Axon angle is significantly reduced in double knock-outs (p < 0.0001) and with Pa-en1 RNAi (p = 0.03). Pa-en2 dsRNA gave variable, bimodally distributed results. E, Anterolateral branches are significantly increased by all dsRNA treatments (Pa-en2: p = 0.03; Pa-en1: p < 0.0001; both: p < 0.0001). Pa-en2 dsRNA alone is significantly less effective than the double knock-out (p = 0.03). F, Dorsal branches are significantly decreased by all dsRNA treatments (Pa-en2: p = 0.008; Pa-en1: p < 0.0001; both: p < 0.0001). Pa-en2 or Pa-en1 dsRNA alone is significantly less effective than the double knock-out (Pa-en2: p = 0.008; Pa-en1: p = 0.04).
Quantitative measurements of arborizations such as those shown in Figure 3 showed that Pa-en2 dsRNA did not significantly alter the mean axon angle, although the variance was greatly increased (Fig. 4D), reflecting the 40% chance of the axon choosing the lateral pathway. However, despite the lack of a consistent effect on axon pathway choice, Pa-En2 knock-out did significantly increase the area of branches in the anterolateral corner of the neuropil (Fig. 4E) and decrease the area of branches in the dorsal third of the cercal glomerulus (Fig. 4F).
Injection of Pa-en1 dsRNA did significantly decrease axon angle (Fig. 4D) and increased and decreased anterolateral and dorsal branches, respectively, almost as effectively as did the mixture of both dsRNAs (Fig. 4E,F), although the area of dorsal branches was not increased as much as in double knock-outs (Fig. 4F). Thus, our results indicate that both Engrailed paralogs affect axonal guidance and growth, but Pa-En1 appears to be more effective than Pa-En2.
Knock-out of Pa-En1 or Pa-En2 has different effects on synaptic specificity
Because the paralogs appear to have different effects on axonal arborization, it was of great interest to find out whether they also have differential effects on the choice of synaptic target interneurons. Selective stimulation of 6m allowed us to measure the amplitude of monosynaptic unitary EPSPs recorded intracellularly from a range of potential target interneurons within the terminal ganglion using standard electrophysiological methods (Thompson et al., 1992; Marie et al., 2000, 2002). Normally, 6m forms synaptic contacts with a very characteristic subset of these targets (interneurons cGI5, iGI2, cGI2, cGI1) (Fig. 5A) and avoids others that are normally targeted by En-negative sensory neurons (interneurons cGI6, iGI6, cGI3, iGI3) (Fig. 5A). With Pa-en2 RNAi, we saw no significant changes in the patterns of synaptic outputs of 6m compared with controls (Fig. 5B). However, with Pa-En1 knock-out, there were significant decreases in the amplitude of synaptic connections normally made with interneurons cGI5, iGI2, cGI2, and cGI1, and the de novo appearance of robust connections to cGI6, iGI6, cGI3, iGI3, neurons with which 6m normally never synapses (Fig. 5C). These changes in synaptic connections were not significantly different from those seen with injection of a mixture of Pa-en1 and Pa-en2 dsRNAs (Fig. 5D).
Figure 5.
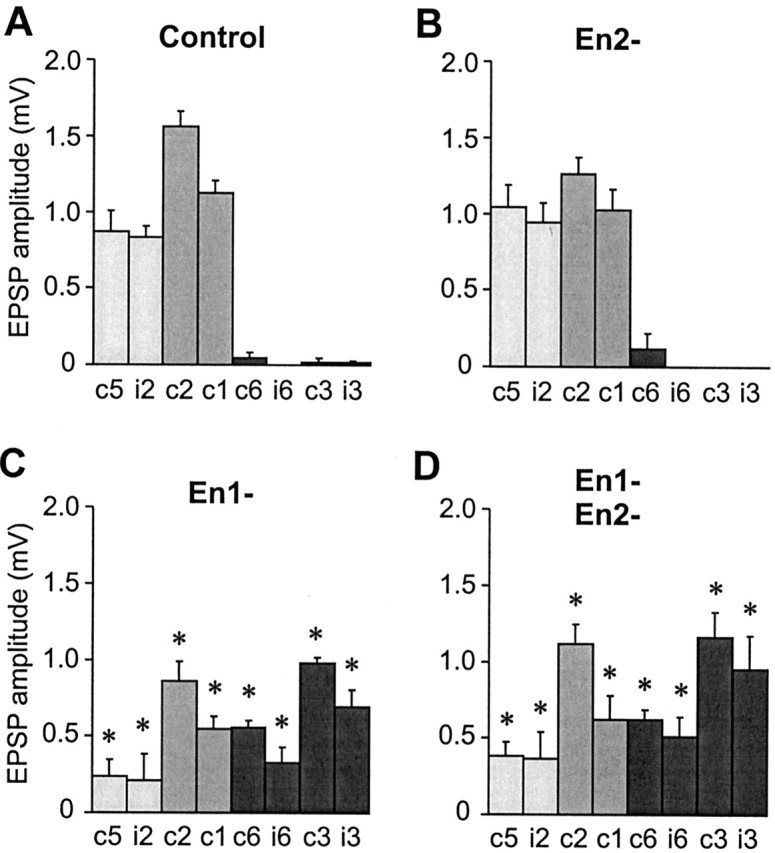
Quantification of the effects of En paralog knock-out on the synaptic connections of neuron 6m. A-D, Bar charts showing the mean amplitudes of monosynaptic unitary EPSPs recorded in eight GIs in response to stimulation of 6m: contralateral GI1 (c1), ipsilateral GI2 (i2), contralateral GI2 (c2), contralateral GI1(c1), contralateral GI6 (c6), ipsilateral Gi6 (i6), contralateral GI3 (c3), and ipsilateral GI3 (i3). Data for ipsilateral GIs 1 and 5 were not shown because they rarely receive synaptic inputs from 6m in control or experimental animals. Light gray bars indicate GIs that, in control animals, receive synaptic inputs from 6m only, medium gray indicates GIs that normally receive inputs both from 6m and from lateral neurons such as 6d (Marie et al., 2000), and dark gray indicates GIs that normally receive inputs only from 6d. Error bars indicate the SEM, and asterisks indicate a significant difference from the control value (p < 0.05). A, Synaptic connections of control 6m (from left to right, n = 13, 16, 15, 13, 16, 16, 16, 17). B, Synaptic connections of 6m after Pa-En2 knock-out (n = 5 for all). No significant differences from control are observed. C, Synaptic connections of 6m after Pa-En1 knock-out (n = 5 for all). There are significant differences from control, with decreases in the amplitude of connections to GI1, GI2, and GI5 and the appearance of de novo connections to GI3 and GI6. There are no significant differences compared with double knock-out. D, Synaptic connections of 6m after double Pa-En1 and Pa-En2 knock-out (n = 8, 8, 7, 6, 9, 8, 8, 8). There are significant differences from control, with decreases in the amplitude of connections to GI1, GI2, and GI5 and the appearance of de novo connections to GI3 and GI6.
Taken together, our results point to the conclusion that the Pa-En1 and Pa-En2 paralogs have different regulatory effects on the genes controlling axon guidance and target recognition. However, because twice as much Pa-En1 is present as Pa-En2, another possible explanation is that the En paralogs have redundant biochemical functions, but transcription of the target genes is differentially affected by different concentrations of total En protein. Thus, expression of the genes controlling the axonal arborization may be altered by a 35% reduction in En activity, whereas expression of the genes controlling synaptic target recognition is affected only by a >35% knockdown of En.
Engrailed dose-response curve
We can test whether the differential effects of single paralog knock-out are simply attributable to dosage effects by titrating the amounts of total En protein, using different dilutions of the equimolar mixture of dsRNAs. If the paralogs have identical functions, then reducing the total amount of En activity in 6m to only 35% of control levels, using an appropriate dilution of the dsRNA mix, should be functionally equivalent to complete abolition of Pa-En1 with an excess of Pa-en1 dsRNA alone. Similarly, reducing total En to 65% of control with a more dilute dsRNA mix should be equivalent to complete knock-out of Pa-En2 using an excess of Pa-en2 dsRNA.
The purpose of these experiments is to compare a partial knockdown of both paralogs to a complete knock-out of one alone. It is not necessary, and may not even be possible, to ensure proportional inhibition of each paralog by adjusting the composition of the dsRNA mixture. Although twice as much Pa-En1 may be present as Pa-En2, suggesting that the latter could be inhibited more strongly by an equimolar mixture, the Pa-en1 dsRNA is longer and could therefore give rise to more active siRNA fragments (Elbashir et al., 2001). In any case, the important point is that any En activity remaining after application of the diluted dsRNA mixture is due to the presence of both Pa-En1 and Pa-En2, irrespective of whether one may be inhibited more than the other.
As a prerequisite for the next experiments, we need to know how the total En protein levels are affected by different dilutions of the mixture of Pa-en1 and Paen2 dsRNAs. To this end, a dose-response curve was constructed, quantifying, as before, the mean percentage intensity of 4D9 staining in the nucleus of 6m (Fig. 6A). From this curve we can calculate the dilutions of the dsRNA mixture necessary to reduce En activity to 35% of the control levels, which, if the paralogs have identical functions, should mimic Pa-En1 knock-out (Fig. 6C,F), and to 65%, equivalent to Pa-En2 knock-out (Fig. 6D,G). These dilutions are 1:30 and 1:200, respectively.
Figure 6.
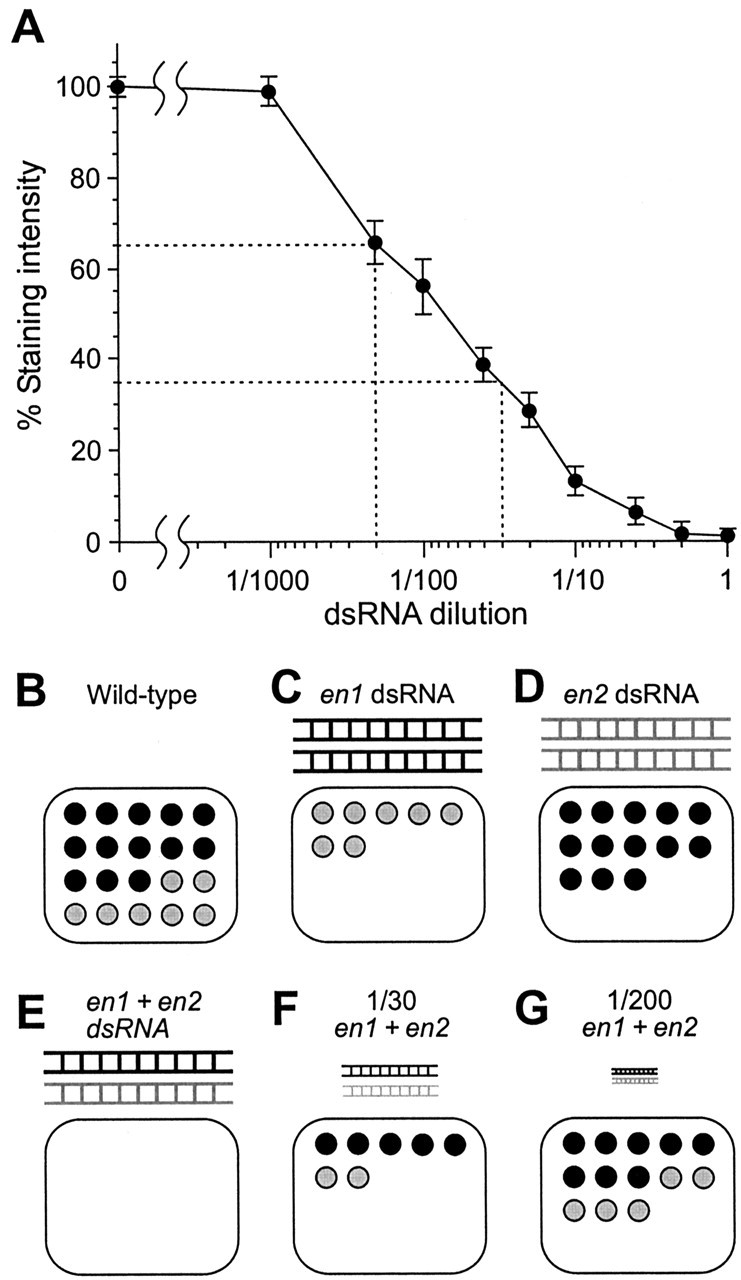
Effects of dsRNA dilution on En levels. A, Dose-response curve of percentage En staining intensity in neuron 6m, plotted against different dilutions of an equimolar mixture of both Pa-en1 and Pa-en2 dsRNAs. From left to right, n = 39, 3, 7, 7, 7, 4, 5, 6, 4, 14. The dashed lines indicate the dilutions of dsRNA required to reduce En staining to 65% of control (1:200) and to 35% of control (1:30). B-G, Diagrams illustrating the effects of different en dsRNAs and different dilutions on the amounts of En proteins. B, Wild-type nucleus, containing 65% Pa-En1 (black circles) and 35% Pa-En2 (gray). C, Application of Pa-en1 dsRNA (black) abolishes Pa-En1, leaving only Pa-En2. D, Application of Pa-en2 dsRNA (gray) abolishes Pa-En2, leaving only Pa-En1. E, Application of an equimolar mixture of the two dsRNAs abolishes all En activity in the nucleus. F, The 1:30 dilution of the dsRNA mixture knocks down total En activity to 35% of control levels, putatively equivalent to Pa-En1 knock-out. In this example, both Pa-En1 and Pa-En2 are reduced proportionately. G, The 1:200 dilution of the dsRNA mixture knocks down total En activity to 65% of control levels, putatively equivalent to Pa-En2 knock-out.
Engrailed dosage effects on axonal arborization
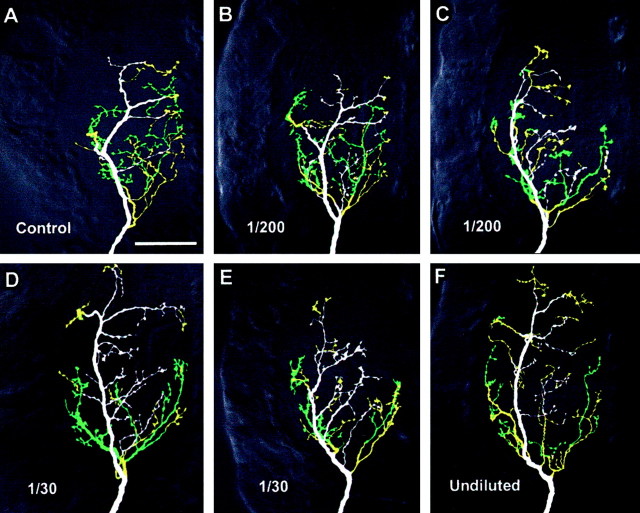
Both 1:200 and 1:30 dilutions of dsRNA had noticeable effects on the arborization of 6m (Fig. 7). As with Pa-En2 knock-out, 1:200 dsRNA had variable qualitative effects on the arbor, having little effect in 43% of the preparations (Fig. 7B) and transforming it to L-type in 57% of the cases (Fig. 7C). Similarly to Pa-En1 knockout, a 1:30 dilution transformed the arbor in 57% of preparations (Fig. 7D) and sometimes resulted in intermediate configurations (Fig. 7E).
Figure 7.
Effects of dsRNA dose on the axonal arbor of neuron 6m. The panels show ventral views of whole-mount terminal ganglia containing antibody-intensified Lucifer Yellow fills of 6m, colored according to branch depth within the neuropil (white represents most ventral, green, most dorsal). The anterior of the ganglion is toward the top and the midline to the right of each panel. A, Typical control medial-type arborization. B, C, Arborizations after treatment with a 1:200 dilution of an equimolar mixture of Pa-en1 and Pa-en2 dsRNAs. The arbor in B appears similar to control, whereas that in C is more similar to lateral-type. D,E, Arborizations of neurons treated with a 1:30 dilution of the dsRNA mix. D illustrates the lateral-type appearance of some of the arbors, whereas E shows an indeterminate arbor type. F, Typical 6m lateral-type arbor after an undiluted application of both en dsRNAs.
Morphological quantification confirmed these results, with the effects of a 1:200 dilution on all three arbor parameters being not significantly different from Pa-En2 knock-out, and a 1:30 dilution mimicking knock-out of Pa-En1 alone (Fig. 8, compare Fig. 4). Taken together, these results are consistent with the hypothesis that the different effects of Pa-En1 and Pa-En2 knockout on axonal arborization can be explained most simply by their differing contributions to the total En activity.
Figure 8.
Quantification of the effects of dsRNA dose on the axonal arbor of neuron 6m. A-C, Histograms of axon angle, anterolateral branches, and dorsal branches, comparing arbors from control 6m (n = 10) and 6d (n = 3) to those from neurons treated with a 1:200 dilution (n = 6) or 1:30 dilution (n = 7) of the mixed dsRNAs, and from neurons treated with the undiluted mixture (1, n = 9). Asterisks indicate significant differences (p < 0.05) from control values. Overall, there are no significant differences between the effects of the 1:200 dilution and Pa-en2 dsRNA alone (compare Fig. 4), or between the 1:30 dilution and Pa-en1 dsRNA alone. A, Axon angle is significantly reduced with a 1:30 dilution (p = 0.01), as in double knock-outs. The reduction with a 1:200 dilution was not significant (p = 0.07) because of the large variance. B, Anterolateral branches are significantly increased by both dsRNA dilutions (1:200: p = 0.03; 1:30: p = 0.0008). C, Dorsal branches are significantly decreased by both dsRNA dilutions (1:200: p = 0.001; 1:30: p < 0.0001). A 1:200 dilution of dsRNA is significantly less effective than the undiluted mixture (p = 0.007).
Engrailed dosage effects on synaptic specificity
In contrast to the dosage effects on arborization, the effects on synaptic connections were not equivalent to those obtained by single paralog knock-out. The 1:200 dilution of the dsRNA mixture, which reduces total En to 65% of control levels (putatively equivalent to Pa-En2 knock-out), did have significant effects on synaptic connections. The amplitude of the synaptic connection to ipsilateral GI2 was reduced and, more importantly, inappropriate connections to contralateral GIs 3 and 6 were formed (Fig. 9B). These effects were significantly different to those of Pa-En2 knock-out alone (compare Figs. 5B,9B). Similarly, the 1:30 dilution of the dsRNA mixture, which reduces En to 35% of control levels (putatively equivalent to Pa-En1 knock-out), did have significant effects on synaptic connections (Fig. 9C) but was significantly less effective than Pa-En1 knock-out alone (compare Figs. 5C, 9C).
Figure 9.
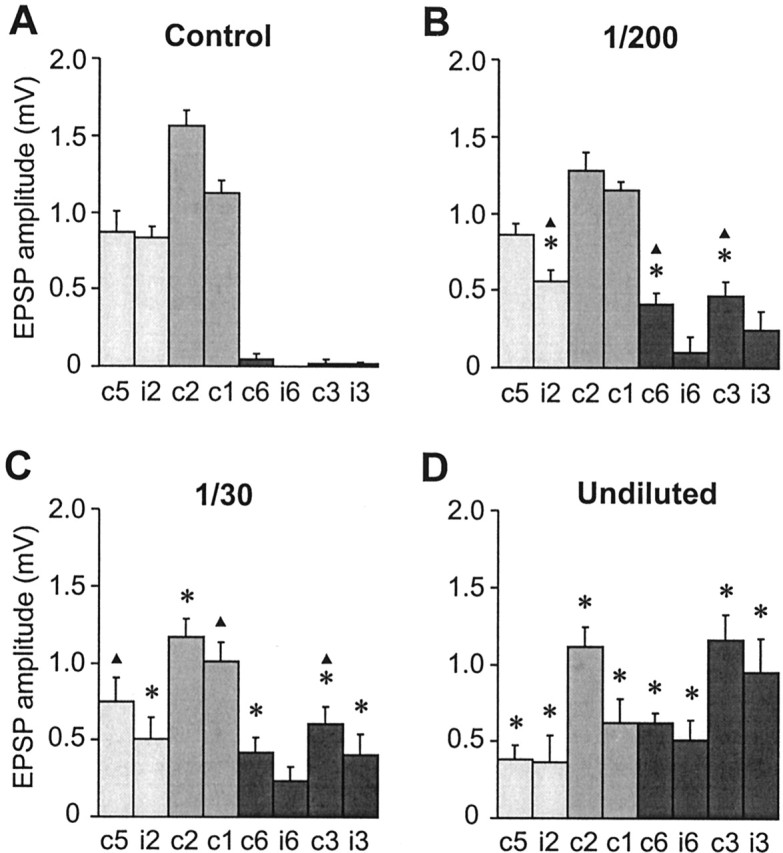
Quantification of the effects of dsRNA dose on the synaptic connections of neuron 6m. A-D, Bar charts showing the mean amplitudes of monosynaptic unitary EPSPs recorded in eight GIs in response to stimulation of 6m. Error bars indicate SEM, and asterisks indicate a significant difference from the control value (p < 0.05). A, Synaptic connections of control 6m (from left to right, n = 13, 16, 15, 13, 16, 16, 16, 17). B, Synaptic connections of 6m after treatment with a 1:200 dilution of the dsRNA mix (n = 6 for all). Significant differences to control are observed, with decreases in the amplitude of connections to iGI2 and the appearance of inappropriate connections to cGI3 and cGI6. Triangles indicate significant differences to Pa-en2 dsRNA alone (Fig. 5B). C, Synaptic connections of 6m after treatment with a 1:30 dilution of dsRNA (n = 6 for all). Significant differences to control are observed, with decreases in the amplitude of connections to GI2 and the appearance of de novo connections to GI3 and GI6. This dilution has significantly less effect than Pa-en1 dsRNA alone (compare Fig. 5C, triangles indicate significant differences). D, Synaptic connections of 6m after treatment with the undiluted dsRNAs (n = 8, 8, 7, 6, 9, 8, 8, 8). There are significant differences to control, with decreases in the amplitude of connections to GI1, GI2, and GI5 and the appearance of de novo connections to GI3 and GI6.
These results indicate that the differential effects of Pa-en1 and Pa-en2 dsRNA on synaptic connections, unlike axon anatomy, are not simply caused by the reduction of the total En protein. On the contrary, the most parsimonious explanation is that the 1:200 dilution of the dsRNA mixture has a significant effect on connectivity (in contrast to Pa-En2 knock-out) because it reduces the levels of Pa-En1. Conversely, the 1:30 dilution is not as effective as Pa-en1 dsRNA alone because, in reducing total En protein to 35% of the original levels, it allows some Pa-En1 activity to remain. Therefore, our results show that Pa-En1 alone is responsible for determining target choice.
Discussion
A biochemical difference between En paralogs
We show here that knock-out of either Pa-En1 or Pa-En2 using RNAi has different effects on axon guidance and synaptic target recognition in an identified sensory neuron. Both Pa-En1 and Pa-En2 have effects on the axon, but only Pa-En1 affects synaptic connections. Quantification of En protein after RNAi showed that Pa-En1 represents 65% of the total Engrailed activity in the cell and Pa-En2 only 35%. We therefore constructed a dose-response curve to enable titration of the amounts of En protein to levels equivalent to single paralog knock-out. RNAi with the equivalent dilutions of the mixed dsRNAs suggested that Pa-En1 and Pa-En2 both contribute toward the control of axonal guidance and confirmed that only Pa-En1 controls synaptic target recognition. We propose that this differential role reflects a biochemical difference between the two paralogs.
What evidence is there for different biochemical functions of Engrailed paralogs in other systems? Spatial and/or temporal separation of En paralog expression appears to be sufficient to account for the different phenotypes of en-1 and en-2 mutations in the mouse. Genetic misexpression experiments have shown that, at least on a gross anatomical level, they appear to have redundancy of function in patterning the CNS (Hanks et al., 1995). In fact, even Drosophila En can substitute for the mouse En-1 (Hanks et al., 1998). However, neither mouse En-2 nor Drosophila En can fulfill the functions of En-1 in patterning the mouse limb (Hanks et al., 1998). In the mouse, En-1 and En-2 compensate for each other in maintaining the differentiation and survival of dopaminergic midbrain neurons (Simon et al., 2001). In Drosophila itself, En and Inv are largely coexpressed, although a few CNS neurons express Inv only (Siegler and Jia, 1999). Mosquito En can substitute for the Drosophila protein during development (Whiteley and Kassis, 1997). However, there is also evidence for separate functions of En and Inv in wing patterning (Simmonds et al., 1995).
What are the domains responsible for the functional difference between Pa-En1 and Pa-En2? Sequence comparison shows that they both retain the known conserved functional domains: the Groucho/TLE corepressor-binding domain, the Exd/Pbx cofactor-binding domain, the homeodomain, and an active repressor domain at the C terminus (Marie and Bacon, 2000). Pa-En2 in addition contains the Invected-specific domain and the RS dipeptide motif (Marie and Bacon, 2000), but there are no conserved domains present in Pa-En1 that are not present in Pa-En2. The function of the Invected domain is unknown but one possibility is that it could be a binding site for an additional cofactor that prevents Pa-En2 from regulating genes that mediate synaptic recognition. It is also possible that differences in function are caused by amino acid substitutions in the less conserved extradomain regions. This would not be unprecedented; it is probably differences in extradomain regions that are responsible for the different effects on limb development of mouse En-1 and En-2 (Hanks et al., 1998). Alternatively, the sequences of these extradomain regions may not be as important as the distances that they interpose between the functional domains themselves. For example, the distance between the Groucho corepressor-interacting domain and the Exd-binding domain is much larger in Pa-En1 than it is in Pa-En2. Also the RS motif present only in En2/Invected extends the distance between the Exd-interacting domain and the homeodomain; this modification has been suggested to affect the homeodomain target specificity (Peltenburg and Murre, 1996; Peterson et al., 1998).
Dose-dependent effects of En
Our results show that the effects of Engrailed are dose-dependent; there is an approximately linear relationship between the amount of En and the morphological characteristics and synaptic connectivity of the neuron. We see no indication that there is a qualitative switch from lateral to medial phenotype governed by a threshold concentration of Engrailed. The axon angle gives the impression of such a switch, however its bimodal distribution rather reflects the binary choice of axon pathways made by the growing axon of 6m (Marie et al., 2002). In one interesting case (Fig. 3E), the axon bifurcated shortly after entering the neuropil and followed both pathways. This dose dependence of the effects of En implies that its target genes bear numerous binding sites of rather low affinity, the progressive occupation of which quantitatively regulates transcription. This has been proposed to be the mechanism by which Ubx suppresses the wing patterning gene sal in Drosophila (Galant et al., 2002).
Engrailed is one of the few neuronal transcription factors for which graded expression patterns and dosage effects are clearly established. It is the graded expression of En across the tectum that sets up the gradient of ephrin-A, providing guidance cues for the topographic mapping of retinal axons (Friedman and O'Leary, 1996; Itasaki and Nakamura, 1996; Logan et al., 1996; Shigetani et al., 1997). Dopaminergic cell survival in the mouse midbrain depends on the dosage of En-1/En-2, with strong, but not weak, expression of a single en-2 allele being sufficient to rescue neurons (Simon et al., 2001). It is perhaps not surprising, therefore, that in this study we also see dosage-dependent effects of En on axon guidance, but at the single cell level. However, we also demonstrate dose-dependent effects of En1 levels on the strength and specificity of synaptic connections. If similar dosage-dependent effects are a common feature in the transcriptional control of phenotype in other neurons, in other systems, this adds an extra dimension of complexity to neuronal determination.
Neofunctionalization or subfunctionalization?
We have shown that the two En paralogs have different functions in the specification of cockroach cercal sensory neurons. How does this result fit with what is known about the phylogeny of these genes and the evolutionary processes underlying their stabilization?
The phylogenetic analysis of conserved domains of Pa-en1 and Pa-en2 (Marie and Bacon, 2000) has shown that the two paralogs strongly group together instead of grouping with their corresponding insect orthologs. This suggests that Pa-en1 and Pa-en2 are the product of a recent duplication within the cockroach lineage. However, coevolution of those domains may have occurred, and the duplication could date back further to the ancestral arthropod (Peterson et al., 1998; Marie and Bacon, 2000). It is clear, however, that the arthropod en duplication occurred independently from the duplications in the vertebrate lineage.
These duplication events have to be considered in the light of two competing, although not necessarily mutually exclusive, models to explain how duplicated genes can become stabilized during evolution. The neofunctionalization model suggests they are permanently preserved by the fixation of rare beneficial mutations that confer a novel function on one of the duplicates (Ohno, 1970; Ohta, 1988; Cooke et al., 1997; Ganfornina and Sanchez, 1999). The more recent Duplication-Degeneration-Complementation (DDC) model (Force et al., 1999) posits complementary restrictions in expression pattern as the most likely initial mechanism for preserving gene duplications, but does not rule out additional biochemical changes in function over the longer term (Force et al., 1999; Gibert, 2002).
The neofunctionalization model would require that one of the duplicated en genes gained a function. In our case, because there is no differential expression, En1 could have been co-opted for the new function of regulating synaptic connectivity. If such a change took place recently in the cockroach lineage, it is hard to understand how it would have been adaptive, because it would have resulted in major re-wiring of the cercal escape circuitry, and of other En-expressing neurons in the CNS. If, in fact, the duplication and co-option occurred in an ancestral insect, we would expect that the role of Engrailed/En1 in other insects is to control both axon guidance and synaptic connections, and that Invected/En2 only affects the former. A comparative study of the neuronal roles of En duplicates in other insects and of the single, invected-like, engrailed genes in the beetle and the grasshopper might answer this question. Whatever the date of the gene duplication, if the function of En1 in controlling synaptic specificity was acquired subsequently, it would imply that En proteins in vertebrates do not necessarily have this role.
In the subfunctionalization, or DDC, model, the ancestral animal had a single En protein that had a role in controlling both axon guidance and synaptic target recognition (as well as the other patterning functions). After the gene duplication, mutations in regulatory sequences separated the expression of En1 and En2 in some non-cercal cells, preserving the duplication, but eventually En2 lost the biochemical function of controlling synaptic specificity. Another possibility with this model is that degenerative mutations in shared regulatory regions would have decreased the expression level of both genes, therefore allowing redundancy. This is not without precedent (Lynch and Force, 2000) and is consistent with the dose-dependent effects of En on arbor shape reported in this study. The DDC model implies that regulation of axon guidance and of synaptic target recognition are both ancestral functions of Engrailed. This opens the possibility that these functions have been conserved throughout the evolution of other phyla, including mammals.
Footnotes
This work was supported by National Institutes of Health (NIH) Grant R01 NS045547 (J.M.B.) with partial support from NIH Research Centers in Minority Institutions award G12 RR-03051.
Correspondence should be addressed to Dr. Jonathan M. Blagburn, Institute of Neurobiology, 201 Boulevard del Valle, San Juan, Puerto Rico 00901. E-mail: jmblagbu@neurobio.upr.clu.edu.
Copyright © 2003 Society for Neuroscience 0270-6474/03/237854-09$15.00/0
References
- Baader SL, Vogel MW, Sanlioglu S, Zhang X, Oberdick J ( 1999) Selective disruption of “late onset” sagittal banding patterns by ectopic expression of Engrailed-2 in cerebellar Purkinje cells. J Neurosci 19: 5370-5379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blagburn JM ( 1989) Synaptic specificity in the first instar cockroach: patterns of monosynaptic input from filiform hair afferents to giant interneurons. J Comp Physiol [A] 166: 133-142. [DOI] [PubMed] [Google Scholar]
- Blagburn JM, Thompson KS ( 1990) Specificity of filiform hair afferent synapses onto giant interneurons in Periplaneta americana: anatomy is not a sufficient determinant. J Comp Neurol 302: 255-271. [DOI] [PubMed] [Google Scholar]
- Blagburn JM, Gibbon CR, Bacon JP ( 1995) Expression of engrailed in an array of identified sensory neurons: comparison with position, axonal arborization, and synaptic connectivity. J Neurobiol 28: 493-505. [DOI] [PubMed] [Google Scholar]
- Condron BG, Patel NH, Zinn K ( 1994) Engrailed controls glial/neuronal cell fate decisions at the midline of the central nervous system. Neuron 13: 541-554. [DOI] [PubMed] [Google Scholar]
- Cooke J, Nowak MA, Boerlijst M, Maynard-Smith J ( 1997) Evolutionary origins and maintenance of redundant gene expression during metazoan development. Trends Genet 13: 360-364. [DOI] [PubMed] [Google Scholar]
- Elbashir SM, Lendeckel W, Tuschl T ( 2001) RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev 15: 188-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC ( 1998) Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans Nature 391: 806-811. [DOI] [PubMed] [Google Scholar]
- Force A, Lynch M, Pickett FB, Amores A, Yan YL, Postlethwait J ( 1999) Preservation of duplicate genes by complementary, degenerative mutations. Genetics 151: 1531-1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman GC, O'Leary DD ( 1996) Retroviral misexpression of engrailed genes in the chick optic tectum perturbs the topographic targeting of retinal axons. J Neurosci 16: 5498-5509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galant R, Walsh CM, Carroll SB ( 2002) Hox repression of a target gene: extradenticle-independent, additive action through multiple monomer binding sites. Development 129: 3115-3126. [DOI] [PubMed] [Google Scholar]
- Ganfornina MD, Sanchez D ( 1999) Generation of evolutionary novelty by functional shift. Bioessays 21: 432-439. [DOI] [PubMed] [Google Scholar]
- Gibert JM ( 2002) The evolution of engrailed genes after duplication and speciation events. Dev Genes Evol 212: 307-318. [DOI] [PubMed] [Google Scholar]
- Hanks M, Wurst W, Anson-Cartwright L, Auerbach AB, Joyner AL ( 1995) Rescue of the En-1 mutant phenotype by replacement of En-1 with En-2. Science 269: 679-682. [DOI] [PubMed] [Google Scholar]
- Hanks MC, Loomis CA, Harris E, Tong CX, Anson-Cartwright L, Auerbach A, Joyner A ( 1998) Drosophila Engrailed can substitute for mouse Engrailed1 function in mid-hindbrain, but not limb development. Development 125: 4521-4530. [DOI] [PubMed] [Google Scholar]
- Itasaki N, Nakamura H ( 1996) A role for gradient En expression in positional specification on the optic tectum. Neuron 16: 55-62. [DOI] [PubMed] [Google Scholar]
- Jia XX, Siegler MV ( 2002) Midline lineages in grasshopper produce neuronal siblings with asymmetric expression of Engrailed. Development 129: 5181-5193. [DOI] [PubMed] [Google Scholar]
- Kennerdell JR, Carthew RW ( 1998) Use of dsRNA-mediated genetic interference to demonstrate that frizzled and frizzled 2 act in the Wingless pathway. Cell 95: 1017-1026. [DOI] [PubMed] [Google Scholar]
- Kornberg T ( 1981) Engrailed: a gene controlling compartment and segment formation in Drosophila Proc Natl Acad Sci USA 78: 1095-1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence PA, Struhl G ( 1982) Further studies of the engrailed phenotype in Drosophila EMBO J 1: 827-833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan C, Wizenmann A, Drescher U, Monschau B, Bonhoeffer F, Lumsden A ( 1996) Rostral optic tectum acquires caudal characteristics following ectopic Engrailed expression. Curr Biol 6: 1006-1014. [DOI] [PubMed] [Google Scholar]
- Lundell MJ, Chu-LaGraff Q, Doe CQ, Hirsh J ( 1996) The engrailed and huckebein genes are essential for development of serotonin neurons in the Drosophila CNS. Mol Cell Neurosci 7: 46-61. [DOI] [PubMed] [Google Scholar]
- Lynch M, Force A ( 2000) The probability of duplicate gene preservation by subfunctionalization. Genetics 154: 459-473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marie B, Bacon JP ( 2000) Two engrailed-related genes in the cockroach: cloning, phylogenetic analysis, expression and isolation of splice variants. Dev Genes Evol 210: 436-448. [DOI] [PubMed] [Google Scholar]
- Marie B, Bacon JP, Blagburn JM ( 2000) Double-stranded RNA interference shows that Engrailed controls the synaptic specificity of identified sensory neurons. Curr Biol 10: 289-292. [DOI] [PubMed] [Google Scholar]
- Marie B, Cruz-Orengo L, Blagburn JM ( 2002) Persistent engrailed expression is required to determine sensory axon trajectory, branching, and target choice. J Neurosci 22: 832-841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morata G, Lawrence PA ( 1975) Control of compartment development by the engrailed gene in Drosophila Nature 255: 614-617. [DOI] [PubMed] [Google Scholar]
- Nibbering PH, Marijnen JG, Raap AK, Leijh PC, van Furth R ( 1986) Quantitative study of enzyme immunocytochemical reactions performed with enzyme conjugates immobilized on nitrocellulose. Histochemistry 84: 538-543. [DOI] [PubMed] [Google Scholar]
- Ohno S ( 1970) Evolution by gene duplication. Heidelberg, Germany: Springer.
- Ohta T ( 1988) Evolution by gene duplication and compensatory advantageous mutations. Genetics 120: 841-847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel NH, Schafer B, Goodman CS, Holmgren R ( 1989) The role of segment polarity genes during Drosophila neurogenesis. Genes Dev 3: 890-904. [DOI] [PubMed] [Google Scholar]
- Peltenburg LT, Murre C ( 1996) Engrailed and Hox homeodomain proteins contain a related Pbx interaction motif that recognizes a common structure present in Pbx. EMBO J 15: 3385-3393. [PMC free article] [PubMed] [Google Scholar]
- Peterson MD, Popadic A, Kaufman TC ( 1998) The expression of two engrailed-related genes in an apterygote insect and a phylogenetic analysis of insect engrailed-related genes. Dev Genes Evol 208: 547-557. [DOI] [PubMed] [Google Scholar]
- Saueressig H, Burrill J, Goulding M ( 1999) Engrailed-1 and netrin-1 regulate axon pathfinding by association interneurons that project to motor neurons. Development 126: 4201-4212. [DOI] [PubMed] [Google Scholar]
- Shigetani Y, Funahashi JI, Nakamura H ( 1997) En-2 regulates the expression of the ligands for Eph type tyrosine kinases in chick embryonic tectum. Neurosci Res 27: 211-217. [DOI] [PubMed] [Google Scholar]
- Siegler MV, Jia XX ( 1999) Engrailed negatively regulates the expression of cell adhesion molecules connectin and neuroglian in embryonic Drosophila nervous system. Neuron 22: 265-276. [DOI] [PubMed] [Google Scholar]
- Siegler MV, Pankhaniya RR, Jia XX ( 2001) Pattern of expression of Engrailed in relation to gamma-aminobutyric acid immunoreactivity in the central nervous system of the adult grasshopper. J Comp Neurol 440: 85-96. [DOI] [PubMed] [Google Scholar]
- Simmonds AJ, Brook WJ, Cohen SM, Bell JB ( 1995) Distinguishable functions for engrailed and invected in anterior-posterior patterning in the Drosophila wing. Nature 376: 424-427. [DOI] [PubMed] [Google Scholar]
- Simon HH, Saueressig H, Wurst W, Goulding MD, O'Leary DD ( 2001) Fate of midbrain dopaminergic neurons controlled by the engrailed genes. J Neurosci 21: 3126-3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson KS, Blagburn JM, Gibbon CR, Bacon JP ( 1992) Correlation of filiform hair position with sensory afferent morphology and synaptic connections in the second instar cockroach. J Comp Neurol 320: 213-227. [DOI] [PubMed] [Google Scholar]
- Webster PJ, Mansour TE ( 1992) Conserved classes of homeodomains in Schistosoma mansoni, an early bilateral metazoan. Mech Dev 38: 25-32. [DOI] [PubMed] [Google Scholar]
- Wenner P, O'Donovan MJ, Matise MP ( 2000) Topographical and physiological characterization of interneurons that express Engrailed-1 in the embryonic chick spinal cord. J Neurophysiol 84: 2651-2657. [DOI] [PubMed] [Google Scholar]
- Whiteley M, Kassis JA ( 1997) Rescue of Drosophila engrailed mutants with a highly divergent mosquito engrailed cDNA using a homing, enhancer-trapping transposon. Development 124: 1531-1541. [DOI] [PubMed] [Google Scholar]
- Wurst W, Auerbach AB, Joyner AL ( 1994) Multiple developmental defects in Engrailed-1 mutant mice: an early mid-hindbrain deletion and patterning defects in forelimbs and sternum. Development 120: 2065-2075. [DOI] [PubMed] [Google Scholar]