Abstract
Despite recent advances in our understanding of lineage of oligodendrocytes, detailed molecular characterization of this lineage in vivo is limited, primarily because of our inability to obtain a pure population of cells in situ. To define the molecular characteristics of oligodendrocyte lineage cells during development and their response to injury, we developed a strategy that uses laser capture microdissection (LCM) to isolate cells from sections and reverse transcription-PCR to determine mRNA expression. As a first step, we examined the expression of myelin-specific protein genes in NG2+ cells in cerebral cortex. We demonstrate that NG2+ cells in both developing and adult mice express NG2 mRNA but not mRNA for proteins specific for astrocytes, neurons, or microglia, indicating that a highly pure population of antigen-specific cells of the oligodendrocyte lineage can be obtained using LCM. Furthermore, we show that NG2+ cells express mRNAs for proteolipid protein (PLP), myelin basic protein, and 2′,3′-cyclic nucleotide 3′-phosphodiesterase, but they dot not express DM-20 mRNA, a PLP mRNA splicing variant. Our data demonstrate that antigen-specific cells of oligodendrocyte lineage differentially express mRNA for myelin-specific proteins and their variants in vivo, partly define the gene expression in NG2+ cells, and raise questions about the cellular sites of DM-20 expression. This work also shows that LCM is a valuable tool to define and analyze gene expression in the cells of the oligodendrocyte lineage.
Keywords: oligodendrocyte precursor, NG2, PLP, DM-20, MBP, gene expression, LCM
Introduction
In the CNS, oligodendrocytes develop from glial progenitors in the ventricular and subventricular zones (Hirano and Goldman, 1988; Levison and Goldman, 1993). Recent developments using molecular biology and immunology techniques have made it possible to identify cells of the oligodendrocyte lineage both in vitro and in vivo. These studies have significantly advanced our knowledge and led to our current understanding of oligodendrocyte development (for review, see Raff, 1989; Woodruff et al., 2001). Despite these advances, detailed molecular characterization of oligodendrocyte lineage cells in vivo during development and after injury is limited, primarily because of our inability to obtain pure populations of cell types in situ. For example, whereas the expression of genes encoding for myelin-specific proteins has been well studied in developing CNS using Northern blot hybridization, RNA protection assay, reverse transcription (RT)-PCR, and Western immunoblot analyses (Morell et al., 1972; Lai et al., 1987; Schindler et al., 1990; Inuzuka et al., 1991;Pedraza et al., 1991), these studies used whole brain or brain regions, making it impossible to identify the cellular origin for each myelin-associated protein. In situ mRNA hybridization analysis, immunohistochemical staining, and combined in situ hybridization and immunostaining also have provided important information about cellular expression of myelin-specific proteins. The unavailability of isoform-specific probes or antibodies, however, has made it difficult to precisely identify the oligodendrocyte lineage cells that express each isoform of the myelin-specific protein genes.
To define the pattern of gene expression in the cells of the oligodendrocyte lineage during development and after injury, we developed a strategy that uses laser capture microdissection (LCM) to remove cells from sections. This allows isolation of RNA from defined in vivo cell populations and use of RT-PCR to precisely identify the mRNAs that they express. Here we report the expression of myelin protein genes in NG2 immunopositive (NG2+) cells in the cerebral cortex (CTX). We provide evidence that the NG2+ cells express mRNAs for proteolipid protein (PLP), MBP, and 2′,3′-cyclic nucleotide 3′-phosphodiesterase (CNP) but not the mRNA for DM-20, a PLP mRNA splicing variant. Our data indicate that NG2 + cells are a unique population of cells in the oligodendrocyte lineage and that LCM is a valuable tool for defining and analyzing the molecular characteristics of antigen-specific cells in oligodendrocyte lineage in vivo.
Materials and Methods
Tissue collection. Under deep anesthesia, brains of mice (C57BL/6) were removed and collected. For whole CTX RNA extraction, CTX were dissected (n = 3) and frozen in liquid nitrogen. For immunohistochemistry and LCM, brains (n = 3) were split near the middle sagittal line, frozen in liquid nitrogen, and sagittally sectioned at a thickness of 8 μm on a cryostat. Both sections and CTX were stored at -80°C until use. All procedures used were approved by the institutional review committee of the University of North Carolina at Chapel Hill.
Immunohistochemistry and cell capture. After thawing at room temperature briefly, sections near the midsagittal line were fixed with 75% ethanol for 20 sec and washed with PBS. In the presence of RNase inhibitor (200 U/ml; Promega, Madison, WI), the sections were immunostained for NG2 + cells for 30 min using an anti-NG2 antibody (1:400; a gift from Dr. Bill Stallcup, The Burnham Institute, La Jolla, CA). Antibody–antigen complexes were detected using peroxidase-conjugated polymers and visualized by incubation with DAB, provided in an EnVision+ immunostaining system (Dakocytomation; Dako, Carpinteria, CA).
After sections were dehydrated via a gradient concentration of ethanol, NG2 + cells (250–500) were microdissected and captured from two sections using a PixCell laser capture microdissector (Arcturus Engineering, Santa Clara, CA). Individual samples (250–500 NG2 + cells) derived from each brain were then transferred to a microcentrifuge tube containing 10 μl of cell lysis buffer provided in an RNA PicoPure kit (Arcturus Engineering).
Total RNA isolation and RT-PCR. Total RNA from NG2 + cells captured with LCM was extracted using the RNA PicoPure kit according the protocol of the manufacturer. Resultant RNA was dissolved in 10 μl of H2O and subjected to RT-PCR. Whole CTX RNA was isolated using the acidic guanidinium thiocyanate–phenol–chloroform method (Chomczynski and Sacchi, 1987) and subjected to RT-PCR. RT was performed using Supercript II reverse transcriptase (Invitrogen, Carlsbad, CA), a 27 base poly deoxythymidine primer, and 10 μl of RNA derived from NG2 + cells or 10 μg of CTX RNA. The resultant cDNA (1 μl of RT reaction derived from CTX RNA and 2.5 μl of RT reaction derived from LCM-captured NG2 + RNA, respectively) was then used as template for PCR amplification (40 cycles, in a 25 μl reaction volume), using specific 20 base oligonucleotide primers. Primers used for PLP–DM-20 amplification were 5′-gaaaagctaattgagaccta-3′ (forward) and 5′-gagcagggaaactagtgtgg-3′ (reverse). The sequences of each primer used and the primer positions for myelin-specific protein genes are provided as supplemental materials at http://www.unc.edu/~pye1/Table1.htm. PCR-amplified DNA fragments were resolved on 1% agarose gel containing ethidium bromide and then photographed. For each gene, the primer pairs are identical or complimentary to sequences that span multiple exons. This strategy allowed amplification of mRNA encoded by several exons, and, therefore, PCR products derived from splicing mRNA variants could be readily identified by size. In addition, this method ensured that the cDNAs obtained could not have been amplified from contaminating genomic DNA [the large DNA intron segment(s) between exons either would not have been amplified because of their size or would have yielded products of sizes that could easily be discriminated from their mRNAs].
Sequence of cDNA derived from RT-PCR. PCR-amplified DNA fragments were resolved on 1% agarose gel. DNA band of interest was cut out and purified using Wizard PCR Preps DNA Purification System (Promega). DNA sequencing was performed at the Automated DNA Sequencing Facility at the University of North Carolina at Chapel Hill.
Southern blot hybridization analysis. PCR-amplified DNA fragments were fractionated on 1% agarose gel, transferred onto a nylon membrane, UV cross-linked, and hybridized with 32P-labeled single-stranded DNA (ssDNA) probe. PLP DNA fragment corresponding to bp 217–903 of mouse PLP cDNA (Hudson et al., 1987) was amplified by PCR and used as a template for hybridization probe. The 32P-labeled ssDNA probe was generated by linear PCR using PLP 3′ end primers and 32P-dCTP (Amersham Biosciences, Arlington Heights, IL), as described previously (Ye et al., 1992, 1995).
Results
Consistent with previous reports (Nishiyama et al., 1997; Reynolds and Hardy, 1997; Diers-Fenger et al., 2001), NG2+ cells in the CTX of adult mice radially extend multiple long processes. To obtain a high pure population of NG2+ cells, soma and theportions of the processes that were proximal to the cell body of each NG2+ cell were made to focally adhere to a membrane by activation with a laser beam (Fig. 1).
Figure 1.
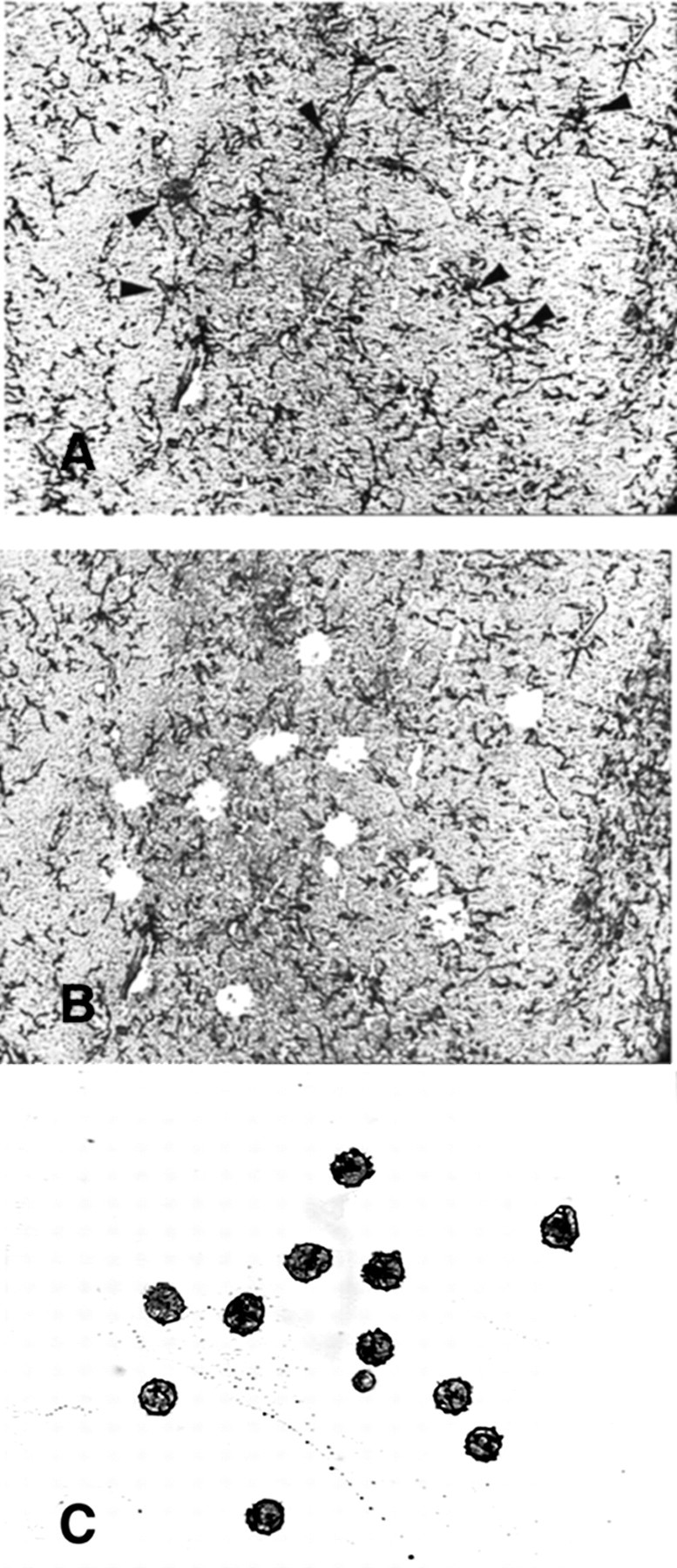
Representative microphotographs of a cerebral cortical section stained for NG2 + precursors derived from a 4-month-old mouse before (A) and after (B) LCM, as well as that of the same captured cells on a membrane (C). Arrows in A indicate NG2 + cells. Note that these cells are absent in B.
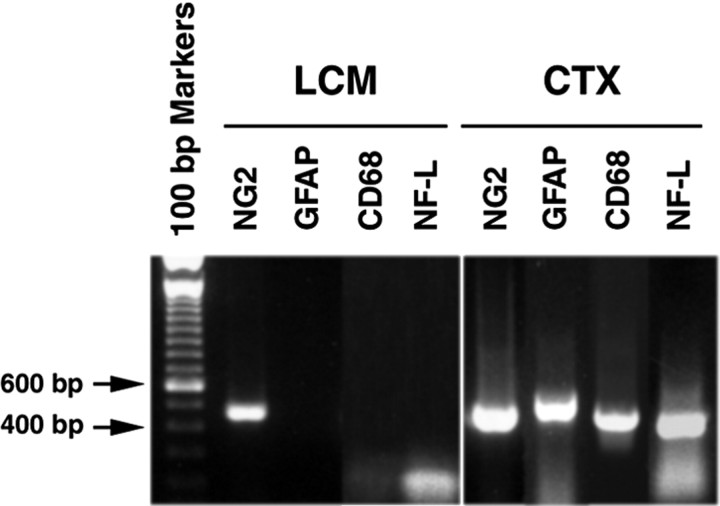
To confirm the identity of LCM-captured NG2+ cells and to determine whether there was contamination of captured NG2+ cells with other cell types, we used RT-PCR to examine the expression of mRNA derived from LCM-captured NG2+ cells derived from 4-month-old mice for the following cell-specific proteins: NG2, glial fibrillary acidic protein (GFAP)(astrocytes), CD68 (microglia–macrophages), and neurofilament light-subunit (NF-L) (neurons). Total RNA derived from whole CTX was used in parallel RT-PCR reactions. When CTX RNA was used, a single DNA band of the expected size was observed for each NG2, GFAP, CD68, and NF-L mRNA, respectively (Fig. 2). In contrast, only NG2 mRNA was detected in the LCM-captured cortical NG2+ cells, and no mRNA for GFAP, CD68, or NF-L mRNA was detected. These data show that LCM can be used to obtain NG2+ cells with little or no contamination of other CNS cell types.
Figure 2.
Expression of NG2, GFAP, CD68, and NF-L mRNAs in cerebral cortical NG2 + cells and whole cerebral cortex. Total RNA was isolated from LCM-captured NG2 + cells (LCM) and from whole cerebral cortex (CTX) of a 4-month-old mouse, and each RNA preparation was subjected to RT-PCR. RT-PCR DNA products were fractionated on 1% agarose gel containing ethidium bromide and photographed under UV light. The mRNA products derived from RT-PCR are indicated on the top of each lane.A DNA ladder of 100 bp markers was loaded in the left lane, and the size of the 400 and 600 bp bands are indicated by arrows. Note that the bands at bottom of the figure are primers used for RT-PCR.
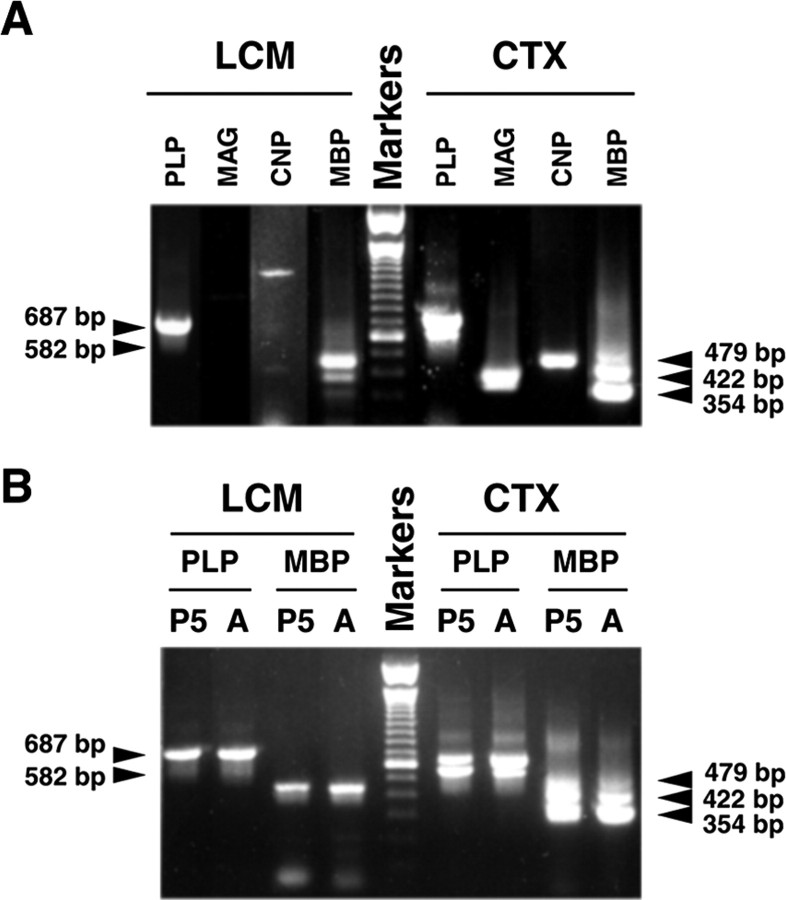
Next, we examined the expression of mRNAs for the four major myelin-specific proteins: PLP, MBP, myelin-associated glycoprotein (MAG), and CNP in NG2+ cells. RNA derived from CTX of adult mice was again used as a control. mRNAs for all four myelin-specific proteins were detected in CTX RNA. As expected, multiple splicing forms of PLP, MBP, and MAG mRNA were clearly distinguished. As Figure 3A shows, two PLP bands of 582 and 687 bp representing DM-20 and PLP mRNA, respectively, were evident. Similarly, three MBP bands of 354, 422–442, and 479 bp (representing mRNA coded for 14, 17, or 18.5 kDa MBP, respectively) and two MAG bands ∼343 and 388 bp (coding for the large and small forms of MAG, MAG-L and MAG-S, respectively) were detected. Among these mRNA variants of each myelin protein gene, mRNA coding for 14 kDa MBP, PLP, and MAG-S was the most abundant, a finding consistent with previous studies showing that 14 kDa MBP, PLP, and MAG-S proteins and their mRNAs are predominately expressed in the CNS of adult mice (Morell et al., 1972; Lai et al., 1987; Campagnoni et al., 1989; Fujita et al., 1989; Schindler et al., 1990; Pedraza et al., 1991; Ikenaka et al., 1992).
Figure 3.
Expression of myelin protein mRNA in cerebral cortical NG2 + cells and whole cerebral cortex. A, Expression of PLP, CNP, MAG, and MBP mRNAs in 4-month-old mice. Total RNA was isolated from LCM-captured NG2 +cells(LCM) and from whole cerebral cortex (CTX) of amouse and was subjected to RT-PCR.RT-PCR DNA products were separated on 1%agarose gel. Each mRNA product derived from RT-PCR is indicated on the top of each lane. B, Expression of PLP–DM-20 and MBP mRNA during development. Total RNA was extracted from cerebral cortical NG2 + cells (LCM) and cerebral cortex (CTX) of a 5-d-old mouse (P5) and a 4-month-old mouse (A), respectively, and subjected to RT-PCR. In both panels, DNA 100 bp markers were loaded in the middle lane.The arrowheads on the left of the panel indicate the expected sizes for RT-PCR amplified DNA derived from PLP (687 bp) and DM-20 (582 bp) mRNA, respectively. The arrowheads on the right indicate the expected sizes for RT-PCR amplified DNA (479, 422, and 354 bp) derived from MBP mRNA variants, respectively.
When RNA derived from LCM NG2+ cells was used, PLP and MBP mRNAs also were readily detected (Fig. 3A). RT-PCR amplification also identified a single faint CNP band and MAG bands. The MAG bands, although clearly visible on the gel under UV light, did not photograph well in Figure 3. In contrast to CTX RNA, which contained both PLP and DM-20 mRNAs, LCM NG2+ cells exhibited only one RT-PCR-amplified DNA band at 687 bp (i.e., the smaller 582 bp DM-20 band was not observed). To be certain that the 687 bp DNA band in LCM samples was derived from PLP mRNA, the DNA band was sequenced after gel purification. DNA sequencing data confirmed that the 687 bp band derived from isolated NG2+ cells corresponded to PLP mRNA (data not shown).
Three MBP 354, 422–442, and 479 bp bands (corresponding to MBP mRNA splicing variants coding for 14, 17, and 18.5 kDa proteins, respectively) also were observed in RNA from NG2+ cells. Unlike adult CTX that expresses predominately mRNA encoded for 14 kDa MBP, however, mRNA for 18.5 kDa MBP (the 479 bp band) was the most abundant form in NG2+ cells.
DM-20 mRNA has been shown to be the predominant form of the two PLP isoforms during early development (Morell et al., 1972; Schindler et al., 1990; Ikenaka et al., 1992). To determine whether DM-20 is expressed in NG2+ cells during early postnatal development, NG2+ cells were isolated from the CTX of mice at postnatal day 5 (P5) and subjected to RT-PCR. RT-PCR examination for NG2, GFAP, CD68, and NF-L mRNAs was first performed to be certain that NG2+ cells were isolated with little or no contamination of other cell types. Similar to the results obtained from the NG2+ cells of adult mice, only NG2 mRNA was detected, and no GFAP, CD68, and NF-L mRNA was detected in the NG2+ cells derived from P5 mice (data not shown). Although abundant DM-20 mRNA was observed in the P5 CTX, no DM-20 mRNA was detected in the NG2+ cells in the CTX of P5 mice (Fig. 3B).
The expression of MBP mRNA in the NG2+ cells of P5 CTX also was examined. Unlike adult CTX, which expresses predominately mRNA coding 14 kDa MBP (354 bp band), mRNAs encoding larger MBP isoforms (17 and 18.5 kDa, i.e., the 422–442 and 479 bp bands, respectively) were more abundant in the NG2+ cells, findings that are consistent with those that we observed in RNA derived from adult NG2+ cells.
To exclude the possibility that DM-20 mRNA is expressed at low levels in NG2+ cells, two rounds of PCR amplification for PLP–DM-20 mRNA from both P5 and adult mice were performed, followed by Southern blot hybridization analysis using a 32P-labeled PLP probe that hybridizes both PLP and DM-20 cDNA. In multiple experiments, we were unable to detect DM-20 mRNA in cortical NG2+ cells derived from P5 or adult mice. A representative PLP Southern hybridization blot is shown in Figure 4.
Figure 4.
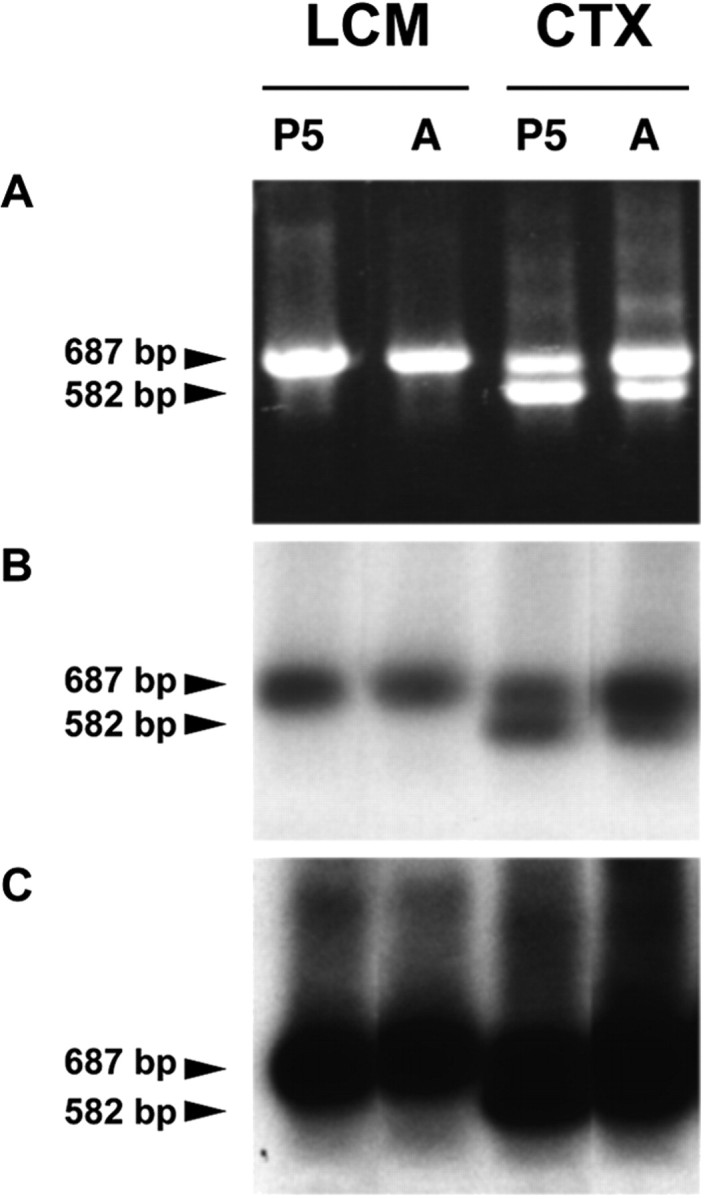
Representative Southern blot hybridization analysis of RT-PCR amplified PLP–DM-20. A, RT-PCR amplified PLP–DM-20 DNA fragments derived from cerebral cortical NG2 + cells (LCM) and cerebral cortex (CTX) of a 5-d-old mouse (P5) and a 4-month-old mouse (A) were fractionated on 1% agarose gel. B, DNA fragments, fractionated on 1% agarose shown in A, were transferred onto a nylon membrane, hybridized with a 32P-labeled PLP ssDNA probe, and exposed to an x-ray film for ∼2 min. C, The blot shown in B was exposed with a film for an extended time (∼30 min). In B and C, arrowheads on the right of each indicate the expected sizes for RT-PCR amplified DNA derived from PLP (687 bp) and DM-20 (582 bp) mRNA, respectively.
Discussion
Using LCM, we demonstrated that it is possible to isolate a population of NG2+ cells from mouse CTX sections with little or no contamination of microglia, astrocytes, or neurons. We showed that NG2+ cell RNA contains mRNAs specific for NG2 and several myelin-specific proteins, including PLP and MBP, but not mRNAs for other cell type-specific proteins, such as GFAP (astrocytes), NF (neurons), and CD68 (microglia). These findings are consistent with previous data showing that NG2+ cells do not react with the antibodies specific for astrocytes, neurons, or microglia (Nishiyama et al., 1997; Reynolds and Hardy, 1997; Keirstead et al., 1998; Diers-Fenger et al., 2001), exhibit immunoreactivity for PLP–DM-20, MBP, and MAG (Trapp et al., 1997), and can drive the expression of a PLP promoter-driven transgene (Mallon et al., 2002). Perhaps more importantly, we demonstrated that NG2+ cells of both developing and adult mice express little or no DM-20 mRNA and that they predominately express 17 and 18.5 kDa MBP mRNA. We believe that this is the first demonstration that antigen-specific cells of oligodendrocyte lineage differentially express mRNA for myelin-specific proteins and their splicing variants in vivo. Our results also demonstrate that LCM is an important and useful tool to define and analyze the molecular characteristics of antigen-specific cells of the oligodendrocyte lineage.
NG2 is a large cell surface proteoglycan that is expressed in the CNS of both developing and adult rodents and humans (for review, see Dawson et al., 2000). In developing rodents, NG2+ cells are widespread in the CNS, and they exhibit many characteristics of oligodendrocyte precursors: they are either bipolar or have few processes and, thus, morphologically resemble oligodendrocyte precursors in culture (Nishiyama et al., 1996; Diers-Fenger et al., 2001; Ye et al., 2002); they express oligodendrocyte precursor markers, such as PDGFαR and A2B5 in vivo and in culture (Stallcup and Beasley, 1987; Nishiyama et al., 1996; Reynolds and Hardy, 1997; Keirstead et al., 1998; Diers-Fenger et al., 2001; Mallon et al., 2002); they proliferate, as judged by their incorporation of bromodeoxyuridine (BrdU) (Mallon et al., 2002); they possess electophysiological properties of glial progenitor cells (Diers-Fenger et al., 2001); and they are capable of developing into mature oligodendrocytes in vivo and in culture (Stallcup and Beasley, 1987; Trapp et al., 1997; Diers-Fenger et al., 2001).
In adult rodents, NG2+ cells also are abundant in the CNS (Nishiyama et al., 1996; Reynolds and Hardy, 1997; Diers-Fenger et al., 2001; Mallon et al., 2002). Unlike during development, however, NG2+ cells in adult rodent brains exhibit large cell bodies with long complex processes (Ye et al., 2000, 2002; Diers-Fenger et al., 2001). Despite the fact that their morphology is not like most precursors, current data indicate that NG2+ cells in adult mice also are oligodendrocyte precursors, as shown by their capability to incorporate BrdU and the fact that they increase in number after injury and in response to growth factor stimulation (Keirstead et al., 1998; Mason et al., 2000; Ye et al., 2000, 2002). Our current data showing NG2+ cells in both developing and adult brains predominately express the 17 and 18.5 kDa MBP isoforms, a characteristic of early developing brain, further support the hypothesis that NG2+ cells are oligodendrocyte precursors.
Our findings that PLP, but not DM-20, mRNA is detected in NG2+ oligodendrocyte precursors during development are consistent with those of Mallon et al. (2002). Using a rat PLP promoter-driven enhanced green fluorescent protein reporter transgene, Mallon et al. demonstrated that PLP gene is actively expressed in CNS NG2+ cells of transgenic mice during development. Our data also are in line with that of Trapp et al. (1997), who reported that some NG2+ cells colocalized with PLP–DM-20-immunoreactive antigen in the developing rodent brain. Our data extends the above-cited reports by demonstrating that NG2+ oligodendrocyte precursors express full-length PLP mRNA but are not a significant site of DM-20 mRNA expression in vivo. The latter finding raises questions about the sites of DM-20 expression.
DM-20 mRNA and its protein, products of an alternatively spliced PLP mRNA that lacks 105 nucleotides in the exon 3 (Nave et al., 1987), are the major forms of PLP expressed during early CNS development (Schindler et al., 1990; Ikenaka et al., 1992). Schindler et al. (1990) reported that DM-20 protein is present before PLP in fetal bovine cerebral cortex. Later, Ikenaka et al. (1992), using combined RT-PCR and Southern blot hybridization analysis, demonstrated that DM-20 mRNA is expressed in the mouse CNS during embryonic development, a time when mature oligodendrocytes are rare. It has been postulated, therefore, that DM-20 is the predominant PLP isoform in oligodendrocyte precursors. These studies, however, used whole brain or brain regions and, thus, could not determine the cell type(s) that express DM-20 mRNA. Although our study clearly demonstrates that NG2+ oligodendrocyte precursors express no or little DM-20 in vivo, our data do not exclude the possibility that there are other oligodendrocyte precursors existing in CNS that are capable of expressing DM-20 mRNA. In other words, it is possible that NG2-negative oligodendrocyte precursors express DM-20 mRNA. Another possible explanation is that, although DM-20 and its mRNA are expressed in myelinating oligodendrocytes, they are mainly synthesized in non-oligodendrocyte cells during embryonic and early postnatal development. The latter speculation is supported by the following findings: DM-20 mRNA is detected in multiple adult mouse organs, including liver, heart, and spleen (Nadon et al., 1997), and in cultured C6 rat astroglioma and several neuronal cell lines (Ikenaka et al., 1992); and premyelinating oligodendrocytes can be immunostained with antibodies that recognize both PLP and DM-20 (Trapp et al., 1997).
In summary, using LCM in combination with RT-PCR, we demonstrated that CNS NG2+ cells express only PLP mRNA and not the DM-20 isoform. These data offer new insights into the gene expression of cells in the oligodendrocyte lineage. Our data also demonstrate that LCM methodology is a powerful tool to investigate the molecular characteristics of single antigenic positive cells in vivo.
Footnotes
This work was supported by National Institutes of Health Grant NS38891 (National Institute of Neurological Disorders and Stroke) to A.J.D. We thank Dr. Pierer Morell for critical reading of this manuscript and helpful discussion.
Correspondence should be addressed to Dr. Ping Ye, Department of Pediatrics, CB 7039, The University of North Carolina at Chapel Hill, Chapel Hill, NC 27599-7220. E-mail: ping_ye@med.unc.edu.
Copyright © 2003 Society for Neuroscience 0270-6474/03/234401-05$15.00/0
References
- Campagnoni AT, Roth HJ, Pretorius PJ, Campagnoni CW ( 1989) Expression of myelin basic protein genes in the developing mouse brain. In: Developmental neurobiology (Evrard P, Minkowski A, eds), pp 95–109.New York: Nestec.
- Chomczynski P, Sacchi N ( 1987) Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem 162: 156–159. [DOI] [PubMed] [Google Scholar]
- Dawson MR, Levine JM, Reynolds R ( 2000) NG2-Expressing cells in the central nervous system: are they oligodendroglial progenitors? J Neurosci Res 61: 471–479. [DOI] [PubMed] [Google Scholar]
- Diers-Fenger M, Kirchhoff F, Kettenmann H, Levine JM, Trotter J ( 2001) AN2/NG2 protein-expressing glial progenitor cells in the murine CNS: isolation, differentiation, and association with radial glia. Glia 34: 213–228. [DOI] [PubMed] [Google Scholar]
- Fujita N, Sato S, Kurihara T, Kuwano R, Sakimura K, Inuzuka T, Takahashi Y, Miyatake T ( 1989) cDNA cloning of mouse myelin-associated glycoprotein: a novel alternative splicing pattern. Biochem Biophys Res Commun 165: 1162–1169. [DOI] [PubMed] [Google Scholar]
- Hirano M, Goldman JE ( 1988) Gliogenesis in rat spinal cord: evidence for origin of astrocytes and oligodendrocytes from radial precursors. J Neurosci Res 21: 155–167. [DOI] [PubMed] [Google Scholar]
- Hudson LD, Berndt JA, Puckett C, Kozak CA, Lazzarini RA ( 1987) Aberrant splicing of proteolipid protein mRNA in the dysmyelinating jimpy mutant mouse. Proc Natl Acad Sci USA 84: 1454–1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikenaka K, Kagawa T, Mikoshiba K ( 1992) Selective expression of DM-20, an alternatively spliced myelin proteolipid protein gene product, in developing nervous system and in nonglial cells. J Neurochem 58: 2248–2253. [DOI] [PubMed] [Google Scholar]
- Inuzuka T, Fujita N, Sato S, Baba H, Nakano R, Ishiguro H, Miyatake T ( 1991) Expression of the large myelin-associated glycoprotein isoform during the development in the mouse peripheral nervous system. Brain Res 562: 173–175. [DOI] [PubMed] [Google Scholar]
- Keirstead HS, Levine JM, Blakemore WF ( 1998) Response of the oligodendrocyte progenitor cell population (defined by NG2 labelling) to demyelination of the adult spinal cord. Glia 22: 161–170. [PubMed] [Google Scholar]
- Lai C, Brow MA, Nave KA, Noronha AB, Quarles R, Bloom FE, Milner RD, Sutcliffe JG ( 1987) Two forms of 1B236/myelin-associated glycoprotein, a cell adhesion molecule for postnatal neural development, are produced by alternative splicing. Proc Natl Acad Sci USA 84: 4337–4341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levison SW, Goldman JJ ( 1993) Both oligodendrocytes and astrocytes develop from progenitors in the subventricular zone of postnatal rat fore-brain. Neuron 10: 201–212. [DOI] [PubMed] [Google Scholar]
- Mallon BS, Shick HE, Kidd GJ, Macklin WB ( 2002) Proteolipid promoter activity distinguishes two populations of NG2-positive cells throughout neonatal cortical development. J Neurosci 22: 876–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason JL, Ye P, Suzuki K, D'Ercole AJ, Matsushima GK ( 2000) Insulin-like growth factor-1 inhibits mature oligodendrocyte apoptosis during primary demyelination. J Neurosci 20: 5703–5708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morell P, Greenfield S, Costantino-Ceccarini E, Wisniewski H ( 1972) Changes in the protein composition of mouse brain myelin during development. J Neurochem 19: 2545–2554. [DOI] [PubMed] [Google Scholar]
- Nadon NL, Miller S, Draeger K, Salvaggio M ( 1997) Myelin proteolipid DM20: evidence for function independent of myelination. Int J Dev Neurosci 15: 285–293. [DOI] [PubMed] [Google Scholar]
- Nave KA, Lai C, Bloom FE, Milner RJ ( 1987) Splice site selection in the proteolipid protein (PLP) gene transcript and primary structure of the DM-20 protein of central nervous system myelin. Proc Natl Acad Sci USA 84: 5665–5669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama A, Lin XH, Giese N, Heldin CH, Stallcup WB ( 1996) Colocalization of NG2 proteoglycan and PDGF α-recepto on O2A progenitor cells in the developing rat brain. J Neurosci Res 43: 299–314. [DOI] [PubMed] [Google Scholar]
- Nishiyama A, Yu M, Drazba JA, Tuohy VK ( 1997) Normal and recative NG2 + glial cells are distinct from resting and activated microglia. J Neurosci Res 48: 299–312. [DOI] [PubMed] [Google Scholar]
- Pedraza L, Frey AB, Hempstead BL, Colman DR, Salzer JL ( 1991) Differential expression of MAG isoforms during development. J Neurosci Res 29: 141–148. [DOI] [PubMed] [Google Scholar]
- Raff MC ( 1989) Glial cell diversification in the rat optic nerve. Science 243: 1450–1455. [DOI] [PubMed] [Google Scholar]
- Reynolds R, Hardy R ( 1997) Oligodendroglial progenitors labeled with the O4 antibody persist in the adult rat cerebral cortex in vivo. J Neurosci Res 47: 455–470. [DOI] [PubMed] [Google Scholar]
- Schindler P, Luu B, Sorokine O, Trifilieff E, Van Dorsselaer A ( 1990) Developmental study of proteolipids in bovine brain: a novel proteolipid and DM-20 appear before proteolipid protein (PLP) during myelination. J Neurochem 55: 2079–2085. [DOI] [PubMed] [Google Scholar]
- Stallcup WB, Beasley L ( 1987) Bipotential glial precursor cells of the optic nerve express the NG2 proteoglycan. J Neurosci 7: 2737–2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapp BD, Nishiyama A, Cheng D, Macklin W ( 1997) Differentiation and death of premyelinating oligodendrocytes in developing rodent brain. J Cell Biol 137: 459–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodruff RH, Tekki-Kessaris N, Stiles CD, Rowitch DH, Richardson WD ( 2001) Oligodendrocyte development in the spinal cord and telencephalon: common themes and new perspectives. Int J Dev Neurosci 19: 379–385. [DOI] [PubMed] [Google Scholar]
- Ye P, Kanoh M, Zhu W, Laszkiewicz I, Royland JE, Wiggins RC, Konat G ( 1992) Cyclic AMP-induced upregulation of proteolipid protein and myelin associated glycoprotein gene expression in C6 cells. J Neurosci Res 37: 683–690. [DOI] [PubMed] [Google Scholar]
- Ye P, Carson J, D'Ercole AJ ( 1995) In vivo actions of insulin-like growth factor-I (IGF-I) on brain myelination: studies of IGF-I and IGF binding protein-1 (IGFBP-1) transgenic mice. J Neurosci 15: 7344–7356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye P, Lee KH, D'Ercole AJ ( 2000) Insulin-like growth factor-I (IGF-I) protects myelination from undernutritional insult: studies of transgenic mice overexpressing IGF-I in brain. J Neurosci Res 62: 700–708. [DOI] [PubMed] [Google Scholar]
- Ye P, Li L, Richards RG, DiAugustine RP, D'Ercole AJ ( 2002) Myelination is altered in insulin-like growth factor-I null mutant mice. J Neurosci 22: 6041–6051. [DOI] [PMC free article] [PubMed] [Google Scholar]