Abstract
The NMDA receptor 3B (NR3B) subunit is the most recently identified member of the NMDA receptor family. In heterologous cells, it has been shown to reduce the Ca2+ permeability of glutamatergic receptor complexes formed together with NR1 and NR2 subunits and to form the unique excitatory glycine receptor complex with the NR1 subunit. However, it is unclear whether NR3B protein is expressed in and exerts similar functions in neurons. In addition, it is not understood how NR3B interacts with NR1 and NR2 and how such an interaction may regulate the membrane trafficking of the NMDA receptor complex. Here we report that our analysis using an antibody specific for NR3B showed that the NR3B protein is selectively expressed in somatic motor neurons in the brainstem of adult mice. Coimmunoprecipitation and electrophysiological analyses demonstrated that NR3B, when exogenously introduced into hippocampal neurons, can coassemble with endogenous NR1 and NR2A and can reduce the Ca2+ permeability of NMDA currents. In contrast, NR3B was not involved in the excitatory glycine response in neurons under our test conditions. Although NR1 or NR3B alone cannot be transported to the cell surface, coexpression of these subunits mutually supported transport of the NMDA receptor complex by interaction involving the specific regions of the C terminus of NR3B. These results indicate that NR3B may modulate the function of NMDA receptors in somatic motor neurons during adulthood by controlling membrane trafficking and by reducing Ca2+ permeability.
Keywords: NMDA, calcium, glutamate, receptor, surface, assembly
Introduction
NMDA receptors play an essential role in many neurodevelopmental, neurophysiological, and neuropathological processes because of their uniquely high Ca2+ permeability (Monaghan et al., 1989; McBain and Mayer, 1994; Dingledine et al., 1999; Cull-Candy et al., 2001). Functional NMDA receptors in the mammalian brain consist of the NMDA receptor 1 (NR1) subunit and one or more of the NR2 subunits: NR2A-NR2D (Forrest et al., 1994; Hollmann and Heinemann, 1994). In addition, NR3A (Ciabarra et al., 1995; Sucher et al., 1995) and NR3B (Nishi et al., 2001; Chatterton et al., 2002; Matsuda et al., 2002) have been recently identified as NMDA receptor subunits. NR1 is ubiquitously expressed in the CNS and provides a binding site for glycine (Hirai et al., 1996), an essential coagonist of NMDA receptors. In contrast, expression of NR2 subunits is spatially and temporally regulated (Monyer et al., 1992; Watanabe et al., 1993; Monyer et al., 1994). NR2 subunits not only provide glutamate-binding sites (Laube et al., 1997) but also modify channel properties such as current kinetics and channel conductance (Monyer et al., 1992). In addition, the C terminus of NR2 controls the cell surface expression and the synaptic localization of the NMDA receptor (Kornau et al., 1995; McIlhinney et al., 1996; Niethammer et al., 1996; Okabe et al., 1999; Steigerwald et al., 2000). The NR3A subunit also modifies channel properties of the NMDA receptor. When coexpressed with NR1 and NR2 in heterologous cells, NR3A decreases channel conductance and Ca2+ permeability of the NMDA receptor (Perez-Otano et al., 2001). Indeed, the NMDA-induced currents were increased in NR3A-/- neurons (Das et al., 1998). Therefore, to understand the functional diversity of native NMDA receptors in the CNS, it is essential to clarify how NMDA receptor subunits are expressed and assembled and how they modify the function of the NMDA receptor complex.
NR3B mRNA is highly expressed in motor neurons in the spinal cord, pons, and medulla. When coexpressed with NR1 and NR2 subunits, NR3B also reduces NMDA-evoked current (Nishi et al., 2001) and Ca2+ permeability (Matsuda et al., 2002) in heterologous cells. Although NR3A mRNA (Ciabarra et al., 1995; Sucher et al., 1995) and protein (Wong et al., 2002) expression drops dramatically after the second postnatal weeks, NR3B mRNA expression persists in these tissues in adult mice. Therefore, expression of NR3B may play a unique role in synaptic plasticity and certain forms of neuronal death in these tissues at later developmental stages and in adulthood. However, it is unclear whether the NR3B protein is expressed in these tissues. In addition, although NR3B coassembles with NR1 to form unique excitatory glycine receptors in Xenopus oocytes (Chatterton et al., 2002), it is not completely clear whether it forms such channels in mammalian neurons. Finally, it is not understood how NR3B interacts with NR1 and NR2 and how such an interaction may regulate the membrane trafficking of the NMDA receptor complex. In this study, we aimed to address these issues by using a specific anti-NR3B antibody and by performing quantitative analysis of NMDA receptor trafficking.
Materials and Methods
Production and affinity purification of polyclonal anti-NR3B antibody production. The synthetic peptide TGPPEGQQERAEQEC, which corresponds to amino acids 885-899, was coupled to keyhole limpet hemocyanin (KLH). Two rabbits were immunized with 0.5 mg of KLH-peptide conjugate in complete Freund's adjuvant and were given booster immunizations three times at 1 week intervals with 0.25 mg of conjugate in incomplete Freund's adjuvant (Rockland, Gilbertsville, PA). Antiserum was further purified by an affinity Sepharose column coupled with the antigenic peptide. The antibody was eluted with 100 mm Gly, pH 2.5, immediately neutralized with Tris-HCl, pH 8.5, and dialyzed in PBS.
Immunohistochemical analysis. The experimental protocol was approved by the Animal Resource Committee of St. Jude Children's Research Hospital, and all procedures related to care and treatment of the animals were performed according to National Institutes of Health guidelines. ICR mice (Harlan, Indianapolis, IN) were anesthetized with tribromoethanol and perfused transcardially with 4% paraformaldehyde. Coronal sections (16 μm thick) were prepared by using a cryostat. Sections were incubated first with PBS containing 4% goat serum, 1% bovine serum albumin (BSA), and 0.4% Triton X-100 and were then subsequently incubated with a 1:250 dilution of rabbit affinity-purified anti-NR3B antibody and a 1:1000 dilution of mouse anti-pan NR1 antibody (Chemicon, Temecula, CA) overnight at 4°C. Sections were further incubated with a 1:2000 dilution of anti-rabbit Ig antibody labeled with Alexa 546 and anti-mouse Ig antibody labeled with Alexa 488 (Molecular Probes, Eugene, OR). Bound antibody was visualized with a microscope (BX60; Olympus Optical, Tokyo, Japan) equipped with a CCD camera (C5810; Hamamatsu Photonics, Shizuoka, Japan).
Construction of expression vectors and cell transfection. NR3B cDNA was cloned into the expression vector pTracer-enhanced green fluorescent protein (EGFP) (Kohda et al., 2000) or pCAGGS (provided by Dr. J. Miyazaki, Tohoku University, Sendai, Japan). In some experiments, cDNA that encoded hemagglutinin (HA) was added just upstream of the stop codon of the NR3B coding region; the resulting plasmid was designated pTracer-NR3B-HA-EGFP or pCAGGS-NR3B-HA. For the cell surface staining analysis, HA cDNA was added immediately downstream of the signal sequence of NR3B and subcloned into pTracer-nucYFP vector (Kohda et al., 2003), which expresses yellow fluorescent protein (YFP) in the nucleus of transfected cells. This plasmid was referred to as pTracer-HA-NR3B-nucYFP. Fluorescent protein and the cloned cDNA in each pTracer-based vector were under the control of two different promoters. The NR1-1a (provided by Dr. J. Boulter, University of California, Los Angeles, CA) and NR2A (from Dr. S. Nakanishi, Kyoto University, Kyoto, Japan) cDNAs were subcloned into pCAGGS. In some experiments, GFP was fused to the extracellular domain of NR1-1a (pCAGGS-EGFP-NR1-1a) to increase the protein molecular weight for better separation from the NR3B protein.
Human embryonic kidney 293 (HEK293) cells (CRL-1573; American Type Culture Collection, Manassas, VA) were maintained in 10% CO2 at 37°C in DMEM (Invitrogen, Rockville, MD) supplemented with 10% fetal bovine serum (Hyclone, Logan, UT) in 6 cm dishes. A total of 10 μg of plasmids (the ratio of pCAGGS-NR1-1a to pCAGGS-NR2A to pTracer-NR3B-HA-EGFP DNA was 3:3:2) were transfected into HEK293 cells by using LipofectAMINE 2000 (Invitrogen, Carlsbad, CA). Cells were protected from NMDA receptor-mediated toxicity by the addition of (5S, 10R)-(+)-5-methyl-10,11-dihydro-5H-dibenzo[a,d]cyclo-hepten-5,10-imine (MK-801) (5 μm; Sigma, St. Louis, MO), d,l-2-amino-5-phosphonovalerate (100 μm), and 7-Cl-kynurenate (10 μm; Tocris, Ballwin, MO) to the medium.
Immunoblotting, cell surface biotinylation, and immunoprecipitation assays. HEK293 cells 36-48 hr after transfection or whole brains were solubilized in TNE buffer (50 mm NaF, 1% NP-40, 20 mm EDTA, 1 μm pepstatin A, 2 μg/ml leupeptin, 10 μg/ml aprotinin, and 50 mm Tris-HCl, pH 8.0) with 0.1% SDS, and the soluble fractions were analyzed by immunoblotting with anti-NR3B or anti-HA antibody (Roche Applied Science, Indianapolis, IN), in which a horseradish peroxidase (HRP)-conjugated secondary antibody (Amersham Biosciences, Buckinghamshire, UK) and an ECL kit (Amersham Biosciences) were used.
For biotinylation of surface proteins, transfected cells were biotinylated for 15 min at 4°C with 0.5 mg/mL EZ-link Sulfo-NHS-LC-biotin or EZ-link Sulfo-NHS-SS-biotin (Pierce, Rockford, IL) in 115 mm NaCl containing borate, pH 8.6. The reaction was terminated by adding 50 mm Tris-HCl, pH 7.5, and washing with 115 mm NaCl containing 50 mm Tris-HCl, pH 7.5. For immunoprecipitation assays followed by detection with HRP-conjugated streptavidin (Amersham Biosciences), cells were first lysed in 1% SDS and incubated for 5 min at 95°C to avoid coimmunoprecipitation of NR1 and NR3B subunits because these subunits have similar molecular weights. The cell lysate was diluted in TNE buffer and incubated with 1 μg of rat anti-HA antibody (Roche Applied Science) or 2.5 μg of rabbit anti-NR1 antibody (Chemicon) for 2 hr and subsequently with 50 μl of protein G-conjugated agarose (Roche Applied Science) for 1 hr. For the avidin precipitation or coimmunoprecipitation assays, cells were directly lysed in TNE buffer with 0.1% SDS, and the cell lysate was incubated with 60 μl of immobilized NeutrAvidin (Pierce), 1 μg of rat anti-HA antibody, 2.5 μg of mouse anti-pan NR1 antibody (Chemicon), or 3 μg of rabbit anti-NR2A antibody (Upstate Biotechnology, Waltham, MA) for 2 hr at 4°C and subsequently with 50 μl of protein G-conjugated agarose (Roche Applied Science) for 1 hr. In all cases, after the precipitates were washed four times with TNE buffer, we added SDS-PAGE sample buffer and incubated the samples for 5 min at 95°C. After centrifugation, the supernatant, which corresponded to 10% of the total cell lysate, was subjected to immunoblot analysis.
Glycosidase digestion. HEK293 cells transfected with pTracer-NR3B-HA-EGFP plus pCAGGS-NR1-1a or pTracer-NR3B-HA-EGFP plus pCAGGS-NR2A were lysed in TNE buffer with 0.1% SDS, followed by immunoprecipitation with anti-HA antibody as described above. Immunoprecipitated samples were denatured at 95°C for 5 min in a solution of 100 mm sodium acetate, pH 5.5, 1% SDS, and 4% β-mercaptoethanol. After 10-fold dilution in 100 mm sodium acetate, pH 5.5, samples were incubated for 16 hr at 37°C in the presence or absence of endoglycosidase H (Endo H; Roche Applied Science).
Immunocytochemical analysis of transfected cells. To confirm the specificity of the anti-NR3B antibody, HEK293 cells transfected with pTracer-NR3A-HA-EGFP or pTracer-NR3B-HA-EGFP were fixed with 4% paraformaldehyde for 10 min at room temperature and incubated with a blocking solution (2% goat serum, 2% BSA, and 0.4% Triton X-100 in PBS) for 1 hr. Cells were then incubated with a dilution of 1:1000 of mouse anti-HA antibody (Berkeley Antibody Co., Inc., Richmond, CA) and anti-NR3B antibody for 1 hr. To visualize the bound primary antibody, the cells were incubated with 1:2000 of Alexa 546-conjugated anti-rabbit Ig and Alexa 350-conjugated anti-mouse Ig antibodies (Molecular Probes).
To investigate the subcellular localization of the NMDA receptor, we cotransfected COS-7 cells with 3 μg of pECFP-ER (Clontech, Palo Alto, CA), which encoded cyan fluorescence protein (CFP) that included an endoplasmic reticulum (ER) retention signal, and a total of 7 μg of pCAGGS-NR1-1a plus pCAGGS-NR3B-HA or pCAGGS-NR2A plus pCAGGS-NR3B-HA (for each combination, the DNA ratio was 4:1, respectively). After cells were fixed and blocked as described above, they were incubated with primary antibodies (anti-GFP and anti-HA antibodies) and finally with secondary antibodies (anti-chicken Ig antibody conjugated to Alexa 488 and anti-mouse Ig antibody conjugated to Alexa 546; Molecular Probes). The ECFP-ER signal completely colocalized with the ER-specific markers (Matsuda and Yuzaki, 2002).
For cell surface staining, HEK293 cells were transfected with pTracer-HA-NR3B-nucYFP with or without pCAGGS-NR1-1 or pCAGGS-NR2A and fixed as described above. Cells were then incubated with PBS containing 2% goat serum and 2% BSA without Triton X-100 (a nonpermeabilizing condition) for 1 hr at room temperature. Cells were incubated with a dilution of 1:1000 of mouse anti-HA antibody for 1 hr to stain N-terminal-tagged HA-NR3B on the cell surface. After permealization with 0.4% Triton X-100 for 30 min, cells were incubated for 1 hr with a dilution of 1:2000 of anti-mouse Ig antibody conjugated to Alexa 546 (Molecular Probes). The average background red fluorescence was determined by using HEK293 cells incubated without the primary antibody but with the secondary antibody. Cells whose surface displayed red fluorescence at a level greater than the background level were counted as HA+. The proportion of HA+ cells among the transfected (YFP-positive) cells was calculated by determining the number of positive cells among 400 cells from three independent experiments.
Primary culture, Sindbis virus infection, and transfection. A primary culture of hippocampal neurons was prepared as previously described (Forrest et al., 1994) and kept in Neurobasal medium with B-27 (Invitrogen). Cortical neurons were prepared from wild-type and NR1 knock-out (NR1-/-) mice (Forrest et al., 1994) at postnatal day 0 as described previously (Chatterton et al., 2002). Whole-cell voltage-clamp recordings were performed on days 21-30 in vitro.
An additional subgenomic promoter and a farnesylated EGFP (Clontech) cDNA were inserted between SphI and StuI sites, and NR3B or HA-tagged NR3B cDNA was inserted into the Pml I site of the pSinRep5 vector (Invitrogen). Sindbis virus particles were prepared according to the manufacturer's protocol. Primary neurons were infected on days 10-14 in vitro. After 24 hr, cells were harvested and subjected to the immunoprecipitation assay with anti-pan NR1 antibody (Chemicon), anti-NR2A antibody (Upstate Biotechnology), and anti-GluR1 antibody (EMD Biosciences, San Diego, CA).
Electrophysiological studies. Hippocampal neurons were transfected with pCAGGS-NR3B-HA or an empty vector together with pCAGGS-EGFP (DNA ratio was 6:1) by using Effectene (Qiagen, Valencia, CA) at day 3 in vitro; the transfected cells were cultured for another 7-10 d. Neurons were voltage-clamped at -60 mV by using Axopatch 200B (Axon Instruments, Foster City, CA). Thin-wall borosilicate glass pipettes (TW150F; World Precision Instruments, Sarasota, FL) had resistance of 4-7 MΩ when filled with an intracellular solution that contained (in mm): 130 CsMeSO4, 10 CsCl, 2 MgCl2, 0.5 CaCl2, 10 HEPES, 2 Na2-ATP, and 5.5 Cs3-EGTA, pH 7.3. The extracellular solution contained (in mm): 150 NaCl, 5 KCl, 2 CaCl2, 20 HEPES, and 20 d-glucose, pH 7.3. For Ca2+ permeability experiments, the Na+ concentration in the bath solution was reduced to 80 mm, and glucose was used to adjust the osmolarity. Cells were exposed to the test solutions containing either 1 or 20 mm Ca2+ at a series of membrane potentials. Ca2+ permeability was calculated as described previously (Yuzaki et al., 1996a). All drugs were dissolved in the recording solution and applied by the Y-tube method (Yuzaki et al., 1996b) for neurons or by the θ-tube method (Kohda et al., 2000) with a Piezo translator (LSS-3100; Burleigh Instruments, Fishers, NY) for HEK293 cells. In electrophysiological studies on hippocampal neurons, tetrodotoxin (1 μm) and picrotoxin (100 μm) were added to the solution to block action potentials and inhibitory glycine and GABA channels. Glycine-evoked spiking activities in cultured cortical neurons were recorded in the presence of strychnine (10 μm). Responses were filtered at 1 kHz with an eight-pole Bessel filter, digitized at 3 kHz. Data acquisition and analysis were done by using pCLAMP 8.0 software (Axon Instruments). Membrane potential was corrected for the liquid junction potential. Statistical analysis was performed by using ANOVA.
Results
Specific expression of NR3B protein in motor neurons in the brainstem To investigate expression of the NR3B protein in the brain, we generated a specific anti-NR3B antibody. Although 62% of the amino acid sequence of NR3B is similar to that of NR3A (Matsuda et al., 2002), the C-terminal region of NR3B, especially its middle portion, is considerably different from that of NR3A (supplementary figure, available at www.jneurosci.org). Therefore, we raised an antibody against the synthetic peptide TGPPEGQQERAEQEC corresponding to the unique region of NR3B. Immunoblot analysis of HEK293 cells expressing NR3A or NR3B showed that this anti-NR3B antibody recognized a protein of ∼100 kDa, which is in agreement with the predicted molecular weight of NR3B (Fig. 1A). In contrast, the antibody did not react with NR3A protein (Fig. 1A), NR1, or NR2A (data not shown). In addition, anti-NR3B antibody interaction with NR3B protein was completely blocked by preabsorption of the antibody with the antigenic peptide (Fig. 1A). Immunohistochemical analysis also showed that the anti-NR3B antibody specifically recognized NR3B but not NR3A in HEK293 cells expressing these proteins (Fig. 1B). These results confirmed the specificity of the anti-NR3B antibody.
Figure 1.
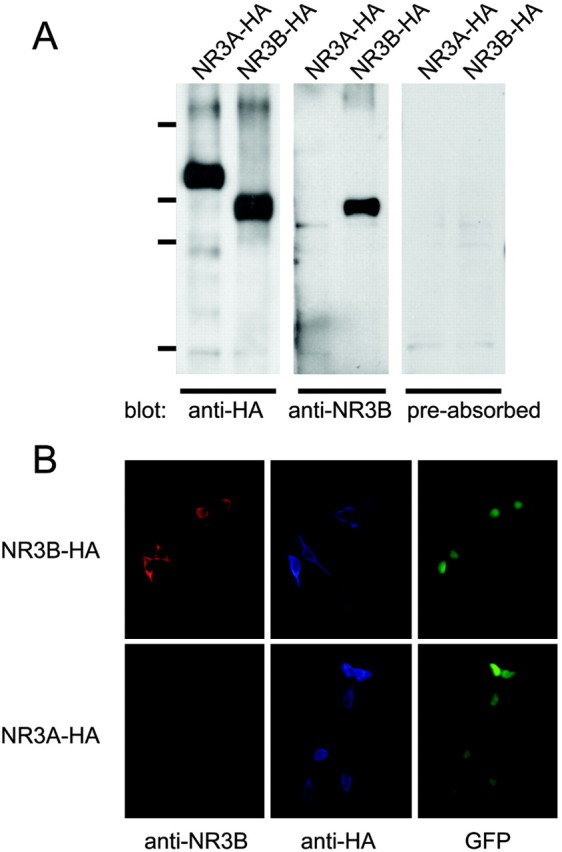
Specificity of anti-NR3B antibody. A, Immunoblot analysis. Lysates of HEK293 cells expressing HA-tagged NR3A or HA-tagged NR3B were subjected to immunoblot analysis using anti-HA antibody (left panel), anti-NR3B antibody (middle panel), or anti-NR3B antibody preabsorbed with the peptide used to immunize the rabbit (right panel). Molecular weight markers correspond to 203, 120, 90, and 51 kDa. B, Immunocytochemical analysis. HEK293 cells expressing HA-tagged NR3A or HA-tagged NR3B were incubated with anti-NR3B and anti-HA antibodies and detected by Alexa 546-conjugated anti-rabbit Ig and Alexa 350-conjugated anti-mouse Ig antibodies. Transfected cells exhibited EGFP fluorescence.
Using this antibody, we conducted immunohistochemical analysis to determine whether NR3B is expressed in adult mouse brain. First, by immunoblot analysis on mice brain homogenates, we confirmed that this antibody recognized a protein of ∼100 kDa (Fig. 2A). The interaction with NR3B protein was completely blocked by preabsorption of the antibody with the antigenic peptide (Fig. 2A), a result suggesting that this antibody specifically recognized endogenous NR3B protein. Strong immunoreactivity was observed in the facial nucleus (Fig. 2B,D,F) and the ambiguous nucleus (Fig. 2C,G) of the brainstem. Motor neurons in the abducent nucleus in the same section were not, or only very weakly, immunoreactive (Fig. 2E); this finding is consistent with the localization of NR3B mRNA (Nishi et al., 2001). In addition, the immunoreactivity of these cells was completely blocked by preabsorption of the antibody with the antigenic peptide (Fig. 2F). We also observed immunoreactivity in motor neurons in the trigeminal motor nucleus and motor neurons in the spinal anterior horn (data not shown). These results clearly indicate that NR3B protein is indeed translated from mRNA in adult mouse brain.
Figure 2.
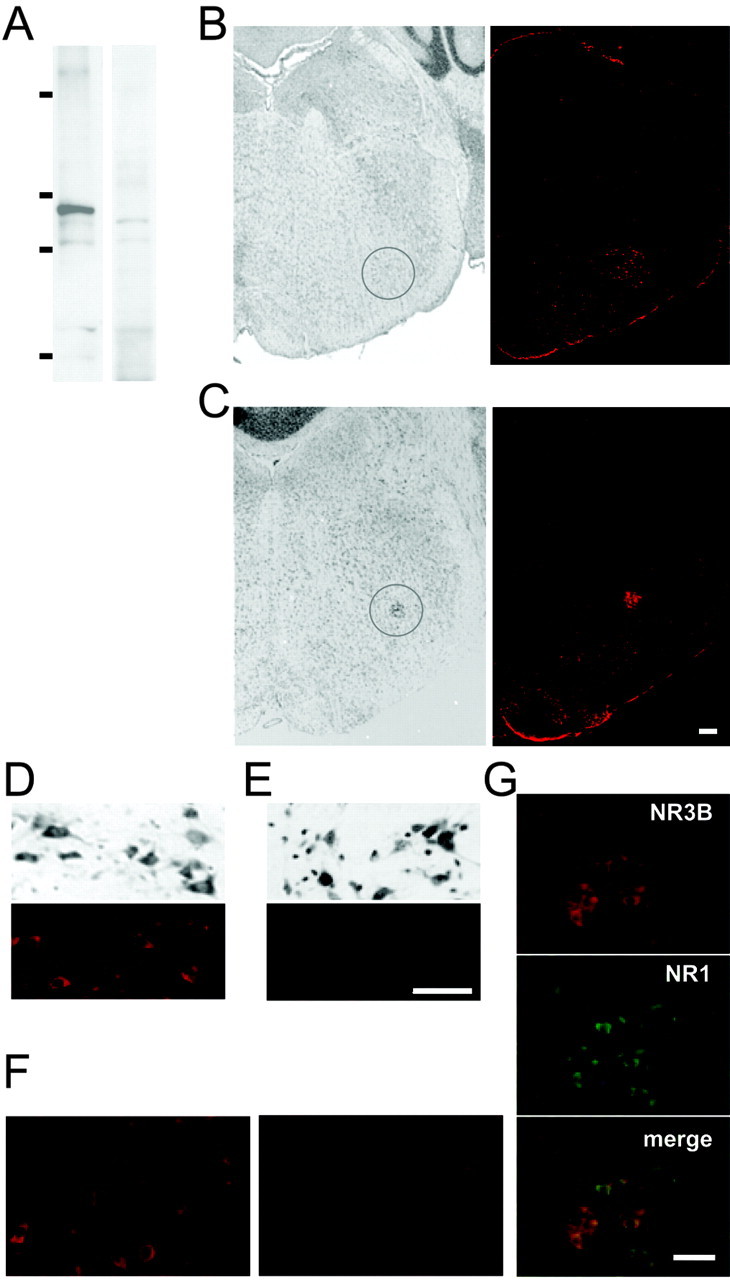
NR3B immunoreactivity in motor neurons in the brainstem. A, Immunoblot analysis. Lysates of mice whole brain were subjected to immunoblot analysis using anti-NR3B antibody (left panel) or anti-NR3B antibody preabsorbed with the peptide used to immunize the rabbit (right panel). Molecular weight markers correspond to 203, 120, 90, and 51 kDa. B, C, Coronal sections of adult mouse brainstem were stained with cresyl violet (left panels), and adjacent sections were stained with anti-NR3B antibody and visualized by Alexa 546-conjugated secondary antibody (right panels). The circled region corresponds to the facial nucleus (B) or the ambiguous nucleus (C). D, E, Enlarged view of the facial nucleus (D) and the abducent nucleus (E) in the same section. Top panels, Corresponding cresyl violet staining. F, Before (left panel) and after (right panel) absorption of anti-NR3B antibody with the peptide. These fields correspond to the facial nucleus. G, Double immunostaining of NR3B (top panel) and NR1 (middle panel) in the ambiguous nucleus. Fluorescent images were overlaid (bottom panel) to show the colocalization. Scale bars: C (also applies to B, C), E, (also applies to D-F), G, 100 μm.
Next, our immunohistochemical analysis (Fig. 2G) determined whether NR3B protein colocalized with the NR1 protein in the ambiguous nucleus. Interestingly, although almost all neurons were immunopositive for NR1, approximately half of them were negative for NR3B. Because anti-NR2 antibodies were also produced in rabbits, double immunostaining of NR3B and NR2s was not performed; however, the patterns of NR1 and NR2 expression in these neurons completely overlapped (Petralia et al., 1994a,b). These results indicate that the NR3B subunit is coexpressed together with the NR1 and NR2 subunits, at least in some neurons in the ambiguous nucleus, and this distinct pattern of NR3B expression may contribute to the functional heterogeneity of native NMDA receptors in these neurons.
Functionality of NR3B in neurons: dominant effects on Ca2+ permeability
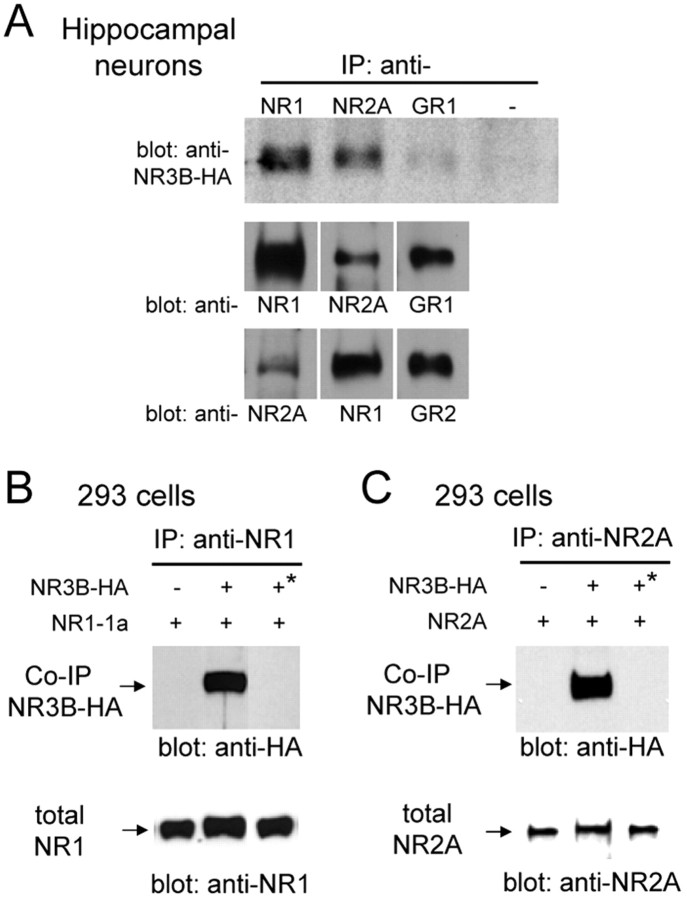
When coexpressed with NR1 and NR2A in heterologous HEK293 cells, the NR3B subunit reduces NMDA-evoked current amplitudes (Nishi et al., 2001) and lowers the Ca2+ permeability of the NMDA receptor complex (Matsuda et al., 2002). However, it is unclear whether NR3B can be incorporated into the native NMDA receptor complex and whether it exerts similar functions in neurons. To test the function of NR3B subunits in neurons, we used the Sindbis virus vector to express HA-tagged NR3B in cultured mouse hippocampal neurons. These neurons mainly express NR1, NR2A, and NR2B subunits (Monyer et al., 1994), but not NR3B (Nishi et al., 2001; Matsuda et al., 2002). Twenty-four hours after infection, proteins were immunoprecipitated from cell lysates by anti-NR1, anti-NR2A, or anti-GluR1 antibody and immunoblotted with anti-HA antibody. Exogenous NR3B subunits were clearly coimmunoprecipitated by either anti-NR1 or anti-NR2A antibody (Fig. 3A). In contrast, little or no NR3B subunits were coimmunoprecipitated by anti-GluR1 antibody, whereas it effectively coimmunoprecipitated AMPA receptor GluR2 (Fig. 3A). We also performed coimmunoprecipitation assays using HEK293 cells coexpressing HA-tagged NR3B and either NR1-1a or NR2A. NR3B subunits were coimmunoprecipitated by anti-NR1 or anti-NR2A antibody (Fig. 3B,C), a result that was consistent with the results seen in hippocampal neurons. When lysates from cells expressing NR3B, NR1-1a, or NR2A alone were mixed together after transfection, coimmunoprecipitation of NR3B was not observed (Fig. 3B,C); this finding confirms the specificity of antibodies and coimmunoprecipitation. These results indicate that exogenous NR3B subunits were incorporated into the endogenous NMDA receptor complex in hippocampal neurons.
Figure 3.
Assembly of complexes containing NR3B with NR1 and NR2 subunits. A, Interaction of NR3B subunit with endogenous NMDA receptors in hippocampal neurons. Lysates of hippocampal neurons infected with Sindbis virus expressing HA-tagged NR3B were immunoprecipitated (IP) by anti-NR1, anti-NR2A, anti-GluR1, or no antibody (negative control) and subjected to the immunoblotting using an anti-HA antibody (top panel). The quantity of each precipitated protein was confirmed by immunoblotting with corresponding antibodies used for immunoprecipitation (middle panel). To confirm coimmunoprecipitation efficiency, same immunoprecipitants were immunoblotted with indicated antibodies (bottom panel). B, Interaction of NR3B with NR1 in HEK293 cells. Lysates of HEK293 cells coexpressing HA-tagged NR3B and NR1-1a were immunoprecipitated by anti-NR1 antibody and subjected to immunoblot analysis using anti-HA antibody. When lysates of cells singly expressing NR1-1a or NR3B were mixed together, coimmunoprecipitation was not observed (third lane, *). The quantity of the precipitated NR1 subunit was confirmed by immunoblotting with an anti-NR1 antibody (bottom panel). C, Interaction of NR3B with NR2A in HEK293 cells. Lysates of HEK293 cells coexpressing HA-tagged NR3B and NR2A were immunoprecipitated by anti-NR2A antibody and subjected to immunoblot analysis using an anti-HA antibody. When lysates of cells singly expressing NR2A or NR3B were mixed together, coimmunoprecipitation was not observed (third lane, *). The quantity of the precipitated NR2A subunit was confirmed by immunoblotting with an anti-NR2A antibody (bottom panel).
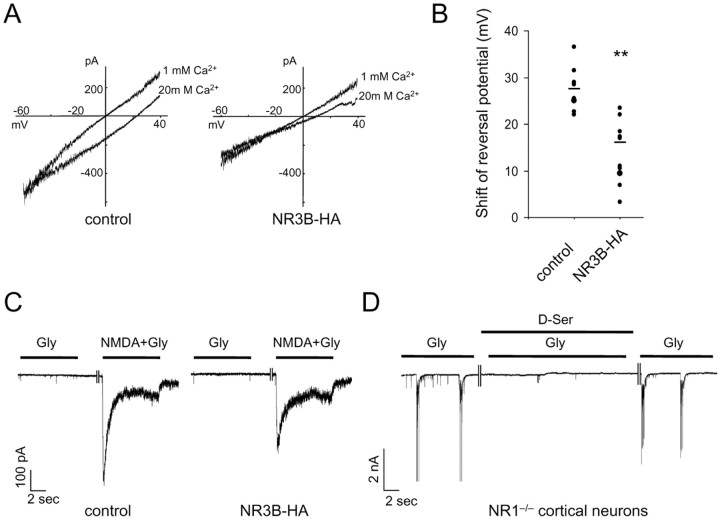
We next examined the function of this newly formed NMDA receptor complex by whole-cell recording of currents evoked by coapplication of NMDA (300 μm) and glycine (10 μm) to neurons. To evaluate the Ca2+ permeability of the newly formed NMDA receptor, we measured the reversal potential of NMDA-induced currents in 1 and 20 mm extracellular Ca2+ concentrations (Fig. 4A). The average shift in reversal potential in control neurons was 27.2 ± 1.7 mV (n = 8; Fig. 4B). According to the Goldman-Hodgkin-Katz equation modified to include divalent cations, assuming no anion permeability and equal permeability for monovalent cations (Mayer and Westbrook, 1987), this shift in reversal potential corresponds to PCa/Pmono = 7.7. When NR3B was expressed, the shift in reversal potential was 16.1 ± 2.0 mV (n = 13), corresponding to PCa/Pmono = 2.2. Together with the results of the coimmunoprecipitation experiments (Fig. 3A), these results suggest that NR3B is incorporated into the endogenous NMDA receptor, is transported to the cell surface, and modifies the function of the NMDA receptor in neurons.
Figure 4.
Functional effects of NR3B expression in neurons. A, Representative current-voltage curves of NMDA-induced currents in neurons expressing only EGFP (left) or NR3B (right). Current responses to the ramp voltage pulses (-60 to 40 mV) during NMDA (300 μm) plus glycine (10 μm) application in solutions containing 1 and 20 mm Ca2+ were superimposed. Changing from 1 to 20 mm Ca2+ resulted in the reversal potential shifting from 0 to 23.5 mV for the control cells and from 0.5 to 11 mV for NR3B-expressing neurons. B, Summary of the Ca2+-induced shift in reversal potential of NMDA-induced currents. The difference in reversal potential of NMDA-induced currents in 1 and 20 mm Ca2+ was plotted in hippocampal neurons expressing only EGFP (n = 8, control) or NR3B (n = 13). **Statistically significant differences (p < 0.005) between the shift of reversal potential of control and NR3B-expressing neurons. C, Responses to glycine in hippocampal neurons. Glycine (10 μm) or NMDA (300 μm) plus glycine (10 μm) was rapidly applied to neurons (voltage-clamped at -60 mV) expressing EGFP or NR3B. Tetrodotoxin (1 μm) and picrotoxin (100 μm) were included in the extracellular solution. Each trace represents recordings from 7 to 11 neurons. D, Glycine-evoked burst of action currents in cultured cortical neurons of NR1-/- mice. Glycine (10 μm) evoked spiking activity in the presence of strychnine (10 μm); however, this activity was completely blocked by d-serine (100 μm). The trace represents recordings from seven neurons.
Although the current amplitude in neurons expressing NR3B (-758 ± 65 pA; n = 25) was smaller than that in neurons expressing a mock vector (-922 ± 88 pA at -60 mV; n = 20), the difference was not significant (p = 0.13). In previous coexpression studies using HEK293 cells (Nishi et al., 2001; Matsuda et al., 2002), NR3B was expressed together with NR1 and NR2. In contrast, in the present study, NR3B was introduced into neurons that were already expressing native NMDA receptors. Thus, depending on the turnover rate of the preexisting NMDA receptor complex, the actual effect of NR3B expression may have been masked. Therefore, the Ca2+ permeability of NMDA receptors containing NR3B may be even lower than the measured value in the current analysis.
We examined whether the NR3B subunit is responsible for the unique excitatory glycine response in mammalian cells. The glycine response was evoked by glycine alone and inhibited by d-serine but not by AP-5, picrotoxin, or strychnine in Xenopus oocytes coexpressing NR1 and NR3B (Chatterton et al., 2002). However, hippocampal neurons expressing NR3B did not show a response to glycine (10 μm) in the presence of tetrodotoxin (1 μm) and picrotoxin (100 μm), which is a blocker of the conventional inhibitory glycine receptor (Fig. 4C). To exclude the possibility that the glycine response was desensitized very fast, we used the Piezo device-based method (Kohda et al., 2000) of fast drug application to apply glycine (10 μm) to lifted HEK293 cells coexpressing NR1 and NR3B. Although the drug exchange rate around the cell was 3-4 msec, no current was evoked (data not shown). It was suggested that glycine-evoked spiking activities originating from out-of-voltage-clamp regions reflected the excitatory glycine receptor in cortical neurons (Chatterton et al., 2002). Thus, to test the possibility that the excitatory glycine response is located at the synaptic, not the extrasynaptic, site, we examined glycine-evoked spiking activities in cortical neurons. We observed similar glycine-evoked spiking activities that were inhibited by the NMDA receptor coagonist d-serine in the presence of strychnine, an antagonist of the inhibitory glycine receptor; however, we also observed this phenomenon in cortical neurons prepared from NR1-/- mice (Fig. 4D). Because the excitatory glycine response absolutely requires the NR1 subunit (Chatterton et al., 2002), this phenomenon may not necessarily reflect the excitatory glycine receptor involving NR3A or NR3B. Although we cannot rule out the possibility that NR3B can participate in the excitatory glycine response under certain conditions, we hypothesize that in neurons, NR3B mainly functions as a modulatory subunit of the NMDA receptor complex by changing the Ca2+ permeability and perhaps also the current amplitude.
Surface transport of NR3B requires coexpression of NR1
In heterologous cells, expression of NR3B alone, NR3B plus NR1, or NR3B plus NR2A does not result in the formation of functional NMDA receptors (Nishi et al., 2001; Matsuda et al., 2002). However, it is unclear whether the failure to detect any channel activity in these cells reflected the nonfunctional channels formed by these subunits or the controlled trafficking of a specific combination of subunits. Indeed, the C-terminal region of NR3B contains many ER retention signal-like motifs (supplementary figure). Therefore, we examined how the cell surface export of NMDA receptors containing NR3B is controlled in HEK293 cells.
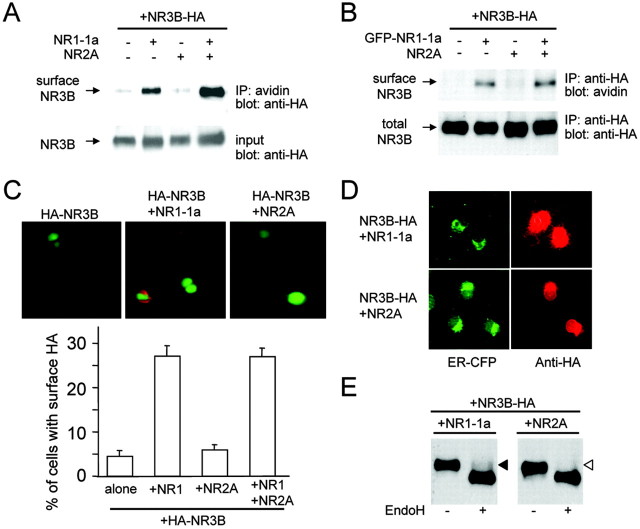
HA-tagged NR3B was expressed in HEK293 cells, and surface proteins were biotinylated and precipitated by streptavidin-conjugated agarose. Immunoblot analysis with anti-HA antibody indicated that biotinylated HA-NR3B was only observed when coexpressed with NR1-1a or NR1-1a plus NR2A (Fig. 5A). We obtained similar results when cell lysates were subjected first to immunoprecipitation with anti-HA antibody and detection by HRP-conjugated streptavidin (Fig. 5B). To further confirm the presence of NR3B on the plasma membrane, we expressed NR3B with an N-terminal HA tag in HEK293 cells. Under the nonpermeabilizing condition, the anti-HA antibody specifically detected HA-tagged NR3B on the surface of HEK293 cells when HA-NR3B was coexpressed with NR1-1a or NR1-1a plus NR2A (Fig. 5C). No staining was detected with the anti-HA antibody when NR3B with a C-terminal HA tag was expressed in HEK293 cells, a result confirming the specific binding of the antibody to surface proteins under this condition (data not shown). These results indicate that, like NR3A (Perez-Otano et al., 2001), NR3B requires the NR1 subunit for transport to the cell surface.
Figure 5.
Surface expression of NR3B requires NR1 coexpression. A, Results of a cell surface biotinylation assay using the avidin precipitation method. HA-tagged NR3B together with certain combinations of NR1-1a and NR2A was expressed in HEK293 cells. The cell surface biotinylation assay was performed as described in Materials and Methods. Cell lysates were precipitated by avidin and subjected to the immunoblot analysis using an anti-HA antibody (top panel). Expression of NR3B was confirmed by immunoblotting of the cell lysate (bottom panel). The results represent those of three independent experiments. B, Cell surface biotinylation assay using immunoprecipitated (IP) NR3B. Cell surface biotinylation of HEK293 cells was performed as described above. NR3B protein was immunoprecipitated from cell lysates by an anti-HA antibody and subjected to immunoblot analysis using HRP-conjugated avidin (top panel). The quantity of precipitated NR3B was confirmed by immunoblotting of the cell lysate (bottom panel). The results represent those of four independent experiments. C, Immunocytochemical analysis of the cell surface NR3B. An HA tag was attached immediately downstream of the signal sequence of NR3B, and the fusion protein was coexpressed with NR1-1a or NR2A in HEK293 cells. Expression of NR3B on the cell surface was examined by using an anti-HA antibody (red) under nonpermeabilizing conditions. The proportion of HA+ cells among the transfected cells, which were identified by the fluorescence of YFP in the nucleus, is summarized. Results are from cells expressing NR3B (n = 439), NR3B plus NR1-1a (n = 389), NR3B plus NR2A (n = 452), and NR3B plus NR1-1a plus NR2A (n = 390). D, Immunocytochemical analysis of NR3B in the ER. HA-tagged NR3B together with CFP-ER was coexpressed with NR1-1a (top row) or NR2A (bottom row) in COS-7 cells. Red signals indicate the presence of an anti-HA (NR3B) antibody. Green signals indicate the presence of ER-CFP. E, Sensitivity to Endo H digestion. HA-tagged NR3B was coexpressed with NR1-1a or NR2A in HEK293 cells. NR3B Protein was immunoprecipitated from cell lysates by an anti-HA antibody, treated with Endo H, and subjected to immunoblot analysis using anti-HA antibody. The filled triangle indicates the Endo H-resistant component of NR3B protein. Results shown in D and E represent those of three or four independent experiments.
To further examine the intracellular trafficking of the NMDA receptor, we coexpressed CFP targeted to the ER (ER-CFP) and HA-tagged NR3B in COS-7 cells with NR1-1a or NR2A. HA-NR3B immunoreactivity was observed beyond the ER-CFP signals when HA-NR3B was coexpressed with NR1-1a. In contrast, the pattern of HA-NR3B immunoreactivity completely overlapped that of ER-CFP signals when HA-NR3B was expressed with NR2A (Fig. 5D), a result suggesting retention of HA-NR3B in the ER. Retention of HA-NR3B in the ER was further confirmed by its sensitivity to Endo H, which removes unprocessed high-mannose oligosaccharides from proteins that reside in the ER and cis-Golgi. HA-NR3B, expressed alone (data not shown) or coexpressed with NR2A, was completely digested by Endo H, whereas HA-NR3B coexpressed with NR1-1a was at least partially resistant to Endo H (Fig. 5E). These results are consistent with those of the cell surface biotinylation (Fig. 5A,B) and surface staining (Fig. 5C) analyses and indicate that association with the NR1-1a subunit is crucial for the NR3B subunit to exit from the ER and to be transported to the cell surface.
The C-terminal region of NR3B controls the cell surface transport of NMDA receptors
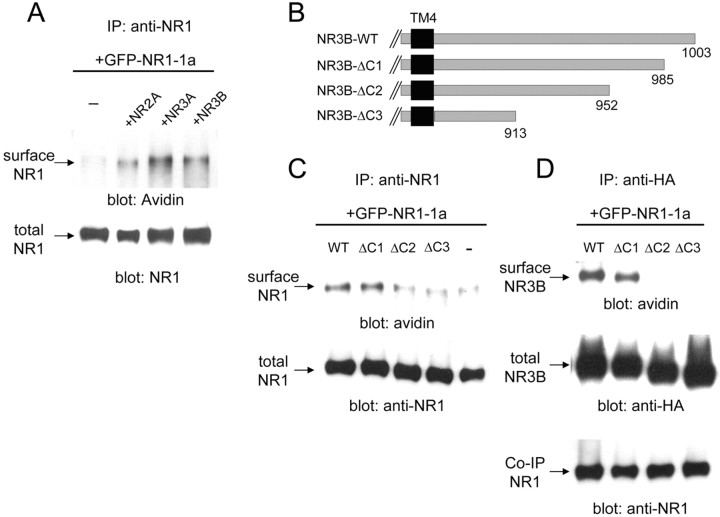
The C-terminal region of the NR1-1 subunit contains the ER retention signal, which must be masked by NR2 or other proteins before the NMDA receptor complex can be transported to the cell surface (Okabe et al., 1999; Standley et al., 2000; Scott et al., 2001). A cell surface biotinylation assay indicated that coexpression of NR3A or NR3B could supplant NR2A in allowing cell surface transport of NR1-1a (Fig. 6A).
Figure 6.
Interaction of the C-terminal region of NR3B with NR1-1a is responsible for cell surface trafficking of the NMDA receptor complex. A, Cell surface biotinylation assay of cells expressing various NMDA receptors. GFP-tagged NR1-1a was coexpressed with NR2A, NR3A, or NR3B in HEK293 cells. After the cell surface biotinylation, NR1 protein was precipitated from cell lysates by an anti-NR1 antibody and subjected to immunoblot analysis using HRP-conjugated avidin (top panel). The quantity of precipitated NR1 was confirmed by immunoblotting with an anti-NR1 antibody (bottom panel). B, Deletion mutants of NR3B. Residues distal to 986 (NR3B-ΔC1), 953 (NR3B-ΔC2), or 914 (NR3B-ΔC3) were deleted (for sequences of these regions in NR3B and NR3A, see supplementary figure). C, Effects of NR3B deletion mutants on the cell surface expression of NR1. NR1-1 and the indicated HA-tagged NR3B deletion mutants were coexpressed in HEK293 cells, and cell surface biotinylation was performed. Proteins were immunoprecipitated (IP) from cell lysates by an anti-NR1 antibody and subjected to immunoblotting using HRP-conjugated avidin. The quantity of precipitated NR1-1a was confirmed by immunoblotting with an anti-NR1 antibody. D, Cell surface expression of NR3B mutants coexpressed with NR1. NR1-1 and the indicated HA-tagged NR3B deletion mutants were coexpressed in HEK293 cells, and cell surface biotinylation was performed. Cell lysates were immunoprecipitated with an anti-HA antibody and subjected to immunoblotting analysis using HRP-conjugated avidin (top panel), anti-HA antibody (middle panel), and anti-NR1 antibody (bottom panel). Results shown in A, C, and D represent those of three or four experiments. WT, Wild type.
To narrow down the region of the NR3B subunit involved in this trafficking effect, we introduced several deletions in the C-terminal region of NR3B (Fig. 6B). When amino acids distal to position 914 (NR3B-ΔC3) or position 953 (NR3B-ΔC2) were removed, mutant NR3Bs no longer promoted the cell surface transport of coexpressed NR1-1a (Fig. 6B,C). Similarly, these mutant NR3Bs failed to go to the cell surface even when they were coexpressed with NR1-1a (Fig. 6D). In contrast, mutant NR3Bs whose amino acids distal to position 986 were deleted (NR3B-ΔC1) reached the cell surface together with coexpressed NR1-1a (Fig. 6C,D). These results indicate that amino acids 952-985 of NR3B play a crucial role in controlling the trafficking of the NMDA receptor complex. Results of coimmunoprecipitation assays indicated that these NR3B deletion mutants interacted with the NR1 subunit in a manner similar to that of wild-type NR3B (Fig. 6D). Therefore, although the C-terminal region of NR3B is not responsible for coassembly with NR1, the region between amino acids 952 and 985 may mask the ER retention signal of the NR1 subunit, which then brings the NR3B subunit to the cell surface as a complex.
Discussion
Expression of NR3B protein in motor neurons
In the present study, immunohistochemical analysis revealed that the newly identified member of the NMDA receptor, NR3B, is expressed as a protein in somatic motor neurons in the brainstem of adult mice. This result is consistent with the pattern of NR3B mRNA expression (Nishi et al., 2001) and indicates that NR3B mRNA is indeed translated in these neurons.
As with NR3A (Wong et al., 2002), NR3B immunoreactivity was mainly observed in neuronal cell bodies. This somatic staining pattern may reflect the intracellular pool of NR3B (Fig. 5) or extrasynaptic NMDA receptors. Similarly, NR1 was often observed in cell bodies (Hall and Soderling, 1997) (Fig. 2G). Extrasynaptic NMDA receptors are activated under certain conditions, such as after repeated synaptic stimulation (Clark and Cull-Candy, 2002), and may play certain roles (see below). It is also possible that trafficking of the intracellular pool of NR3A (Perez-Otano et al., 2001) or NR3B (Fig. 5) to the cell surface may be controlled by certain mechanisms, similar to those reported for NR1 subunits (Lan et al., 2001a,b). Alternatively, the weak immunoreactivity in dendrites may simply be caused by masking of the NR3B epitope because the C termini of membrane receptors are often masked by intracellular proteins in dendrites (Yamada et al., 2001). In this case, NMDA receptors containing NR3B may also act as synaptic receptors. The precise intracellular localization of endogenous NR3B should be investigated further by using antibodies that recognize different epitopes of NR3B.
Dominant modulatory function of NR3B in neurons
Coimmunoprecipitation and electrophysiological analyses demonstrated that NR3B can coassemble with endogenous NR1 and NR2A in hippocampal neurons and can reduce the Ca2+ permeability of the NMDA receptor. Ca2+ influx through NMDAreceptors is thought to initiate processes leading to synaptic plasticity and, if the receptors are overloaded, to certain forms of neuronal death. Properties of NMDA receptors observed in this study mainly reflect those of extrasynaptic receptors; analysis of synaptic NMDA receptors containing NR3B is not straightforward because the current amplitude and Ca2+ permeability are influenced by the electrotonic distance of the synapse and the release probability of the presynaptic sites. However, synaptically and extrasynaptically activated NMDA receptors are equally capable of excitotoxicity (Sattler et al., 2000) and induce distinct transcriptional programs (Li et al., 2002). Although NR3A mRNA (Ciabarra et al., 1995; Sucher et al., 1995) and protein expression (Wong et al., 2002) decreases dramatically after the second postnatal week, NR3B immunoreactivity (Fig. 2) and mRNA expression (Matsuda et al., 2002) persist in adult mice. Therefore, expression ofNR3B, which alters the Ca2+ permeability of the NMDA receptor complex, may play an important role in modulating synaptic plasticity and neuronal death in somatic motor neurons at later developmental stages and in adulthood.
Interestingly, NR3B immunoreactivity (Fig. 2E) and mRNA expression (Nishi et al., 2001) was very low in motor neurons controlling extraocular muscles, which are usually spared in amyotrophic lateral sclerosis (ALS). This finding suggests that NR3B may not be a critical factor determining susceptibility to degeneration of motor neurons in ALS. It is possible, however, that expression levels of NR3B in the spinal cord may modify the course of neuronal cell death via glutamate excitotoxicity in certain neuronal disorders, such as ALS (Rothstein, 1996) and ischemic spinal injuries (Lee et al., 1999).
We could not find evidence supporting the involvement of NR3B in the unique excitatory glycine response (Chatterton et al., 2002) in HEK293 cells and neurons. One possible explanation is that this response may only be observed in Xenopus oocytes, not in mammalian neurons or other cells. For example, in these mammalian cells, NR3B may be associated with a protein that inhibits or masks the expression of the excitatory glycine receptor. Alternatively, it is also possible that the excitatory glycine receptor is only formed in synaptic sites, not in extrasynaptic sites, which we studied. However, although it was suggested that glycine-evoked spiking activity in cortical neurons reflect the synaptic excitatory glycine response (Chatterton et al., 2002), we observed a similar phenomenon in cortical neurons prepared from NR1-/- mice. Because the excitatory glycine response should not occur in the absence of the NR1 subunit, other mechanisms may cause the glycine-evoked spiking activity in cortical neurons. Therefore, although NR3B may be part of the excitatory glycine receptor under certain circumstances, we propose that NR3B mainly functions as a modulatory subunit of the NMDA receptor complex by changing the current amplitude and the Ca2+ permeability in neurons.
Role of the NR1 in the cell surface targeting of NMDA receptors
Many multimeric receptor channels, such as K+ channels, GABAA receptors, and acetylcholine receptors, are preassembled in the ER, where assembly is tightly coupled to the trafficking of individual components to prevent monomers and incompletely assembled complexes from reaching the cell surface (Griffon et al., 1999; Zerangue et al., 1999; Wang et al., 2002). In conventional NMDA receptors, cell surface transport of the NMDA receptor complex is precisely controlled by masking of the ER retention signal (RXR) in the C-terminal region of the NR1 subunit by NR2 or other proteins (Okabe et al., 1999; Standley et al., 2000; Scott et al., 2001). We confirmed that export of NR1-1a is facilitated when NR3B, as well as NR2 or NR3A, is coexpressed in HEK293 cells (Fig. 6A). Because this effect required the specific region between positions 952 and 985 of the C-terminal region of NR3B (Fig. 6C), our results suggest that this region masks the retention signal of the NR1 subunit. Interestingly, although the C-terminal sequence of NR3B is considerably different from that of NR3A, the region between positions 952 and 985 shares similar amino acid sequences with NR3A (supplementary figure), a finding suggesting a common mechanism for masking the retention signal.
There are multiple RXR motifs in the C-terminal region of the NR3B; NR1-1a and NR3A contain only one motif, and NR2A has three motifs. However, in addition to NR3B-ΔC3 and NR3B-ΔC2 (Fig. 6D), NR3B without any retention signals failed to reach the cell surface with or without NR1 (data not shown). In contrast, the NR1 subunit lacking the C terminus (Krupp et al., 1999) or the retention signal (Standley et al., 2000; Scott et al., 2001) can exit from the ER to reach the cell surface. Therefore, we propose that the NR1 subunit contains forward-trafficking signals; NR3B does not contain the forward-trafficking signals, but when assembled with NR1, it can mask the ER retention signals in the C-terminal region of NR1 and exit from the ER together with NR1. Alternatively, through association with the NR1 subunit, the C-terminal region of NR3B may undergo conformational changes and bind to proteins, such as the band 4.1 protein (Walensky et al., 1999; Coleman et al., 2003), which may provide a forward trafficking signal and override the retention signal in the C-terminal portion of NR1.
Results of coimmunoprecipitation assays indicated that the C-terminal region of the NR3B subunit is not responsible for the association with NR1 (Fig. 6D). In contrast, the N-terminal leucine-isoleucine-valine-binding protein (LIVBP)-like domain was previously shown to be important in the subtype-specific assembly of the GluR1 (Leuschner and Hoch, 1999; Ayalon and Stern-Bach, 2001). This LIVBP-like domain is also important in the association between NR1 and NR2A (Meddows et al., 2001). Similarly, small deletions in the LIVBP-like domain of the glutamate receptor δ2 significantly reduce its participation in intersubunit assembly (Matsuda and Yuzaki, 2002; Wang et al., 2003). Therefore, the N-terminal LIVBP-like domain controls the assembly of NMDA receptor subunits, whereas the interaction at the C terminus of the NR1 subunit may determine the membrane-trafficking process. It has become increasingly clear that glutamatergic signaling is modulated in vivo by membrane-trafficking processes that control the expression of receptors on the cell surface (Malinow and Malenka, 2002). Further studies are warranted to better characterize the mechanisms by which the C-terminal region of NR3B controls the trafficking of NMDA receptors.
Footnotes
This work was supported by the Uehara Memorial Foundation (K.M.), the Japan Society for the Promotion of Science (Y.K.), National Institutes of Health Grant NS36925, Cancer Center Support Grant CA21765, and the American Lebanese Syrian Associated Charities (M.Y.). We thank Dr. N. Yamada, T. Torashima, and Y. Wang for technical assistance and Dr. H. Hirai and Dr. S. Matsuda for useful discussions.
Correspondence should be addressed to Michisuke Yuzaki, Department of Physiology, Keio University School of Medicine, 35 Shinano-machi, Shinjuku-ku, Tokyo 160-8582, Japan. E-mail: myuzaki@sc.itc.keio.ac.jp.
Copyright © 2003 Society for Neuroscience 0270-6474/03/2310064-10$15.00/0
References
- Ayalon G, Stern-Bach Y ( 2001) Functional assembly of AMPA and kainate receptors is mediated by several discrete protein-protein interactions. Neuron 31: 103-113. [DOI] [PubMed] [Google Scholar]
- Chatterton JE, Awobuluyi M, Premkumar LS, Takahashi H, Talantova M, Shin Y, Cui J, Tu S, Sevarino KA, Nakanishi N, Tong G, Lipton SA, Zhang D ( 2002) Excitatory glycine receptors containing the NR3 family of NMDA receptor subunits. Nature 415: 793-798. [DOI] [PubMed] [Google Scholar]
- Ciabarra AM, Sullivan JM, Gahn LG, Pecht G, Heinemann S, Sevarino KA ( 1995) Cloning and characterization of chi-1: a developmentally regulated member of a novel class of the ionotropic glutamate receptor family. J Neurosci 15: 6498-6508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark BA, Cull-Candy SG ( 2002) Activity-dependent recruitment of extrasynaptic NMDA receptor activation at an AMPA receptor-only synapse. J Neurosci 22: 4428-4436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman SK, Cai C, Mottershead DG, Haapalahti JP, Keinanen K ( 2003) Surface expression of GluR-D AMPA receptor is dependent on an interaction between its C-terminal domain and a 4.1 protein. J Neurosci 23: 798-806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cull-Candy S, Brickley S, Farrant M ( 2001) NMDA receptor subunits: diversity, development and disease. Curr Opin Neurobiol 11: 327-335. [DOI] [PubMed] [Google Scholar]
- Das S, Sasaki YF, Rothe T, Premkumar LS, Takasu M, Crandall JE, Dikkes P, Conner DA, Rayudu PV, Cheung W, Chen HS, Lipton SA, Nakanishi N ( 1998) Increased NMDA current and spine density in mice lacking the NMDA receptor subunit NR3A. Nature 393: 377-381. [DOI] [PubMed] [Google Scholar]
- Dingledine R, Borges K, Bowie D, Traynelis SF ( 1999) The glutamate receptor ion channels. Pharmacol Rev 51: 7-61. [PubMed] [Google Scholar]
- Forrest D, Yuzaki M, Soares HD, Ng L, Luk DC, Sheng M, Stewart CL, Morgan JI, Connor JA, Curran T ( 1994) Targeted disruption of NMDA receptor 1 gene abolishes NMDA response and results in neonatal death. Neuron 13: 325-338. [DOI] [PubMed] [Google Scholar]
- Griffon N, Buttner C, Nicke A, Kuhse J, Schmalzing G, Betz H ( 1999) Molecular determinants of glycine receptor subunit assembly. EMBO J 18: 4711-4721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall RA, Soderling TR ( 1997) Differential surface expression and phosphorylation of the N-methyl-d-aspartate receptor subunits NR1 and NR2 in cultured hippocampal neurons. J Biol Chem 272: 4135-4140. [DOI] [PubMed] [Google Scholar]
- Hirai H, Kirsch J, Laube B, Betz H, Kuhse J ( 1996) The glycine binding site of the N-methyl-d-aspartate receptor subunit NR1: identification of novel determinants of co-agonist potentiation in the extracellular M3-M4 loop region. Proc Natl Acad Sci USA 93: 6031-6036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollmann M, Heinemann S ( 1994) Cloned glutamate receptors. Annu Rev Neurosci 17: 31-108. [DOI] [PubMed] [Google Scholar]
- Kohda K, Wang Y, Yuzaki M ( 2000) Mutation of a glutamate receptor motif reveals its role in gating and delta2 receptor channel properties. Nat Neurosci 3: 315-322. [DOI] [PubMed] [Google Scholar]
- Kohda K, Kamiya Y, Matsuda S, Kato K, Umemori H, Yuzaki M ( 2003) Heteromer formation of delta2 glutamate receptors with AMPA or kainate receptors. Brain Res Mol Brain Res 110: 27-37. [DOI] [PubMed] [Google Scholar]
- Kornau HC, Schenker LT, Kennedy MB, Seeburg PH ( 1995) Domain interaction between NMDA receptor subunits and the postsynaptic density protein PSD-95. Science 269: 1737-1740. [DOI] [PubMed] [Google Scholar]
- Krupp JJ, Vissel B, Thomas CG, Heinemann SF, Westbrook GL ( 1999) Interactions of calmodulin and alpha-actinin with the NR1 subunit modulate Ca2+-dependent inactivation of NMDA receptors. J Neurosci 19: 1165-1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan JY, Skeberdis VA, Jover T, Grooms SY, Lin Y, Araneda RC, Zheng X, Bennett MV, Zukin RS ( 2001a) Protein kinase C modulates NMDA receptor trafficking and gating. Nat Neurosci 4: 382-390. [DOI] [PubMed] [Google Scholar]
- Lan JY, Skeberdis VA, Jover T, Zheng X, Bennett MV, Zukin RS ( 2001b) Activation of metabotropic glutamate receptor 1 accelerates NMDA receptor trafficking. J Neurosci 21: 6058-6068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laube B, Hirai H, Sturgess M, Betz H, Kuhse J ( 1997) Molecular determinants of agonist discrimination by NMDA receptor subunits: analysis of the glutamate binding site on the NR2B subunit. Neuron 18: 493-503. [DOI] [PubMed] [Google Scholar]
- Lee JM, Zipfel GJ, Choi DW ( 1999) The changing landscape of ischaemic brain injury mechanisms. Nature 399: A7-A14. [DOI] [PubMed] [Google Scholar]
- Leuschner WD, Hoch W ( 1999) Subtype-specific assembly of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionic acid receptor subunits is mediated by their n-terminal domains. J Biol Chem 274: 16907-16916. [DOI] [PubMed] [Google Scholar]
- Li B, Chen N, Luo T, Otsu Y, Murphy TH, Raymond LA ( 2002) Differential regulation of synaptic and extra-synaptic NMDA receptors. Nat Neurosci 5: 833-834. [DOI] [PubMed] [Google Scholar]
- Malinow R, Malenka RC ( 2002) AMPA receptor trafficking and synaptic plasticity. Annu Rev Neurosci 25: 103-126. [DOI] [PubMed] [Google Scholar]
- Matsuda S, Yuzaki M ( 2002) Mutation in hotfoot-4J mice results in retention of delta2 glutamate receptors in ER. Eur J Neurosci 16: 1507-1516. [DOI] [PubMed] [Google Scholar]
- Matsuda K, Kamiya Y, Matsuda S, Yuzaki M ( 2002) Cloning and characterization of a novel NMDA receptor subunit NR3B: a dominant subunit that reduces calcium permeability. Brain Res Mol Brain Res 100: 43-52. [DOI] [PubMed] [Google Scholar]
- Mayer ML, Westbrook GL ( 1987) Permeation and block of N-methyl-d-aspartic acid receptor channels by divalent cations in mouse cultured central neurones. J Physiol (Lond) 394: 501-527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBain CJ, Mayer ML ( 1994) N-Methyl-d-aspartic acid receptor structure and function. Physiol Rev 74: 723-760. [DOI] [PubMed] [Google Scholar]
- McIlhinney RA, Molnar E, Atack JR, Whiting PJ ( 1996) Cell surface expression of the human N-methyl-d-aspartate receptor subunit 1a requires the co-expression of the NR2A subunit in transfected cells. Neuroscience 70: 989-997. [DOI] [PubMed] [Google Scholar]
- Meddows E, Le Bourdelles B, Grimwood S, Wafford K, Sandhu S, Whiting P, McIlhinney RA ( 2001) Identification of molecular determinants that are important in the assembly of N-methyl-d-aspartate receptors. J Biol Chem 276: 18795-18803. [DOI] [PubMed] [Google Scholar]
- Monaghan DT, Bridges RJ, Cotman CW ( 1989) The excitatory amino acid receptors: their classes, pharmacology, and distinct properties in the function of the central nervous system. Annu Rev Pharmacol Toxicol 29: 365-402. [DOI] [PubMed] [Google Scholar]
- Monyer H, Sprengel R, Schoepfer R, Herb A, Higuchi M, Lomeli H, Burnashev N, Sakmann B, Seeburg PH ( 1992) Heteromeric NMDA receptors: molecular and functional distinction of subtypes. Science 256: 1217-1221. [DOI] [PubMed] [Google Scholar]
- Monyer H, Burnashev N, Laurie DJ, Sakmann B, Seeburg PH ( 1994) Developmental and regional expression in the rat brain and functional properties of four NMDA receptors. Neuron 12: 529-540. [DOI] [PubMed] [Google Scholar]
- Niethammer M, Kim E, Sheng M ( 1996) Interaction between the C terminus of NMDA receptor subunits and multiple members of the PSD-95 family of membrane-associated guanylate kinases. J Neurosci 16: 2157-2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishi M, Hinds H, Lu HP, Kawata M, Hayashi Y ( 2001) Motoneuron-specific expression of NR3B, a novel NMDA-type glutamate receptor subunit that works in a dominant-negative manner. J Neurosci 21: RC185(1-6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okabe S, Miwa A, Okado H ( 1999) Alternative splicing of the C-terminal domain regulates cell surface expression of the NMDA receptor NR1 subunit. J Neurosci 19: 7781-7792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Otano I, Schulteis CT, Contractor A, Lipton SA, Trimmer JS, Sucher NJ, Heinemann SF ( 2001) Assembly with the NR1 subunit is required for surface expression of NR3A-containing NMDA receptors. J Neurosci 21: 1228-1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petralia RS, Yokotani N, Wenthold RJ ( 1994a) Light and electron microscope distribution of the NMDA receptor subunit NMDAR1 in the rat nervous system using a selective anti-peptide antibody. J Neurosci 14: 667-696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petralia RS, Wang YX, Wenthold RJ ( 1994b) The NMDA receptor subunits NR2A and NR2B show histological and ultrastructural localization patterns similar to those of NR1. J Neurosci 14: 6102-6120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothstein JD ( 1996) Excitotoxicity hypothesis. Neurology 47: S19-S26. [DOI] [PubMed] [Google Scholar]
- Sattler R, Xiong Z, Lu WY, MacDonald JF, Tymianski M ( 2000) Distinct roles of synaptic and extrasynaptic NMDA receptors in excitotoxicity. J Neurosci 20: 22-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott DB, Blanpied TA, Swanson GT, Zhang C, Ehlers MD ( 2001) An NMDA receptor ER retention signal regulated by phosphorylation and alternative splicing. J Neurosci 21: 3063-3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Standley S, Roche KW, McCallum J, Sans N, Wenthold RJ ( 2000) PDZ domain suppression of an ER retention signal in NMDA receptor NR1 splice variants. Neuron 28: 887-898. [DOI] [PubMed] [Google Scholar]
- Steigerwald F, Schulz TW, Schenker LT, Kennedy MB, Seeburg PH, Kohr G ( 2000) C-terminal truncation of NR2A subunits impairs synaptic but not extrasynaptic localization of NMDA receptors. J Neurosci 20: 4573-4581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sucher NJ, Akbarian S, Chi CL, Leclerc CL, Awobuluyi M, Deitcher DL, Wu MK, Yuan JP, Jones EG, Lipton SA ( 1995) Developmental and regional expression pattern of a novel NMDA receptor-like subunit (NMDAR-L) in the rodent brain. J Neurosci 15: 6509-6520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walensky LD, Blackshaw S, Liao D, Watkins CC, Weier HU, Parra M, Huganir RL, Conboy JG, Mohandas N, Snyder SH ( 1999) A novel neuron-enriched homolog of the erythrocyte membrane cytoskeletal protein 4.1. J Neurosci 19: 6457-6467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JM, Zhang L, Yao Y, Viroonchatapan N, Rothe E, Wang ZZ ( 2002) A transmembrane motif governs the surface trafficking of nicotinic acetylcholine receptors. Nat Neurosci 5: 963-970. [DOI] [PubMed] [Google Scholar]
- Wang Y, Matsuda S, Drews V, Torashima T, Meisler MH, Yuzaki M ( 2003) A hot spot for hotfoot mutations in the gene encoding the 2 glutamate receptor. Eur J Neurosci 17: 1581-1590. [DOI] [PubMed] [Google Scholar]
- Watanabe M, Inoue Y, Sakimura K, Mishina M ( 1993) Distinct spatio-temporal distributions of the NMDA receptor channel subunit mRNAs in the brain. Ann NY Acad Sci 707: 463-466. [DOI] [PubMed] [Google Scholar]
- Wong HK, Liu XB, Matos MF, Chan SF, Perez-Otano I, Boysen M, Cui J, Nakanishi N, Trimmer JS, Jones EG, Lipton SA, Sucher NJ ( 2002) Temporal and regional expression of NMDA receptor subunit NR3A in the mammalian brain. J Comp Neurol 450: 303-317. [DOI] [PubMed] [Google Scholar]
- Yamada K, Fukaya M, Shimizu H, Sakimura K, Watanabe M ( 2001) NMDA receptor subunits GluRepsilon1, GluRepsilon3 and GluRzeta1 are enriched at the mossy fibre-granule cell synapse in the adult mouse cerebellum. Eur J Neurosci 13: 2025-2036. [DOI] [PubMed] [Google Scholar]
- Yuzaki M, Forrest D, Curran T, Connor JA ( 1996a) Selective activation of calcium permeability by aspartate in Purkinje cells. Science 273: 1112-1114. [DOI] [PubMed] [Google Scholar]
- Yuzaki M, Forrest D, Verselis LM, Sun SC, Curran T, Connor JA ( 1996b) Functional NMDA receptors are transiently active and support the survival of Purkinje cells in culture. J Neurosci 16: 4651-4661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerangue N, Schwappach B, Jan YN, Jan LY ( 1999) A new ER trafficking signal regulates the subunit stoichiometry of plasma membrane K(ATP) channels. Neuron 22: 537-548. [DOI] [PubMed] [Google Scholar]