Abstract
In vertebrates, individual olfactory neurons are thought to express a single odorant receptor (Or) gene, but it is not clear that all odor-evoked activity in each neuron is exclusively dependent on an individual odorant receptor. In Drosophila, little is known about what receptors impart odor sensitivity to particular olfactory neurons. Here, we demonstrate the use of gene targeting to produce a null mutant of the putative odorant receptor Or43b and find that the mutant is defective for odor-evoked activity in ab8A neurons, a single functional class of olfactory neurons in Drosophila. ab8A neurons lacking Or43b are still present in the mutants and display spontaneous activity but are insensitive to odor stimulation. Therefore, Or43b is required for odor responsiveness in these olfactory neurons in vivo. Or83b, a receptor expressed in a large fraction of olfactory neurons including Or43b neurons, does not confer odor responsiveness in the absence of Or43b. Olfactory behavior elicited by odorants that activate the ab8A neurons is indistinguishable between Or43b mutants and controls, demonstrating a surprising degree of functional redundancy among the limited odor receptor repertoire in this species. These studies demonstrate that a reverse genetic approach can be used to correlate specific olfactory receptors with odor specificity of functional classes of olfactory neurons.
Keywords: behavior, Drosophila, odor, olfactory, receptor, tuning
Introduction
All animals have the ability to detect and interpret relevant biological information from their environment. Discrimination of sensory cues is achieved by integration of inputs from cells tuned to specific subsets of sensory space. For example, color vision is achieved by weighing the relative inputs of neurons expressing distinct opsins tuned to different wavelengths of light (Nathans, 1999). Similarly, odor discrimination is thought to result from integration of the activity of specific subsets of olfactory neurons activated by specific odors. Although the spectral tuning of visual pigments is well established, the tuning of olfactory neurons and odorant receptors has not been extensively analyzed.
Vertebrate animals have millions of olfactory neurons expressing hundreds of odorant receptor (Or) genes, but each receptor neuron is thought to express a single allele of one receptor gene (Ressler et al., 1993; Vasser et al., 1993; Chess et al., 1994; Malnic et al., 1999). Heterologous expression of odorant receptors suggests these receptors confer responsiveness to discrete sets of odorants (Krautwurst et al., 1998; Zhou et al., 1998; Bozza et al., 2002; Gaillard et al., 2002), and that odor specificity is at least in part a function of the odorant receptors expressed in these neurons. It is not clear whether odor sensitivity of olfactory neurons in vivo results exclusively from a single odorant receptor gene product.
The Drosophila olfactory system is anatomically similar to that of vertebrates but is numerically simpler. There are ∼60 odor receptors expressed in ∼1200 olfactory neurons located in the third antennal segment (Stocker, 1994; Clyne et al., 1999; Vosshall et al., 1999, 2000). Most olfactory receptor neurons in Drosophila express at least two putative odorant receptors. Or83b is expressed in approximately two-thirds of all olfactory receptor neurons (Vosshall et al., 1999; Kalidas and Smith, 2002; Ng et al., 2002). All other Or genes are expressed in a much more limited number of cells (Clyne et al., 1999; Gao et al., 1999; Vosshall et al., 1999; Elmore et al., 2001). Despite the relatively limited repertoire of odor receptor genes, flies are capable of odorant discrimination and can be trained to avoid specific odorants paired with aversive stimuli (Tully and Quinn, 1985). How does this small number of odorant receptor genes and expression of more than one receptor per cell relate to odor discrimination in the fly? What receptors are actually required for odorant sensitivity? What contribution does a single receptor gene make to olfactory behavior?
To address these issues, we used gene targeting to create mutants defective for expression of a Drosophila odor receptor gene, Or43b. This reverse genetic approach allowed us to determine the in vivo odor specificity of this odor receptor in its normal cellular environment. Our results indicate that a single receptor confers odor responsiveness to a single functional group of olfactory neurons, and that the Drosophila olfactory system is functionally redundant, such that loss of a receptor, Or43b, has little effect on the olfactory behavior elicited by the odorants it detects.
Materials and Methods
Drosophila stocks. w1118; 70FLP, 70I-SceI, Sco/S2,CyO;+/+ containing FLP recombinase and I-Sce1 restriction enzyme regulated by a heat shock promoter, and w1118; FLP/FLP;+/+ flies were kindly provided by K. Golic (University of Kansas, Lawerence, KS). Df3364 was obtained from the Bloomington stock center as B#3364 df(2R)CA53/CyO. Wild-type Oregon R flies with the X chromosome from w1118 (w1118;OrR) were generated for the homologous recombination screen to minimize DNA polymorphisms between endogenous and targeted Or43b genes. Transgenic animals were made in the w1118 background with w+ transformation vectors by standard transformation protocols (Spradling and Rubin, 1982). For electrophysiological and behavior assays, Or43b1 was back-crossed to isogenic w1118 flies for 10 generations, selecting single red-eyed males each generation. White-eyed siblings from the progeny of the last back-cross were used as controls in electrophysiological and behavioral experiments to ensure control and knock-out mutants had identical genetic backgrounds.
DNA constructs for homologous recombination. Two PCR fragments corresponding to nucleotides 54299-57700 and 57262-60074 from AC005463 were amplified from w1118;OrR. PCR mutagenesis was used to replace A→T at 58402 and C→A at 57163. The 18 base pair I-SceI restriction sequence (GGCCGCTAGGGATAACAGGGTAAGTAC) was inserted into the Cla-I site at position 57279, joining the two PCR fragments. The 6 kb-targeting construct was cloned as a Not-I fragment into the pTV vector that contained the white gene and FLP recombination site (kindly provided by K. Golic). Drosophila transformations of the plasmid P[>whs, Or43b Δ], were performed by standard methods; two lines were generated on chromosome 3.
Genetics for homologous recombination. Virgin females carrying P[>whs, Or43b Δ] on the third chromosome were crossed to w1118; 70FLP, 70I-SceI, Sco/S2,CyO;+/+ males. Zero- to three-day-old progeny were heat shocked at 37° for 1 hr. Virgin Sco females from this cross were crossed 20 in a bottle to 10 w1118; FLP/FLP;+/+ males. Adults were discarded after 1 week, and the progeny were screened for red eyes. Single red-eyed flies were selected and crossed to Df3364/CyO flies to establish lines. Red-eyed non-Cy progeny were screened for targeting.
Analysis of targeted events. PCR primers were designed to independently amplify each copy of Or43b from recombinant lines. Genomic DNA was obtained from flies homozygous for the homologous recombination chromosome or heterozygous over a deficiency lacking Or43b (Df3364). The upstream copy of each mutant allele was amplified with F1 primer (ACAAGTGTCTGTTCCCACCGAAAGAAG) and R1 primer (CCCATACGGCTGTAATATGCTG). The downstream copy was amplified with F2 primer (CATGGCAAACTGTCTCACGACGTTTTG) and R2 primer (AACTTTTGGTCCAGGTTCATACCGCTC). PCR fragments were cloned into pCR 2.1 (Invitrogen, San Diego, CA) and sequenced. The high frequency that we recovered the Or43b alleles with multiple mutations and Sce1 sites suggests there was gene conversion occurring during the targeting.
In situ hybridization. Polytene chromosomes were prepared from salivary glands of late third instar larvae and hybridized as described by Langer-Sofer et al. (1982). A 2.7 kb BcgI fragment containing the Or43b gene was labeled with biotin-16-deoxyuridine triphosphate (Roche Products, Hertforshire, UK) and detected using a streptavidin-coupled peroxidase (Jackson ImmunoResearch, West Grove, PA) and diaminobenzidine (Sigma, St. Louis, MO). In situ hybridization was performed on control and Or43b mutant larval salivary gland polytene chromosome squashes using a probe encoding a fragment of the white gene. We found that all five mutant lines had white DNA sequences present at chromosomal position 43b (data not shown). For in situ hybridization to olfactory neurons, antisense riboprobes were hybridized to frozen Drosophila head tissue sections and detected as described by Vosshall et al. (1999).
Immunohistochemistry. Antibody generation of Or43b was described by Elmore et al. (2001). Twelve micrometer sections were transferred to ProbeOn Plus slides (Fisher Scientific, Springfield, NJ) and air dried for 1 hr, fixed for 5′ 100% methanol and 5′ 100% acetone, and both solutions were prechilled in –20°C and then washed three times in 1× PBS. The sections were then blocked for 30′ in 3% normal goat serum, 0.1% Triton X-100, and 0.1% bovine serum albumin in 1× PBS and then incubated overnight at 4°C with a 1:100 dilution of affinity-purified Or43b antibody in blocking buffer. Sections were washed three times in 1× PBS and 0.03% Triton X-100, incubated for 2 hr with a Cy3-conjugated secondary goat-anti-rabbit antibody (Jackson ImmunoResearch), washed, mounted in 90% glycerol, 1× PBS, and 0.01% phenylenediamine, and examined with a laser-scanning microscope (MRC1024; Bio-Rad, Hercules, CA).
Behavior assays. The T-maze assay was performed as described previously (Tully and Quinn, 1985). The performance index was calculated as [(number of flies attracted by odor/total number flies) × 2 – 1] × 100 (Wang et al., 2001). Twenty flies (10 mutant, 10 controls) were tested together for each trial of 2 min. A minimum of 15 trials was made for each genotype at each concentration of odorant. All odorants were obtained from Aldrich (Milwaukee, WI), and the maximum purity available was used.
For population cage assays, ∼10,000 Or43b mutants (red eyes) and an equal number of control flies with white eyes with the same genetic background were added to a humidified 24 by 24 inch Plexiglas population cage and starved for 12 hr. One hundred to three hundred odorant traps (each consisting of a borosilicate test tube, with a defined dilution of odorant mixed in agarose in the bottom, and an ependorf tube with a one-sixteenth inch hole drilled through the bottom) were placed in the cage for 12–24 hr. Flies in each trap were sorted by eye color and counted. Consistent differences of twofold or greater were considered significant. lush mutants typically were five times more abundant than controls in traps containing 25% v/v ethanol. A complete list of screened odorants is presented as supplemental data (available at www.jneurosci.org).
Single-sensillum recordings. Extracellular electrophysiological recordings were generally performed according to de Bruyne et al. (2001). Briefly, a 4- to 6-d-old fly was restrained in a truncated pipette tip with the head protruding. The antenna was placed on an elevated coverslip platform and stabilized with a glass micropipette. Action potentials were recorded by inserting a tungsten wire electrode (World Precision Instruments, Sarasota, FL) in the shaft of a sensillum. Signals were amplified 1000× (Iso-Dam; World Precision Instruments) and fed into a computer via a 16 bit analog-to-digital converter to be analyzed off-line with AUTOSPIKE software (universal serial BUS–intelligent data aquisition controller system; Syntech, Hilversum, The Netherlands). Analysis of data was performed according to de Bruyne et al. (2001). The odor stimulation system was calibrated to give levels of responses similar to those reported by de Bruyne et al. (2001), facilitating direct comparisons of physiological data.
Results
Generation of Or43b null mutants by gene targeting
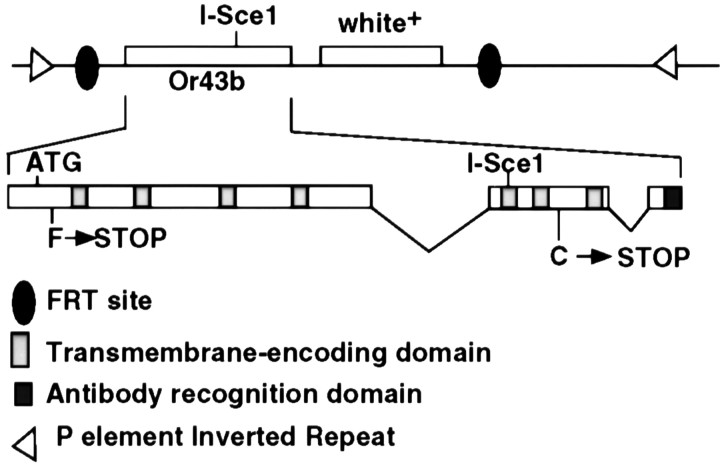
To generate mutants specifically defective for the Or43b odorant receptor, we targeted mutations to the Or43b gene in living flies using homologous recombination (Rong and Golic, 2000, 2001). Figure 1 shows the structure of the Or43b construct that we used to introduce stop codons into the endogenous receptor gene.
Figure 1.
Targeting construct to induce null mutations in Or43b. The Or43b gene was modified in vitro to introduce two stop mutations into the open reading frame. The first mutation (F→STOP) was introduced at codon 5, and the second (C→STOP) was introduced at codon 320, just before the DNA encoding the seventh transmembrane domain. A restriction site for the rare cutting restriction enzyme I-Sce1 was introduced into the Or43b gene between the two stop mutations. This construct was then cloned into the homologous recombination targeting P element vector pTV2 that contains the white eye color marker and FRT sites (Rong et al., 2002). Transgenic flies were created carrying the integrated construct on the third chromosome.
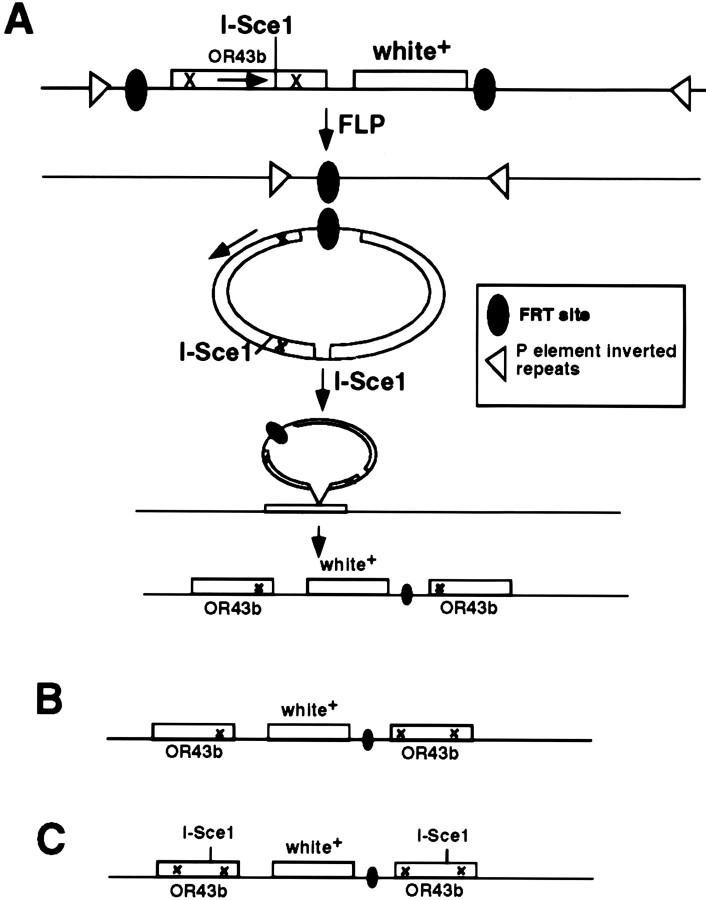
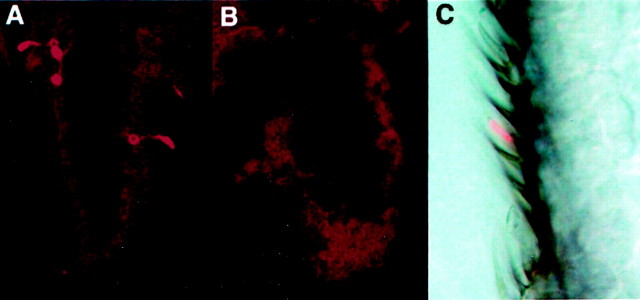
We generated 14,000 mosaic females and recovered 60 candidate Or43b targeted alleles on the basis of translocation of the white+ eye color marker from the third chromosome to the endogenous Or43b gene located on the second chromosome (see Materials and Methods). The homologous recombination approach is expected to produce a duplication of the Or43b gene with each copy carrying a single nonsense mutation (Rong and Golic, 2000). Figure 2A shows the events and expected gene structure of a targeted allele of Or43b. We screened our pool of candidates for Or43b expression in the antenna using anti-Or43b antiserum (Elmore and Smith, 2001). We identified five independent Or43b mutants defective for expression of the Or43b receptor on the basis of loss of antigen in the antenna (Fig. 3, compare A and B). Figure 2, B and C, depicts the structure of the five Or43b alleles that we recovered. Or43b mutant flies are healthy and fertile with no gross behavioral or morphological phenotypes.
Figure 2.
Molecular structure of expected and recovered Or43b mutants. A, Flies carrying the targeting construct were crossed to transgenic flies carrying the genes encoding FLP recombinase and I-Sce1 restriction enzyme regulated by heat-shock promoters (Rong et al., 2002). A illustrates the events leading to Or43b gene targeting after heat shock of the progeny of this cross. FLP expression results in excision of a closed circle of DNA containing all sequences between the FRT sites. This circle is linearized by the restriction enzyme I-Sce1, which is also expressed in the flies under heat-shock control. This triggers DNA repair mechanisms to integrate the free DNA into the endogenous Or43b locus. The expected result is a duplication of Or43b with each copy containing one of the two engineered termination codons and the white eye color marker gene located between the two Or43b genes. X denotes introduced stop codon within the coding sequence of Or43b. B, C, Depiction of the molecular events recovered. Of the five independent Or43b mutant alleles recovered, two had the structure depicted in B, and three had the structure depicted in C. None of the alleles expressed detectable Or43b. The structure of these mutants suggests that gene conversion and DNA repair may be important for the ultimate molecular structure of the targeted mutants. Or43b1 and Or43b2 correspond to the structure in B.
Figure 3.
Loss of Or43b antigen in Or43b mutants and localization to basiconic sensilla. A, Frozen tissue section through a wild-type antenna. Or43b receptor is detected with antiserum against Or43b. The majority of the Or43b receptor was localized to the dendrites, in which olfactory signal transduction is thought to occur. B, Frozen tissue section through Or43b mutant antenna. No Or43b antigen is detectable. Identical results were obtained for all five mutant alleles. C, High-power image of a frozen tissue section through a wild-type antenna reacted with Or43b antiserum. Overlaying the light and fluorescent images revealed that Or43b expression is restricted to basiconic sensilla.
Or43b is expressed in a subset of basiconic sensilla
On the basis of RNA in situ hybridization (Clyne et al., 1999; Gao and Chess, 1999; Vosshall et al., 2000) and, more recently, protein localization studies (Elmore and Smith, 2001), Or43b expression is restricted exclusively to ∼15 olfactory neurons in each antenna. However, the morphological type of sensillum that houses the Or43b-expressing neurons has not been determined. Sensilla are hair-like structures on the surface of the olfactory organs. Each sensillum houses the dendrites of one to four olfactory neurons. Several classes of sensilla have been described on the basis of morphology (Stocker, 1994; Shanbhag et al., 1999). We wished to identify the morphological class of sensillum in which Or43b is expressed, so we could focus our electrophysiological analysis on those sensilla in our mutants.
To identify the morphological class of sensillum expressing Or43b, we used antiserum specific for this receptor on frozen sections of Drosophila antennae. Figure 3C shows that Or43b localization is specifically restricted to a small subset of basiconic sensilla. Most of the Or43b-positive sensilla were located on the posterior surface of the antenna. Or43b-expressing neurons were not present in other classes of sensilla. Therefore, we conclude that Or43b is expressed exclusively in a small subset of olfactory neurons localized to basiconic sensilla that are located primarily on the posterior surface of the antenna.
Identification of ab8, a novel class of sensillum
The olfactory neurons in basiconic sensilla can be functionally subdivided into at least 16 classes on the basis of sensitivity to a defined odorant panel (de Bruyne et al., 2001). If Or43b functions to determine part or all of the odor sensitivity of the olfactory neurons in which it is expressed, we predicted that a subset of basiconic neurons would have defective odor responses in our mutants. We used single sensillum recording techniques to systematically characterize the olfactory responses of the neurons within basiconic sensilla in the region of the antenna where Or43b is expressed.
Systematic studies have defined the odor response profiles from basiconic sensilla on the maxillary palps and antenna (de Bruyne et al., 1999, 2001). The responses of the olfactory neurons within the large basiconic sensilla and most of the small basiconic sensilla in the Or43b mutant were indistinguishable from those observed in previous studies (de Bruyne et al., 2001). Our studies confirmed the presence of seven functional classes of basiconic sensilla, ab1–7, that contain neurons with characteristic odorant sensitivities. However, we did observe a subset of basiconic sensilla in the general region where Or43b is normally expressed in wild-type flies that do not match any previously defined functional class. We call this novel sensillum class ab8.
Wild-type ab8 sensilla contain two neurons, A and B, that are distinguishable by the shape and amplitude of their action potentials in single-unit electrophysiological recordings (Fig. 4). The A neuron produces larger amplitude action potentials and is sensitive to pentyl acetate and several other odorants (Figs. 4, 5). The B neuron gives the greatest mean response to ethyl butyrate and 2,3-dibutanedione. Surveys of the basiconic sensilla on the antenna in wild-type controls revealed that most ab8 sensilla are localized to the posterior surface of the antenna, in agreement with the expression of Or43b receptor protein (Elmore and Smith, 2001).
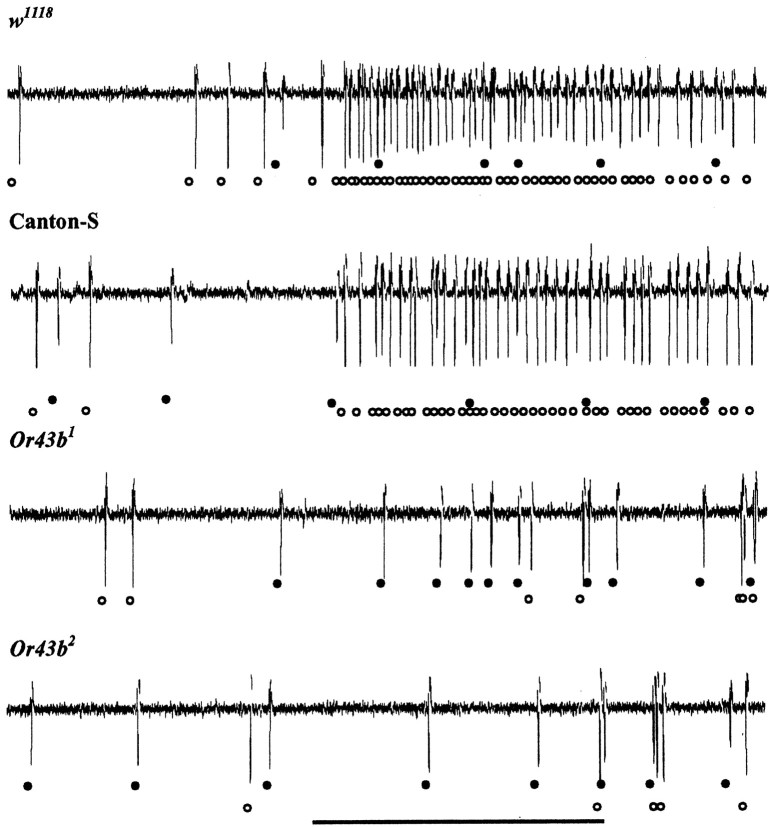
Figure 4.
Single-sensillum recordings from control and Or43b mutant ab8A neurons. Extracellular recordings from ab8 sensilla in control w1118 flies, Canton-S flies, and the two mutant strains Or43b1 and Or43b2. The traces show the activity of the ab8A (open circles) and ab8B (filled circles) neurons after stimulation with 1-hexanol at a dilution of 10–3. In w1118 and Canton-S flies, this concentration of 1-hexanol elicits a strong excitatory response of the A neuron but not the B neuron. Although ab8A neurons display spontaneous activity, they do not show a normal excitatory response to odors in Or43b mutants. Spontaneous activity in ab8A neurons in Or43b mutants often occurs in short bursts of two to three action potentials. The thick horizontal bar indicates the stimulation period (500 msec). Spike classification was performed as discussed previously (de Bruyne et al., 2001).
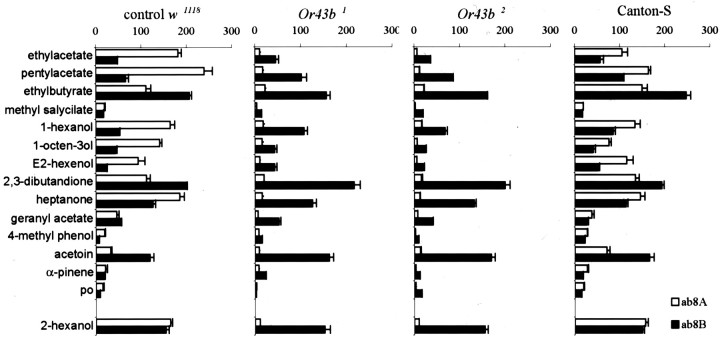
Figure 5.
Odor sensitivity of ab8 sensilla in Or43b mutants and controls. Response profiles of olfactory neurons housed in ab8 sensilla for control w1118, Canton-S flies, and two mutant strains, Or43b1 and Or43b2, to a diagnostic set of 14 odors diluted 10–2 with the solvent control paraffin oil (po). Each odorant was spotted on filter paper, and an air stream was diverted over the paper to apply the odor stimulus. The actual concentration reaching the antenna is not known but is considerably diluted using this method. Each bar represents the increase in spike frequency (spikes per second ± SEM) after odorant stimulation with a 10–2 dilution of the indicated odorant (12≤ n ≤15 recordings).
Or43b mutants lack odor-evoked electrical activity in ab8A neurons
We compared odor-evoked responses from sensilla in control and Or43b mutant antennae. No differences were detected in large basiconic sensilla or most types of small basiconic sensilla. However, we did note a striking defect in the ab8 basiconic sensilla in the Or43b mutants (Figs. 4, 5). Odor-evoked responses in the ab8A neurons are virtually absent in the Or43b mutants. However, there was little if any effect on odor-evoked activity in the B neurons in ab8 sensilla, confirming that we were recording from ab8 sensilla. Figure 4 shows examples of recordings from ab8 sensilla in response to 1-hexanol. Application of odorants failed to produce high frequency action potential trains in the Or43b mutants characteristic of the A neuron in genetically matched controls. 1-hexanol normally activates the A but not the B neuron in wild-type ab8 sensilla at the tested concentration. Figure 4 shows that in wild-type flies, 1-hexanol normally triggers a train of large amplitude action potentials in the ab8A neurons. The ab8A neurons are not activated by 1-hexanol in the Or43b mutants (Figs. 4, 5). Indeed, the ab8A neuron in Or43b mutants was not activated by any of the 55 odorants applied in these studies (Fig. 5) (data not shown). Recordings from a second independent allele of Or43b mutant revealed that ab8A neurons also lack odor-evoked potentials in this mutant allele (Fig. 4, Or43b2). This confirms that these defects arise from the lack of Or43b expression and not a random spontaneous mutation affecting ab8A function in Or43b1. These data establish that Or43b is required for odor sensitivity of ab8A neurons, consistent with a role as a functional odorant receptor.
Although the odor-evoked responses of the ab8A neurons are absent in Or43b mutants, spontaneous activity from these neurons is still present in the ab8 sensilla (Fig. 4); they have similar amplitude and shape as in control traces. Interestingly, the activity of the ab8A neurons in Or43b mutants is often observed in short bursts of two to three action potentials. A similar bursting phenotype is observed in mutant neurons that lack expression of Or22a (Dobritsa et al., 2003). The presence of action potentials characteristic of the ab8A neuron demonstrates that the A neuron is still present in Or43b mutants and is capable of firing action potentials. We conclude that the ab8A neuron normally expresses the Or43b odorant receptor, and this odorant receptor is required for odor-evoked activity in these neurons.
Odor specificity of ab8A neurons
Having established that Or43b is required for odorant responses of ab8A neurons, we explored the range of odors that activate ab8A neurons. We tested 55 odorants and found that ab8A neurons respond strongly to chemically diverse odorants, including ethyl acetate, pentyl acetate, heptanone, 1-hexanol, and 2-hexanol (Fig. 5). Weaker responses were observed for 1-octen-3-ol, 2,3-dibutanedione, ethyl butyrate, E2-hexenal, and geranyl acetate. The 45 other odorants tested produced little or no response above the spontaneous action potential firing rate. These results indicate that ab8A neurons are odor selective but can be stimulated by a wide range of chemically diverse odorants. None of these odorants evoked activity in the ab8A neurons in Or43b mutants, indicating that these neurons lack functional odorant receptors.
Or83b does not function as an independent receptor for any tested odorant
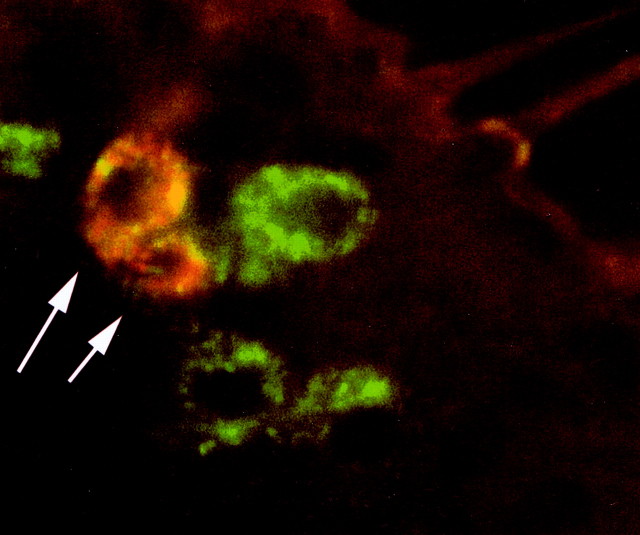
Or83b is a putative Drosophila odorant receptor expressed in a large fraction of olfactory neurons. The function of this receptor is unknown, but it could function as an independent odor receptor or as a coreceptor that functions with other Drosophila odor receptors. We used in situ hybridization in frozen tissue sections through the antenna to colocalize Or43b and Or83b expression. Figure 6 shows that olfactory neurons expressing Or43b also express Or83b. We did not detect any Or43b-positive neurons that were not Or83b positive. The simplest explanation of these results is that in ab8A, the Or83b receptor alone does not confer odor sensitivity to any of the 55 odorants tested.
Figure 6.
Or43b and Or83b are coexpressed. Antisense probes to Or43b (red) and Or83b (green) hybridized to olfactory neurons in frozen Drosophila tissue sections through the antenna. Or83b is expressed in a large fraction of olfactory neurons, including those expressing Or43b. Arrows indicate olfactory neurons that are positive for both Or43b and Or83b probes.
Olfactory behavioral responses of Or43b mutants
Or43b mutants are defective for odor-induced activation of ab8A neurons. What is the contribution of ab8A neurons to chemosensory behavior? We tested the olfactory behavioral responses of Or43b mutants and controls to determine whether loss of ab8A activity results in alterations in olfactory behavior. We performed T-maze assays (Tully and Quinn, 1985) on Or43b mutant and control flies using serial dilutions of odorants that activate the ab8A neuron in wild-type flies. For these odorants, we observed no significant differences in chemosensory behavior between controls and Or43b mutants (data not shown). We screened ∼200 additional compounds or complex odor mixtures for their ability to induce abnormal chemosensory behavior compared with control flies using a population cage assay (complete list of odorants, available at www.jneurosci.org). This assay successfully identified an increased affinity of LUSH mutants (Kim et al., 1998) for high concentrations of ethanol compared with controls (data not shown). We detected no significant differences in olfactory behavior between control and Or43b mutants. These results suggest that the loss of Or43b odorant receptor and corresponding loss of odor-evoked responses in the ab8A neurons have negligible effects on olfactory behavior.
Discussion
Or43b is required for odor-evoked activity in ab8A neurons
We demonstrated that mutant flies defective for expression of odorant receptor Or43b lack odor-evoked activity in ab8A olfactory neurons. Two independent alleles of Or43b mutants displayed the same phenotype. This defect is specific to ab8A olfactory neurons, because other classes of olfactory neurons located within basiconic sensilla are not affected in the mutants. Or43b expression is localized to the region of the antenna where ab8 basiconic sensilla are located (Elmore and Smith, 2001). These data provide strong support for the idea that Or43b is normally expressed in ab8A neurons, and this receptor is required for odor-evoked activity in these neurons. Immunolocalization of Or43b protein to the dendrites also supports the idea that Or43b functions directly as an odorant receptor (Elmore and Smith, 2001).
Odor specificity of ab8A neurons
In wild-type flies, ab8A neurons respond to a wide range of chemically diverse odorants, including ethyl acetate, pentyl acetate, ethyl butyrate, 1-hexanol, 2-hexanol, 1-octene-3-ol, 2,3-dibutanedione, heptanone, and E2-hexenal. This list includes long- and short-chain acetates, alcohols, ketones, an acid, and an aldehyde. Therefore, this olfactory neuron is broadly tuned to a chemically diverse set of odorants. However, a large number of other odorants that we applied does not activate ab8A neurons, demonstrating that although broadly tuned, these neurons are still chemically selective.
Most olfactory neurons in Drosophila, including the ab8A neurons, are tuned to a chemically diverse set of odorants (Clyne et al., 1997; de Bruyne et al., 1999, 2001). Our studies and those of Dobritsa et al. (2003) suggest this tuning likely reflects the chemical specificity of the odorant receptors expressed in these neurons, and that generally these receptors are broadly tuned. Some insect olfactory neurons are tuned to specific odorant molecules. Pheromone-responsive neurons in male moths are exquisitely sensitive to female pheromones and are not activated by structurally similar molecules (for review, see Kaissling, 1997). However, the tuning of pheromone-responsive receptors is under great selective pressure for chemical specificity and may not reflect tuning of other insect odorant receptors. Or43b appears broadly tuned, suggesting overlap in odor specificity among different receptors. This is in contrast to our current view of tuning of vertebrate olfactory neurons that are thought to be narrowly tuned. For example, individual olfactory neurons in the mouse are activated by structurally related odor molecules with similar chain lengths and functional groups (Malnic et al., 1999; Araneda et al., 2000).
Why would flies have broadly tuned olfactory neurons? The Drosophila odorant receptor gene repertoire is an order of magnitude smaller than that of mice or worms. There are ∼60 odorant receptor genes in the Drosophila melanogaster genome-encoding receptors that must detect and discriminate a large number of odorants relevant to the survival of this species. One possibility is that broadly tuned receptors compensate for a limited receptor set. Using combinatorial coding, discrimination would still be possible among a vast array of odorants if the subset of receptors activated by each odorant still produced a unique spatial and temporal pattern of activity in the glomeruli of the antennal lobes.
Why do flies have such a limited receptor set in the first place? The answer to this question is unknown, but one possibility is that this may reflect the challenges of wiring the peripheral olfactory system without using receptor gene products to guide axonal pathfinding (Elmore and Smith, 2001, Dobritsa et al., 2003).
Or83b is unlikely to function as an independent odorant receptor
ab8A expresses at least two members of the Or family, Or43b and Or83b. When Or43b is deleted, the affected neuron fails to show a strong response to any tested odorant. Flies lacking Or43b lack odor-evoked responses despite expression of Or83b in the same neurons. This suggests that Or83b alone is not an independent odorant receptor. It is possible that both Or43b and Or83b can function as odor receptors but are obligate chaperones for each other. For example, GABAB receptors are dimers that include one subunit that passes ions and another subunit required for plasma membrane localization (White et al., 1998). However, misexpression of Or43b in neurons lacking Or22a confers an odor response profile similar to ab8A neurons (Hallem and Carlson, unpublished results). This implicates Or43b as the primary, if not the exclusive, functional odor receptor expressed in ab8A neurons. However, Or83b could still function as a coreceptor. Alternatively, Or83b may not function directly in olfaction at all and could perform some unknown homeostatic role. Finally, we cannot rule out the possibility that Or83b is an odorant receptor but is tuned to a chemical structure not tested in our studies. Genetic studies of Or83b will be required to clearly define the role of this unique receptor. However, at present, Or83b mutants are not available.
Or43b and olfactory behavior
The loss of Or43b had a dramatic effect on ab8A neuronal activity but, surprisingly, it had no detectable effect on olfactory behavior. Indeed, for all odorants tested, no effect on chemosensory behavior was observed with the T-maze or population cage assays. We could detect no differences in quantitative behavior or sensitivity to any of the odorants that we have shown to activate ab8A neurons. This could reflect limitations of these particular behavioral assays to detect the abnormal behavior induced by the loss of this receptor. Alternatively, perhaps some odorant not tested here can induce differential olfactory behavior in control and Or43b mutants. However, the lack of observable behavioral defects for odors that activate ab8A neurons suggests that there is functional redundancy between odorant receptors in Drosophila that can compensate for the loss of ab8A neuron function. Indeed, most of the odorants that activate ab8A are known to activate other classes of olfactory neurons (de Bruyne et al., 2001). Functional redundancy among olfactory receptors could also explain why mutant screens performed over the last 25 years to identify mutants with defective chemosensory behavior have recovered a variety of signal transduction components but failed to recover Drosophila odorant receptor mutants.
A particular odorant at a particular concentration is thought to activate a specific subset of receptors and thus a specific subset of glomeruli. The pattern of glomeruli activated is thought to be unique to each odorant, thus defining an odorant “code” in the brain. The loss of ab8A olfactory neuron activity failed to produce defective olfactory behavior, suggesting that odor coding is a robust phenomenon in Drosophila. If a pattern of glomerular activity underlies odor discrimination that, in turn, leads to odor-evoked behavior, and if ab8A neurons project to a single glomerulus (Vosshall et al., 2000; Dobritsa et al., 2003), our results suggest that the loss of one glomerular component does not appreciably influence the sensitivity or the quality of the behavioral response. It is not clear from these studies whether odor discrimination is affected in these mutants. In future studies, it will be interesting to determine whether odorants that activate ab8A neurons reveal abnormalities in the ability of these mutants to discriminate among these and closely related odorants, and to evaluate whether these odorants induce different neuronal activity in the higher olfactory-processing centers in Or43b mutants.
Reverse genetics and analysis of olfactory behavior
The numerical simplicity of the Drosophila olfactory system, combined with the available genetic and imaging tools available, make flies an attractive model system to dissect olfactory function. Functional maps of the Drosophila peripheral olfactory system that define the odorant specificity of specific functional classes of olfactory neurons are being developed (de Bruyne et al., 1999, 2001). Analysis of receptor gene expression patterns is concurrently elucidating anatomic maps (Clyne et al., 1999; Vosshall et al., 1999, 2000). Integration of these maps (Dobritsa et al., 2003) is useful in formulating models of odor coding, but ultimately these models must be tested. We demonstrated here that a reverse genetic approach on the basis of gene targeting can be used to explore the link between selected olfactory gene products of interest and chemosensory behavior, and we have correlated expression of a specific receptor with a functional class of olfactory neurons. A similar analysis of related gene products using homologous recombination or RNA interference (Kalidas and Smith, 2002) will expand our knowledge of the contributions of specific chemosensory receptors to peripheral and central odor-coding mechanisms.
Footnotes
This work was supported by American Heart Association Grant EI 0040029N (D.P.S.) and National Institutes of Health Grants DC02539 (D.P.S.), DC04729, and DC02174 (J.R.C.). We thank Ashley Nevin and John Morse for technical assistance and Helmut Kramer for helpful comments on this manuscript.
Correspondence should be addressed to Dean P. Smith, Department of Pharmacology and Center for Basic Neuroscience, University of Texas Southwestern Medical Center, Dallas, TX 75390-9111. E-mail: dean.smith@utsouthwestern.edu.
Copyright © 2003 Society for Neuroscience 0270-6474/03/239906-07$15.00/0
References
- Araneda RC, Kini AD, Firestein S ( 2000) The molecular receptive fields of an odorant receptor. Nat Neurosci 3: 1248–1255. [DOI] [PubMed] [Google Scholar]
- Bozza T, Teinstein P, Zheng C, Mombaerts P ( 2002) Odorant receptor expression defines functional units in the mouse olfactory system. J Neurosci 22: 3033–3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chess A, Simon I, Cedar H, Axel R ( 1994) Allelic inactivation regulates olfactory receptor gene expression. Cell 78: 823–834. [DOI] [PubMed] [Google Scholar]
- Clyne P, Grant A, O'Connell R, Carlson JR ( 1997) Odorant response of individual sensilla on the Drosophila antenna. Invert Neurosci 3: 127–135. [DOI] [PubMed] [Google Scholar]
- Clyne PJ, Warr CG, Freeman MR, Lessing D, Kim J, Carlson JR ( 1999) A novel family of divergent seven-transmembrane proteins: candidate odorant receptors in Drosophila Neuron 22: 327–338. [DOI] [PubMed] [Google Scholar]
- de Bruyne M, Clyne P, Carlson JR ( 1999) Odor coding in a model olfactory organ: the Drosophila maxillary palp. J Neurosci 19: 4520–4532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bruyne M, Foster K, Carlson JR ( 2001) Odor coding in the Drosophila antenna. Neuron 30: 537–552. [DOI] [PubMed] [Google Scholar]
- Dobritsa AA, van der Goes van Naters W, Warr CG, Steinbrecht RA, Carlson JR ( 2003) Integrating the molecular and cellular basis of odor coding in the Drosophila antenna. Neuron 37: 827–841. [DOI] [PubMed] [Google Scholar]
- Elmore T, Smith DP ( 2001) Putative Drosophila odor receptor OR43b localizes to dendrites of olfactory neurons. Insect Biochem Mol Biol 31: 791–798. [DOI] [PubMed] [Google Scholar]
- Gaillard I, Rouquier S, Pin J-P, Mollard P, Richard S, Barnabe C, Demaille J, Giorgi D ( 2002) A single olfactory recceptor specifically binds a set of odorant molecules. Eur J Neurosci 15: 409–418. [DOI] [PubMed] [Google Scholar]
- Gao Q, Chess A ( 1999) Identification of candidate olfactory receptors from genomic DNA sequence. Genomics 60: 31–39. [DOI] [PubMed] [Google Scholar]
- Kaissling K-E ( 1997) Olfactory transduction in moths: 1. Generation of receptor potentials and nerve impulses. In: From structure to information in the nervous system (Taddei-Ferretti C, Musio C, eds). Singapore: World Scientific.
- Kalidas S, Smith DP ( 2002) Novel genomic cDNA hybrids produce effective RNA interference in adult Drosophila Neuron 33: 177–184. [DOI] [PubMed] [Google Scholar]
- Kim M-S, Repp A, Smith DP ( 1998) LUSH odorant binding protein mediates chemosensory responses to alcohols in Drosophila melanogaster Genetics 150: 711–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krautwurst D, Yau KW, Reed RR ( 1998) Identification of ligands for olfactory receptors by functional expression of a receptor library. Cell 23: 917–926. [DOI] [PubMed] [Google Scholar]
- Langer-Sofer PR, Levine M, Ward DC ( 1982) Immunological method for mapping genes on Drosophila polytene chromosomes. Proc Natl Acad Sci USA 79: 4381–4385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malnic B, Hirono J, Sato T, Buck LB ( 1999) Combinatorial receptor codes for odors. Cell 96: 713–723. [DOI] [PubMed] [Google Scholar]
- Nathans J ( 1999) The evolution and physiology of human color vision: insights from molecular genetic studies of visual pigments. Neuron 24: 299–312. [DOI] [PubMed] [Google Scholar]
- Ng M, Roorda RD, Lima SQ, Zeleman BV, Morcillo P, Meisenbock G ( 2002) Transmission of olfactory information between three populations of neurons in the antenna lobe of the fly. Neuron 36: 463–474. [DOI] [PubMed] [Google Scholar]
- Ressler KJ, Sullivan SL, Buck LB ( 1993) A zonal organization of odorant gene expression in the olfactory epithelium. Cell 73: 596–609. [DOI] [PubMed] [Google Scholar]
- Rong YS, Golic KG ( 2000) Gene targeting by homologous recombination in Drosophila Science 288: 2013–2018. [DOI] [PubMed] [Google Scholar]
- Rong YS, Golic KG ( 2001) A targeted gene knockout in Drosophila Genetics 157: 1307–1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong YS, Titen SW, Xie HB, Golic MM, Bastiani M, Bandyopadhyay P, Brodsky M, Rubin GM, Golic KG ( 2002) Targeted mutagenesis by homologous recombination in D. melanogaster Genes Dev 16: 1568–1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanbhag SR, Müller B, Steinbrecht RA ( 1999) Atlas of olfactory organs of Drosophila melanogaster 1. Types, external organization, innervation and distribution of olfactory sensilla. Int J Insect Morphol Embryol 28: 377–397. [Google Scholar]
- Spradling AC, Rubin GM ( 1982) Transposition of cloned P elements into Drosophila germline chromosomes. Science 218: 341–347. [DOI] [PubMed] [Google Scholar]
- Stocker RF ( 1994) The organization of the chemosensory system in Drosophila melanogaster: a review. Cell Tissue Res 275: 3–26. [DOI] [PubMed] [Google Scholar]
- Tully T, Quinn WT ( 1985) Classical conditioning and retention in normal and mutant Drosophila melanogaster J Comp Physiol 157: 263–277. [DOI] [PubMed] [Google Scholar]
- Vasser R, Ngai J, Axel R ( 1993) Spatial segregation of odorant receptor expression in the mammalian olfactory epithelium. Cell 74: 309–318. [DOI] [PubMed] [Google Scholar]
- Vosshall LB, Amrein H, Morozov PS, Rzhetsky A, Axel R ( 1999) A spatial map of olfactory receptor expression in the Drosophila antenna. Cell 96: 725–736. [DOI] [PubMed] [Google Scholar]
- Vosshall LB, Wong AM, Axel R ( 2000) An olfactory sensory map in the fly brain. Cell 102: 147–159. [DOI] [PubMed] [Google Scholar]
- Wang Y, Wright NJD, Guo H-F, Xie Z, Svoboda K, Malinow R, Smith DP, Zhong Y ( 2001) Genetic manipulation of the odor-evoked distributed neural activity in the Drosophila mushroom body. Neuron 29: 267–276. [DOI] [PubMed] [Google Scholar]
- White JH, Wise A, Main MJ, Green A, Fraser NJ, Disney GH, Barnes AA, Emson P, Foord SM, Marshall FH ( 1998) Heterodimerization is required for the formation of a functional GABA(B) receptor. Science 396: 679–682. [DOI] [PubMed] [Google Scholar]
- Zhou H, Ivic L, Otaki JM, Hashimoto M, Mikoshiba K, Firestein SJ ( 1998) Functional expression of a mammalian odorant receptor. Science 279: 237–242. [DOI] [PubMed] [Google Scholar]