Abstract
The dendritic transport and local translation of mRNA may be an essential mechanism to regulate synaptic growth and plasticity. We investigated the molecular mechanism and function of β-actin mRNA localization in dendrites of cultured hippocampal neurons. Previous studies have shown that β-actin mRNA localization to the leading edge of fibroblasts or the growth cones of developing neurites involved a specific interaction between a zipcode sequence in the 3′ untranslated region and the mRNA-binding protein zipcode-binding protein-1 (ZBP1). Here, we show that ZBP1 is required for the localization of β-actin mRNA to dendrites. Knock-down of ZBP1 using morpholino antisense oligonucleotides reduced dendritic levels of ZBP1 and β-actin mRNA and impaired growth of dendritic filopodia in response to BDNF treatment. Transfection of an enhanced green fluorescent protein (EGFP)-β-actin construct, which contained the zipcode, increased the density of dendritic filopodia and filopodial synapses. Transfection of an EGFP construct, also with the zipcode, resulted in recruitment of endogenous ZBP1 and β-actin mRNA into dendrites and similarly increased the density of dendritic filopodia. However, the β-actin zipcode did not affect filopodial length or the density of mature spines. These results reveal a novel function for an mRNA localization element and its binding protein in the regulation of dendritic morphology and synaptic growth via dendritic filopodia.
Keywords: mRNA localization, zipcode-binding protein, β-actin, β-actin mRNA, dendritic filopodia, filopodial synapse, BDNF
Introduction
Although there has been extensive study of the active role that axonal growth cones play to initiate physical contacts with dendrites of target cells, there has also been evidence of an active role for dendritic filopodia in seeking out contacts with navigating axons (Cooper and Smith, 1992; Ziv and Smith, 1996). Dendritic filopodia are actin-rich protrusions from the cell membrane but are distinguished from dendritic spines by their long thin morphology, the absence of a stable synaptic connection, and their ability to undergo more robust motility characterized by dynamic cycles of extension and retraction (Cailliau and Yuste, 2001). It has been proposed that the dynamic contacts made between dendritic filopodia and axons may play a key role in synaptogenesis, and possibly spine maturation, during development (Ziv et al., 1996). An alternative view arises from observations that filopodia are not prerequisites for spine formation, but rather that they recruit shaft-synapses, which may later become spines (Fiala et al., 1998). Regardless of which model is correct, it becomes essential to understand the cellular and molecular mechanisms that regulate the formation and density of dendritic filopodia during neuronal development, because these are likely to be intimately connected to the process of synaptogenesis.
Despite our understanding of the importance of F-actin dynamics in the morphological changes of dendritic filopodia and spines during development and plasticity (Fischer et al., 1998, 2000; Halpain et al., 1998; Dunaevsky et al., 1999; Zhang and Benson, 2001), very little is known about the molecular mechanisms involved in the sorting of distinct actin isoforms and the spatial regulation of their synthesis which could, in principle, play a major regulatory role in actin-based plasticity in dendrites during periods of synaptic growth. A number of studies have shown that the β-actin isoform is highly enriched in actin structures undergoing dynamic polymerization and remodeling of actin, such as the fibroblast leading edge (Kislauskis et al., 1997), the growth cone of developing neurites (Bassell et al., 1998), and dendritic spines of adult cerebellar neurons (Micheva et al., 1998). The unique amino terminus of the β-actin isoform may favor interactions with specific proteins involved in regulation of actin polymerization at submembranous sites (Shuster et al., 1995; Zhang et al., 1999).
The targeting of β-actin to submembranous sites in some polarized cells has been shown to be mediated by a 54 nucleotides (nt) sequence in the 3′ untranslated region (UTR), which positions β-actin mRNA for local translation. In chick fibroblasts and neurons, the zipcode was bound by the mRNA-binding protein zipcode-binding protein-1 (cZBP1) (Ross et al., 1997; Zhang et al., 2001; Farina et al., 2003). The interaction between the zipcode and cZBP1 was important for normal motility of the fibroblast leading edge or neurite growth cone (Shestakova et al., 2001; Zhang et al., 2001). We have shown that β-actin mRNA is localized to dendrites, filopodia, and spines in cultured rat hippocampal neurons (Tiruchinapalli et al., 2003) and hypothesized that there may be a critical period during development when β-actin mRNA localization may provide an important function for synaptogenesis and developmental plasticity. In this study, we cloned the rat orthologue of cZBP1 (rZBP1) from embryonic brain and show that it is required for the localization of β-actin mRNA to dendrites of cultured hippocampal neurons. Modulation of rZBP1 levels using morpholino antisense oligonucleotides or overexpression of zipcode-containing constructs had opposing effects on filopodial density. We also observed a zipcode-dependent effect on the formation of filopodial synapses. These results reveal a novel function for the localization of β-actin mRNA in dendritic morphology and synaptic growth via dendritic filopodia.
Materials and Methods
Disociated hippocampal cultures. Following the method of neuronal culture for embryonic rat hippocampus [embryonic day (E) 18] detailed previously (Goslin et al., 1998), cells were dissociated with trypsin and plated at low density (120,000 cells/cm2) on poly-l-lysine-coated (1.0 mg/ml) coverslips. The coverslips were inverted on a plate of astrocytes and grown as a coculture with defined media containing N2 supplements. After 10-12 d in vitro (DIV), neurons were fixed in paraformaldehyde (4% in 1 × PBS with 5 mm MgCl2) for 20 min at room temperature.
In situ hybridization with digoxigenin-labeled probes. Four amino-modified oligonucleotide probes (∼50 nt each), complementary either to rat β-actin mRNA or calcium/calmodulin-dependent protein kinase II (CaMKIIα) mRNA, were synthesized and chemically labeled with digoxigenin succinamide ester at five amino-modified thymidine residues (Roche Applied Science, Indianapolis, IN) as described previously (Zhang et al., 2001). Rat β-actin mRNA probes corresponded to the coding region and 3′UTR nucleotide positions: 166-218, 400-453, 1132-1181, 1440-1489. Oligo 1132-1181 was excluded for the experiment shown in Figure 8 because of complementarity to the β-actin zipcode. CaMKIIα mRNA probes corresponded to 3′UTR nucleotide positions: 1671-1720, 2261-2310, 2692-2741 (Mori et al., 2000). In situ hybridization was completed as described previously (Bassell et al., 1998). As negative controls, digoxigenin-labeled oligonucleotide probes were hyrbidized with 100 × excess unlabeled oligos. Alternatively, oligos for β-galactosidase were used. These negative controls did not reveal either fluorescent signal or granules. The digoxigenin-labeled oligonucleotide probes were detected by immunofluorescence using a Cy3-conjugated monoclonal antibody to digoxigenin and a Cy3-conjugated anti-mouse antibody (Jackson ImmunoResearch, West Grove, PA). Coverslips were mounted with gelvatol with n-propyl gallate (6 mg/ml) as an anti-bleaching agent as described previously (Zhang et al., 1999).
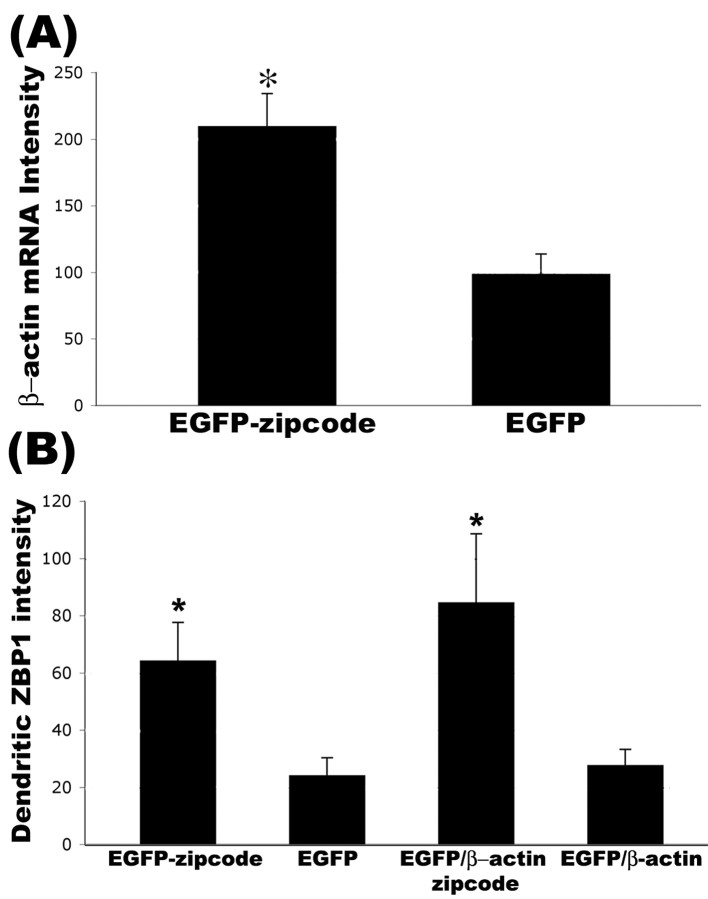
Figure 8.
Overexpression of EGFP constructs with the β-actin zipcode resulted in recruitment of endogenous β-actin mRNA and ZBP1. Cultured rat hippocampal neurons were transfected with EGFP, EGFP-zipcode, EGFP-β-actin, and EGFP-β-actin-zipcode. All constructs were expressed for 48 hr. A, Fluorescence in situ hybridization to detect endogenous β-actin mRNA was performed using oligonucleotide probes that excluded the region containing the zipcode. Measurement of average β-actin mRNA levels in dendrites using digital imaging analysis showed an increase in neurons transfected with EGFP-zipcode compared with EGFP alone (*p < 0.05; Student's t test). B, Quantitative immunofluorescence of ZBP1 levels in dendrites showed an increase in both zipcode-containing constructs compared with constructs lacking the zipcode (*p < 0.05; Student's t test; histogram indicates mean values ± SEM).
Immunofluorescence. For detection of endogenous proteins, we used rabbit anti-ZBP1 antibody (Farina et al., 2003), mouse anti-microtubule associated protein-2 (MAP2) (Roche Molecular Biochemicals) and rabbit anti-synapsin antibodies (Chemicon, Temecula, CA). All secondary antibodies were affinity-purified donkey antibodies to mouse or rabbit IgG conjugated to a fluorochrome (Jackson ImmunoResearch). Incubations for antibodies were for 1 hr at room temperature in Tris-buffered saline with 1% bovine serum albumin (BSA) (Roche Molecular Biochemicals) and 0.1% Triton X-100. Coverslips were also mounted with gelvatol with n-propyl gallate as mentioned above (6 mg/ml).
Fluorescence microscopy and quantitative digital imaging. Neurons were viewed under a Nikon (Tokyo, Japan) Eclipse inverted microscope equipped with a 60 × Plan-Neofluar objective, phase optics, a 100 W mercury arc lamp, and HiQ bandpass filters (Chroma Technology, Brattleboro, VT), and images were acquired by a cooled CCD camera (Quantix; PhotoMetrics, Huntington Beach, CA) using a 35 mm shutter and processed using IP Lab Spectrum (Scanalytics, Fairfax, VA) as described previously (Zhang et al., 2001). Exposure times were kept constant in the analysis of each experiment. Dendritic morphology was imaged using anti-MAP2 or phase images, such that the user was blind to the signal for rZBP1 or mRNA, respectively. The filter set was then changed, and images of β-actin mRNA (in situ hybridization) or ZBP1 (immunofluorescence) were acquired. Quantitative analysis of mRNA or protein localization within dendrites was calculated as mean fluorescence intensity per unit area within a region of interest. Each histogram shown in Figure 3 reflects the combined quantitative analysis of many dendrites (1-3 dendrites per cell, 10-15 dendrites per experiment with duplicate coverslips, and 3 experiments from separate cultures).
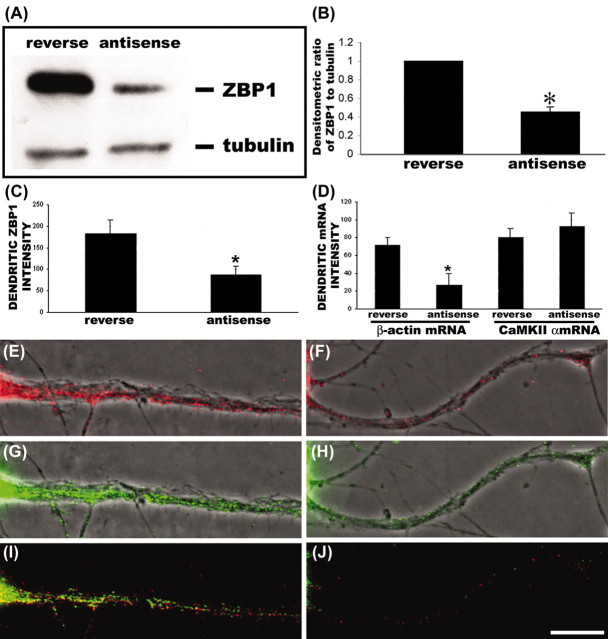
Figure 3.
Reduced ZBP1 and β-actin mRNA localization in dendrites after antisense knock-down of rZBP1. A, Western blot analysis with anti-ZBP1 and antitubulin antibodies from extracts of antisense and reverse antisense-treated neurons. Note that the tubulin band is weaker than ZBP because of excessive dilution of antitubulin (1:4000). This prevented oversaturation of the tubulin signal so it could be used for normalization. rZBP1 levels were decreased after 48 hr of antisense treatment, although no changes in tubulin levels were noted. B, Ratio analysis of ZBP1/tubulin band intensities. In antisense-treated cells, the relative intensity of rZBP1 (compared with tubulin) was decreased compared with reverse antisense-treated cells. C, Antisense treatment decreased rZBP1 levels in dendrites, as measured using quantitative immunofluorescence. Cultured rat hippocampal neurons were incubated with either antisense or reverse antisense oligonucleotides to rZBP1 for 48 hr and processed for immunofluorescence. D, Quantitative FISH analysis showed that rZBP1 antisense treatment decreased β-actin mRNA levels in dendrites but not CaMKIIα mRNA levels. E, Example of β-actin mRNA signal in reverse antisense-treated neuron. G, Corresponding signal for ZBP1 immunofluorescence in the same neuron. I, Merge of both β-actin mRNA and ZBP1 showing partial pixel overlap. F, Example of β-actin mRNA signal in rZBP1 antisense-treated neuron. H, Corresponding signal for ZBP1 immunofluorescence in the same neuron. J, Merge of both β-actin mRNA and ZBP1 signal. B, C, D, *, Significant difference in mean values ± SEM; p < 0.05; Student's t test. Scale bar, (E-J) 10 μm.
For quantitative analysis of density of filopodia and spine density (see Figs. 5, 6, 7), a visual scoring method was used from randomly acquired images. Neurons were fixed after 48 hr of construct expression. At least 10 neurons per construct were randomly selected using phase or differential interference contrast, followed by acquisition of phalloidin (see Fig. 5) or EGFP (see Figs. 6, 7) fluorescence at constant exposure times. A quantitative morphometric analysis was performed using image analysis software (IP Lab; Scanalytics) to measure the average density of total surface protrusions and the average length of each protrusion. The density of protrusions was obtained by dividing their total number by the total length of dendritic segments that were analyzed. A few dendrites from each neuron were analyzed, and each experiment was repeated using separate cultures. This resulted in several hundred to often thousands of protrusions being quantified for each construct. The majority of surface protrusions at this stage in culture were filopodia, which are long and thin and not colocalized with synapsin puncta. Filopodial-spine synapses were defined as long, thin, nonbulbous protrusions in contact with a synapsin puncta. Spines were defined as a dendritic protrusion with a broad or bulbous morphology, which was in contact with synapsin puncta, as detected by immunofluorescence with anti-synapsin antibodies. Spines that were unclear as to their distinct morphology were not analyzed.
Figure 5.
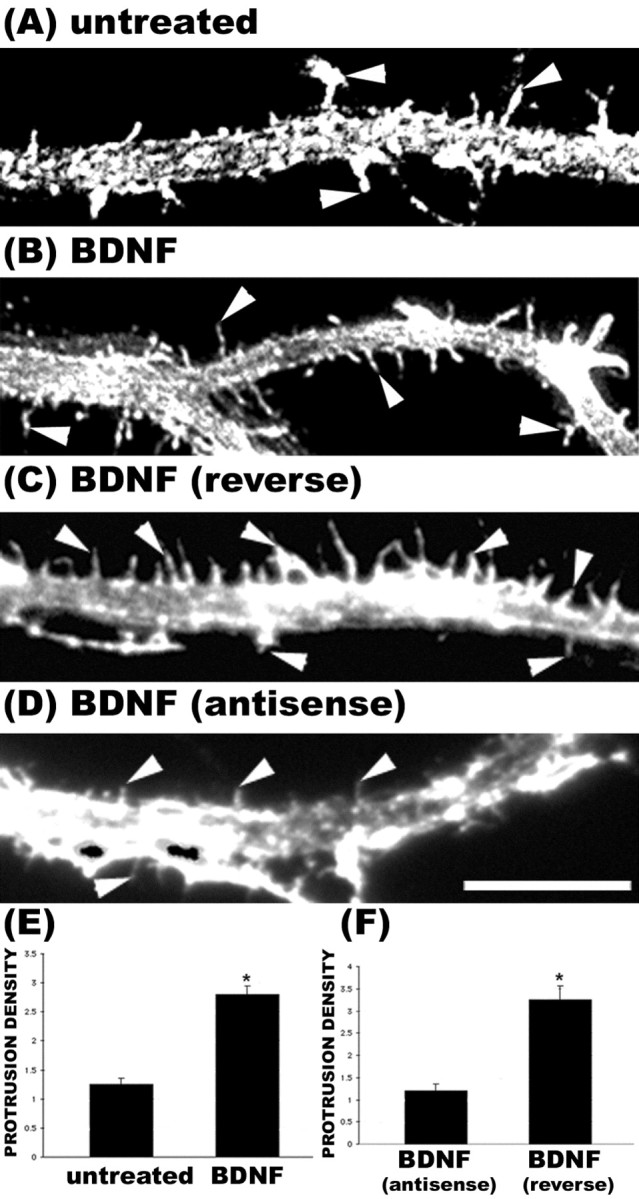
Knock-down of rZBP1 inhibited the growth of dendritic protrusions in response to BDNF stimulation. A, Phalloidin stain of an untreated neuron. B, Phalloidin labeling of a neuron treated with BDNF (25 ng/ml) for 2 hr before fixation. An increase in the density of filopodial-like protrusions was observed (arrowheads). C, Control neurons treated with reverse antisense oligonucleotides to rZBP1 (48 hr) and then stimulated with BDNF (2 hr) also showed an increased density of protrusions (arrowheads). D, In contrast, treatment of neurons with antisense oligonucleotides to rZBP1 resulted in markedly fewer filopodial protrusions after BDNF treatment (arrowheads). E, Quantitation of the density of total F-actin protrusions in untreated and BDNF-treated cultures showed a significant increase after BDNF (p < 0.05). These neurons were not exposed to any morpholino oligonucleotides. F, Quantitation of the density of F-actin protrusions in BDNF-treated cultures exposed to reverse antisense or antisense morpholino oligonucleotides; note the significantly reduced density in antisense-treated neurons (*p < 0.05; Student's t test; histogram indicates mean value ± SEM). Scale bar, 10 μm.
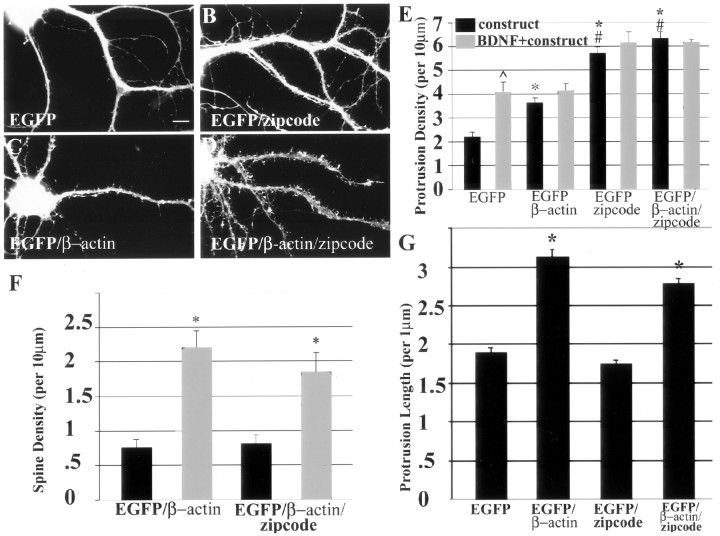
Figure 6.
Morphologic analysis of dendritic protrusions in neurons transfected with EGFP constructs. Cultured rat hippocampal neurons were transfected with the following cDNAs: EGFP (A), EGFP-zipcode with the β-actin zipcode in the 3′UTR (B), EGFP-β-actin (C), and EGFP-β-actin-zipcode with the β-actin zipcode in the 3′UTR (D). Neurons were fixed after 48 hr of construct expression. A quantitative morphometric analysis was performed using image analysis software (IP Lab; Scanalytics) to measure mean protrusion density (E), mean protrusion length (G), and mean spine density (F). Spines were defined as a dendritic protrusion with a broad or bulbous morphology in contact with synpasin puncta, which was detected by immunofluorescence. At least 10 neurons per construct were randomly selected, followed by acquisition of GFP fluorescence at constant exposure times. A few dendrites from each neuron were analyzed, and each experiment was repeated using separate cultures. This resulted in several hundred to often thousands of protrusions being quantified for each construct. E, A significant increase in protrusion density was observed after overexpression of EGFP-β-actin without the zipcode when compared with EGFP alone (*p < 0.05; ANOVA). Highly significant increases in protrusion density were observed after overexpression of EGFP-β-actin and EGFP constructs containing the zipcode (*p < 0.001; ANOVA). Both zipcode-containing constructs resulted in highly significant increases in protrusion density when compared with β-actin lacking the zipcode (#p < 0.01; ANOVA). A significant increase in protrusion density was observed in EGFP-transfected cells after BDNF stimulation (^p < 0.001). Zipcode-containing constructs seemed to occlude any additional increase in protrusion density by BDNF treatment. F, BDNF treatment significantly increased spine density in neurons transfected with EGFP-β-actin with (*p < 0.01; Student's t test) or without the zipcode (*p < 0.001; Student's t test). G, Overexpression of actin resulted in highly significant increases in the mean length of protrusions, which were not affected by the zipcode (*p < 0.01; Student's t test). Error bars indicate SEM.
Figure 7.
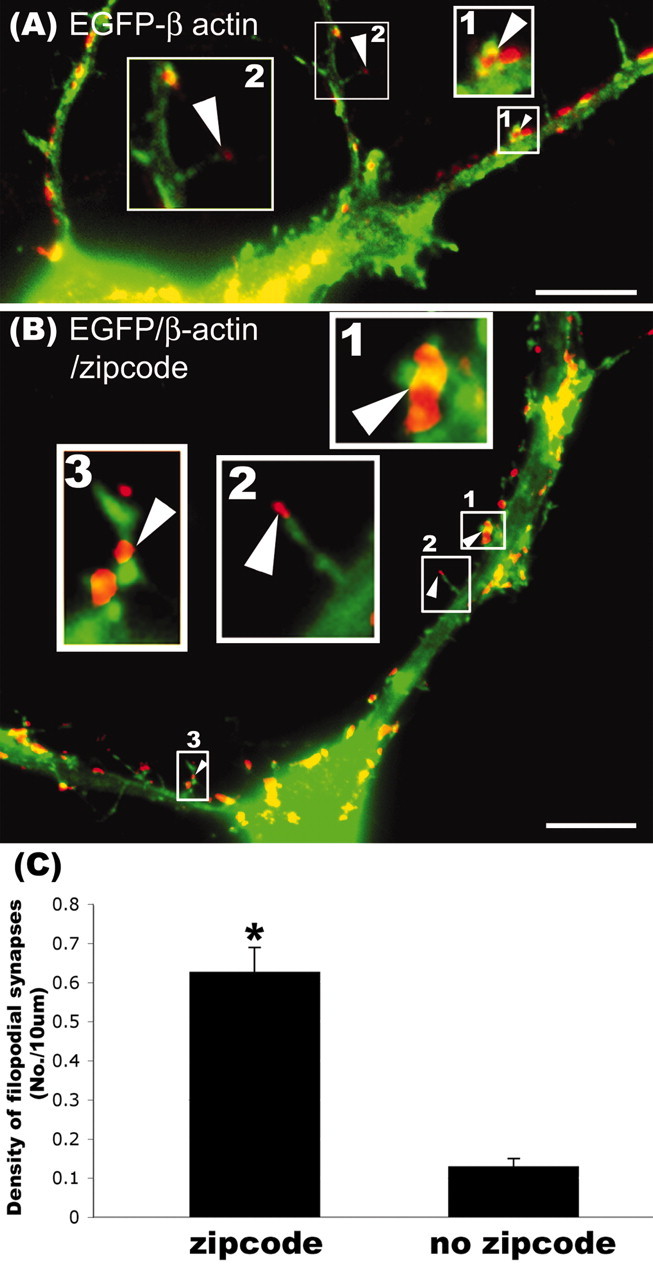
Overexpression of EGFP-β-actin increased the density of filopodial synapses in a zipcode-dependent manner. Cultured rat hippocampal neurons were transfected with either EGFP-β-actin or EGFP-β-actin-zipcode. Neurons were fixed and stained with antisynapsin antibody (red) as a presynaptic marker. A, EGFP-β-actin (no zipcode): the enlarged insets illustrate and contrast spines with a bulbous or headed morphology (box 1, arrowhead) with those having a long, thin filopodial morphology (box 2, arrowhead). Both types of spines were in contact with synapsin puncta (red). We referred to the filopodial spines (box 2) as filopodial synapses in the text. B, Expression of EGFP-β-actin-zipcode increased the density of filopodial synapses (insets 2, 3, arrowheads). A bulbous headed spine is also depicted in this dendrite for comparison (box 1, arrowhead). C, Quantitation of the average density of filopodial synapses showed that the zipcode significantly increased their density after transfection of EGFP-β-actin (*p < 0.05; Student's t test). Scale bar, 10 μm.
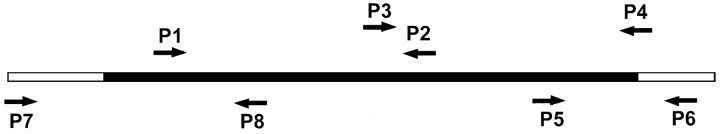
RNA isolation and reverse transcriptase-PCR. Using a reverse transcriptase (RT)-PCR approach, we cloned the rat orthologue of chicken ZBP1 from E18 rat brain (see diagram below for primer locations). Two overlapping sets of primers (P1/P2 and P3/P4) were first used to sequence 1642 nt, which corresponded to most of the coding region of rat ZBP1 but excluded the 5′ end. Sequence analysis of both fragments confirmed a single open-reading frame. To obtain partial 5′ and 3′UTR sequences, primers were designed to rat expression sequence tag (EST) sequences that corresponded to putative upstream and downstream UTR sequences and were used in combination with primers to the rat ZBP1-coding sequence obtained above. PCR performed with the primer to the 5′UTR EST sequence (P7), and a coding region primer (P8) allowed sequence analysis of the 5′ end that contained 222 nt upstream of the start codon. Similarly, PCR performed with the primer to the 3′UTR EST sequence (P6) and a primer to the coding region (P5) allowed sequence analysis of the 3′ end that included 149 nt of 3′UTR downstream of the stop codon. Full-length amplification of the coding region (P7/P4 primers) has also been performed with primers flanking both start and stop codons, confirming a coding region of 1734 nt. A schematic of the primers used is shown below (open bar, UTRs; black bar, coding region). Total RNA from rat forebrain of E18 embryos was isolated using Tri Reagent, following product instructions (Molecular Research Center, Cincinnati, OH) as described previously (Zhang et al., 2001). Total RNA was dissolved in DEPC-treated distilled water. RT-PCR was performed with the following primers as depicted above. P1, sense: 5′TCTT GGTCAAATCCGGCTA3′ [coding region determinant binding protein (CRDBP) nt positions, 228-246] (Doyle et al., 1998); P2, antisense: 5′CGGCCACGTCGTT CTCGTAA3′ (CRDBP, 1180-1199); P3, sense: 5′CAGGATC TCACGCTCTATAAC3′ (CRDBP, 1076-1096); P4, antisense: 5′CGG GGTCACTTCCTCCGA GC3′ (CRDB, 1850-1869). To obtain the 3′UTR sequence of rZBP1, the primers were P5, sense (on the basis of our rZBP1 sequence; 5′CAAGTTAAGCAGCAGC ACCA3′), and P6, antisense [EST sequence gene identification (GI): 14921191; 5′TGAGGTTTTATAAAAC CTGAAGG3′]. To obtain the 5′UTR sequence of rZBP1, the primers were P8, antisense (on the basis of our rZBP1 sequence; 5′TGGTTC TCCA GTT GATGGCC3′), and P7, sense (EST sequence GI: 4278139;5′TCGTCAGC CTCCC TACAC TT3′). The full-length cloned rZBP1 sequence (GenBank accession number AF541940) has 577 aa and shares 95% identity with chick ZBP1, differing in 32 aa.
Western blot analysis. Protein extracts (10 μl) were resolved by 12% SDS-PAGE, and fractionated proteins were transferred to a Hybond ECL nitrocellulose membrane (Amersham Biosciences, Arlington Heights, IL) at 4°C overnight. rZBP1 from brain extract and recombinant rZBP1 was detected with a rabbit anti-ZBP1 antibody (Farina et al., 2003), and α-tubulin was detected with a mouse anti-α-tubulin antibody (Sigma, St. Louis, MO) at 1:1000 or 1:5000 in PBS with 1% BSA. The membrane was washed and incubated with peroxidase-conjugated goat anti-rabbit IgG (Jackson ImmunoResearch). The membrane was exposed to x-ray film (Kodak, Rochester, NY) by using ECL detection reagents (Amersham Biosciences).
Expression and purification of recombinant rZBP1. Recombinant rZBP1 protein was purified using the IMPACT-CN system (New England Biolabs, Beverly, MA). rZBP1 cDNA was inserted into the Ndel/SapI sites of the pTYB11 vector. pTYB11-rZBP1 was expressed in E. coli BL21-Codon Plus(DE3)-RP strain cells (Stratagene, La Jolla, CA). Recombinant rZBP1 was cleaved from an intein tag in 1,4-dithiothreitol (DTT) and eluted through a chitin column and used for electrophoretic mobility shift assay (EMSA).
EMSA. The rat β-actin sequence (GI: 55574) corresponding to the zipcode was obtained by RT-PCR from E18 rat brain and subcloned into RNA expression vector pGEM3Zf(-). A PCR product (88 nt) containing the zipcode was inserted into the blunt end/XbaI sites of the pGEM3Zf(-) vector (Promega, Hawthorne, Australia). Primers used were sense 5′ACAAGATCTGCGGACTGTTA3′ and antisense 5′ACATCTAGAGCCATGCCAA3′. This 88 nt sequence is located just downstream of the stop codon in the 3′UTR and contains the ACACCC consensus motif for rZBP1 binding analogous to the 54 nt chick β-actin zipcode (Ross et al., 1997).
32P-labeled RNA probes were transcribed in vitro from XbaI-linearized pGEM-zipcode construct and from nonrelated sequences as a negative control using SP6 RNA polymerase (Promega) and 50 μCi of [α-32P] CTP (Amersham Biosciences) for 1 hr at 37°C. The synthesized labeled RNA probes were resolved in 5% urea-polyacrylamide gels and purified. Radiolabeled RNA probes (50,000 cpm/μl) were incubated with pooled protein extracts for 20 min at room temperature in binding solution (20 mm HEPES, pH 7.6, 3 mm MgCl2, 100 mm KCl, 2 mm DTT, 5% glycerol). Proteins were extracted from rat brain (E18) as described previously (Zhang et al., 2001). Unbound RNA was digested with RNase T1 (1 U) (Invitrogen, San Diego, CA), and nonspecific RNA-protein interactions were minimized by incubation with heparin (5 mg/ml). RNA protein complexes were resolved in a nondenaturing 4% polyacrylamide native gel and visualized by autoradiography. Competition assays were performed by preincubation of the protein extract with unlabeled RNA competitors for 10 min at room temperature before the radiolabeled RNA was added.
EGFP-rZBP1 construct. For transfection of rZBP1, pEGFP-rZBP1 was generated by inserting the open-reading frame of rZBP1 into the EcoRI site of the pEGFP-C1 vector (BD Biosciences). For transfection of the pEGFP-zipcode and the pEGFP-β-actin-zipcode constructs, a fragment of amplified pGEM-3Zf(-)-zipcode (88 nt) was inserted into the EcoRI/SalI sites of pEGFP-C1. To add a stop codon before the zipcode, a fragment which has a stop codon (5′AGATCTC GGATCAAAAAAGAAGAGAAAGGTAGATATCTCGAGCTC3′) was added into BglII/SacI sites of pEGFP-C1. A BglII/StuI cleaved fragment of pEGFP-zipcode was inserted into the BamHI/StuI sites of the pEGFP-β-actin vector (BD Biosciences Clontech, Palo Alto, CA).
Transfection of cDNA or morpholino antisense oligonucleotides. Constructs were transfected when neurons were 9 DIV as described above. Coverslips with attached neurons were transferred into a new 35 mm culture dish and equilibrated with 2 ml of 1-d-old N2-conditioned medium at 5% CO2 at 37°C for 30 min. DNA (3-5 μg) was dissolved in 60 μl of 250 mm CaCl2, and 60 μl of 2 × N,N-bis[2-hydroxyethyl]-2-aminoethane sulfonic acid-buffered saline was added to the reaction tube. Cells were incubated with transfection mixture at 2.5% CO2 and 37°C until the proper size of precipitate was observed. Transfected cells were fixed after 48 hr. For antisense treatment, morpholino oligonucleotides were transfected instead of DNA. Sequences for antisense and reverse antisense oligonucleotides (Gene-Tools, Philomath, OR) were antisense 5′AGCTTGTTTATGGTGGCGGTGGCGG3′ and reverse antisense 5′GGCGGT GGCGGTGGTATTTGTTCGA3′. After 48 hr of construct expression or antisense exposure, neurons were fixed as described above. Some samples were exposed to BDNF by bath application (25 ng/ml for 1-2 hr before fixation).
Results
Molecular characterization of rZBP1 and its association with the β-actin zipcode in embryonic rat brain
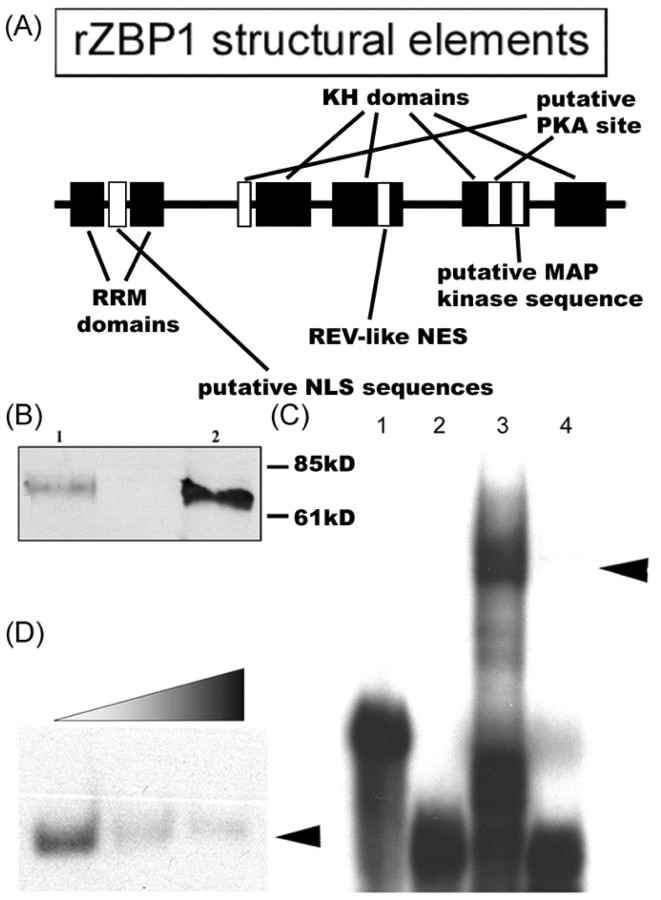
The mRNA-binding protein cZBP1 was originally isolated and cloned from chick embryos and shown to exhibit sequence-specific binding to the 54 nt β-actin zipcode in vitro but not after mutation of a critical ACACCC sequence, which was required for localization of β-actin mRNA reporter constructs in transfected fibroblasts (Ross et al., 1997). We recently showed that ZBP1 is expressed in rat hippocampal neurons on the basis of immunofluorescence and Western blot analysis with a polyclonal antibody to chicken ZBP1 (Tiruchinapalli et al., 2003). We also showed that ZBP1 colocalized with β-actin mRNA in dendrites and spines using double-label immunofluorescence and in situ hybridization (Tiruchinapalli et al., 2003). In this study, we investigated whether ZBP1 forms a biochemical complex with the zipcode that is required for the dendritic localization of β-actin mRNA in cultured hippocampal neurons. This first necessitated that we clone the rat orthologue of ZBP1 (designated rZBP1). A PCR-based approach was used from extracts of E18 embryonic rat brain (see Materials and Methods). The rZBP1 sequence (GenBank accession number AF541940) has 577 amino acids and shares 95% identity with chick ZBP1, differing in 32 aa. Like chick ZBP1, rat ZBP1 also contains several RNA-binding domains, including two RNA recognition motifs (RRM) and four heterogeneous ribonucleoprotein (hnRNP) K homology (KH) domains (Fig. 1A). In addition, there are putative nuclear localization signal (NLS) sites, putative nuclear export signal (NES) sites, and phosphorylation consensus sites for MAP kinase and PKA.
Figure 1.
Molecular analysis of rZBP1. rZBP1 was cloned from embryonic rat hippocampus using PCR (see Materials and Methods), and its sequence has been entered into GenBank (accession number AF541940). A, rZBP1 structural elements. B, Western blot analysis of recombinant rZBP1 (lane 1) and endogenous rZBP1 from E18 rat hippocampus (lane 2) showed a single band at ∼69 kDa using a rabbit polyclonal antibody to cZBP1. C, An mRNA protein complex was indicated between the β-actin zipcode and rat brain protein extracts using EMSA (lane 3), which was not observed in the absence of extract (lane 1). The control probe migrated to the bottom of the gel in the presence (lane 4) or absence of protein extract (lane 2). D, Formation of an mRNA protein complex between zipcode and recombinant rZBP1 was significantly inhibited by increasing amounts of unlabeled probes.
Western blot analysis from E18 rat hippocampal extracts, using a rabbit polyclonal antibody to chick ZBP1, recognized a 69 kDa protein (Fig. 1B, lane 2) analogous to the molecular weight of chick ZBP1. Similarly, recombinant rZBP1 expressed in bacteria also showed a single band of 69 kDa (Fig. 1B, lane 1). To demonstrate that rZBP1 formed a sequence-specific mRNP complex with the rat β-actin zipcode in vitro, EMSAs were performed with both brain extracts and recombinant rZBP1. When the labeled zipcode was used as a probe, a distinct complex was observed (Fig. 1C, lane 3), which migrated significantly more slowly than radiolabeled probes incubated without protein extract (lanes 1, 2) or brain extract incubated with probe to a mutated sequence (lane 4). A similar band-shift complex was observed with recombinant rZBP1 that could be competed by excess amount of unlabeled zipcode (Fig. 1D). These results indicate that rZBP1 can form a complex with the β-actin zipcode in vitro.
Localization of rZBP1 granules in dendrites and actin-rich protrusions of cultured hippocampal neurons
A previous immunofluorescence study in rat hippocampal neurons has shown that ZBP1 was distributed in dendrites in the form of granules, which were prominent in both the microtubule-rich dendritic shaft and F-actin-rich protrusions, both filopodia and spines (Tiruchinapalli et al., 2003) (Fig. 2A). To specifically investigate the localization of the cloned rZBP1, we fused it to EGFP and transiently transfected cultured hippocampal neurons. EGFP-rZBP1 was localized in granules that were distributed throughout dendrites and into F-actin-rich protrusions (Fig. 2B; a, b, insets, higher magnification). In the same cell, immunofluorescence localization of MAP2 (Fig. 2C, blue) was used as a marker for the dendritic shaft, and F-actin protrusions were identified using tetramethylrhodamine isothiocyanate (TRITC)-phalloidin (Fig. 2C, red). Higher magnification of two dendritic regions of interest (Fig. 2B,C; a, b, insets, arrows) showed EGFP-rZBP1 granules within F-actin-rich protrusions that extended beyond the microtubule-rich dendritic shaft.
Figure 2.
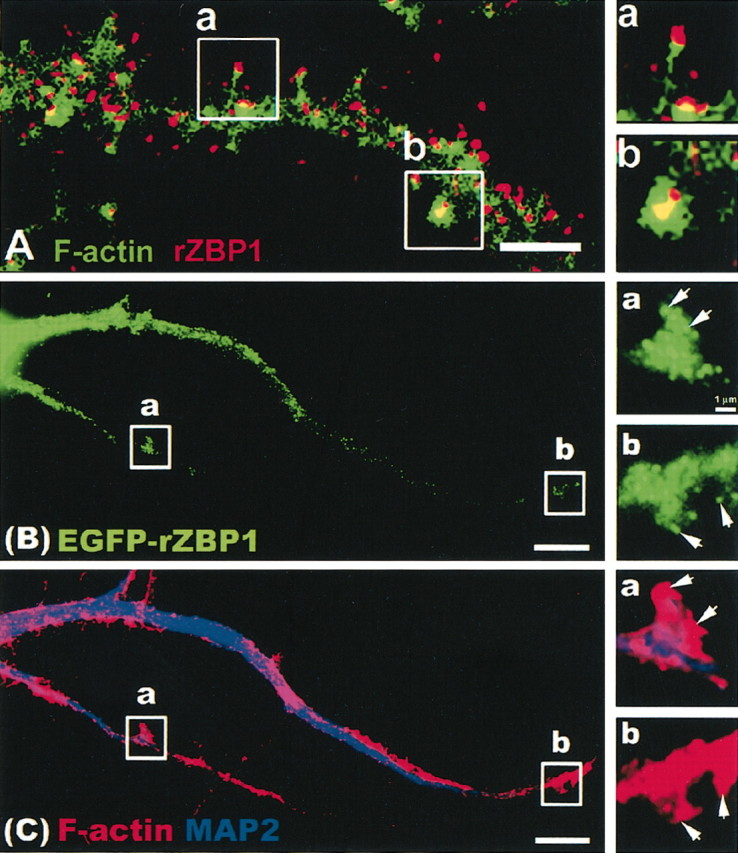
Dendritic localization of endogenous rZBP1 and EGFP-rZBP1. A, Immunofluorescence detection of ZBP1 (red) using a polyclonal antibody. F-actin was detected in the same neuron using phalloidin (Alexa 488), and images were superimposed. Two segments of interest (a,b) were enlarged (right) to show ZBP1 granules in the tip of a long, thin filopodial protrusion (a) and a spine-like, bulbous or headed protrusion (b). Scale bar, 2.5 μm. B, Transfection of EGFP-rZBP1 in cultured hippocampal neurons followed by immunofluorescence detection (C) of MAP2 (blue) and F-actin using TRITC-labeled phalloidin (red). B, C, Two dendritic segments (a,b) are enlarged (right) showing the EGFP-rZBP1 signal (B) and MAP2 and phalloidin signals (C). Arrows denote localization of EGFP-rZBP1 granules in F-actin-rich structures that protrude from the dendritic shaft. Scale bar, 10 μm.
rZBP1 is necessary for β-actin mRNA localization in dendrites of cultured neurons
To determine whether rZBP1 was required for β-actin mRNA localization in dendrites, morpholino antisense oligonucleotides were used to knock-down rZBP1, and then β-actin mRNA localization was analyzed by in situ hybridization. Western blot analysis with anti-ZBP1 antibody normalized to tubulin levels (as a loading control) demonstrated a 54% decrease in rZBP1 levels after antisense treatment (Fig. 3A,B). To show that rZBP levels were also reduced in dendrites, rZBP1 immunofluorescence intensities were measured by quantitative digital imaging microscopy. A 52% decrease in mean pixel intensities for ZBP1 was observed by immunofluorescence after antisense treatment (Fig. 3C,G,H). Fluorescence in situ hybridization (FISH) to β-actin mRNA showed a 63% decrease in mean pixel intensities after antisense treatment (Fig. 3D,E,F). As a control, CaMKIIα mRNA levels in dendrites were not reduced (Fig. 3D), consistent with the absence of homology between the β-actin zipcode and CaMKIIα mRNA localization sequences (Mori et al., 2000). These data indicate that rZBP1 is necessary for β-actin mRNA localization in dendrites and suggest that one level of regulation may be ZBP1 expression or availability.
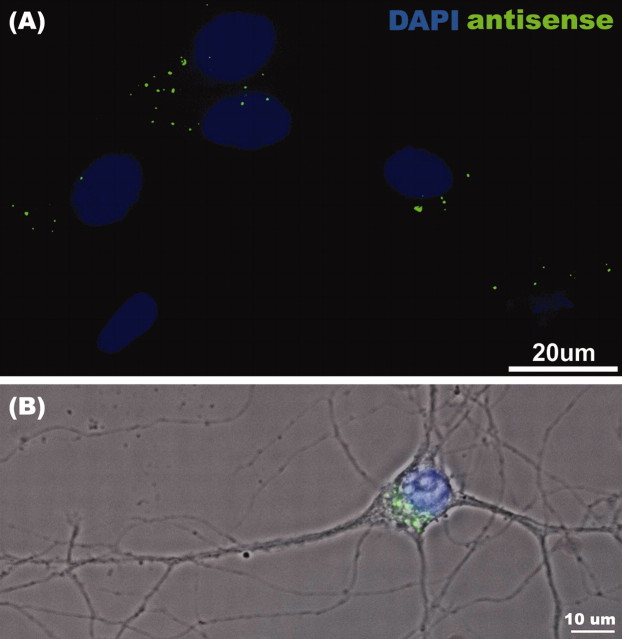
To confirm that the morpholino antisense oligonucleotides were efficient in the entry of most cells, we cotransfected morpholino antisense, which was conjugated with FITC as a tracer (∼10% of total antisense). More than 80% of the analyzed neurons demonstrated the presence of morpholiono oligonucleotides in the cytoplasm, as judged by random scoring of several hundred cells for the presence or absence of FITC signal (Fig. 4A,B). These results show that morpholino antisense is an effective tool to knock-down rZBP1 levels in most cultured cells and is consistent with our ability to detect a significant reduction in ZBP1 expression by Western blot analysis.
Figure 4.
Efficient entry of morpholino antisense oligonucleotides into cultured hippocampal neurons. Cultured neurons were transfected with morpholino oligonucleotides (as described in Materials and Methods) with the inclusion of an FITC-conjugated tracer oligonucleotide. A, Cells were fixed and stained with 4′,6′-diamidino-2-phenylindole (DAPI) to reveal nuclei; note five of six cells with FITC labeling in the cytoplasm. B, Higher magnification reveals the presence of FITC-morpholino antisense oligonucleotide in the perinuclear cytoplasm by overlay of the fluorescence and phase images. DAPI-stained nucleus is shown in blue.
rZBP1-mediated localization of β-actin mRNA in dendrites facilitates filopodial growth
Antisense-treated cells (48 hr) were examined morphologically to determine whether there was a phenotype that resulted from a knock-down of rZBP1. Phalloidin labeling suggested a reduction in the density of actin-rich filopodia in neurons treated with antisense, although these results were not statistically significant (data not shown). We then asked whether antisense-treated cells would show impaired growth of filopodia in response to neurotrophin exposure. It has been well established that neurotrophin signaling through tyrosine receptor kinase receptors can stimulate actin polymerization in cultured forebrain neurons (Bassell et al., 1999) and actin-dependent growth of dendritic filopodia and spines in cultured cerebellar neurons (Shimada et al., 1998). In cultured hippocampal neurons, we observed that 2 hr of exposure to BDNF resulted in a 127% increase in the density of F-actin-rich protrusions along dendrites (Fig. 5A,B,E). However, in cultures that were first exposed to rZBP1 antisense oligonucleotides for 48 hr, the BDNF-induced increase in the density of F-actin-rich protrusions was completely inhibited (Fig. 5D,F). In contrast, neurons treated with control oligonucleotides (reverse sequence) exhibited the normal BDNF-induced increase in the density of F-actin protrusions (Fig. 5C,F). These results suggest that under limiting amounts of ZBP1 and β-actin mRNA in dendrites, neurons have impaired ability to elicit robust actin growth in response to BDNF.
We next investigated whether overexpression of β-actin mRNA could stimulate growth of filopodia and spines. The strategy was to transfect EGFP-β-actin and determine whether the presence of the β-actin zipcode could affect the density of actin-rich protrusions along dendrites. We hypothesized that although overexpression of EGFP-β-actin would result in more β-actin mRNA within cells, only by insertion of the zipcode would there be more localized β-actin mRNA. We predicted that the endogenous rZBP1 would bind not only endogenous β-actin mRNA but also the EGFP-β-actin fusion mRNA promoting its localization. Such overexpression of β-actin mRNA (endogenous plus EGFP fusion mRNA) would be evident by increased levels of rZBP1 in dendrites.
Insertion of the β-actin zipcode into the 3′UTR of the EGFP-β-actin mRNA, after the stop codon, resulted in a marked increase in the total density of protrusions along dendrites (Fig. 6D), compared with neurons transfected with EGFP-β-actin lacking the zipcode (Fig. 6C). The vast majority of protrusions at this stage in culture are filopodia. Quantitative analysis of the density of total surface protrusions showed a 75% increase in their density when EGFP-β-actin with the zipcode was compared with the construct lacking the zipcode (Fig. 6E). Overexpression of EGFP-β-actin without the zipcode was also observed to promote an increase in protrusion density compared with EGFP alone. However, this increase was significantly less than that observed when the zipcode was present (Fig. 6C,E).
We also examined the effects of an EGFP construct with the β-actin zipcode subcloned into the 3′UTR of the EGFP vector. Because this construct encodes only EGFP, it might not be expected to result in a morphologic phenotype and perhaps might be expected to produce a dominant negative effect. However, because the insertion of the β-actin zipcode was sufficient to localize an EGFP reporter mRNA (Kislauskis et al., 1994), one might also envision that such localized EGFP mRNAs could recruit other endogenous mRNAs and proteins that could produce a morphologic effect. Here, we observed that EGFP containing the zipcode resulted in a 158% increase in the density of protrusions compared with EGFP alone (Fig. 6A,B,E). These results demonstrate the dramatic effect of a single cis-acting, untranslated element on dendritic morphology.
Although an increased protrusion density was observed with either the EGFP-zipcode or EGFP-β-actin-zipcode constructs, we note that the protrusions were significantly longer when actin was present (Fig. 6B,D,G). We observed a 65% increase in protrusion length after overexpression of actin, compared with EGFP-transfected neurons (Fig. 6G). However, an analysis of protrusion length did not reveal any role for the zipcode. Protrusion length was similar in neurons transfected with EGFP-β-actin, with or without the zipcode (Fig. 6G). Protrusion length was also similar in neurons transfected with EGFP, with or without the zipcode (Fig. 6G). These data show that overexpression of actin can increase both protrusion density and length, whereas the zipcode can act only to increase the density of filopodial protrusions.
BDNF stimulation has been shown to increase the density of dendritic filopodia and spines (Shimada et al., 1998). Here we show that neurons transfected with EGFP exhibited an 84% increase in protrusion density after BDNF treatment (Fig. 6E). However, neurons that were transfected with actin, which also resulted in increased protrusion density (Fig. 6C,E), did not show any additional increase in their density after BDNF treatment (Fig. 6E). This suggests an occlusion whereby BDNF exhibits a weakened response in neurons already having very high levels of actin. Because BDNF is known to enhance actin expression, this is consistent with the cell being saturated after overexpression of actin and unable to exhibit a response to BDNF. Expression of zipcode-containing constructs (EGFP or EGFP-β-actin) also did not show an additional increase in protrusion density after BDNF treatment. Hence, overexpression of localized β-actin mRNA may occlude any additional additive effects by BDNF, possibly because there may be common pathways involved, because BDNF can enhance actin expression and promote endogenous β-actin mRNA localization.
Analysis of both the density and morphology of dendritic spines revealed a zipcode-dependent increase in the density of filopodial protrusions that were in contact with synapsin-labeled terminals (Fig. 7). We refer to these synapses as filopodial synapses (Fiala et al., 1998), which have also been referred to as filopodia spines (Okabe et al., 2001) or headless spines (Papa et al., 1995). Using this morphologic criteria (Fig. 7A,B, insets), the zipcode significantly increased the density of filopodial synapses after transfection of EGFP-β-actin (Fig. 7B,C). Although filopodial synapses were noted in both constructs (Fig. 7A,B), the zipcode containing EGFP-β-actin construct resulted in a marked increase in their density (Fig. 7C).
In contrast, the zipcode did not have an effect on the density of mature spines having a bulbous or headed morphology (Fig. 6F). An increased density of headed spines was observed after BDNF treatment; however, insertion of the zipcode did not result in any additional increase in their density (Fig. 6F). In neurons transfected with actin, BDNF treatment increased the density of headed spines (Fig. 6F), a result that contrasted with the occlusion of a BDNF response in the modulation of filopodial density (Fig. 6E). These results suggest separate pathways involved in modulation of filopodia from those that regulate formation of a mature headed spine.
Overexpression of the β-actin zipcode promoted localization of endogenous ZBP1 and β-actin mRNA into dendrites
We investigated how ovexpression of an EGFP reporter with the 3′UTR β-actin zipcode increased the density of filopodial protrusions, compared with EGFP alone (Fig. 6A,B,E). We hypothesized that ZBP1 trafficking to processes might be enhanced by overexpression of zipcode-containing mRNAs. Because the localization machinery would not discriminate between endogenous β-actin mRNA and EGFP mRNA containing the zipcode, both mRNAs would be packaged into granules and localized. To test this hypothesis, we designed oligonucleotide probes to β-actin mRNA at regions outside the zipcode for in situ hybridization to neurons that were transfected with either EGFP or EGFP-zipcode (Fig. 8A). Because the probe cannot hybridize to the EGFP-zipcode mRNA, an increased signal by FISH would indicate enhanced localization of endogenous β-actin mRNA. Quantitative digital imaging analysis revealed a 112% increase in the mean fluorescence intensity for β-actin mRNA in neurons transfected with EGFP-zipcode compared with EGFP alone (Fig. 8A). We also observed increased levels of endogenous ZBP1 within dendrites after overexpression of zipcode-containing EGFP constructs. A 167% increase in ZBP1 levels in dendrites was observed when EGFP-zipcode was compared with EGFP (Fig. 8B). A 206% increase in ZBP1 levels in dendrites was observed when EGFP-β-actin-zipcode was compared with EGFP-β-actin (Fig. 8B). Such observations of a more robust localization response with ZBP1 than β-actin mRNA suggest the possible recruitment of other mRNAs with ZBP1, which could also contribute to any morphologic effect involving overexpression of zipcode-containing mRNAs.
Western blot analysis did not reveal any increases in EGFP-β-actin levels from the insertion of the zipcode (Fig. 9A). This provides additional support for our conclusion that the zipcode-dependent increase in protrusion density was attributable to ZBP1-mediated localization of β-actin mRNA, rather than increased production of β-actin protein. In addition, there was no evidence of an increase in total levels of ZBP1 (Fig. 9B) after overexpression of the zipcode, suggesting that the increase in ZBP1 immunofluorescence intensity in dendrites (Fig. 8B) was attributable to localization of pre-existing ZBP1 from the cell body after expression of EGFP-β-actin fusion mRNA that contained the zipcode.
Figure 9.
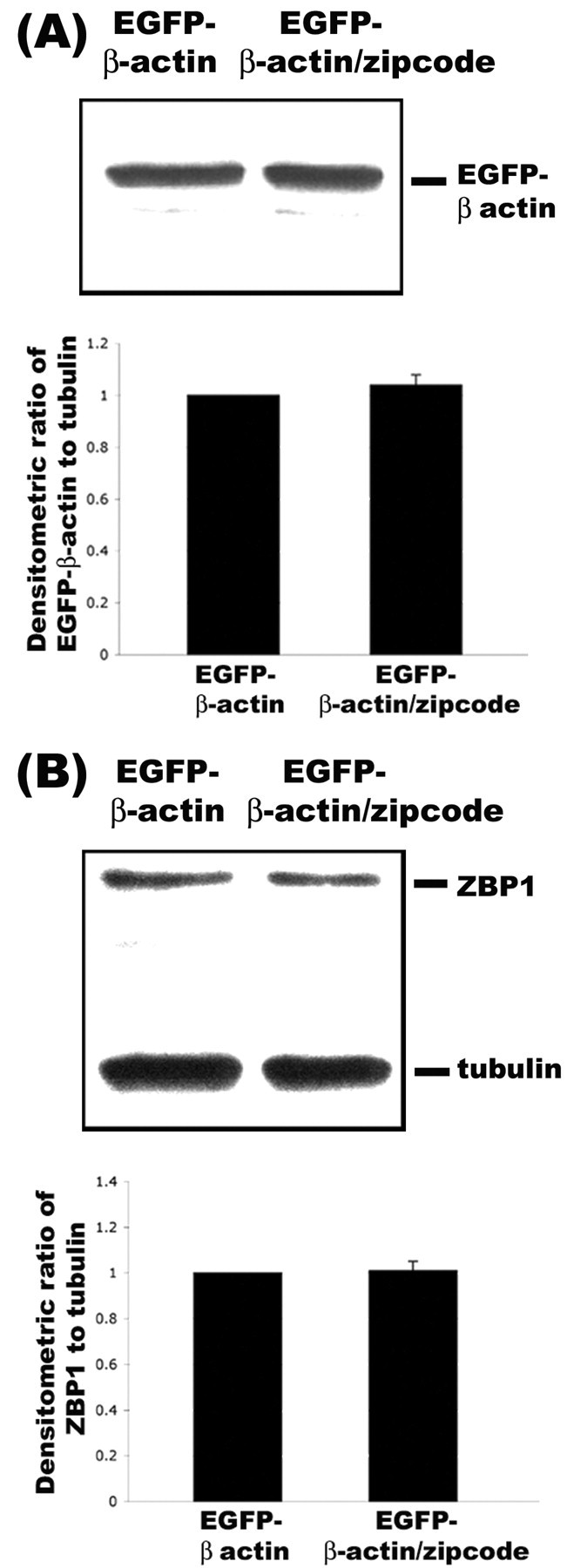
Expression levels of EGFP-β-actin were not dependent on the presence of zipcode. A, EGFP-β-actin constructs without the zipcode (lane 1) and with the zipcode (lane 2) were transfected into human embryonic kidney 293 cells and processed by Western blot analyses with antibody to EGFP. Quantitative analysis of EGFP levels relative to tubulin using densitometry did not reveal any differences in their levels (bottom). B, From the same lysates, antibodies to ZBP1 were also used for Western blot analysis. No changes in ZBP1 levels were observed relative to tubulin as a loading control (bottom).
Discussion
Requirement of specific mRNA-binding proteins in mRNA localization
The molecular mechanism of mRNA localization in many polarized cells, such as oocytes and neurons, appears to involve the recognition of cis-acting sequences by mRNA-binding proteins, which may link the mRNA with cytoskeletal-based transport machinery and direct their targeting to distinct intracellular compartments (Kloc et al., 2002). Several proteins have been shown to bind localization sequences within the 3′UTR. These include ZBP1 (Ross et al., 1997; Zhang et al., 2001; Farina et al., 2003) and ZBP2 (Gu et al., 2002) binding to the 54 nt β-actin zipcode, Vg1 RNA binding protein (VgRBP)-VgLE binding and endoplasmic reticulum association (VERA) binding to the localization element of Vg1 RNA (Deshler et al., 1998; Havin et al., 1998), hnRNP A2 binding to the 21 nt RNA transport sequence of MBP mRNA (Hoek et al., 1998), and MAP2 RNA transacting factor binding to the dendritic targeting element of MAP2 mRNA (Rehbein et al., 2002). Mutations in cis-acting elements that disrupt protein binding have also been shown to impair mRNA localization in fibroblasts (Ross et al., 1997), oligodendrocytes (Munro et al., 1999), and Xenopus oocytes (Cote et al., 1999). Such correlative analysis suggests that these interactions may be required for mRNA localization. Antisense oligonucleotides directed to the β-actin zipcode impaired the formation of the zipcode-ZBP1 complex in vitro and also β-actin mRNA localization when applied to cultured chick forebrain neurons (Zhang et al., 2001). Transfection of truncated forms of ZBPs that possess the minimal KH domains required for zipcode binding were shown to act as a dominant negative and inhibit β-actin mRNA localization in fibroblasts (Gu et al., 2002; Farina et al., 2003). One interpretation of these data is that the KH domains sequester the endogenous β-actin mRNAs and prevent their localization, because KH domains may be necessary for zipcode binding but not sufficient for localization. However, it is also possible that these truncated proteins interfere with many other proteins that interact with endogenous ZBP1, and observations of impaired β-actin mRNA localization may be indirect and not unequivocally indicative of a direct role for ZBP1.
A direct approach for demonstrating the requirement for ZBP1 in β-actin mRNA localization is to investigate the effects of ZBP1 knock-down using antisense oligonucleotides. Here, we provide quantitative evidence for knock-down of rZBP1 levels by Western blot and immunofluorescence. This treatment reduced β-actin mRNA localization in dendrites but did not affect CaMKIIα mRNA, whose localization sequences lack homology to the zipcode. In addition, we showed that overexpression of zipcode-containing constructs increased the dendritic localization of rZBP1. The combined use of these approaches demonstrated a role for a zipcode-ZBP1 interaction in β-actin mRNA localization in neurons. These studies have important implications for the study of other dendritically localized mRNAs. Several mRNAs have been localized in dendrites, which include MAP2 (Garner et al., 1988; Kleiman et al., 1990), the α subunit of CaMKIIα (Burgin et al., 1990; Mayford et al., 1996), and the activity-regulated cytoskeletal-associated protein (Lyford et al., 1995). Although the molecular mechanism of dendritic mRNA localization has been shown to involve cis-acting sequences in the 3′UTR (Mayford et al., 1996; Blichenberg et al., 1999, 2001; Mori et al., 2000), the requirement of specific mRNA-binding proteins in facilitating the localization of these mRNAs is unclear. Recent work indicated that the mRNA-binding protein, CPEB, can facilitate the localization of mRNAs containing the cytoplasmic polyadenylation sequence, such as that found in CaMKIIα mRNA (Huang et al., 2003).
The ZBP1 family of KH domain-containing RNA-binding proteins
cZBP1 was originally isolated from chick fibroblasts by using its affinity to the β-actin zipcode (Ross et al., 1997). Here, we cloned the rat orthologue of ZBP1 that has >95% identity to cZBP1. ZBP1 is a member of a family of KH domain-containing proteins that has been shown in other cell types to be involved in mRNA localization, translation, and stability (Farina and Singer, 2002). The Xenopus homologs, VgRBP (Havin et al., 1998) or Vera (Deshler et al., 1998), bind to the Vg1 mRNA localization sequence required for targeting to the vegetal pole of oocytes. These proteins share 75.9% identity to ZBP1. ZBP1 is also highly related to CRDBP, which binds a coding region determinant involved in stabilization of c-myc mRNA (Doyle et al., 1998), and also IGF-II mRNA-binding proteins involved in translational repression (Nielson et al., 1999). All ZBP1 homologs share two KH didomains, two RRM didomains, and nuclear localization and export sequences. These proteins are believed to act as shuttling proteins by first forming mRNP complexes in the nucleus that are then exported to specific cytoplasmic sites in granules (Farina et al., 2002; Oleynikov and Singer, 2003).
Function of β-actin mRNA localization to influence actin morphology and dynamics during neuronal development
Disruption of the interaction between cZBP1 and the β-actin zipcode using antisense decreased the number of actin nucleation sites at the fibroblast leading edge and impaired cell motility (Shestakova et al., 2001). Antisense to the zipcode also prevented localization of β-actin mRNA in response to neurotrophin-3 (NT-3), which resulted in reduced β-actin protein levels in growth cones and impaired growth cone extension (Zhang et al., 2001). These results point to a specific role for β-actin mRNA localization in localized protein synthesis that affects actin polymerization and cell motility.
The present study provides new insight into the function of β-actin mRNA localization to dendrites. We show that the density of filopodia and filopodial synapses were modulated by ZBP1-mediated localization of β-actin mRNA. The ability of a growth factor, BDNF, to stimulate filopodial growth was dramatically impaired in cells that had reduced dendritic levels of ZBP1. We speculate that ZBP1 antisense-treated cells have reduced levels of G-actin pools in dendrites that are available to sustain F-actin polymerization and assembly needed to grow new filopodia and maintain an increased density of these protrusions in response to stimulation by growth factors. Neurons may be especially dependent on a local supply of newly synthesized β-actin monomers because of the long distances that actin would have to be transported from the cell body. Although it has been established that actin is a component of slow axonal transport (and possibly in analogous manner in dendrites) (Mills et al., 1996), this does not a priori imply that actin delivered in this manner should be readily recruitable throughout the dendritic arbor in response to signals that elicit robust nucleation of actin filament assembly in filopodia.
One way to distinguish the affect of somatic transport versus local synthesis would be to compare β-actin-EGFP with and without the mRNA localization sequence. Previous work has shown that expression of β-actin-EGFP, lacking the complete β-actin 3′UTR, did not impair the localization of β-actin-EGFP protein to dendritic spines (Kaech et al., 1997). Because this construct lacked the reported β-actin mRNA localization sequence, actin monomers must have been delivered into dendrites and spines post-translationally. Here, we show that insertion of the 54 nt β-actin zipcode markedly increased the density of filopodial protrusions in neurons transfected with EGFP-β-actin. Furthermore, our data also show that the total amount of actin in the neuron may not be the only driving force behind the regulation of filopodial density. It was striking that overexpression of just the 3′UTR zipcode in a plasmid encoding only EGFP protein was able to increase filopodial density more than achieved by overexpression of EGFP-β-actin itself (without zipcode). These results demonstrate that a single untranslated element can have a marked influence on filopodial density. Our results suggest that this response was mediated in part by the recruitment of endogenous β-actin mRNA into dendrites. Additional work is needed to determine how overexpression of EGFP-zipcode mRNA might also promote the recruitment of other mRNAs that may be localized by rZBP1. It is possible that the mechanism of ZBP1 facilitated mRNA localization from the cell body into the process may be regulated by the amount or availability of zipcode-containing mRNAs. Overexpression of EGFP mRNAs that harbor a zipcode sequence might shift a population of unbound ZBP1 to bind mRNAs and promote their localization. ZBP1 localization machinery would not discriminate between endogenous β-actin mRNA and EGFP-zipcode mRNAs; both mRNAs would be packaged into granules and targeted into the process.
Developmental functions for zipcode-binding protein
Our studies on ZBP1 suggest a broad range of functions for β-actin mRNA localization in the regulation of actin morphology during neuronal development. During the first week of neurite outgrowth in culture, localization of ZBP1 and β-actin mRNA was strongly correlated with the enrichment of β-actin protein within growth cones after NT-3 stimulation and contributed to the regulated dynamics and motility of growth cones (Zhang et al., 1999, 2001). ZBP1 and β-actin mRNA were localized to growth cones of both the minor neurites and incipient axon, with the later showing a more prominent enrichment. In this study, we demonstrated that rZBP1 is required for the dendritic localization of β-actin mRNA and elucidated its function in regulation of the density of filopodial protrusions and filopodial synapses. Regulation of β-actin mRNA localization and local translation may be an important mechanism during development to influence synaptic growth via the formation of dendritic filopodia. Although β-actin mRNA localization has not yet been detected in dendrites in vivo, we note that most in situ hybridization studies have used sections from adult brain to reveal dendritic mRNAs (Steward and Schuman, 2001). It has long been known that polyribosomes are enriched at postsynaptic sites during synaptogenesis in vivo (Steward and Falk, 1986). It will be of interest to know whether local synthesis of β-actin protein occurs in dendrites during this developmental period. Our findings of a role for β-actin mRNA localization, in modulation of dendritic filopodia and synaptic growth in a cultured neuron model, provide motivation to look for these and other mRNAs in dendrites at earlier developmental stages in vivo. We suggest that zipcode-binding protein may play a key role in the developmental regulation of many localized mRNAs, the local translation of which may collectively impact F-actin cytoskeletal dynamics essential for process growth, synaptogenesis, and developmental plasticity.
Footnotes
This work was supported by National Institutes of Health RO1 Grants HD46368-06 and NS39641 to G.J.B. and AR41480 to R.H.S. We thank Joel Yisraeli for suggestions on the use of morpholino antisense, Kim Farina for antibody to zipcode-binding protein, and Stefan Huttelmaier for helpful discussion.
Correspondence should be addressed to Dr. Gary Bassell, Department of Neuroscience, Albert Einstein College of Medicine, 1410 Pelham Parkway, Bronx, NY 10461. E-mail: bassell@aecom.yu.edu.
T. Eom's present address: Laboratory of Molecular Neurooncology, Rockefeller University, New York, NY 10021.
Copyright © 2003 Society for Neuroscience 0270-6474/03/2310433-12$15.00/0
References
- Bassell GJ, Zhang HL, Byrd AL, Femino AM, Singer RH, Taneja KL, Lifshitz LM, Herman IM, Kosik KS ( 1998) Sorting of beta actin mRNA and protein to neurites and growth cones in culture. J Neurosci 18: 251-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassell GJ, Oleynikov Y, Singer RH ( 1999) The travels of mRNAs through all cells large and small. FASEB J 13: 447-454. [DOI] [PubMed] [Google Scholar]
- Blichenberg A, Schwanke B, Rehbein M, Garner CC, Richter D, Kindler S ( 1999) Identification of a cis-acting dendritic targeting element in MAP2 mRNAs. J Neurosci 19: 8818-8829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blichenberg A, Rehbein M, Muller R, Garner CC, Richter D, Kindler S ( 2001) Identification of a cis-acting dendritic targeting element in the mRNA encoding the alpha subunit of CaMKII. Eur J Neurosci 13: 1-9. [DOI] [PubMed] [Google Scholar]
- Burgin KE, Waxham MN, Rickling S, Westgate SA, Mobley WC, Kelly PT ( 1990) In situ hybridization histochemistry of Ca2+/Calmodulin-dependent protein kinase in developing rat brain. J Neurosci 10: 1788-1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cailliau P, Yuste R ( 2001) On the function of dendritic filopodia. Rev Neurol 33: 1158-1166. [PubMed] [Google Scholar]
- Cooper MW, Smith SJ ( 1992) A real time analysis of growth cone target cell interactions during the formation of stable contacts between hippocampal neurons in culture. J Neurobiol 23: 814-828. [DOI] [PubMed] [Google Scholar]
- Cote CA, Gautreau D, Denegre JM, Kress TL, Mowry KL ( 1999) A Xenopus protein related to hnRNP I has a role in cytoplasmic RNA localization. Mol Cell 4: 431-437. [DOI] [PubMed] [Google Scholar]
- Deshler JO, Highett MI, Schnapp BJ ( 1998) A highly conserved RNA binding protein for cytoplasmic mRNA localization in vertebrates. Curr Biol 8: 489-496. [DOI] [PubMed] [Google Scholar]
- Doyle GA, Betz NA, Leeds PF, Fleisig AJ, Prokipcak RD, Ross J ( 1998) The c-myc coding region determinant-binding protein: a member of a familiy of KH domain RNA-binding proteins. Nucleic Acid Res 26: 5036-5044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunaevsky A, Tashiro A, Majewska A, Mason C, Yuste R ( 1999) Developmental regulation of spine motility in the mammalian central nervous system. Proc Natl Acad Sci USA 96: 13438-13443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farina KL, Singer RH ( 2002) The nuclear connection in RNA transport and localization. Trends Cell Biol 12: 466-472. [DOI] [PubMed] [Google Scholar]
- Farina K, Huttelmaier S, Musunuru K, Darnell R, Singer RH ( 2003) Two ZBP1 KH domains facilitate β-actin mRNA localization, granule formation and cytoskeletal attachment. J Cell Biol 160: 77-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiala JC, Feinberg M, Popov V, Harris KM ( 1998) Synaptogenesis via dendritic filopodia in developing hippocampal area CA1. J Neurosci 18: 8900-8911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer M, Kaech S, Knutti D, Matus A ( 1998) Rapid actin based plasticity in dendritic spines. Neuron 20: 847-854. [DOI] [PubMed] [Google Scholar]
- Fischer M, Kaech S, Wagner U, Brinkhaus H, Matus A ( 2000) Glutamate receptors regulate actin-based plasticity in dendritic spines. Nat Neurosci 3: 887-894. [DOI] [PubMed] [Google Scholar]
- Garner CC, Tucker RP, Matus A ( 1988) Selective localization of mRNA for cytoskeletal protein MAP2 in dendrites. Nature 336: 674-679. [DOI] [PubMed] [Google Scholar]
- Goslin K, Asmussen H, Banker G ( 1998) Rat hippocampal neurons in low density culture. In: Culturing nerve cells (Banker G, Golsin K, eds), pp 339-371. Cambridge, MA: MIT.
- Gu W, Pan F, Zhang HL, Bassell GJ, Singer RH ( 2002) A predominantly nuclear protein affecting cytoplasmic localization of b-actin mRNA in fibrobalsts and neurons. J Cell Biol 156: 41-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halpain S, Hipolito A, Saffer L ( 1998) Regulation of F-actin stability in dendritic spines by glutamate receptors and calcineurin. J Neurosci 18: 9835-9844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havin L, Git A, Elisha Z, Oberman F, Yaniv K, Schwartz SP, Standart N, Yisraeli JK ( 1998) RNA binding protein conserved in both microtubule and microfilament based RNA localization. Genes Dev 12: 1593-1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoek KS, Kidd GJ, Carson JH, Smith R ( 1998) hnRNPA2 selectively binds the cytoplasmic transport sequence of MBP mRNA. Biochemistry 37: 7021-7029. [DOI] [PubMed] [Google Scholar]
- Huang YS, Carson JH, Barbarese E, Richter JD ( 2003) Facilitation of dendritic mRNA transport by CPEB. Genes Dev 17: 638-653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaech S, Fischer M, Doll T, Matus A ( 1997) Isoform specificity in the relationship of actin to dendritic spines. J Neurosci 17: 9565-9572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kislauskis E, Zhu X, Singer RH ( 1997) β-actin messenger RNA localization and protein sythesis augment cell motility. Cell Biol 136: 1263-1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleiman R, Banker G, Steward O ( 1990) Differential subcellular localization of particular mRNAs in hippocampal neurons in culture. Neuron 5: 821-830. [DOI] [PubMed] [Google Scholar]
- Kloc MK, Zearfoss NR, Etkin LD ( 2002) Mechanisms of subcellular mRNA localization. Cell 108: 533-544. [DOI] [PubMed] [Google Scholar]
- Lyford GL, Yamagata K, Kaufman WE, Barnes CA, Sanders LK, Copeland NG, Gilbert DJ, Jenkins NA, Lanahan AA, Worley PF ( 1995) ArC, a growth factor and activity related gene encodes a novel cytoskeletal protein that is enriched in neuronal dendrites. Neuron 14: 433-445. [DOI] [PubMed] [Google Scholar]
- Mayford M, Baranes D, Podsypanina K, Kandel ER ( 1996) The 3′-untranslated region of CaMKIIα is a cis acting signal for the localization and translation of mRNA in dendrites. Proc Natl Acad Sci USA 93: 13250-13255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micheva KD, Vallee A, Beaulieu C, Herman IM, Leclerc N ( 1998) Beta-actin is confined to structures having high capacity of remodelling in developing and adult rat cerebellum. Eur J Neurosci 10: 3785-3798. [DOI] [PubMed] [Google Scholar]
- Mills RG, Minamide LS, Yuan A, Bamburg JR, Bray JJ ( 1996) Slow axonal transport of soluble actin with ADF, cofilin and profilin suggest actin moves in an unassembled form. J Neurochem 67: 1225-1234. [DOI] [PubMed] [Google Scholar]
- Mori Y, Imaizumi K, Katayama T, Yoneda T, Tohyama M ( 2000) Two cis-acting elements in the 3′UTR of CaMKIIa regulate its dendritic targeting. Nat Neurosci 3: 1079-1084. [DOI] [PubMed] [Google Scholar]
- Munro TP, Magee RJ, Kidd GJ, Carson JH, Barbarese E, Smith LM, Smith R ( 1999) Mutational analysis of a heterogeneous nuclear ribonucleoprotein A2 response element for RNA trafficking. J Biol Chem 274: 34389-34395. [DOI] [PubMed] [Google Scholar]
- Nielson J, Wewer UM, Nielson FC ( 1999) A family of insulin like growth factor mRNA binding proteins repress translation late in development. Mol Cell Biol 19: 1262-1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okabe S, Miwa A, Okado H ( 2001) Spine formation and correlated assembly of presynaptic and postsynaptic molecules. J Neurosci 21: 6105-6114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oleynikov Y, Singer RH ( 2003) Real-time visualization of ZBP1 association with β-actin mRNA during transcription and localization. Curr Biol 13: 199-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papa M, Bundman MC, Greenberger V, Segal M ( 1995) Morphological analysis of dendritic spine development in primary cultures of hippocampal neurons. J Neurosci 15: 1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehbein M, Wege K, Buck F, Schweizer M, Richter D, Kindler S ( 2002) Molecular characterization of MARTA1, a protein interacting with the dendritic targeting element of MAP2 mRNAs. J Neurochem 82: 1039-1046. [DOI] [PubMed] [Google Scholar]
- Ross A, Oleynikov Y, Kislauskis E, Taneja K, Singer R ( 1997) Characterization of a β-actin mRNA zipcode-binding protein. Mol Cell Biol 17: 2158-2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shestakova EA, Singer RH, Condeelis J ( 2001) The physiological significance of b-actin mRNA localization in determining cell polarity and directional motility. Proc Natl Acad Sci USA 98: 7045-7050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada A, Mason CA, Morrison ME ( 1998) TrkB signaling modulates spine density and morphology independent of dendrite structure in cultured Purkinje cells. J Neurosci 18: 8559-8570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuster CB, Herman IM ( 1995) Indirect association of ezrin with F-actin: isoform specificity and calcium sensitivity. J Cell Biol 128: 837-848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steward O, Schuman EM ( 2001) Protein synthesis at synaptic sites on dendrites. Annu Rev Neurosci 24: 299-325. [DOI] [PubMed] [Google Scholar]
- Tiruchinapalli DM, Oleynikov Y, Kelic S, Shenoy SM, Hartley A, Stanton PK, Singer RH, Bassell GJ ( 2003) Activity-dependent trafficking and dynamic localization of zipcode-binding protein 1 and beta-actin mRNA in dendrites and spines of hippocampal neurons. J Neurosci 23: 3251-3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang HL, Singer RH, Bassell GJ ( 1999) Neurotrophin regulation of beta-actin mRNA and protein localization within growth cones. J Cell Biol 147: 59-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang HL, Eom T, Oleynikov Y, Shenoy SM, Lieblet DA, Dictenberg JB, Singer RH, Bassell GJ ( 2001) Neurotrophin induced transport of b-actin mRNP complex increases β-actin levels and stimulates growth cone motility. Neuron 31: 261-275. [DOI] [PubMed] [Google Scholar]
- Zhang W, Benson DL ( 2001) Stages of synapse development defined by dependence on F-actin. J Neurosci 21: 5169-5181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziv NE, Smith SJ ( 1996) Evidence for a role of dendritic filopodia in synaptogenesis and spine formation. Neuron 17: 91-102. [DOI] [PubMed] [Google Scholar]