Abstract
Rho family of small GTPases are key regulators for morphological changes of neurons on the basis of reorganization of the actin cytoskeleton. Rnd1, a novel member of this family, is predominantly expressed in neurons in brain; however, the neuronal functions of Rnd1 are not known. Here we investigated the effect of Rnd1 on neuronal morphology. Northern blot analysis of Rnd1 expression in rat brain showed that Rnd1 mRNA was highly expressed during early postnatal period, the synaptogenic stage. In situ hybridization analysis at this period revealed that Rnd1 mRNA was strongly expressed in neurons, including the hippocampal pyramidal neurons. Furthermore, immunoblot analysis showed that Rnd1 protein was localized in synaptosomal membrane fraction. Ectopical overexpression of Rnd1 in cultured rat hippocampal neurons promoted the elongation of dendritic spines. On the other hand, suppression of endogenous Rnd1 level by antisense oligonucleotide of Rnd1 caused the increase in the percentage of headless protrusions accompanied by the reduction in the spine number and spine width and shortened the length of the headless protrusions. These results indicate that Rnd1 plays a role in spine formation in the developmental synaptogenic stage.
Keywords: Rnd, Rho, dendritic spines, actin, cytoskeleton, hippocampal neurons, synaptogenesis
Introduction
The organization of the nervous system is a complex and orchestrated process. Neurons migrate to their characteristic locations, extend axons and dendrites toward proper target regions, and induce spine formation and synaptic connections with appropriate partners. These dynamic morphological changes of neurons are primarily decided by the cytoskeletal organization.
The Rho family of small GTPases has been implicated in the reorganization of the actin cytoskeleton and subsequent morphological changes in various cells (Hall, 1998). Presently, at least 14 mammalian Rho family proteins have been identified: Rho (A, B, and C), Rac (1, 2, and 3), Cdc42, RhoD, RhoG, RhoH/TTF, TC10, and Rnd (1, 2, and 3). Among them, the functions of Rho, Rac, and Cdc42 have been characterized extensively. In fibroblasts, the activation of Rho leads to formation of actin stress fibers and assembly of focal adhesions (Ridley and Hall, 1992), whereas the activation of Rac and Cdc42 induces formation of lamellipodia and filopodia, respectively (Ridley et al., 1992; Nobes and Hall, 1995). In neuronal cells, Rho family GTPases have been shown to be involved in the regulation of neuronal cell morphology, and activation of Rac and Cdc42 induces the formation of lamellipodia and filopodia of the growth cone, respectively, whereas activation of Rho causes the collapse of the growth cone and neurite retraction (Negishi and Katoh, 2002). The less studied members of Rho family GTPases, such as RhoG and TC10, are also involved in the regulation of neuritogenesis (Katoh et al., 2000; Tanabe et al., 2000), suggesting that a variety of Rho family GTPases participate in the regulation of neuronal morphology. Dendritic spines are tiny protrusions that receive excitatory synaptic input and compartmentalize postsynaptic response (Hering and Sheng, 2001). Rho family GTPases are also involved in spine morphogenesis (Luo et al., 1996; Nakayama et al., 2000; Tashiro et al., 2000; Penzes et al., 2001, 2003; Irie and Yamaguchi, 2002), and they are considered as a common denominator controlling spine morphology in that several genes for the molecules of Rho family GTPase signaling pathways are involved in X-linked mental retardation, which is associated with an immature morphology of synaptic spines (Ramakers, 2000).
Recently, a new branch of Rho family GTPases, the Rnd subfamily, consisting of Rnd1, Rnd2, and Rnd3, has been identified (Foster et al., 1996; Guasch et al., 1998; Nobes et al., 1998). Rnd1 and Rnd2 are mainly expressed in brain, whereas Rnd3 is expressed ubiquitously. Unlike other Rho family GTPases, Rnd1 and Rnd3 possess very low intrinsic GTPase activity and constitutively bind to GTP, indicating that they are constitutively active. In fibroblasts, transient expression of Rnd1 or Rnd3 leads to loss of stress fibers, retraction and rounding of the cell body, and protrusion of extensively branching processes, and a part of these morphological effects of Rnd1 or Rnd3 is mediated by a novel Rnd GTPase-interacting protein, Socius (Katoh et al., 2002). In neuronal PC12 cells, expression of Rnd1 induces the formation of many neuritic processes from the cell body, with disruption of the cortical actin filaments that is probably attributable to the inhibition of Rho signaling pathway (Aoki et al., 2000). It appears, therefore, that Rnd1 and Rnd3 possess antagonistic effects on the action of RhoA in various cell types. However, the precise role of Rnd1 in the nervous system remains unknown. Here we demonstrate that Rnd1 is highly expressed in the cerebral cortex and hippocampus during the stage of synapse formation and plays a role in spine formation.
Materials and Methods
Plasmid constructions. The cording sequence for rat Rnd1 was obtained from adult rat brain by reverse transcription-PCR. The PCR product was cloned into pCR2.1 vector (Invitrogen, Carlsbad, CA), sequenced completely, and was subcloned into pBluescript SK(+) (Stratagene, La Jolla, CA). For expression in cultured rat hippocampal neurons, human Rnd1 cDNA (Aoki et al., 2000) was fused in frame with a sequence in pcDNA3 (Invitrogen) encoding an initiating methionine, followed by the Myc tag sequence at the N terminus by using KpnI/EcoRI sites. We also subcloned cDNA encoding enhanced green fluorescent protein (EGFP) into pcDNA3.
Northern blot analysis. Timed Wistar rats were ether anesthetized deeply and decapitated. Whole brains from embryonic or postnatal rats were removed, frozen immediately in liquid nitrogen, and then stored at -80°C until use. Purification of total RNA and Northern blotting were performed as described previously (Fujita et al., 2002). The membrane was hybridized with a 32P-labeled cDNA probe for rat Rnd1. For normalization, the membrane was rehybridized with a cDNA probe for the mouse glyceraldehyde 3-phosphate dehydrogenase (GAPDH).
In situ hybridization. Antisense and sense riboprobes of the sequence of rat Rnd1 were synthesized and digoxigenin (DIG) labeled by in vitro transcription with T7 and T3 RNA polymerases and DIG RNA labeling mix (Roche, Basel, Switzerland) from the BamHI- and SalI-digested plasmid. In situ hybridization was performed as described previously (Ishikawa et al., 2002). Briefly, 40-μm-thick coronal sections of the postnatal day 12 (P12) rat brains were acetylated and treated with 2.5 μg/ml proteinase K (Roche) for 10 min at room temperature before hybridization. After prehybridization, the sections were incubated overnight at 55°C with 500 ng/ml DIG-labeled sense or antisense probe. The sections were then washed in 2× SSC at 55°C three times for 1 hr each. DIG-labeled probes were immunodetected using alkaline phosphatase-conjugated anti-DIG antibody (1:1000 dilution; Roche) and then reacted with chromogenic substrates 5-bromo-4-chloro-3-indolylphosphate and nitroblue tetrazolium.
Subcellular fractionation of rat brains. The brains of P12 Wistar rats were homogenized by 10 strokes with a Teflon homogenizer in homogenizing buffer (0.32 m sucrose, 20 mm Tris-HCl, pH 7.4, 10 mm MgCl2, 1 μg/ml aprotinin, 1 μg/ml leupeptin, 0.1 mm benzamidin, 0.2 mm PMSF, and 1 mm EDTA). The homogenate was centrifuged at 800 × g to remove nuclei and large debris, and the supernatant was further centrifuged at 100,000 × g to obtain the cytosol (S100) and crude membrane (P100) fractions. Preparation of synaptosomal membrane fraction (LP1) was performed as described previously (Dunah and Standaert, 2001) with some modification. Briefly, homogenate of P12 rat brains in the homogenizing buffer was centrifuged at 800 × g, and the supernatant (S1) was further centrifuged at 13,800 × g to obtain crude synaptosomal fraction (P2). P2 fraction was then lysed hypo-osmotically and centrifuged at 25,000 × g to precipitate a LP1 fraction. Protein concentrations were determined by Bradford's method.
Immunoblotting. Proteins were separated by 7.5% SDS or 12.5% PAGE and were electrophoretically transferred onto a polyvinylidene difluoride membrane (Millipore, Billerica, MA). The membrane was blocked with 3% low-fat milk in Tris-buffered saline and then incubated with primary antibodies. The primary antibodies were detected by using horseradish peroxidase-conjugated secondary antibodies (Dako, Glostrup, Denmark) and an ECL detection kit (Amersham Biosciences, Piscataway, NJ). Antibody for Rnd1 was raised against bacterially expressed glutathione S-transferase-fused peptide, corresponding to the residues 53-61 (CLETEEQRV) of Rnd1, and the specific antibody was purified with the peptide-conjugated affinity column. The antibody was used for the detection of Rnd1 at a concentration of 2 μg/ml. Anti-hemagglutinin (HA) rabbit polyclonal antibody (Y-11; Santa Cruz Biotechnology, Santa Cruz, CA), anti-PSD-95 (postsynaptic density 95) mouse monoclonal IgG (clone K28/43; Upstate Biotechnology, Lake Placid, NY), and anti-α-tubulin mouse monoclonal IgG (B-5-1-2; Sigma, St. Louis, MO) were used at 1:100, 1:5000, and 1:200 dilutions, respectively.
Hippocampal cultures and transfection. Hippocampal cultures were prepared from the hippocampi of embryonic day 19 (E19) rats as described previously (Goslin et al., 1998). Briefly, embryos were removed from timed pregnant Wistar rats after the animals were killed, and their hippocampi were dissected in ice-cold calcium- and magnesium-free HBSS. The hippocampi were washed in HBSS and incubated in HBSS with 0.25% trypsin and 0.1% DNase for 15 min at 37°C. After the incubation, the hippocampi were washed three times in HBSS, followed by trituration with fire-polished Pasteur pipette in the culture medium [Neurobasal medium (Invitrogen, Grand Island, NY) with 2% B27 supplement (Invitrogen), 0.5 mm l-glutamine, and 100 U/ml penicillin]. The cells were seeded onto round 13 mm coverslips (Matsunami, Osaka, Japan) coated with 0.05 mg/ml poly-d-lysine (Sigma) in culture plates (24-wells) at a density of 2 × 104 cells and cultured in the culture medium under the humidified conditions in 95% air and 5% CO2 at 37°C. For immunoblot analysis, cells were cultured on poly-d-lysine-coated 60 mm culture dishes. One-half the medium was changed at 4 d in vitro (4 DIV) and then once per week. Hippocampal neurons were transfected with test plasmids using Lipofectamine 2000 (Invitrogen) according to the instructions of the manufacturer.
Antisense oligonucleotide treatment. Phosphorothioate oligonucleotides were used in this study. We constructed an antisense oligonucleotide (5′-TCTCTCCTTCATGGCTGCAGCCTA-3′) consisting of the complement sequence of nucleotides -12 to +12 of the sequence of rat Rnd1. The corresponding sense oligonucleotide was also constructed as a control. Hippocampal neurons were transfected with pEGFP-C1 (Clontech, Palo Alto, CA) at 8 DIV to visualize spine morphology, oligonucleotides were added to the cultures at a concentration of 2 μm every 24 hr from 15 to 16 DIV, and the cells were fixed at 17 DIV.
Immunofluorescence microscopy. Immunofluorescence was performed as described previously (Katoh et al., 2002). Hippocampal neurons on coverslips were fixed with 4% paraformaldehyde in PBS for 15 min. After residual formaldehyde had been quenched with 50 mm NH4Cl in PBS for 10 min, cells were permeabilized with 0.2% Triton X-100 in PBS for 10 min and incubated with 10% fetal bovine serum in PBS for 30 min to block nonspecific antibody binding. For the detection of cells expressing Myc-Rnd1, cells were incubated with anti-Myc mouse monoclonal IgG (9E10, 1:500 dilution; Santa Cruz Biotechnology) in PBS for 1 hr, followed by incubation with Alexa 594-conjugated anti-mouse IgG (1:1000 dilution ; Molecular Probes, Eugene, OR). For PSD-95 detection, anti-PSD-95 mouse monoclonal IgG was used at 1:250 dilution. For visualizing endogenous Rnd1 in cultured hippocampal neurons, neurons were fixed with methanol for 10 min at -20°C. After incubation with 10% fetal bovine serum in PBS, cells were incubated with anti-Rnd1 antibody at 5 μg/ml, followed by incubation with Alexa 594-conjugated anti-rabbit IgG (1:1500 dilution ; Molecular Probes). Morphology of neurons was visualized by transfection with pEGFP-F (Clontech), which effectively outlines the morphology of methanol-fixed neurons. Cells on coverslips were mounted in 90% glycerol containing 0.1% p-phenylenediamine dihydrochloride in PBS. Fluorescence microscopic images were gained by using an MRC-1024 laser scanning confocal imaging system (Bio-Rad, Hercules, CA) equipped with a Nikon Eclipse E800 microscope and a Nikon Plan Apo 60-by-1.4 oil immersion objective lens (Nikon, Tokyo, Japan).
Data analysis. For quantification of various parameters of spine morphology, confocal z-series EGFP image stacks of the proximal segments of dendrites were taken, and their morphological measurements were performed using Image Pro 4.5 software (Media Cybernetics, Silver Spring, MD). Spines were defined as <6 μm dendritic protrusions with rounded heads whose edges were clearly defined by EGFP fluorescence. Approximately 15 neurons were collected per construct from two or three experiments, and spines were analyzed within dendritic segments of 30-50 μm/cell (for analysis of overexpression experiments, 345 spines from 16 cells for control and 257 spines from 14 cells; for analysis of antisense oligonucleotide experiments, 415 spines from 17 cells for sense treatment and 352 spines from 17 cells for antisense treatment). Headless protrusions were defined as <10 μm dendritic protrusions without head region and without branch, which were distinguished from spines. For quantification of headless protrusion length, 135 or 151 protrusions were collected in sense- or antisense-treated neurons (17 cells each), respectively. For counting PSD-95-positive protrusions among all protrusions (= spines + headless protrusions) or headless protrusions, immunofluorescence images of PSD-95 were converted to binary images by thresholding intensity to clarify PSD-95-immunopositive puncta. PSD-95-positive protrusions were defined as protrusions in which the immunopositive puncta were observed. Statistical difference was determined by Student's t test.
Results
Rnd1 is strongly expressed in the neurons in developing rat brain
To identify developmental changes of expression of Rnd1, Northern blot analysis was performed for rat brain RNA from E14 to the adult (Fig. 1A). Rnd1 mRNA expression was poor during embryonic stage but gradually increased during early postnatal days toward the peak of the expression level at P14, the level declining in adult.
Figure 1.
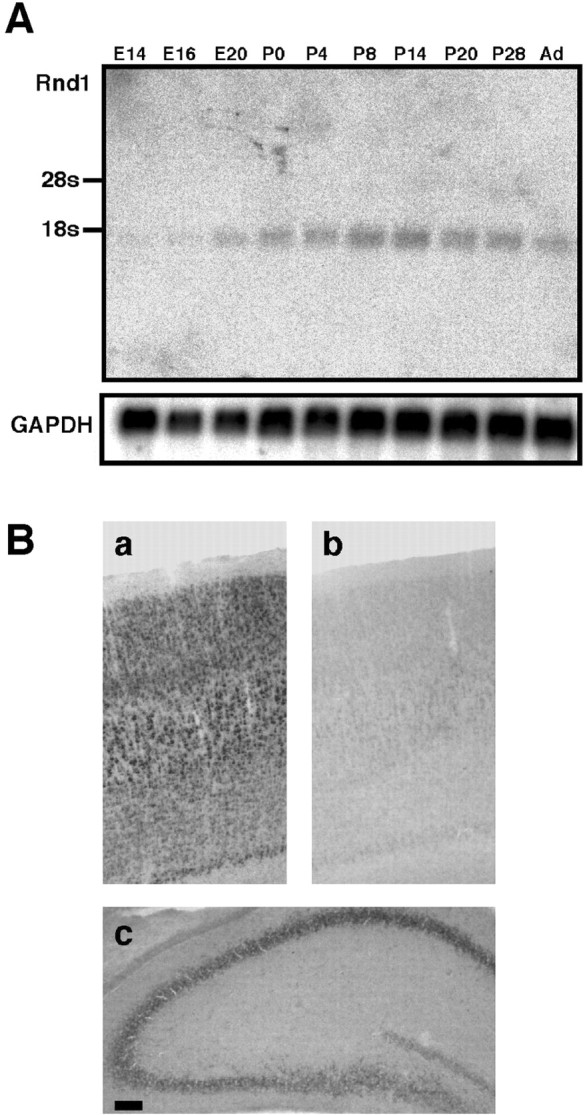
Expression of Rnd1 mRNA in developing rat brain. A, Total RNA (20 μg) isolated from embryonic (E14, E16, and E20) or postnatal (P0, P4, P8, P14, P20, P28, and adult) rat brain was subjected to Northern blot analysis with a cDNA probe for Rnd1. The same membrane rehybridized with GAPDH cDNA probe is shown below as the internal control. B, Distribution of Rnd1 in the P12 rat brain. In situ hybridization was performed with antisense Rnd1 riboprobe (a, c) and the corresponding sense riboprobe (b) for coronally sectioned P12 rat cortex (a, b) and hippocampus (c). Scale bar, 100 μm.
To reveal the distribution of Rnd1 mRNA in the rat brain at P12, we performed in situ hybridization using a DIG-labeled Rnd1 antisense riboprobe for the brain section prepared from P12 rat (Fig. 1B). Expression of Rnd1 mRNA was observed in cortical neurons (Fig. 1Ba; control sense strand hybridization shown in Fig. 1Bb) and hippocampal pyramidal neurons (Fig. 1Bc) with high intensity. Rnd1 mRNA-expressing neurons were also found in other brain regions, including the piriform cortex and cerebellum (data not shown).
Subcellular localization of Rnd1 in early postnatal brain
We raised an antibody against a peptide, corresponding to the amino acid residues 53-61 of Rnd1, which is a unique sequence of Rnd1 among Rnd subfamily as well as other Rho family GTPases. The antibody specifically recognized Rnd1 expressed in COS-7 cells among three Rnd proteins, Rnd1, Rnd2, and Rnd3 (Fig. 2A). We then investigated subcellular localization of endogenous Rnd1 protein in P12 rat brain by immunoblotting (Fig. 2B). Rnd1 was highly localized in the particulate fraction (P100 fraction) but not in the cytosolic fraction (S100 fraction), and further subcellular fractionation revealed that Rnd1 protein was localized to a synaptosomal membrane fraction (LP1 fraction). Effectiveness of the subcellular fractionation was evaluated by using a synapse marker protein, PSD-95. We showed in Figure 1Bc that Rnd1 mRNA was highly expressed in hippocampal neurons. We then prepared cultured rat hippocampal neurons and analyzed the expression of Rnd1 protein by immunoblotting. As shown in Figure 2C, Rnd1 protein was clearly detected in the hippocampal neurons cultured for 17 d. Distribution of endogenous Rnd1 protein in 17 DIV hippocampal neurons was studied by immunofluorescence (Fig. 2D). Morphology of neurons was visualized by ectopical expression of EGFP-F, which was localized at membrane fraction by farnesylation. Immunofluorescence showed that endogenous Rnd1 protein was detected as punctuated structures in dendritic shafts, including some spine-like structures (Fig. 2Da). Double immunolabeling with PSD-95 revealed that Rnd1-immunopositive clusters were in part colocalized with PSD-95 puncta (Fig. 2Db). These data suggest that Rnd1 is involved in spine formation near or in dendritic spines.
Figure 2.

Subcellular distribution of Rnd1 protein in rat brain. A, Specificity of an anti-Rnd1 antibody. COS-7 cells were transiently transfected with expression vectors encoding HA-tagged Rnd1, Rnd2, Rnd3, or an empty vector. Cell lysates were immunoblotted with antibody against Rnd1 (top) or HA epitope (bottom). B, Each subcellular fraction of P12 rat brain (80 μg of total protein) immunoblotted with the anti-Rnd1 antibody (top) or anti-PSD-95 antibody (bottom). S100, Cytosol; P100, crude membrane; P2, crude synaptosome; LP1, synaptosomal membrane; S1, homogenate without nuclei and large debris; S2, supernatant above P2 fraction. C, Endogenous Rnd1 protein in hippocampal cell cultures. COS-7 cell lysate expressing HA-Rnd1 and P100 fractions (45 μg of total protein) of dissociated hippocampal cell cultures at 17 and 31 DIV were immunoblotted with the anti-Rnd1 antibody. D, Immunofluorescence microscopy of dendrites of cultured hippocampal neurons. Cells at 17 DIV were fixed with methanol and immunostained. a, Cells were transfected with EGFP-F vector at 6 DIV. EGFP-F and Rnd1 labeling are shown in the top and middle panels, respectively. Bottom panels (Merge) show the superposition of the top and middle images. Rnd1 immunostains are observed in the dendritic shaft regions (arrows) including spine-like structures (arrowheads). b, PSD-95 and Rnd1 labeling were shown in the top and middle panels, respectively. Bottom panels (Merge) show the superposition of the top and middle images. Arrowheads or arrows indicate the regions in which Rnd1 immunostains are localized with or without PSD-95-immunopositive puncta, respectively. Scale bar, 5 μm.
Overexpression of Rnd1 in hippocampal neurons causes elongation of spines
To study the effect of Rnd1 on the morphology of hippocampal neurons, the 13 DIV cultured hippocampal neurons were transfected with plasmids encoding Myc-Rnd1 and EGFP to visualize neuronal morphology and fixed 48 hr later (15 DIV) (Fig. 3). At 15 DIV, a period characterized by dendritic maturation, including spine stabilization and synapse formation (Ziv and Smith, 1996), spines in Myc-Rnd1-transfected neurons frequently had longer necks than mock-transfected neurons (Fig. 3a′, b′), although the dendrite patterning did not seem to be changed (Fig. 3a,b). On the other hand, a negative form of Rnd1, Rnd1N27, and an effector mutant of Rnd1, Rnd1A45, failed to induce spine elongation, suggesting that the activity of Rnd1 is required for the spine elongation (data not shown).
Figure 3.
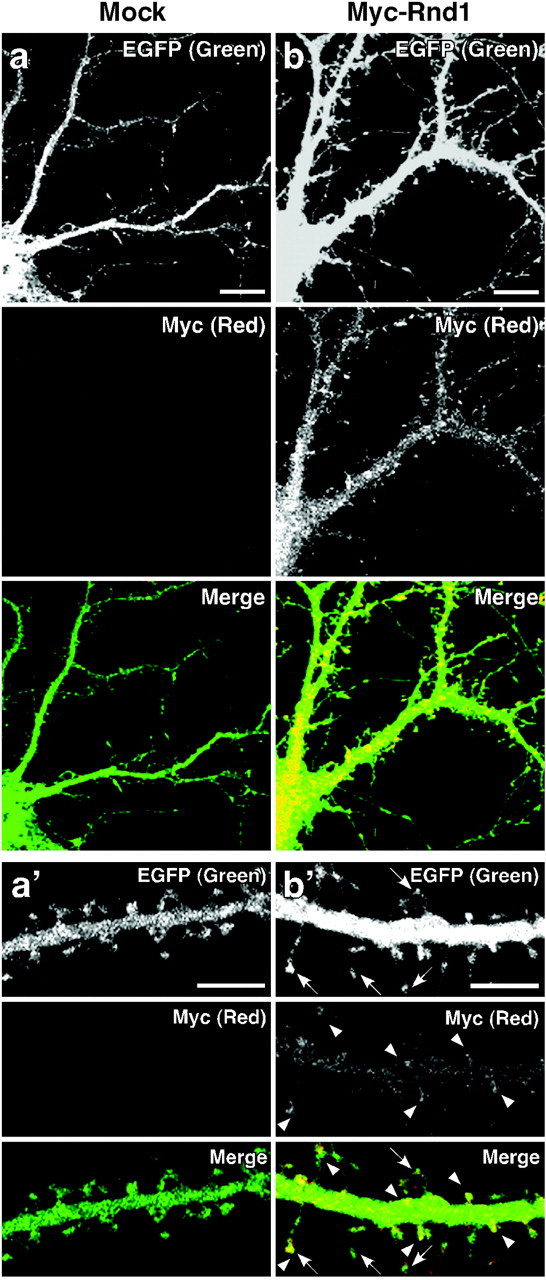
The effect of overexpression of Rnd1 on the morphology of hippocampal neurons. After 13 DIV, hippocampal neurons were transfected with a vector for EGFP alone or cotransfected with vectors for EGFP and Myc-Rnd1, and cells at 15 DIV were fixed and immunostained with anti-Myc antibody. Immunofluorescence microscopy was shown for neuron expressing EGFP alone (a) or EGFP and Myc-Rnd1 (b). Magnified images of dendrites of neurons in a and b are shown in a′ and b′, respectively. EGFP and Myc-Rnd1 labeling were shown in the top and middle panels, respectively. Bottom panels (Merge) show the superposition of the top and middle images. Arrowheads indicate Myc-Rnd1 enriched in dendritic spines, and arrows indicate elongated spines. Scale bars: a, b, 10 μm; a′, b′, 5 μm.
The effect of overexpression of Rnd1 on the spine morphology was quantified by measuring the length and width of spine heads and spine density (Fig. 4). Cumulative distributions of spine length and head width showed that spines in Myc-Rnd1-transfected neurons were longer than those in mock-transfected neurons (Fig. 4a), without significant difference in head width (Fig. 4c). There was no statistical difference in spine density (Fig. 4e) (4.45 ± 0.42 spines/10 μm dendrite for Myc-Rnd1-transfected neurons vs 4.85 ± 0.38 spines/10 μm dendrite for mock-transfected neurons; p > 0.05). Counting the percentage of >2-μm-length spines showed that spines of Myc-Rnd1-transfected neurons were significantly longer than those of mock-transfected neurons (Fig. 4b) (53.6 ± 4.3% for Myc-Rnd1-transfected neurons vs 22.1 ± 3.5% for mock-transfected neurons; p < 0.0005). We also counted the percentage of >0.8 μm head width spines, but significant difference was not determined (Fig. 4d) (22.7 ± 3.4% for Myc-Rnd1-transfected neurons vs 32.5 ± 5.0% for mock-transfected neurons; p > 0.05). These data suggest that Rnd1 promotes the elongation of spines.
Figure 4.
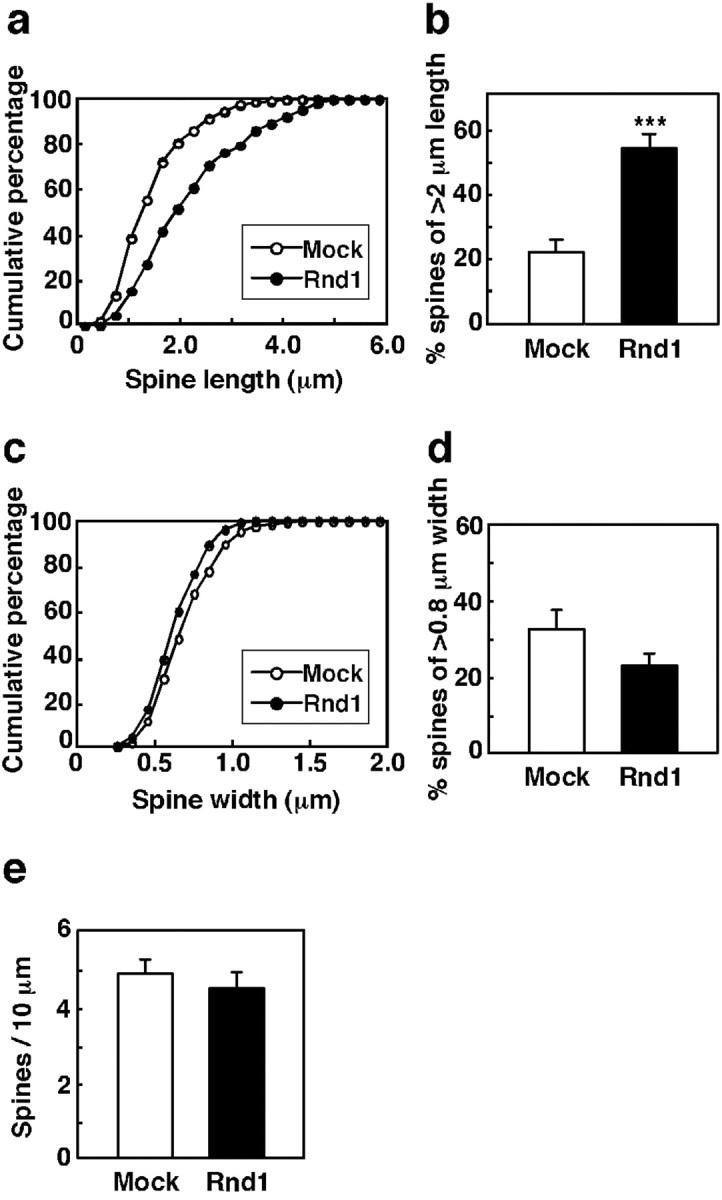
Quantification of dendritic spine morphology of transfected neurons. a, b, Cumulative distribution of spine length of mock- (○) or Myc-Rnd1- (•) transfected neurons (a) and the percentage of spines of >2 μm length in the same population (mock, □; Myc-Rnd1, ▪) (b). c, d, Cumulative distribution of head width of mock- (○) or Myc-Rnd1- (•) transfected neurons (c) and the percentage of spines of >0.8 μm width in the same population (mock, □; Myc-Rnd1, ▪) (d). e, The number of spines of mock- (□) or Myc-Rnd1- (▪) transfected neurons was counted, and the spine density was calculated (spines/10 μm dendrite). Myc-Rnd1-transfected neurons had significantly longer spines than mock-transfected neurons (***p < 0.0005; Student's t test), but there were not significant differences in spine width and density (p > 0.05; Student's t test). The data are the means ± SE.
Suppression of Rnd1 expression results in immaturation of spine shape
Immunoblotting and immunocytochemistry showed that Rnd1 was expressed endogenously in cultured hippocampal neurons during the stage of spine formation (Fig. 2C,D). To examine the significance of Rnd1 in the spine formation, we set out to suppress the expression of endogenous Rnd1 in hippocampal neurons by using an antisense oligonucleotide. The antisense oligonucleotide corresponding to the nucleotides -12 to +12 of the sequence of rat Rnd1 significantly reduced Rnd1 protein expression in cultured hippocampal neurons at 17 DIV (Fig. 5A).
Figure 5.

The effect of Rnd1 antisense treatment on the morphology of hippocampal neurons. Hippocampal cultures at 15 DIV were treated for 2 d with sense or antisense oligonucleotide and harvested (Aa, Ab) or fixed (Ac, Ad, B) at 17 DIV. Aa, P100 fraction (20 μg of total protein) of harvested cells was immunoblotted with anti-Rnd1 antibody (top) or anti-α-tubulin antibody (bottom). Ab, Quantification of Rnd1 protein level of sense- or antisense-treated neurons. The level of Rnd1 protein was normalized to that of α-tubulin and expressed as fold of the value of sense-treated neurons. Ac, Ad, Immunofluorescence microscopy was shown for EGFP-F-expressing hippocampal neuron treated with sense (c) or antisense (d) oligonucleotide. EGFP-F and Rnd1 labeling were shown in the top and middle panels, respectively. Bottom panels (Merge) show the superposition of the top and middle images. Note that Rnd1 immunostain is faint in antisense-treated neuron but is obvious in sense-treated neuron. B, Immunofluorescence microscopy was shown for EGFP-expressing hippocampal neuron treated with sense (a) or antisense (b) oligonucleotide. EGFP and PSD-95 labeling were shown in the top and middle panels, respectively. Bottom panels (Merge) show the superposition of the top and middle images. Arrows indicate mature-shaped spines, and arrowheads indicate headless protrusions. Another example is shown in a′ for sense-treated neuron or in b′ for antisense-treated neuron. Scale bars, 5 μm.
Antisense-treated neurons visualized by EGFP expression at 17 DIV had less rounded-head spines and alternatively more tiny headless protrusions or slimmer spines (Fig. 5Bb,b′) compared with sense-treated neurons, which had stubby- or mushroom-shaped spines, indicating mature ones (Fig. 5Ba,a′). Immunostains for PSD-95, a spine marker, were hardly detectable in the headless protrusions of antisense-treated neurons (Fig. 5B, arrowheads), although PSD-95 was easily detectable in mature spines of sense-treated neurons (Fig. 5B, arrows). Quantitative analysis showed that the percentage of headless protrusions among all protrusions was higher in antisense-treated neurons than that in sense-treated neurons (Fig. 6Aa) (25.1 ± 4.3% for antisense-treated neurons vs 16.3 ± 2.7% for sense-treated neurons; p < 0.05). Furthermore, the cumulative percentage of headless protrusion length showed that length of headless protrusions was shorter in antisense-treated neurons than those in sense-treated neurons (Fig. 6Ac). The antisense treatment decreased the percentage of >2-μm-length of headless protrusions (Fig. 6Ad) (37.1 ± 4.1% for antisense-treated neurons vs 62.1 ± 4.5% for sense-treated neurons; p < 0.0005). Quantification of PSD-95-positive protrusions in all dendritic protrusions showed that less PSD-95-positive protrusions existed in antisense-treated neurons than those in sense-treated neurons (Fig. 6Ab, left) (67.8 ± 4.3% for antisense-treated neurons vs 85.5 ± 2.0% for sense-treated neurons; p < 0.005). On the other hand, there was no significant difference in the number of PSD-95-positive headless protrusions between sense and antisense treatments (Fig. 6Ab, right) (15.3 ± 3.1% for antisense-treated neurons vs 19.4 ± 3.2% for sense-treated neurons; p > 0.05), indicating that the antisense treatment decreases synapse formation by inhibiting spine formation. We then measured spine length, width, and density (Fig. 6B). The antisense treatment induced marked reduction of head width (Fig. 6Bd) (18.4 ± 5.1% of >0.8 μm head width for antisense-treated neurons vs 40.1 ± 5.3% for sense-treated neurons; p < 0.005) and slight but significant reduction of spine density (Fig. 6Be) (4.91 ± 0.48 spines/10 μm dendrite for antisense-treated neurons vs 6.03 ± 0.41 spines/10 μm dendrite for sense-treated neurons; p < 0.05) without any difference in the length (Fig. 6Ba,b). These results suggest that suppression of Rnd1 expression inhibits elongation of headless protrusions, spine precursors, and then subsequently causes spine immaturation.
Figure 6.

A, Quantification of dendritic protrusions. Aa, Percentage of headless protrusions among all dendritic protrusions (= spines + headless protrusions) of sense- (□) or antisense- (▪) treated neurons. Ab, Percentage of PSD-95-positive protrusions among all protrusions (left) or among headless protrusions (right) of sense- (□) or antisense- (▪) treated neurons. Ac, Ad, Cumulative distribution of headless protrusion length of sense- (○) or antisense- (•) treated neurons (c) and the percentage of >2-μm-length protrusions in the same population (sense, □; antisense, ▪) (d). B, Quantification of dendritic spines. Ba, Bb, Cumulative distribution of spine length of sense- (○) or antisense- (•) treated neurons (a) and the percentage of spines of >2 μm length in the same population (sense, □; antisense, ▪) (b). Bc, Bd, Cumulative distribution of head width of sense- (○) or antisense- (•) treated neurons (c) and the percentage of spines of >0.8μm width in the same population (sense, □; antisense, ▪) (d). Be, The number of spines of sense- (□) or antisense- (▪) treated neurons was counted, and the density was calculated (spines/10 μm dendrite). Statistical difference was determined by Student's t test (*p < 0.05; **p < 0.005; ***p < 0.0005). The data are the means ± SE.
Discussion
The focus of this study has been an analysis of the role of Rnd1, a novel Rho family GTPase predominantly expressed in brain, in regulating neuronal development. Here we show that Rnd1 is highly expressed in the cerebral cortex and hippocampus during the stage of synapse formation and that overexpression of Rnd1 induces elongation of spine length, but suppression of endogenous Rnd1 expression disturbs the spine maturation in cultured hippocampal neurons. Our findings demonstrate that Rnd1 plays a role in spine formation.
In contrast to RhoA, Rac1, and Cdc42, which are expressed continuously throughout development (Threadgill et al., 1997), Rnd1 is absent in early embryonic period but highly expressed during the synaptogenic period of approximately P14-P20 after the birth, and then the expression level gradually decreases in adult. Rho, Rac, and Cdc42 are usually localized in cytosol as a complex with Rho-GDI (GDP dissociation inhibitor), and they are recruited to membranes and activated by GEF (GDP/GTP exchange factor) in response to various extracellular stimuli (Etienne-Manneville and Hall, 2002). In contrast to these well known Rho family GTPases, Rnd1 is constitutively GTP-bound active form and localized in plasma membranes in fibroblasts, suggesting that Rnd1 activity is probably determined by expression (Nobes et al., 1998). We here showed that, in brain, Rnd1 is mostly found in the membrane fraction, especially in the synaptosomal fraction, with little or no existence in the cytosol. Although at present Rnd1-associated protein(s) or receptor(s) is not identified in the dendritic spines, Rnd1 expressed in the synaptogenic stage is constitutively active and may play a role in regulation of spine morphology.
Dendritic filopodia are widely accepted to be the precursors of dendritic spines. Dendritic filopodia protrude from dendrites during the stages of synaptogenesis and contact with nearby axons and thereafter develop mushroom-shaped spines. We here showed that the overexpression of Rnd1 causes spine elongation without any change in the number of spines in cultured hippocampal neurons, suggesting that Rnd1 promotes elongation of preexisting protrusions but does not initiate new protrusions. Spine formation and maturation are mediated by various steps, including head formation and neck retraction. Overexpression of Rnd1 promotes continuously neck elongation, and this strong activity may overwhelm neck retraction. On the other hand, we showed that the reduction of Rnd1 level by the antisense treatment causes the increase in the percentage of headless protrusions accompanied by the reduction in the spine number and spine width and shortens the length of the headless protrusions, suggesting that suppression of Rnd1 activity results in the inhibition of elongation of dendritic filopodia, which reduces the opportunity to contact nearby axons, eventually causing immaturation of spine formation. Considering that Rnd1 is localized near dendritic spines, Rnd1 therefore appears to play a crucial role in the protrusion of dendritic filopodia.
F-actin is highly concentrated in dendritic spines, and regulation of the actin cytoskeleton is crucial for the development and stability of spines. A recent study (Zhang and Benson, 2001) showed that an actin polymerization inhibitor, latrunculin, treatment of neurons transformed spines into filopodia-like processes, suggesting that actin polymerization is essential for elongation of filopodia and subsequent spine formation. Rho family GTPases are known to be important regulators for the actin cytoskeleton, and a growing number of evidence supports that Rho, Rac, and Cdc42 are involved in regulation of spine morphology. It has been reported that dominant-negative Rac1 causes a progressive reduction in spine number (Nakayama et al., 2000), whereas constitutively active Rac1 increases spine density (Luo et al., 1996), indicating that Rac1 activity is important for the maintenance of spine density. Loss-of-function mutation of Cdc42 was also reported to show a reduction in dendritic spine density of vertical system neurons in Drosophila (Scott et al., 2003). Rac and Cdc42 may participate in the promotion of dendritic filopodia and subsequent mature spine formation, because Rac and Cdc42 are known to direct outgrowth of the actin cytoskeleton through extension of filopodia and lamellipodia in various cell lines, including neurons. In contrast, constitutively active Rho has been shown to strongly induce a loss of mature-shaped spines, whereas inhibition of Rho activity by C3 transferase resulted in elongated spine necks (Nakayama et al., 2000; Tashiro et al., 2000), suggesting that Rho is a negative regulator for spine formation. Therefore, Rho and Rac/Cdc42 signaling act antagonistically in spine formation and growth, and the balance between Rho and Rac/Cdc42 activities is likely to be crucial point for the regulation of spine morphology (Yamaguchi et al., 2001). Rnd1 possesses the inhibitory effects on Rho signaling pathway, such as inhibition of Rho-mediated actin stress fiber formation and the assembly of focal adhesions in fibroblasts (Nobes et al., 1998) and inhibition of Rho-mediated Ca2+ sensitization of rat smooth muscle (Loirand et al., 1999). In addition, we showed that Rnd1 promoted filopodial protrusion from the cell body and subsequent formation of neuritic processes attributable to the inhibition of Rho signaling pathway in PC12 cells (Aoki et al., 2000). In the light of this antagonistic action of Rnd1, it is plausible that Rnd1 promotes protrusion of dendritic filopodia through inhibition of Rho signaling. We recently identified Socius as a novel Rnd1-interacting protein, which mediates the Rnd1-induced disassembly of actin stress fibers (Katoh et al., 2002). Socius is an effector of Rnd1, displaying inhibitory action for Rho signaling. Although we do not know the expression of Socius in the hippocampal neurons, Socius might be involved in the Rnd1-induced filopodial protrusion and spine maturation.
It has been reported recently that Rnd1 interacts with cytoplasmic domain of Plexin-A1, a receptor for semaphorin 3A, and induces semaphorin 3A-mediated cell collapse in COS-7 cells (Zanata et al., 2002). Semaphorin 3A is known to act as a repulsive factor for axon guidance through the binding to its receptor complex, neuropilin-1 and Plexin-A (Polleux et al., 2000). In sharp contrast to other Rho family GTPases, Rnd1 is constitutively active and exists as a membrane-bound form. In the case of Plexin-A1, Rnd1 stably associates with Plexin-A1 and transduces the signal of semaphorin 3A for growth cone collapse and axonal retraction. In addition to this regulation of axon guidance, our present work proposes a new possible function of Rnd1, regulation of spine morphology.
In conclusion, we demonstrated that Rnd1 is highly expressed in the cerebral cortex and hippocampus during the stage of synapse formation and plays a role in spine formation. However, many questions have not yet been elucidated about Rnd1 signaling, for example, a receptor(s) or adaptor molecules associated with Rnd1 on the spine membranes. Additional studies focusing on the identification of Rnd1-interacting proteins will contribute to understanding the Rnd1-siganling pathway for the regulation of spine morphology.
Footnotes
This work was supported in part by Grants-in-Aid for Scientific Research from the Ministry of Education, Science, Sports, and Culture of Japan and a grant from the Takeda Science Foundation.
Correspondence should be addressed to Manabu Negishi, Laboratory of Molecular Neurobiology, Graduate School of Biostudies, Kyoto University, Sakyo-ku, Kyoto 606-8502, Japan. E-mail address: mnegishi@pharm.kyoto-u.ac.jp.
Copyright © 2003 Society for Neuroscience 0270-6474/03/2311065-08$15.00/0
References
- Aoki J, Katoh H, Mori K, Negishi M ( 2000) Rnd1, a novel Rho family GTPase, induces the formation of neuritic processes in PC12 cells. Biochem Biophys Res Commun 278: 604-608. [DOI] [PubMed] [Google Scholar]
- Dunah AW, Standaert DG ( 2001) Dopamine D1 receptor-dependent trafficking of striatal NMDA glutamate receptors to the postsynaptic membrane. J Neurosci 21: 5546-5558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etienne-Manneville S, Hall A ( 2002) Rho GTPases in cell biology. Nature 420: 629-635. [DOI] [PubMed] [Google Scholar]
- Foster R, Hu KQ, Lu Y, Nolan KM, Thissen J, Settleman J ( 1996) Identification of a novel human Rho protein with unusual properties: GTPase deficiency and in vivo farnesylation. Mol Cell Biol 16: 2689-2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita H, Katoh H, Ishikawa Y, Mori K, Negishi M ( 2002) Rapostlin is a novel effector of Rnd2 GTPase inducing neurite branching. J Biol Chem 47: 45428-45434. [DOI] [PubMed] [Google Scholar]
- Goslin K, Asmussen H, Banker G ( 1998) Rat hippocampal neurons in low-density culture. In: Culturing nerve cells, Ed 2 (Banker G, Goslin K, eds), pp 339-370. Cambridge, MA: MIT.
- Guasch RM, Scambler P, Jones GE, Ridley AJ ( 1998) RhoE regulates actin cytoskeleton organization and cell migration. Mol Cell Biol 18: 4761-4771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall A ( 1998) Rho GTPases and the actin cytoskeleton. Science 279: 509-514. [DOI] [PubMed] [Google Scholar]
- Hering H, Sheng M ( 2001) Dendritic spines: structure, dynamics and regulation. Nat Rev Neurosci 2: 880-888. [DOI] [PubMed] [Google Scholar]
- Irie F, Yamaguchi Y ( 2002) EphB receptors regulate dendritic spine development via intersectin, Cdc42 and N-WASP. Nat Neurosci 5: 1117-1118. [DOI] [PubMed] [Google Scholar]
- Ishikawa Y, Katoh H, Nakamura K, Mori K, Negishi M ( 2002) Developmental changes in expression of small GTPase RhoG mRNA in the rat brain. Brain Res Mol Brain Res 106: 145-150. [DOI] [PubMed] [Google Scholar]
- Katoh H, Yasui H, Yamaguchi Y, Aoki J, Fujita H, Mori K, Negishi M ( 2000) Small GTPase RhoG is a key regulator for neurite outgrowth in PC12 Cells. Mol Cell Biol 20: 7378-7387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh H, Harada A, Mori K, Negishi M ( 2002) Socius is a novel Rnd GTPase-interacting protein involved in disassembly of actin stress fibers. Mol Cell Biol 22: 2952-2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loirand G, Cario-Toumaniantz C, Chardin P, Pacaud P ( 1999) The Rho-related protein Rnd1 inhibits Ca2+ sensitization of rat smooth muscle. J Physiol (Lond) 516: 825-834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo L, Hensch TK, Ackerman L, Barbel S, Jan LY, Jan YN ( 1996) Differential effects of the Rac GTPase on Purkinje cell axons and dendritic trunks and spines. Nature 379: 837-840. [DOI] [PubMed] [Google Scholar]
- Nakayama AY, Harms MB, Luo L ( 2000) Small GTPases Rac and Rho in the maintenance of dendritic spines and branches in hippocampal pyramidal neurons. J Neurosci 20: 5329-5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negishi M, Katoh H ( 2002) Rho family GTPases as key regulators of neuronal network formation. J Biochem 132: 157-166. [DOI] [PubMed] [Google Scholar]
- Nobes CD, Hall A ( 1995) Rho, rac, and cdc42 GTPases regulate the assembly of multimolecular focal complexes associated with actin stress fibers, lamellipodia, and filopodia. Cell 81: 53-62. [DOI] [PubMed] [Google Scholar]
- Nobes CD, Lauritzen I, Mattei MG, Paris S, Hall A, Chardin P ( 1998) A new member of the Rho family, Rnd1, promotes disassembly of actin filament structures and loss of cell adhesion. J Cell Biol 141: 187-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penzes P, Johnson RC, Sattler R, Zhang X, Huganir RL, Kambampati V, Mains RE, Eipper BA ( 2001) The neuronal Rho-GEF Kalirin-7 interacts with PDZ domain-containing proteins and regulates dendritic morphogenesis. Neuron 29: 229-242. [DOI] [PubMed] [Google Scholar]
- Penzes P, Beeser A, Chernoff J, Schiller MR, Eipper BA, Mains RE, Huganir RL ( 2003) Rapid induction of dendritic spine morphogenesis by trans-synaptic ephrinB-EphB receptor activation of the Rho-GEF Kalirin. Neuron 37: 263-274. [DOI] [PubMed] [Google Scholar]
- Polleux F, Morrow T, Ghosh A ( 2000) Semaphorin 3A is a chemoattractant for cortical apical dendrites. Nature 404: 567-573. [DOI] [PubMed] [Google Scholar]
- Ramakers GJA ( 2000) Rho proteins and the cellular mechanisms of mental retardation. Am J Med Genet 94: 367-371. [DOI] [PubMed] [Google Scholar]
- Ridley AJ, Hall A ( 1992) The small GTP-binding protein rho regulates the assembly of focal adhesions and actin stress fibers in response to growth factors. Cell 70: 389-399. [DOI] [PubMed] [Google Scholar]
- Scott EK, Reuter JE, Luo L ( 2003) Small GTPase Cdc42 is required for multiple aspects of dendritic morphogenesis. J Neurosci 23: 3118-3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanabe K, Tachibana T, Yamashita T, Che YH, Yoneda Y, Ochi T, Tohyama M, Yoshikawa H, Kiyama H ( 2000) The small GTP-binding protein TC10 promotes nerve elongation in neuronal cells, and its expression is induced during nerve regeneration in rats. J Neurosci 20: 4138-4144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tashiro A, Minden A, Yuste R ( 2000) Regulation of dendritic spine morphology by the Rho family small GTPases: antagonistic roles of Rac and Rho. Cereb Cortex 10: 927-938. [DOI] [PubMed] [Google Scholar]
- Threadgill R, Bobb K, Ghosh A ( 1997) Regulation of dendritic growth and remodeling by Rho, Rac, and Cdc42. Neuron 19: 625-634. [DOI] [PubMed] [Google Scholar]
- Yamaguchi Y, Katoh H, Yasui H, Mori K, Negishi M ( 2001) RhoA inhibits the nerve growth factor-induced Rac1 activation through Rho-associated kinase-dependent pathway. J Biol Chem 276: 18977-18983. [DOI] [PubMed] [Google Scholar]
- Zanata SM, Hovatta I, Rohm B, Püschel AW ( 2002) Antagonistic effects of Rnd1 and RhoD GTPases regulate receptor activity in Semaphorin 3A-induced cytoskeletal collapse. J Neurosci 22: 471-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Benson DL ( 2001) Stages of synapse development defined by dependence on F-actin. J Neurosci 21: 5169-5181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziv NE, Smith SJ ( 1996) Evidence for a role of dendritic filopodia in synaptogenesis and spine formation. Neuron 17: 91-102. [DOI] [PubMed] [Google Scholar]


