Abstract
Small Src homology domain 2 (SH2) and 3 (SH3) adapter proteins regulate cell fate and behavior by mediating interactions between cell surface receptors and downstream signaling effectors in many signal transduction pathways. The CT10 regulator of kinase (Crk) family has tissue-specific roles in phagocytosis, cell migration, and neuronal development and mediates oncogenic signaling in pathways like that of Abelson kinase. However, redundancy among the two mammalian family members and the position of the Drosophila gene on the fourth chromosome precluded assessment of Crk’s full role in embryogenesis. We circumvented these limitations with short hairpin RNA and CRISPR technology to assess Crk’s function in Drosophila morphogenesis. We found that Crk is essential beginning in the first few hours of development, where it ensures accurate mitosis by regulating orchestrated dynamics of the actin cytoskeleton to keep mitotic spindles in syncytial embryos from colliding. In this role, it positively regulates cortical localization of the actin-related protein 2/3 complex (Arp2/3), its regulator suppressor of cAMP receptor (SCAR), and filamentous actin to actin caps and pseudocleavage furrows. Crk loss leads to the loss of nuclei and formation of multinucleate cells. We also found roles for Crk in embryonic wound healing and in axon patterning in the nervous system, where it localizes to the axons and midline glia. Thus, Crk regulates diverse events in embryogenesis that require orchestrated cytoskeletal dynamics.
INTRODUCTION
Cells have an incredible ability to receive environmental inputs and respond in context-dependent manners during embryogenesis and adult homeostasis. Diverse signals, including growth factors, adhesion to neighbors, extracellular matrix engagement, or bioactive lipids, lead to diverse cellular responses, including changes in transcription, proliferation, cell shape, motility, or cell death. Despite the diversity of inputs and the responses they evoke, pathways often share an underlying logic. Signals are usually received by transmembrane receptors (e.g., receptor tyrosine kinases [RTKs], integrins, or cadherins), and signal transduction is often initiated by the formation of multiprotein complexes associated with receptor cytoplasmic tails. Small adapter proteins such as Grb2, Crk, or Nck play critical roles in both signal amplification and specificity (e.g., Lettau et al., 2009; Belov and Mohammadi, 2012; Martinez-Quiles et al., 2014). They confer signal specificity by using Src homology domain 2 (SH2) and 3 (SH3) or other protein interaction domains to physically link activated receptors and downstream effectors. Particular small adapter proteins only interact with select receptors and effectors, ensuring specificity.
CT10 regulator of kinase (Crk) proteins are a conserved family of small modular adapters shared by diverse animal lineages (reviewed in Feller, 2001; Birge et al., 2009; and Hossain et al., 2012). In vertebrates, two genes encode three protein isoforms: CrkI, CrkII (alternative splice forms of crk), and Crk-like (CrkL). An N-terminal SH2 domain allows them to bind tyrosine phosphorylated proteins, while a canonical SH3 domain binds proline-rich motifs (“PXXP”). CrkII and CrkL have an additional, noncanonical C-terminal SH3 domain that does not bind PXXP motifs, but serves important regulatory roles. This organization allows Crk family adapters to link PXXP-containing downstream effectors to upstream proteins containing phosphorylated tyrosines, such as activated RTKs.
Our interest in Crk was initiated by our work on the developmental roles of the non-RTK Abelson tyrosine kinase (Abl; reviewed in Khatri et al., 2016). Abl is the oncogene activated in chronic myelogenous leukemia and the target of the clinically effective kinase inhibitor imatinib (Greuber et al., 2013). Drosophila Abl plays important roles in many processes, ranging from the cytoskeletal rearrangements of syncytial development and cellularization (Grevengoed et al., 2003), to cell shape changes during gastrulation and dorsal closure (Grevengoed et al., 2001; Fox and Peifer, 2007), and to axon outgrowth and guidance in the nervous system (e.g., Gertler et al., 1989; Bashaw et al., 2000; Kannan and Giniger, 2017). While Abl kinase activity is important for function, an even more essential determinant of function is a short, conserved PXXP motif in the Abl linker known in mammals to bind Crk, Nck, and Abi (Rogers et al., 2016). Crk is a known Abl substrate and a potential upstream activator (Hossain et al., 2012), and Crk knockdown alleviates the effects of Bcr-Abl transformation (e.g., Seo et al., 2010). This stimulated our interest in Crk as a potential Abl regulator or effector in Drosophila.
Crk adapters modulate a wide range of cell behaviors in cultured cells, including cell growth, migration, adhesion, and immune function (reviewed in Feller, 2001; Birge et al., 2009; Hossain et al., 2012). For example, in fibroblasts CrkI/II and CrkL play redundant roles in regulating cell growth, proliferation (Park et al., 2016), morphology, motility (Park and Curran, 2014; Park et al., 2016), and oncogene-mediated cell transformation (Koptyra et al., 2016). Crk proteins localize to focal adhesions to promote their stability (Senechal et al., 1996; Li et al., 2003; Antoku and Mayer, 2009; Watanabe et al., 2009; Park and Curran, 2014), thus promoting cell spreading and motility (Chodniewicz and Klemke, 2004), and work downstream of integrins to coordinate motility and mechanosensing in T-cells (Roy et al., 2018). Not surprisingly, Crk proteins also can play important roles in cancer, promoting cell transformation and tumor growth (reviewed in Sriram and Birge, 2010; Kumar et al., 2014).
Crk’s roles in normal development are less well understood. CrkII is dispensable (Imaizumi et al., 1999). Mice lacking both CrkI and CrkII die as embryos with multiple defects in cardiovascular and craniofacial development (Park et al., 2006). CrkL knockout mice have reduced viability and multiple defects associated with cardiac and neural crest derivatives (Guris et al., 2001), at least in part via effects on FGF signaling (Moon et al., 2006). Interestingly, CrkL maps to a chromosomal region commonly deleted in DiGeorge syndrome patients, and phenotypic similarities suggest its loss may contribute to this disease. Tissue-specific knockouts revealed additional roles for Crk or CrkL in places ranging from T-cell migration (Huang et al., 2015; Roy et al., 2018) to Bcr-Abl transformation of mouse hematopoietic progenitors (Seo et al., 2010). The two mouse Crk family members have redundant roles in many tissues. Simultaneous neural-specific knockout of Crk and CrkL revealed essential overlapping roles downstream of Disabled-1 in the Reelin pathway regulating neuronal positioning (Park and Curran, 2008). Similar overlapping roles were seen in the kidney (George et al., 2014) and lens (Collins et al., 2018). However, to our knowledge, there has been no analysis of embryos lacking both Crk and CrkL from the onset of development. In Caenorhabditis elegans and zebrafish, Crk adapters play important developmental roles in cell migration, axon outgrowth, phagocytosis, and myoblast fusion. C. elegans Ced-2 regulates phagocytic clearance of apoptotic cells, cell migration, and motor neuron development as part of a Crk/DOCK180/Rac pathway (Reddien and Horvitz, 2000; Gumienny et al., 2001; Wu et al., 2001, 2002). In zebrafish, morpholino-mediated knockdown of either Crk or CrkL impairs fusion of fast-twitch myoblasts (Moore et al., 2007), while overexpressing Crk or CrkL enhances fusion.
Drosophila has a single Crk family member, thus providing a simplified model to define core Crk functions. Crk can directly bind potential upstream regulators such as the RTK PDGF- and VEGF-receptor related (PVR) (Ishimaru et al., 2004) and the Ig-family receptor Sticks and Stones (Kim et al., 2007), as well as potential downstream effectors such as the Rac Guanine-nucleotide exchange factor Myoblast City (Mbc, Drosophila DOCK180; Galletta et al., 1999; Balagopalan et al., 2006), Verprolin 1 (Drosophila Wiskott-Aldrich syndrome protein (WASP)-interacting protein; Kim et al., 2007), and its regulator Blown fuse (Jin et al., 2011). Much of this analysis has focused on roles of Crk in mediating Dock180/ELMO/Rac1 signaling to the actin cytoskeleton via WASP, suppressor of cAMP receptor (SCAR), and the actin-related protein 2/3 (Arp2/3) complex (reviewed in Deng et al., 2017). Tissue-specific RNA interference (RNAi) revealed Crk acts upstream of Mbc/Elmo/Rac1 signaling in phagocytic clearance of axonal debris and pathogenic protein aggregates by glial cells (Ziegenfuss et al., 2012; Pearce et al., 2015), developmental pruning of larval neurites (Tasdemir-Yilmaz and Freeman, 2014), removal of apoptotic neurons (Nakano et al., 2019), and salivary gland clearance by autophagy (McPhee et al., 2010). Fly Crk also regulates phagocytic uptake of pathogenic bacteria (Elwell et al., 2008; Pielage et al., 2008), as does mammalian Crk (reviewed in Martinez-Quiles et al., 2014). Dock180/ELMO/Rac1 signaling acts downstream of the RTK PVR during thorax closure and border cell migration, and use of tissue-specific RNAi and a hypomorphic crk allele supports roles for Crk in both processes (Ishimaru et al., 2004; Geisbrecht et al., 2008). Another DOCK family GEF, the DOCK3 relative Sponge, acts in parallel to Mbc in both processes (Bianco et al., 2007; Morishita et al., 2014). Dock180/ELMO/Rac1 signaling also regulates the actin cytoskeleton during myoblast fusion, but despite clear evidence that Crk binds proteins essential for myoblast fusion, it is unclear whether Crk is required. Overexpressing a membrane-targeted form of Crk disrupts myoblast fusion (Abmayr et al., 2003), but Mbc mutants lacking all Crk-binding sites fully restore fusion (Balagopalan et al., 2006). However, muscle-specific RNAi led to a flightless phenotype with missing indirect flight muscles (Schnorrer et al., 2010). Thus, tissue-specific Crk knockdown revealed important developmental roles, most involving DOCK/ELMO/Rac regulation of actin.
While informative, analyses via tissue-specific RNAi do not reveal the core, conserved roles for Crk adapters during embryogenesis. We sought to generate embryos devoid of Crk and thus analyze its full set of functions. Despite its identification nearly 20 yr ago, there has been surprisingly little genetic analysis of Drosophila crk, largely due to the fact that it maps to the fourth chromosome, which was less genetically tractable. The only allele was a hypomorphic mobile element insertion (Ishimaru et al., 2004). Modern tools allowed us to circumvent this. We used second-generation short hairpin RNA (shRNA) lines and developed a suite of genetic tools, including a null allele, a gene replacement platform, and overexpression constructs. These allowed us to manipulate Crk levels in many ways, revealing that Crk is required for embryonic viability and plays critical roles during actin-dependent morphogenic events.
RESULTS
Crk is required for embryonic viability
Despite Crk’s importance in oncogenic signaling and the known tissue-specific roles of the two mammalian family members, its full importance in normal development remains unclear. Drosophila provides a superb model to examine this role, as there is only a single family member, thus avoiding the partial redundancy of the mammalian proteins. However, the position of crk on the fourth chromosome hindered the ability to examine its function, with previous studies limited to the use of a probable hypomorphic allele derived from P-element insertion (Ishimaru et al., 2004) or tissue-specific RNAi. New tools offered the opportunity to develop ways to reduce or eliminate Crk function.
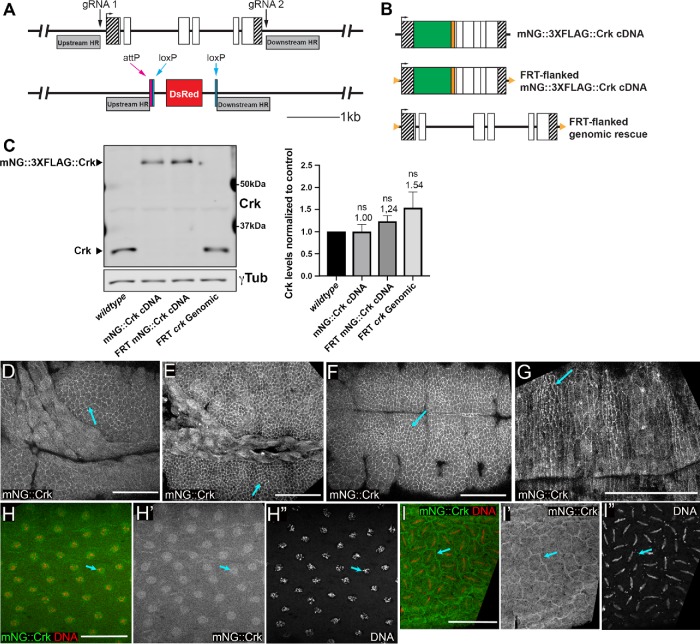
We took two approaches: using CRISPR to generate tagged and null alleles and RNAi to knockdown function. We generated a crk null allele, crkΔattP, using CRISPR to replace the entire crk locus with a 50 base-pair attP phage recombination site and positive selectable marker (Figure 1A). This allele was verified by PCR (Supplemental Figure 1, A and B). Consistent with the previously characterized allele crkKG00336 (Ishimaru et al., 2004), homozygous zygotic crk∆attP mutants were embryonic viable but died as pupae, revealing crk to be an essential gene. We next created a series of rescue constructs, in which either the complete genomic region or an monomeric NeonGreen (mNG)-tagged cDNA was targeted back into the locus at the attP site (Figure 1B). The genomic rescue construct and one version of the cDNA rescue constructed were flanked by FRT sites (Figure 1B; full descriptions are in Supplemental Figure 1C) allowing deletion via the FLP recombinase to facilitate production of females whose germlines were homozygous mutant. We also generated a Crk antibody using the full-length protein as the antigen, which recognize a single protein of molecular weight ∼31 kDa (Figure 1C), consistent with the predicted size of Crk (31 kDa). All three rescue constructs are expressed at essentially normal levels (Figure 1C), and all rescued viability and fertility.
FIGURE 1:
Generation of a crk null allele and a gene replacement platform. (A) Schematic representation: crk locus before and after CRISPR/Cas9-mediated deletion of crk to generate crkΔattP. We used two gRNAs to introduce double-strand breaks 110 base pairs upstream of the transcription start site and 99 base pairs downstream of the end of the 3′UTR. Using a double-stranded donor repair template containing flanking homology regions (indicated by shaded gray bars), we replaced the crk coding sequence with an attP recombination site and a loxP-flanked, positive visual marker (3xP3-DsRed expressed in the eye). (B) Schematic representation: rescue alleles targeted into crkΔattP. The three constructs include cDNA-based constructs containing a tandem N-terminal tag (mNG; green, and a 3XFLAG tag; orange), with or without flanking FRT sites (orange triangles), and an FRT-flanked genomic rescue construct carrying the exact sequence removed in generating crkΔattP. (C) Representative Western blot and quantification of Crk levels in wild type and crkΔattP-targeted rescue lines. Rescue constructs restore endogenous levels of Crk; n = 4, error bars = SEM, ns = not statistically significant. (D–I) Representative images: Crk localization throughout embryonic development. Embryos expressing mNG-tagged Crk (mNG::Crk) inserted at the endogenous locus. The tagged protein was imaged using mNG fluorescence. H and I were also stained to reveal DNA. (D) Stage 9. (E) Stage 10. (F) Stage 11. (G) Stage 14. After gastrulation Crk localized to a cytoplasmic pool with enrichment at the cell cortex (arrows). (H, I) Syncytial blastoderm. Crk is enriched in actin caps that overlie nuclei (H, arrow) and at pseudocleavage furrows (I, arrow). Scale bars = 50 µm.
While zygotic mutants were embryonic viable, we suspected this was due to maternally contributed Crk, as crk mRNA is maternally loaded and broadly expressed (Galletta et al., 1999). We hypothesized Crk would be broadly expressed and essential for embryonic morphogenesis; consistent with this, our mNG-tagged construct targeted into the endogenous locus was expressed throughout embryogenesis (Figure 1, D-I). After gastrulation, we observed cortical enrichment in the ectoderm and developing epidermis, along with a cytoplasmic pool (Figure 1, D–G, arrows)–-this continued until at least the completion of dorsal closure (Figure 1G).
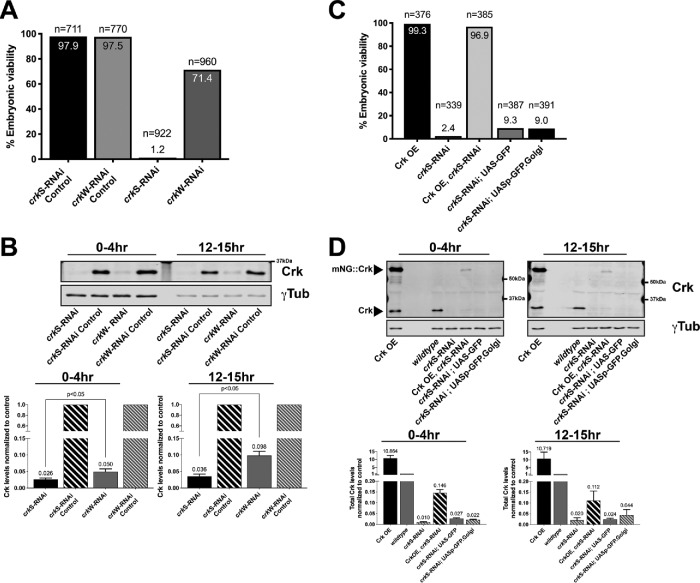
To directly test the hypothesis that Crk is essential for embryonic development, we used RNAi to maternally and zygotically deplete crk. We used two independent UAS-driven shRNA lines targeting different regions of crk developed by the Transgenic RNAi Project (TRiP; Perkins et al., 2015). shRNA constructs can mimic maternal/zygotic loss of gene products when expressed using germline drivers (Staller et al., 2013). We drove expression of shRNAs targeting crk using two copies of a strong germline driver (maternal αtubulin-GAL4; matα-GAL4) to knock down maternal crk. Both RNAi lines impair embryonic viability, but to differing extents. The stronger of the two lines, referred to below as strong crk RNAi (crkS-RNAi), severely impaired embryonic viability, reducing viability to 1.2% (Figure 2A; n = 922), while the second line, referred to below as weak crk RNAi (crkW-RNAi), resulted in weaker effects, reducing viability to 71.4% (Figure 2A; n = 960), compared with UAS–shRNA-only controls, which exhibited 97.9% (Figure 2A; n = 711) and 97.5% (Figure 2A; n = 770) viability, respectively (Supplemental Table S1 includes details of the number of biological replicates and other quantitative information for this and subsequent figures).
FIGURE 2:
Crk is required for embryonic viability. (A) Quantification of embryonic viability following shRNA-mediated knockdown of Crk. Compared to shRNA-only controls (crkS-RNAi Control and crkW-RNAi Control), Crk knockdown using either of two shRNA lines driven by a pair of strong maternal GAL4 drivers reduces embryonic viability. (B) Representative Western blot and quantification of Crk levels in Crk knockdown embryos compared with controls. crkS-RNAi embryos have a stronger reduction in Crk levels compared with crkW-RNAi embryos; n = 4, error bars = SEM. (C) Quantification of embryonic viability following overexpression of Crk or simultaneous knockdown/overexpression of Crk. Simultaneous overexpression of Crk fully restores embryonic viability in crkS-RNAi embryos. Crk overexpression (Crk OE; ∼10-fold higher than endogenous) alone has no impact on embryonic viability. Expressing two different extraneous GFP-tagged proteins to control for effects of two UAS promotors does not substantially rescue lethality. (D) Representative Western blot and quantification of Crk levels in embryos overexpressing Crk (lane one), wild type (lane four, note lanes two and three intentionally left empty), crkS-RNAi embryos (lane five), embryos undergoing simultaneous knockdown/overexpression (lane six), or in crkS-RNAi embryos simultaneously expressing GFP-tagged proteins to control for effects of two UAS promotors (lanes seven and eight), n = 3, error bars = SEM.
To verify that the shRNA constructs knocked down crk, and, in parallel, to test specificity of our antibody, we examined Crk accumulation at two stages of embryogenesis by immunoblotting. Both shRNAs strongly reduced Crk protein levels during early embryonic development, with crkS-RNAi dropping levels to 2.6% and crkW-RNAi to 5.0% at 0–4 h (Figure 2B; left), consistent with their differential effects on embryonic lethality. Expression remained strongly reduced at 12–15 h (3.6% and 9.8%, respectively; Figure 2B, right). We next tested whether simultaneous overexpression of Crk, using UAS-driven mNG-tagged Crk, rescued effects of knockdown. Crk overexpression alone, even at ∼10-fold endogenous levels (Figure 2D), had no effect on embryonic viability (99.3%, n = 376; Figure 2C). Simultaneously overexpressing Crk in the crkS-RNAi background restored viability to wild-type levels (96.9% viability, n = 385; Figure 2C), supporting the idea that embryonic lethality of Crk knockdown was due to on-target effects. Immunoblotting revealed that this restored total Crk levels (tagged plus endogenous) to 11–15% of normal endogenous levels (Figure 2D). To verify that this rescue of viability was due to Crk overexpression and not due to titration of GAL4 by the presence of additional UAS-driven elements, we overexpressed either GFP (Valium10 GFP) or a Golgi marker (UASp-GFP.Golgi) in the crkS-RNAi background. Neither substantially rescued lethality (9.3% viability, n = 387 and 9.0% viability, n = 391, respectively; Figure 2C). We further verified the essential role of Crk in embryonic development using our FRT-flanked allele by expressing FLP recombinase in the female germline to delete the FRT-flanked crk gene, creating females lacking maternally contributed Crk. When these females were crossed to crk heterozygous males, 78% of the embryos died (n = 698), consistent with fully penetrant maternal-zygotic lethality and only partial ability of the paternal zygotic gene to rescue viability. Thus, Crk is essential for embryonic viability.
Loss of Crk disrupts embryonic morphogenesis and leads to formation of multinucleate cells
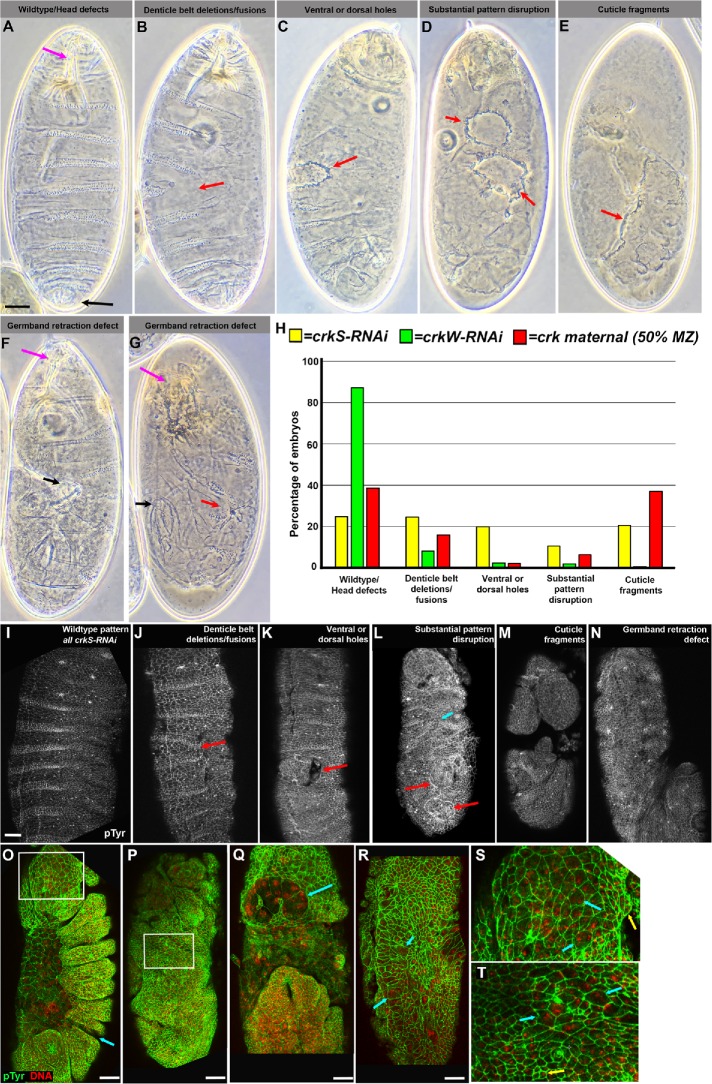
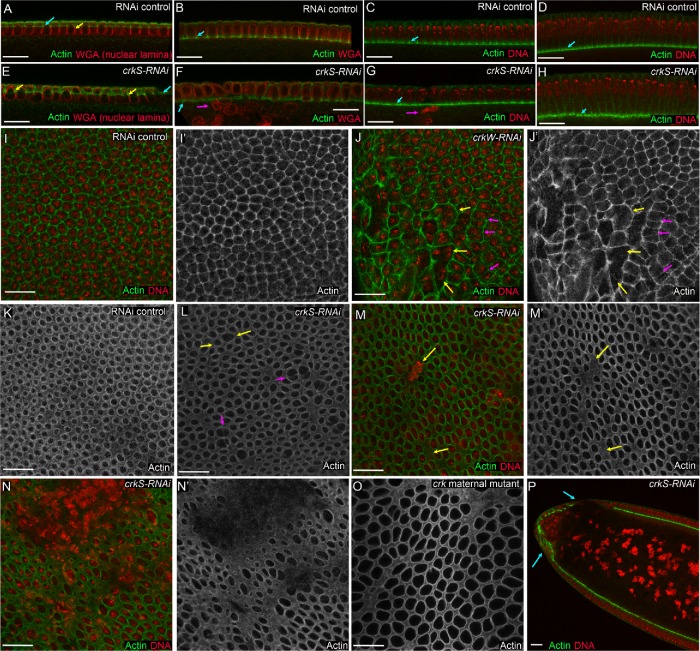
As a first assessment of Crk’s role in embryonic development, we examined the cuticle secreted by the epidermis to ask whether cell fates were properly determined, major morphogenetic movements such as head involution and dorsal closure were successfully completed, and whether the epidermis remained intact (Figure 3, A–G). crkS-RNAi and crkW-RNAi embryos exhibited certain characteristic defects, though at different frequencies (Figure 3, A–H). crkS-RNAi embryos had stronger phenotypes, with substantial disruption of epidermal integrity in most embryos. In 25% apparent cell loss occurred, leading to deletion or fusion of denticle belts (Figure 3, B and G, red arrows, and H). Twenty percent had holes in the head, ventral, or dorsal cuticle (Figure 3, C, arrow, and H), while 11% had more substantial pattern disruption (Figure 3, D, arrows, and H), and 21% retained only fragments of cuticle (Figure 3, E, arrow, and H). In embryos in the first three categories, we also noted mild to complete failure to complete germband retraction in 41% (Figure 3, F and G vs. A, magenta to black arrows). We suspect the phenotypic spectrum reflects the fact that embryos can inherit one or two shRNA constructs. After crkW-RNAi most embryos secreted largely wild-type cuticles (87% including the 71% that hatched; Figure 3H). However, in 8%, deletion or fusion of denticle belts was observed (Figure 3H). Twenty percent of the lethal embryos were also defective in germband retraction. We next generated embryos maternally mutant for Crk using FLP recombinase to excise the genomic rescue construct in the female germline–-half of these were also zygotically mutant. The spectrum of defects was quite similar to those seen in crkS-RNAi. Roughly half, potentially those receiving a wild-type gene zygotically, had wild-type cuticles or deletion or fusion of denticle belts (Figure 3H). The other half had more severe disruptions of the embryonic pattern or only fragments of cuticle (Figure 3H). Defects in head involution, germband retraction, and epidermal integrity are all shared with abl maternal/zygotic mutants (Grevengoed et al., 2001; Rogers et al., 2016). The more severe pattern disruptions were similar to those observed in embryos maternally mutant for the septin protein Peanut (Adam et al., 2000), or the actin-regulatory protein Bottleneck (Schejter and Wieschaus, 1993), suggesting the possibility that, as in peanut or bottleneck mutants, the underlying defects might occur very early, when many cells were disrupted at or right after cellularization.
FIGURE 3:
Crk loss disrupts embryonic morphogenesis and leads to the formation of multinucleate cells. (A–G) Embryonic cuticles, anterior at top. Range of phenotypes in crk maternal-zygotic mutants or after crkS-RNAi or crkW-RNAi. (A) Wild type. Note intact, correctly patterned epidermis, and correct completion of germband retraction with head skeleton at anterior (magenta arrow) and spiracles at posterior (black arrow). (B) Some mutant or knockdown embryos had deletion or fusion of denticle belts (arrow), although the cuticle remained intact. (C) Others had holes in the anterior, dorsal, or ventral cuticle (arrow). (D) In severe cases, there were multiple segmental deletions or epidermal holes (arrows). (E) In the most severe cases, only fragments of cuticle remained (arrow). (F) Embryo with a wild-type pattern but in which germband retraction failed, leaving the spiracles (black arrow) curled around the dorsal side. (G) Embryo combining denticle belt deletion (red arrow) and germband retraction failure (black arrow). (H) Frequencies of defects illustrated; n = 439 crkS-RNAi, 168 crkW-RNAi, and 286 crk maternal cuticles scored. crk maternal-zygotic mutants and crkS-RNAi lead to a similar range of defects while those caused by crkW-RNAi are generally milder. (I–N) Stage 15 embryos stained with an antibody to pTyr revealing cell outlines. Defects in morphogenesis mirror those seen later in the cuticle, including denticle belt fusions (J, arrow) and epidermal holes (K, L, red arrows). (O–T) Embryos stained for pTyr and a DNA dye. (O, P) Stages 13 and 14. Note pattern disruptions or epidermal holes (arrow in O). Boxed areas are enlarged in S and T. (Q) Stage 11. Note large multinucleate cells in the head (arrow). (R) Stage 9. Many large multinucleate cells are seen in the ectoderm (arrows). (S, T) Closeups from embryos in O and P, contrasting normal cells (yellow arrows) with larger multinucleate cells (cyan arrows). Scale bars = 30 µm.
We extended this analysis by staining embryos with antibodies to phosphotyrosine (pTyr) to outline cells, allowing rapid assessment of morphogenesis. We first examined embryos after morphogenesis should largely be complete, at dorsal closure, or later. We observed embryos with a range of defects reflecting those seen in the cuticles (Figure 3, I–N). Some were wild type (Figure 3I), while others had mild disruptions of the epidermal pattern or small holes in the epidermis (Figure 3, J and K, arrows). In others, morphogenesis was more severely disrupted, with defects in head involution or more severe disruptions in epidermal patterning (Figure 3L, cyan arrow) or integrity (Figure 3, L, red arrows, and M). Embryos with germband retraction defects were also observed (Figure 3N). Earlier stage mutants similarly exhibited gaps in the embryonic pattern and holes in the ectoderm (Figure 3, O and P). Most striking, we noted frequent examples of multinucleate cells in the developing head (Figure 3O, box enlarged in Q and S) and at a lower frequency in the thorax and abdomen (Figure 3P, box enlarged in T). Once again, these were defects we had previously observed in both peanut (Adam et al., 2000) and abl maternal/zygotic mutants (Grevengoed et al., 2003), both of which have defects in syncytial development and/or cellularization. These results prompted us to begin a detailed analysis at these stages.
Loss of Crk disrupts the cytoskeletal events driving syncytial development
Our data suggested that Crk knockdown or loss causes defects that precede the onset of morphogenesis at gastrulation. Drosophila begins development as a syncytium with a series of nuclear divisions without cytokinesis, and thus we examined these syncytial divisions to define the earliest role of Crk. The last four rounds of nuclear division occur after nuclei have migrated to the embryo cortex, thus setting the initial membrane and cytoskeletal polarity of the system (Mavrakis et al., 2009). The nuclei become increasingly closely packed through these four rounds of mitosis, and there is a dynamic cycle of rearrangement of the actin and myosin cytoskeletons that prevents collision of adjacent mitotic spindles, which could lead to aneuploidy. During interphase, actin forms “caps” at the cortex above each nucleus. As spindle assembly begins, these actin caps expand, and when adjacent caps meet, the cytoskeleton is driven inward into transient furrows that surround each mitotic spindle and insulate it from its neighbors (Zhang et al., 2018). These pseudocleavage furrows recede as mitosis is completed and caps reform, beginning a new cycle. In mutants in which furrow formation is blocked or reduced (e.g., Sullivan et al., 1993; Afshar et al., 2000; Zallen et al., 2002; Webb et al., 2009), spindles often collide, leading to DNA damage and/or aneuploidy. This triggers a Chk2-mediated response by which aneuploid nuclei are removed from the embryo surface by “dropping” into the interior of the egg during the next mitotic cycle (Takada et al., 2003). At the end of nuclear cycle 13, furrows form around each nucleus and pull membranes downward, enclosing each nucleus in a cell in a process called cellularization. Actin rings stabilize yolk channels at the bottom of each new cell, connecting it to the underlying yolk.
In wild type, the dynamic cycle of actin rearrangements ensures that most syncytial mitoses occur without errors. Nuclei are well spaced at cycle 10, not all caps encounter a neighbor, and furrows are relatively shallow. Spindles assemble, and chromosomes segregate without errors (Figure 4A). As each cycle proceeds, nuclear density increases (Figure 4, C, E, G, and J); however, pseudocleavage furrows generally succeed in maintaining spindle separation. In wild type, occasional defects in spindle separation and chromosome segregation occur in later cycles (1/8 and 4/19 cycle 11 and 12 embryos had one to two spindle collisions per field of view; Figure 4P). Defective nuclei are removed into the yolk in the next cycle. This affects 2–3% of nuclei in wild type (Sullivan et al., 1993). Centrosomes remain behind and continue to organize the actin cytoskeleton into smaller caps and furrows (Figure 4G, arrow). Nuclear loss tends to affect small groups of neighboring nuclei. In wild type, increasing numbers of cycle 11 or 12 embryos had small patches of nuclear loss (4/8 and 13/19 at cycles 11 and 12 relative to 1/10 in cycle 10), but there were only 1–4 patches per embryo and these included only a relatively small number of nuclei (e.g., Figure 4G, arrow).
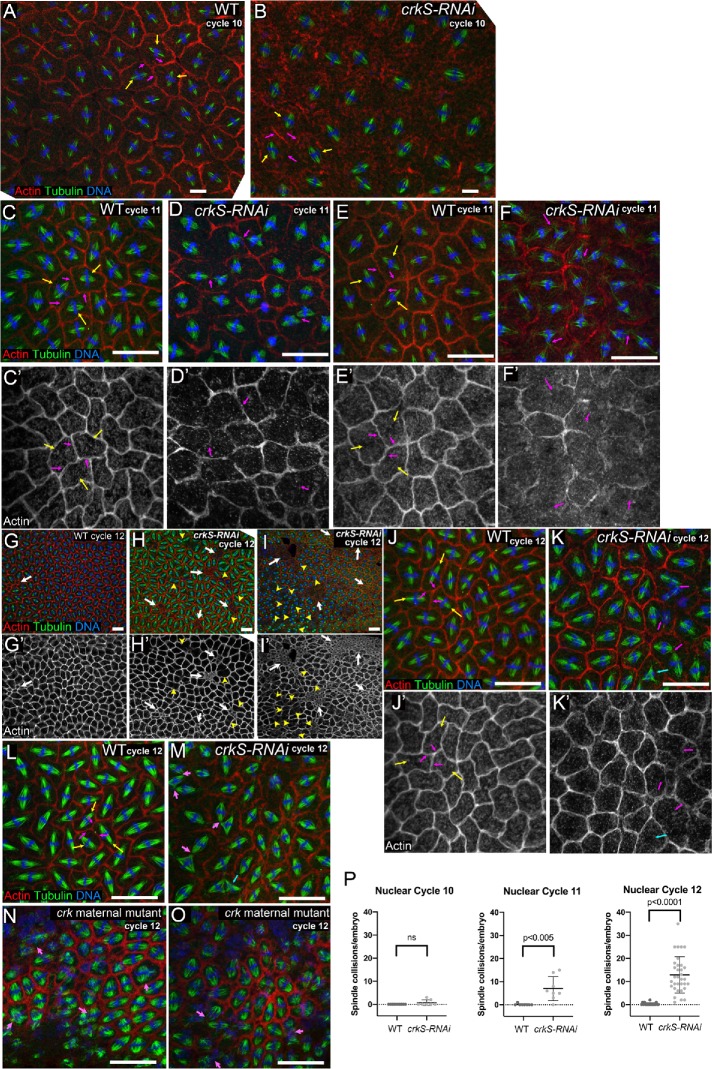
FIGURE 4:
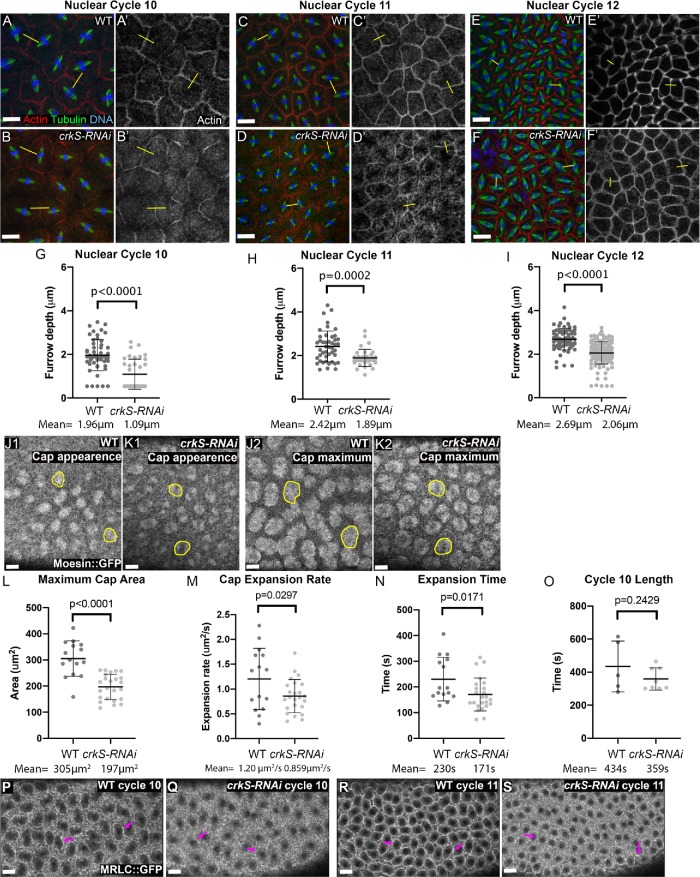
Crk knockdown disrupts the cytoskeletal events driving syncytial development. (A–O) Embryos in metaphase to anaphase of indicated syncytial nuclear cycle, stained to visualize F-actin, tubulin, and DNA. (A, B) Nuclear cycle 10. (A) Wild type. Each spindle and its associated chromosomes (yellow arrows) are surrounded by a pseudocleavage furrow (magenta arrows), separating nuclear compartments and preventing spindle collisions. (B) crkS-RNAi. Spindles remain separated from one another (yellow arrows), although pseudocleavage furrows were often less apparent (magenta arrows). (C–F) Nuclear cycle 11. (C, E) Wild type. Spindles remain well separated (yellow arrows) and pseudocleavage furrows continue to separate nuclear compartments (magenta arrows). (D, F) crkS-RNAi. Most embryos have multiple spindle collisions (magenta arrows). These usually occur in places where pseudocleavage furrows are discontinuous or absent. (G–O) Nuclear cycle 12. (G) Most wild-type embryos have a small amount of nuclear loss, revealed by small actin rings, usually only involving a few nuclei (arrow). Spindle collisions remain very rare. (H, I) crkS-RNAi. Most embryos have multiple regions of nuclear loss (white arrows), and in severe cases these occupy a significant fraction of the embryo surface (I, arrows). Spindle collisions (yellow arrowheads) occur in regions adjacent to and distant from regions of nuclear loss. (J, L) Wild type. Spindles (yellow arrows) remain separated by pseudocleavage furrows (magenta arrows). (K, M) crkS-RNAi. All embryos have multiple spindle collisions (magenta arrows), sometimes leading to tripolar spindles (cyan arrows). Spindle collisions occur in regions where pseudocleavage furrows are weak or absent. (N, O) crk maternal mutants exhibiting spindle collisions (arrows) and nuclear loss. (P) Quantification, number of spindle collisions per embryo in wild type and crkS-RNAi in nuclear cycles 10–12; n = 10 wild type/7 crkS-RNAi, 8 wild type/8 crkS-RNAi, and 19 wild type/33 crkS-RNAi embryos scored for nuclear cycles 10, 11, and 12, respectively. Spindle collisions are elevated in cycle 11 and highly elevated in cycle 12. Scale bars = 20 µm.
We first examined Crk localization during syncytial divisions, using our mNG-tagged construct. Strikingly, Crk is enriched in both actin caps (Figure 1H) and metaphase furrows (Figure 1I), consistent with the possibility that Crk plays a role in cytoskeletal regulation. We next examined the effect of Crk knockdown. In crkS-RNAi embryos, nuclear cycle 10 proceeded relatively normally (Figure 4B), though pseudocleavage furrows appeared less continuous (see below), and 2/7 embryos observed had individual examples of spindles that had collided (quantified in Figure 4P). However, by nuclear cycle 11, spindle defects became more frequent: in 7/8 embryos examined neighboring spindles collided (Figure 4, D and F, magenta arrows), often in places where pseudocleavage furrows were reduced or absent. Embryos often had multiple examples in a single field of view (quantified in Figure 4P). At cycle 11, the frequency of embryos with regions of nuclear loss remained similar to that in wild type (4/8 embryos observed). However, by nuclear cycle 12 all crkS-RNAi embryos (33/33) had multiple spindle collisions, and in the most severely affected, these included a substantial fraction of the spindles in the field of view (Figure 4, H and I, arrowheads; K and M, magenta arrows, quantified in P). The incidence of nuclear loss increased (91% affected vs. 68% in wild type), and the fraction of nuclei involved was much higher, with 71% having multiple groups of nuclei lost, including embryos in which this affected much of the embryo surface (Figure 4, H and I, white arrows). We think that the extensive nuclear loss likely reflects the frequency of spindle collisions in the previous cycle. Spindle collisions and nuclear loss were also observed in crk maternal mutants generated using our FRT flanked genomic constructs (Figure 4, N and O). These data reveal that Crk plays an important role in regulating the intricate cytoskeletal rearrangements of syncytial development.
At the end of the syncytial stage, cellularization forms the first cells (Lee and Harris, 2014; Schmidt and Grosshans, 2018). An actomyosin ring assembles at the cortex above each nucleus–-the rest of the cortex is covered with actin-rich microvilli. Cofilin, Anillin, Septins, Rho-driven myosin contractility, regulated actin assembly involving the formin Diaphanous, and regulation of membrane dynamics all play roles (e.g., Grosshans et al., 2005; Padash Barmchi et al., 2005; Figard et al., 2016; Xue and Sokac, 2016), driving the movement of these rings inward, enclosing each nucleus in a plasma membrane (Figure 5, A–D and I). Both actin and myosin localize to the leading edge during this event (Figure 5, A–D, cyan arrows). At the end of cellularization, the actomyosin rings constrict to partially close off each cell, but this halts before completion, leaving an actin-lined “yolk channel” at the basal end of each cell (Figure 5K). Abl plays a role in this process (Grevengoed et al., 2003), and many genes involved in syncytial divisions also play a role in cellularization. We thus examined cellularization after crk RNAi.
FIGURE 5:
Crk knockdown leads to defects in cellularization. Cellularizing control, crkS-RNAi and crk maternal mutant embryos stained with phalloidin to visualize F-actin and either WGA to visualize the nuclear lamina or a DNA dye. (A–H) Cross-section views during early (A, E), mid- (B vs. F, C vs. G), and late (D, H) cellularization. (A–D) In controls, as in wild type, actin localizes to the cellularization front (cyan arrows) moving basally as a ring around each nucleus and then partially closing, leaving the yolk channel. Apical actin-rich microvilli gradually disappear during this process. Nuclei are uniform in shape (e.g., A, yellow arrow) and positioned in a regular row. (E–H) crkS-RNAi. Actin can assemble at the front and move basally (cyan arrows). However, the nuclear shape is irregular (E, yellow arrows; F), possibly reflecting premature ring closure. Some nuclei reside below the cortex (F, G, magenta arrows), likely a result of nuclear loss in syncytial cycles. (I, J) En face views, midcellularization. (I) Wild type. Actin rings are uniform in size and actin intensity and enclose single nuclei. (J) crkS-RNAi. Note multinucleate actin compartments (yellow arrows) and examples of weak actin localization to a subset of compartment boundaries (magenta arrows). (K–O). Late cellularization at the level of forming yolk channels. (K) Wild type. Rings partially close, leaving actin-lined yolk channels of uniform size. (L, M) Examples of milder defects after crkS-RNAi. Yolk channels are uneven in size (L, yellow arrows) and sometimes two channels are separated only by a weak actin border (L, magenta arrows). Regions without yolk channels are also seen (M, yellow arrows), some of which coincide with places where nuclei lie below the cortex. (N) Severe example of crkS-RNAi defects with a large region of nuclear loss. (O) crk maternal mutant exhibiting similar variability in yolk channel size. (P) Cross-section: crkS-RNAi embryo at gastrulation onset. Many embryos have severe cellularization defects in the presumptive head (arrows). Scale bars = 20 µm.
Several defects were readily apparent in most crkS-RNAi embryos. Nuclear shape was less uniform than in controls (Figure 5, E, yellow arrows vs. A and B). Nuclei often appeared “bottle-shaped” rather than oval, a phenotype that can reflect premature closure of the actomyosin ring before it passes the bottom of the nucleus, thus squeezing the nuclei and resulting in the so-called bottleneck phenotype (Schejter and Wieschaus, 1993). In controls, nuclei form a uniform row of similar shape (Figure 5, A–C), whereas in crkS-RNAi embryos, nuclei were seen below this row (Figure 5, F and G, magenta arrows); these may include both nuclei removed from the surface in the previous syncytial division and nuclei forced downward by premature actin ring closure. Viewed en face, actin rings in wild-type embryos are largely uniform in shape and intensity, and each encloses a single nucleus (Figure 5I). In contrast, after crk RNAi, actin rings were less uniform, the actin staining at the border between two nuclear compartments was sometimes less intense (Figure 5J, magenta arrows), and in other cases it was absent, leaving multiple nuclei in a single membrane compartment (Figure 5J, yellow arrows). This disruption of nuclear compartmentalization is a likely mechanism for the formation of multinucleate cells we observed above. We also observed striking defects in yolk channels in Crk-depleted embryos. While these are uniform in size and shape in wild type (Figure 5K), they varied dramatically in size and shape following crkS-RNAi (Figure 5, L–N). Yolk channels were uneven in size, with some channels abnormally enlarged (Figure 5L, yellow arrows), and others appeared to be separated by only a weak actin border (Figure 5L, magenta arrows). Other regions were devoid of yolk channels altogether–-these often overlaid nuclei that had been removed from the cortex (Figure 5M, arrows). In the most severe cases, large regions of the embryo were affected (Figure 5N), as we had observed at the syncytial stages–-these were prominent in the presumptive head as gastrulation began (Figure 5P, arrows). Defects in cellularization leading to variable size yolk channels were also observed in crk maternal mutants generated using our FRT flanked genomic construct (Figure 5O). Intriguingly, live imaging revealed the ability of embryos to “repair” the regions devoid of nuclei (Supplemental Movie S1), helping explain the embryos with pattern deletions we observed at later stages. Together, these data reveal that Crk is important for proper regulation of the dynamic behavior of the actomyosin cytoskeleton during syncytial divisions and cellularization.
Movie S1.
Crk knockdown often leads to wide-spread nuclear loss but embryos can largely repair the epithelium before gastrulation. Time-lapse movies showing actin dynamics in control and crkS-RNAi embryos expressing Moesin::GFP to label F-actin. The Movie begins during the last syncytial nuclear division and extends until the onset of gastrulation. In the crkS-RNAi embryo, large patches of nuclear loss are apparent following the last nuclear division. During cellularization, cells at the margins of these patches can be seen exhibiting protrusive cell behavior and migrating into areas of cell loss to enclose the blastoderm embryo prior to the onset of gastrulation. Images acquired at 10 s intervals and displayed at 10 frames per second. Time between frames varies between genotypes due to different sizes of region scanned. Scan time was 2.82 seconds for the control embryo and 2.30 seconds for the crkS-RNAi embryo.
Crk is important for actin cap expansion and the resulting establishment of pseudocleavage furrows
In some mutants with similar phenotypes, the primary defect is in the proper formation of pseudocleavage furrows, which form as expanding actomyosin caps collide (Zhang et al., 2018). We thus focused our attention on this event, measuring maximum furrow depth at metaphase. We drew lines across furrows separating neighboring nuclei (e.g., Figure 6, A–F), reconstructed z-axis projections from z stacks, and used these to measure furrow depth. In wild type, pseudocleavage furrows first form during cycle 10 and then become sequentially deeper in the subsequent cycles. We compared furrow depths in wild type to those in crkS-RNAi embryos, choosing pairs of nuclei without spindle collisions to avoid events biased by earlier furrow failure. As our en face views had suggested (Figure 6, A vs. B), pseudocleavage furrow invagination was significantly shallower at cycle 10 (Figure 6G, mean 1.96 µm in wild type vs. 1.09 µm in crkS-RNAi). This defect remained significant at cycles 11 (Figure 6H, mean 2.42 µm in wild type vs. 1.89 µm in crkS-RNAi) and 12 (Figure 6I, mean 2.69 µm in wild type vs. 2.06 µm in crkS-RNAi). The remaining furrows were similar in depth at nuclear cycle 13 (mean 3.18 µm in wild type vs. 3.26 µm in crkS-RNAi), but by that stage spindle collisions were so frequent in crkS-RNAi embryos that we could only measure furrows in the least affected embryos.
FIGURE 6:
Crk knockdown reduces pseudocleavage furrow depth and cap expansion. (A–F) Representative images of wild type and crkS-RNAi embryos at nuclear cycles 10, 11, and 12. Embryos were fixed and stained to visualize actin, tubulin, and DNA; selected for those in metaphase to anaphase; and binned by nuclear density into the respective cycles. Furrows were selected, avoiding those involved in apparent mitotic spindle collisions. Lines were drawn perpendicular to the spindle (examples in yellow), z-axis projections were made, and metaphase furrow depth was measured (see Materials and Methods for details). (G–-I) Quantification of furrow depth; n = 11 wild type/7 crkS-RNAi embryos, nuclear cycle 10; 9 wild type/8 crkS-RNAi embryos, nuclear cycle 11; and 14 wild type/ 28 crkS-RNAi embryos, nuclear cycle 12. Furrows were significantly less deep in nuclear cycles 10–12 in crkS-RNAi embryos. (J, K) Movie stills illustrating examples of wild-type embryos at first appearance of actin caps and at the point when caps were maximally expanded; outlines are examples of caps measured for the analysis. Full movie can be seen in Supplemental Movie S2. (L–O) Quantification of cap dynamics (see Materials and Methods for definitions); n = 5 wild type/8 crkS-RNAi embryos. (P–S) Stills from movies of wild type and crkS-RNAi embryos expressing MRLC::GFP visualized at the peak of cap expansion in nuclear cycles 10 and 11. Note the reduction in myosin accumulation at cap margins after crkS-RNAi. Full movie can be seen in Supplemental Movie S3. Scale bars = 20 µm.
We also examined cap expansion directly by using imaging of GFP fused to the filamentous actin (F-actin) binding domain of Moesin (sGMCA; Edwards et al., 1997) to visualize the actin cytoskeleton in living embryos. Our movies suggested cap expansion was less extensive in mutants (Supplemental Movie S2); this was particularly striking in the early cycles in crkS-RNAi embryos. To quantify cap expansion, we focused on cycle 10 when increased cap spacing allowed more accurate measurement of individual cap area. We measured cap area both when caps first appeared at the surface of the embryo (Figure 6, J1 and K1) and again when they reached maximal expansion (Figure 6, J2 and K2; see Materials and Methods for details). While starting cap area was similar between wild type and crkS-RNAi embryos, maximum cap area was significantly reduced after crkS-RNAi (Figure 6, J2 vs. K2, and L). We also calculated the cap expansion rate by plotting the change in cap area over time and found it was significantly reduced following crkS-RNAi (Figure 6M), partially due to premature termination of cap expansion (Figure 6N). However, overall length of nuclear cycle 10 (measured from first cap appearance to the next cap appearance) was not significantly changed (Figure 6O).
Movie S2.
Crk knockdown leads to reduced actin cap expansion and less apparent pseudocleavage furrows during syncytial nuclear cycles. Time-lapse movies showing actin dynamics in control and crkS-RNAi embryos expressing Moesin::GFP to label F-actin during nuclear cycles 10-14. In both control and crkS-RNAi embryos, actin caps form, expand, and merge into pseudocleavage furrows. However, in crkS-RNAi conditions, actin cap expansion is reduced compared to controls, leading to impaired pseudocleavage furrow formation. Images acquired every 10 s and displayed at 10 frames per second. Time between frames varies between genotypes due to different sizes of region scanned. Scan time was 2.30 seconds for the control embryo and 2.93 seconds for crkS-RNAi embryo. Focal plane was occasionally adjusted to maintain a clear view of the furrows. This video is related to figure 6K,L.
As actin caps collide, myosin becomes enriched at the cap borders and furrows (Zhang et al., 2018). We visualized myosin live through the syncytial cycles using a GFP-tagged form of Myosin Regulatory Light Chain (MRLC = Sqh::GFP; Supplemental Movie S3). Stills from the end of cap expansion during nuclear cycles 10 and 11 are shown in Figure 6, P–S. In wild-type myosin is clearly enriched at cap margins at both nuclear cycles (Figure 6, P and R, arrows). In contrast, myosin enrichment at cap margins appeared substantially reduced in crkS-RNAi embryos (Figure 6, Q and S, arrows). Together, these data suggest the defects in syncytial divisions in crkS-RNAi embryos result from defects in actin dynamics, with reduced cap expansion leading to shorter pseudocleavage furrows and subsequent spindle collisions and nuclear loss.
Movie S3.
Crk loss impairs actin cap expansion and myosin remodeling during syncytial nuclear cycles. Time-lapse movies showing myosin dynamics in control and crkS-RNAi embryos during nuclear cycles 10-14, via MRLC::GFP (=Sqh). In control embryos, inter-cap myosin zones are remodeled during actin cap expansion, resulting in an enrichment of myosin in pseudocleavage furrows. In crkS-RNAi embryos, this remodeling is impaired, resulting in broader inter-cap myosin zones and reduced enrichment of myosin in pseudocleavage furrows. Images acquired every 10 s and displayed at 10 frames per second. Time between frames varies between genotypes due to different sizes of region scanned. Scan time was 2.01 seconds for the control embryo and 2.43 seconds for the crkS-RNAi embryo. Focal plane was occasionally adjusted to maintain a clear view of the furrows. This video is related to figure 6Q-T.
Crk knockdown reduces localization of Arp2/3 and its activator SCAR to actin caps and pseudocleavage furrows
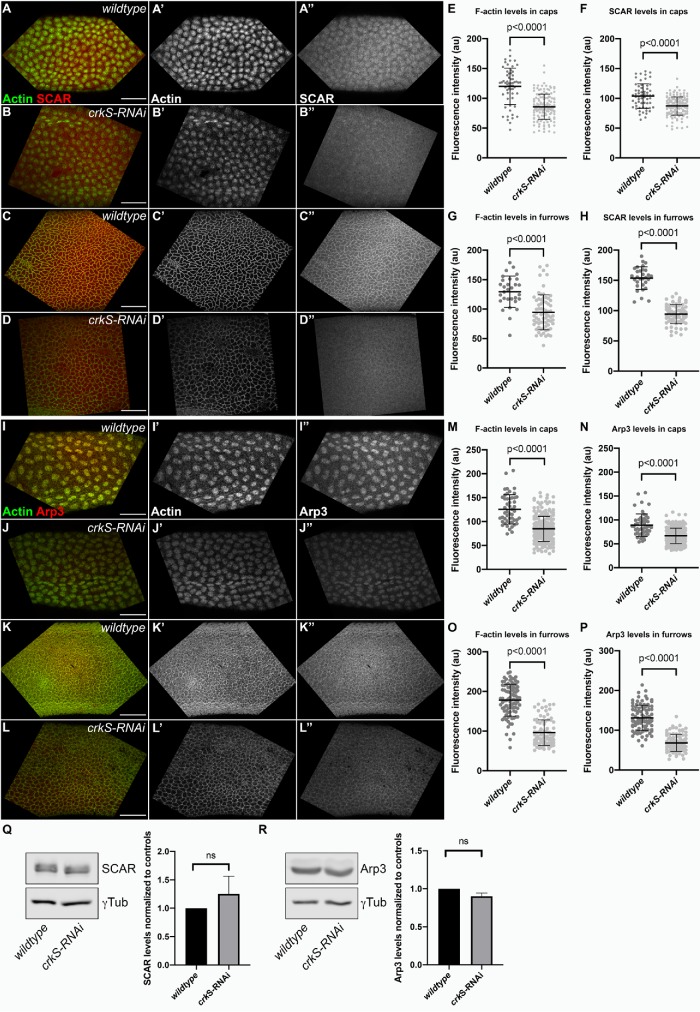
The reduced expansion of actin caps and impaired establishment of pseudocleavage furrows suggested the possibility that the localization and/or activity of actin cytoskeletal regulators was altered by loss of Crk. The Arp2/3 complex (Stevenson et al., 2002) and its upstream activator SCAR (Zallen et al., 2002) are both required for actin cap expansion and pseudocleavage furrow formation during syncytial development. Since Crk regulates Arp2/3-dependent actin remodeling in other cellular contexts (e.g., myoblast fusion, phagocytosis, and immune synapse maturation), we hypothesized that localization and/or activity of the Arp2/3 complex or SCAR might be altered by loss of Crk. We tested this hypothesis using quantitative imaging of Arp3 and SCAR localization to actin caps and pseudocleavage furrows. To directly compare fluorescence intensity between genotypes, we stained crkS-RNAi embryos together with embryos marked with Histone::RFP as internal, wild-type controls. Compared to controls, SCAR intensity in both actin caps (Figure 7, B compared with A) and pseudocleavage furrows (Figure 7, D compared with C) was significantly decreased following crkS-RNAi (Figure 7, F and H). Arp3 localization to both actin caps (Figure 7, J compared with I) and pseudocleavage furrows (Figure 7, L compared with K) was also significantly reduced by crkS-RNAi (Figure 7, N and P). To rule out the possibility that loss of Crk may reduce overall levels of either SCAR or Arp3, we performed Western blot analysis on extracts from 0- to 4-h embryos and quantified SCAR and Arp levels in crkS-RNAi compared with wild-type controls. This analysis revealed that total SCAR (Figure 7Q) and Arp3 (Figure 7R) levels are unchanged in crkS-RNAi embryos. Quantification of phalloidin intensity revealed F-actin levels were, on average, also significantly reduced in both actin caps (Figure 7, B and J compared with A and I, respectively; quantified in E and M) and pseudocleavage furrows (Figure 7, D and L compared with C and K, respectively; quantified in G and O). Taken together, these data suggest the altered actin cap dynamics and impaired pseudocleavage furrow formation observed after crkS-RNAi result from altered localization and thus activity of the Arp2/3 complex and its upstream activator, SCAR.
FIGURE 7:
Crk knockdown leads to reduced localization of the Arp2/3 activator SCAR, Arp3, and F-actin to caps and pseudocleavage furrows. (A–D) Representative images of wild type and crkS-RNAi embryos stained with phalloidin (visualizing F-actin) and an antibody against SCAR. (E–H) Quantification of fluorescence intensity of F-actin or SCAR in actin caps (E, F; n = 6 wild type/14 crkS-RNAi embryos) and pseudocleavage furrows (G, H; n = 3 wild type/7 crkS-RNAi embryos). (Q) Representative Western blot and quantification of total SCAR levels in 0–4 h crkS-RNAi embryos compared with controls; n = 4, error bars = SEM. In wild type, SCAR localizes to both actin caps (A, A’) and pseudocleavage furrows (C, C’). SCAR still localizes to actin caps (B, B”) and pseudocleavage furrows (D, D”) following Crk knockdown, but localization of F-actin and SCAR to both structures is significantly reduced after crkS-RNAi (E–H) while total SCAR levels are unchanged (Q). (I–L) Representative images of wild type and crkS-RNAi embryos stained with phalloidin and an antibody against Arp3. (M–P) Quantification of fluorescence intensity of F-actin and Arp3 in actin caps (M, N; n = 5 wild type/22 crkS-RNAi embryos) and pseudocleavage furrows (O, P; n = 9 wild type/9 crkS-RNAi embryos). (R) Representative Western blot and quantification of total Arp3 levels in 0–4 h crkS-RNAi embryos compared with controls; n = 4, error bars = SEM. Arp3 also localizes to actin caps and pseudocleavage furrows in both wild type (I–I’’, K–K’’) and crkS-RNAi (J–J’’, L–L’’) embryos. Like SCAR, Arp3 localization to both actin caps (N) and pseudocleavage furrows (P) is significantly reduced after crkS-RNAi while total Arp3 levels are unchanged (R). F-actin localization to actin caps (M) and pseudocleavage furrows (O) is also significantly reduced after crkS-RNAi. In E–H and M–P, mean = black bar; error bars = SD. All scale bars = 50 µm.
Loss of Crk leads to defects in axon patterning in the CNS similar to those seen in abl mutants
On the basis of Crk’s multiple roles in mammalian cells, we suspected Crk likely played additional roles in Drosophila embryogenesis. We initiated work on Crk because we suspected it was a regulator or effector of Abl kinase, as the PXXP motif within Abl that can bind Crk is essential for Abl function (Rogers et al., 2016). Abl plays many roles in embryonic morphogenesis, but the best characterized are its roles in the CNS, where it regulates axon outgrowth and pathfinding (e.g., Gertler et al., 1989; Wills et al., 1999a,b; Bashaw et al., 2000; Liebl et al., 2000; Crowner et al., 2003; Forsthoefel et al., 2005). We first examined whether Crk was expressed in the CNS using our mNG-tagged rescue construct. Crk localized to the axon tracks of the CNS (Figure 8A, cyan arrow), as well as to midline glia (Figure 8A, magenta arrow), suggesting that Crk may play a role in axon patterning. We thus examined the nervous systems of Crk knockdown embryos using both our strong and weak shRNA lines. We visualized axons using antibodies to BP102, which labels all CNS axons (Seeger et al., 1993), and Fasciclin II (FasII), an adhesion molecule expressed on a subset of CNS axons (Grenningloh et al., 1991).
FIGURE 8:
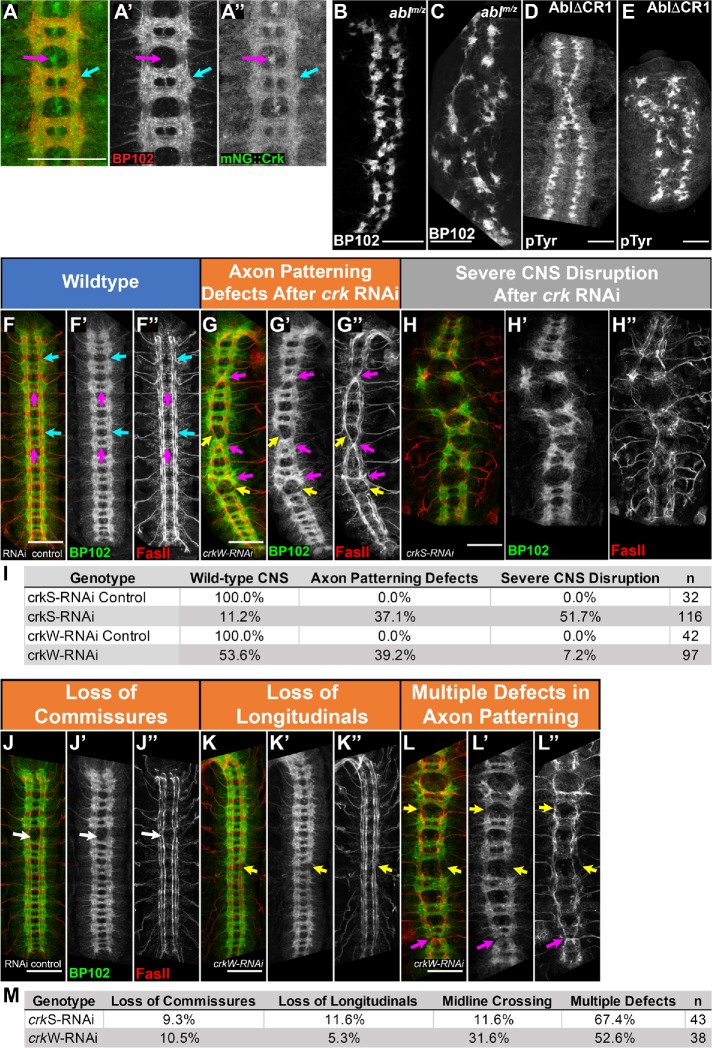
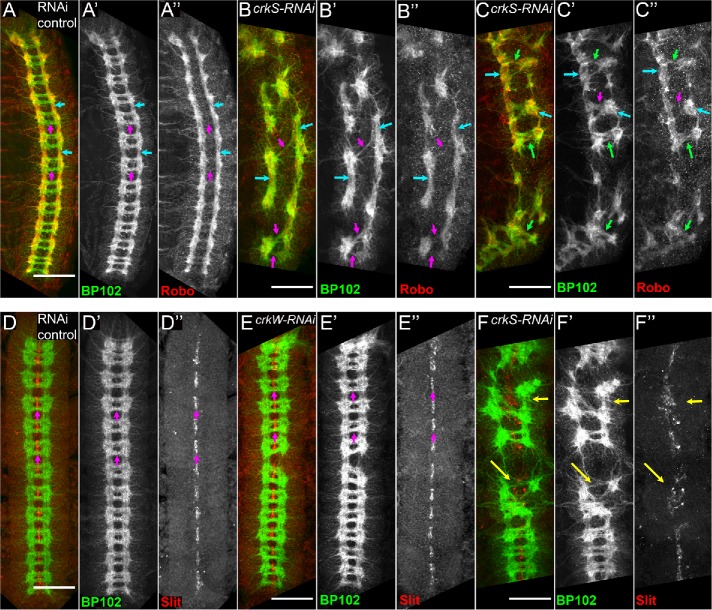
Crk is required for proper patterning of the embryonic CNS. (A–H, J–L) Stage 15 embryos, antigens and genotypes indicated. (I, M) Quantification of penetrance of phenotypes depicted. (A) mNG::Crk expressed at endogenous levels. Crk is enriched in CNS axons (cyan arrow) and midline glia (magenta arrow), suggesting Crk may play a role in axon pathfinding or function. (B, C) Representative images of CNS phenotypes, abl maternal, and zygotic null mutants. (D, E) CNS phenotypes of abl mutants expressing a form of Abl (AblΔCR1) that cannot bind to PXXP-dependent partners. Both can lead to the loss of commissures (B, D) or severe disruption of CNS patterning (C, E). (F–I) Representative images of range of CNS phenotypes in crkS-RNAi and crkW-RNAi embryos and quantification of their prevalence (I). In wild type, axons form a stereotypical ladder-like pattern with longitudinal axon bundles running anterior to posterior (cyan arrows, F–F’’) and commissural axon bundles crossing the midline at defined points in each body segment (magenta arrows, F–F’’). FasII-positive axons run in three parallel bundles on either side of the midline and do not normally cross the midline (F’’). Crk knockdown results in a range of CNS phenotypes including milder axon patterning defects and severe disruption of CNS patterning (G, H’’, I). (J–M) Representative images, different axon patterning defects observed after Crk knockdown and quantification of their prevalence. These include aberrant midline crossing (magenta arrows, G–G’’, L–L”), loss of longitudinal axons (yellow arrows, G–G’’, K–K”, L–L”), and loss of commissural axon bundles (white arrow, J–J’’). All scale bars = 50 µm.
In wild type, CNS axons form a ladder-like array (Figure 8F), with axons extending longitudinally on either side of the midline (Figure 8F, cyan arrows) and crossing the midline at two commissures per body segment (Figure 8F, magenta arrows). FasII-positive axons follow three parallel longitudinal tracks on each side of the midline and do not normally cross the midline (Figure 8F”). Embryos maternally and zygotically mutant for null alleles of abl exhibit a range of CNS defects (Rogers et al., 2016). Roughly half have moderate defects in axon patterning, with frequent loss of commissures (Figure 8B). The other half have more severe defects in axon patterning (Figure 8C). Embryos mutant for abl∆CR1, in which the PXXP motif is deleted, exhibit a similar range of defects (Figure 8, D and E), though the penetrance is less complete–-22% are wild type while 70% have commissural defects (Rogers et al., 2016). We thus compared these defects to the effects of knocking down Crk.
crkS-RNAi led to substantial and highly penetrant defects in axon pattern (Figure 8, F–H, quantified in I). In 52% of embryos, there was severe disruption of nervous system patterning (e.g., Figure 8H). Thirty-seven percent of embryos had milder defects in axon patterning (Figure 8G), with inappropriate midline crossing of FasII-positive axons (Figure 8, G and L, magenta arrows), as well as loss of commissural (Figure 8J, arrow) or longitudinal (Figure 8, G, K, and L, yellow arrows) axons in some segments (quantified in Figure 8M). Eleven percent of embryos had a normal CNS, consistent with the embryonic viability data after strong knockdown. This range of defects was similar to what we previously observed in abl maternal/zygotic mutants or after mutation of Abl’s PXXP motif (Figure 8, B–E; Grevengoed et al., 2001; Rogers et al., 2016), though total loss of commissures was less frequent after Crk knockdown. crkW-RNAi produced weaker and less penetrant defects (quantified in Figure 8, I and M). Only 7% of embryos had severe CNS disruption, once again consistent with the reduced embryonic lethality. However, 39% had milder but still significant CNS defects, once again ranging from loss of longitudinal or commissural axons in some segments to inappropriate midline crossing of FasII-positive axons.
Because crkS-RNAi leads to major defects in cellularization, including cell loss at gastrulation onset, we suspected that many of the severe CNS defects might be a secondary consequence of these earlier defects, with loss of ectodermal cells that should have become neuroblasts. However, crkW-RNAi results in fewer major morphogenesis defects (Figure 3), suggesting some of the milder defects we observed might be due to effects of Crk knockdown in the CNS itself. To assess this, we examined epidermal integrity and CNS patterning in the same embryos using an antibody to pTyr to outline epidermal cells and antibodies to BP102 and FasII to examine the underlying nervous system (Supplemental Figure 3). As we suspected, most embryos with strong defects in the CNS also had strong defects in the overlying epidermis, ranging from epidermal holes to strong disruptions to the epidermal pattern (Supplemental Figure 3, A and B, arrows), consistent with early cell loss. In other cases, more subtle epidermal defects, such as denticle belt fusions, also correlated with underlying CNS defects (Supplemental Figure 3C, arrow). However, we also observed mild CNS defects in some embryos in which the overlying epidermis was intact Supplemental Figure 3, D and E, arrows), keeping open the possibility that Crk also plays important CNS-intrinsic roles.
Crk is not essential for the patterns of expression of the guidance cues Slit and Robo, but may play more subtle roles in Slit/Robo signaling
A variety of axon guidance cues shape the pattern of the CNS. Among these is Slit/Robo signaling. The Robo receptor is expressed on a subset of CNS axons (Figure 9A, cyan arrows). Slit is expressed by midline glial cells (Figure 9D, magenta arrows), and acts as a repulsive ligand for the Robo receptor, so that axons expressing Robo strictly follow longitudinal axon pathways (Figure 9A, cyan arrows) and do not cross the midline (Figure 9A, magenta arrows; Blockus and Chedotal, 2016). Slit/Robo signaling requires Abl function (Bashaw et al., 2000). The midline crossing defects seen following Crk knockdown could reflect defects in Slit or Robo expression or function. We assessed this by examining expression patterns of both proteins. In wild type, Robo is expressed in a subset of axons that all follow the longitudinal axon tracks (Figure 9A, cyan arrows) and is not expressed on axons that cross the midline (Figure 9A, magenta arrows). Slit is expressed in glial cells along the midline (Figure 9D, magenta arrows), where the commissural axons cross. We examined Slit and Robo expression patterns in a subset of Crk knockdown embryos in which the CNS was not totally disrupted. Most Crk knockdown embryos retained clear enrichment of Robo expression in the longitudinal tracts (Figure 9, B and C, cyan arrows), though these were often displaced laterally. The fact that most Robo-positive axons remained restricted to longitudinal tracks suggested midline repulsion was not completely inactivated. However, while some axons crossing the midline were Robo-negative (Figure 9, B and C, magenta arrows), a subset of Robo-expressing axons did cross the midline (Figure 9C, green arrows), suggesting that Slit/Robo signaling may be impaired following Crk loss. In Crk knockdown embryos in which the nervous system was only mildly disrupted, Slit expression remained confined to the midline glia (Figure 9E, magenta arrows). However, in embryos with more severely disrupted nervous systems, Slit-expressing cells were sometimes displaced (Figure 9F, yellow arrows). Thus, Crk does not appear to be required for the basic expression pattern of Robo or Slit, and its knockdown does not fully abrogate midline repulsion. However, the severity of the CNS disruptions observed made it impossible to definitively determine whether Crk plays more subtle roles in Slit/Robo signaling. Together, this work suggests Crk may play two roles in CNS patterning: one indirect, via maintaining the survival of neural precursors just following cellularization, and one CNS intrinsic in axon patterning. However, this latter role remains speculative.
FIGURE 9:
Crk is not required for proper expression of Robo and Slit but may play a more subtle role in Slit/Robo signaling. (A–F) Stage 15 embryos, antigens, and genotypes indicated. (A–C) Representative images of Robo localization in control and Crk knockdown embryos. (A) In wild type, Robo is strongly enriched in longitudinal axon bundles (A–A”, cyan arrows) and excluded from commissural axon bundles crossing the midline (A–A”, magenta arrows). (B, C) Following Crk knockdown, Robo expression is still largely restricted to longitudinal axon tracks (B–C’’, cyan arrows). In most cases, midline-crossing axon bundles are Robo-negative (B–C’’, magenta arrows); however, a subset of Robo-positive axons appear to cross the midline in Crk knockdown embryos (C–C’’, green arrows), suggesting Slit/Robo signaling may be impaired following Crk loss. (D–F) Representative images of Slit localization in control and Crk knockdown embryos. (D) In wild type, Slit expression is confined to the midline glia along the ventral midline (magenta arrows). (E, F) In Crk knockdown embryos, Slit expression remains confined to midline glia, generally forming a stereotypical line along the ventral midline (E–E”, magenta arrows). However, Crk knockdown embryos with more severely disrupted CNS patterning defects can have mispositioned Slit-expressing midline glia (F–F”, yellow arrows). Scale bars = 50 µm.
Crk is required for timely wound healing in the embryonic epidermis
Another embryonic process requiring coordinated regulation of adhesion and actin dynamics is embryonic wound healing (e.g., Martin and Lewis, 1992; Wood et al., 2002; Abreu-Blanco et al., 2012; Fernandez-Gonzalez and Zallen, 2013). Embryonic wounds are repaired through the collective movements of the cells around the wound. Upon wounding, the cells immediately adjacent to the wound polarize their actin cytoskeleton and assemble a supracellular actin cable that coordinates cell movements. This process is tightly coordinated with dynamic changes in cell–cell adhesion (reviewed in Hunter and Fernandez-Gonzalez, 2017). Abl is required for efficient wound closure via effects on the organization of F-actin and the reorganization of cell–cell junctions at the wound margin (Zulueta-Coarasa et al., 2014). We thus investigated whether Crk plays a role in the collective movements that drive embryonic wound closure
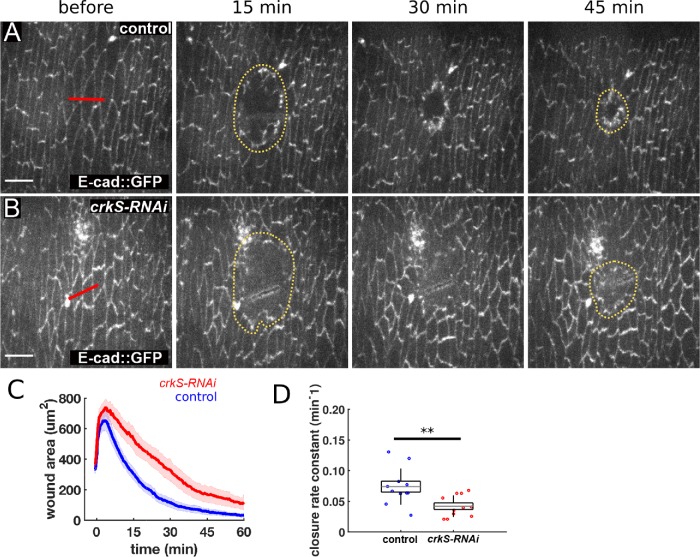
To do so, we quantified wound healing dynamics in control vs. crkS-RNAi embryos (Figure 10, A and B). We used laser ablation to induce wounds in the ventral epidermis of stage 14 and 15 embryos expressing E-cadherin::GFP, which outlines cells and allows us to visualize cell behavior during wound closure. In wild-type embryos, wounds close rapidly, with almost complete closure after 45 min (Figure 10A). Wounds closed more slowly in crkS-RNAi embryos (Figure 10, B, quantified in C). The rate of wound closure (measured as the decay constant of an exponential fit to the wound area vs. time curve of each wound) decreased by 43% in crkS-RNAi embryos with respect to controls (2.5 ± 0.3 h-1 vs. 4.4 ± 0.5 h-1, respectively) (mean ± SEM). Consistent with this, when we previously measured the rate of closure during the fast phase of wound repair in abl maternal/zygotic mutants, it was 37% slower than in wild-type embryos (Zulueta-Coarasa et al., 2014). Thus, wound closure provides another example of a morphogenetic process requiring both Abl and Crk.
FIGURE 10:
Crk knockdown slows embryonic wound healing. (A, B) Wound closure in control (A) and crkS-RNAi (B) embryos expressing endo-DE-cadherin::GFP. Red lines indicate wound sites; yellow dotted lines outline the wounds. Anterior, left; ventral, down. Scale bar = 10 μm. (C, D) Mean wound area over time (C) and closure rate constant (D) for controls (blue, n = 11 wounds) and crkS-RNAi embryos (red, n = 11). **P < 0.01 (Mann-Whitney test).
DISCUSSION
Small adapter proteins play key roles in assembling multiprotein signaling complexes mediating the interface between cell surface receptors and downstream effectors. The Crk family provides an example. We developed new genetic tools that allowed us to fully assess the role of Crk in embryonic development, revealing novel roles in the cytoskeletal events of syncytial development and cellularization, in CNS development, and in embryonic wound healing.
Crk is an important regulator of actin dynamics during syncytial development and cellularization, acting via SCAR and the Arp2/3 complex
The Drosophila syncytial and cellularizing blastoderm provides a superb model for studying dynamic cytoskeletal rearrangements. During each of the last four nuclear cycles, the actin cytoskeleton undergoes a dynamic set of rearrangements. The sequential formation of actin caps and pseudocleavage furrows are critical to allow the underlying nuclei to accurately segregate chromosomes. The subsequent coordinated movement of actin rings around each nucleus and their incomplete closure during cellularization leads to the formation of the initial polarized epithelium. Our data reveal that these coordinated events require Crk function and suggest that defects in actin cap expansion and pseudocleavage furrow invagination after Crk knockdown lead to defects in separation of mitotic spindles and subsequent nuclear loss.
During the normal syncytial division cycles, actin caps expand and collide, with collision driving furrow ingression (Zhang et al., 2018). Growth of the actin caps requires the DOCK-family guanine nuclear exchange factor Sponge (Postner et al., 1992), the Arp2/3 regulator SCAR (Zallen et al., 2002), and the Arp2/3 complex (Stevenson et al., 2002). Data also support a role for the small GTPase Rac downstream of Sponge and upstream of SCAR (Zhang et al., 2018). The known physical interactions of Crk with DOCK family members and its role in DOCK/ELMO/Rac1 pathways in phagocytic clearance, nervous system remodeling, thoracic closure, and border cell migration are thus intriguing in this regard. The roles for Crk we documented in enhancing cap expansion and furrow invagination are consistent with it playing a role in this pathway, as are the effects of Crk knockdown on cortical localization of SCAR, Arp3, and branched F-actin to actin caps and furrows. Thus, Crk may act upstream of SCAR, although roles in regulating other important players such as the formin Diaphanous, Anillin, or the septin Peanut have not been ruled out (Fares et al., 1995; Field and Alberts, 1995; Adam et al., 2000; Afshar et al., 2000; Field et al., 2005). Our data are also consistent with a role for Crk in cellularization, although at this stage the exact mechanisms involved and the extent to which this is a primary role or a secondary effect of defects in syncytial divisions remain unclear.
Much work remains to clearly define the pathways and mechanistic connections driving actin rearrangements during syncytial development. For example, the upstream, initiating regulators of this process have yet to be identified. Unlike in myoblast fusion or in RTK-dependent actin remodeling, there is no known role for transmembrane receptors in actin remodeling during syncytial development. Centrosomes play an important role and can continue to organize actin cap and furrow dynamics even in the absence of nuclei (Raff and Glover, 1989; Stevenson et al., 2001), but the mechanisms by which they communicate with the DOCK-family GEF Sponge and ultimately activate SCAR remain unknown. While recent data are consistent with Rac being a downstream effector (Zhang et al., 2018), other data suggest that Rap1 acts downstream of Sponge during cellularization (Schmidt et al., 2018), as it does in later embryonic events (Biersmith et al., 2015). Intriguingly, C3G, a Rap1 guanine-nucleotide exchange factor, was the first Crk SH3-domain interacting protein identified (Knudsen et al., 1994; Tanaka et al., 1994). In mammals Src/Crk/C3G/Rap1 signaling is involved in activation of Cdc42 during nectin-induced adherens junction formation (Fukuyama et al., 2005). In T-cells, Rap1 activation and integrin-dependent adhesion downstream of T-cell receptor engagement are dependent on both Abl activity and CrkL/C3G recruitment to the plasma membrane (Nolz et al., 2008). It will be exciting to explore these mechanistic connections further and to examine potential roles of other pathway members such as Verprolin 1 (Kim et al., 2007) and its upstream regulator Blown fuse (Jin et al., 2011).
Crk plays additional roles in the CNS and in wound healing
We suspected Crk’s roles would not be restricted to early events, but the major defects in syncytial development and cellularization posed a challenge in determining whether defects in later events were a primary or secondary consequence. For example, we suspect the major disruption of the CNS seen in a subset of Crk knockdown embryos reflects early cell loss of both epidermal and neural precursors. This is supported by the strong, although not complete, correlation between defects in the CNS and defects in the overlying epidermis. In the future it will be important to directly examine stages when neuroblasts invaginate to test this hypothesis. However, our range of knockout and RNAi tools helped circumvent this difficulty. Another subset of embryos, particularly those in the crkW-RNAi background, had more mild CNS defects even when the overlying epidermis was intact. These data are consistent with Crk playing a more direct role in axon guidance. Intriguingly, abl (Rogers et al., 2016), SCAR (Zallen et al., 2002), and sponge/mbc loss (Biersmith et al., 2011) all can lead to CNS defects similar to those seen after Crk knockdown. Crk knockdown also modifies photoreceptor axon targeting defects arising from overexpression of Eyes absent (Hoi et al., 2016), an event also regulated by Abl (Xiong et al., 2009). Our data also suggest that not all Robo-positive axons respect the normal midline repulsive signals after Crk knockdown, although basic Robo and Slit expression patterns are intact. Future work should directly explore the role of Crk in axon guidance and examine genetic interactions between crk and known players in this process. Development of tools to target Crk for proteasomal destruction in specific tissues (e.g., Caussinus and Affolter, 2016) would be particularly valuable. For example, tissue-specific degradation of Crk could provide new insight into its potential roles in myoblast fusion or axon patterning.
We also examined embryonic wound healing, which revealed a second place where Crk plays a role postgastrulation. Wound healing requires a complex interplay between cell adhesion and the actomyosin cytoskeleton (reviewed in Hunter and Fernandez-Gonzalez, 2017). Actin and myosin rapidly accumulate at the wound edge forming a supracellular contractile cable that coordinates cell movements to help drive rapid wound closure. In parallel, cell–cell junctions are remodeled, with concentration of the cadherin–catenin complex at tricellular junctions along the wound edge. Our analysis revealed that Crk knockdown slows wound healing. Intriguingly, Abl loss has a similar effect, and in the case of Abl, its loss impairs both actin organization at the wound edge and enrichment of the cadherin–catenin complex at tricellular junctions (Zulueta-Coarasa et al., 2014). SCAR is also critical for embryonic wound healing (Matsubayashi et al., 2015). In the future it will be important to explore which cell biological events required for wound healing are regulated by Crk and compare these to those affected by loss of Abl or SCAR. Germband retraction is a third postgastrulation event our data suggest requires Crk. Cell–matrix adhesion plays a key role in this process, in which different tissues exert mechanical force on one another (reviewed in Lacy and Hutson, 2016). Once again, Abl loss has a parallel effect (Grevengoed et al., 2001) to that of loss of Crk. This will provide another place where the cell biological roles of Crk can be explored.
More broadly, our data suggest potential functional cooperation between Abl and Crk, but this remains speculative, based on their known physical interaction in mammals and their phenotypic overlap during syncytial development and cellularization, CNS patterning, and wound healing. We think it is most likely that Crk has both Abl-dependent and Abl-independent functions. The effects of Crk knockdown or loss on actin dynamics during syncytial development and the overall effects of Crk knockdown on epithelial integrity, as assessed by cuticle pattern, are more severe than those of Abl mutants lacking the PXXP motif to which Crk can bind,and, in fact, more severe than those of complete loss of Abl (Rogers et al., 2016). In the future, it will be important to examine double mutants and explore genetic interactions, along with extending analysis of the biochemical and cell biological interactions between Crk and Abl.
MATERIALS AND METHODS
Fly husbandry and cuticle preparation
All stocks were maintained on standard cornmeal agar media at room temperature or 25°C. All RNAi crosses and controls were maintained at 25°C. Stocks used are described in Table 1. We used RNAi to generate embryos with reduced maternal and zygotic Crk as follows. We used two independent UAS-driven shRNA lines targeting different regions of crk developed by TRiP (Perkins et al., 2015)–-HMC03964 (use of which is referred to as crkS-RNAi) and HMJ22995 (use of which is referred to as crkW-RNAi). Both shRNA constructs are in a VALIUM20 backbone and are inserted in the same genomic locus (attP40) using PhiC31-mediated integration, making them directly comparable. We generated females carrying two copies of matα-tubulin-GAL4 (one copy on the second and one on the third chromosome) and a single copy of the indicated UAS-shRNA construct. These females were set up in cups with males either homozygous (crkS-RNAi) or heterozygous (crkW-RNAi) for the indicated UAS-shRNA and allowed to lay eggs for the appropriate period. For mNG::3XFLAG::Crk overexpression, we generated females carrying two copies of matα-tubulin GAL4 and a single copy of UASp-mNG::3XFLAG::dCrk. These females were mated to homozygous UASp-mNG::3XFLAG::dCrk males and allowed to lay eggs for the appropriate period. For simultaneous knockdown/overexpression, we generated a second chromosome bearing both UASp-mNG::3XFLAG::dCrk and UAS-crkS-RNAi using standard recombination and generated embryos as described above. To generate crk germline clones, we generated females that were heterozygous for the indicated FRT-flanked rescue construct over crkΔattP and were simultaneously carrying a single copy of ovo-FLP. These females were crossed to y, w; crkΔattP/In(4) ciD ciD panciD males in cups and allowed to lay eggs for the appropriate period. Cuticle preparations were performed as described in Wieschaus and Nüsslein-Volhard (1986).
TABLE 1:
Description of the Drosophila melanogaster stocks used in this study and their sources.
| Fly stocks | Use/description | Source |
|---|---|---|
| y[1], sc[*], v[1]; P{y[+t7.7] v[+t1.8]=TRiP.HMC03964}attP40 (stock #55277) | crkS-RNAi | Bloomington Drosophila Stock Center (Bloomington, IN) |
| y[1], sc[*], v[1]; P{y[+t7.7] v[+t1.8]=TRiP.HMJ2295}attP40/CyO | crkW-RNAi | National Institute of Genetics (Mishima, Japan) |
| w; P(mat-tub-Gal4)mat67; P(mat-tub-Gal4)mat15 (referred to as matII; matIII) | Double-copy matGAL4 | Peifer lab stocks |
| y,w | Wild-type control | Peifer lab stocks |
| w[*]; P{w[+mC]=His2Av-mRFP1}II.2 (stock #23651) | Internal wild-type control | Bloomington Drosophila Stock Center (Bloomington, IN) |
| w[*]; P{w[+mC]=His2Av-mRFP1}III.1, P{w[+mC]=sGMCA}3.1 (stock #59023) | Live imaging of F-actin dynamics (Moesin::GFP) | Bloomington Drosophila Stock Center (Bloomington, IN) |
| w[1118]; P{w[+mC]=sqh-GFP.RLC}3 (stock #57145) | Live imaging of myosin dynamics (MRLC::GFP) | Bloomington Drosophila Stock Center (Bloomington, IN) |
| P{w[+mC]=ovo-FLP.R}M1A, w[*] (stock #8727) | Germline FLPase for making germline clones | Bloomington Drosophila Stock Center (Bloomington, IN) |
| y,w; crk[ΔattP]/In(4) ci[D] ci[D] pan[ciD] | crk null allele | See Materials and Methods |
| y,w; P{w[+]=mNG::3XFLAG::Crk}crk[ΔattP] | cDNA-based rescue construct at crk locus | See Materials and Methods |
| y,w; P{w[+]=FRT-mNG::3XFLAG::Crk-FRT}crk[ΔattP] | FRT-flanked cDNA-based rescue construct at crk locus | See Materials and Methods |
| y,w; P{w[+]=FRT-crk genomic-FRT}crk[ΔattP] | FRT-flanked genomic rescue construct at crk locus | See Materials and Methods |
| w; P{w[+]=UASp-mNG::3XFLAG::Crk}attP16 | mNG::3XFLAG::Crk overexpression | See Materials and Methods |
Generation of Crk antibody
The ORF from crk isoform RA was PCR-amplified from a gBlocks Gene Fragment (IDT, Coralville, IA) using the primers listed below and cloned into the NdeI/EcoRI sites of pET28P (6XHis-dCrk) or the BamHI/EcoRI sites of pGEX-6P-2 (GST-dCrk). Resulting clones were screened by restriction digest and verified using Sanger sequencing.
PCR primers (restriction sites are in bold; target sequences are underlined):
-
pET28P Forward: 5′-ACTGCATATGGATACATTTGACGTTTCTGA-3′
Reverse: 5′-ACTGGAATTCTTAGCATATTTCTGTGGAGTTTTG-3′
-
pGEX-6-P2 Forward: 5′-ACTGGGATCCATGGATACATTTGACGTTTCTGA-3′
Reverse: 5′-ACTGGAATTCTTAGCATATTTCTGTGGAGTTTTTG-3′
Recombinant protein was expressed in BL21 Star (DE3) cells and purified using standard affinity chromatography techniques. Pocono Rabbit Farm & Laboratory (Canadensis, PA) raised antisera against 6XHIS-dCrk in two rabbits. Antisera were affinity-purified against GST-dCrk using standard techniques prior to use for Western blot analysis.
Generation of a crk null allele
The crk null allele, crkΔattP, was generated by using CRISPR to replace the entire crk locus (beginning 110 base pairs upstream of transcription start site and ending 99 base pairs beyond the 3′UTR) with a 50–base pair attP phage recombination site and positive marker (a loxP-flanked 3xP3-DsRed cassette) as described in Gratz et al. (2014). Briefly, guide RNAs (gRNAs ) flanking the crk locus were selected using flyCRISPR Target Finder (http://tools.flycrispr.molbio.wisc.edu/targetFinder/) to identify 19- to 20–base pair CRISPR targets with NGG PAM sites and zero predicted off-target effects using maximum stringency. Annealed gRNA oligo nucleotides were cloned into the BbsI site of pU6-BbsI-chiRNA. We generated a dsDNA donor template for homology-directed repair by cloning upstream and downstream flanking regions of the crk locus into the AarI and SapI sites of pHD-DsRed-attP, respectively. An 815–base pair upstream homology region, extending from 5′-CTTGAGCATGCAAAGGAATG-… to …GCTGACAGTACGTCCT-3′ and flanked by AarI sites, was synthesized as a gBlocks Gene Fragment (IDT, Coralville, IA). A 1-kb downstream homology region was PCR-amplified from genomic DNA using the primers described below. An injection mixture containing both U6-gRNA plasmids (75 ng/µl each) and the dsDNA template (250 ng/µl) was injected into y1, M{vas-Cas9.RFP-}ZH-2A, w1118 flies. Injections were performed by BestGene (Chino Hills, CA). Surviving G0 animals were outcrossed to y, w and CRISPR-edited lines were identified by the presence of DsRed eye fluorescence. DsRed-positive G1 animals were further outcrossed to y, w to remove M{vas-Cas9.RFP-}ZH-2A and subsequently established as stocks over In(4) ciD, ciD, panciD. Deletion of the crk locus was verified by PCR using the primers described below.
Oligo nucleotides for gRNA1:
Target (PAM underlined): GCTGACAGTACGTCCTAAAGGG
Sense oligo: 5′-CTTCGCTGACAGTACGTCCTAAA-3′
Antisense oligo: 5′-AAACTTTAGGACGTACTGTCAGC -3′
Oligo nucleotides for gRNA2:
Target (PAM underlined): GAATTGTGTAACTAAACGGTAGG
Sense oligo: 5′-CTTCGAATTGTGTAACTAAACGGT-3′
Antisense oligo: 5′-AAACACCGTTTAGTTACACAATTC-3′
Primers for PCR amplification of the downstream homology region (Sap1 sites are in bold; target sequences are underlined):
Forward: 5′-GACTGCTCTTCATATTGCTACAGAAAACCCTTTGG -3′
Reverse: 5′-GACTGCTCTTCAGACGAACATAGACAGCACAAGTTTGTTG -3′
Primers for PCR verification of crk locus deletion:
Detection of DsRed:
Forward: 5′-CGAGGACGTCATCAAGGAGT-3′
Reverse: 5′-GGTGATGTCCAGCTTGGAGT-3′
Flanking upstream HR:
Forward: 5′-AATTGCGCGCTGTTAAAAAT-3′
Reverse: 5′-GCTTCGAGCCGATTGTTTAG-3′
Flanking downstream HR:
Forward: 5′-GTGGACTCCAAGCTGGACAT-3′
Reverse: 5′-TAACAGGCAAAAACGCTTCC-3′
Generation of rescue constructs for PhiC31 integrase-mediated targeting into crk∆attP
The following rescue constructs were generated by introducing the indicated PCR-amplified fragments into EcoRI/KpnI-linearized pGE-attBGMR (Huang et al., 2009) using NEBuilder HiFi DNA Assembly (New England Biolabs). We generated a genomic rescue construct by PCR amplifying a 3151–base pair fragment of the crk locus extending from 5′-AAAGGGGTTTACCACCACAG-3′… to …5′-TTGTGTAACTAAACGGTAGG-3′ from genomic DNA–-this rescue construct corresponds to the exact sequence deleted in generation of crk∆attP.
Primers used (overlap with pGE-attBGMR in lowercase, target sequence underlined):
Forward: 5′-gctccccgggcgcgtactccacgaattcAAAGGGGTTTACCACCAC-3′
Reverse: 5′-attatacgaagttatggtacCCTACCGTTTAGTTACACAATTC-3′
We generated a cDNA-based rescue construct using the ORF from crk isoform RA by assembling the following PCR-amplified fragments into EcoRI/KpnI linearized pGE-attBGMR:
5′ deleted region (includes 5′UTR and 110 base pairs upstream of the transcription start site):
Primers used (overlap with neighboring fragments are in lowercase; target sequences are underlined):
Forward: 5′-gggctccccgggcgcgtactccacgaattcAAAGGGGTTTACCACCAC-3′
Reverse: 5′-tgtcttcttcacctttggaaaccatTTCCGTTTAAGGTTTATCTTAGAC-3′
mNG::3XFLAG::dCrk (synthesized as a gBlocks Gene Fragment [IDT, Coralville, IA]; 3XFLAG sequences are in bold; dCrk-RA sequences are underlined):
ATGGTGAGCAAGGGCGAGGAGGATAACATGGCCTCTCTCCCAGCGACACATGAGTTACACAT
CTTTGGCTCCATCAACGGTGTGGACTTTGACATGGTGGGTCAGGGCACCGGCAATCCAAATGATG
GTTATGAGGAGTTAAACCTGAAGTCCACCAAGGGTGACCTCCAGTTCTCCCCCTGGATTCTGGTCCCTCATATCGGGTATGGCTTCCATCAGTACCTGCCCTACCCTGACGGGATGTCGCCTTTCCAGGCCGCCATGGTAGATGGCTCCGGCTACCAAGTCCATCGCACAATGCAGTTTGAAGATGGTGCCTCCCTTACTGTTAACTACCGCTACACCTACGAGGGAAGCCACATCAAAGGAGAGGCCCAGGTGAAGGGGACTGGTTTCCCTGCTGACGGTCCTGTGATGACCAACTCGCTGACCGCTGCGGACTGGTGCAGGTCGAAGAAGACTTACCCCAACGACAAAACCATCATCAGTACCTTTAAGTGGAGTTACACCACTGGAAATGGCAAGCGCTACCGGAGCACTGCGCGGACCACCTACACCTTTGCCAAGCCAATGGCGGCTAACTATCTGAAGAACCAGCCGATGTACGTGTTCCGTAAGACGGAGCTCAAGCACTCCAAGACCGAGCTCAACTTCAAGGAGTGGCAAAAGGCCTTTACCGATGTGATGGGCATGGACGAGCTGTACAAGATGGACTACAAAGACCATGACGGTGATTATAAAGATCATGACATCGATTACAAGGATGACGATGACAAGATGGATACATTTGACGTTTCTGATAGGAACAGCTGGTACTTTGGTCCCATGTCTAGACAGGATGCTACTGAAGTTTTGATGAACGAACGCGAGCGGGGAGTGTTTTTAGTCCGTGATAGTAACTCGATAGCAGGGGATTATGTACTTTGTGTAAGAGAAGATACAAAAGTTAGCAACTACATCATTAACAAAGTTCAACAACAGGATCAAATCGTTTACCGCATTGGGGATCAGTCTTTTGACAATCTACCGAAACTCTTAACTTTTTACACTCTTCATTATTTGGATACAACCCCTTTAAAACGGCCTGCGTGTAGAAGGGTGGAAAAAGTAATAGGAAAGTTCGATTTCGTTGGCAGCGATCAAGATGATTTACCTTTTCAAAGAGGTGAAGTTTTAACAATAGTTCGAAAAGACGAGGATCAATGGTGGACTGCGCGTAACTCCTCGGGGAAAATTGGTCAAATACCGGTTCCCTATATACAACAGTATGACGATTATATGGATGAAGATGCTATTGATAAAAACGAACCTTCCATTTCGGGATCTAGCAATGTATTTGAAAGTACTCTTAAAAGGACAGATTTAAATCGAAAACTACCTGCATACGCCCGCGTAAAACAGTCAAGGGTCCCTAACGCATACGATAAGACTGCATTAAAATTGGAAATAGGTGACATTATTAAAGTCACTAAAACAAACATTAATGGGCAATGGGAGGGAGAATTAAATGGAAAAAATGGTCATTTTCCCTTCACGCACGTTGAATTTGTCGATGATTGTGATTTAAGCAAAAACTCCACAGAAATATGCTAA
3′ deleted region (includes 3′UTR and 99 base pairs downstream):
Primers used (overlap with neighboring fragments are in lowercase; target sequences are underlined):
Forward: 5′-caaaaactccacagaaatatgctaaATAGGAAGGAATGGAAGG-3′
Reverse: 5′-catacattatacgaagttatggtacCCTACCGTTTAGTTACACAATTC-3′
For our FRT-flanked rescue constructs, we introduced flanking FRT sites 104 base pairs upstream of the transcription start site and 99 base pairs downstream of the end of the 3′UTR using a Q5 Site-Directed Mutagenesis kit (New England Biolabs) and the following primers:
Upstream FRT (FRT sequences are in lowercase; target sequences are underlined):
Forward: 5′-tagagaataggaacttcGTTTACCACCACAGAGCAG-3′
Reverse: 5′-gaaagtataggaacttcaCCCTTTGAATTCGTGGAG-3′
Downstream FRT (FRT sequences are in lowercase; target sequences are underlined):
Forward: 5′-tagagaataggaacttcGTACCATAACTTCGTATAATG-3′
Reverse: 5′-gaaagtataggaaCTTCACCTACCGTTTAGTTACAC-3′
All rescue constructs were verified by a combination of PCR, restriction digest, and Sanger sequencing prior to injection. Transgenic lines were generated by coinjecting rescue constructs and PhiC31 helper plasmid into y, w; crkΔattP/ In(4) ciD, ciD panciD embryos. Injections were performed by BestGene (Chino Hills, CA).
Generation of Crk overexpression constructs
We generated two constructs for overexpressing fluorescent protein-tagged Crk using the UAS/GAL4 system–-mNG-3XFLAG-Crk and tdTomato-3XFLAG-Crk. mNG-3XFLAG-Crk was generated by cloning PCR-amplified mNG-3XFLAG-dCrk into the KpnI/SphI sites of pTIGER (Ferguson et al., 2012). Similarly, tdTomato-3XFLAG-Crk was generated by sequentially cloning PCR-amplified tdTomato and 3XFLAG-Crk sequences into the KpnI/NotI and NotI/SphI sites of pTIGER, respectively. Constructs were verified using both restriction digest and Sanger sequencing. Transgenic lines were generated using PhiC31 integrase-mediated site-specific integration into attP16 on 2R (Bloomington Stock #53C4; y1, w67c23; P{CaryP}attP16). Injections were performed by BestGene (Chino Hills, CA).
Primers used
mNG::3XFLAG::Crk (restriction sites are in bold; Kozac consensus sequences are in lowercase; target sequences are underlined):
Forward: 5′-GACTGGTACCcaccATGGTTTCCAAAGGTGAAGAA-3′
Reverse: 5′-ACTGGCATGCTTAGCATATTTCTGTGGAGTTTTTG-3′
3XFLAG::Crk (restriction sites in bold, target sequence underlined)
Forward: 5′-GACTGCGGCCGCAATGGACTACAAGGATCATGACGG-3′
Reverse: 5′-ACTGGCATGCTTAGCATATTTCTGTGGAGTTTTTG-3′
tdTomato (restriction sites in bold, Kozac consensus sequence in lowercase, target sequence underlined):
Forward: 5′-GACTGGTACCcaccATGGTGAGCAAGGGCGAG -3′
Reverse: 5′-ACTGGCGGCCGCCTTGTACAGCTCGTCCATGC -3′
Western blotting
Flies were allowed to lay eggs on apple juice agar plates with yeast paste for the indicated time. Dechorionated embryos were lysed by manual homogenization in ice-cold RIPA lysis buffer (1% NP-40; 0.5% Na deoxycholate; 0.1% SDS; 50 mM Tris, pH 8; 300 mM NaCl; 1× Halt protease and phosphatase inhibitor cocktail [Thermo Scientific]). Lysates were cleared by centrifugation at 21,000 × g at 4°C for 1 min and protein concentration was determined using Bio-Rad Protein Assay Dye. Normalized samples were mixed 1:1 with 2× SDS sample buffer and boiled for 5 min. Samples were either run immediately or stored at –20°C and boiled 5 min prior to running on 10% SDS–PAGE and transferring to nitrocellulose. Blots were blocked in 5% milk in TBST (0.1% Tween20 in Tris-buffered saline, pH 7.6). Western blots were probed with mouse anti-γTubulin (1:2500; GTU-88, Sigma-Aldrich), rabbit anti-dCrk (1:10,000), rat anti-Arp3 (1:1000; Stevenson et al., 2002), and mouse anti-SCAR (1:10; Developmental Studies Hybridoma Bank, Iowa City, IA). Secondary antibodies used were IRDye–conjugated donkey anti-mouse (1:5000), goat anti-rabbit (1:5000), and goat anti-rat (1:5000). Blots were imaged using Odyssey CLx infrared imaging system (LI-COR Biosciences). Band densitometry was performed using ImageStudio software version 4.0.21 (LI-COR Biosciences). Statistical comparisons were made using unpaired t tests in Prim8 (GraphPad).
Immunofluorescence and image analysis
Flies were allowed to lay eggs for the appropriate time for the desired stages of development on apple juice agar plates with yeast paste. Embryos were dechorionated in 50% bleach, washed 3× in 0.1% Triton-X, and fixed at room temperature in 1:1 heptane/formaldehyde diluted in phosphate-buffered saline (PBS). For phalloidin staining, syncytial embryos were fixed for 30 min using 18.5% formaldehyde and devitellinized by either hand-peeling or shaking in 1:1 heptane/90% ethanol. In all other instances, embryos were fixed for 20 min using 4% formaldehyde and devitellinized by shaking in 1:1 heptane/methanol. Embryos were blocked in PNT (PBS, 0.1% Triton X-100, and 1% normal goat serum) for ≥30 min followed by incubation with primary antibodies diluted in PNT at room temperature for 4 h or overnight at 4°C. Following washing 3× with PBT (PBS, 0.1% Triton X-100), embryos were incubated in secondary antibodies diluted in PNT at room temperature for 2 h or overnight at 4°C. Embryos were washed 3× with PBT and mounted on glass slides using Aquapolymount (Polysciences, Warrington, PA). Imaging was done on a Zeiss (Oberkochen, Germany) LSM-5 Pascal, a Zeiss 710, or a Zeiss 880 scanning confocal microscope. Primary antibodies used were: anti-BP102 (1:200), anti-FasII (1:100), anti-Robo (1:100), anti-SCAR (1:50), anti-Slit (1:10; all from the Developmental Studies Hybridoma Bank, Iowa City, IA); anti-Arp3 (1:500; Stevenson et al., 2002), anti-pTyr (1:1000; 4G10 clone; EMD Millipore); and anti-αTubulin (1:2000; DM1A clone; Sigma). Secondary antibodies used were: Alexa Fluor 488, 568, and 647 conjugated Goat anti-mouse (pan or isotype-specific as required), Goat anti-rat, or Goat anti-rabbit secondaries (1:500; Life Technologies). Other stains used: Alexa Fluor 488–, 568–, or 647–conjugated phalloidin (1:500; Life Technologies); DRAQ7 (1:500; BD Biosciences); and AF647 wheat germ agglutinin (1:1,000; Life Technologies).
Analysis of SCAR and Arp3 localization
To be able to directly compare fluorescence intensity levels, wild-type control (Histone::RFP) and crkS-RNAi embryos were collected and processed in the same tube. Embryo fixation and staining were carried out as described above in Immunofluoresence and image analysis. Embryos were fixed in 18.5% formaldehyde diluted in PBS for 30 min at room temperature and devitellinized using 1:1 heptane/90% ethanol. Images were acquired as 8-bit, single-plane confocal sections using a Zeiss (Oberkochen, Germany) LSM-5 Pascal. All images within a given experiment were acquired using the same settings (i.e., laser power, gain, offset, etc.). These confocal sections were taken at the level in which actin caps or pseudocleavage furrows appeared most obvious for each embryo. To measure fluorescence intensity in caps, we used the Fiji distribution of ImageJ (version 1.52N; Schindelin et al., 2012; Rueden et al., 2017) to draw 30-pixel-diameter circles centered over 10 randomly selected caps for each embryo analyzed and recorded mean gray values at each position. To measure fluorescence intensity in pseudocleavage furrows, we freehand drew 3-pixel-wide lines over 10 randomly selected pseudocleavage furrows and recorded mean gray values at each position. These are illustrated in Supplemental Figure 2. We compared fluorescence intensity levels for SCAR, Arp3, and phalloidin between genotypes by plotting the mean gray values for each individual cap or pseudocleavage furrow measured. Statistical comparisons made using unpaired t tests in Prism8 (GraphPad).
Wound healing assays
Crk knockdown was achieved by crossing mat67, endo-D-E-cadherin:GFP; mat15 virgins to crkS-RNAi/CyO; Pri/TM3 males. The mat67, endo-DE-cadherin:GFP/crkS-RNAi; mat15/(Pri or TM3) virgins were subsequently crossed to crkS-RNAi males, and their progeny were wounded and imaged. The progeny of mat67, endo-DE-cadherin:GFP/CyO; mat15/(Pri or TM3) virgins crossed to y, w males were used as controls. For wound healing assays, stage 14 and 15 embryos were dechorionated for 2 min in 50% bleach. Embryos were arranged on an apple juice agar pad with the ventral side down and transferred to a coverslip coated with heptane glue. Embryos were then covered with a 1:1 mix of halocarbon oil 27:700 (Sigma-Aldrich). Embryos were imaged using a Revolution XD spinning disk confocal microscope (Andor Technology) with an iXon Ultra 897 camera (Andor Technology), a 60× (NA 1.35; Olympus) oil-immersion lens, and Metamorph software (Molecular Devices). Wounding was performed using a Micropoint nitrogen dye cell laser (Andor Technology) tuned to 365 nm. To wound the embryos, 10 laser pulses were delivered at spots 2 μm apart along a 14-μm line. Sixteen-bit z stacks consisting of 16 slices at 0.2-μm steps were acquired every 30 s after wounding. Maximum intensity projections of time-lapse images were analyzed using SIESTA (Fernandez-Gonzalez and Zallen, 2011), custom software written in MATLAB (Mathworks) using the DIPImage toolbox (Delft University of Technology). The wound margin was segmented using the semiautomated LiveWire tool at every time point within the time-lapse image. A two-parameter exponential function was fitted to individual wound healing curves:
where A(t) represents the area of the wound at time t, A0 is the maximum wound area, and k is the decay constant of the exponential. A least-squares fit by the Gauss-Newton method was done starting at the point of maximum wound area. The decay constant of the exponential was used to quantify the rate of wound closure.
Live imaging
Embryos with Crk knockdown and carrying live imaging constructs were produced by crossing either crkS-RNAi/CyO; His2av mRFP, sGMCA or crkS-RNAi/CyO; MRLC::GFP males to matα tub-GAL4 II; matα tub-GAL4 III females. crkS-RNAi/ matα tub-GAL4 II; His2av mRFP, sGMCA/matα tub GAL4 III or crkS-RNAi/ matα tub-GAL4 II; MRLC::GFP/matα tub GAL4 III female progeny were then crossed to crkS-RNAi males and the progeny were imaged. CyO/matα tub -GAL4 II; His2av mRFP, sGMCA/matα tub-GAL4 III females crossed to y, w males were used as controls. Flies were allowed to lay on apple juice agar plates with yeast for 2 h. Embryos were dechorionated in 50% bleach for 3 min, washed 3× with 0.01% Triton-X, and transferred to a fresh apple juice agar plate where they were immersed in halocarbon oil 27(Apex Bioresearch Products). Seven to 10 embryos were then transferred to halocarbon oil on the gas-permeable membrane of a Lumox dish (Sarstedt) and covered with a glass coverslip. Imaging was done on a Zeiss LSM-5 Pascal scanning confocal microscope (Oberkochen, Germany). Embryos were imaged at the apical cortex where the tops of actin caps were visible. Frames were taken at 10-s intervals for ∼2 h or until cellularization began.
Furrow analysis
The length of five metaphase furrows per embryo was measured using the Fiji distribution of ImageJ (version 1.52N; Schindelin et al., 2012; Rueden et al., 2017). Furrows were selected by drawing a perpendicular line between two unique spindles that had not collided with another, such that the line passed through the middle of the dividing furrow. The ImageJ Stacks>Reslice function was used to construct a z-axis projection along that line. The resulting projected image was binarized using the Max Entropy Auto Threshold feature (Kapur et al., 1985) to improve visibility of the furrow. Furrows were then measured by drawing a line from the top of the furrow to the bottom. In some cases when not enough of a furrow could be discerned to be effectively measured but some pixels were still visible, a value of 0.54 μm was recorded as this is the step length between slices of the z stack.
Cap expansion analysis
Cap expansion during nuclear cycle 10 was quantified in three nonadjacent caps per embryo live imaged as above. ImageJ was used for the analysis. Videos were binarized using the Default Auto Threshold feature and processed to remove some noise using Process>Noise> Despeckle to improve visibility of cap borders. Cap area was measured by freehand-outlining the cap at each frame from when they first appear at the cortex to when they began to break up to enter the next nuclear cycle. Expansion time was reported as the time in seconds between when the cap first was measurable to when it was maximally expanded. Maximum expansion was defined as the maximum area of the cap before expansion plateaued or decreased. Expansion rate was calculated as the difference in area between when the cap was first measured to when it was maximally expanded divided by the expansion time. Cycle length was measured as the seconds between when caps first reappear to when they appear in the next cycle.
Supplementary Material
Acknowledgments
We are grateful to Victoria Deneke and Stefano Di Talia for advice and guidance in live imaging; Yixie Zhang, Tony Harris, Steve Rogers, and Kevin Slep for advice and helpful discussions; the Bloomington Drosophila Stock Center, the TRiP, and the Developmental Studies Hybridoma Bank for reagents; Andrew Hudson and Lynn Cooley for the Arp3 antisera; Scott Ferguson for pTIGER vectors; Tony Perdue of the Biology Imaging Center; and members of the Peifer lab for helpful discussions and reading the manuscript. The work was supported by National Institutes of Health R35 GM118096 to M.P. A.S. was supported by National Institutes of Health T32 CA009156 and F32 GM117803. A.B. was supported by the UNC (University of North Carolina) Science and Math Achievement and Resourcefulness Track (SMART), in partnership with the North Carolina Louis Stokes Alliance for Minority Participation (NC-SLSAMP; supported by National Science Foundation HRD-1202467), and by the William W. and Ida W. Taylor fellowship through the UNC Summer Undergraduate Research Fellowship (SURF) program. R.F.G. and A.F. are supported by grants from the Canada Foundation for Innovation (#30279) and the Canadian Institutes for Health Research (#156279). R.F.-G. is the Tier II Canada Research Chair in Quantitative Cell Biology and Morphogenesis.
Abbreviations used:
- Abl
Abelson tyrosine kinase
- Arp2/3
actin-related protein 2/3
- Crk
CT10 regulator of kinase
- CrkL
Crk-like
- crkS-RNAi
strong crk RNAi
- crkW-RNAi
weak crk RNAi
- F-actin
filamentous actin
- FasII
Fasciclin II
- gRNA
guide RNA
- Mbc
Myoblast City
- mNG
monomeric NeonGreen
- MRLC
myosin regulatory light chain
- PBS
phosphate-buffered saline
- pTyr
phosphotyrosine
- PVR
PDGF- and VEGF-receptor related
- RTK
receptor tyrosine kinase
- SCAR
suppressor of cAMP receptor
- sGMCA
GFP fused to the F-actin binding domain of Moesin
- SH2
Src homology domain 2
- SH3
Src homology domain 3
- WASP
Wiskott-Aldrich syndrome protein.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E19-05-0302) on July 18, 2019.
REFERENCES
- Abmayr SM, Balagopalan L, Galletta BJ, Hong SJ. (2003). Cell and molecular biology of myoblast fusion. Int Rev Cytol , 33–89. [DOI] [PubMed] [Google Scholar]
- Abreu-Blanco MT, Verboon JM, Liu R, Watts JJ, Parkhurst SM. (2012). Drosophila embryos close epithelial wounds using a combination of cellular protrusions and an actomyosin purse string. J Cell Sci , 5984–5997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adam JC, Pringle JR, Peifer M. (2000). Evidence for functional differentiation among Drosophila septins in cytokinesis and cellularization. Mol Biol Cell , 3123–3135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afshar K, Stuart B, Wasserman SA. (2000). Functional analysis of the Drosophila diaphanous FH protein in early embryonic development. Development , 1887–1897. [DOI] [PubMed] [Google Scholar]
- Antoku S, Mayer BJ. (2009). Distinct roles for Crk adaptor isoforms in actin reorganization induced by extracellular signals. J Cell Sci , 4228–4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balagopalan L, Chen MH, Geisbrecht ER, Abmayr SM. (2006). The CDM superfamily protein MBC directs myoblast fusion through a mechanism that requires phosphatidylinositol 3,4,5-triphosphate binding but is independent of direct interaction with DCrk. Mol Cell Biol , 9442–9455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashaw GJ, Kidd T, Murray D, Pawson T, Goodman CS. (2000). Repulsive axon guidance: Abelson and Enabled play opposing roles downstream of the roundabout receptor. Cell , 703–715. [DOI] [PubMed] [Google Scholar]
- Belov AA, Mohammadi M. (2012). Grb2, a double-edged sword of receptor tyrosine kinase signaling. Sci Signal , pe49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianco A, Poukkula M, Cliffe A, Mathieu J, Luque CM, Fulga TA, Rorth P. (2007). Two distinct modes of guidance signalling during collective migration of border cells. Nature , 362–365. [DOI] [PubMed] [Google Scholar]
- Biersmith B, Liu ZC, Bauman K, Geisbrecht ER. (2011). The DOCK protein sponge binds to ELMO and functions in Drosophila embryonic CNS development. PLoS One , e16120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biersmith B, Wang ZH, Geisbrecht ER. (2015). Fine-tuning of the actin cytoskeleton and cell adhesion during drosophila development by the unconventional guanine nucleotide exchange factors myoblast city and sponge. Genetics , 551–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birge RB, Kalodimos C, Inagaki F, Tanaka S. (2009). Crk and CrkL adaptor proteins: networks for physiological and pathological signaling. Cell Commun Signal , 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blockus H, Chedotal A. (2016). Slit-Robo signaling. Development , 3037–3044. [DOI] [PubMed] [Google Scholar]
- Caussinus E, Affolter M. (2016). deGradFP: A system to knockdown GFP-tagged proteins. Methods Mol Biol , 177–187. [DOI] [PubMed] [Google Scholar]
- Chodniewicz D, Klemke RL. (2004). Regulation of integrin-mediated cellular responses through assembly of a CAS/Crk scaffold. Biochim Biophys Acta , 63–76. [DOI] [PubMed] [Google Scholar]
- Collins TN, Mao Y, Li H, Bouaziz M, Hong A, Feng GS, Wang F, Quilliam LA, Chen L, Park T, et al (2018). Crk proteins transduce FGF signaling to promote lens fiber cell elongation. eLife , e32586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowner D, Le Gall M, Gates MA, Giniger E. (2003). Notch steers Drosophila ISNb motor axons by regulating the Abl signaling pathway. Curr Biol , 967–972. [DOI] [PubMed] [Google Scholar]
- Deng S, Azevedo M, Baylies M. (2017). Acting on identity: Myoblast fusion and the formation of the syncytial muscle fiber. Semin Cell Dev Biol , 45–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards KA, Demsky M, Montague RA, Weymouth N, Kiehart DP. (1997). GFP-moesin illuminates actin cytoskeleton dynamics in living tissue and demonstrates cell shape changes during morphogenesis in Drosophila. Dev Biol , 103–117. [DOI] [PubMed] [Google Scholar]
- Elwell CA, Ceesay A, Kim JH, Kalman D, Engel JN. (2008). RNA interference screen identifies Abl kinase and PDGFR signaling in Chlamydia trachomatis entry. PLoS Pathog , e1000021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fares H, Peifer M, Pringle JR. (1995). Loclaization and possible functions of Drosophila septins. Mol Biol Cell , 1843–1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feller SM. (2001). Crk family adaptors-signalling complex formation and biological roles. Oncogene , 6348–6371. [DOI] [PubMed] [Google Scholar]
- Ferguson SB, Blundon MA, Klovstad MS, Schupbach T. (2012). Modulation of gurken translation by insulin and TOR signaling in Drosophila. J Cell Sci , 1407–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Gonzalez R, Zallen JA. (2011). Oscillatory behaviors and hierarchical assembly of contractile structures in intercalating cells. Phys Biol , 045005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Gonzalez R, Zallen JA. (2013). Wounded cells drive rapid epidermal repair in the early Drosophila embryo. Mol Biol Cell , 3227–3237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field CM, Alberts BM. (1995). Anillin, a contractile ring protein that cycles from the nucleus to the cell cortex. J Cell Biol , 165–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field CM, Coughlin M, Doberstein S, Marty T, Sullivan W. (2005). Characterization of anillin mutants reveals essential roles in septin localization and plasma membrane integrity. Development , 2849–2860. [DOI] [PubMed] [Google Scholar]
- Figard L, Wang M, Zheng L, Golding I, Sokac AM. (2016). Membrane supply and demand regulates F-actin in a cell surface reservoir. Dev Cell , 267–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsthoefel DJ, Liebl EC, Kolodziej PA, Seeger MA. (2005). The Abelson tyrosine kinase, the Trio GEF and Enabled interact with the Netrin receptor Frazzled in Drosophila. Development , 1983–1994. [DOI] [PubMed] [Google Scholar]
- Fox DT, Peifer M. (2007). Abelson kinase (Abl) and RhoGEF2 regulate actin organization during cell constriction in Drosophila. Development , 567–578. [DOI] [PubMed] [Google Scholar]
- Fukuyama T, Ogita H, Kawakatsu T, Fukuhara T, Yamada T, Sato T, Shimizu K, Nakamura T, Matsuda M, Takai Y. (2005). Involvement of the c-Src-Crk-C3G-Rap1 signaling in the nectin-induced activation of Cdc42 and formation of adherens junctions. J Biol Chem , 815–825. [DOI] [PubMed] [Google Scholar]
- Galletta BJ, Niu XP, Erickson MR, Abmayr SM. (1999). Identification of a Drosophila homologue to vertebrate Crk by interaction with MBC. Gene , 243–252. [DOI] [PubMed] [Google Scholar]
- Geisbrecht ER, Haralalka S, Swanson SK, Florens L, Washburn MP, Abmayr SM. (2008). Drosophila ELMO/CED-12 interacts with Myoblast city to direct myoblast fusion and ommatidial organization. Dev Biol , 137–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George B, Fan Q, Dlugos CP, Soofi AA, Zhang J, Verma R, Park TJ, Wong H, Curran T, Nihalani D, Holzman LB. (2014). Crk1/2 and CrkL form a hetero-oligomer and functionally complement each other during podocyte morphogenesis. Kidney Int , 1382–1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gertler F, Bennett R, Clark M, Hoffmann F. (1989). Drosophila abl tyrosine kinase in embryonic CNS axons: a role in axonogenesis is revealed through dosage-sensitive interactions with disabled. Cell , 103–113. [DOI] [PubMed] [Google Scholar]
- Gratz SJ, Ukken FP, Rubinstein CD, Thiede G, Donohue LK, Cummings AM, O’Connor-Giles KM. (2014). Highly specific and efficient CRISPR/Cas9-catalyzed homology-directed repair in Drosophila. Genetics , 961–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grenningloh G, Rehm EJ, Goodman CS. (1991). Genetic analysis of growth cone guidance in Drosophila: fasciclin II functions as a neuronal recognition molecule. Cell , 45–57. [DOI] [PubMed] [Google Scholar]
- Greuber EK, Smith-Pearson P, Wang J, Pendergast AM. (2013). Role of ABL family kinases in cancer: from leukaemia to solid tumours. Nat Rev Cancer , 559–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grevengoed EE, Fox DT, Gates J, Peifer M. (2003). Balancing different types of actin polymerization at distinct sites: roles for Abelson kinase and Enabled. J Cell Biol. , 1267–1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grevengoed EE, Loureiro JJ, Jesse TL, Peifer M. (2001). Abelson kinase regulates epithelial morphogenesis in Drosophila. J Cell Biol , 1185–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosshans J, Wenzl C, Herz HM, Bartoszewski S, Schnorrer F, Vogt N, Schwarz H, Muller HA. (2005). RhoGEF2 and the formin Dia control the formation of the furrow canal by directed actin assembly during Drosophila cellularisation. Development , 1009–1020. [DOI] [PubMed] [Google Scholar]
- Gumienny TL, Brugnera E, Tosello-Trampont AC, Kinchen JM, Haney LB, Nishiwaki K, Walk SF, Nemergut ME, Macara IG, Francis R. (2001). CED-12/ELMO, a novel member of the CrkII/Dock180/Rac pathway, is required for phagocytosis and cell migration. Cell , 27–41. [DOI] [PubMed] [Google Scholar]
- Guris DL, Fantes J, Tara D, Druker BJ, Imamoto A. (2001). Mice lacking the homologue of the human 22q11.2 gene CRKL phenocopy neurocristopathies of DiGeorge syndrome. Nat Genet , 293–298. [DOI] [PubMed] [Google Scholar]
- Hoi CS, Xiong W, Rebay I. (2016). Retinal axon guidance requires integration of Eya and the Jak/Stat pathway into phosphotyrosine-based signaling circuitries in Drosophila. Genetics , 1283–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossain S, Dubielecka PM, Sikorski AF, Birge RB, Kotula L. (2012). Crk and ABI1: binary molecular switches that regulate abl tyrosine kinase and signaling to the cytoskeleton. Genes Cancer , 402–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Clarke F, Karimi M, Roy NH, Williamson EK, Okumura M, Mochizuki K, Chen EJ, Park TJ, Debes GF. (2015). CRK proteins selectively regulate T cell migration into inflamed tissues. J Clin Invest. , 1019–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Zhou W, Dong W, Watson AM, Hong Y. (2009). From the cover: directed, efficient, and versatile modifications of the Drosophila genome by genomic engineering. Proc Natl Acad Sci USA , 8284–8289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter MV, Fernandez-Gonzalez R. (2017). Coordinating cell movements in vivo: junctional and cytoskeletal dynamics lead the way. Curr Opin Cell Biol , 54–62. [DOI] [PubMed] [Google Scholar]
- Imaizumi T, Araki K, Miura K, Araki M, Suzuki M, Terasaki H, Yamamura K. (1999). Mutant mice lacking Crk-II caused by the gene trap insertional mutagenesis: Crk-II is not essential for embryonic development. Biochem Biophys Res Commun , 569–574. [DOI] [PubMed] [Google Scholar]
- Ishimaru S, Ueda R, Hinohara Y, Ohtani M, Hanafusa H. (2004). PVR plays a critical role via JNK activation in thorax closure during Drosophila metamorphosis. EMBO J , 3984–3994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin P, Duan R, Luo F, Zhang G, Hong SN, Chen EH. (2011). Competition between Blown fuse and WASP for WIP binding regulates the dynamics of WASP-dependent actin polymerization in vivo. Dev Cell , 623–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannan R, Giniger E. (2017). New perspectives on the roles of Abl tyrosine kinase in axon patterning. Fly (Austin) , 260–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapur JN, Sahoo PK, Wong AKC. (1985). A new method for gray-level picture thresholding using the entropy of the histogram. Comput Vision Graph , 273–285. [Google Scholar]
- Khatri A, Wang J, Pendergast AM. (2016). Multifunctional Abl kinases in health and disease. J Cell Sci , 9–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Shilagardi K, Zhang S, Hong SN, Sens KL, Bo J, Gonzalez GA, Chen EH. (2007). A critical function for the actin cytoskeleton in targeted exocytosis of prefusion vesicles during myoblast fusion. Dev Cell , 571–586. [DOI] [PubMed] [Google Scholar]
- Knudsen BS, Feller SM, Hanafusa H. (1994). Four proline-rich sequences of the guanine-nucleotide exchange factor C3G bind with unique specificity to the first Src homology 3 domain of Crk. J Biol Chem , 32781–32787. [PubMed] [Google Scholar]
- Koptyra M, Park TJ, Curran T. (2016). Crk and CrkL are required for cell transformation by v-fos and v-ras. Mol Carcinog , 97–104. [DOI] [PubMed] [Google Scholar]
- Kumar S, Fajardo JE, Birge RB, Sriram G. (2014). Crk at the quarter century mark: perspectives in signaling and cancer. J Cell Biochem , 819–825. [DOI] [PubMed] [Google Scholar]
- Lacy ME, Hutson MS. (2016). Amnioserosa development and function in Drosophila embryogenesis: Critical mechanical roles for an extraembryonic tissue. Dev Dyn , 558–568. [DOI] [PubMed] [Google Scholar]
- Lee DM, Harris TJ. (2014). Coordinating the cytoskeleton and endocytosis for regulated plasma membrane growth in the early Drosophila embryo. Bioarchitecture , 68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lettau M, Pieper J, Janssen O. (2009). Nck adapter proteins: functional versatility in T cells. Cell Commun Signal , 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Guris DL, Okura M, Imamoto A. (2003). Translocation of CrkL to focal adhesions mediates integrin-induced migration downstream of Src family kinases. Mol Cell Biol , 2883–2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebl EC, Forsthoefel DJ, Franco LS, Sample SH, Hess JE, Cowger JA, Chandler MP, Shupert AM, Seeger MA. (2000). Dosage-sensitive, reciprocal genetic interactions between the Abl tyrosine kinase and the putative GEF trio reveal trio’s role in axon pathfinding. Neuron , 107–118. [DOI] [PubMed] [Google Scholar]
- Martin P, Lewis J. (1992). Actin cables and epidermal movement in embryonic wound healing. Nature , 179–183. [DOI] [PubMed] [Google Scholar]
- Martinez-Quiles N, Feuerbacher LA, Benito-Leon M, Hardwidge PR. (2014). Contribution of Crk adaptor proteins to host cell and bacteria interactions. Biomed Res Int , 372901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubayashi Y, Coulson-Gilmer C, Millard TH. (2015). Endocytosis-dependent coordination of multiple actin regulators is required for wound healing. J Cell Biol , 677–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavrakis M, Rikhy R, Lippincott-Schwartz J. (2009). Plasma membrane polarity and compartmentalization are established before cellularization in the fly embryo. Dev Cell , 93–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPhee CK, Logan MA, Freeman MR, Baehrecke EH. (2010). Activation of autophagy during cell death requires the engulfment receptor Draper. Nature , 1093–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon AM, Guris DL, Seo JH, Li L, Hammond J, Talbot A, Imamoto A. (2006). Crkl deficiency disrupts Fgf8 signaling in a mouse model of 22q11 deletion syndromes. Dev Cell , 71–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore CA, Parkin CA, Bidet Y, Ingham PW. (2007). A role for the Myoblast city homologues Dock1 and Dock5 and the adaptor proteins Crk and Crk-like in zebrafish myoblast fusion. Development , 3145–3153. [DOI] [PubMed] [Google Scholar]
- Morishita K, Ozasa F, Eguchi K, Yoshioka Y, Yoshida H, Hiai H, Yamaguchi M. (2014). Drosophila DOCK family protein sponge regulates the JNK pathway during thorax development. Cell Struct Funct , 113–124. [DOI] [PubMed] [Google Scholar]
- Nakano R, Iwamura M, Obikawa A, Togane Y, Hara Y, Fukuhara T, Tomaru M, Takano-Shimizu T, Tsujimura H. (2019). Cortex glia clear dead young neurons via Drpr/dCed-6/Shark and Crk/Mbc/dCed-12 signaling pathways in the developing Drosophila optic lobe. Dev Biol (in press). [DOI] [PubMed] [Google Scholar]
- Nolz JC, Nacusi LP, Segovis CM, Medeiros RB, Mitchell JS, Shimizu Y, Billadeau DD. (2008). The WAVE2 complex regulates T cell receptor signaling to integrins via Abl- and CrkL-C3G-mediated activation of Rap1. J Cell Biol , 1231–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padash Barmchi M, Rogers S, Hacker U. (2005). DRhoGEF2 regulates actin organization and contractility in the Drosophila blastoderm embryo. J Cell Biol , 575–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park T, Koptyra M, Curran T. (2016). Fibroblast Growth Requires CT10 Regulator of Kinase (Crk) and Crk-like (CrkL). J Biol Chem , 26273–26290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park TJ, Boyd K, Curran T. (2006). Cardiovascular and craniofacial defects in Crk-null mice. Mol Cell Biol , 6272–6282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park TJ, Curran T. (2008). Crk and Crk-like play essential overlapping roles downstream of disabled-1 in the Reelin pathway. J Neurosci , 13551–13562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park TJ, Curran T. (2014). Essential roles of Crk and CrkL in fibroblast structure and motility. Oncogene , 5121–5132. [DOI] [PubMed] [Google Scholar]
- Pearce MMP, Spartz EJ, Hong W, Luo L, Kopito RR. (2015). Prion-like transmission of neuronal huntingtin aggregates to phagocytic glia in the Drosophila brain. Nat Commun , 6768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins LA, Holderbaum L, Tao R, Hu Y, Sopko R, McCall K, Yang-Zhou D, Flockhart I, Binari R, Shim HS. (2015). The Transgenic RNAi Project at Harvard Medical School: Resources and validation. Genetics , 843–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pielage JF, Powell KR, Kalman D, Engel JN. (2008). RNAi screen reveals an Abl kinase-dependent host cell pathway involved in Pseudomonas aeruginosa internalization. PLoS Pathog , e1000031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postner MA, Miller KG, Wieschaus EF. (1992). Maternal effect mutations of the sponge locus affect actin cytoskeletal rearrangements in Drosophila melanogaster embryos. J Cell Biol , 1205–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raff JW, Glover DM. (1989). Centrosomes, and not nuclei, initiate pole cell formation in Drosophila embryos. Cell , 611–619. [DOI] [PubMed] [Google Scholar]
- Reddien PW, Horvitz HR. (2000). CED-2/CrkII and CED-10/Rac control phagocytosis and cell migration in Caenorhabditis elegans. Nat Cell Biol , 131–136. [DOI] [PubMed] [Google Scholar]
- Rogers EM, Spracklen AJ, Bilancia CG, Sumigray KD, Allred SC, Nowotarski SH, Schaefer KN, Ritchie BJ, Peifer M. (2016). Abelson kinase acts as a robust, multifunctional scaffold in regulating embryonic morphogenesis. Mol Biol Cell , 2613–2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy NH, MacKay JL, Robertson TF, Hammer DA, Burkhardt JK. (2018). Crk adaptor proteins mediate actin-dependent T cell migration and mechanosensing induced by the integrin LFA-1. Sci Signal , eaat3178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rueden CT, Schindelin J, Hiner MC, DeZonia BE, Walter AE, Arena ET, Eliceiri KW. (2017). ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinformatics , 529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schejter ED, Wieschaus E. (1993). bottleneck acts as a regulator of the microfilament network governing cellularization of the Drosophila embryo. Cell , 373–385. [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B. (2012). Fiji: an open-source platform for biological-image analysis. Nat Methods , 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt A, Grosshans J. (2018). Dynamics of cortical domains in early Drosophila development. J Cell Sci , jcs212795. [DOI] [PubMed] [Google Scholar]
- Schmidt A, Lv Z, Grosshans J. (2018). ELMO and Sponge specify subapical restriction of Canoe and formation of the subapical domain in early Drosophila embryos. Development , dev157909. [DOI] [PubMed] [Google Scholar]
- Schnorrer F, Schonbauer C, Langer CC, Dietzl G, Novatchkova M, Schernhuber K, Fellner M, Azaryan A, Radolf M, Stark A. (2010). Systematic genetic analysis of muscle morphogenesis and function in Drosophila. Nature , 287–291. [DOI] [PubMed] [Google Scholar]
- Seeger M, Tear G, Ferres-Marco D, Goodman CS. (1993). Mutations affecting the growth cone guidance in Drosophila: genes necessary for guidance toward or away from the midline. Neuron , 409–426. [DOI] [PubMed] [Google Scholar]
- Senechal K, Halpern J, Sawyers CL. (1996). The CRKL adaptor protein transforms fibroblasts and functions in transformation by the BCR-ABL oncogene. J Biol Chem , 23255–23261. [DOI] [PubMed] [Google Scholar]
- Seo JH, Wood LJ, Agarwal A, O’Hare T, Elsea CR, Griswold IJ, Deininger MW, Imamoto A, Druker BJ. (2010). A specific need for CRKL in p210BCR-ABL-induced transformation of mouse hematopoietic progenitors. Cancer Res , 7325–7335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sriram G, Birge RB. (2010). Emerging roles for crk in human cancer. Genes Cancer , 1132–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staller MV, Yan D, Randklev S, Bragdon MD, Wunderlich ZB, Tao R, Perkins LA, Depace AH, Perrimon N. (2013). Depleting gene activities in early Drosophila embryos with the “maternal-Gal4-shRNA” system. Genetics , 51–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson V, Hudson A, Cooley L, Theurkauf WE. (2002). Arp2/3-dependent pseudocleavage furrow assembly in syncytial Drosophila embryos. Curr Biol , 705–711. [DOI] [PubMed] [Google Scholar]
- Stevenson VA, Kramer J, Kuhn J, Theurkauf WE. (2001). Centrosomes and the Scrambled protein coordinate microtubule- independent actin reorganization. Nat Cell Biol , 68–75. [DOI] [PubMed] [Google Scholar]
- Sullivan W, Fogarty P, Theurkauf W. (1993). Mutations affecting the cytoskeletal organization of syncytial Drosophila embryos. Development , 1245–1254. [DOI] [PubMed] [Google Scholar]
- Takada S, Kelkar A, Theurkauf WE. (2003). Drosophila checkpoint kinase 2 couples centrosome function and spindle assembly to genomic integrity. Cell , 87–99. [DOI] [PubMed] [Google Scholar]
- Tanaka S, Morishita T, Hashimoto Y, Hattori S, Nakamura S, Shibuya M, Matuoka K, Takenawa T, Kurata T, Nagashima K, et al. (1994). C3G, a guanine nucleotide-releasing protein expressed ubiquitously, binds to the Src homology 3 domains of CRK and GRB2/ASH proteins. Proc Natl Acad Sci USA , 3443–3447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasdemir-Yilmaz OE, Freeman MR. (2014). Astrocytes engage unique molecular programs to engulf pruned neuronal debris from distinct subsets of neurons. Genes Dev , 20–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Tsuda M, Makino Y, Konstantinou T, Nishihara H, Majima T, Minami A, Feller SM, Tanaka S. (2009). Crk adaptor protein-induced phosphorylation of Gab1 on tyrosine 307 via Src is important for organization of focal adhesions and enhanced cell migration. Cell Res , 638–650. [DOI] [PubMed] [Google Scholar]
- Webb RL, Zhou MN, McCartney BM. (2009). A novel role for an APC2-Diaphanous complex in regulating actin organization in Drosophila. Development , 1283–1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieschaus E, Nüsslein-Volhard C. (1986). Looking at embryos. In: Drosophila, A Practical Approach, ed. Roberts DB, Washington, DC: IRL Press, 199–228. [Google Scholar]
- Wills Z, Bateman J, Korey CA, Comer A, Van Vactor D. (1999a). The tyrosine kinase Abl and its substrate enabled collaborate with the receptor phosphatase Dlar to control motor axon guidance. Neuron , 301–312. [DOI] [PubMed] [Google Scholar]
- Wills Z, Marr L, Zinn K, Goodman CS, Van Vactor D. (1999b). Profilin and the Abl tyrosine kinase are required for motor axon outgrowth in the Drosophila embryo. Neuron , 291–299. [DOI] [PubMed] [Google Scholar]
- Wood W, Jacinto A, Grose R, Woolner S, Gale J, Wilson C, Martin P. (2002). Wound healing recapitulates morphogenesis in Drosophila embryos. Nat Cell Biol , 907–912. [DOI] [PubMed] [Google Scholar]
- Wu YC, Cheng TW, Lee MC, Weng NY. (2002). Distinct rac activation pathways control Caenorhabditis elegans cell migration and axon outgrowth. Dev Biol , 145–155. [DOI] [PubMed] [Google Scholar]
- Wu YC, Tsai MC, Cheng LC, Chou CJ, Weng NY. (2001). C. elegans CED-12 acts in the conserved crkII/DOCK180/Rac pathway to control cell migration and cell corpse engulfment. Dev Cell , 491–502. [DOI] [PubMed] [Google Scholar]
- Xiong W, Dabbouseh NM, Rebay I. (2009). Interactions with the Abelson tyrosine kinase reveal compartmentalization of eyes absent function between nucleus and cytoplasm. Dev Cell , 271–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Z, Sokac AM. (2016). Back-to-back mechanisms drive actomyosin ring closure during Drosophila embryo cleavage. J Cell Biol , 335–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zallen JA, Cohen Y, Hudson AM, Cooley L, Wieschaus E, Schejter ED. (2002). SCAR is a primary regulator of Arp2/3-dependent morphological events in Drosophila. J Cell Biol , 689–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Yu JC, Jiang T, Fernandez-Gonzalez R, Harris TJC. (2018). Collision of expanding actin caps with actomyosin borders for cortical bending and mitotic rounding in a syncytium. Dev Cell , 551–564.e554. [DOI] [PubMed] [Google Scholar]
- Ziegenfuss JS, Doherty J, Freeman MR. (2012). Distinct molecular pathways mediate glial activation and engulfment of axonal debris after axotomy. Nat Neurosci , 979–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zulueta-Coarasa T, Tamada M, Lee EJ, Fernandez-Gonzalez R. (2014). Automated multidimensional image analysis reveals a role for Abl in embryonic wound repair. Development , 2901–2911. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.