Abstract
It has been widely accepted that there is a distance‐decay pattern in the soil microbiome. However, few studies have attempted to interpret the microbial distance‐decay pattern from the perspective of quantifying underlying processes. In this study, we examined the processes governing bacterial community assembly at multiple spatial scales in maize fields of Northeast China using Illumina MiSeq sequencing. Results showed that the processes governing spatial turnover in bacterial community composition shifted regularly with spatial scale, with homogenizing dispersal dominating at small spatial scales and variable selection dominating at larger scales, which in turn explained the distance‐decay pattern that closer located sites tended to have higher community similarity. Together, homogenizing dispersal and dispersal limitation resulting from geographic factors governed about 33% of spatial turnover in bacterial community composition. Deterministic selection processes had the strongest influence, at 57%, with biotic factors and abiotic environmental filtering (mainly imposed by soil pH) respectively contributing about 37% and 63% of variation. Our results provided a novel and comprehensive way to explain the distance‐decay pattern of soil microbiome via quantifying the assembly processes at multiple spatial scales, as well as the method to quantify the influence of abiotic, biotic, and geographic factors in shaping microbial community structure, thus enabling understanding of widely acknowledged microbial biogeographic patterns and microbial ecology.
Keywords: distance‐decay pattern, maize field, Northeast China, soil bacteria
1. INTRODUCTION
The distance‐decay pattern refers to the spatial pattern in which community similarity decreases as geographical distance increases (Morlon et al., 2008; Soininen, McDonald, & Hillebrand, 2007). In other words, ecological communities that are geographically close to one another tend to have more similar species composition than geologically distant ones. Many studies have confirmed the existence of distance‐decay pattern in soil microbial communities (Chemidlin et al., 2014; Chu et al., 2016; Durrer et al., 2017; Fan et al., 2017; Martiny, Eisen, Penn, Allison, & Horner‐Devine, 2011; Shi et al., 2018; Terrat et al., 2015; Tuomisto, Ruokolainen, & Yli‐Halla, 2003; Wang, Lu, et al., 2017; Yang et al., 2017; Zhang et al., 2017). Such studies have also disentangled the contribution of habitat heterogeneity and biogeographic differences to distance‐decay patterns (Fan et al., 2017; Langenheder & Ragnarsson, 2007; Ramette & Tiedje, 2006; Ranjard et al., 2013; Shi et al., 2018; Zhang et al., 2017). Recently, the development of methods for understanding community assembly processes has enabled us to explore the underlying processes that result in the observed microbial biogeographic patterns (Hanson, Fuhrman, Horner‐Devine, & Martiny, 2012; Shi et al., 2018; Stegen et al., 2013; Stegen, Lin, Fredrickson, & Konopka, 2015; Stegen, Lin, Konopka, & Fredrickson, 2012; Wang, Li, et al., 2017). However, there have been few studies on how these assembly processes vary at different spatial scales.
According to the framework developed by Stegen et al. (2013) and modified in Stegen et al. (2015) and Dini‐Andreote et al. (2015), community assembly processes can be divided into five categories: variable selection (VS), homogeneous selection (HS), dispersal limitation (coupled with drift) (DL), homogenizing dispersal (HD), and undominated. VS (HS) refers to the scenario where high (or conversely, low) community turnover results from selective pressure imposed by variable (consistent) abiotic and biotic environmental factors (Dini‐Andreote et al., 2015; Stegen et al., 2015; Vellend, 2010). When environmental selection is weak, the effect of extreme high (low) dispersal can result in communities with low (high) turnover, and the communities are said to be dominated by HD (DL) (Stegen et al., 2013, 2015). Dispersal is generally considered to be the effect of geographical factors on the microbial community (Bahram et al., 2015; Tuomisto et al., 2003; Wang et al., 2013). ”Undominated” refers to a scenario where no single process dominates (Stegen et al., 2015). These processes have various effects on microbial turnover across space: HD and HS can lead to low community turnover while DL and VS can result in high community turnover (Hanson et al., 2012; Stegen et al., 2015). The relative importance of community assembly processes can vary across space. Shi et al.’s (2018) study of wheat fields of the North China Plain found that stochastic processes governed bacterial communities at small scales (<900 km), while deterministic processes governed at larger scales (>900 km). Additionally, the prevailing role of stochastic processes was also observed on small soil eukaryotes at the local scale in temperate forests (Bahram et al., 2015). However, these studies did not provide any direct explanation for the decreased community similarity at larger scales, as the deterministic process—including VS and HS, and stochastic processes including HD, DL and drift—can have divergent influences on spatial turnover of the microbiome (Hanson et al., 2012; Stegen et al., 2015). Hence, quantitatively discerning the relative importance of these assembly processes at different spatial scales is necessary in order to provide direct and exact explanation for microbial distance‐decay pattern.
Northeast China is the main commercial grain producing area of China and maize is its main grain crop, which amounting to about 30% of the nation's gross maize production. This area is known as the Golden‐Maize‐Belt in China (Liu et al., 2014; Liu, Yang, Hubbard, & Lin, 2012; Zhang, Sui, Zhang, Meng, & Herbert, 2007). Here, we chose to study maize field monoculture in Northeast China—a relatively simple and homogenous ecosystem—to examine the underlying mechanisms of the bacterial distance‐decay pattern, as well as to quantify the importance of abiotic, biotic, and geographical factors in structuring bacterial communities. We hypothesized that the greater community similarity between closer sites would mainly be due to greater relative importance of HD, as the greater importance of stochastic processes has been reported at smaller spatial scales (Shi et al., 2018). Among these, HD would be more likely to dominate due to high dispersal rate across closely located sites (Bahram et al., 2015; Stegen et al., 2015). By contrast, the lower community similarity between relatively more distant sites would be mainly caused by VS, as increased geographical distance is nearly always associated with greater environmental difference, which would deterministically select for microbiomes with different adaptive strategies (Caruso et al., 2011; Shi et al., 2018). To test our hypothesis, we studied the relative importance of different processes across space according to Stegen et al.’s (2015) framework to see their variation tendency and the dominant process at each specific spatial scale. To test the wider applicability of our hypothesis, the result from our study on maize fields is further compared with that in wheat fields of the North China Plain, which have a significant distance‐decay pattern as well as the dominate role of stochasticity at small spatial scale (<900 km) and determinacy at larger spatial scale (>900 km) for bacterial community (Shi et al., 2018).
2. MATERIALS AND METHODS
2.1. Soil location description and sampling method
Our sampling region ranges from 122°E to 131°E and 43°N to 48°N in Northeast China (Appendix Figure A1). The region has a monsoon‐influenced humid continental climate (https://en.wikipedia.org/wiki/Köppen_climate_classification), with the mean annual temperature ranging from 1°C to 7°C, and mean annual precipitation ranging from 392 to 657 mm (http://www.worldclim.org/) for the years 1970 to 2000. The soils were classified as dark brown soil (Heilongjiang) and Baijiang soil (Jilin) (Appendix Figure A1) (http://www.soil.csdb.cn/).
To test our hypothesis, we collected 81 soil samples from nine sites during the elongation stage of maize (15th–25th June, 2017). The nine sites were relatively uniformly distributed in the major maize fields of Northeast China. In each site, nine replicates were sampled in a ~100 km2 apart from their adjacent replicates (Appendix Figure A1). Within each replicate, nine soil cores were taken from 0 to 10 cm soil using an alcohol‐wiped auger, across the quadrat following an s‐shaped course, then the nine soil cores were combined as a composite soil sample. After sampling, the soil samples were packed into polyethylene bags and transported on ice to laboratory as soon as possible. The soils were then sieved through a 2‐mm mesh and divided into two parts: one kept at 4°C for soil properties analysis and the other placed at −40°C for DNA extraction.
2.2. Soil properties analysis
To identify the factors that might govern the spatial distribution of soil bacteria, 22 soil properties were measured, including soil pH, soil moisture (SM), nitrate (NO3 ‐), ammonium (NH4 +), total C (TC), total N (TN), total P (TP), total K (TK), dissolved organic carbon (DOC), dissolved organic nitrogen (DON), available P (AP), available K (AK), total Al, Fe, Ca, Mg, Mn, Cd, Cr, Pb,Zn, Cu. Soil pH was measured using a pH monitor (Thermo 0rion‐868, MA, USA) with a fresh soil to water ratio of 1:5. Soil moisture was measured gravimetrically by drying the soil to constant weight at 105°C. Soil NH4 + and NO3 ‐ content were measured by automated segmented flow analysis (AAIII; Bran and Luebbe, Germany). TC, TN, TP, TK content were determined by combustion using air dried (2 mm mesh) soil samples (2,400 II CHNS/0 Elemental l Analyzer, Perkin‐Elmer, Boston, MA). DOC and DTN were determined using a total organic carbon‐total nitrogen (TOC‐TN) analyzer (Shimadzu, Kyoto, Japan). AK was extracted by 1 M ammonium acetate extracts and determined by flame photometry (FP640, INASA, China). AP was determined in 0.5 M NaHCO3 extracts using the molybdenum blue method. Cu, Pb, Zn, Cd, Cr were determined using HPLC‐ICP‐MS. Ca, Mg, Al, Fe, Mn were determined using ICP‐AES Optima 8,000 (Perkin‐Elmer, Waltham, MA). In addition, mean annual precipitation (MAP) and mean annual temperature (MAT) of each sample site was the average value of the years 1970 to 2000 in http://www.worldclim.org/.
2.3. DNA extraction, PCR amplification and bioinformatics analysis
0.5g soil of each sample was used for DNA extraction using Power Soil DNA kit (MO BIO Laboratories, Carlsbad, CA, USA) by following the manufacturer's instructions. The V4‐V5 region of bacterial 16S rRNA was amplified using primers 515F (5′‐GTGCCAGCMGCCGCGGTAA‐3′) and 907R (5′‐CCGTCAATTCCTTTGAGTTT‐3′) (Biddle, Fitzgibbon, Schuster, Brenchley, & House, 2008). The 20‐µl PCR reaction included 4‐uL 5 × FastPfu buffer, 2‐µl 2.5‐mM dNTPs, 0.8‐µl forward primer(5 μM), 0.8‐µl reverse primer(5 μM), 0.4‐µl FastPfu polymerase, 0.2‐µl BSA, 10‐ng template DNA with the rest volume filled with ddH2O. PCR reaction were performed under the procedures of 95°C for 3 min, 35 cycles of 95°C for 30 s, 55 °C for 30 s, and 72°C for 45 s. The final extension was performed at 72°C for 10 min. The reaction was then held at 10°C until sequencing on the Illumina MiSeq PE250 platform. The sequence data were available in the National Center for Biotechnology Information with the accession number PRJNA508421.
After sequencing, the 2,961,808 raw sequences (30,020−44,899 for each sample) were processed and analyzed using QIIME software (http://qiime.sourceforge.net/) (Caporaso, Kuczynski, et al., 2010). The sequences were re‐assigned to each sample using the multiple_split_libraries_fastq.py command. The sequences with quality score <30 or length out the range of 380–440, or contained chimera sequences were discarded, resulting in 1,454,129 high quality sequences (14,617–18,014 for each sample). These high quality sequences were then clustered into OTUs at a 97% similarity using parallel_pick_otus_uclust_ref.py command (Edgar, 2010). The chimera sequences were identified using Userach61 against Greengenes database (http://greengenes.lbl.gov/) and filtered out using filter_fasta.py command. Greengenes database was used for taxonomy assignment using parallel_assign_taxonomy_uclust.py command. The final OTU table was constructed based on a randomly selected subset of 14,617 sequences which represented the lowest sequence number yielded by all samples. The aligned sequences of the represent OTUs using PyNAST algorithm(Caporaso, Bittinger, et al., 2010) were then used to construct phylogenetic tree using FastTree for phylogenetic analysis (Price, Dehal, & Arkin, 2010).
2.4. Distance‐decay analysis
To test whether bacterial community in our study has a significant distance‐decay pattern, the turnover rates (Z value) of bacterial community composition across spatial distance was tested following the formula: log10 (χd) = (–2z)*log10 (d)+b, where χd is the pairwise Sørensen similarity(presence/absence data) calculated using dsvdis function in labdsv package (R 3.4.4); d is between‐site distance (meter), and b is the intercept of the linear relationship (Ranjard et al., 2013). Between‐site distance was transformed from geographic coordinates in PASSaGE 2.
2.5. Phylogenetic analysis
2.5.1. Quantify the community assembly processes at different spatial scales
To quantify the community assembly processes at different spatial scales, we first utilized the combination of our sampling characteristics and “mantel.correlog” function (“vegan” package, R 3.1.4) which tests the relationship between community dissimilarity based on Bray–Curtis distance and geographic distance using default parameters to separate the whole distance into 15 spatial scales, then the processes were quantified in each of the 15 spatial scales separately.
The importance of the five processes was identified by using the combination of β‐nearest taxon index (βNTI) and Bray‐Curtis‐based Raup‐Crick metrics (RCbray) (Stegen et al., 2013, 2015). βNTI measuring the deviation of observed β‐ mean nearest taxon distance (βMNTD) from βMNTD in null model, in which taxa labels were shuffled across the tips of the phylogeny, were calculated in Phylocom 4.2 (Hardy, 2008). Using phylogenetic metrics such as βNTI to infer community assembly processes requires significant phylogenetic signal (the traits that regulate assembly processes should be phylogenetically conserved) (Stegen et al., 2012). To test for the phylogenetic signal, the relationship between phylogenetic distance of pairwise OTUs and their environmental optimal differences was tested using “mantel.correlog” (Stegen et al., 2012; Tripathi et al., 2018; Wang, Li, et al., 2017). Phylogenetic distance of pairwise OTUs was calculated using ”cophenetic” function in ”picante” package (R 3.1.4). The environmental differences of pairwise OTUs were calculated as euclidean distance of their environmental optima, for which abundance‐weighted mean value was calculated for each environmental variable. The significant relationship across short phylogenetic distance indicated that phylogenetic signal were observed in this study (Appendix Figure A2). On this occasion, significant deviation of observed βMNTD from null model (|βNTI| >2) indicates the dominant role of deterministic selection imposed by abiotic factors such as environmental filtering and biotic factors such as interactions among species (Stegen et al., 2012; Wiens, 2011). Among them, the fraction of |βNTI| <2 includes DL, HD and undominated categories, which were differentiated using the RCbray (see also https://github.com/stegen/Stegen_etal_ISME_2013) (Stegen et al., 2013).
A combination of |βNTI| <2 (thus the community is not dominated by selection) and RCbray > 0.95 indicates the dominant role of DL. |βNTI| <2 and RCbray <−0.95 indicates the dominant role HD. |βNTI| <2 and |RCbray| <0.95 indicates that no single process dominates (undominated). Detailed logical explanation of the framework can be referred to Stegen et al. (2013, 2015). Bacterial OTUs with a relative abundance higher than 0.001% were included in calculation of βNTI and RCbray matrices in maize field of Northeast China (about 8,200 OTUs, and the chosen OTUs were of the highest relative abundance). To find the law that each process changed across space, the relative importance of each process (percentage) at multiple spatial scales were fitted to linear, quadratic, cubic, logarithmic, and reciprocal models in SPSS Statistics 22. For the bacterial community in the wheat field in the North China Plain, βNTI and RCbray matrices based on 5,000 and 1,000 OTUs were both calculated by Shi et al. (2018). As the variation tendency of each process across space can be better demonstrated when 5,000 OTUs data‐sets were used (the relative importance of HD and HS were nearly 0 at all spatial scales in 1,000 OTUs data‐sets), we only reported the results based on 5,000 OTUs data‐sets in our study. Of note, Shi et al.’s (2018) results were based on 1,000 OTUs.
2.5.2. Quantify the abiotic, biotic, and geographic factors that shape bacterial community
Previous study of the geographical factors influencing community structure was often focused on the effect of DL (Blois et al., 2014; Li & Waller, 2016; Zhang et al., 2017). In this article, we defined the effect of geographical factors as those operating through mechanisms other than deterministic selection which cause the variation in community structure—either HD or DL. To further quantify the abiotic and biotic factors that impose deterministic selection, a forward selection model using βNTI as response variable was used.
Briefly, the method followed two steps. First, the geographical distances of 81 samples were transformed into spatial eigenvectors (PCNM axes) using function “pcnm” in “vegan” package (R 3.1.4). Second, the significant environmental variables and PCNM axes retained by forward selection (“ordiR2step” in package “vegan”, R 3.1.4) were fitted to βNTI using distance‐based redundancy analysis (Blanchet, Legendre, & Borcard, 2008; Legendre & Anderson, 1999) (“capscale” function in package “vegan”, R 3.1.4) to discern the factors that impose selection. Of note, βNTI was rescaled between 0 and 1 by first plus the absolute magnitude of the minimum (negative) and then divided by the maximum before distance‐based redundancy analysis was performed. Any variables that were retained by forward selection using βNTI as response variable reflected environmental variables that imposed selection. These retained PCNM axes reflected the selection imposed by unmeasured spatially auto‐correlated environmental variables. Here, we defined the part that βNTI cannot be explained by environmental variables and PCNM axes as the effect of biotic factors that impose selection, which is analogous to Blois et al.’s (2014) approach that the nonrandom part of community structure that cannot be explained by geographical or environmental differences is defined as the effect of biotic factors, as deterministic selection could only be imposed by biotic and abiotic variables.
3. RESULTS
3.1. The overall bacterial community composition in maize fields of Northeast China
The soil and climate properties varied significantly across the maize field of Northeast China (Appendix Table A1). After annotation, we found that there were totally 13 phyla/classes yielding the average relative abundance higher than 1% across the 81 samples: Acidobacteria (23.3%), Thermoleophilia (12.2%), Alphaproteobacteria (10.6%), Betaproteobacteria (9.59%), Gammaproteobacteria (8.79%), Bacteroidetes (7.69%), Chloroflexi (7.17%), Planctomycetes (4.87%), Gemmatimonadetes (4.58%), Deltaproteobacteria (3.01%), Nitrospirae (2.05%), Firmicutes (1.24%), and Fimbriimonadia (1.02%), which accounted for more than 96% of all the sentences (Appendix Figure A3). Mantel test showed that among the measured 24 soil and climate properties, 14 of them had a significant relationship with bacterial community, in which soil pH had the highest correlation (r = 0.64) (Appendix Table A2).
3.2. The distance‐decay pattern of bacterial community
The linear regression between log‐transformed bacterial community similarity and geographic distance indicated that there was a significant distance‐decay pattern for bacterial community (p < 0.01) (Figure 1), and the turnover rate (Z value) of bacterial community composition was 0.05. So spatially closely located samples had more similar community structure, while samples that were further apart had more different community structure.
Figure 1.
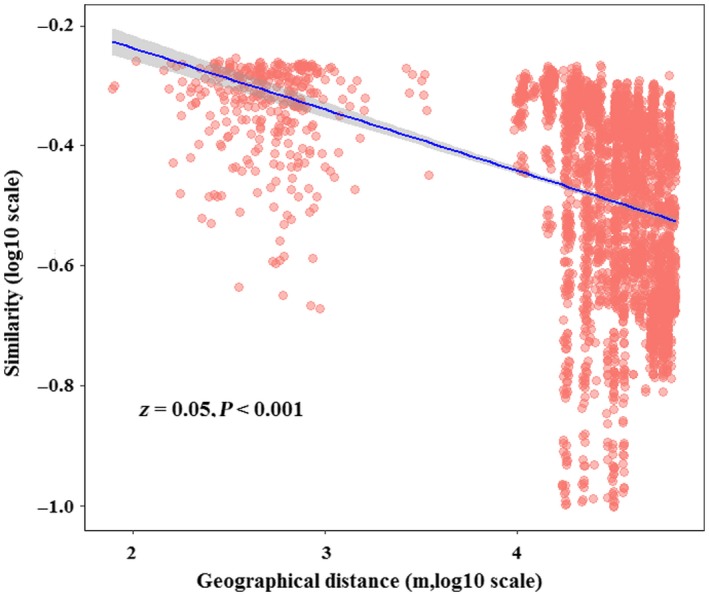
The distance‐decay pattern of bacterial community based on Sørensen similarity in the maize field of Northeast China
3.3. The processes varied at different spatial scales
As a whole, the spatial turnover of bacterial community composition was dominated by VS (~52.87%) (Table 1). When looking into the processes at different spatial scales, we found that, at relatively smaller spatial scales (scales 1‐5), the bacterial community was dominated by HD (Table 1). At medium spatial scales (scales 6‐8), deterministic processes (VS and HS) tended to overtake stochastic processes (DL, HD, and undominated). Additionally, DL tended to exceed HD above scale 7(>294 km). At larger spatial scales (scales 8‐15), the bacterial community was dominated by VS (above 50%) (Figure 2A). By fitting the relative importance of each process to existing mathematical models, we found that the importance of VS could be significantly fitted to linear (R 2 = 0.76, p < 0.001), logarithmic (R 2 = 0.81, p < 0.001), and quadratic (R 2 = 0.81, p < 0.001) models, all of which indicated the increasing importance of VS with the increasing spatial distance (Figure 2B). The importance of HS can be only fitted to a reciprocal model (R 2 = 0.40, p = 0.02), indicating that there was a sharp decrease of its importance followed by a slower decline after scale 2 (>133 km) (Figure 2C). As for HD, linear (R 2 = 0.77, p < 0.001), logarithmic (R 2 = 0.89, p < 0.001), and cubic models (R 2 = 0.94, p < 0.001) could all be significantly fitted. Though the correlation of the cubic model was the highest, we believed that it was impossible for the importance to increase after scale 12, thus overall the effect of HD would decrease when the scale became larger (Figure 2D). None of the mathematical models could be fitted to the variation in importance of DL in the maize field of Northeast China (Figure 2E).
Table 1.
Relative importance (%) of different assembly processes across whole region and at different spatial scales in the maize field of Northeast China
| Spatial scale | Spatial distance(km) | VS | HS | DL | HD | UD |
|---|---|---|---|---|---|---|
| whole | whole | 52.87 | 4.17 | 16.14 | 16.82 | 10.00 |
| 1 | 0–3 | 22.64 | 16.98 | 7.55 | 45.28 | 7.55 |
| 2 | 3–10 | 15.16 | 9.02 | 13.52 | 51.23 | 11.06 |
| 3 | 10–80.27 | 15.38 | 11.54 | 15.38 | 38.46 | 19.23 |
| 4 | 80.27–133.79 | 33.33 | 1.23 | 1.23 | 51.85 | 12.34 |
| 5 | 133.79–187.31 | 36.42 | 2.24 | 13.74 | 40.89 | 6.71 |
| 6 | 187.31–240.82 | 53.40 | 4.36 | 7.63 | 24.25 | 10.35 |
| 7 | 240.82–294.34 | 29.15 | 5.83 | 34.98 | 17.49 | 12.56 |
| 8 | 294.34–347.85 | 60.80 | 3.52 | 13.32 | 8.54 | 13.82 |
| 9 | 347.85–401.37 | 61.85 | 2.59 | 18.89 | 7.04 | 9.63 |
| 10 | 401.37–454.89 | 53.62 | 4.02 | 20.11 | 5.63 | 16.62 |
| 11 | 454.89–508.40 | 60.64 | 3.72 | 22.87 | 0.00 | 12.76 |
| 12 | 508.40–561.92 | 70.72 | 5.32 | 13.31 | 4.18 | 6.46 |
| 13 | 561.92–615.43 | 87.5 | 0.00 | 10.65 | 0.46 | 1.39 |
| 14 | 615.43–668.95 | 71.63 | 3.26 | 22.32 | 0.93 | 1.86 |
| 15 | 668.95–670.57 | 66.67 | 0.00 | 33.33 | 0.00 | 0.00 |
The scales 1–15 indicated the spatial distance between two samples, the percentage of the five processes indicated the relative importance of these processes in dominating the community dissimilarity between two samples within each specific spatial scale.
Abbreviations: Whole: the whole region; VS: variable selection; HS: homogeneous selection; DL: dispersal limitation; HD: homogenizing dispersal; UD: undominated.
Figure 2.

The importance of the four assembly processes at different spatial scales in shaping bacterial community composition in the maize field of Northeast China. (a) The dots that represent the importance of the four assembly processes are connected with smoothed curve. The significant mathematical models that could be fitted to the importance of variable selection (b), homogeneous selection (c), homogenizing dispersal (d) and dispersal limitation (e).We did not include the scenario “undominated” because of its low importance and ambiguous role in shaping bacterial community structure. When there were more than three models that were significantly fitted to the data, we plotted only three models with relatively higher correlation values. The numbers on the X axis indicates the spatial scales. VS: variable selection; HS: homogeneous selection; HD: homogenizing dispersal; DL: dispersal limitation
The spatial trends in the various factors structuring communities were further tested in the bacterial community in wheat field of North China Plain. We found that, although VS dominated at all spatial scales (Appendix Table A3, Appendix Figure A4A), the general trends in the importance of VS, HS, and HD across spatial scales were consistent with that in the maize field of Northeast China (Appendix Figure A4B–D). The difference lies in the importance of DL in shaping bacterial community in the North China Plain: it could be significantly fitted to the quadratic model (R 2 = 0.42, p = 0.03), which suggested that its importance would first increase to its peak at about scale 9 and then declined steadily (Appendix Figure A4E).
3.4. Abiotic, biotic, and geographic factors combined in shaping bacterial community
Homogenizing dispersal and DL respectively explained 16.14% and 16.82% of spatial turnover in bacterial community composition (Table 1). We thus concluded that geographical factors contributed about 33% to spatial variation of bacterial community composition in the maize fields of Northeast China.
Deterministic processes (VS and HS) could explain about 57% of bacterial community spatial variation in the maize fields of Northeast China (Table 1). There were five significant explanatory environmental variables and three PCNM axes that were significantly retained in the forward selection model using βNTI as the response variable. All variables totally explained about 63% of βNTI (Table 2). Among the five environmental variables, soil pH had the biggest adjusted R 2, followed by MAT, MAP, SM, and TP (Table 2). The three significant PCNM axes (PCNM 3, 6, 32) represented unmeasured, spatially auto‐correlated environmental variables, which can impose deterministic selection on soil bacterial community (Table 2). Thus, we inferred that abiotic factors accounted for about 63% of deterministic selection on bacterial community, and biotic factors contributed about 37%.
Table 2.
Summary of PCA axes that had significant effect on βNTI of bacterial community in maize field of Northeast China
| PCA | Highest loading | R 2.adj | Cum.R 2.adj | p |
|---|---|---|---|---|
| 4 | AP | 0.30 | 0.30 | 0.002** |
| 11 | MAP | 0.057 | 0.36 | 0.002** |
| 7 | Ca | 0.055 | 0.41 | 0.002** |
| 6 | DOC | 0.036 | 0.45 | 0.002** |
| 15 | MAT | 0.028 | 0.48 | 0.002** |
| 1 | AK | 0.027 | 0.50 | 0.004** |
| 17 | pH | 0.022 | 0.52 | 0.002** |
| 2 | NH4 + | 0.023 | 0.55 | 0.008** |
| 3 | NO3 ‐ | 0.021 | 0.57 | 0.008** |
| 12 | Pb | 0.0098 | 0.58 | 0.024* |
| 5 | DOC | 0.0096 | 0.59 | 0.03* |
| 21 | spatial | 0.0077 | 0.60 | 0.028* |
| 34 | spatial | 0.0065 | 0.60 | 0.04* |
The PCA axes are ordered according to their adjusted coefficients of determination (R 2.adj). Items in the “Highest loading” column are the variables that have the biggest loading on each PCA axes, “spatial” means that the highest loading is imposed by PCNM eigenvector; R 2.adj: the adjusted R 2 for βNTI in the forward selection model; Cum.R 2.adj:the cumulative adjusted R 2 for βNTI in the forward selection model.
Indicates p < 0.05,
Indicates p < 0.01.
4. DISCUSSION
4.1. Significant distance‐decay relationship
In our study, we found significant distance‐decay pattern of the bacterial community in maize field of Northeast China (Figure 1). The turnover rate (Z value) in our study was 0.05, which was in the range of the estimated microbial community turnover rate from 0.002 to 0.26 in other studies (Green & Bohannan, 2006; Horner‐Devine, Lage, Hughes, & Bohannan, 2004; Woodcock, Curtis, Head, Lunn, & Sloan, 2006). Our results suggested that relatively nearby bacterial communities would have more similar composition, which was in accordance with previous research (Hanson et al., 2012; Martiny et al., 2011).
4.2. Scale‐dependent shifts in dominant processes governing spatial turnover in bacteria community composition
The dominant processes that governed spatial turnover in bacterial community composition changed with increasing spatial scale, from HD at the smallest scales to VS at the largest scales (Table 1; Figure 2A). Such results were consistent with our hypothesis that greater community similarity at closer sites would be mainly caused by HD, with the lower community similarity at more distant sites being brought about by VS. In addition, the result resembled that of Stegen et al.’s (2015) simulation experiment, in which HD dominated in communities which were closely located and where the environmental selection was weak, whereas distantly located communities where the environmental selection was strong were mainly dominated by VS. It was also consistent with Caruso et al.’s (2011) finding that differences between desert microbial communities within continents with more similarity were dominated by stochastic processes, and those across continents with more dissimilarities were dominated by deterministic processes. The reason may be that, the stronger role of homogeneous dispersal at smaller scales was caused by the high microbial exchange rate (high dispersal rate) and a low level of selection due to greater environmental similarity at closer sites (Bahram et al., 2015; Figure 1; Appendix Figure A5). High dispersal rates can homogenize communities across environments through the mass effect (Evans, Martiny, & Allison, 2017). As geographical distance increased, the failure of movement and establishment of some microbial species as well as the increased environmental heterogeneity should increase the effect of DL and selection for species with variable survival strategies. Finally, when the distance exceeded a certain threshold, the community variation caused by deterministic processes surpassed stochastic processes and a dominant role of VS overtook HD and DL.
As the effect of soil pH was most significantly correlated with bacterial community structure in our dataset (Appendix Table A2), we also examined the relationship between soil pH difference and geographical distance, as well as the variation tendency of different processes at different soil pH scales. The results supported our explanation that soil pH difference generally increased significantly with increasing geographical distance (Appendix Figure A5). Besides, as the soil pH difference increased, the importance of the four processes shifted similarly in their relationship to increasing geographical distance (Appendix Table A4; Appendix Figure A6). Our results were in agreement with previous findings showing that the assembly processes of soil bacteria could be significantly influenced by soil pH, with stochastic processes dominated in neutral conditions and deterministic processes dominated in acid and alkaline conditions (Tripathi et al., 2018). To our knowledge, this is the first study that explains the distance‐decay pattern of the microbial community from the perspective of discerning the assembly processes at multiple spatial scales, though previous research has evaluated the influence of spatial scale on phylogenetic community structure, phylogenetic relatedness and the assembly processes (Cavender‐Bares, Keen, & Miles, 2006; Kembel & Hubbell, 2006; Kraft, Cornwell, Webb, & Ackerly, 2007; Shi et al., 2018).
Though the trends in the four processes governing bacterial community across space in the North China Plain resembled those in Northeast China (Table 1; Figure 2; Appendix Table A3; Appendix Figure A4), the dominant role of HD was not observed at the smallest spatial scales in the North China Plain. However, we suspected that HD would dominate microbial community assembly at even smaller spatial scales if we had sampled at those scales, just as Bahram et al. (2015) concluded that both DL and deterministic selection had a weak effect on eukaryotes at local scales (<2 m). The spatial scale beyond which HD is overtaken by DL and VS may depend on the particular environment gradient and the species’ dispersal ability (Graham & Fine, 2008; Hanson et al., 2012). The threshold scale should be larger if environmental selection is weak and species have high dispersal ability. Conversely, if environmental selection is strong and species have poor dispersal ability, the threshold scale should be smaller. Hence, the smallest spatial scales we sampled in the North China Plain (35.58 km) may be too large for HD to dominate.
To explain the discrepancy in importance of community structuring factors between Northeast China and the North China Plain, the correlations among community similarity, soil pH difference and geographical distance were examined. We found that, although the spatial turnover of soil bacterial community in Northeast China (z = 0.05) were larger than on the North China Plain (z = 0.018), the soil pH difference and bacterial community similarity in Northeast China were not significantly correlated with geographical distance at small spatial scales (<80.27 km), and such nonsignificant relationships were not observed in the North China Plain when corresponding geographical distances were used (Appendix Table A5). The results suggested that, compared with the North China Plain, the bacterial community and soil pH in Northeast China were more homogenous on small spatial scales. This can in turn explain the dominance of HD at small spatial scales in Northeast China rather than the North China Plain.
The results in Northeast China and the North China Plain lead us to propose a conceptual model (Figure 3). We hypothesize that the trends in HD and HS are similar, both falling gradually to nearly 0. However, the importance of HD may be constantly higher than HS, as the homogeneous environmental conditions which are crucial for HS to dominate may be rare in the natural ecosystem. The case for DL may be a little more complicated: though no model fitted significantly to it in the Northeast China, we still think it should first reach to its peak at a certain distance and then decrease to a relatively stable value. Not only because it is the pattern that DL shows in the North China Plain, but also we believe that its importance would peak at some spatial distance before the growth rate of the effect of VS caused by increasing geographical distance outweighted that of DL. With increasing spatial scale, the peak importance of DL may not exceed the importance of VS or exceed VS after/before VS exceeds HD (scenario 1, 2 and 3 respectively for DL in Figure 3), it may depend on the relative growing rate of VS and DL across space. The influence of VS would steadily increase with the increasing geographical distance, which would exceed HD and end up being the most dominant assembly process at larger spatial scales. As for the dominant processes at different spatial scales, the microbial community will first be dominated by HD (d<d1), and finally VS (d>d2). Among the distance that d1<d<d2, the effect of deterministic processes will exceed stochastic processes, and dispersal will become limited (Figure 3).
Figure 3.
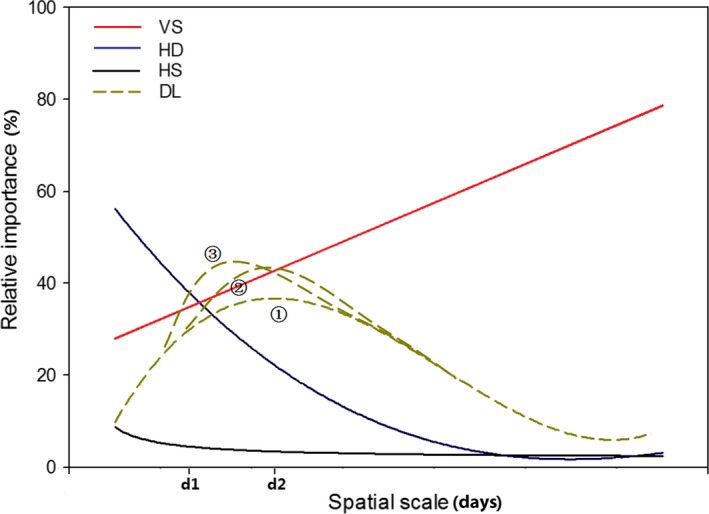
The conceptual model showing how the relative importance of each assembly process changes across space. For DL, there are three possibilities. Scenario ①: the relative importance of DL is always smaller than that of VS; scenario ②: the maximum relative importance of DL exceeds VS after VS exceeds HD; scenario ③: the maximum relative importance of DL exceeds VS before VS exceeds HD. “d1”: the spatial scale before which HD dominates; “d2”: the spatial scale after which VS dominates. For abbreviation, see legend of Figure 2.
4.3. The abiotic and biotic variables that impose deterministic selection
Among the eight environmental variables and PCNM axes retained in the forward selection model using βNTI as responsive variables, soil pH had the largest adjusted coefficients of determination (Table 2). This agreed with the result of the Mantel test, that soil pH had the biggest correlation of bacterial community composition (r = 0.64, p < 0.001 in Appendix Table A2). Many studies have identified soil pH as the most important variable influencing bacterial community in both natural and agricultural ecosystems (Fan et al., 2017; Fierer & Jackson, 2006; Liu et al., 2014; Shen et al., 2013; Sun, Zhang, Guo, Wang, & Chu, 2015; Tripathi et al., 2018). The reason for this could be that bacterial species or strains are capable of optimal growth only within a narrow pH range (Lauber, Hamady, Knight, & Fierer, 2009; Ramirez, Craine, & Fierer, 2010), such that changes in environmental pH would directly deterministically select for species with different pH optima. Besides, soil pH usually covaries with other environmental variables such as climate, soil nutrients, plant properties, and the content or valence state of heavy metal elements (Van Nostrand, Sowder, Bertsch, & Morris, 2005; Williams, Jangid, Shanmugam, & Whitman, 2013; Wang, Zheng, et al., 2017; Lammel et al., 2018; Appendix Table A6). Hence, increased soil pH difference could result in the shift of the assembly processes, especially for VS, which increased steadily with spatial scale (Appendix Figure A6). In addition, we also identified three PCNM axes (Table 2) that was retained in the forward‐selection model, hence they represented the unmeasured, spatially auto‐correlated environmental variables that imposed deterministic selection.
All the measured and unmeasured, spatially auto‐correlated environmental variables explained 63% of deterministic selection, and we thus inferred that the remaining 37% was caused by biotic factors. The high proportion of biotic factors that imposed selection indicated the necessity of taking biotic factors into account when seeking for the factors that structured microbial communities. However, although we have measured a wide range of environmental variables (24 in total and contributed 58.6% in the totally identified 62.1% abiotic influences among the deterministic selection) and taken the unmeasured, spatially auto‐correlated environmental factors into consideration, there should still be a caveat that we may have overlooked the deterministic selection imposed by unmeasured environmental variables that are not spatially auto‐correlated. These would by default be added to the biotic factors in our study, possibly erroneously: thus, the actual biotic fraction may be smaller than 37%.
CONFLICT OF INTERESTS
The authors declare no conflict of interest.
AUTHOR CONTRIBUTIONS
HC, YS and YGZ designed the study. MF and YS collected soil samples. MF conducted the experiment and analyzed data. HC, BMT and JMA evaluated the data. MF, BMT, JMA and HC wrote the manuscript.
ETHICS STATEMENT
None required.
DATA ACCESSIBILITY
The raw data were uploaded to the National Center for Biotechnology Information SRA database under the accession number PRJNA508421.
ACKNOWLEDGEMENTS
We are grateful to Liyan Zhang, Jianmei Wang for assistance in soil sampling and laboratory assays. This work was supported by the Strategic Priority Research Program (XDB15010101) of the Chinese Academy of Sciences, the National Key Research and Development Program of China (2017YFC0504002, 2017YFD0200604), and the China Biodiversity Observation Networks (Sino BON).
APPENDIX 1.
Figure A1.

Locations of sampling region in Northeast China and quadrat sets demonstration at one site
Figure A2.

The Mantel's correlogram about the relationship between phylogenetic distances of two OTUs and their niche difference in the maize field of Northeast China. Solid and open symbols represent significant and nonsignificant correlations, respectively
Figure A3.

The relative abundance of the dominant bacterial phyla/classes (>1%) in the nine sites in the maize field of Northeast China
Figure A4.

The importance of the four processes at different spatial scales in shaping the spatial turnover of bacterial community composition in the wheat field of the North China Plain. (a) The dots that represent the importance of the four processes are connected with smoothed curve. The significant mathematical models that could be fitted to the importance of variable selection (b), homogeneous selection (c), homogenizing dispersal, (d) and dispersal limitation (e). We did not include the scenario “undominated” because of its low importance and ambiguous role in shaping bacterial community structure. When there were more than three models that were significantly fitted to the data, we plotted only three models with relatively higher correlation values. The numbers on the X axis indicates the spatial scales. VS: variable selection; HS: homogeneous selection; HD: homogenizing dispersal; DL: dispersal limitation
Figure A5.
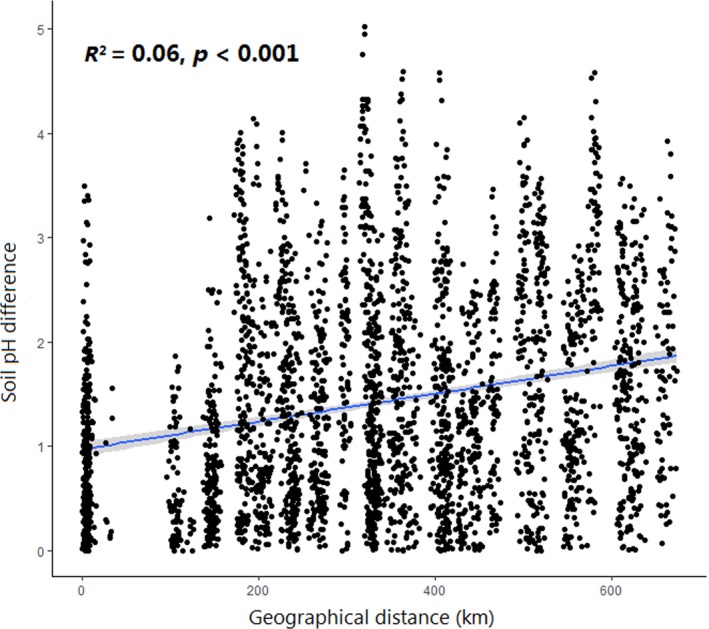
The relationship between soil pH difference and geographical distance in the maize field of Northeast China
Figure A6.

The importance of the four processes in different soil pH scales in governing spatial turnover of bacterial community composition in the maize field of Northeast China. (a) The dots that represent the importance of the four processes are connected with smoothed curve. The significant mathematical models that could be fitted to the importance of variable selection (b), homogeneous selection (c), homogeneous dispersal (d) and dispersal limitation (e). We did not include the scenario “undominated” because of its low importance and ambiguous role in shaping bacterial community structure. When there were more than three models that were significantly fitted to the data, we plotted only three models with relatively higher correlation values. The numbers on the X axis indicates soil pH difference. For abbreviation, see Figure A4.
Table A1.
The comparison of the soil and climate properties among the nine sites in the maize field of the Northeast China
| SL | CL | TN | KS | SH | TL | TY | JX | JL | |
|---|---|---|---|---|---|---|---|---|---|
| SM | 0.135 (0.029)d | 0.110 (0.102)d | 0.104 (0.020)d | 0.290 (0.038)a | 0.276 (0.024)a | 0.271 (0.029)ab | 0.231 (0.034)bc | 0.207 (0.038)c | 0.209 (0.032)c |
| pH | 7.67 (0.64)ab | 7.97 (0.46)a | 8.42 (0.62)a | 6.73 (0.72)c | 7.01 (0.69)bc | 8.05 (0.62)a | 5.71 (1.06)d | 6.76 (1.11)c | 5.37 (0.77)d |
| NO3 − (µg/g) | 159.6 (107.0)a | 62.3 (80.4)b | 28.4 (19.6)b | 32.2 (22.2)b | 63.4 (26.8)b | 42.7 (49.3)b | 56.5 (36.4)b | 37.0 (12.6)b | 45.1 (36.5)b |
| NH4 + (g/g) | 6.13 (0.32)a | 6.35 (0.68)a | 6.76 (0.36)a | 7.81 (2.58)a | 9.12 (1.64)a | 8.31 (0.32)a | 11.0 (6.6)a | 7.67 (1.04)a | 9.38 (1.18)a |
| DON(µg/g) | 9.15 (10.4)a | 12.3 (4.84)a | 10.2 (4.4)a | 11.4 (7.1)a | 13.9 (4.0)a | 6.85 (1.6)a | 17.8 (9.1)a | 11.2 (4.2)a | 15.3 (6.9)a |
| DOC (µg/g) | 47.9 (2.470d | 58.0 (5.63)cd | 76.6 (15.3)b | 98.1 (33.5)a | 68.6 (15.5)bc | 66.7 (12.8)bc | 64.5 (9.8)bcd | 74.8 (18.8)b | 49.9 (7.21)d |
| TP (mg/g) | 0.69 (0.09)cd | 0.53 (0.07)d | 0.33 (0.07)e | 1.07 (0.25)a | 0.87 (0.09)bc | 0.55 (0.09)d | 0.95 (0.17)ab | 0.94 (0.39)ab | 0.79 (0.18)bc |
| TK (mg/g) | 21.3 (1.1)ab | 22.2 (0.8)a | 21.6 (0.7)ab | 20.3 (1.26)b | 20.7 (0.58)ab | 22.1 (2.44)a | 20.8 (0.87)ab | 21.6 (2.8)ab | 20.2 (0.65)b |
| AP (µg/g) | 30.4 (12.8)bc | 18.3 (9.0)c | 10.2 (4.07)c | 76.6 (72.7)a | 59.0 (20.7)ab | 23.2 (12.0)c | 65.6 (24.8)a | 89.2 (43.7)a | 73.4 (37.7)a |
| AK (µg/g) | 182.8 (49.3)bc | 172.2 (74.3)bc | 130.6 (37.8)c | 289.1 (116.8)a | 256.7 (64.4)a | 156.9 (21.6)bc | 221.7 (70.6)ab | 221.2 (81.4)ab | 167.5 (80.8)bc |
| TN (%) | 0.11 (0.03)b | 0.09 (0.02)b | 0.08 (0.01)b | 0.10 (0.03)b | 0.09 (0.02)b | 0.10 (0.02)b | 0.10 (0.03)b | 0.14 (0.03)a | 0.10 (0.02)b |
| TC (%) | 1.24 (0.34)cd | 1.33 (0.28)bcd | 1.68 (0.20)b | 1.36 (0.30)bcd | 1.56 (0.36)bc | 1.20 (0.23)cd | 1.57 (0.30)bc | 2.26 (0.71)a | 1.16 (0.26)d |
| MAT (°C) | 6.86 (0.1 1)a | 5.55 (0.056)c | 5.80 (0.049)b | 1.52 (0.15)g | 2.92 (0.021)h | 4.67 (0.24)e | 2.64 (0.054)i | 4.16 (0.084)f | 4.83 (0.044)d |
| MAP (mm) | 490 (10.3)f | 468 (3.32)g | 415 (3.26)h | 448 (152)e | 518 (4.O0)d | 395 (1.83)i | 590 (7.11)b | 556 (3.68)c | 650 (5.00)a |
| Al (mg/g) | 62.7 (6.08)ab | 58.2 (2.25)b | 51.8 (1.56)c | 66.2 (3.61)a | 65.8 (2.10)a | 65.1 (5.39)a | 65.7 (3.66)a | 62.3 (9.50)ab | 65.4 (3.86)a |
| Fe (mg/g) | 21.6 (5.19)bc | 15.9 (1.13)d | 12.2 (0.85)e | 28.3 (1.O0)a | 28.1 (1.28)a | 21.1 (2.35)c | 24.8 (1.78)ab | 24.7 (8.18)ab | 26.4 (2.95)a |
| Ca (mg/g) | 15.5 (5.51)c | 23.4 (5.36)b | 34.2 (6.59)b | 9.24 (1.38)d | 10.7 (1.99)cd | 26.7 (14.3)a | 6.07 (0.508)d | 6.74 (1.24)d | 6.83 (0.75)d |
| Mg (mg/g) | 6.75 (1.26)ab | 5.15 (0.62)c | 4.20 (0.46)d | 6.9O (0.584)ab | 7.24 (0.309)a | 6.17 (1.14)b | 5.02 (0.36)cd | 4.66 (1.28)cd | 6.31 (0.91)b |
| Mn (mg/g) | 0.45 (0.099)c | 0.38 (0.042)cd | 0.29 (0.034)d | 0.67 (0.094)b | 0.65 (0.12)b | 0.38 (0.060)cd | 0.68 (0.078)b | 0.88 (0.326)a | 0.69 (0.08 7)b |
| Cr (µg/g) | 46.1 (10.4)ab | 26.8 (3.28)c | 18.3 (2.66)d | 52.3 (1.69)a | 51.3 (2.92)a | 39.0 (4.92)b | 44.9 (2.90)ab | 42.0 (20.3)b | 47.4 (6.19)ab |
| Zn (µg/g) | 61.9 (13.2)c | 42.5 (2.97)d | 32.4 (2.23)e | 65.5 (5.92)c | 69.6 (3.38)abc | 63.0 (7.02)c | 67.4 (8.42)bc | 78.4 (19.3)a | 75.6 (9.04)ab |
| PK (µg/g) | 22.8 (2.44)de | 21.3 (0.75)e | 18.4 (0.45)f | 25.5 (1.60)bc | 25.6 (1.33)bc | 24.2 (2.62)cd | 27.4 (1.93)ab | 27.5 (3.18)ab | 28.3 (2.90)a |
| Cu (µg/g) | 18.9 (4.70)bc | 13.2 (1.10)d | 10.9 (1.50)d | 21.8 (2.39)ab | 20.8 (1.36)ab | 17.2 (2.04)c | 20.8 (2.11)ab | 22.5 (6.05)a | 22.2 (3.50)a |
| Cd (µg/g) | 0.186 (0.050)a | 0.167 (0.043)ab | 0.098 (0.018)b | 0.126 (0.028)ab | 0.146 (0.019)ab | 0.0156 (0.057)ab | 0.161 (0.131)ab | 0.18 (0.116)a | 0.181 (0.051)a |
Abbreviations: SM: soil moisture; DON: dissolved organic nitrogen; DOC: dissolved organic carbon; TP: total phosphorus; TK: total potassium; AP: available phosphorus; AK: available potassium; TN: total nitrogen; TC: total carbon; MAT: mean annual temperature; MAP: mean annual precipitation
Table A2.
The soil and climate properties that had significant (p < 0.05) influence on bacterial community structure in the maize field of Northeast China based on Bray–Curtis distance
| pH | MAP | Pb | Mn | Ca | MAT | Fe | AP | Cu | Zn | SM | Cr | TP | Al | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| r | 0.64** | 0.60** | 0.38** | 0.36** | 0.33** | 0.31** | 0.27** | 0.24** | 0.23** | 0.20** | 0.15** | 0.14** | 0.14** | 0.13** |
The results were determined by Mantel test (package “vegan” in R 3.1.4).
p < 0.01. For abbreviations, see Table A1.
Table A3.
Relative importance (%) of different processes in shaping the spatial turnover of soil bacterial community composition of the whole region and at different spatial scales in the wheat field of the North China Plain
| Spatial scale | Spatial distance(km) | VS | HS | DL | HD | UD |
|---|---|---|---|---|---|---|
| whole | whole | 63.53 | 0.03 | 21.74 | 7.38 | 7.32 |
| 1 | <35.58 | 49.44 | 0.31 | 6.67 | 38.97 | 4.61 |
| 2 | 35.58–106.16 | 54.8 | 0.00 | 17.20 | 18.17 | 9.83 |
| 3 | 106.16–176.75 | 60.16 | 0.00 | 22.11 | 8.77 | 8.95 |
| 4 | 176.75–247.33 | 61.26 | 0.00 | 23.62 | 8.06 | 7.05 |
| 5 | 247.33–317.91 | 67.19 | 0.02 | 16.53 | 8.82 | 7.43 |
| 6 | 317.91–388.5 | 69.65 | 0.03 | 17.20 | 5.64 | 7.47 |
| 7 | 388.5–459.08 | 63.40 | 0.03 | 23.62 | 5.69 | 7.26 |
| 8 | 459.08–529.66 | 65.39 | 0.06 | 23.94 | 3.14 | 7.46 |
| 9 | 529.66–600.25 | 56.58 | 0.00 | 34.24 | 2.69 | 6.49 |
| 10 | 600.25–670.83 | 60.36 | 0.00 | 27.28 | 2.81 | 9.55 |
| 11 | 670.83–741.42 | 71.46 | 0.00 | 23.62 | 0.34 | 4.57 |
| 12 | 741.42–812.00 | 71.98 | 0.00 | 22.52 | 0.35 | 5.14 |
| 13 | 812.00–882.58 | 56.05 | 0.25 | 38.27 | 0.00 | 5.43 |
| 14 | 882.58–953.17 | 86.58 | 0.00 | 11.58 | 0.00 | 1.83 |
| 15 | 953.17‐1,023.8 | 68.12 | 0.00 | 25.00 | 0.00 | 6.88 |
| 16 | 1,023.80‐1,094.30 | 95.06 | 0.00 | 4.32 | 0.00 | 0.62 |
The scales 1–16 indicated the spatial distance between two samples, the percentage of the five processes indicated the relative importance of these processes in dominating the community dissimilarity between two samples within each specific spatial scale.
Abbreviations: Whole: the whole region; VS: variable selection; HS: homogenous selection; DL: dispersal limitation; HD: homogenizing dispersal; UD: undominated.
Table A4.
Relative importance (%) of different processes at different soil pH scales in the maize field of Northeast China
| pH difference | VS | HS | DL | HD | UD |
|---|---|---|---|---|---|
| [0,0.1) | 29.41 | 4.41 | 11.03 | 37.50 | 17.65 |
| [0.1,0.2) | 31.43 | 6.43 | 12.86 | 30.00 | 19.28 |
| [0.2,0.3) | 29.58 | 9.86 | 9.86 | 36.62 | 14.08 |
| [0.3,0.4) | 29.30 | 6.37 | 12.10 | 34.39 | 17.83 |
| [0.4,0.5) | 31.65 | 8.63 | 17.27 | 30.22 | 12.23 |
| [0.5,1) | 36.55 | 5.91 | 17.57 | 26.44 | 13.53 |
| [1,1.5) | 47.41 | 5.01 | 22.36 | 14.13 | 11.09 |
| [1.5,2) | 57.80 | 2.44 | 24.88 | 6.58 | 8.29 |
| [2,2.5) | 76.94 | 0.56 | 15.56 | 3.06 | 3.89 |
| [2.5,3) | 83.21 | 1.14 | 10.69 | 1.91 | 3.05 |
| [3,3.5) | 89.60 | 0.58 | 4.62 | 4.62 | 0.58 |
| [3.5,4) | 89.02 | 2.44 | 1.22 | 4.88 | 2.44 |
| [4,5.02) | 100 | 0 | 0 | 0 | 0 |
For abbreviations, please see Table A3
Table A5.
The slope, intercept, and adjusted coefficients of determination (R 2.adj) of linear regression model and correlation value (Cor) of the relationships between community similarity and soil pH difference (otu‐pH), soil pH difference and geographical distance (pH‐geo) and community similarity and geographical distance (otu‐geo) in both Northeast China and North China Plain
| Northeast China | Cor | slope | North China Plain | |||||
|---|---|---|---|---|---|---|---|---|
| slope | intercept | R 2.adj | intercept | R 2.adj | Cor | |||
| whole | ||||||||
| otu‐pH | −0.097 | 0.49 | 0.41*** | −0.64*** | −0.065 | 0.54 | 0.64*** | −0.80*** |
| pH‐geo | 0.0013 | 0.97 | 0.056*** | 0.24*** | 0.0009 | 0.91 | 0.02*** | 0.14*** |
| otu‐geo | −0.00042 | 0.5 | 0.24*** | −0.50*** | −0.00015 | 0.51 | 0.08*** | −0.29*** |
| small | ||||||||
| otu‐pH | −0.068 | 0.56 | 0.0001*** | −0.38*** | −0.075 | 0.6 | 0.28*** | −0.53*** |
| pH‐geo | −0.0016 | 0.88 | 0.0001 | −0.01 | 0.035 | 0.16 | 0.034*** | 0.19*** |
| otu‐geo | −0.0007 | 0.51 | −0.002 | −0.03 | −0.003 | 0.59 | 0.01*** | −0.12*** |
The nonsignificant relationships are marked in bold.
Indicates p < 0.001.
Table A6.
The correlation between the total 24 environmental variables
| Fe | Ca | Mg | Mn | Cr | Zn | Pb | Cu | Cd | SM | pH | NO3 − | NH4 + | DON | DOC | TP | TK | AP | AK | MAT | MAP | TN | TC | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Al | 0.61** | −0.54** | 0.47** | 0.36** | 0.54** | 0.58** | 0.68** | 0.54** | 0.11 | 0.5** | −0.42** | 0.03 | −0.12 | −0.06 | −0.08 | 0.43** | 0.02 | 0.33** | 0.2 | −0.41** | 0.35** | 0.2 | −0.03 |
| Fe | −0.65** | 0.68** | 0.76** | 0.94** | 0.86** | 0.78** | 0.91** | 0.18 | 0.68** | −0.59** | −0.05 | −0.17 | −0.06 | 0.12 | 0.75 | −0.52** | 0.55** | 0.46** | −0.59** | 0.52** | 0.31** | 0.18 | |
| Ca | −0.11 | −0.61** | −0.59** | −0.64** | −0.71** | −0.63** | −0.18 | −0.34** | 0.67** | −0.03 | 0.35** | 0.16 | −0.02 | −0.58** | 0.08 | −0.52** | −0.26* | 0.47** | −0.75** | −0.14 | −0.02 | ||
| Mg | 0.26* | 0.73** | 0.5** | 0.32** | 0.56** | 0.13 | 0.49** | −0.18 | 0.15 | −0.07 | −0.08 | 0.01 | 0.41** | −0.48** | 0.21 | 0.31** | −0.22* | 0.08 | 0.16 | −0.08 | |||
| Mn | 0.62** | 0.71** | 0.76** | 0.73** | 0.13 | 0.41** | −0.62** | −0.12 | −0.14 | −0.05 | 0.09 | 0.61** | −0.37** | 0.51** | 0.34** | −0.5** | 0.64** | 0.3** | 0.32** | ||||
| Cr | 0.84** | 0.66*8 | 0.89** | 0.25* | 0.61** | −0.52** | 0.04 | −0.17 | −0.12 | 0.09 | 0.73** | −0.56** | 0.48** | 0.46** | −0.46** | 0.44** | 0.3** | 0.14 | |||||
| Zn | 0.81** | 0.9** | 0.4** | 0.58** | −0.57** | −0.03 | −0.16 | −0.05 | 0.06 | 0.74** | −0.33** | 0.66** | 0.47** | −0.38** | 0.57** | 0.4** | 0.29** | ||||||
| Pb | 0.79** | 0.23* | 0.54** | −0.7** | −0.12 | −0.18 | −0.06 | −0.07 | 0.56** | −0.27* | 0.51** | 0.21 | −0.47** | 0.66** | 0.22* | 0.09 | |||||||
| Cu | 0.36** | 0.6** | −0.57** | −0.04 | −0.13 | −0.07 | 0.09 | 0.78** | −0.5** | 0.63** | 0.5** | −0.45** | 0.57** | 0.41** | 0.28* | ||||||||
| Cd | 0 | −0.16 | 0.13 | −0.09 | −0.06 | −0.11 | 0.28* | 0.02 | 0.28* | 0.17 | 0.11 | 0.19 | 0.22* | 0.22* | |||||||||
| SM | −0.31** | −0.05 | −0.06 | −0.06 | 0.3** | 0.56** | −0.35** | 0.37** | 0.34** | −0.69** | 0.17 | 0.24* | 0.09 | ||||||||||
| Ph | 0.05 | 0.19 | 0 | 0.1 | −0.43** | 0.31** | −0.42** | −0.08 | 0.42** | −0.77** | −0.18 | 0.01 | |||||||||||
| NO3 − | 0.07 | 0.08 | −0.26* | 0.01 | 0.14 | −0.02 | 0.02 | 0.3** | 0 | 0.1 | −0.16 | ||||||||||||
| NH4 + | 0.82 | −0.1 | −0.18 | 0.04 | −0.1 | −0.05 | 0.09 | −0.18 | 0.02 | 0.08 | |||||||||||||
| DON | −0.07 | −0.06 | 0.14 | 0.06 | 0.06 | −0.03 | −0.02 | 0.01 | 0.03 | ||||||||||||||
| DOC | 0.38** | −0.09 | 0.48** | 0.62** | −0.46** | −0.19 | 0.02 | 0.23* | |||||||||||||||
| TP | −0.39** | 0.84** | 0.77** | −0.61** | 0.5** | 0.32** | 0.33** | ||||||||||||||||
| TK | −0.16 | −0.18 | 0.27* | −0.3** | −0.2 | −0.17 | |||||||||||||||||
| AP | 0.79** | −0.44** | 0.51** | 0.26* | 0.31** | ||||||||||||||||||
| AK | −0.39** | 0.13 | 0.28* | 0.39** | |||||||||||||||||||
| MAT | −0.3** | −0.02 | −0.12 | ||||||||||||||||||||
| MAP | 0.14 | 0.07 | |||||||||||||||||||||
| TN | 0.63** |
p < 0.01,
p < 0.05.
Feng M, Tripathi BM, Shi Y, Adams JM, Zhu Y‐G, Chu H. Interpreting distance‐decay pattern of soil bacteria via quantifying the assembly processes at multiple spatial scales. MicrobiologyOpen. 2019;8:e851 10.1002/mbo3.851
Data Availability Statement: The raw data were uploaded to the National Center for Biotechnology Information SRA database under the accession number PRJNA508421.
REFERENCES
- Bahram, M. , Kohout, P. , Anslan, S. , Harend, H. , Abarenkov, K. , & Tedersoo, L. (2015). Stochastic distribution of small soil eukaryotes resulting from high dispersal and drift in a local environment. ISME Journal, 10, 885–896. 10.1038/ismej.2015.164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biddle, J. F. , Fitzgibbon, S. , Schuster, S. C. , Brenchley, J. E. , & House, C. H. (2008). Metagenomic signatures of the peru margin subseafloor biosphere show a genetically distinct environment. Proceedings of the National Academy of Sciences of the United States of America, 105, 10583–10588. 10.1073/pnas.0709942105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchet, F. G. , Legendre, P. , & Borcard, D. (2008). Forward selection of explanatory variables. Ecology, 89, 2623–2632. 10.1890/07-0986.1 [DOI] [PubMed] [Google Scholar]
- Blois, J. L. , Gotelli, N. J. , Behrensmeyer, A. K. , Faith, J. T. , Lyons, S. K. , Williams, J. W. , … Wing, S. (2014). A framework for evaluating the influence of climate, dispersal limitation, and biotic interactions using fossil pollen associations across the late Quaternary. Ecography, 37, 1095–1108. 10.1111/ecog.00779 [DOI] [Google Scholar]
- Caporaso, J. G. , Bittinger, K. , Bushman, F. D. , Desantis, T. Z. , Andersen, G. L. , & Knight, R. (2010). Pynast: A flexible tool for aligning sequences to a template alignment. Bioinformatics, 26, 266–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso, J. G. , Kuczynski, J. , Stombaugh, J. , Bittinger, K. , Bushman, F. D. , Costello, E. K. , … Knight, R. (2010). QIIME allows analysis of high‐throughput community sequencing data. Nature Methods, 7, 335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caruso, T. , Chan, Y. , Lacap, D. C. , Lau, M. C. , McKay, C. P. , & Pointing, S. B. (2011). Stochastic and deterministic processes interact in the assembly of desert microbial communities on a global scale. ISME Journal, 5, 1406–1413. 10.1038/ismej.2011.21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavender‐Bares, J. , Keen, A. , & Miles, B. (2006). Phylogenetic structure of floridian plant communities depends on taxonomic and spatial scale. Ecology, 87, 109–122. 10.1890/0012-9658(2006)87[109:PSOFPC]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Chemidlin Prévost‐Bouré, N. , Dequiedt, S. , Thioulouse, J. , Lelièvre, M. , Saby, N. P. A. , Jolivet, C. , … Ranjard, L. (2014). Similar processes but different environmental filters for soil bacterial and fungal community composition turnover on a broad spatial scale. PLoS ONE, 9, e111667 10.1371/journal.pone.0111667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu, H. Y. , Sun, H. B. , Tripathi, B. M. , Adams, J. M. , Huang, R. , Zhang, Y. J. , & Shi, Y. (2016). Bacterial community dissimilarity between the surface and subsurface soils equals horizontal differences over several kilometers in the western Tibetan Plateau. Environmental Microbiology, 18, 1523–1533. 10.1111/1462-2920.13236 [DOI] [PubMed] [Google Scholar]
- Dini‐Andreote, F. , de Cássia Pereira e Silva, M. , Triadó‐Margarit, X. , Casamayor, E. O. , van Elsas, J. D. , & Salles, J. F. (2015). Dynamics of bacterial community succession in a salt marsh chronosequence: Evidences for temporal niche partitioning. ISME Journal, 8, 1989– 2001. 10.1038/ismej.2014.54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durrer, A. , Gumiere, T. , Taketani, R. G. , Costa, D. P. D. , Pereira e Silva, M. D. C. , & Andreote, F. D. (2017). The drivers underlying biogeographical patterns of bacterial communities in soils under sugarcane cultivation. Applied Soil Ecology, 110, 12–20. 10.1016/j.apsoil.2016.11.005 [DOI] [Google Scholar]
- Edgar, R. C. (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics, 26, 2460e2461 10.1093/bioinformatics/btq461 [DOI] [PubMed] [Google Scholar]
- Evans, S. , Martiny, J. B. , & Allison, S. D. (2017). Effects of dispersal and selection on stochastic assembly in microbial communities. ISME Journal, 11, 176–185. 10.1038/ismej.2016.96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan, K. , Cardona, C. , Li, Y. , Shi, Y. U. , Xiang, X. , Shen, C. , … Chu, H. (2017). Rhizosphere‐associated bacterial network structure and spatial distribution differ significantly from bulk soil in wheat crop fields. Soil Biology & Biochemistry, 113, 275–284. 10.1016/j.soilbio.2017.06.020 [DOI] [Google Scholar]
- Fierer, N. , & Jackson, R. B. (2006). The diversity and biogeography of soil bacterial communities. Proceedings of the National Academy of Sciences of the United States of America, 103, 626–631. 10.1073/pnas.0507535103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham, C. H. , & Fine, P. V. (2008). Phylogenetic beta diversity: Linking ecological and evolutionary processes across space in time. Ecology Letters, 11, 1265–1277. 10.1111/j.1461-0248.2008.01256.x [DOI] [PubMed] [Google Scholar]
- Green, J. , & Bohannan, B. J. M. (2006). Spatial scaling of microbial biodiversity. Trends in Ecology & Evolution, 21, 501–507. 10.1016/j.tree.2006.06.012 [DOI] [PubMed] [Google Scholar]
- Hanson, C. A. , Fuhrman, J. A. , Horner‐Devine, M. C. , & Martiny, J. B. (2012). Beyond biogeographic patterns: Processes shaping the microbial landscape. Nature Reviews Microbiology, 10, 497–506. 10.1038/nrmicro2795 [DOI] [PubMed] [Google Scholar]
- Hardy, O. J. (2008). Testing the spatial phylogenetic structure of local communities: Statistical performances of different null models and test statistics on a locally neutral community. Journal of Ecology, 96, 914–926. 10.1111/j.1365-2745.2008.01421.x [DOI] [Google Scholar]
- Horner‐Devine, M. C. , Lage, M. , Hughes, J. B. , & Bohannan, B. J. M. (2004). A taxa‐area relationship for bacteria. Nature, 432, 750–753. 10.1038/nature03073 [DOI] [PubMed] [Google Scholar]
- Kembel, S. W. , & Hubbell, S. P. (2006). The phylogenetic structure of a neotropical forest tree community. Ecology, 87, 86–99. 10.1890/0012-9658(2006)87[86:TPSOAN]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Kraft, N. J. B. , Cornwell, W. K. , Webb, C. O. , & Ackerly, D. D. (2007). Trait evolution, community assembly, and the phylogenetic structure of ecological communities. American Naturalist, 170, 271–283. 10.1086/519400 [DOI] [PubMed] [Google Scholar]
- Lammel, D. R. , Barth, G. , Ovaskainen, O. , Cruz, L. M. , Zanatta, J. A. , Ryo, M. , … Pedrosa, F. O. (2018). Direct and indirect effects of a pH gradient bring insights into the mechanisms driving prokaryotic community structures. Microbiome, 6, 106 10.1186/s40168-018-0482-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langenheder, S. , & Ragnarsson, H. (2007). The role of environmental and spatial factors for the composition of aquatic bacterial communities. Ecology, 88, 2154–2161. 10.1890/06-2098.1 [DOI] [PubMed] [Google Scholar]
- Lauber, C. L. , Hamady, M. , Knight, R. , & Fierer, N. (2009). Pyrosequencing‐based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Applied and Environmental Microbiology, 75, 5111–5120. 10.1128/AEM.00335-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legendre, P. , & Anderson, M. J. (1999). Distance‐based redundancy analysis: Testing multispecies responses in multifactorial ecological experiments. Ecological Monographs, 69, 1–24. 10.1890/0012-9615(1999)069[0001:DBRATM]2.0.CO;2 [DOI] [Google Scholar]
- Li, D. J. , & Waller, D. (2016). Long‐term shifts in the patterns and underlying processes of plant associations in Wisconsin forests. Global Ecology and Biogeography, 25, 516–526. 10.1111/geb.12432 [DOI] [Google Scholar]
- Liu, J. , Sui, Y. , Yu, Z. , Shi, Y. U. , Chu, H. , Jin, J. , … Wang, G. (2014). High throughput sequencing analysis of biogeographical distribution of bacterial communities in the black soils of northeast China. Soil Biology and Biochemistry, 70, 113–122. 10.1016/j.soilbio.2013.12.014 [DOI] [Google Scholar]
- Liu, Z. J. , Yang, X. G. , Hubbard, K. G. , & Lin, X. M. (2012). Maize potential yields and yield gaps in the changing climate of northeast China. Global Change Biology, 18, 3441–3454. 10.1111/j.1365-2486.2012.02774.x [DOI] [PubMed] [Google Scholar]
- Martiny, J. B. H. , Eisen, J. A. , Penn, K. , Allison, S. D. , & Horner‐Devine, M. C. (2011). Drivers of bacterial β‐diversity depend on spatial scale. Proceedings of the National Academy of Sciences of the United States of America, 108, 7850–7854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morlon, H. , Chuyong, G. , Condit, R. , Hubbell, S. , Kenfack, D. , Thomas, D. , … Green, J. L. (2008). A general framework for the distance–decay of similarity in ecological communities. Ecology Letters, 11, 904–917. 10.1111/j.1461-0248.2008.01202.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price, M. N. , Dehal, P. S. , & Arkin, A. P. (2010). FastTree 2–approximately maximum‐likelihood trees for large alignments. PLoS ONE, 5, e9490 10.1371/journal.pone.0009490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramette, A. , & Tiedje, J. M. (2006). Multiscale responses of microbial life to spatial distance and environmental heterogeneity in a patchy ecosystem. Proceedings of the National Academy of Sciences of the United States of America, 104, 2761–2766. 10.1073/pnas.0610671104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez, K. S. , Craine, J. M. , & Fierer, N. (2010). Nitrogen fertilization inhibits soil microbial respiration regardless of the form of nitrogen applied. Soil Biology and Biochemistry, 42, 2336–2338. [Google Scholar]
- Ranjard, L. , Dequiedt, S. , Chemidlin Prévost‐Bouré, N. , Thioulouse, J. , Saby, N. , Lelievre, M. , … Lemanceau, P. (2013). Turnover of soil bacterial diversity driven by wide‐scale environmental heterogeneity. Nature Communications, 4, 1434 10.1038/ncomms2431 [DOI] [PubMed] [Google Scholar]
- Shen, C. , Xiong, J. , Zhang, H. , Feng, Y. , Lin, X. , Li, X. , … Chu, H. (2013). Soil pH drives the spatial distribution of bacterial communities along elevation on Changbai Mountain. Soil Biology and Biochemistry, 57, 204–211. 10.1016/j.soilbio.2012.07.013 [DOI] [Google Scholar]
- Shi, Y. U. , Li, Y. , Xiang, X. , Sun, R. , Yang, T. , He, D. , … Chu, H. (2018). Spatial scale affects the relative role of stochasticity versus determinism in soil bacterial communities in wheat fields across the North China Plain. Microbiome, 6, 27 10.1186/s40168-018-0409-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soininen, J. , McDonald, R. , & Hillebrand, H. (2007). The distance decay of similarity in ecological communities. Ecography, 30, 3–12. 10.1111/j.0906-7590.2007.04817.x [DOI] [Google Scholar]
- Stegen, J. C. , Lin, X. , Fredrickson, J. K. , Chen, X. , Kennedy, D. W. , Murray, C. J. , … Konopka, A. (2013). Quantifying community assembly processes and identifying features that impose them. ISME Journal, 7, 2069–2079. 10.1038/ismej.2013.93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegen, J. C. , Lin, X. J. , Fredrickson, J. K. , & Konopka, A. E. (2015). Estimating and mapping ecological processes influencing microbial community assembly. Frontiers in Microbiology, 6, 370 10.3389/fmicb.2015.00370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegen, J. C. , Lin, X. J. , Konopka, A. E. , & Fredrickson, J. K. (2012). Stochastic and deterministic assembly processes in subsurface microbial communities. ISME Journal, 6, 1653–1664. 10.1038/ismej.2012.22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, R. B. , Zhang, X. X. , Guo, X. S. , Wang, D. Z. , & Chu, H. Y. (2015). Bacterial diversity in soils subjected to long‐term chemical fertilization can be more stably maintained with the addition of livestock manure than wheat straw. Soil Biology and Biochemistry, 88, 9–18. 10.1016/j.soilbio.2015.05.007 [DOI] [Google Scholar]
- Terrat, S. , Dequiedt, S. , Horrigue, W. , Lelievre, M. , Cruaud, C. , Saby, N. P. A. , … Chemidlin Prévost‐Bouré, N. (2015). Improving soil bacterial taxa–area relationships assessment using DNA meta‐barcoding. Heredity, 114, 468–475. 10.1038/hdy.2014.91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi, B. M. , Stegen, J. C. , Kim, M. , Dong, K. , Adams, J. M. , & Lee, Y. K. (2018). Soil pH mediates the balance between stochastic and deterministic assembly of bacteria. ISME Journal, 12, 1072–1083. 10.1038/s41396-018-0082-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuomisto, H. , Ruokolainen, K. , & Yli‐Halla, M. (2003). Dispersal, environment, and floristic variation of western Amazonian forests. Science, 299, 241–244. 10.1126/science.1078037 [DOI] [PubMed] [Google Scholar]
- Van Nostrand, J. D. , Sowder, A. G. , Bertsch, P. M. , & Morris, P. J. (2005). Effect of pH on the toxicity of nickel and other divalent metals to Burkholderia cepacia pr1301. Environmental Toxicology and Chemistry, 24(11), 2742–2750. 10.1897/04-335R.1 [DOI] [PubMed] [Google Scholar]
- Vellend, M. (2010). Conceptual synthesis in community ecology. The Quarterly Review of Biology, 85, 183–206. 10.1086/652373 [DOI] [PubMed] [Google Scholar]
- Wang, C. , Zheng, M. M. , Song, W. F. , Wen, S. L. , Wang, B. R. , Zhu, C. Q. , & Shen, R. F. (2017). Impact of 25 years of inorganic fertilization on diazotrophic abundance and community structure in an acidic soil in southern China. Soil Biology and Biochemistry, 113, 240–249. [Google Scholar]
- Wang, J. , Shen, J. I. , Wu, Y. , Tu, C. , Soininen, J. , Stegen, J. C. , … Zhang, E. (2013). Phylogenetic beta diversity in bacterial assemblages across ecosystems: Deterministic versus stochastic processes. ISME Journal, 7, 1310–1321. 10.1038/ismej.2013.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X. B. , Lu, X. T. , Yao, J. , Wang, Z. W. , Deng, Y. , Cheng, W. X. , … Han, X. G. (2017). Habitat‐specific patterns and drivers of bacterial beta‐diversity in China's drylands. ISME Journal, 11, 1345–1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y. S. , Li, C. N. , Kou, Y. P. , Wang, J. J. , Tu, B. , Li, H. , … Yao, M. (2017). Soil pH is a major driver of soil diazotrophic community assembly in Qinghai‐Tibet alpine meadows. Soil Biology and Biochemistry, 115, 547–555. [Google Scholar]
- Wiens, J. J. (2011). The niche, biogeography and species interactions. Philosophical Transactions of the Royal Society B: Biological Sciences, 366, 2336–2350. 10.1098/rstb.2011.0059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, M. A. , Jangid, K. , Shanmugam, S. G. , & Whitman, W. B. (2013). Bacterial communities in soil mimic patterns of vegetative succession and ecosystem climax but are resilient to change between seasons. Soil Biology and Biochemistry, 57, 749–757. 10.1016/j.soilbio.2012.08.023 [DOI] [Google Scholar]
- Woodcock, S. , Curtis, T. P. , Head, I. M. , Lunn, M. , & Sloan, W. T. (2006). Taxa‐area relationships for microbes: The unsampled and the unseen. Ecology Letters, 9, 805–812. 10.1111/j.1461-0248.2006.00929.x [DOI] [PubMed] [Google Scholar]
- Yang, T. , Adams, J. M. , Shi, Y. , Sun, H. B. , Cheng, L. , Zhang, Y. Z. , & Chu, H. Y. (2017). Fungal community assemblages in a high elevation desert environment: Absence of dispersal limitation and edaphic effects in surface soil. Soil Biology and Biochemistry, 115, 393–402. 10.1016/j.soilbio.2017.09.013 [DOI] [Google Scholar]
- Zhang, K. , Adams, J. M. , Shi, Y. U. , Yang, T. , Sun, R. , He, D. , … Chu, H. (2017). Environment and geographic distance differ in relative importance for determining fungal community of rhizosphere and bulk soil. Environmental Microbiology, 19, 3649–3659. 10.1111/1462-2920.13865 [DOI] [PubMed] [Google Scholar]
- Zhang, X. Y. , Sui, Y. Y. , Zhang, X. D. , Meng, K. , & Herbert, S. J. (2007). Spatial variability of nutrient properties in black soil of northeast China. Pedosphere, 17, 19–29. 10.1016/S1002-0160(07)60003-4 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The raw data were uploaded to the National Center for Biotechnology Information SRA database under the accession number PRJNA508421.


