Abstract
The mouse homeodomain protein, Engrailed-1, is generally viewed as an essential player in the early establishment and maintenance of the midbrain/hindbrain region that gives rise to the cerebellum and midbrain. In keeping with this, engineered null mutations at this locus have been reported to lead to perinatal lethality accompanied by near-total absence of cerebellar and caudal midbrain structures. We report here that these cerebellar phenotypes are nearly completely suppressed on a C57BL/6J genetic background. All cell types are present and arranged properly in both the cortex and the deep nuclei, and cell counts reveal no significant absence of cerebellar Purkinje cells. Folial patterns are nearly normal, although an apparent fusion of lobules IV and V is consistently noted. Significantly, no change in the Engrailed-2 mutant phenotype occurs after a similar background switch, and whole-mount in situ hybridization reveals identical En2 expression patterns in wild-type C57BL/6J and 129/Sv mice. One likely mechanism for the En1-/- phenotype suppression is a temporal and/or spatial change in the pattern of Engrailed-2 expression apparent only in the absence of Engrailed-1. In support of this, C57BL/6—En1-/- embryos that are also En2+/- lack a cerebellum and caudal midbrain: a phenotype identical to 129/Sv—En1-/- mice.
Keywords: Engrailed-1, cerebellar development, Engrailed-2, genetic background, Purkinje cells, midbrain/hindbrain
Introduction
The vertebrate cerebellum develops from a specialized region of the neural tube located at the midbrain/hindbrain border (MHB) (for review, see Liu and Joyner, 2001). As part of this process, the two mouse Engrailed genes (En1 and En2) appear at the MHB in a broad transverse domain. En1 is expressed first, at the 1-somite stage (Davis et al., 1988; McMahon et al., 1992) followed by En2 at the 5-somite stage (Davis and Joyner, 1988; Davis et al., 1988). Later in development, En1 is also expressed in regions of the spinal cord and limb buds, whereas En2 expression remains restricted to the brain. Expression of both genes continues in the adult, in whom their functions are unknown (Davis and Joyner, 1988).
Null alleles of both Engrailed genes have been created. The analysis of mutations of Engrailed-1 on a 129/Sv inbred or C57BL/6J × 129/Sv hybrid background support the view that En1 is critical for midbrain/hindbrain development (Wurst et al., 1994). Homozygous En1 mutants die at birth, with a near-total absence of cerebellum and dorsal midbrain. In addition, En1-/- mutants have limb and skeletal abnormalities that include defects of paws, sternum, and ribs (Wurst et al., 1994; Loomis et al., 1996). In contrast, En2 homozygous mutants on similar genetic backgrounds exhibit only a mild phenotype that appears restricted to cerebellum (Joyner et al., 1989, 1991). There is a decrease in cerebellar size, minor abnormalities of folial patterning (Millen et al., 1994) plus a 35% reduction in Purkinje, granule, deep nuclear, and inferior olive neuron number (Kuemerle et al., 1997). As a likely consequence, En2 mutants have mild behavioral problems (Gerlai et al., 1996). In the CNS, the Engrailed genes appear to be interchangeable. When the En1 gene is replaced by either En2 (Hanks et al., 1995) or the Drosophila en gene (Hanks et al., 1998) the resulting homozygous mice show virtually no brain defects. However, such substitutions do not rescue the skeletal abnormalities (Loomis et al., 1996). This separation of brain and limb phenotypes is not unexpected, given the absence of En2 expression in the developing limb field, but the significance of the separation may run deeper, as the results of the current work suggest.
We report here that transferring the original En1 null allele (En1Hd) (Wurst et al., 1994) from the inbred 129/Sv strain to the C57BL/6J background results in a near-total suppression of the cerebellar phenotype. On a C57BL/6J background, mutant mice undergo a relatively normal cerebellar development and are born alive with mild folial pattern abnormalities of the cerebellum. Despite these dramatic effects, the 129/Sv-to-C57BL/6 background shift results in little or no rescue of limb and sternum defects. Our findings emphasize the fact that the cerebellar field is created by a complex genetic network that is capable of extensive compensatory changes.
Materials and Methods
Mouse strains. Mice carrying the En1Hd allele (Wurst et al., 1994) on the 129/Sv inbred background were provided by Dr. A. Joyner (Skirball Institute, New York). The original animals were mated to wild-type C57BL/6J mice to transfer the mutation to the new background. Offspring of these matings were genotyped via PCR, and the mice carrying the En1Hd allele were backcrossed to C57BL/6J mates. This breeding scheme was maintained for 10 generations to establish a congenic line: C57BL/6J—En1. For genotyping, three sets of PCR primers were used. To detect the neomycin gene in the En1Hd allele, primers 1 (5′-GCTTGGGTGGAGAGGCTATTC-3′) and 2 (5′-CAAGGTGAGATGACAGGAGATC-3′) amplify a 280-bp sequence, whereas primers 3 (5′-TACTTTCTCGGCAGGAGCAAGGTG-3′) and 4 (TGGATTGCACGCAGGTTCTC) amplify a 325-bp sequence. The wild-type En1 allele was detected by amplification using primers 5 (5′-GCAAACCGCTATATCACGGA-3′) and 6 (5′-CACGGTCGTAAGCAGTTTGG-3′), which amplify a 528 bp sequence. Although suppression of the mutant phenotype was first noted at backcross generation N3, all mice used in this study ranged from generation N6 to N10 on the C57BL/6J background and no phenotypic differences in rescue animals were detected regardless of generation number.
Mice carrying the En2Hd allele (Joyner et al., 1991) on the 129/Sv inbred background were maintained and genotyped as described previously (Kuemerle et al., 1997). To transfer this mutation to the C57BL/6J background, heterozygous En2+/Hd mice were mated with wild-type C57BL/6J mice; offspring carrying the En2Hd allele (neo-positive by PCR) were backcrossed to C57BL/6J mates. All C57BL/6J En2Hd mice used in this study were N5 animals.
Histology. For embryonic analysis of mouse cerebella and limbs, pregnant mothers were anesthetized with ether and killed by cervical dislocation. Pups were dissected from the uterus into ice-cold PBS, decapitated, and then fixed by immersion overnight in 4% paraformaldehyde and 0.1 m phosphate buffer (PB) at 4°C. After fixation, a midsagittal cut was made in each head to determine the presence or absence of a cerebellum. For postnatal analysis, the animals were anesthetized with Avertin (0.02 cc/gm of body weight) and transcardially perfused with PBS, followed by 4% paraformaldehyde/PB. Brains were dissected into fresh fixative overnight at 4°C, then transferred to PBS. For paraffin embedding, brains were dehydrated through 50, 70, 80, 95, and 100% ethanol, followed by xylenes, before being placed in paraffin at 56°C. A midsagittal or coronal cut was then made in the brains before they were embedded in fresh paraffin overnight. Sections were cut at 10 μm on a rotary microtome and rehydrated before staining with cresyl violet.
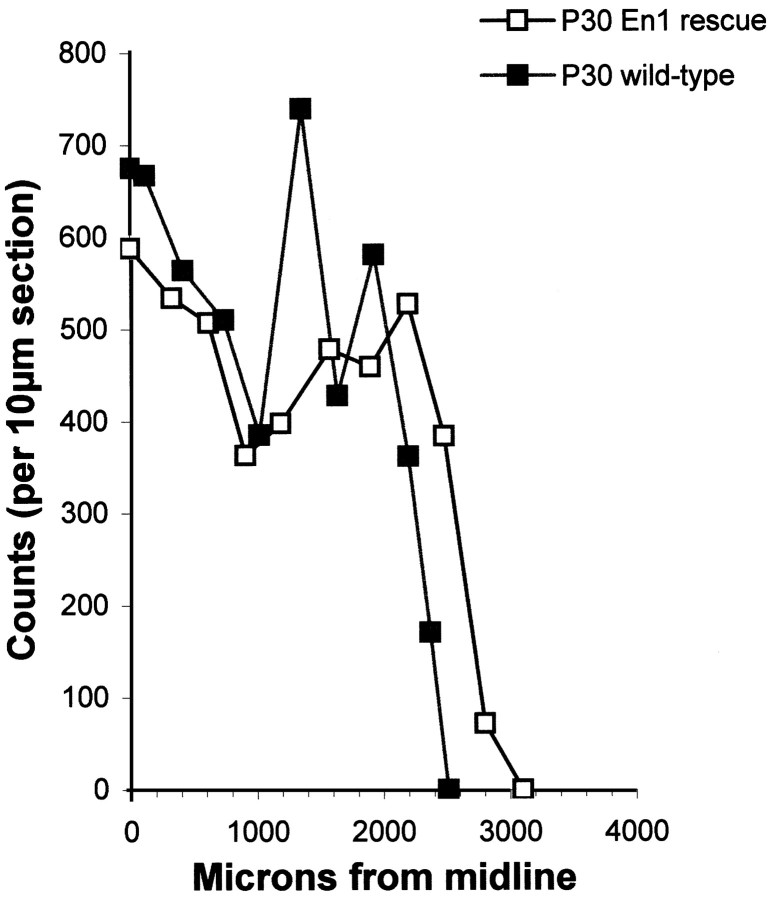
Cell counts. To estimate differences in the Purkinje cell number between wild-type C57BL/6J and C57BL/6J—En1Hd/Hd, two postnatal day 30 (P30) sagittally sectioned cerebella of each genotype were analyzed. Purkinje cells whose nuclear outline could be seen were counted in approximately every 30th section, starting at the midline. After plotting the cell number as a function of distance from the midline, the area under the curve was calculated to give an estimate of total Purkinje cell number. Student's one-tailed t test was used to determine statistical significance.
Immunohistochemistry. Cryostat sections (10 μm) of embryonic brains were rehydrated in three changes of 1× PBS for 5 min each, blocked in PBS with 10% goat serum and 0.5% Triton X-100 for 45 min at room temperature (RT), rinsed three times in PBS for 5 min each, and incubated at 4°C overnight with anti-calbindin antibody in a 1:1000 dilution (Santa Cruz Biotechnology, Santa Cruz, CA). Slides were then rinsed three times in PBS, incubated in a 1:250 dilution of Cy3-labeled goat anti-mouse IgG (Jackson ImmunoResearch, West Grove, PA) for 2 hr at RT in the dark, rinsed again in PBS, and mounted in 1:1 PBS/glycerol for viewing.
Whole-mount in situ hybridization. Embryos were harvested as above in ice-cold PBS then transferred to a 4% paraformaldehyde/PB solution at 4°C for 2 hr to overnight. Tissue was washed twice with PBS/0.1% Tween 20 (PBST), followed by dehydration through 50 and 75% methanol in PBST. The embryos were then rinsed twice with 100% methanol and stored in 100% methanol at -20°C for up to 1 month. A 1.7 kb En1 probe, subcloned into pBluescript, was provided by A. Joyner. All remaining probes were generated by reverse transcription PCR using RNA from an embryonic day 9.5 (E9.5) head for the Pax2 and Wnt1 probes and adult brain RNA for the En2 probe. The primers contained restriction enzyme sequences for directional cloning into pBluescript. For Pax2 a 554 bp fragment was amplified from the 3′-untranslated region using the following primers: 5′-CCGCCAGTGGGATCCTACTCCAT-3′ (forward) and 5′-CGCTCCCAGGCACCATTC-3′ (reverse). A 432 bp Wnt1 fragment was generated from the coding region using the following primers: 5′-AGTGGCTGCTTCAGCCCA-3′ (forward) and 5′-CTCACGCTGTGCAGGATC-3′ (reverse). Finally, a 458 bp En2 fragment was amplified from the 3′-untranslated region using 5′-GGCTCTGCCGCTTGCTTT-3′ (forward) and 5′-TGCGTGCACATGCATACA-3′ (reverse). All fragments were then subcloned into pBluescript and linearized for T7 (antisense) transcription. RNA probes were transcribed using the RNA Transcription Kit from Stratagene (La Jolla, CA) and digoxigenin-UTP from Roche (Indianapolis, IN). After transcription, the reactions were treated with RNase-free DNase I at 37°C for 15 min, stopped with 2 μl of 0.5 m EDTA and precipitated with ammonium acetate and 10 μg of tRNA overnight at -20°C. The probes were spun down and washed with 70% ethanol and allowed to air dry. The pellets were resuspended in 50–70 μl of DEPC water, aliquoted, and stored at -20°C. Each probe was tested to determine the optimal concentration for hybridization.
Embryos were rehydrated through 75, 50, and 25% methanol in PBST followed by one rinse in PBST. Embryos were then digested with proteinase K (20 μg/ml) at RT for 20 min, followed by two 10 min washes in PBST containing 2 mg/ml of glycine and two additional 5 min washes with PBST alone. The embryos were then fixed in 4% paraformaldehyde/0.1% glutaraldehyde/PBST for 20 min at RT, followed by four washes in PBST for 10 min each. Half of the final wash was removed and replaced with prewarmed (65°C) hybridization solution (50% formamide, 5× SSC, 50 μg/ml yeast RNA, 50 μg/ml heparin, and 0.2% Tween 20) for 5–10 min at 65°C. The embryos were then rinsed twice in prewarmed hybridization solution for 10 min each, followed by incubation in hybridization solution at 65°C for 1 hr. The hybridization solution was then replaced with fresh solution containing digoxigenin-labeled probe at 1 μg/ml (1:200 dilution) and incubated at 65°C overnight. The embryos were washed twice with Solution I (2× SSC, 0.5% SDS, and 25% formamide) for 30 min at 70°C; once with a mixture of Solution I and Solution II (0.5 m NaCl, 10 mm Tris-HCl, pH 7.5, and 0.1% Tween 20) (1:1) for 10 min at 70°C; three times in Solution II alone for 5 min at RT; twice in Solution II/RNase A (100 μg/ml) for 30 min at 37°C; once in Solution II for 5 min at RT; once in Solution III (50% formamide, 2× SSC) for 5 min at RT; twice in Solution III for 30 min at 65°C; and three times in TBS/0.1% Tween 20 (TBST)/2 mm levamisole for 5 min at RT. The embryos were then incubated in blocking solution containing 2 mm levamisole, 2% blocking reagent (Roche) and 10% heat-inactivated goat serum in TBST for 1–2 hr at RT. The blocking solution was then replaced with fresh blocking solution containing antidigoxigenin antibody conjugated with alkaline phosphatase (Roche) at 1:2000 and incubated overnight at 4°C. The embryos were then washed three times with TBST/2 mm levamisole for 5 min each; three times with NTMT (100 mm NaCl, 100 mm Tris-HCl, pH 9.5, 50 mm MgCl2, and 1% Tween 20) then developed in NTMT containing 4.5 μl/ml nitroblue-tetrazolium-chloride and 3.5 μl/ml 5-bromo-4-chlor0-indolyl-phosphate at RT for 3 hr overnight in the dark. Final washes were done in TBST three times for 3 min, followed by fixation in 4% paraformaldehyde/0.1% glutaraldehyde overnight at 4°C. Embryos were then rinsed in PBST and stored at 4°C in PBST containing 0.1% sodium azide.
Results
Rescue of En1Hd/Hd cerebellar phenotype on C57BL/6J background
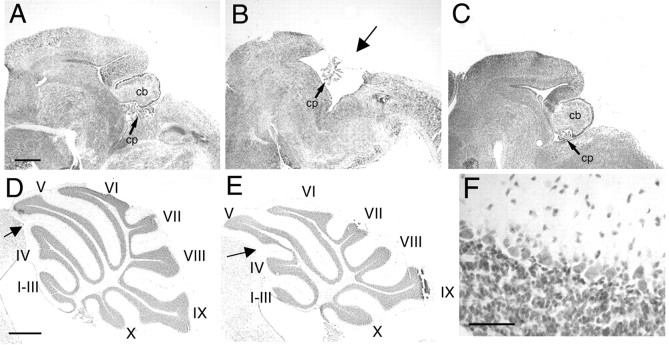
Targeted disruption of the En1 locus leads to perinatal lethality on the 129/Sv strain background. Mutant mice display numerous abnormalities, including a deletion of the cerebellum and truncated colliculi, particularly in posterior regions (Wurst et al., 1994) (Fig. 1B). To determine whether the En1Hd mutation was sensitive to changes in the genetic background, 129/Sv mice heterozygous for the En1Hd allele (a functional null allele) were mated to wild-type C57BL/6J mice; progeny were then backcrossed to C57BL/6J to establish a congenic line. At the N3 backcross generation heterozygous animals were intercrossed. When litters were examined at E17—E18 approximately one-fourth of the embryos were homozygous En1Hd/Hd (Fig. 1C) yet displayed a phenotypically normal cerebellar structure (Fig. 1A). Thus, factors in the C57BL/6J strain background resulted in suppression of the cerebellar phenotypes of the En1Hd mutation. The rescue of the midbrain was less complete. In virtually every animal we examined, we found evidence of a mild truncation of the posterior midbrain, the region destined to give rise to the inferior colliculus. The full description of this phenotype will be the subject of a separate study.
Figure 1.
Cerebellar phenotype of En1Hd/Hd mutants on 129/Sv and C57BL/6J backgrounds. Cresyl violet-stained sagittal cryostat sections of E17.5 midbrain/hindbrain region of C57BL/6J wild-type (A), 129/Sv—En1Hd/Hd (B), and C57BL/6J—En1Hd/Hd (C). The arrow in B indicates where the cerebellum would normally appear in the 129/Sv mutant. Comparison between adult C57BL/6J wild-type cerebellum (D) and C57BL/6J—En1Hd/Hd rescue cerebellum (E) reveals a partial fusion of vermal lobes IV and V, but no other abnormalities. At higher magnification (F), the microarchitecture of the cerebellar cortex is indistinguishable from normal. cb, Cerebellum; cp, choroid plexus. Scale bars: (in D), A—E, 500 μm; F, 25 μm.
To explore the cerebellar phenomenon further, pregnant females from C57BL/6—En1+/Hd intercrosses were allowed to deliver and rear their young. Within a few days after birth, the “rescued” En1Hd/Hd mice can be identified because they are smaller than their normal littermates. Furthermore, they exhibit a reduced vitality that frequently results in their death before weaning. Despite this postnatal fragility, there is no apparent loss of En1Hd/Hd mice prenatally. We have analyzed 75 mice from litters ranging in age from E12 to P0 and from 3 to 10 C57BL/6 backcross generations. Seventeen of these embryos (23%) were genotyped as En1Hd/Hd, close to the expected ratio of 25%. Examination of the brains of these animals revealed that 16 of the 17 genetically mutant embryos were phenotypically normal: a well formed cerebellar and midbrain architecture were readily visible. Cresyl violet-stained sections of E18.5 brains show that the cerebella of C57BL/6J—En1Hd/Hd rescue animals are indistinguishable from those of wild-type littermates (Fig. 1A,C). For comparison, a typical cerebellar remnant of a 129/Sv—En1Hd/Hd embryo is shown in Figure 1B. We have examined C57BL/6J—En1Hd/Hd animals as old as 10 months of age, and this rescue is apparently maintained throughout the entire developmental process. In all homozygous animals (from the N3 generation onward) the overall size and foliation pattern of the cerebellum closely resembles that of wild-type C57BL/6J mice. The close resemblance of the foliation of the rescue to that of the C57BL/6J mouse is particularly noteworthy because there are distinct differences between the patterns of the 129/Sv and C57BL/6J strains. However, the pattern rescue is not complete because we detected a consistent fusion of lobules IV and V in the vermis of rescue animals (Fig. 1D,E, arrows). In every case the defect appeared as a small sulcus appended to the anterior face of lobule V (culmen). The source of this modest difference is unclear, because it is not accompanied by any change in cell number (see below).
The microarchitecture of the C57BL/6J—En1Hd/Hd cerebellar cortex is also normal. A well defined single layer of Purkinje cells separates a normal-looking internal granule cell layer and a cell-sparse molecular layer (Fig. 1F). To examine the dendritic structure of the cerebellar Purkinje cells in more detail, we stained both wild-type (Fig. 2A,B) and C57BL/6J—En1Hd/Hd (Fig. 2C,D) cerebella with antibodies directed against the Purkinje cell-specific protein calbindin. In all lobules, healthy Purkinje cells with extensive richly branched dendritic arbors were readily visible. Each Purkinje cell dendrite was covered with the characteristic postsynaptic spines that are the site of synapse between the parallel fiber axon and the Purkinje cell dendrite (Fig. 2D).
Figure 2.
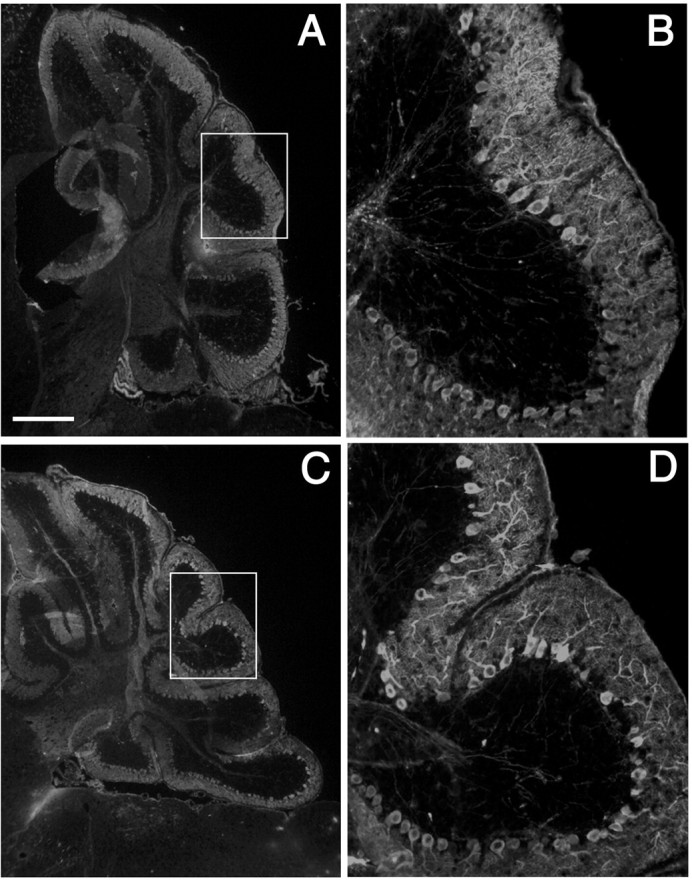
Purkinje cell morphology in adult C57BL/6J—En1Hd/Hd cerebella are indistinguishable from normal. Sagittal cryostat sections immunostained for calbindin, a Purkinje cell-specific calcium-binding protein; the C57BL/6J wild-type [A, B (magnification of inset in A)] and C57BL/6J—En1Hd/Hd cerebella [C, D (magnification of inset in C)] show no difference in either the density of Purkinje cell bodies or the complexity of their dendritic arborizations. Scale bar: (in A) A, C, 400 μm; B, D, 100 μm.
The normal appearance of the cells of the cerebellar cortex is extended to the more quantitative features of the structure as well. We estimated the Purkinje cell number in two C57BL/6—En1Hd/Hd animals and found no significant differences (p = 0.3; one-tailed Student's t test) compared with wild-type controls (Fig. 3). We did not apply any geometric or stereologic correction to our counts; thus, our analysis is not sufficiently accurate to rule out changes of ±10%. Nonetheless, the counts offer clear semiquantitative evidence that the C57BL/6J—En1Hd/Hd animals have cerebella that differ little from their wild-type counterparts. Thus, on the C57BL/6J background, most aspects of cerebellar development appear to proceed normally even in the total absence of Engrailed-1 protein.
Figure 3.
No detectable Purkinje cell loss occurs in the cerebella of rescue animals. Two P30 C57BL/6J animals of each genotype (En1+/+ and En1Hd/Hd) were analyzed. There is no significant difference in Purkinje cell number between the two genotypes (p = 0.29; one-tailed Student's t test). The raw counts per 10 μm section plotted as a function of distance from the midline is shown.
Limb defects in En1Hd/Hd mutants are not suppressed on the C57BL/6J background
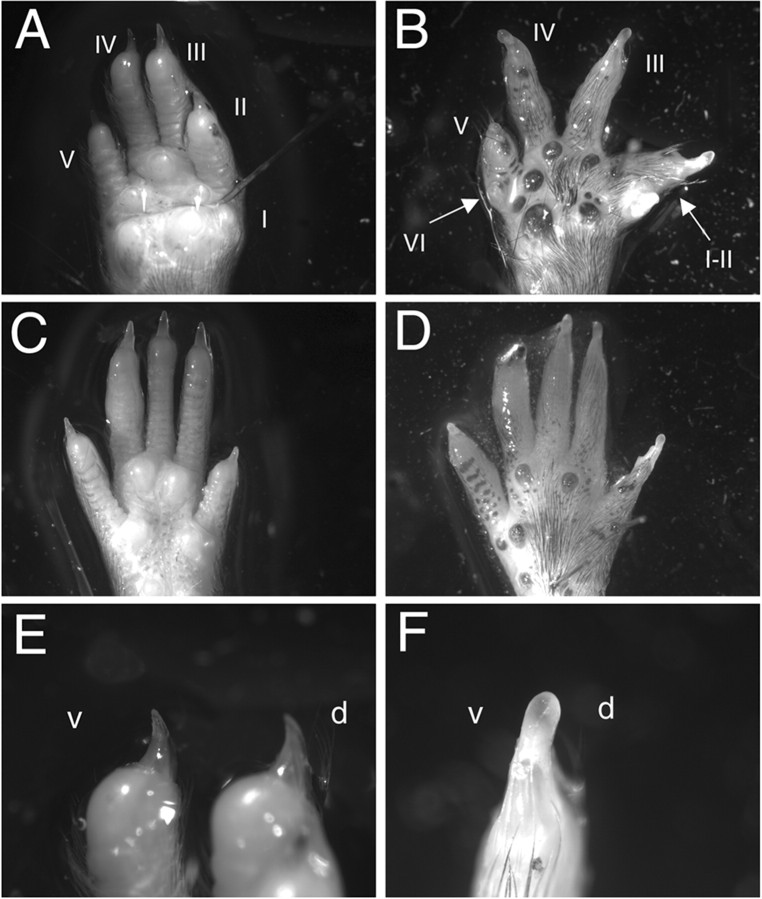
Embryonic En1Hd/Hd mice on the 129/Sv background have severe deformities in their forelimbs, including polydactyly, syndactyly, and shortened and/or outwardly splayed digits (Wurst et al., 1994). Therefore, we examined the forelimbs and hindlimbs of the embryonic C57BL/6J—En1Hd/Hd mice. Contrary to our findings in the cerebellum, all C57BL/6J—En1Hd/Hd mice exhibited forelimb defects similar to those in their 129/Sv—En1Hd/Hd counterparts. Multiple abnormalities and variability of the phenotypes between the right and left paws can often be found in the same embryo. As has been reported previously (Wurst et al., 1994), the severity of the forelimb defects is greater than that of the hindlimbs.
The paw defects develop postnatally and thus become more apparent with age. At weaning, the animals display ectopic footpads, fused digits, ectopic digits, and extra postaxial digits (Fig. 4B,D) compared with their wild-type littermates (Fig. 4A,C). An apparent duplication of dorsal paw phenotype was also seen, manifested as fur growth on the ventral surface of the paws (Fig. 4B,D) and circumferential nails on all digits of affected limbs (Fig. 4F), a condition that was never observed in the wild-type animals (Fig. 4E). Variations in phenotype were found from animal to animal and on different limbs of the same animal, but all C57BL/6J—En1Hd/Hd mice exhibited one or more limb abnormalities. It is also worth noting that although there are clear differences in the detailed structure of the mutant paws on the two genetic backgrounds, the broad categories of developmental failures are the same.
Figure 4.
Mutant limb abnormalities are not suppressed in C57BL/6J—En1Hd/Hd animals; they are identical to those seen on the 129/Sv background. A—F, Adult paws of C57BL/6J wild-type (A, C, E) and C57BL/6J—En1Hd/Hd (B, D, F) animals. Forelimbs are shown in A, B, E, and F; hindlimbs in C and D. Arrows in B denote a sixth postaxial digit (left arrow) and a fusion of digits I and II (right arrow). Ectopic footpads and hair growth are also found on the ventral surface of rescue limbs (B). The right hindlimb of a rescue mutant (D) showed similar abnormalities, but generally hindlimb defects were less severe. The nails of the wild-type digits (E) are hooked, with a clear dorsal (d)/ventral (v) asymmetry. In contrast, the nails of the C57BL/6J—En1Hd/Hd paw (F) are symmetrical and straight.
En2Hd/Hd mutant phenotype is unchanged on C57BL/6J background
Based on their overlapping spatiotemporal patterns of expression and the ability of Engrailed-2 to substitute for Engrailed-1 in “knock-in” experiments, it would appear that the functions of Engrailed-1 and Engrailed-2 overlap. Therefore, one plausible model to explain our findings would be that on the C57BL/6J background, the role of Engrailed-2 becomes predominant, with Engrailed-1 playing a more minor role. According to this hypothesis, if the En2Hd mutation were transferred to the C57BL/6J background, the phenotype should worsen considerably. Mice carrying the En2Hd allele (Joyner et al., 1991) on the 129/Sv inbred background were mated with wild-type C57BL/6J mice for several generations to transfer the mutation to the new strain. We analyzed three N5 C57BL/6J—En2Hd/Hd mice from three separate litters (from heterozygous parents). The cerebellum of 129/Sv—En2Hd/Hd mice, as reported previously, is smaller and has reduced cell numbers in every cell population examined as well as a distorted foliation pattern, particularly in the posterior cerebellar lobules, VI to IX. As with their 129/Sv counterparts, the C57BL/6J—En2Hd/Hd mice survive and appear identical to wild-type littermates in outward appearance. In general, the cerebella of the mutants are reduced in size, but are well foliated with normal molecular, Purkinje cell, and granule cell layers. Closer examination of cresyl violet-stained paraffin sections reveals defects in the posterior lobes (Fig. 5D). As described previously on the 129/Sv genetic background, lobules VIII and IX appear distorted and fused with one another (Fig. 5B) compared with wild-type (Fig. 5A). The same description applies broadly to the C57BL/6J—En2Hd/Hd cerebellum (Fig. 5, compare C with D). Exact comparisons are difficult because the wild-type C57BL/6J cerebellum has a folial pattern that is distinct from its 129/Sv cousin. For example, if one examines Figure 5, comparing A with C, it is plain that in the 129/Sv mouse (Fig. 5A) there is a reduction of lobule VI and a relative enlargement in the size of lobule IX compared with C57BL/6J (Fig. 5C). Thus, although the exact lobulation pattern of the C57BL/6J—En2Hd/Hd cerebellum is different from the 129/Sv—En2Hd/Hd, the alterations seem well within the variation expected given the morphological differences between wild-type animals of the two strains.
Figure 5.
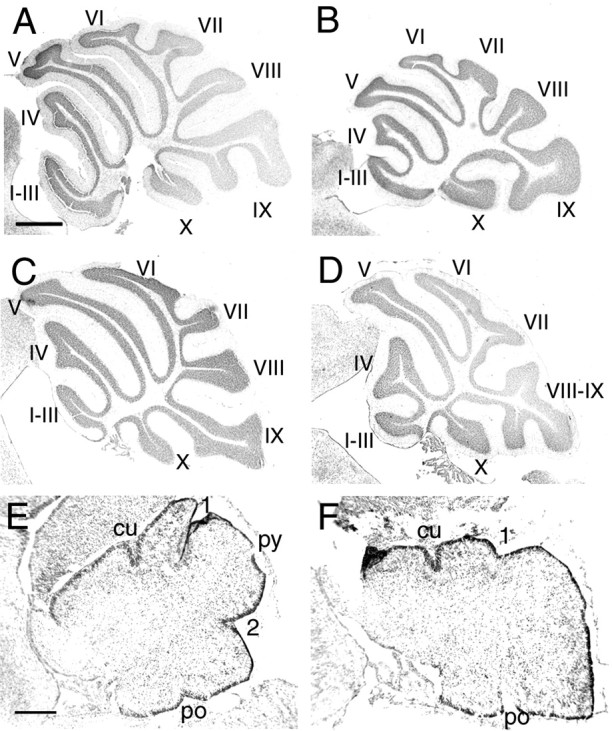
Strain background does not alter the En2Hd/Hd phenotype. Cresyl violet-stained sagittal paraffin sections of 129/Sv—En2Hd/Hd adults (B) revealed a posterior lobe defect in which the VIIIth and IXth vermal lobes are fused, unlike in the 129/Sv wild-type (A). Sagittal sections of C57BL/6J—En2Hd/Hd adults (D) display abnormal vermal lobe branching, similar to that of the 129/Sv—En2Hd/Hd mutants, but quite different from the branching seen in the C57BL/6J wild-type (C). Cresyl violet-stained sagittal cryostat sections of newborn C57BL/6J—En2Hd/Hd mice (F) reveal abnormal fissure formation in the posterior region of the cerebellum compared with wild-type C57BL/6J (E). The secondary (2) and prepyramidal (py) fissures are less obvious in the developing mutants, whereas the anterior fissures appear normal. cu, Preculminate; 1, primary; po, posterolateral. Scale bars: (in A) A—D, 500 μm; (in E) E, F, 200 μm.
To emphasize this point further, we showed that the developing C57BL/6J—En2Hd/Hd cerebellum exhibits the same temporal delay in foliation as that reported by Millen et al. (1994) in 129/Sv—En2Hd/Hd mice. When examined at birth, cresyl violet-stained cryostat sections of newborn C57BL/6J—En2Hd/Hd mutants reveal abnormalities in the cerebellar structure consistent with those seen in the original mutant (Fig. 5E,F). Compared with wild-type cerebella (Fig. 5E), few differences in the rate of formation of the anterior fissures (the preculminate and primary fissures) were observed in the mutants (Fig. 5F). However, in the posterior region of the cerebellum, the depths of the developing secondary and prepyramidal fissures were substantially reduced. In summary, the C57BL/6J—En2Hd/Hd animals displayed cerebellar abnormalities that are nearly identical to those seen in 129/Sv—En2Hd/Hd mice. Thus, the shift in genetic background has virtually no effect on the expression of this mutation, either during development or in the adult, and argues against the hypothesis that the background effect is achieved by a swap in the relative prominence of Engrailed-1 and Engrailed-2.
En1, En2, Pax2, and Wnt1 expression patterns are equivalent in wild-type 129/Sv and C57BL/6J mice
The data above illustrate that Engrailed-1 and Engrailed-2 have not switched in their developmental roles between the two inbred strains. However, it is still possible that the explanation for the mutant suppression seen on the C57BL/6J background is that the regulation of the network of genes that is necessary for establishing the cerebellar primordium is somehow different between C57BL/6J and 129/Sv. It has been shown that Engrailed-2 can functionally substitute for Engrailed-1 in the developing midbrain/hindbrain; thus, an expansion of either the temporal or spatial En2 domain in C57BL/6J mice could explain the rescue phenotype. Alternatively, differences in the expression patterns of either the En gene or other key genes important for establishing the cerebellar field could also be responsible for the CNS rescue. Therefore, we compared the expression patterns of En1, En2, Pax2, and Wnt1 in C57BL/6J and 129/Sv wild-type embryos using whole-mount in situ hybridization. We examined embryos ranging in age from E8 to E11. No difference in the expression patterns of the four genes was detected at any of the embryonic ages examined. The results from the E9.5 embryos are shown in Figure 6. These images make it clear that simple differences between the C57BL/6J and 129/Sv strains in the wild-type spatial patterns and/or the timing of expression (for these genes at least) are not responsible for the rescue phenotype. We paid particular attention to the timing of En2 appearance in the two strains and found that in both C57BL/6J and 129/Sv En2 the mRNA first appears at about the 5-somite stage (Fig. 7A—D).
Figure 6.
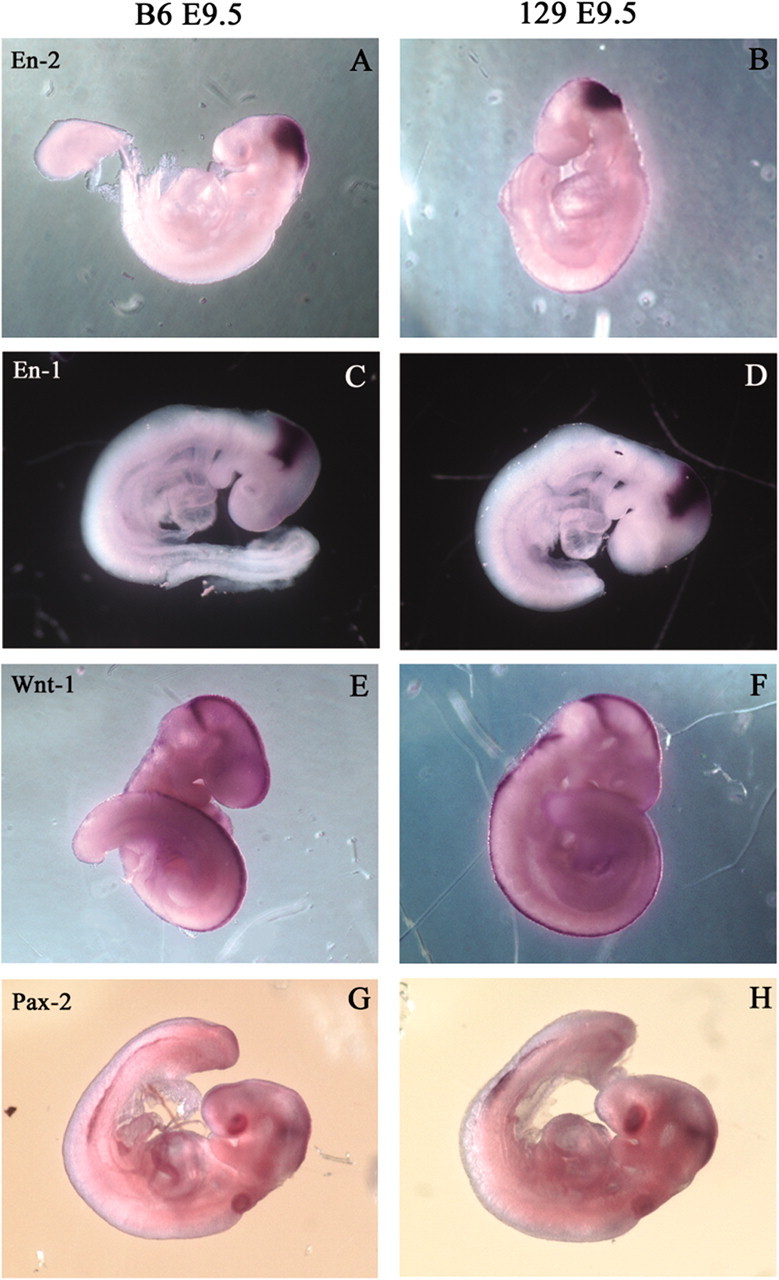
Expression patterns of early pattern formation genes are not affected by strain background. Whole-mount in situ hybridization of E9.5 embryos for En2 (A, B), En1 (C, D), Wnt1 (E, F), and Pax2 (G, H). Two different strains were used, C57BL/6J (A, C, E, G) and 129/Sv (B, D, F, H). No differences are seen between the two strains.
Figure 7.
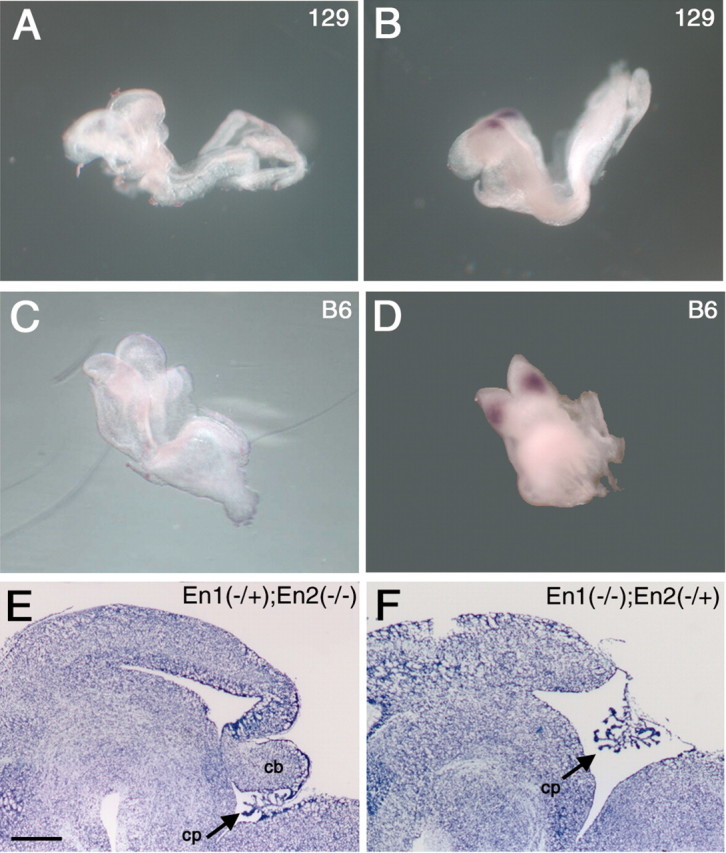
There is no difference in onset of En2 expression between 129/Sv (A, B) and C57BL/6J (C, D) wild-type mice. Whole-mount in situ hybridization of the 4- to 5-somite stage (A, C) shows no expression in either strain, whereas at the 6-somite stage (B, D), expression is first detected. Despite this, in the absence of En1, development of the cerebellum depends critically on En2 expression. Cresyl violet-stained sagittal cryostat sections of E17.5 (En1Hd/Hd; En2Hd/+) mutants (F) reveal a lack of cerebellum, whereas the(En1Hd/+; En2Hd/Hd) mutants (E) show no such loss. Both E and F are on a C57BL/6J background. Scale bar: (in E) E, F, 500 μm.
To further explore the role of Engrailed-2 in the rescue phenomenon, we crossed congenic C57BL/6—En1Hd/+ and C57BL/6—En2Hd/+ heterozygotes with each other. We then identified and intercrossed the resulting first filial generation progeny (F1) double heterozygotes and examined the F2 progeny at E17.5. Five litters were analyzed, but none of the resulting progeny were double mutants. Despite this, we have recovered En1 mutant embryos that were missing one copy of the wild-type En2 gene (En1Hd/Hd;En2Hd/+); all embryos of this genotype are missing their cerebellum (Fig. 7F). In contrast, En2 mutant embryos missing one copy of En1 (En1Hd/+;En2Hd/Hd) (Fig. 7E) were indistinguishable from single mutants (En1+/+;En2Hd/Hd). This demonstrates that in the absence of Engrailed-1, an adequate level of Engrailed-2 is crucial to establishing a normal cerebellar field.
Discussion
We have shown that shifting the mouse En1 mutation from the 129/Sv to the C57BL/6J genetic background has a profound effect on the observed phenotype. The En1Hd mutation, which leads to major malformations of the mesencephalic and metencephalic derivatives on 129/Sv, is transformed into a condition that is virtually free of cerebellar defects on the C57BL/6J background. That factors in the genetic background can affect the expression of a given mutation is not a novel finding. Indeed, our observations add to a growing literature concerning the importance of genetic background in the evaluation and interpretation of genetic mutations. In the mouse alone, the list of affected genes is diverse, ranging from those encoding p53 (Ikeda et al., 1999) and TGF-β (Bonyadi et al., 1997) to the p75NGF receptor gene (Greferath et al., 2000). Two specific examples of background effects that involve the strains used in our study are Fmr1 and Pax2. In studies of the mouse model for human fragile X mental retardation, in which the Fmr1 gene is disrupted, learning deficits arise on the 129/Sv background but not on the C57BL/6J background. A more dramatic difference is observed with Pax2. The Pax2 gene encodes a transcription factor in the same genetic network as the En genes and is known to be important for the specification and subsequent development of the midbrain/hindbrain region. On the 129/Sv background, 100% of the Pax-2-deficient mice display exencephaly in this region, whereas on a C57BL/6J background the frequency of this phenotype is reduced to 30% (Schwarz et al., 1997). As in our study, there is no effect on the expression patterns of other genes important for the establishment of the midbrain/hindbrain region in C57BL/6J animals.
The effect of genetic background on the En1 gene is unusual by virtue of its magnitude and the completeness of the rescue. Although shifting the genetic background of an engineered epidermal growth factor receptor mutation from 129 to CD1 produces major effects on phenotypic expression (Threadgill et al., 1995), the mildest configuration still leads to a CNS that is badly deformed. In contrast to this partial effect, En1-/- mice on a C57BL/6J background develop a virtually normal cerebellum and midbrain (Fig. 1), despite the fact that on a 129/Sv background only thin remnants of these structures remain. This rescue has a number of important implications. First, the data show that in the absence of Engrailed-1, the brain can develop the entire complement of cerebellar cell types. This implies that none of the cells in the cerebellum depends exclusively on En1 for their identity. Furthermore, the developmental interactions that regulate cell number are also completely restored. Purkinje cell counts reveal no hint of a deficit in the Engrailed-1-deficient animals. Coupled with the near-normal structure of the granule cell layer and inferior olive and the well documented developmental dependency of these two populations on the Purkinje cell (Herrup and Sunter, 1987; Vogel et al., 1989; Herrup et al., 1996) it is likely that all populations are present in normal numbers. Also noteworthy is the extent of the restoration of patterning in the region. With the exception of a curious malformation in the anterior lobes, the complex morphological details of the C57BL/6J cerebellum are rendered in nearly perfect detail despite the absence of the En1 gene product. The cerebellar cortex of each inbred mouse strain has a signature pattern of folding at the midline (Inouye and Oda, 1980). A sense of this can be gained by comparing the shapes and size of lobules VI and IX in the two wild-type animals illustrated in Figure 5, A and C. Therefore, it is noteworthy that from lobule V through lobule X in the C57BL/6J—En1-/- cerebellum, no significant difference appears in the shapes and relative sizes of these cortical folds. In other words, not only is a cerebellum restored on the C57BL/6J background, but its finest details are quintessentially C57BL/6J in character. The anomalies in the more anterior lobes, although minor, are a consistent finding that varies in severity. These observations hint that subtle (perhaps regionally variable) forces that require En1 function still lie beneath the surface, a suggestion that is supported by the minor reduction in caudal midbrain. One implication of the fact that the lobule distortion is similar in kind but different in severity and location from that observed in En2-/- mice may be that different anteroposterior domains of the cerebellum rely to different extents on the two Engrailed proteins.
Insight into the possible mechanisms mediating the Engrailed-1 rescue comes from our analysis of the gene expression patterns illustrated in Figure 6. The near identity of the patterns in wild-type mice from the two different strains suggests that the En1Hd rescue does not occur by virtue of a fundamental difference in the 129/Sv and C57BL/6J developmental programs. Rather, the explanation for the different En1Hd/Hd phenotypes in the two backgrounds must lie in the response of the genetic network to the mutation. This interpretation is further supported by the demonstration by Schwarz et al. (1997) of a partial CNS phenotypic rescue in Pax2 knock-out animals maintained on the C57BL/6J background. We suggest that there is a fundamental resiliency of the developmental program responsible for the establishment of the early midbrain/hindbrain region in the C57BL/6J strain context. Furthermore, the background differences that lead to this resiliency are only revealed in the absence of key proteins (in this case, Engrailed-1 or Pax-2) and their functions. This explanation is in keeping with the ease with which wild-type C57BL/6J and 129/Sv mice can be interbred and with the vitality of the hybrids.
Thus, we propose that the most plausible explanation for our results is that, in the C57BL/6J strain context, the normal functions of En1 can be subsumed by the actions of En2. In support of this hypothesis, the Joyner laboratory has shown that the two Engrailed proteins are functionally equivalent in the CNS (Hanks et al., 1995). The implication is that the genetic network responsible for establishing the field properties of the midbrain/hindbrain region contains enough plasticity to allow for a compensatory response in the timing and/or spatial pattern of En2 expression. Indeed, we have shown that C57BL/6J compound mutants (En1Hd/Hd;En2Hd/+) lack a cerebellum; directly demonstrating that Engrailed-2 substitution for Engrailed-1 must be the final common pathway for the rescue and that a minimum amount of functional protein must be present for normal development.
In all of the animals we have examined, no intermediate phenotype has been observed; this finding has implications for the mechanism involved in the rescue phenomenon. The apparent absence of any intermediate phenotypes means that this shift is not a gradually changing quantitative trait, but instead represents an all-or-none shift in the cross regulations of the genes in the network that specifies the midbrain/hindbrain pattern. Although this would suggest a single gene effect, preliminary genetic analyses suggest the involvement of multiple modifier loci because fewer than 5% of the En1Hd/Hd progeny of a C57BL/6J × 129/Sv—En1Hd/+ intercross demonstrate the rescue phenotype.
The lack of change in the phenotype of the En2Hd mutation after a similar genetic background shift should also be considered in this context. The developmental genetic expressions of En2 in C57BL/6J and 129/Sv are nearly identical; thus, the loss of En2 function has the same consequence in both. The finding that the En2 cerebellar phenotype is unchanged in both backgrounds whereas an En1 cerebellar phenotype is lost on C57BL/6J may result from the later times of Engrailed-2 protein action in the midbrain/hindbrain region. En1 expression decreases substantially after E12, and it persists in only a small number of regions, none of them in the cerebellum. The suggestion is that there is no workaround in the regulatory network that can compensate for the absence of this apparently vital component. This interpretation is also consistent with the finding that limb abnormalities associated with the En1Hd/Hd phenotype persist on the C57BL/6J background, because En2 is never expressed outside of the CNS.
The specific genotypic differences between C57BL/6J and 129/Sv that are responsible for altering the En1 mutant phenotype remain to be determined. Whether because of alterations in the identified players such as Pax2, Fgf8, Wnt1, etc., or because of additional genes heretofore uncharacterized, it is likely that the identification of these differences will lend additional insight into the regulatory pathways that direct the formation of the midbrain/hindbrain region.
Footnotes
This work was supported by National Institutes of Health Grant NS18381, by the National Alliance for Autism Research (K.H.), and by National Research Service Award NS43844 (C.L.M.). We thank Dr. Alexandra Joyner for providing the original Engrailed-1 mutation.
Correspondence should be addressed to Dr. Karl Herrup, Alzheimer Research Laboratory, Case Western Reserve University, E 504 School of Medicine, 10900 Euclid Avenue, Cleveland, OH 44106. E-mail: kxh26@po.cwru.edu.
S. Maricich's present address: Department of Child Neurology, Texas Children's Hospital, Houston, TX 77030-2399.
Copyright © 2003 Society for Neuroscience 0270-6474/03/235105-08$15.00/0
N.A.B. and R.R.R.-D. contributed equally to this work.
References
- Bonyadi M, Rusholme SA, Cousins FM, Su HC, Biron CA, Farrall M, Akhurst RJ ( 1997) Mapping of a major genetic modifier of embryonic lethality in TGF β1 knockout mice. Nat Genet 15: 207–211. [DOI] [PubMed] [Google Scholar]
- Davis C, Joyner A ( 1988) Expression patterns of the homeobox-containing genes En-1 and En-2 and the proto-oncogene int-1 diverge during mouse development. Genes Dev 2: 1736–1744. [DOI] [PubMed] [Google Scholar]
- Davis C, Noble-Topham S, Rossant J, Joyner A ( 1988) Expression of the homeobox-containing gene En-2 delineates a specific region of the developing mouse brain. Genes Dev 2: 361–371. [DOI] [PubMed] [Google Scholar]
- Gerlai R, Millen KJ, Herrup K, Fabien K, Joyner AL, Roder J ( 1996) Impaired motor learning performance in cerebellar En-2 mutant mice. Behav Neurosci 110: 126–133. [DOI] [PubMed] [Google Scholar]
- Greferath U, Bennie A, Kourakis A, Bartlett PF, Murphy M, Barrett GL ( 2000) Enlarged cholinergic forebrain neurons and improved spatial learning in p75 knockout mice. Eur J Neurosci 12: 885–893. [DOI] [PubMed] [Google Scholar]
- Hanks M, Wurst W, Anson-Cartwright L, Auerbach A, Joyner A ( 1995) Rescue of the En-1 mutant phenotype by replacement of En-1 with. En-2 Science 269: 679–682. [DOI] [PubMed] [Google Scholar]
- Hanks MC, Loomis CA, Harris E, Tong CX, Anson-Cartwright L, Auerbach A, Joyner A ( 1998) Drosophila engrailed can substitute for mouse Engrailed1 function in mid-hindbrain, but not limb development. Development 125: 4521–4530. [DOI] [PubMed] [Google Scholar]
- Herrup K, Sunter K ( 1987) Numerical matching during cerebellar development: quantitative analysis of granule cell death in staggerer mouse chimeras. J Neurosci 7: 829–836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrup K, Shojaeian-Zanjani H, Panzini L, Sunter K, Mariani J ( 1996) The numerical matching of source and target populations in the CNS: the inferior olive to Purkinje cell projection. Brain Res Dev Brain Res 96: 28–35. [DOI] [PubMed] [Google Scholar]
- Ikeda S, Hawes NL, Chang B, Avery CS, Smith RS, Nishina PM ( 1999) Severe ocular abnormalities in C57BL/6 but not in 129/Sv p53-deficient mice. Invest Ophthalmol Vis Sci 40: 1874–1878. [PubMed] [Google Scholar]
- Inouye N, Oda S ( 1980) Strain-specific variations in the folial pattern of the mouse cerebellum. J Comp Neurol 190: 357–362. [DOI] [PubMed] [Google Scholar]
- Joyner A, Auerbach B, Davis C, Herrup K, Rossant J ( 1991) Subtle cerebellar phenotype in mice homozygous for a targeted deletion of the En-2 homeobox. Science 259: 1239–1243. [DOI] [PubMed] [Google Scholar]
- Joyner AL, Skarnes WC, Rossant J ( 1989) Production of a mutation in mouse En-2 gene by homologous recombination in embryonic stem cells. Nature 338: 153–156. [DOI] [PubMed] [Google Scholar]
- Kuemerle B, Zanjani H, Joyner A, Herrup K ( 1997) Pattern deformities and cell loss in Engrailed-2 mutant mice suggest two separate patterning events during cerebellar development. J Neurosci 17: 7881–7889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu A, Joyner AL ( 2001) Early anterior/posterior patterning of the midbrain and cerebellum. Annu Rev Neurosci 24: 869–896. [DOI] [PubMed] [Google Scholar]
- Loomis CA, Harris E, Michaud J, Wurst W, Hanks M, Joyner AL ( 1996) The mouse Engrailed-1 gene and ventral limb patterning. Nature 382: 360–363. [DOI] [PubMed] [Google Scholar]
- McMahon A, Joyner A, Bradley A, McMahon J ( 1992) The midbrainhindbrain phenotype of Wnt-1-/Wnt-1- mice results from stepwise deletion of engrailed-expressing cells by 9.5 days post coitum Cell 69: 581–595. [DOI] [PubMed] [Google Scholar]
- Millen KJ, Wurst W, Herrup K, Joyner AL ( 1994) Abnormal embryonic cerebellar development and patterning of postnatal foliation in two mouse Engrailed-2 mutants. Development 120: 695–706. [DOI] [PubMed] [Google Scholar]
- Schwarz M, Alvarez-Bolado G, Urbanek P, Busslinger M, Gruss P ( 1997) Conserved biological function between Pax-2 and Pax-5 in midbrain and cerebellum development: evidence from targeted mutations. Proc Natl Acad Sci USA 94: 14518–14523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Threadgill DW, Dlugosz AA, Hansen LA, Tennenbaum T, Lichti U, Yee D, LaMantia C, Mourton T, Herrup K, Harris RC, Barnard JA, Yuspa SH, Coffey RJ, Magnuson T ( 1995) Targeted disruption of mouse EGF receptor: effect of genetic background on mutant phenotype. Science 269: 230–234. [DOI] [PubMed] [Google Scholar]
- Vogel MW, Sunter K, Herrup K ( 1989) Numerical matching between granule and Purkinje cells in lurcher chimeric mice: a hypothesis for the trophic rescue of granule cells from target-related cell death. J Neurosci 9: 3454–3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurst W, Auerbach AB, Joyner AL ( 1994) Multiple developmental defects in Engrailed-1 mutant mice: an early mid-hindbrain deletion and patterning defects in forelimbs and sternum. Development 120: 2065–2075. [DOI] [PubMed] [Google Scholar]