Abstract
Neurons require Ca2+-dependent gene transcription for their activity-dependent survival, the mechanisms of which have not been fully elucidated yet. Here, we demonstrate that a novel primary response gene, alivin 1 (ali1), is an activity-dependent gene and promotes survival of neurons. Sequence analyses reveal that rat, mouse, and human Ali1 proteins contain seven leucine-rich repeats, one IgC2-like loop and a transmembrane domain, and display homology to Kek and Trk families. Expression of ali1 mRNA in cultured cerebellar granule neurons is rigidly regulated by KCl and/or NMDA concentrations in the culture medium and tightly correlated to depolarization-dependent survival and/or NMDA-dependent survival of the granule neuron. ali1 mRNA expression was regulated at the transcriptional step by the Ca2+ influx through voltage-dependent L-type Ca2+ channels when the cells were stimulated by 25 mm KCl. Expression of ali1 mRNA in cultured cortical neurons was inhibited when their spontaneous electrical activity was blocked by tetrodotoxin. Thus, the expression is neuronal activity dependent. Overexpression of Ali1 in cerebellar granule neurons inhibited apoptosis that was induced by the medium containing 5 mm KCl. The addition of anti-Ali1 antiserum or the soluble putative extracellular Ali1 domain to the 25 mm KCl-supported culture inhibited the survival of the granule neuron. These results suggest that expression of ali1 promotes depolarization-dependent survival of the granule neuron. Mouse ali1 was mapped to a locus ∼55.3 cM from the centromere on chromosome 15 that is syntenic to positional candidate loci for familial Alzheimer's disease type 5 and Parkinson's disease 8 on human chromosome 12.
Keywords: alivin, leucine-rich repeat, Ig superfamily, apoptosis, survival, activity-dependent, NMDA, depolarization, Alzheimer's disease, Parkinson's disease
Introduction
Neuronal activity regulates a wide variety of functions, including memory, synaptic transmission, neurite outgrowth, differentiation, synaptogenesis, cell death, and survival. During brain development, neuronal activity suppresses apoptosis and promotes survival of neurons, thus adjusting cell numbers and their connection patterns (Oppenheim, 1991). To date, a number of in vivo and in vitro studies report that neurons suppress apoptosis and promote cell survival in an activity-dependent manner (Gallo et al., 1987; Ono et al., 1997a; Ikonomidou et al., 1999; Monti and Contestabile, 2000). Stimulation of neurons by high concentrations of KCl or glutamate receptor agonists leads to an influx of Ca2+ through voltage-dependent Ca2+ channels or glutamate receptor channels, respectively, and thus enhances Ca2+ turnover (Ono et al., 1997a; Kohara et al., 1998). This Ca2+ signal activates Ca2+/calmodulin-dependent protein kinases (See et al., 2001), protein kinase B/Akt (Yano et al., 1998), and RAS/MEK/MAPK cascades that suppress apoptosis and promote neuronal survival through both transcription-dependent and -independent pathways (Bonni et al., 1999). cAMP response element-binding protein (CREB) and/or MEF2C-mediated gene transcription processes are indispensable for the activity-dependent survival of neurons (Bonni et al., 1999; Mao et al., 1999; Walton and Dragunow, 2000). However, their downstream targets that mediate activity-dependent survival are yet to be fully elucidated. Several secretory proteins, including BDNF, PACAP, and parathyroid hormone-related protein, that are expressed in an activity-dependent and/or Ca2+-dependent manner promote cell survival or possess neuroprotective effects (Ghosh et al., 1994; Ono et al., 1997b; Tabuchi et al., 2001). These proteins are secreted from neurons and act in an autocrine and/or paracrine manner to promote survival. For instance, a number of studies show that BDNF is expressed through CREB and/or calcium-responsive transcription factor-dependent transcription(s) (Shieh et al., 1998; Tao et al., 1998, 2002) and facilitates the survival of neurons (Ghosh et al., 1994). However, BDNF alone is not enough to fully support neuronal existence (Ichikawa et al., 1998), and therefore, multiple signal factors may be involved in activity-dependent survival (Franke et al., 2000). To comprehensively identify the factors involved in this process, we used mRNA differential display to screen genes expressed in cerebellar granule neurons cultured specifically under survival-promoting (i.e., depolarizing or NMDA supported) conditions. This useful technique has revealed a number of candidate genes that could be involved in activity-dependent neuronal survival (T. Ono, unpublished data). Consequently, we successfully identified both known and novel factors by selecting genes displaying good correlation between expression patterns and depolarization-dependent and/or NMDA-dependent survival (Ono et al., 1997b, present study).
Here, we show a novel gene that plays a crucial role(s) in the depolarization-dependent survival of cerebellar granule neurons, designated “alivin 1” (ali1) after “alive” and “activity-dependent leucine-rich-repeat and Ig superfamily survival-related protein.” The gene encodes a putative transmembrane protein that is a member of a novel class of leucine-rich repeat (LRR) and Ig superfamily proteins. The putative extracellular domain of Ali1 displays striking similarity to the human Trk and Drosophila Kek family, suggesting that the protein plays a critical role(s) in signal transduction of neuronal survival.
Materials and Methods
Primary culture of cerebellar granule neurons and cell survival assays. Cerebellar granule neurons were cultured according to the method of Levi et al. (1989) with some modifications, as described previously (Ono et al., 1997b). In some cases, 30 μm (±)-2-amino-5-phosphonopentanoic acid (AP-5) were added to the culture medium simultaneously with KCl for prevention of potential effects of glutamate, which might have been present in the FCS on the survival of granule neurons. Cell survival was assayed as described by Mosmann (1983), similar to earlier reports (Ono et al., 1997b).
Primary culture of cortical neurons. Cortical neurons were cultured by the method of Ogura et al. (1987) with some modifications. Briefly, cerebra were dissected from 20-d-old embryos of the Wister rat. Cortical neurons were prepared by using dissociation solution (MB-X9901; Sumitomo Bakelite Co. Ltd.). The dissociated cells were plated at a density of 5 × 10 6 cells per 3.5-cm dish containing 2 ml of DMEM supplemented with 5% of FCS and 5% of horse serum.
RNA isolation and Northern blotting. Total cellular RNA was isolated by the method of Chomczynski and Sacchi (1987) with some modifications. Cells were lysed with a 1:1 (v/v) mixture of solution A and water-saturated phenol. RNA was separated on a 1% formaldehyde agarose gel and transferred to Biodyne B membrane (Pall). A 0.24–9.5 kb RNA ladder (Invitrogen, Carlsbad, CA) was used as a size marker. cDNA probes were labeled with random 9-mers (Takara) and [α-32P] dCTP. Hybridization was performed at 65°C in rapid hybridization buffer (Amersham), following the manufacturer's instructions. After hybridization, the membrane was washed with 2× SSC containing 0.1% SDS at room temperature, followed by washing with 0.1× SSC containing 0.1% SDS at 55°C. Membranes were reprobed with mouse glucose-6-phosphate dehydrogenase cDNA (G6PDH) to confirm equivalent loading of RNA samples (Inokuchi et al., 1996). Levels of mRNA were normalized to that of G6PDH mRNA by hybridization signal analyses using a FUJIX BAS2000 Bioimage analyzer (Fuji Photo film).
Differential display and cDNA cloning. Differential display was performed as described previously (Ono et al., 1997b). The primer pair used included 5′-ACACGCTCAC-3′ and 5′-T12VA-3′ (the anchor primer, whereby V is a mixture of A, G, and C). The resulting PCR product was cloned into the pCR-Script Amp SK(+) vector (Stratagene, La Jolla, CA). Positive clones were selected using Northern blotting analyses and sequenced with the Thermo Sequenase fluorescent-labeled primer cycle kit (Amersham).
Molecular cloning of mouse and human ali1 cDNA. Mouse ali1 cDNA was cloned by PCR, using LA-TaqDNA polymerase (Takara) and mouse brain cDNA as a template. The cycling conditions were: 94°C for 1 min, 35 cycles of 94°C for 30 sec, 66°C for 30 sec, and 68°C for 3 min, followed by extension at 68°C for 3 min. The primers used were: 5′-GCTAAGTATTTCCGTTCAATTGTTT-3′ (sense) and 5′-GGTATTCAGGACAGCGAGGACGGAG-3′ (antisense). Human ali1 cDNA was obtained with two subsets of PCRs, using human brain cDNA as a template. The primers used were 5′-GCTGGTCTTTTGGTATCGTAGGC-3′ (sense), 5′-TCCCCCTCGTGGACTTTAGG-3′ (antisense), 5′-GAGGGCATCCTAAAGTCCACG-3′ (sense), and 5′-GTGTAGTCTATTCATTCTGGTCTAAAACGG-3′ (antisense).
Competitive reverse transcriptase-PCR for rat ali1 mRNA expression. Total RNA (1 μg) was treated with DNase I (Invitrogen) and converted into cDNA with Superscript II (Invitrogen), using the manufacturer's instructions. The competitor was constructed using a competitive DNA construction kit (Takara). The sequences used for competitive PCR analysis of rat ali1 included sense primer (1992–2014), 5′-AAACGCACTGAACCAAAGCAACG-3′; antisense primer (2266–2291), 5′-AATCACGACACCAGACGAACACAGG-3′; competitive sense primer, 5′-AAACGCACTGAACCAAAGCAACGTACGGTCATCATCTGACAC-3′; and competitive antisense primer, 5′-AATCACGACACCAGACGAACACAGGCGGTGAGTATCTGCATATGAT-3′. The sense and antisense primer pair generated a 300-bp product, whereas PCR using the competitive primer pair led to the amplification of a 199-bp fragment. The competitive PCR was performed with 10 4 copies of the competitor and 50 ng of the cDNA.
Bacterial expression of rat Ali1. The coding sequence of rat ali1 cDNA was amplified by PCR with Pfu polymerase (Stratagene). Appropriate restriction sites were incorporated in both primers, and the His-tag sequence was included in the antisense primer. The primers used for raising the specific antibody to rat Ali1 (57–520) were 5′-GGAATTCCATATGAACCTGTCTAAGGTGCCTGGG-3′ and 5′-CCGCTCGAGAGTGGATGCCACGAAGGGG-3′. Primers used to amplify the putative extracellular domain [Ali1 (57–402)] were 5′-GGAATTCCATATGAACCTGTCTAAGGTGCCTGGG-3′ and 5′-CCGCTCGAGGGTGGTGAAAGC-3′. The resulting PCR products were cloned in-frame into pET22b vector. Bacterially expressed proteins were induced by 1 mm isopropyl-β-d-thiogalactoside for 3 hr and purified by Ni 2+-NTA column (Qiagen, Hilden, Germany) chromatography. The bacterially expressed Ali1 (57–520) was further purified by SDS gel electrophoresis and used for immunizing rabbits to raise the anti-Ali1 antibody.
Subcellular fractionation of the rat brain. Brains were dissected from male rats, 10 weeks of age. Subcellular components were fractionated according to the method described by Gordon-Weeks (1987). Plasma membrane-rich fraction and synaptosomal and mitochondrial fractions were obtained by Ficoll density gradient centrifugation, using a procedure described by the same authors. A marker enzyme for plasma membrane, 5′-nucleotidase, was assayed as described by Graham et al. (1968).
Western blotting analysis. Cell extract was applied on 10% SDS gel electrophoresis. After proteins were transferred onto polyvinylidene difluoride membranes, Ali1 immunoreactivity was detected with 3000-fold diluted anti-Ali1 antiserum, either with ECL or ECL-plus reagents (Amersham).
Overexpression of ali1 in cerebellar granule neurons. A coding sequence of Flag epitope was fused to the coding sequence of rat ali1 cDNA in its 3′ end by PCR. The amplified cDNA was cloned into pTriEx-1.1 vector (Novagen, Madison, WI). Transfection was performed by using Lipofectamine 2000 reagent (Invitrogen) at the same time when the cells were plated at 0 d in vitro (DIV). Either the ali1 expression vector or the control vector was mixed with enhanced green fluorescent protein (EGFP) expression vector pEGFP-N3 (Clontech) in a ratio of 4:1, and then the mixture was diluted in basal medium Eagle (BME). An equal amount of Lipofectamine 2000 reagent (Invitrogen) was mixed with BME and incubated for 5 min at room temperature. The diluted Lipofectamine 2000 solution was mixed with the diluted DNA and incubated for 20 min at room temperature. The DNA-Lipofectamine 2000 complex was mixed with the dissociated cell suspension in BME supplemented with 10% FCS, in a ratio of 1:4. Aliquots of 0.5 ml, each containing 2.5 × 10 6 cells, were dispensed onto polyethyleneimine-coated glass coverslips in 12-well plates. Each culture well received ∼1.25 μg of plasmid DNAs. The medium was replaced with the fresh one after 5 hr. Twenty four hours after plating the cells, AraC and AP-5 were added to the cultures as described previously (Ono et al., 1997b). At 3.0 DIV, neurons were fixed with 4% formaldehyde and stained with anti-Flag antibody M2 (Sigma, St. Louis, MO). Apoptotic nuclei were visualized by staining with Hoechst 33342 (See et al., 2001).
Radiation hybrid mapping. The chromosomal location of mouse ali1 was determined by PCR analyses that were typed in duplicate on 94 DNA samples (omitting typing cell lines 12, 25, 61,62, 99, and 100) from the T31 radiation hybrid (RH) panel (Research Genetics/Invitrogen) (McCarthy et al., 1997). The PCR was performed in a total volume of 15 μl comprising 20 ng of genomic DNA, 0.66 μm of each primer, 0.15 U of AmpliTaq Gold (Applied Biosystems, Foster City, CA), 200 μm dNTP, 1.5 mm MgCl2, and 1× Gold buffer. The following cycle conditions were used: 95°C for 5 min (enzyme activation), 94°C for 30 sec, 55°C for 40 sec, and 72°C for 1 min for a total 40 cycles. PCR products were electrophoresed on a 4% agarose gel (3% NuSieve agarose and 1% SeaKem ME agarose) in Tris–acetate–EDTA electrophoresis buffer and visualized by staining with ethidium bromide. Data were recorded from gel images, using a specific scoring system (0 = negative, 1 = positive, and 2 = line not tested or not reproducible). The obtained data were used to establish a RH framework map. After mapping of the locus, the flanking markers were retyped using a cell line from the RH panel. Distances in cR3000 were determined by analysis of RH mapping data in Map Manager QT (Manly and Olson, 1999).
Immunohistochemistry for Ali1. Frozen sections (15 μm thick) of adult rat brain were fixed with 100% methanol at -20°C for 10 min. Sections were blocked with Dulbecco's PBS containing 5% BSA and 5% goat serum. Then, the sections were reacted with rabbit anti-Ali1 antiserum, followed by anti-rabbit IgG conjugated to FITC. Nonimmune serum was used for staining negative control preparations.
Results
Identification of ali1 as a depolarization- and/or NMDA-induced gene
Cerebellar granule neurons survived extremely well when cultured in medium containing either 25 mm KCl or both 150 μm NMDA and 15 mm KCl (Fig. 1). Conversely, these neurons underwent apoptosis (Ono et al., 1997a) and gradually degenerated on culturing in medium containing 5 or 15 mm KCl, the latter of which had a better survival rate than the former. The neuron was also degenerated, and approximately half of the cells died in 36 hr when the cells were cultured in medium containing both 150 μm NMDA and 15 mm KCl for 3.5 DIV, and then 300 μm AP-5 were added to the culture to block NMDA receptor. This cell death was also apoptosis (data not shown). To identify the genes involved in depolarization-dependent survival and/or NMDA-dependent survival of neurons, we initially determined the time required for granule neurons to be committed to apoptosis as 3 DIV (Ono et al., 1997b). Accordingly, we extracted RNA at 3 DIV from sets of cells cultured under both survival-promoting conditions (medium containing 150 μm NMDA and 15 mm KCl) and apoptosis-inducing conditions (medium containing 5 mm KCl and 30 μm AP-5). These RNA samples were subjected to differential display analysis. A pair of bands that had ∼150 bases in size were detected in lanes loaded with PCR products derived from cells cultured under survival-promoting conditions, but not in those loaded with PCR products of cells cultured under apoptosis-inducing conditions (data not shown). These bands were excised from the gel and reamplified, as described previously (Ono et al., 1997b). PCR products were cloned into pCR-Script Amp SK(+) vector (Strategene) and sequenced. The cDNA obtained was identified as a nucleotide fragment (2957–3095 bp) of rat ali1 cDNA (GenBank accession number AB078879), which is the most downstream region of the 3′-untranstrated sequence.
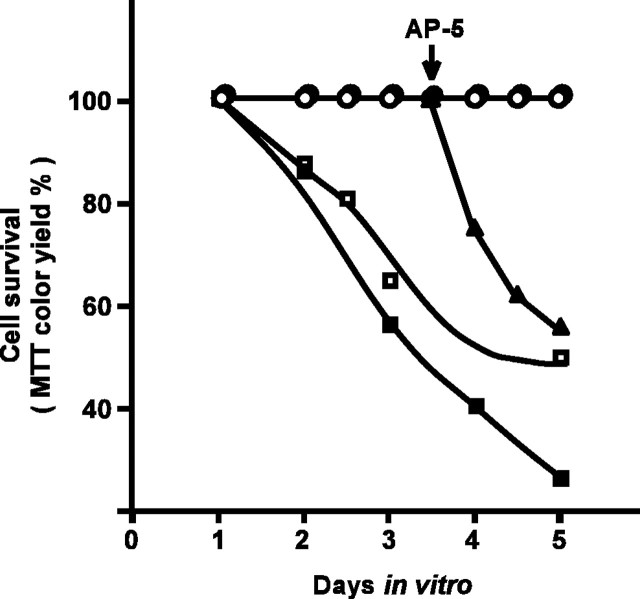
Figure 1.
Time course of depolarization-dependent survival and/or NMDA-dependent survival of cerebellar granule neurons. Cerebellar granule neurons were cultured in medium containing 5 mm KCl and 30 μm AP-5 (▪), 15 mm KCl and 30 μm AP-5 (□), 25 mm KCl and 30 μm AP-5 (○), or 150μm NMDA and 15 mm KCl(•). At 3.5 DIV, 300μm AP-5 were added to some of the NMDA-supported cultures and apoptosis was induced (▴). Relative cell survival rate (MTT color yield) of each culture was plotted against the survival rate in the culture medium containing 25 mm KCl and 30 μm AP-5.
Molecular cloning and sequence analysis of ali1 cDNA
The cloned cDNA fragment was further used to screen a λZap II library constructed from RNA extracted from rat cerebellar granule neurons cultured for 5.0 DIV in medium containing 15 mm KCl and 150 μm NMDA (Ono et al., 1997b). Consequently, several overlapping clones were obtained. Rescreening the same library using the most upstream regions of the sequences obtained yielded several clones that had 5′ extended sequences of ∼500 bp. Full-length rat ali1 cDNA comprises 3095 bp (GenBank accession number AB078879). GenBank searches revealed significant homology of multiple mouse and human expressed sequence tag (EST) clones to the 3′-region of rat ali1 cDNA. Moreover, the entire rat ali1 cDNA sequence displayed considerable identity (72% identity without gaps of >16 consecutive bases) to part of a bacterial artificial chromosome clone harboring a human chromosomal DNA (GenBank accession number AC004010), suggesting that full-length mRNA sequence of the human homolog of rat ali1 is encoded in a single exon. We compared the sequence of rat ali1 cDNA and the genomic sequence of human ali1 for cloning purposes. A 5′-primer was designed, based on the most upstream region of the consensus of the two sequences (see Materials and Methods). Specific 3′-primers were designed from the 3′-untranslated sequences of mouse and human EST clones. Full-length mouse ali1 cDNA (GenBank accession number AB078880) was obtained by PCR, using the 5′-primer and the 3′-primer specific for mouse EST clones. Initial attempts to clone human ali1 cDNA using the same 5′-primer and a 3′-primer specific for human EST clones were unsuccessful. Consequently, several other 5′-primers were designed, based on the genomic sequence of human ali1 (as described in Materials and Methods), and partial human ali1 cDNA was successfully obtained (GenBank accession number AB079074). A human EST clone (GenBank accession number BG724413) that overlapped this partial human ali1 cDNA was identified. These two sequences were ligated to construct full-length human ali1 cDNA (accession number AB080610). The EST clone included amino acids 1–239, whereas human ali1 cDNA obtained by PCR encompassed amino acids 240–522.
The ORFs of rat, mouse, and human ali1 cDNA encode proteins with 520, 519, and 522 aa, respectively. As shown in Figure 2, each protein has a putative signal peptide sequence at the NH2 terminus, seven LRRs flanked by two cysteine-rich domains, one IgC2-like domain, and a putative transmembrane domain. Signal peptide cleavage sites were predicted between residues 38 and 39 for Ali1 of all species, using the method of Nielson et al. (1997). Notably, the second half of the fifth LRR (amino acids 169–190 for rat and mouse; 170–191 for human) is 100% compatible with the consensus leucine zipper sequence (LxxxxxxLxxxxxxLxxxxxxL). Each protein comprises a putative intracellular domain with ∼100 aa at the COOH-terminal region, which does not display homology to other proteins with known functions. A search of the databases revealed other members of the ali family, specifically ali2 and ali3. The structural characteristics of the ali family are described in Discussion.
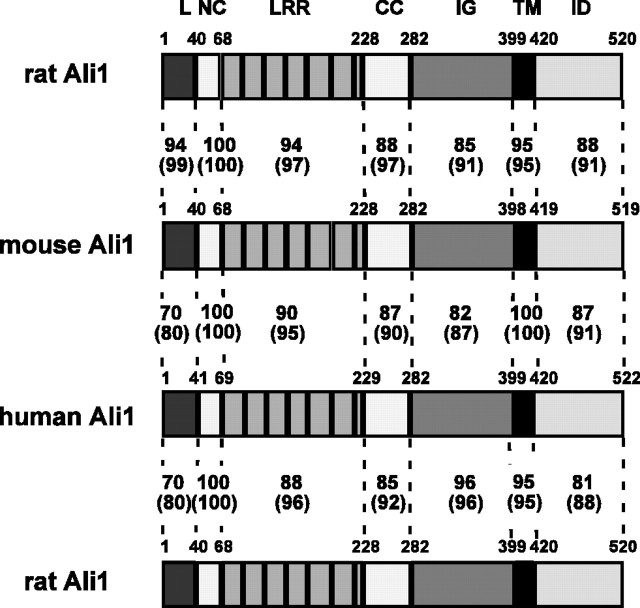
Figure2.
Structure of Alivin 1 proteins. The domain organizations of the deduced amino acid sequence of Ali1 proteins are shown schematically. The numbers on the Ali1 structures indicate positions of amino acid residues. The numbers between Ali1 structures indicate homology: numbers outside the parentheses indicate percentage identity, whereas numbers inside represent percentage similarity. L, Leader sequence including the signal peptide sequence; NC, NH2-terminal flanking cysteine-rich domain; CC, COOH-terminal flanking cysteine-rich domain; IG, IgC2-like loop; TM, transmembrane domain; ID, intracellular domain.
Expression of ali1 mRNA is tightly associated with depolarization-dependent survival and/or NMDA-dependent survival of cerebellar granule neurons
To clarify the relationship between granule neuronal survival and expression of ali1 mRNA, a time course of the expression pattern was assayed by Northern blotting. As shown in Figure 3A, ali1 mRNA was 3.3 kb in size, and an additional minor transcript of 8.5 kb was detected. The expression of ali1 mRNA increased rapidly during the culture period and was confined to cells cultured under survival-promoting conditions (i.e., medium containing 25 mm KCl, 30 μm AP-5 or 150 μm NMDA, and 15 mm KCl). In contrast, ali1 mRNA was barely detected under apoptosis-inducing conditions (i.e., medium containing 5 mm KCl, 30 μm AP-5 or 15 mm KCl, and 30 μm AP-5) (Fig. 3A,B). When AP-5, a specific NMDA receptor antagonist, was added at a final concentration of 300 μm to the NMDA-supported culture, ali1 mRNA expression was rapidly downregulated (t1/2 = 1.4 hr) (Fig. 3C, D). Downregulation of ali1 mRNA was additionally observed when the constituent of the culture medium was changed from 25 mm KCl, 30 μm AP-5, to 5 mm KCl (data not shown). After downregulation, neurons were restimulated with either 25 mm KCl, 30 μm AP-5 or 150 μm NMDA, or 15 mm KCl (Ono et al., 1997b; Ichikawa et al., 1998). ali1 mRNA was upregulated, but recovery to original levels took between 5 and 24 hr (data not shown). To investigate the mechanism of upregulation of ali1 mRNA, various inhibitors were added to the culture medium before stimulation by KCl and/or NMDA, and expression of the transcript was assayed at 24 hr after stimulation (Fig. 3E,F). Expression of ali1 mRNA was completely inhibited by actinomycin D and partially inhibited by cycloheximide (43% of the control level for 25 mm KCl stimulation and 49% of the control level for NMDA stimulation, respectively). Moreover, upregulation of ali1 mRNA was inhibited by nifedipine, a specific antagonist for voltage-dependent L-type Ca2+ channels. Our results collectively suggest that ali1 is a primary response gene, the expression of which is regulated at the transcriptional step by Ca2+ influx through voltage-dependent L-type Ca2+ channels.
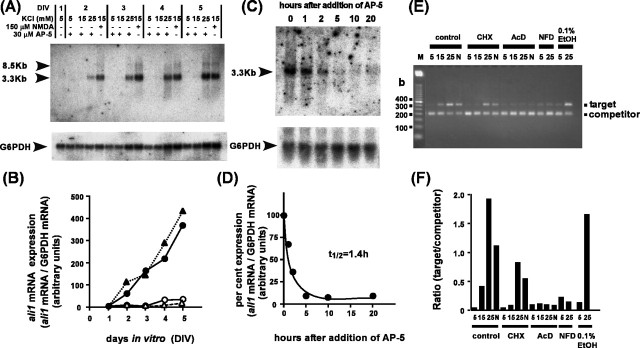
Figure3.
Expression of ali1 mRNA is tightly associated with depolarization-dependent survival and/or NMDA-dependent survival of cerebellar granule neurons.A,Northern blotting of ali1 mRNA expression in granule neurons. Total RNA samples (10μg) extracted from granule neurons cultured in the presence of 5 mm KCl, 30μm AP-5; 15 mm KCl, 30μm AP-5; 25 mm KCl, 30μm AP-5; or 150 μm NMDA, 15 mm KCl were subjected to Northern blotting. The cDNA obtained by differential display analysis (which corresponds to the 3′-untranslated region of rat ali1 cDNA) was used as a probe. The bottom panel depicts expression levels of G6PDH mRNA. B, Quantification of data shown in A. Granule neurons were cultured in the presence of 5 mm KCl,30μm AP-5(▵); 15 mm KCl,30μm AP-5 (○); 25 mm KCl, 30μm AP-5 (▴); or 15 mm KCl, 150μm NMDA (•). C, Rapid downregulation of ali1 mRNA expression by blocking the NMDA receptor with AP-5. Granule neurons were cultured for 4.5 d in medium containing 15 mm KCland150μm NMDA, and the NMDA receptor was blocked by the addition of AP-5 to a final concentration of 300μm. Total RNA (10μg) extracted at the indicated times was subjected to Northern blotting analyses. The bottom panel displays G6PDH mRNA expression levels. D, The data shown in C were quantified. E, Effects of various inhibitors on ali1 mRNA expression. Cerebellar granule neurons were cultured for 4 d in medium containing 25 mm KCl, as described in Materials and Methods. The medium was replaced with one containing 5 mm KCl, and the culture was continued for another 24 hr, after which cells were stimulated with 5 mm KCl, 30μm AP-5 (5); 15 mm KCl, 30μm AP-5 (15); 25 mm KCl, 30μm AP-5 (25); or 15 mm KCl, 150μm NMDA (N). Inhibitors were added to the culture medium 30 min before stimulation with KCl and/or NMDA. After stimulation (24 hr), total RNA was extracted and subjected to competitive PCR analysis. The inhibitors used included 35 μm cycloheximide (CHX), 4.0 μm actinomycin D (AcD), and 0.2 μm nifedipine (NFD). Ethanol was used as vehicle for nifedipine. F, The data shown in E were quantified.
Neuronal activity-dependent expression of ali1 mRNA
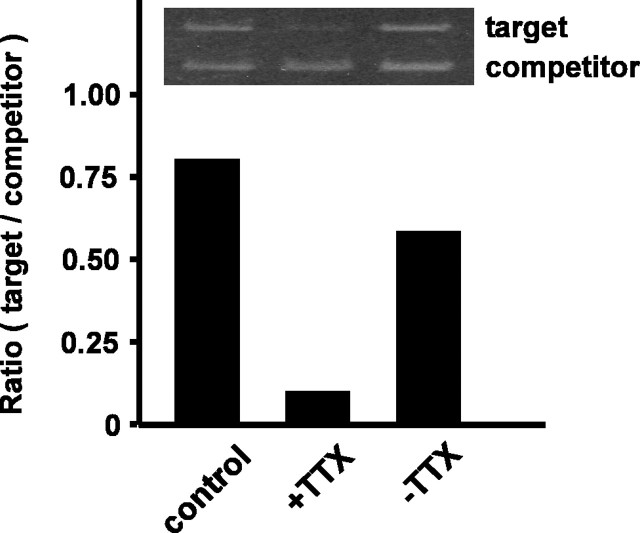
Although most studies that have examined activity-dependent genes have used chronic depolarization induced by high concentration of KCl, Brosenitsch and Katz (2001) reported that chronic depolarization regulates genes in a different way(s) from physiological pattern of electrical impulses. This prompted us to examine whether ali1 mRNA expression was regulated by synaptic excitation. Ogura et al. (1987) and Kuroda et al. (1992) reported that primary cultured hippocampal and cortical neurons formed functional synapses and spontaneously developed periodical synaptic excitation that was synchronized among the cells. They showed that the addition of TTX to the culture medium completely inhibited the spontaneous excitation of neurons and removal of TTX from the culture restored the excitation of the cells. We used primary culture of cortical neurons to see whether the expression of ali1 mRNA depended on electrical activity of neurons. Cortical neurons were prepared from rat embryos and cultured for 2 weeks, by which time the cells matured and formed synapses (Kuroda et al., 1992). These cells expressed ali1 mRNA (Fig. 4). The expression of ali1 mRNA was significantly reduced when Na+ channels were blocked by TTX. Removal of TTX from the culture restored ali1 mRNA to comparable with its original level. These results suggest that the expression of ali1 mRNA depends on the electrical activity of neurons and, hence, ali1 mRNA expression is neuronal activity dependent.
Figure 4.
Expression of ali1 is activity dependent. Rat cortical neurons were cultured for 14 d (control). TTX (1μm) was added to the culture medium, and the cells were continued to culture for another 30 hr (+TTX). Then, the medium was replaced with fresh one and the cells were continued to culture for another 30 hr(-TTX). Total RNA was extracted from each experimental group and subjected to competitive PCR analysis, as described in Materials and Methods. The data shown are representative results of two experiments.
Tissue distribution and developmental regulation of ali1 mRNA
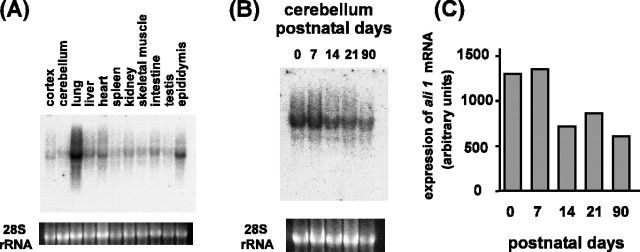
ali1 mRNA was expressed most abundantly in the lung (Fig. 5A). Low ali1 mRNA levels were observed throughout the tissues examined, indicating that expression is not specific to the CNS. Although no results clarifying the roles of ali1 in the lung or other tissues other than brain are currently available, our data suggest that the gene has a function(s) specific to the lung and/or common to all tissue.
Figure 5.
Expression of ali1 mRNA in rat tissue. A, Tissue distribution of ali1 mRNA. Total RNA (10 μg) extracted from various tissues of adult rat was subjected to Northern blotting. The bottom panel shows ethidium bromide staining of 28S rRNA. B, Developmental expression of ali1 mRNA in the cerebellum. Total RNA was extracted from the cerebellum on specified postnatal days, and 10μg of RNA were subjected to Northern blotting analysis. The bottom panel shows ethidium bromide staining of 28S rRNA. C, The data shown in B were quantified.
The expression of ali1 mRNA in the cerebellum is subject to developmental regulation. The rat cerebellum develops after birth, and developmental morphogenesis is complete by postnatal day 21. As shown in Figure 5, B and C, ali1 mRNA in the cerebellum was downregulated to approximately half that in newborn rats between postnatal days 7 and 14, during which cerebellar granule neurons migrate from the external to the internal granule layer. This reduced level of ali1 mRNA was maintained up to postnatal day 90. Our data suggest that ali1 has a developmental role in the cerebellum.
Immunohistochemical localization of Ali1 in rat brain
To assess the distribution of Ali1 in rat brain, specific antibodies against the protein were raised in rabbit. On a Western blot, the antibody cross-reacted with 66K and 63K proteins in the rat brain (Fig. 6A). Immunohistochemical analyses of the rat brain using this specific antibody were performed (Fig. 6B,C). In rat cerebellum, the majority of Ali1 immunoreactivity was localized in the somata of cerebellar granule neurons and Purkinje cells, whereas in the rat hippocampus Ali1 immunoreactivity was localized in the somata of pyramidal cells between CA1 and CA3 regions and those of granule cells of the dentate gyrus. No staining was obtained in the negative control preparations (Fig. 6D,E).
Figure 6.
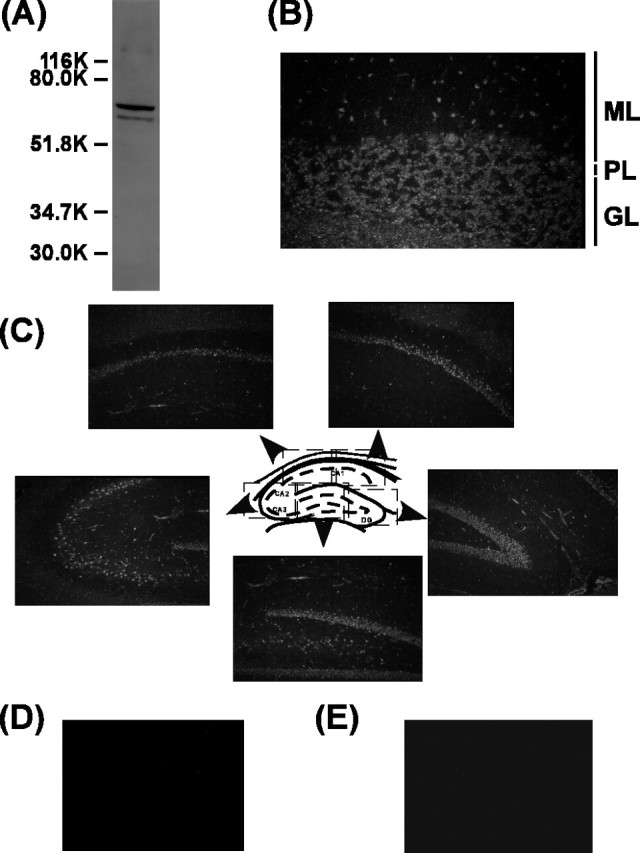
Immunohistochemical localization of Ali1 in brain. A, Total brain extract (100μg) was subjected to 10% SDS-PAGE, and Western blotting was performed with anti-rat Ali1 antiserum. B, Immunohistochemical localization of Ali1 in the rat cerebellum. ML, Molecular layer; PL, Purkinje cell layer; GL, granule cell layer. C, Immunohistochemical localization of Ali1 in the rat hippocampus. D, Negative control staining of the rat cerebellum. E, Negative control staining of the rat hippocampus.
Subcellular localization of Ali1 in rat brain
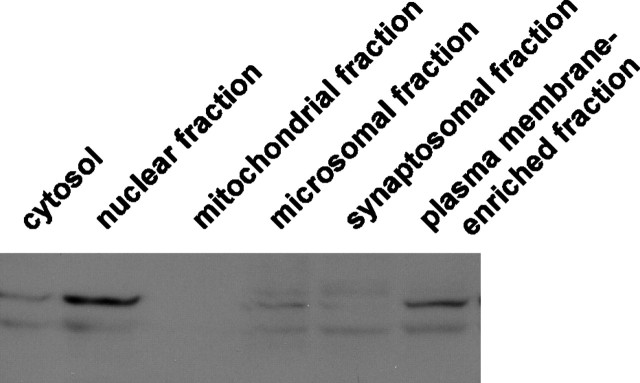
Whole rat brain was fractionated into various subcellular fractions by differential and density gradient centrifugation, as described in Materials and Methods. Individual fractions were subjected to Western blotting analysis with anti-Ali1 antibody. The majority of Ali1 immunoreactivity was localized in the nuclear fraction (Fig. 7, lane 2) and the plasma membrane-enriched fraction (Fig. 7, lane 6), the latter of which displayed the highest 5′-nucleotidase activity that is the marker enzyme for the plasma membrane (data not shown). Therefore, our data suggest that a portion of Ali1 is localized in the plasma membrane.
Figure 7.
Subcellular distribution of Ali1 in brain. Whole brains of rats, 10 weeks of age, were separated into various subcellular fractions. The protein (100 μg) was subjected to 10% SDS-PAGE. Western blotting was performed with anti-rat Ali1 antiserum.
Ali1 promotes depolarization-dependent survival of cerebellar granule neurons
We next examined whether Ali1 regulates survival of neurons. The expression vector of Ali1-Flag fusion protein and the EGFP expression vector were cotransfected to cerebellar granule neurons, and the effect on apoptosis was examined by scoring apoptotic cells in the transfected cell population (Flag-positive and EGFP-positive cells). Control experiments were performed by transfecting the control vector and the EGFP expression vector, and transfected cells were monitored by EGFP fluorescence. As shown in Figure 8, A and B, overexpression of Ali1-Flag fusion protein in the cells cultured in the medium containing 5 mm KCl and 30 μm AP-5 showed that 47% of the transfected cells were apoptotic, whereas the control experiments showed that 75% of the transfected cells were apoptotic. These results indicate that Ali1 inhibits apoptosis and promotes survival of cerebellar granule neurons.
Figure 8.
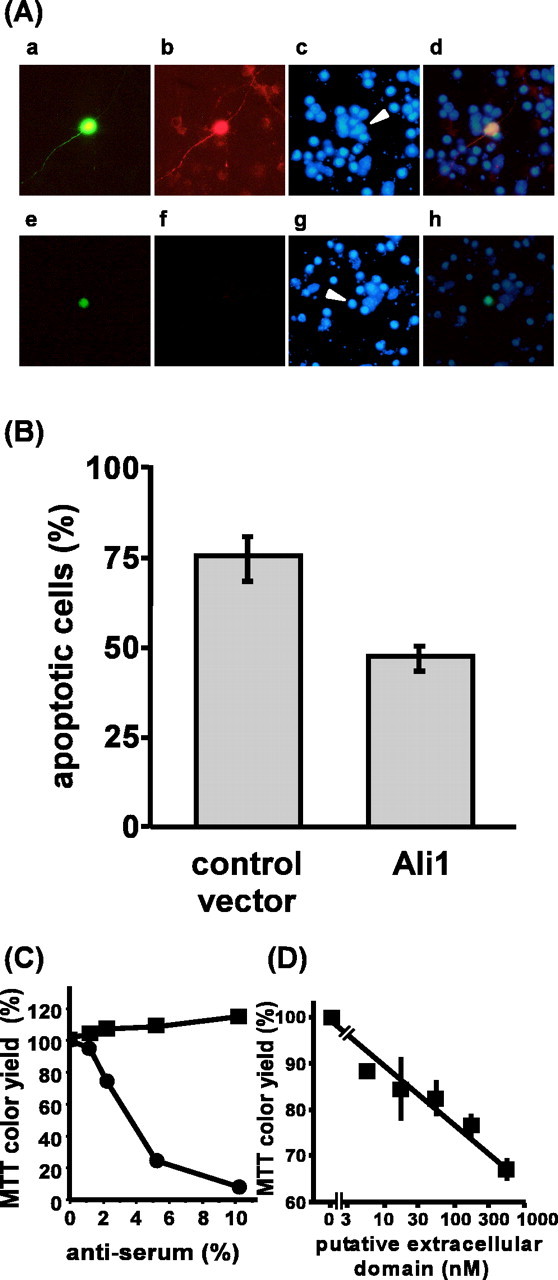
Ali1 is involved in depolarization-dependent survival of cerebellar granule neurons. A, Ali1 promotes survival of neurons. Expression vectors were transfected to cells at 0 DIV, and the cells were cultured in medium containing 5 mm KCl and 30μm AP-5. At 3.0 DIV, the cells were fixed with 4% paraformaldehyde. Transfected cells were monitored by staining with anti-Flag antibody and EGFP fluorescence. Apoptotic cells were monitored by condensed and/or fragmented nuclei by staining with Hoechst dye. a–d, Ali1-Flag expression vector; e–h, control vector; a, e, EGFP fluorescence; b, f, staining with anti-Flag antibody and Alexa Flour 594-coupled secondary antibody; c, g, staining with Hoechst dye; d, h, overlay of EGFP fluorescence, staining with anti-Flag antibody, and Hoechst staining. Arrowheads indicate transfected cells. B, Quantification of the effect of ali1 transfection on apoptosis in cerebellar granule neurons. Results are presented as mean ± SD. The cultures transfected with ali1 expression vector contained a significantly reduced rate of apoptotic cells (Student's t test; p < 0.001; n = 4). C, Effect of anti-Ali1 antiserum on cerebellar granule neuron survival. Anti-Ali1 antiserum was dialyzed extensively against BME and sterilized by filtering through a 0.22 μm membrane (Millipore, Billerica, MA). Granule neurons were cultured in medium containing 25 mm KCl and 30 μm AP-5, as described in Materials and Methods. Antiserum or nonimmune serum was added to the culture at 1.0 DIV, and survival was assayed on 5.0 DIV. The data shown are representative of two independent experiments. Each value is the mean ± SD of duplicate analyses. The horizontal axis (anti-serum %) indicates that percentage of the volume of the antiserum or control serum in the total volume of the culture medium. •, Anti-Ali1 antiserum; ▪, nonimmune serum. D, Effect of Ali1 (57–402) on the survival of cerebellar granule neurons. Rat Ali1 (52–402) was dialyzed extensively against BME and sterilized by filtering through a 0.22 μm membrane (Millipore). Granule neurons were cultured in medium containing 25 mm KCl and 30 μm AP-5, as described in Materials and Methods. Ali1 (57–402) was added to the culture at 2.0 DIV, and neuronal survival was assayed at 6.0 DIV. The data shown are representative of two independent experiments. Each value is the mean ± SD of duplicate analyses.
Because the results shown in Figure 7 indicated that a portion of Ali1 is localized in the plasma membrane, we examined whether the Ali1 present on the surface of the cell was involved in the function of Ali1. As shown in Figure 8C, the addition of the anti-Ali1 antibody to culture medium inhibited survival of granule neurons in a dose-dependent manner when the cells were cultured in the medium containing 25 mm KCl and 30 μm AP-5. However, the antiserum did not affect the survival of neurons when they were cultured in the medium containing 5 mm KCl and 30 μm AP-5 (data not shown). These results suggest that Ali1 expressed on the surface of cells is involved in depolarization-dependent survival of cerebellar granule neurons. To further confirm the involvement of the Ali1 expressed on the surface of cells in depolarization-dependent survival, a bacterially expressed extracelluar segment of Ali1 was additionally used. The extracellular segment may work as a competitive inhibitor to Ali1 because the extracellular segment has protein-to-protein interaction motifs through which it could bind to a ligand(s) for Ali1. Similar experimental design has been reported already on the successful use of soluble extracellular domains of TGF-β receptor and Trk A to inhibit receptor functions in similar experiments (Robertson et al., 2001; Rowland-Goldsmith et al., 2001). The addition of the extracellular segment of rat Ali1, Ali1 (52–402), to the culture medium containing 25 mm KCl and 30 μm AP-5 inhibited survival of cerebellar granule neurons by 65% in a dose-dependent manner (Fig. 8D). All these results indicate that Ali1 on the surface of cell is involved in depolarization-dependent survival of cerebellar granule neurons. However, these results do not rule out the possibility that Ali1 present in the nucleus plays roles for survival of neurons.
Chromosomal mapping of mouse ali1
To determine the chromosomal localization of ali1 on mouse chromosomes, we performed RH mapping. The mouse ali1 locus was mapped to ∼55.3 cM from the centromere of chromosome 15, compared with the Mouse Genomic Database (MGD2002: http://www.informatics.jax.org/mgihome/).
Discussion
It is well established that Ca2+-dependent gene transcription is required for activity-dependent neuronal survival (Ichikawa et al., 1998; Shieh et al., 1998; Bonni et al., 1999; Mao et al., 1999). However, genes that play crucial roles for activity-dependent survival of neurons have not been well characterized. In this study, we compared gene expression between cerebellar granule neurons cultured in survival-promoting conditions and those cultured in apoptosis-inducing conditions. Our analyses led to the identification of a survival-promoting gene, ali1, which is a downstream target of Ca2+-dependent survival signals. The expression of ali1 mRNA is rigidly regulated by depolarization at the transcriptional step and tightly associated with depolarization-dependent survival and/or NMDA-dependent survival of cerebellar granule neurons. Our results also show that expression of ali1 mRNA is regulated by synaptic excitation; thus ali1 mRNA expression is neuronal activity dependent.
The Ali1 protein is a member of a novel class of LRR and Ig superfamily. A search of the databases revealed other members of the ali family, i.e., mouse ali2 (GenBank accession number BC010598), human ali2 (GenBank accession number LOC127001 and AB032989), and human ali3 (GenBank accession number AB058754). As shown in Figures 2, 9A, a striking sequence similarity is observed among members of the Ali family. The similarity within the Ali1 orthologs is between 92% and 95%, whereas that between paralogs is between 46% and 56%. Moreover, the structural organization of Ali proteins is strikingly conserved among family members (Fig. 9B). Each Ali member contains a signal peptide sequence at the NH2-terminal end, seven LRRs flanked by a cysteine-rich sequence at both NH2-terminal and COOH-terminal ends, one IgC2-like domain, one transmembrane domain, and an intracellular domain of ∼100 aa. Apart from the transmembrane domains, homology between the domain structures of human paralogs is the highest in the N-terminal flanking cysteine-rich and LRR domains. The consensus sequence of LRR domains of Ali family proteins is LxxLxxLxLxxNxaxxaxxxa, which is a typical extracellular LRR sequence in animal cells (Kobe and Deisenhofer, 1994; Kajava, 1998).
Figure 9.
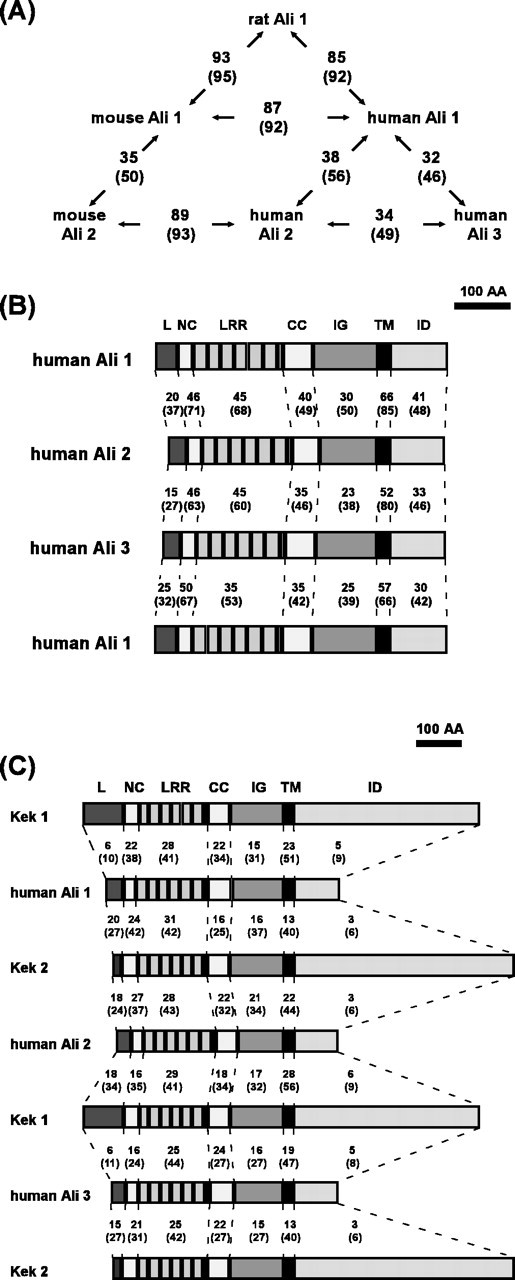
Homology comparisons.A, The homology between each Ali family member is shown in the schematic diagram. The numbers outside the parentheses indicate percentage identity, whereas the numbers inside the parentheses represent percentage similarity. B, The domain organization of members of the human Ali family are shown schematically, and the homology between each homologous domain is compared. The numbers outside the parentheses indicate percentage identity, whereas the numbers inside represent percentage similarity. L, Leader sequence including the signal peptide sequence; NC, NH2-terminal flanking cysteine-rich domain; CC, COOH-terminal flanking cysteine-rich domain; IG, IgC2-like loop; TM, transmembrane domain; ID, intracellular domain. C, Domain organization of human Ali1 and the Drosophila Kek family is shown schematically, and the homology between the each domain is compared as in B.
To date, a number of LRR family members have been identified (Kobe and Deisenhofer, 1994). However, only a limited number of proteins with both LRRs and Ig-like domains have been reported. These proteins include Kek 1/2 (Musacchio and Perrimon, 1996; Ghiglione et al., 1999), Trk A/B/C (Schneider and Schweiger, 1991), ISLR (Nagasawa et al., 1997), NLRR 1/2/3 (Taguchi et al., 1996; Taniguchi et al., 1996), Pal (Gomi et al., 2000), LIG-1 (Suzuki et al., 1996), and peroxidasin (Nelson et al., 1994). In addition, NLRR 1/2/3 and Pal contain fibronectin type 3 domains, whereas peroxidasin possesses peroxidase and thrombospondin/procollagen domains. Among proteins containing both LRRs and Ig-like domains, the putative extracellular domains of the Ali family display significant similarity to those of Drosophila Kek1 and Kek2, the former of which binds the epidermal growth factor receptor (EGFR) in Drosophila embryos and inhibits the Grk/EGFR pathway that controls dorsoventral patterning by restricting a ventralizing signal (Ghiglione et al., 1999). Overall, structural organization is highly conserved between the Kek and Ali families (Fig. 9C). The Kek family has one incomplete LRR and six complete LRRs flanked by two cysteine-rich sequences, one IgC2-like loop, and one single-pass transmembrane domain (Musacchio and Perrimon, 1996). A high degree of homology between the Ali and Kek families is observed in the putative extracellular domains and the transmembrane domains, but not in the putative intracellular domains. The extracellular domains of the Ali family are additionally homologous to those of the Trk family. Trk proteins are neurotrophin receptors that comprise a signal peptide sequence, three LRRs flanked by cysteine-rich sequences, two IgC2-like loops, a single-pass transmembrane domain, and an intracellular tyrosine-kinase domain (Schneider and Schweiger, 1991). Human Trk C exhibits highest homology to human Ali1. The extracellular domain of human Trk C displays 21% amino acid identity and 37% similarity to that of human Ali1.
Our results (shown in Fig. 8) clearly demonstrated that Ali1 is a positive regulator of depolarization-dependent survival of neurons. However, mechanisms of how it promotes neuronal survival remains to be elucidated. Many proteins that contain LRR and/or Ig-like domains are involved in cell adhesion and/or signal transduction through protein-to-protein interactions on cell surface (Kobe and Deisenhofer, 1994). Accordingly, we hypothesize that Ali1 is involved in signal transduction and/or cell adhesion via protein-to-protein interactions. Our results that anti-Ali1 antibody and the extracellular segment of Ali1 inhibited the survival of neurons indicate that the Ali1 on the cell surface plays crucial roles for survival of neurons. We currently assume that Ali1 is a receptor or a modulator molecule of a surviving signal(s) because of its structural similarities to Trk families and Kek families. Additional studies are required to clarify this issue.
Here, we showed that mouse ali1 locus was mapped to a region on chromosome 15. This region is syntenic to human chromosome 12. According to the database, human ali1 is mapped to 12q13.11 (GenBank accession number AC004010). This locus is mapped to within the region in which familial Alzheimer's disease type 5 [AD5; 12p11.23–12q13.12; Online Mendelian Inheritance in Man (OMIM) number 602096] has been mapped (Pericak-Vance et al., 1997). AD5 is a late-onset type (at least 60 years of age) Alzheimer's disease that differs from the disorders caused by mutations in the amyloid precursor, presenilin 1/2, and apolipoprotein E genes (OMIM number 104300). The causative gene for AD5 has not been identified yet. Another gene mapped to the same region is Parkinson's disease 8 (PARK8; 12p11.23–12q13.11; OMIM number 607060). PARK8 was reported in a large Japanese family with autosomal dominant parkinsonism (Funayama et al., 2002). Several causative genes of Parkinson's disease including a-synuclein, UCHL1, parkin, and DJ1 have been identified (OMIM number 168600). However, the causative gene for PARK8 has not been identified yet. Our findings showed that ali1 promotes survival of neurons and suggest that mutations of ali1 could cause serious neurodegenerative diseases like Alzheimer's disease and Parkinson's disease. Taking into account our findings, we propose that ali1 is one of the positional candidate genes of AD5 and/or PARK8.
Subcellular and immunohistochemical localization studies on Ali1 suggest that the majority of the protein is contained in the nucleus and plasma membrane-enriched fraction. Several studies show examples of signal-transducing receptors that are believed to be localized in the cell membrane but are actually present in the nucleus. These include EGFR, FGF receptor, growth factor receptor, and c-erb-4/HER-4 growth factor receptor (Lobie et al., 1994; Xie and Hung, 1994; Maher, 1996; Srinivasan et al., 2000; Brosenitsch and Katz, 2001; Marti et al., 2001). A recent study reported that the EGFR displays transactivator activity and may function as a transcription factor (Brosenitsch and Katz, 2001). Although at present no results are available demonstrating that Ali1 is a transcription factor, the localization of this protein in the nucleus indicates involvement in nuclear roles, in addition to functions on the cell surface.
We showed that ali1 is a primary response gene upregulated by the Ca2+ signal and rapidly downregulated on blocking of the survival signal (Fig. 3). This downregulation occurs before the death of the majority of cells, suggesting that ali1 plays a crucial role(s) in neuronal survival. The ali1 mRNA sequence has a number of features in common with early response gene family members. Immediate early genes, such as c-fos and c-myc, have multiple copies of AUUUA motifs in their 3′-untranslated regions. This motif contributes to the short life of the mRNA (Jones and Cole, 1987; Wilson and Treisman, 1988). Rat, mouse, and human ali1 mRNA have two, three, and four copies of AUUUA motifs, respectively, and one copy of the UACAAAA motif [another common feature shared by early response gene family members that is essential for mRNA trafficking (Freter et al., 1992)] in their 3′-untranslated regions.
The functions of the other members of the ali family remain to be elucidated. However, human Ali3/KIAA1851 (GenBank accession number AB058754) has been suggested in the database to code for GDP-mannose pyrophosphorylase B (GenBank accession number NP 037466). Human Ali3 exhibits only 11% identity and 24% similarity to the amino acid sequence of GDP-mannose pyrophosphorylase B. Notably, GDP-mannose pyrophosphorylase B does not contain the LRR, Ig-like, or transmembrane domains, which are characteristic of Ali protein structure, and, thus, it is presently unclear whether Ali3 has GDP-mannose pyrophosphorylase activity. Additional experimental evidence is required to clarify the function of Ali3.
In conclusion, our findings indicate that ali1 plays a crucial role(s) in the depolarization-dependent survival of cerebellar granule neurons. To elucidate the molecular mechanism by which ali1 regulates the survival of neurons, additional studies are required to characterize the specific ligands, association molecules, and diseases caused by mutation of this gene, especially in relation to Alzheimer's disease and Parkinson's disease. Experiments along these lines are currently in progress in our laboratory.
Footnotes
We thank Drs. Y. Kanaho and K. Itoh of The Tokyo Metropolitan Institute of Medical Science for comments on a previous version of this manuscript.
Correspondence should be addressed to Dr. Tomio Ono, Department of Laboratory Animal Science, The Tokyo Metropolitan Institute of Medical Science, Tokyo Metropolitan Organization for Medical Research, 3-18-22 Honkomagome, Bunkyo-ku, Tokyo 113-8613, Japan. E-mail ono@rinshoken.or.jp.
Copyright © 2003 Society for Neuroscience 0270-6474/03/235887-10$15.00/0
References
- Bonni A, Brunet A, West AE, Datta SR, Takasu MA, Greenberg ME ( 1999) Cell survival promoted by the Ras-MAPK signaling pathway by transcription-dependent and -independent mechanisms. Science 286: 1358-1362. [DOI] [PubMed] [Google Scholar]
- Brosenitsch TA, Katz DM ( 2001) Physiological patterns of electrical stimulation can induce neuronal gene expression by activating N-type calcium channels. J Neurosci 21: 2571-2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N ( 1987) Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem 162: 156-159. [DOI] [PubMed] [Google Scholar]
- Franke B, Bayatti N, Engele J ( 2000) Neurotrophins require distinct extracellular signals to promote the survival of CNS neurons in vitro Exp Neurol 165: 125-135. [DOI] [PubMed] [Google Scholar]
- Freter RR, Irminger JC, Porter JA, Jones SD, Stiles CD ( 1992) A novel 7-nucleotide motif located in 3′ untranslated sequences of the immediate-early gene set mediates platelet-derived growth factor induction of the JE gene. Mol Cell Biol 12: 5288-5300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funayama M, Hasegawa K, Kowa H, Saito M, Tsuji S, Obata F ( 2002) A new locus for Parkinson's disease (PARK8) maps to chromosome 12p11.2-q13.1. Ann Neurol 51: 296-301. [DOI] [PubMed] [Google Scholar]
- Gallo V, Kingsbury A, Balazs R, Jorgensen OS ( 1987) The role of depolarization in the survival and differentiation of cerebellar granule cells in culture. J Neurosci 7: 2203-2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghiglione C, Carraway III KL, Amundadottir LT, Boswell RE, Perrimon N, Duffy JB ( 1999) The transmembrane molecule kekkon 1 acts in a feedback loop to negatively regulate the activity of the Drosophila EGF receptor during oogenesis. Cell 96: 847-856. [DOI] [PubMed] [Google Scholar]
- Ghosh A, Carnahan J, Greenberg ME ( 1994) Requirement for BDNF in activity-dependent survival of cortical neurons. Science 263: 1618-1623. [DOI] [PubMed] [Google Scholar]
- Gomi F, Imaizumi K, Yoneda T, Taniguchi M, Mori Y, Miyoshi K, Hitomi J, Fujikado T, Tano Y, Tohyama M ( 2000) Molecular cloning of a novel membrane glycoprotein, pal, specifically expressed in photoreceptor cells of the retina and containing leucine-rich repeat. J Neurosci 20: 3206-3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon-Weeks PR ( 1987) Isolation of synaptosomes, growth cones and their subcellular components. In: Neurochemistry: a practical approach (Turner AJ, Bachelard HS, eds), pp 1-26. Oxford: IRL.
- Graham JM, Higgins JA, Green C ( 1968) The isolation of rat liver plasma membrane fragments. Biochim Biophys Acta 150: 303-305. [DOI] [PubMed] [Google Scholar]
- Ichikawa D, Tabuchi A, Taoka A, Tsuchiya T, Tsuda M ( 1998) Attenuation of cell death mediated by membrane depolarization different from that by exogenous BDNF in cultured mouse cerebellar granule cells. Brain Res Mol Brain Res 56: 218-226. [DOI] [PubMed] [Google Scholar]
- Ikonomidou C, Bosch F, Miksa M, Bittigau P, Vockler J, Dikranian K, Tenkova TI, Stefovska V, Turski L, Olney JW ( 1999) Blockade of NMDA receptors and apoptotic neurodegeneration in the developing brain. Science 283: 70-74. [DOI] [PubMed] [Google Scholar]
- Inokuchi K, Kato A, Hiraia K, Hishinuma F, Inoue M, Ozawa F ( 1996) Increase in activin β A mRNA in rat hippocampus during long-term potentiation. FEBS Lett 382: 48-52. [DOI] [PubMed] [Google Scholar]
- Jones TR, Cole MD ( 1987) Rapid cytoplasmic turnover of c-myc mRNA: requirement of the 3′ untranslated sequences. Mol Cell Biol 7: 4513-4521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajava AV ( 1998) Structural diversity of leucine-rich repeat proteins. J Mol Biol 277: 519-527. [DOI] [PubMed] [Google Scholar]
- Kobe B, Deisenhofer J ( 1994) The leucine-rich repeat: a versatile binding motif. Trends Biochem Sci 19: 415-421. [DOI] [PubMed] [Google Scholar]
- Kohara K, Ono T, Tominaga-Yoshino K, Shimonaga T, Kawashima S, Ogura A ( 1998) Activity-dependent survival and enhanced turnover of calcium in cultured rat cerebellar granule neurons. Brain Res 809: 231-237. [DOI] [PubMed] [Google Scholar]
- Kuroda Y, Ichikawa M, Muramoto K, Kobayashi K, Matsuda Y, Ogura A, Kudo Y ( 1992) Block of synapse formation between cerebral cortical neurons by a protein kinase inhibitor. Neurosci Lett 135: 255-258. [DOI] [PubMed] [Google Scholar]
- Levi G, Aloisi F, Ciotti MT, Thangnipon W, Kingsbury A, Balazs R ( 1989) Preparation of 98% pure cerebellar granule cell cultures. In: A dissection and tissue culture manual of the nervous system (Shahar A, de Vellis J, Vernadakis A, Haken B, eds), pp 211-214. New York: Liss.
- Lobie PE, Wood TJ, Chen CM, Waters MJ, Norstedt G ( 1994) Nuclear translocation and anchorage of the growth hormone receptor. J Biol Chem 269: 31735-31746. [PubMed] [Google Scholar]
- Maher PA ( 1996) Nuclear translocation of fibroblast growth factor (FGF) receptors in response to FGF-2. J Cell Biol 134: 529-536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manly KF, Olson JM ( 1999) Overview of QTL mapping software and introduction to map manager QT. Mamm Genome 10: 327-334. [DOI] [PubMed] [Google Scholar]
- Mao Z, Bonni A, Xia F, Nadal-Vicens M, Greenberg ME ( 1999) Neuronal activity-dependent cell survival mediated by transcription factor MEF2. Science 286: 785-790. [DOI] [PubMed] [Google Scholar]
- Marti U, Ruchti C, Kampf J, Thomas GA, Williams ED, Peter HJ, Gerber H, Burgi U ( 2001) Nuclear localization of epidermal growth factor and epidermal growth factor receptors in human thyroid tissues. Thyroid 11: 137-145. [DOI] [PubMed] [Google Scholar]
- McCarthy LC, Terrett J, Davis ME, Knights CJ, Smith AL, Critcher R, Schmitt K, Hudson J, Spurr NK, Goodfellow PN ( 1997) A first-generation whole genome-radiation hybrid map spanning the mouse genome. Genome Res 7: 1153-1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monti B, Contestabile A ( 2000) Blockade of the NMDA receptor increases developmental apoptotic elimination of granule neurons and activates caspases in the rat cerebellum. Eur J Neurosci 12: 3117-3123. [DOI] [PubMed] [Google Scholar]
- Mosmann T ( 1983) Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods 65: 55-63. [DOI] [PubMed] [Google Scholar]
- Musacchio M, Perrimon N ( 1996) The Drosophila kekkon genes: novel members of both the leucine-rich repeat and immunoglobulin superfamilies expressed in the CNS. Dev Biol 178: 63-76. [DOI] [PubMed] [Google Scholar]
- Nagasawa A, Kubota R, Imamura Y, Nagamine K, Wang Y, Asakawa S, Kudoh J, Minoshima S, Mashima Y, Oguchi Y, Shimizu N ( 1997) Cloning of the cDNA for a new member of the immunoglobulin superfamily (ISLR) containing leucine-rich repeat (LRR). Genomics 44: 273-279. [DOI] [PubMed] [Google Scholar]
- Nelson RE, Fessler LI, Takagi Y, Blumberg B, Keene DR, Olson PF, Parker CG, Fessler JH ( 1994) Peroxidasin: a novel enzyme-matrix protein of Drosophila development. EMBO J 13: 3438-3447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen H, Engelbrecht J, Brunak S, von Heijne G ( 1997) Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng 10: 1-6. [DOI] [PubMed] [Google Scholar]
- Ogura A, Iijima T, Amano T, Kudo Y ( 1987) Optical monitoring of excitatory synaptic activity between cultured hippocampal neurons by a multisite Ca 2+ fluorometry. Neurosci Lett 78: 69-74. [DOI] [PubMed] [Google Scholar]
- Ono T, Kudo Y, Kohara K, Kawashima S, Ogura A ( 1997a) Activity-dependent survival of rat cerebellar granule neurons is not associated with sustained elevation of intracellular Ca 2+ Neurosci Lett 228: 123-126. [DOI] [PubMed] [Google Scholar]
- Ono T, Inokuchi K, Ogura A, Ikawa Y, Kudo Y, Kawashima S ( 1997b) Activity-dependent expression of parathyroid hormone-related protein (PTHrP) in rat cerebellar granule neurons. Requirement of PTHrP for the activity-dependent survival of granule neurons. J Biol Chem 272: 14404-14411. [DOI] [PubMed] [Google Scholar]
- Oppenheim RW ( 1991) Cell death during development of the nervous system. Annu Rev Neurosci 14: 453-501. [DOI] [PubMed] [Google Scholar]
- Pericak-Vance MA, Bass MP, Yamaoka LH, Gaskell PC, Scott WK, Terwedow HA, Menold MM, Conneally PM, Small GW, Vance JM, Saunders AM, Roses AD, Haines JL ( 1997) Complete genomic screen in late-onset familial Alzheimer disease. Evidence for a new locus on chromosome 12. JAMA 278: 1237-1241. [PubMed] [Google Scholar]
- Robertson AG, Banfield MJ, Allen SJ, Dando JA, Mason GG, Tyler SJ, Bennett GS, Brain SD, Clarke AR, Naylor RL, Wilcock GK, Brady RL, Dawbarn D ( 2001) Identification and structure of the nerve growth factor binding site on TrkA. Biochem Biophys Res Commun 282: 131-141. [DOI] [PubMed] [Google Scholar]
- Rowland-Goldsmith MA, Maruyama H, Kusama T, Ralli S, Korc M ( 2001) Soluble type II transforming growth factor-β (TGF-β) receptor inhibits TGF-beta signaling in COLO-357 pancreatic cancer cells in vitro and attenuates tumor formation. Clin Cancer Res 7: 2931-2940. [PubMed] [Google Scholar]
- Schneider R, Schweiger M ( 1991) A novel modular mosaic of cell adhesion motifs in the extracellular domains of the neurogenic trk and trkB tyrosine kinase receptors. Oncogene 6: 1807-1811. [PubMed] [Google Scholar]
- See V, Boutillier AL, Bito H, Loeffler JP ( 2001) Calcium/calmodulin-dependent protein kinase type IV (CaMKIV) inhibits apoptosis induced by potassium deprivation in cerebellar granule neurons. FASEB J 15: 134-144. [DOI] [PubMed] [Google Scholar]
- Shieh PB, Hu SC, Bobb K, Timmusk T, Ghosh A ( 1998) Identification of a signaling pathway involved in calcium regulation of BDNF expression. Neuron 20: 727-740. [DOI] [PubMed] [Google Scholar]
- Srinivasan R, Gillett CE, Barnes DM, Gullick WJ ( 2000) Nuclear expression of the c-erbB-4/HER-4 growth factor receptor in invasive breast cancers. Cancer Res 60: 1483-1487. [PubMed] [Google Scholar]
- Suzuki Y, Sato N, Tohyama M, Wanaka A, Takagi T ( 1996) cDNA cloning of a novel membrane glycoprotein that is expressed specifically in glial cells in the mouse brain. LIG-1, a protein with leucine-rich repeats and immunoglobulin-like domains. J Biol Chem 271: 22522-22527. [DOI] [PubMed] [Google Scholar]
- Tabuchi A, Koizumi M, Nakatsubo J, Yaguchi T, Tsuda M ( 2001) Involvement of endogenous PACAP expression in the activity-dependent survival of mouse cerebellar granule cells. Neurosci Res 39: 85-93. [DOI] [PubMed] [Google Scholar]
- Taguchi A, Wanaka A, Mori T, Matsumoto K, Imai Y, Tagaki T, Tohyama M ( 1996) Molecular cloning of novel leucine-rich repeat proteins and their expression in the developing mouse nervous system. Brain Res Mol Brain Res 35: 31-40. [DOI] [PubMed] [Google Scholar]
- Taniguchi H, Tohyama M, Takagi T ( 1996) Cloning and expression of a novel gene for a protein with leucine-rich repeats in the developing mouse nervous system. Brain Res Mol Brain Res 36: 45-52. [DOI] [PubMed] [Google Scholar]
- Tao X, Finkbeiner S, Arnold DB, Shaywitz AJ, Greenberg ME ( 1998) Ca 2+ influx regulates BDNF transcription by a CREB family transcription factor-dependent mechanism. Neuron 20: 709-726. [DOI] [PubMed] [Google Scholar]
- Tao X, West AE, Chen WG, Corfas G, Greenberg ME ( 2002) A calcium-responsive transcription factor, CaRF, that regulates neuronal activity-dependent expression of BDNF. Neuron 33: 383-395. [DOI] [PubMed] [Google Scholar]
- Walton MR, Dragunow I ( 2000) Is CREB a key to neuronal survival? Trends Neurosci 23: 48-53. [DOI] [PubMed] [Google Scholar]
- Wilson T, Treisman R ( 1988) Removal of poly(A) and consequent degradation of c-fos mRNA facilitated by 3′ AU-rich sequences. Nature 336: 396-399. [DOI] [PubMed] [Google Scholar]
- Xie Y, Hung MC ( 1994) Nuclear localization of p185neu tyrosine kinase and its association with transcriptional transactivation. Biochem Biophys Res Commun 203: 1589-1598. [DOI] [PubMed] [Google Scholar]
- Yano S, Tokumitsu H, Soderling TR ( 1998) Calcium promotes cell survival through CaM-K kinase activation of the protein-kinase-B pathway. Nature 396: 584-587. [DOI] [PubMed] [Google Scholar]