Abstract
Staufen1, the mammalian homolog of Drosophila Staufen, assembles into ribonucleoprotein particles (RNPs), which are thought to transport and localize RNA into dendrites of mature hippocampal neurons. We therefore investigated whether additional components of the RNA localization complex besides Staufen are conserved. One candidate is the mammalian homolog of Drosophila Barentsz (Btz), which is essential for the localization of oskar mRNA to the posterior pole of the Drosophila oocyte and is a component of the oskar RNA localization complex along with Staufen. In this study, we report the characterization of mammalian Btz, which behaves like a nucleocytoplasmic shuttling protein. When expressed in the Drosophila egg chamber, mammalian Btz is still able to interact with Drosophila Staufen and reach the posterior pole in the wild-type oocyte, but does not rescue the btz mutant phenotype. Most interestingly, we show by immunoprecipitation assays that Btz interacts with mammalian Staufen in an RNA-dependent manner through a conserved domain, which encompasses the region of homology to the Drosophila Btz protein and contains a novel conserved motif. One candidate for an RNA that mediates this interaction is the dendritically localized brain cytoplasmic 1 transcript. In addition, Btz and Staufen1 colocalize within particles in the cell body and, to a more variable extent, in dendrites of mature hippocampal neurons. Together, our data suggest that the mRNA transport machinery is conserved during evolution, and that mammalian Btz is an additional component of the dendritic RNPs in hippocampal neurons.
Keywords: MLN51, Barentsz, Staufen, ribonucleoprotein particles, RNA transport, hippocampal neurons, BC1 RNA
Introduction
The asymmetric distribution and localization of mRNA have been described in many organisms and different cell types (Bashirullah et al., 1998; Bassell et al., 1999; Palacios and St. Johnston, 2001). For example, in Drosophila melanogaster, the localization of oskar and bicoid mRNAs is required to establish the embryonic body axes. In neurons, translation of dendritically localized mRNAs might be involved in mechanisms such as synaptic plasticity and memory (Kiebler and DesGroseillers, 2000; Smith et al., 2001; Steward and Schuman, 2001). Drosophila mutants in kinesin heavy chain, mago nashi, staufen, cytoplasmic tropomyosin II, Y14, and barentsz (btz1) are all defective in the posterior localization of oskar mRNA (Ephrussi et al., 1991; Erdelyi et al., 1991; Newmark and Boswell, 1994; Kim-Ha et al., 1995; Tetzlaff et al., 1996; van Eeden et al., 2001). The clearest link between any of these proteins and their effects on the localization of oskar mRNA has been in the case of Staufen protein. The double-stranded RNA binding protein Staufen is essential for the cytoplasmic localization of oskar mRNA and colocalizes with the oskar transcript to the posterior pole of the oocyte. In Drosophila, oskar mRNA assembles into ribonucleoprotein particles (RNPs) together with Mago nashi, Y14, Barentsz (Btz), and Staufen proteins (van Eeden et al., 2001). Btz thereby transiently colocalizes with oskar mRNA and Staufen to the posterior pole. Until now, only a few components of such RNP complexes, which are involved in RNA transport and localization in mammalian cells and, in particular, primary neurons, have been identified (Krichevsky and Kosik, 2001; Ohashi et al., 2002). The mammalian homolog of Drosophila Staufen has been implicated in RNA localization into dendrites of hippocampal neurons (Kiebler et al., 1999; Köhrmann et al., 1999a; Tang et al., 2001), and the Staufen-containing RNPs are thought to be the active transport units for dendritically localized mRNAs (Köhrmann et al., 1999a; Mallardo et al., 2003; Roegiers, 2003). Recently, protein phosphatase 1 was identified as a Staufen1 (Stau1)-interacting protein; this suggested that both proteins might be recruited into the same RNPs (Monshausen et al., 2002). An attractive candidate for an additional component of such Staufen-containing RNPs is the mammalian homolog of Drosophila Btz (van Eeden et al., 2001), which shares restricted homology with the human nucleocytoplasmic shuttling protein malignant lymph node 51 (MLN51) (Tomasetto et al., 1995; Degot et al., 2002). We show in this study that mammalian Btz (mBtz) does not rescue the btz1 mutant phenotype when exogenously expressed in the Drosophila oocyte. mBtz maintains the capacity, however, to interact with Drosophila Staufen and reach the posterior pole in the wild-type oocyte, suggesting a conserved function of this mBtz–Staufen complex. In hippocampal neurons, overexpressed mBtz- and Staufen-tagged proteins colocalize in granular structures in the dendritic compartment. Whereas the degree of colocalization is highest within the cell body, it gradually decreases with progressive distance from the cell body toward distal dendrites. Most importantly, immunoprecipitation (IP) assays reveal that this interaction is RNA dependent. Therefore, mBtz may represent an important component of the dendritic mRNA transport particles in hippocampal neurons.
Materials and Methods
Animals. For cell culture, pregnant Sprague Dawley rats were used. For tissue extracts, 4- to 6-week-old Sprague Dawley rats (<250 gm) were anesthetized with ether and decapitated in accordance with the state law. All steps were performed under strictly RNase-free conditions at 4°C or on ice.
Cloning. Using information on expressed sequence tags (ESTs) and 5′-rapid amplification of cDNA ends, the cDNAs encoding both the rat and mouse homologs of MLN51–Btz were cloned and sequenced. The rat and mouse Btz cDNAs were cloned either in frame with the pd2EGFP-N1 (Clontech, Heidelberg, Germany) or in the same vector without enhanced green fluorescent protein (EGFP) to express untagged Btz protein.
For the expression of mBtz in Drosophila, the mBtz coding region was cloned into the BamHI–SpeI sites of pD277-green fluorescent protein (GFP)6 vector (Micklem et al., 1997; van Eeden et al., 2001) to generate a transformation construct in which the α4-tubulin promoter drives the germline-specific expression of an mBTZ–GFP6 fusion protein. The construct to express Staufen1–hemagglutinin (HA) was described previously by Duchaîne et al. (2000).
For Btz1–158–GFP, Btz152–359–GFP, and Btz1–359–GFP, cDNAs from rat brain were amplified by PCR using the Herculase HotStart DNA polymerase (Stratagene, Heidelberg, Germany) with the following primers: 5′- GCTAGCATGGCGGACCGGCGGCGGC (NheI, 1), 5′- ACCGGTTTGTTCTCCACAGGCTCTGTG (AgeI, reverse 158), 5′- GCTAGCATGACAGAGCCTGTGGAGAACAAAG (NheI, 152), and 5′- ACCGGTTTAGCTTCATGCTTAAGAGTCTC (AgeI, reverse 359). (Underlining indicates the sequence recognized by the corresponding restriction enzymes.) The PCR products were sequenced and cloned in frame with the d2EGFP. The L465A point mutation was created using the QuickChange site-directed mutagenesis kit (Stratagene) according to the manufacturer's protocol with the following primers: 5′-GAGCAAGATGTGGCACAG GCGAATATAGCAGAACAAAATTGGAGT and 5′-ACTCCAATTTTGTTCTGCTATATT CGCCTGTGCCACATCTTGCTC.
Computer analysis. Sequences related to mouse MLN51 were identified in the nonredundant database at National Center for Biotechnology Information using Blast (http://www.ncbi.nlm.nih.gov/BLAST/) and retrieved using Entrez (http://http://www.ncbi.nlm.nih.gov/Entrez/). The relationship between MLN51 and thyroid hormone receptor-associated protein 150 kDa (TRAP150) sequences was detected through pattern searches at Pôle Bio-Informatique Lyonnais using Pattinprot [http://npsa-pbil.ibcp.fr/cgi-bin/npsa_automat.pl?page=/NPSA/npsa_pattinprot.html]. Alignments were made in Macaw (Schuler et al., 1991).
Northern blotting and in situ hybridization. Northern analysis was performed using commercial mouse mRNA blots (Clontech). The cDNA encoding the full-length mouse Btz protein was labeled with [α-32P]deoxycytidine triphosphate using Prime-It RmT random primer labeling kit (Stratagene); prehybridization and hybridization steps were performed according to the manufacturer's protocol (Clontech). The in situ hybridization was performed as described previously by Knoell et al. (2001). Antisense and sense RNA probes labeled with digoxigenin were synthesized according to the manufacturer's protocol (Roche, Mannheim, Germany). Postnatal day 14 (P14) mouse brains were snap-frozen, cryosectioned, and then fixed in 4% paraformadehyde for 20 min.
Antibody production. Amino acids 175–317 and 356–527 of the rat Btz gene and amino acids 109–353 from the mouse Btz gene were expressed in Escherichia coli using the pPROEX-T vector (Invitrogen, Karlsruhe, Germany). The first fusion protein was purified under denaturing conditions, and the others were purified under native conditions using nickel-nitrilotriacetic acid chromatography (Qiagen, Hilden, Germany). For immunization in rabbits, the first two antigens were used, whereas the third one was injected into mice. Antigen 2 was then used for affinity purification.
In vitro transcription–translation. The cDNA of mBtz cloned in Bluescript KS (Stratagene) was transcribed and translated using the TNT quick-coupled transcription–translation system (Promega, Mannheim, Germany) according to the manufacturer's instructions in the presence of [35S]methionine. Samples were run on SDS-polyacrylamide gels and then exposed for 1–6 hr.
Immunocytochemistry. Immunocytochemistry on mature primary hippocampal neurons in culture [16 d in vitro (DIV)] was performed as described previously (Duchaîne et al., 2002). The following primary antibodies (incubated overnight at 4°C) were used: affinity-purified polyclonal rabbit anti-Btz (dilution, 1:50) and mouse polyclonal anti-Staufen 1 antibodies (1:200). As secondary antibodies, Alexa Fluor 488 goat anti-rabbit IgG [heavy and light (H+L)] conjugated (1:500; Molecular Probes, Leiden, The Netherlands) and Cy3-conjugated goat anti-mouse IgG (H+L) (1:500; Dianova, Hamburg, Germany) were used. Fluorescence and confocal microscopy were performed as described previously (Kiebler et al., 1999; Macchi et al., 2003). The Metamorph 5.0 software package was used to quantify the degree of colocalization between mBtz and Staufen1 in hippocampal neurons. Confocal pictures of hippocampal neurons immunostained with anti-mBtz and anti-Stau1 polyclonal antibodies were analyzed in detail by choosing random areas from three different compartments (cell body, proximal dendrite, or distal dendrite), and the percentage of colocalization (mBtz vs Staufen1) was measured. The mean and SD were then calculated using Microsoft Excel software.
Cell culture, transient transfection, and LMB treatment. Baby hamster kidney (BHK) cells were grown and transfected as described previously by Macchi et al. (2003). For leptomycin B (LMB) experiments, BHK cells were treated (16 hr after transfection) with 50 nm LMB for 3 hr before fixation. Adult primary hippocampal neurons were transfected as described previously by Köhrmann et al. (1999b). Cells were analyzed by fluorescence microscopy as described previously by Köhrmann et al. (1999a).
Fly stocks and Staufen antibody stainings. The barentsz mutant allele used was w;e btz1 ca/TM3 Sb (van Eeden et al., 2001). The mBtz–GFP construct was introduced into flies using standard methods. The transgenic used was w; mBtzGFP/CyO. Antibody stainings were performed as described previously by Palacios and St. Johnston (2002), and the anti-Staufen antibody was used at 1:1000 (St. Johnston et al., 1991).
Preparation of cell and tissue extracts. BHK cells were washed with PBS, lysed in lysis buffer [0.1% Triton X-100, 150 mm KCl, 50 mm HEPES, pH 7.4, and 1 mm DTT plus proteinase inhibitor mixture (Roche), including 1 mm phenylmethylsulfonyl fluoride (PMSF)]. Extracts (50 μg) from brain tissue as well as extracts from cultured glia or hippocampal neurons were prepared. Equal amounts of proteins were separated by 8% SDS-PAGE and blotted onto nitrocellulose (Schleicher & Schuell, Dassel, Germany). Western blot analysis was performed as described previously by Macchi et al. (2003).
For both the DNase treatment and the RNase treatment, two rat forebrains were homogenized in a 5 ml Dounce homogenizer on ice with 4 ml of lysis buffer using 10 manual strokes, followed by 10 strokes in a motordriven Dounce. The homogenate was centrifuged for 10 min at 21,000 × g [supernatant at 14,000 × rpm (S14)]. For RNase treatment, 5 mm EDTA was added to the supernatant, and the sample was divided in half. One sample (+RNase, DNase free; Sigma, Munich, Germany) was incubated for 1 hr at 30°C with 40 μg/ml RNase A in 10 mm Tris-Cl, pH 7.5, and 15 mm NaCl, and the other one-half, as control, was incubated in an equal amount of buffer (-RNase). For DNase (MBI Fermentas, St. Leon-Rot, Germany) treatment, the S14 was incubated with DNase buffer alone (control) or together with 2 U of DNase at 37°C for 30 min, and after 15 min, an additional 2 U of the enzyme were added. After incubation, samples were centrifuged for 20 min at 100,000 × g to collect the pellet at 100,000 × g (P100) and supernatant at 100,000 × g (S100), respectively. The pellets were then processed as described previously by Mallardo et al. (2003).
Immunoprecipitations. BHK cells grown in 5 cm Petri dishes were transfected, washed in PBS at 14–18 hr posttransfection, and then harvested in 500 μl of lysis buffer [0.5% Triton X-100, 50 mm Tris-Cl, pH 7.5, 100 mm KCl, 1 mm PMSF, and protease inhibitor mixture (Roche)]. The lysate was centrifuged at 14,000 × g for 10 min at 4°C. For immunoprecipitations, 500 μg of total protein were preincubated in a total volume of 1 ml, either with anti-HA ascites fluid (1:333) or affinity-purified rabbit polyclonal anti-GFP antibodies (1:500, Molecular Probes) and 30 μl of 50% protein A-Sepharose slurry (Amersham Biosciences, Freiburg, Germany) equilibrated in lysis buffer for at least 1 hr at 4°C and then centrifuged at 500 × g for 10 sec at 4°C. The pellet was washed four times with washing buffer (0.1% Triton X-100, 50 mm Tris-Cl, pH 7.5, 100 mm KCl, and 1 mm PMSF), resuspended in reducing (DTT-containing) sample buffer, and analyzed by SDS-PAGE and Western blotting using polyclonal rabbit anti-GFP (1:2000; Molecular Probes), monoclonal mouse anti-GFP (1:2000; Clontech) or monoclonal anti-HA (1:1000; Roche) antibodies. The RNase treatment was performed as described previously. For the immunoprecipitations of the endogenous complexes, embryonic day 17 rat forebrains were lysed in lysis buffer. The preclearing was performed by incubation of the lysate (2 mg of protein in total in each sample) with preimmune serum and protein A for 1 hr at 4°C. After centrifugation, the supernatants were immunoprecipitated with the indicated antibodies and protein A. The immunocomplexes were collected by centrifugation and washed. Samples were then processed as described above.
RNA isolation and radioactive reverse transcription (RT)-PCR from immunoprecipitates were performed as described previously by Mallardo et al. (2003) with the following modifications. MgCl2 (1 mm) was added to both the lysis and the washing buffer. The latter also contained 0.5% Triton X-100 (instead of 0.1%). To avoid a possible DNA contamination, extracts were incubated with RNase-free DNase (MBI Fermentas). First-strand cDNA synthesis was performed with random hexamers by using Moloney murine leukemia virus reverse transcriptase (Genaxxon, Stafflangen, Germany). A semiquantitative PCR was then achieved using the following primers: 5′-GGGGTTGGGGATTTAGCTC (forward) and 5′-GGTTGTGTGTGCCAGTTACC (reverse) for brain cytoplasmic 1 (BC1); 5′-CATGCCCCCATTCCATCTG (forward) and 5′-GGACCCACTCCACAAACTC (reverse) for microtuble-associated protein 2 (MAP2); TTCGCGGGCGACGATGCTCC (forward) and 5′-CAGGTCCAGACGCAGGATGG (reverse) for β-actin; and 5′-CTCCGGTTAAATGGGAGTGG and 5′-CCCGGCACTAGTTACACTTG (reverse) for Histone H1.
Results
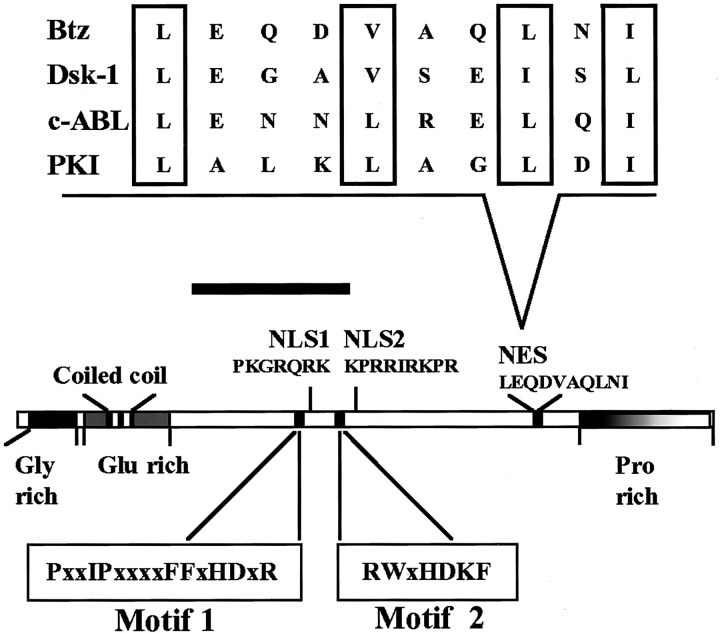
A recent study identified a novel protein called Barentsz (Btz), which is involved in the posterior localization of oskar mRNA in Drosophila and is a component of the oskar RNA transport machinery together with Staufen protein (van Eeden et al., 2001). In an attempt to further characterize the function and composition of the Staufen-containing RNPs in vertebrate neurons (Köhrmann et al., 1999a; Kiebler and DesGroseillers, 2000; Krichevsky and Kosik, 2001; Ohashi et al., 2002; Mallardo et al., 2003), we examined whether a mammalian homolog of Drosophila Barentsz (dBtz) has been conserved during evolution. The N-terminal part of dBtz protein showed significant homology to the human MLN51 protein whose mRNA is overexpressed in breast cancer cell lines (Tomasetto et al., 1995) and in gastric cancer (Varis et al., 2002). Using the EST data bank, we cloned the full-length mouse (accession number AF526276) and rat (accession number AF525467) homologs of dBtz. The mouse Btz transcript shows an open reading frame of 2094 nucleotides that encodes a protein of 698 aa and a predicted molecular mass of ∼76 kDa. The rat Btz protein consists of 699 aa and shares 97% of identity with its mouse homolog. A schematic representation of the rat Btz protein is given in Figure 1. The protein contains a glycine-rich region, a glutamine-rich region, a coiled-coil domain at the N terminal and a proline-rich region at the C terminal as in Drosophila. Four putative Src homology (SH)3 domain binding sites, one putative SH2 domain binding site, and two potential monopartite nuclear localization signals (NLSs) were predicted by Degot et al. (2002). In addition, one nuclear export signal (NES) consensus sequence is present at the C terminal (Fig. 1). Interestingly, one consensus sequence for the NLS appears to be conserved in Drosophila. However, the functionality of the NLS within dBtz has not yet been experimentally verified. The NES, in contrast, is present only in mBtz. Additionally, a thorough computer analysis revealed the existence of two conserved motifs that are present in the N terminal of MLN51 proteins and at the very C-terminal end of TRAP150 proteins (Fig. 1, supplementary data; available at www.jneurosci.org). No additional conserved domains were identified.
Figure 1.
Domain structure of mammalian Barentsz. Schematic representation of rat Btz protein. The different domains are indicated. Two putative NLSs and an NES have been predicted. The black bar indicates the region of homology to Drosophila Btz. A comparison of the NES sequences of mBtz, drosulfakinin (Dsk1), Abelson murine leukemia viral oncogene (c-ABL), and protein kinase inhibitor (PKI) is shown on the top of the figure. Hydrophobic residues important for NES activity are indicated by open boxes.
Expression of mBtz in different tissues
The expression of the mammalian Btz transcript was then tested by Northern blotting of different mouse tissues (Fig. 2A). A single transcript of ∼4.4 kb was found to be expressed at a high level in heart, brain, liver, kidney, and testis. Lower expression was detected in muscle, lung, and spleen. This fits well with the identification of a single 4.2 kb transcript of MLN51 in all of the breast cancer cell lines, many other human cell lines, and a wide variety of human tissues (Degot et al., 2002). Because we were interested in the expression of mBtz in the hippocampus, we performed in situ hybridization on mouse brain sections (Fig. 2B). Btz mRNA was detected in the hippocampus, in particular in the cell body layers of the dentate gyrus and the CA1 and CA3 regions (Fig. 2B1,2). Btz mRNA was also expressed in other regions of the brain, especially in the cerebellum (data not shown). No signal was detected in the hippocampus when the sense probe was used instead (Fig. 2B3).
Figure 2.

Expression of Btz mRNA in different tissues. A, Northern blot analysis of polyA mRNA from the following different mouse tissues: heart (He), brain (Br), spleen (Sp), lung (Lu), liver (Li), skeletal muscle (Mu), kidney (Ki), and testis (Te). Bottom panel,β-Actin control. B, Btz mRNA is expressed in the hippocampus. In situ hybridization on P14 mouse brain sections showing the expression of Btz mRNA. Coronal section of the hippocampus (1), including a high magnification insert (2). 3, In situ hybridization performed with the sense probe in the hippocampus.
mBtz is a nucleocytoplasmic shuttling protein
To determine the subcellular localization of Btz, we cloned the full-length cDNA of mouse Btz in front of the green fluorescent protein reporter (mBtz–GFP). When the resulting construct was transfected into BHK, mBtz–GFP was almost excluded from the nucleus and showed a prominent cytoplasmic distribution, which was often concentrated at the perinuclear region (Fig. 3A, left panel, arrow). A similar expression pattern was observed when yellow fluorescent protein was cloned at the N terminus instead and expressed in BHK cells to exclude the possibility that the position of the added tag influences the intracellular localization of the fusion protein (data not shown). Because the Btz protein contains both predicted NLS and NES domains, we wanted to test whether Btz enters the nucleus or not. LMB specifically inhibits chromosome region maintenance 1 (CRM1)-dependent protein export from the nucleus (Fukuda et al., 1997). LMB treatment led to the accumulation of Btz–GFP in the nucleus (Fig. 3A, right panel). The same results (Fig. 3A, bottom panels) were obtained using axin1 as a positive control, a CRM1-dependent nucleocytoplasmic shuttling protein (F. Fagotto, unpublished observations). These results suggested that both the predicted NLS and the NES in mammalian Btz are indeed functional. To further test this hypothesis, a series of deletion constructs were expressed in BHK cells, and their intracellular localization was analyzed. In contrast to wild-type Btz (Fig. 3B, row i), a truncated Btz–GFP fusion protein that contains the two putative NLSs but lacks the putative NES accumulated within the nucleus (Fig. 3B, row ii). To further map the region of Btz required for nuclear import, we fused GFP to a series of N-terminal fragments. Btz1–158–GFP was not imported into the nucleus (Fig. 3B, row iii); the Btz152–359–GFP, in contrast, ended up exclusively in the nucleus (Fig. 3B, row iv). These results indicate that the two NLSs are functional within Btz. In addition, a signal appears to exist within the N-terminal first 159 aa that could potentially interact with an unknown factor in the cytoplasm partially overriding the exclusive nuclear localization of Btz152–359–GFP; this is currently under investigation.
Figure 3.

Leptomycin B-sensitive export of mBtz protein. A, LMB treatment of BHK cells expressing mBtz–GFP fusion protein (mBtz–GFP). The CRM1-dependent axin1 served as control (F. Fagotto, unpublished observations). Three hour treatment with LMB causes the retention of Btz protein within the nucleus. Arrows indicate the perinuclear and cytoplasmic pattern of Btz-GFP. The experiments were performed either in the absence (-LMB) or in the presence (+LMB) of LMB. Scale bar, 10 μm. B, Mapping of the nuclear localization and nuclear export signal of mammalian Btz. BHK cells transfected with Btz–GFP show cytoplasmic localization of the fusion protein (i). In the subsequent panels, cells were transfected with a vector expressing the N-terminal half of the protein fused with GFP, Btz1–359–GFP (ii), Btz1–158–GFP (iii), or Btz152–359–GFP (iv). The bottom panel (v) shows the nuclear accumulation of BtzL465A–GFP, in which leucine 465 of the putative NES has been mutated to alanine. Scale bar, 10 μm (for all images in B). C, Expression of Btz–GFP in transiently transfected hippocampal neurons confirms the phenotype observed in transfected BHK cells. GFP fluorescence is shown on the left, whereas both the respective 4′,6′-diamidino-2-phenylindole (DAPI) staining and phase contrast (PC) are shown on the right. Transfected glia are shown in the small panels. Arrowheads indicate the particulate Btz–GFP structures observed in dendrites. The inset represents a high magnification with increased exposure time of the area indicated by the dotted square. Scale bar, 10 μm (for all images in C).
To further investigate whether the putative NES is indeed functional, we introduced a point mutation within the NES consensus region (L458xxxV462xxL465xI467) by changing the indicated leucine 465 (Fig. 1) to alanine. The mutant BtzL465A–GFP now accumulated in the nucleus because of the abolished nuclear export (Fig. 3B, row v). To verify that Btz is behaving as a nucleocytoplasmic shuttling protein in nerve cells also, we transiently transfected hippocampal neurons (Köhrmann et al. 1999b) with constructs expressing either mBtz–GFP or N-terminal portions of mBtz tagged with GFP (Fig. 3C). Neurons expressing mBtz–GFP showed perinuclear and cytoplasmic staining (Fig. 3C, first panel); signals were also detected in dendrites as discrete punctate structures when images were acquired at high magnification and increased exposure time (Fig. 3C, top inset). When neurons were transfected with the domain of mBtz containing the two NLSs (Btz152–359–GFP), the recombinant protein accumulated in the nucleus (Fig. 3C, fourth panel). The expression of the N-terminal half of Btz (Btz1–359) led to both cytoplasmic and nuclear localization (Fig. 3C, second panel); in addition, the expression of Btz1–158–GFP caused cytoplasmic localization of the recombinant protein (Fig. 3C, third panel). The same results were observed in transfected glial cells (small insets on the right). Together, these experiments indicate that both the NLS and the NES sequences in Btz are functional and that Btz is a nucleocytoplasmic shuttling protein in mammalian cells.
mBtz interacts with Staufen in the Drosophila oocyte
To test whether mBtz is a functional homolog of the Drosophila protein, we analyzed whether expression of mBtz fused to GFP (mBtz–GFP) in btz1 mutant egg chambers rescued the phenotype. In most btz1 homozygous egg chambers, both Staufen protein and oskar mRNA failed to localize to the posterior pole of the oocyte at stage 9 and remained at the anterior pole instead, until the beginning of stage 10B. However, occasionally, some oskar mRNA may localize to the posterior pole at stage 9 and become anchored there throughout oogenesis (van Eeden et al., 2001). When mBtz–GFP was expressed in wild-type chambers, its localization was indistinguishable from the endogenous dBtz protein (Fig. 4A). mBtz–GFP accumulated in the oocytes in the germarium and to the posterior crescent at stage 5 (Fig. 4A, left panel). Furthermore, mBtz also concentrated around the nuclei of the nurse cells (Fig. 4A, middle panel). Finally, mBtz localized to the posterior pole of the stage 9 oocyte when oskar mRNA is transported to this pole (Fig. 4A, right panel, arrow; B, top panel). In heterozygous flies, mBtz was also detected at the posterior pole, most likely in a complex with Staufen (Fig. 4B, top panels). In the btz1 background (Fig. 4B, middle panels), however, mBtz and Staufen did not localize to the posterior pole and were instead found at the anterior pole of the oocyte. The complex of mouse Btz–Stau and probably oskar mRNA entered the oocyte (they did not stay in the nurse cells), but the final anterior to posterior transport was blocked. Together, all of these results show that mBtz is able to interact in vivo with Drosophila Staufen and is a component of the oskar mRNA complex but is not able to function in the transport of oskar mRNA to the posterior pole of the oocyte.
Figure 4.
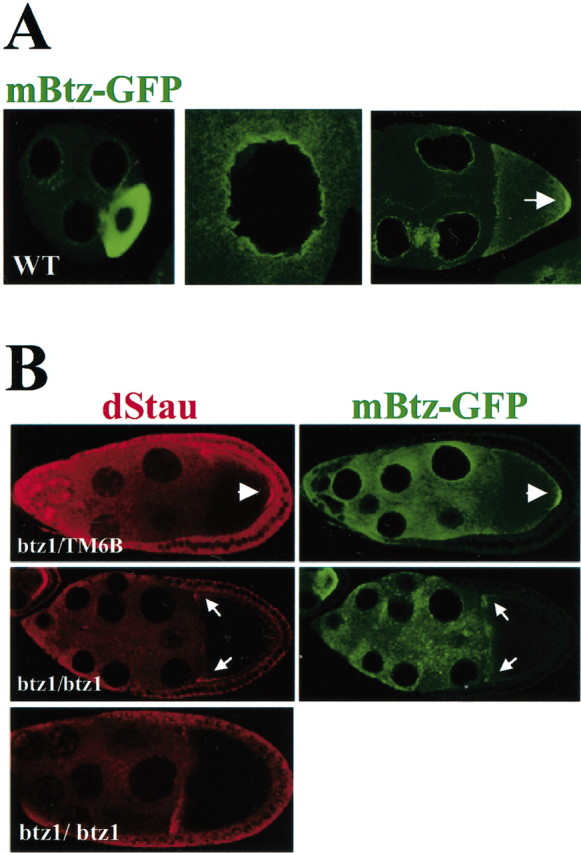
Mammalian Btz colocalizes with Staufen and oskar mRNA in the Drosophila oocyte. A, Localization of mBtz–GFP in the wild-type (WT) Drosophila germ line. Similar to Drosophila Btz, mBtz accumulates in the stage 5 oocyte (left panel), in a rim around the nuclei of the nurse cells (middle panel), and at the posterior pole of the stage 9 oocyte (right panel, arrow), where both Staufen and oskar mRNA localize. B, Localization of Drosophila Staufen (dStau) (red) and mBtz–GFP (green) in btz1/TM6B and btz1 mutant egg chambers expressing mBtz–GFP (top and middle panels, respectively). The arrowheads indicate the colocalization of Drosophila Staufen and mBtz–GFP proteins to the posterior pole. The bottom panel shows Staufen localization in btz1 mutant egg chambers that do not express mBtz–GFP. In the btz1 mutant oocyte, both Staufen and mBtz–GFP do not accumulate at the posterior pole but instead localize at the anterior pole (middle panel, arrows). Thus, mBtz–GFP is unable to rescue the oskar mRNA localization phenotype in barentsz mutant egg chambers.
Btz is present in Staufen1 particles in dendrites of hippocampal neurons
To further investigate the role of mammalian Btz, we raised polyclonal antisera against several domains of the protein in rabbits and mice and tested its expression level in the brain. On Western blots of rat brain extract, the antibodies specifically recognized a band of ∼115 kDa (Fig. 5A, left panel, I), higher then the predicted molecular mass of 76 kDa. This abnormal electrophoretic mobility, however, had also been observed for Drosophila Btz (van Eeden et al., 2001) as well as for human MLN51 protein (Degot et al., 2002). The preimmune serum, in contrast, gave no specific bands (Fig. 5A, left panel, PI). We then went on to affinity-purify the Btz antisera. As indicated in the right lane of Figure 5A (Pur.), these antibodies were monospecific for the 115 kDa band. A second higher band was detected (Fig. 5A, arrowhead); this corresponds to a phosphorylated form of Btz, because the treatment with alkaline phosphatase abolished this band (data not shown). To exclude possible posttranslational modifications of the Btz protein, we performed an in vitro translation and subsequent SDS-PAGE (Fig. 5A, right panel). A single band was observed at 115 kDa, proving that the antibodies specifically recognize full-length Btz protein. Furthermore, on Western blots from BHK cell extracts and different neuronal tissue samples (Fig. 5B), the antibodies recognized in all of the cases a protein with the same molecular mass that has the same electrophoretic mobility as the untagged, full-length Btz protein overexpressed in BHK cells (Fig. 5B, right panel). Together, these experiments indicate that the antibodies specifically identify the 115 kDa protein as mBtz protein. These results encouraged us to investigate whether mBtz and Staufen also interact in mammalian cells. As an experimental system, we chose isolated hippocampal neurons in culture in which Staufen is expressed in RNA-containing particles in the somatodendritic domain (Kiebler et al., 1999). Immunocytochemistry was performed on mature hippocampal neurons using anti-mBtz antibodies. When we focused on the cell body of these cells (Fig. 6A), Btz is predominantly found in the cytoplasm with a strong perinuclear and granular localization pattern (Fig. 6A, top panel). The localization of the endogenous Btz protein was then compared with the expression pattern of Btz–GFP in transfected hippocampal neurons. Both patterns were remarkably similar (Fig. 6A, bottom panel). In particular, mBtz–GFP formed granules in the perinuclear region (Fig. 6A, inset), indicating that our reporter protein actually behaved like the endogenous counterpart. Similar results were obtained by transfecting hippocampal neurons with constructs expressing untagged mBtz protein (data not shown). Confocal microscopy showed that Staufen1 and mBtz colocalized in the perinuclear endoplasmic reticulum (ER) region. A punctate expression pattern of Btz was also detected in the dendritic compartment (Fig. 6 B). High magnifications of two representative dendrites (proximal–top to distal–bottom) are shown in Figure 6C. Interestingly, a nonhomogeneous pattern of colocalization was observed depending on the distance of the dendritic particle from the cell body. A quantitative analysis of the confocal images from three different neurons (Fig. 6D) revealed that an almost complete colocalization between mBtz and Stau1 was observed in the cell body segment (>93 ± 5.54%), in agreement with the nucleocytoplasmic shuttling activity of mBtz. A significant colocalization (38 ± 15.2%; up to 70 ± 17.86%) was still detected in the proximal dendritic compartment (≤1 cell body in distance from the soma) that was decreasing in distal dendrites (32 ± 17.4%; up to 45 ± 22.2%) with increasing distance from the cell body. To prove that mBtz and Staufen1 are indeed interacting in hippocampal neurons, we cotransfected neurons with constructs expressing mBtz and Staufen1–HA (Fig. 6E). Again, both proteins were recruited into the same, discrete particular structures in the cell body as well as in dendrites (Fig. 6E). This suggests that both proteins are actually part of the same transport machinery.
Figure 5.

Mammalian Btz is a 115 kDa protein. A, Western blot of extracts from hippocampal neurons probed with either preimmune (PI) (lane 1) or the corresponding immune antisera. Lane 2, A monospecific mouse polyclonal antisera (I); lane 3, affinity-purified (Pur.), rabbit polyclonal anti-Btz. Our different monospecific polyclonal antisera recognize a 115 kDa protein; this is in good accordance with the observed molecular mass of Drosophila Btz, which runs as a 125 kDa protein. Right panel, In vitro translation experiment showing that mBtz protein migrates as a 115 kDa protein. A negative control (-) was performed without DNA. The arrowhead identifies the phosphorylated form of mBtz. B, Western blot using extracts of different tissues. The antibodies recognize the protein overexpressed in BHK (right panel) but also the endogenous protein from BHK or from different neuronal tissues (left panel). αBtz, Anti-Btz antibodies.
Figure 6.

Intracellular localization of mBtz protein in hippocampal neurons. A, Endogenous mBtz and mBtz–GFP show a remarkably similar localization in the cell body of primary hippocampal neurons (10–12 DIV). Neurons were immunostained for mBtz protein (top panel) or transiently transfected with a construct encoding mBtz–GFP, and fluorescent images were taken from a representative cell (bottom panel). Both proteins yielded a typical punctate, perinuclear localization (see also inset). B, Confocal images from fixed mature hippocampal neurons in culture. Like Staufen1, mBtz forms particles that localize to the dendritic compartment. The immunostaining was performed with affinity-purified rabbit polyclonal anti-mBtz antibodies. C, Higher magnifications of confocal microscopical images. Two different regions of dendrites (proximal–top to distal–bottom) are shown. Hippocampal neurons were labeled with immunopurified rabbit polyclonal anti-mBtz (green) and mouse anti-Stau1 (red). D, Quantitative analysis to evaluate the colocalization between mBtz and Stau1. Three different regions (shown in the schematic drawing) were chosen. The three-column set represents quantifications from three distinct neurons. The number on the top of each column indicates the number of areas analyzed per region. E, In vivo colocalization of Staufen1 and mBtz in transfected hippocampal neurons. Mature neurons were transiently transfected with constructs encoding mBtz and Staufen1–HA, and then immunostaining was performed. Fluorescent images were taken from three different cells. Panels marked Merge in B and E are an overlay of the two images to the left. αBtz, Anti-Btz antibodies; αStau1, anti-Staufen1 antibodies.
mBtz interacts with Staufen in an RNA-dependent manner
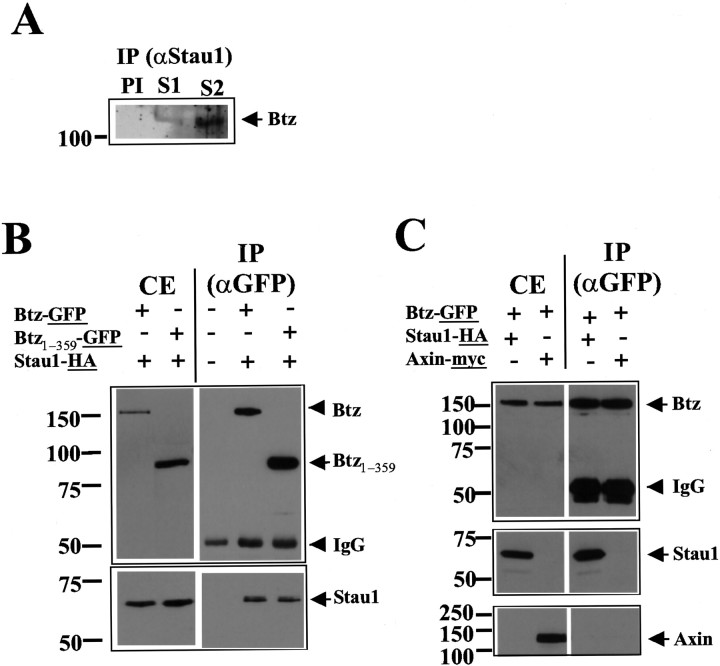
To verify whether Staufen1 and Btz interact, we performed immunoprecipitation experiments on brain extracts using two different anti-Staufen1 polyclonal antibodies (Fig. 7A, S1 and S2). The immunoprecipitates obtained were then subjected to SDS-PAGE followed by Western blot. As shown in Figure 7A, Btz was specifically coimmunoprecipitated with Staufen1; no coimmunoprecipitation was observed when the preimmune (PI) serum was used instead. These results suggest that mBtz is a new component of dendritic RNPs.
Figure 7.
mBtz coimmunoprecipitates with Staufen1. A, Endogenous Staufen1 immunoprecipitates Btz. IPs were performed with two different antisera (S1, S2), and mBtz was detected with a specific antibody. As negative control, the IP was performed with the preimmune serum (PI). B, C, In vivo Staufen1–mBtz interaction. BHK cells were cotransfected with different constructs and lysed, and immunoprecipitations were performed as indicated. B, Btz–GFP and Btz1–359–GFP coimmunoprecipitate with Stau1–HA. The left panel includes the indicated cell extracts (CE) showing the respective molecular masses of mBtz–GFP and Staufen1–HA. IPs (right panel) were performed with anti-GFP antibodies (αGFP), and proteins were visualized by Western blotting (underlined tags indicate the various antibodies). C, The interaction of mBtz is specific for Staufen1. Cotransfection of BHK cells with constructs expressing both mBtz–GFP and axin1–myc did not result in a coimmunoprecipitation of the two recombinant proteins. αStau1, anti-Staufen1 antibodies.
To further investigate the specificity of the molecular interaction of mBtz and Staufen1 in vivo, we performed coimmunoprecipitation assays from BHK cells cotransfected with tagged proteins. In detail, the following constructs coding for Staufen1–HA3 (Stau1–HA) and for either full-length Btz (Btz–GFP or untagged Btz) or truncated Btz (Btz1-359–GFP) were used. The latter construct contains the N-terminal part of mBtz including the homologous domain with Drosophila Btz (van Eeden et al., 2001) (Fig. 1, supplementary data). After cotransfection of BHK cells, IP assays were performed using either anti-GFP (Fig. 7B) or anti-HA antibodies (data not shown), and the immunoprecipitated proteins were visualized by Western blotting using anti-HA, anti-GFP, anti-Btz, or anti c-myc antibodies (Fig. 7C). Stau1–HA interacted with both full-length Btz–GFP and the N-terminal Btz1-359–GFP fusion proteins indicating that both Staufen1 and Btz are components of a common protein complex (Fig. 7B). Similar results were obtained when the coimmunoprecipitation was done using anti-HA antibodies (data not shown); this confirmed that the order of IPs of both proteins was irrelevant in this assay. To investigate whether the observed interaction was specific for the two proteins analyzed, the following additional control IPs were performed. First, no coimmunoprecipitation was observed when the pull-down was performed with monoclonal anti-reticulin antibodies (data not shown). Second, GFP alone did not pull down endogenous Stau1 (data not shown) or Stau1–HA protein (Fig. 8B). Finally, when BHK cells were cotransfected with Btz–GFP and another overexpressed protein (e.g., axin1–myc instead of Stau1–HA), Btz–GFP specifically interacted with Staufen1–HA, but not with axin1–myc, in these coimmunoprecipitation assays (Fig. 7C). Together, these experiments indicate that Staufen1 and Btz proteins specifically interact with each other in mammalian cells.
Figure 8.

The interaction of Btz and Staufen1 is RNA dependent. A, RNA-dependent interaction of mBtz with organelles. RNase treatment of S14 supernatants from 3-week-old rat forebrain extracts releases mBtz from the P100 (P) into the soluble fraction (S). DNase treatment, in contrast, does not change the distribution of mBtz protein. The same membrane was redecorated with monoclonal anti-calnexin antibodies [kind gift of Ari Helenius (Eidgenössische Technische Hochschule, Zürich, Switzerland)] to show that the ER marker calnexin is not removed by RNase treatment. B, mBtz interacts with Staufen1 in an RNA-dependent manner. BHK cells were cotransfected and processed as indicated in Figure 7. Extracts were pretreated with RNase where indicated, and IPs were performed with anti-GFP antibodies. In this experiment, Btz–GFP and Stau1–HA only coimmunoprecipitate in the presence of RNA. Treatment with RNase A before the IP abolishes this interaction. As a control, GFP alone does not interact with Staufen1. The underlining indicates the portion of the antigen recognized by the antibodies. C, The conserved central domain of mammalian Btz is responsible for the RNA-dependent interaction with Staufen1. Different constructs coding for various parts of Btz were expressed together with Stau1–HA in BHK cells, and IPs were performed using anti-GFP antibodies in the presence or absence of RNase. The region from amino acid 152–359 in mouse Btz protein mediates the interaction with Stau1–HA. This domain contains the two identified motifs (1 and 2) (Fig. 1) that are conserved between Drosophila and mBtz proteins. CE, Cell extract; αGFP, Anti-GFP antibodies.
To investigate a possible role of mBtz in RNA transport and whether it is a component of Staufen-containing RNPs, we performed differential centrifugation of rat brain extract. The P100 and S100 derived from rat total brain were run on SDS-PAGE and blotted with anti-mammalian Btz antibodies. The majority of mBtz protein was associated with the pellet, indicating that it may be associated with intracellular membranes. Similar results have been obtained for Staufen proteins, because they are also primarily found in the P100-containing intracellular membranes and ribosomes (Ohashi et al., 2002; Mallardo et al., 2003). Interestingly, RNase treatment before differential centrifugation caused a significant release of both Staufen proteins as well as mBtz protein from the pellet into the S100 fraction (Fig. 8A). DNase treatment, in contrast, had no effect. These data strongly suggest that both Staufen1 and mBtz proteins may be components of the same particles that are associated with intracellular membranes or ribosomes, most likely ER, and that RNase treatment releases these particles into the supernatant. Calnexin, a known ER marker, served as membrane marker (Ou et al., 1993).
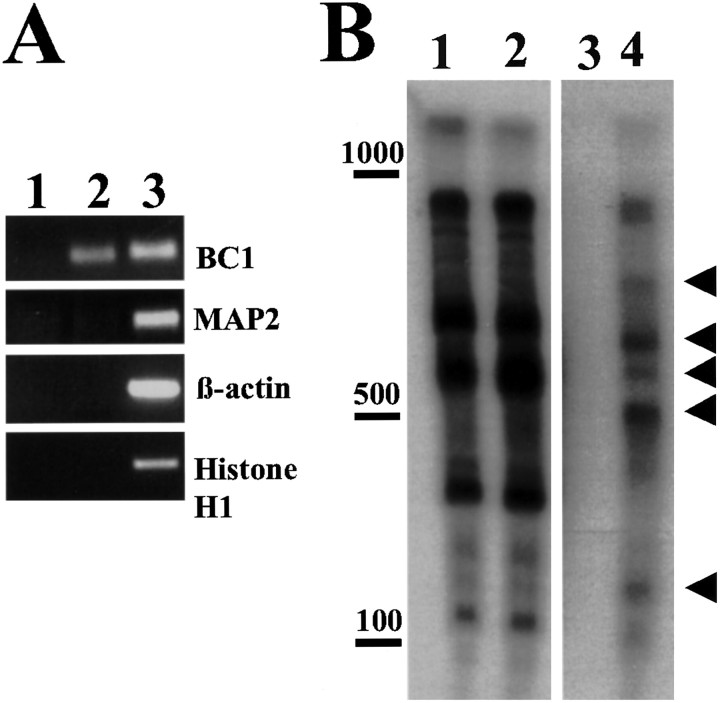
We therefore asked the question whether the interaction of mBtz with Staufen1 in our coimmunoprecipitation assays might also be mediated by RNA. Interestingly, RNase treatment abolished Btz and Staufen1 interaction (Fig. 8B), confirming that RNA plays a role in the interaction. In contrast, DNase treatment did not significantly affect the Btz and Staufen1 interaction (data not shown). Because homology between Drosophila and mammalian Btz proteins is restricted to the N-terminal domain (Fig. 1, supplementary data), and the interaction of mBtz with Staufen1 appears to be conserved, we went on to analyze whether the N-terminal conserved region of mBtz was indeed responsible for the interaction with Staufen1. Coimmunoprecipitation assays with Staufen1–HA and several N-terminal domains of mBtz tagged with GFP (Fig. 8C) clearly showed that both Btz1–359 and Btz152–359–GFP fusion proteins still have the ability, as does full-length Btz–GFP, to interact with Stau1–HA in an RNA-dependent manner. In contrast, the Btz fragment encompassing the first 158 aa (Btz1–158–GFP), which does not contain the homology domain, did not coimmunoprecipitate with Staufen1 (Fig. 8C). Together, several conclusions can be drawn from these experiments. First, the interaction of mBtz with Staufen is mediated by the region homologous to Drosophila Btz, confirming that the interaction domain has been conserved during evolution. Second, our experiments suggest that mBtz selectively binds to Staufen1 in the RNA-bound conformation and then possibly forms RNA-containing Staufen1 complexes in mammalian cells. To determine whether localized mRNAs were physically associated with the endogenous mBtz complex, as in the case of Staufen1 (Mallardo et al., 2003), we performed RT-PCR on immunoprecipitates from rat brain extracts using anti-mBtz antibodies. Primers specific for localized RNAs [e.g., BC1, MAP2, or Ca2+-calmodulin kinase IIα (CaMKIIα) (data not shown)] were used. As negative controls, primers were designed to detect both β-actin and Histone H1 mRNAs (Fig. 9A). The IPs that were performed either in the absence of the primary immunopurified antibodies or in the presence of the preimmune rabbit serum (data not shown) did not reveal any PCR-amplified cDNA product (lane 1). In contrast, BC1 transcript (lane 2) was clearly detected in the mBtz immunoprecipitates. In contrast, MAP2 was not detected (Fig. 9A). Both β-actin and Histone H1 transcripts were also not amplified from the immunoprecipitates (Fig. 9A). To check for the presence of additional mRNAs in the mBtz immunoprecipitates, radioactive RT-PCR was performed (Mallardo et al., 2003). The first two lanes of Figure 9B represent mRNAs derived from S100 supernatants from either protein A-Sepharose alone (lane 1) or mBtz antibodies bound to protein A-Sepharose (lane 2). The observed pattern represents cytosolic mRNAs. In contrast, a distinct set of bands was detected in the Btz immunoprecipitate (Fig. 9B, lane 4, arrowheads). The control lane 3 did not yield any band, indicating that no RNA binding protein binds to protein A-Sepharose. Whether these unknown bands actually represent potential cargo RNAs for the Btz–Staufen RNPs will be the subject of additional investigation.
Figure 9.
Characterization of RNA in mBtz immunoprecipitates. A, The dendritically localized BC1 transcript is present in the mBtz IPs. RNA isolated from the mBtz IPs was retrotranscribed, and the resulting cDNA was amplified using primers specific for either dendritically localized (BC1, MAP2) or nonlocalized (β-actin, Histone H1) transcripts. RNA was collected from IPs performed either in absence of antibodies (lane 1) or with immunopurified anti-mBtz polyclonal antibodies (lane 2) and then retrotranscribed, and the indicated cDNAs were detected by PCR. As positive controls, PCRs were also performed on cDNA from S100 (lane 3). B, Radioactive labeling of mRNAs presentin the mBtz IPs. Radioactive RT-PCR was performed on RNA extracted from either mBtz immunoprecipitates (lane 4) or the corresponding supernatant (lane 2). As negative control, the anti-mBtz antibodies were omitted for the IPs (lane3) to detect for mRNAs unspecifically bound to protein A-Sepharose beads. Lane 1 represents the corresponding supernatant. Arrowheads label a distinct set of bands that were specifically enriched in the mBtz IPs (lane 4).
Discussion
The purpose of this study was to identify additional proteins that interact with mammalian Staufen1. An attractive candidate for such an interactor is the mammalian homolog of Drosophila Btz, a protein component of the oskar RNPs that also contain Staufen, Mago nashi, Y14, and kinesin heavy chain (KHC) protein (van Eeden et al., 2001, and references therein). In btz1 homozygous mutants, these RNPs got stuck on their way to the posterior pole in the Drosophila oocyte. van Eeden et al. (2001) first pointed out the partial homology of Drosophila Btz with a previously identified mouse protein called MLN51. Its mRNA was found to be upregulated in cell lines derived from human metastatic breast and gastric cancer (Tomasetto et al., 1995; Varis et al., 2002). These facts prompted us to investigate whether MLN51 might represent a functional homolog of mBtz protein. Btz mRNA is highly expressed in the hippocampus and cerebellum where Staufen1 is also expressed (Monshausen et al., 2001; Duchaîne et al., 2002). When mBtz protein is made in the wild-type Drosophila egg chamber, it shows identical localization to the Drosophila protein in the female germline; Btz–GFP is detected in the perinuclear region of the nurse cells and localizes, together with Drosophila Staufen, to the posterior pole of the oocyte with the oskar mRNA localization complex. In btz mutant egg chambers, however, the mammalian protein can still enter the oocyte but remains at the anterior pole together with Staufen. This result is the first strong hint that MLN51 is the mammalian homolog of Btz protein, because it can still interact, through its conserved domain, with Drosophila Staufen. On the basis of these data, it is tempting to speculate that the apparent lack of ability to reach the posterior pole in the btz mutant oocyte is attributable to the difference within the C terminal of Btz. This C-terminal region is not conserved in mammals, indicating a significant divergence during evolution.
In Drosophila, most Btz is associated with nuclear envelope membranes of the nurse cells and joins the oskar mRNA–Mago nashi protein complex as it is exported into the cytoplasm. In mammals, in contrast, Btz seems to behave as a nucleocytoplasmic shuttling protein. First, treatment with LMB, a drug that specifically inhibits CRM1-dependent protein export from the nucleus (Fukuda et al., 1997), led to the accumulation of Btz–GFP in the nucleus. Second, the removal of either the two NLSs or the NES inhibited its cytosolic localization in mammalian fibroblasts. In addition to these sequence elements, there may be an additional domain at the very N terminal, located within the first 152 aa, of mammalian Btz protein that may be responsible for its cytoplasmic localization. Only if this domain is missing is the protein found exclusively in the nucleus of mammalian fibroblasts.
As in the case of the Drosophila Btz, the sequence of the mammalian homolog gave few clues to its biochemical functions. We therefore investigated whether mammalian Btz is still able to form a complex with mammalian Staufen1. In mammalian neurons, Staufen1–GFP forms particles in the cytoplasm that are associated with the nuclear–ER membrane system. Some of the smaller particles subsequently move in a saltatory manner along microtubules into dendrites of living hippocampal neurons (Köhrmann et al. 1999a). In this respect, it is of particular interest that both the endogenous and the recombinant Btz protein, as well as Staufen1, formed granules that can be found in the cell body and to a lesser extent in dendrites of hippocampal neurons. Most endogenous Btz protein, in comparison, not only localized in a ring shape around the nuclear membrane, but some staining was also detected in dendrites (Fig. 6). When we carefully evaluated the degree of colocalization between mBtz and Staufen1, we observed a somewhat dynamic pattern. Whereas the degree of colocalization was highest within the cell body, it gradually decreased with progressive distance of the particles from the cell body toward distal dendrites. There could be several reasons for the observed phenomenon. First, the interaction of mBtz with Staufen1 might be highly dynamic and transient as in the case of Drosophila. Here, Drosophila Btz is only transiently associated with the oskar mRNA localization complex and is only found at the posterior pole when oskar mRNA localizes there (van Eeden et al., 2001). In the case of mature hippocampal neurons, this could indicate that mBtz only transiently associates with Staufen RNPs until they reach their final destination in dendrites. Therefore, the degree of colocalization progressively decreases because of the partial disassembly of mBtz from particles in dendrites. Alternatively, Btz could interact additionally with other double-stranded RNA binding proteins (e.g., Staufen2). Interestingly, a drastic increase in the degree of colocalization in Staufen1-containing particles was observed when both mBtz and Staufen1 proteins were overexpressed in hippocampal neurons (Fig. 6E). This apparent difference between endogenous and overexpressed Btz protein could be explained by the fact that excess of both Staufen1 and Btz may yield significantly larger particles that will then be subsequently transported into dendrites. The dynamics of the dendritic transport of Btz-containing particles is currently under investigation.
The mBtz–Staufen1 interaction was then further validated by immunoprecipitations from doubly transfected BHK cells. These experiments showed that mBtz interacts with Staufen1 through its conserved N-terminal domain. A thorough computer analysis revealed the existence of two conserved motifs within this domain, which are also present in other protein families. In this respect, it is of particular interest that mBtz protein does not share any other conserved domain, and that it is characterized by an unusual absence of any significant secondary structure (data not shown). Interestingly, mBtz and Staufen1 do not seem to bind directly to each other, but this interaction appears to be mediated by RNA. This is in good accordance with the situation in Drosophila, in which both proteins assemble with oskar mRNA into RNPs (van Eeden et al., 2001). Preliminary evidence suggests that mBtz does not bind synthetic double-stranded RNA in an in vitro Northwestern assay (data not shown). Therefore, the RNA-dependent association is mediated by either Staufen1 or, alternatively, another RNA binding protein present in the complex. Additional work is required to identify the molecular nature of such additional RNA binding proteins present in Staufen-containing particles.
Unfortunately, the identity of the cargo RNA for mammalian Staufen proteins has not been revealed to date. A recent publication showed that Staufen1 binds to the dendritic targeting element of MAP2 mRNA in a trihybrid system (Monshausen et al., 2001). Whether this interaction actually occurs in vivo has not yet been demonstrated. Additional putative candidates for such cargo RNA (e.g., CaMKIIa, TrkB, and NMDAR1) have been characterized in a recent study by Krichevsky and Kosik (2001). The identification of cargo RNAs in Staufen RNPs, however, has been hampered by the fact that Staufen binds any double-stranded RNA in vitro (M. Kiebler, D. St. Johnston, L. DesGroseillers, unpublished observations). Therefore, we recently established a biochemical isolation procedure to enrich Staufen-containing RNPs from rat brain and to identify the bound RNA (Mallardo et al., 2003). This study showed that several dendritically localized RNAs are indeed enriched in the Staufen particles (e.g., the noncoding RNA BC1, CaMKIIα, as well as 15 novel RNAs) (M. Mallardo and M. Kiebler, unpublished observation). We succeeded in detecting one of these potential Staufen1 cargo RNAs, BC1, in our mBtz immunoprecipitates. Additionally, we present new evidence of the presence of a restricted number of novel, not-yet-characterized mRNAs that are associated with mBtz. The detection of BC1 RNA together with the presence of additional mRNAs within both Staufen1 and mBtz complexes clearly strengthens the hypothesis that mBtz and Staufen may be part of the same particles, and that the observed interaction between these two proteins occurs in vivo. Additional work is needed, however, to characterize the RNAs that mediate the binding of Staufen1 to Btz and to unravel the molecular mechanism of this interaction.
Together, our results suggest the following scenario. Btz is exported from the nucleus into the cytoplasm as a component of the ribonucleoprotein complex. Staufen1 can then be recruited into this complex via its bound cargo RNA. Alternatively, the RNA-dependent interaction of mBtz with Staufen1 could actually be mediated by another RNA binding protein that possibly exists within such an RNP (Wilhelm et al., 2000; Ohashi et al., 2002; Arn et al., 2003). This complex can then be transported into the dendritic compartment along microtubules with the help of KHC (S. Kroening, unpublished observations).
Footnotes
This work was supported by PhD fellowships from the Italian government (S.B.) and the Claussen-Simon-Stiftung (S.K.), a Royal Society Dorothy Hodgkin postdoctoral fellowship (I.M.P.), the Sonderforschungsbereich SFB446 (P.M. and M.K.), and the Human Frontier Science Programme network grant (D.S.J. and M.K.). We thank Drs. L. DesGroseillers, M. Mallardo, and F. van Eeden for their critical comments.
Correspondence should be addressed to Dr. Michael Kiebler, Max-Planck-Institute for Developmental Biology, Spemannstrasse 35, D-72076 Tübingen, Germany. E-mail: michael.kiebler@tuebingen.mpg.de.
S. Baldassa's present address: Università degli Studi di Milano, Dipartimento di Fisiologia e Biochimica Generali, Milano, Italy.
Copyright © 2003 Society for Neuroscience 0270-6474/03/235778-11$15.00/0
References
- Arn EA, Cha BJ, Theurkauf WE, Macdonald PM ( 2003) Recognition of a bicoid mRNA localization signal by a protein complex containing Swallow, Nod, and RNA binding proteins. Dev Cell 4: 41-51. [DOI] [PubMed] [Google Scholar]
- Bashirullah A, Cooperstock RL, Lipshitz HD ( 1998) RNA localization in development. Annu Rev Biochem 67: 335-394. [DOI] [PubMed] [Google Scholar]
- Bassell GJ, Oleynikov Y, Singer RH ( 1999) The travels of mRNAs through all cells large and small. FASEB J 13: 447-454. [DOI] [PubMed] [Google Scholar]
- Degot S, Regnier CH, Wendling C, Chenard MP, Rio MC, Tomasetto C ( 2002) Metastatic Lymph Node 51, a novel nucleo-cytoplasmic protein overexpressed in breast cancer. Oncogene 21: 4422-4434. [DOI] [PubMed] [Google Scholar]
- Duchaîne T, Wang HJ, Luo M, Steinberg SV, Nabi IR, DesGroseillers L ( 2000) A novel murine Staufen isoform modulates the RNA content of Staufen complexes. Mol Cell Biol 20: 5592-5601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duchaîne TF, Hemraj I, Furic L, Deitinghoff A, Kiebler MA, DesGroseillers L ( 2002) Staufen2 isoforms localize to the somatodendritic domain of neurons and interact with different organelles. J Cell Sci 115: 3285-3295. [DOI] [PubMed] [Google Scholar]
- Ephrussi A, Dickinson LK, Lehmann R ( 1991) Oskar organizes the germ plasm and directs localization of the posterior determinant nanos. Cell 66: 37-50. [DOI] [PubMed] [Google Scholar]
- Erdelyi M, Michon AM, Guichet A, Glotzer JB, Ephrussi A ( 1995) Requirement for Drosophila cytoplasmic tropomyosin in oskar mRNA localization. Nature 377: 524-527. [DOI] [PubMed] [Google Scholar]
- Fukuda M, Asano S, Nakamura T, Adachi M, Yoshida M, Yanagida M, Nishida E ( 1997) CRM1 is responsible for intracellular transport mediated by the nuclear export signal. Nature 390: 308-311. [DOI] [PubMed] [Google Scholar]
- Kiebler MA, DesGroseillers L ( 2000) Molecular insights into mRNA transport and local translation in the mammalian nervous system. Neuron 25: 19-28. [DOI] [PubMed] [Google Scholar]
- Kiebler MA, Hemraj I, Verkade P, Köhrmann M, Fortes P, Marión RM, Ortín J, Dotti CG ( 1999) The mammalian Staufen protein localizes to the somatodendritic domain of cultured hippocampal neurons: implications for its involvement in mRNA transport. J Neurosci 19: 288-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim-Ha J, Smith JL, Macdonald PM ( 1991) oskar mRNA is localized to the posterior pole of the Drosophila oocyte. Cell 66: 23-35. [DOI] [PubMed] [Google Scholar]
- Knoell B, Isenmann S, Kilic E, Walkenhorst J, Engel S, Wehinger J, Bähr M, Drescher U ( 2001) Graded expression patterns of ephrin-As in the superior colliculus after lesion of the adult mouse optic nerve. Mech Dev 106: 119-127. [DOI] [PubMed] [Google Scholar]
- Köhrmann M, Luo M, Kaether C, DesGroseillers L, Dotti CG, Kiebler MA ( 1999a) Microtubule-dependent recruitment of Staufen-green fluorescent protein into large RNA-containing granules and subsequent dendritic transport in living hippocampal neurons. Mol Biol Cell 10: 2945-2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhrmann M, Haubensak W, Hemraj I, Kaether C, Lessmann VJ, Kiebler MA ( 1999b) Fast, convenient, and effective method to transiently transfect primary hippocampal neurons. J Neurosci Res 58: 831-835. [PubMed] [Google Scholar]
- Krichevsky AM, Kosik KS ( 2001) Neuronal RNA granules: a link between RNA localization and stimulation-dependent translation. Neuron 32: 683-696. [DOI] [PubMed] [Google Scholar]
- Macchi P, Hemraj I, Goetze B, Grunewald B, Mallardo M, Kiebler MA ( 2003) A GFP-based system to uncouple mRNA transport from translation in a single living neuron. Mol Biol Cell 14: 1570-1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallardo M, Deitinghoff A, Müller J, Goetze B, Macchi P, Peters C, Kiebler MA ( 2003) Isolation and characterization of Staufen-containing ribonucleoprotein particles from rat brain. Proc Natl Acad Sci USA 100: 2100-2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micklem DR, Dasgupta R, Elliott H, Gergely F, Davidson C, Brand A, Gonzalez-Reyes A, St Johnston D ( 1997) The mago nashi gene is required for the polarisation of the oocyte and the formation of perpendicular axes in Drosophila. Curr Biol 7: 468-478. [DOI] [PubMed] [Google Scholar]
- Monshausen M, Putz U, Rehbein M, Schweizer M, DesGroseillers L, Kuhl D, Richter D, Kindler S ( 2001) Two rat brain staufen isoforms differentially bind RNA. J Neurochem 76: 155-165. [DOI] [PubMed] [Google Scholar]
- Monshausen M, Rehbein M, Richter D, Kindler S ( 2002) The RNA-binding protein Staufen from rat brain interacts with protein phosphatase-1. J Neurochem 81: 557-564. [DOI] [PubMed] [Google Scholar]
- Newmark PA, Boswell RE ( 1994) The mago nashi locus encodes an essential product required for germ plasm assembly in Drosophila. Development 120: 1303-1313. [DOI] [PubMed] [Google Scholar]
- Ohashi S, Koike K, Omori A, Ichinose S, Ohara S, Kobayashi S, Sato TA, Anzai K ( 2002) Identification of mRNP complexes containing pur alpha, mStaufen, fragile X protein and myosin Va, and their association with rough endoplasmic reticulum equipped with a kinesin motor. J Biol Chem 277: 37804-37810. [DOI] [PubMed] [Google Scholar]
- Ou WJ, Cameron PH, Thomas DY, Bergeron JJ ( 1993) Association of folding intermediates of glycoproteins with calnexin during protein maturation. Nature 364: 771-776. [DOI] [PubMed] [Google Scholar]
- Palacios IM, St Johnston D ( 2001) Getting the message across: the intracellular localization of mRNAs in higher eukaryotes. Annu Rev Cell Dev Biol 17: 569-614. [DOI] [PubMed] [Google Scholar]
- Palacios IM, St Johnston D ( 2002) Kinesin light chain-independent function of the Kinesin heavy chain in cytoplasmic streaming and posterior localisation in the Drosophila oocyte. Development 129: 5473-5485. [DOI] [PubMed] [Google Scholar]
- Roegiers F ( 2003) Insights into mRNA transport in neurons. Proc Natl Acad Sci USA 100: 1465-1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuler GD, Altschul SF, Lipman DJ ( 1991) A workbench for multiple alignment construction and analysis. Proteins 9: 180-190. [DOI] [PubMed] [Google Scholar]
- Smith WB, Aakalu G, Schuman EM ( 2001) Local protein synthesis in neurons. Curr Biol 11: R901-R903. [DOI] [PubMed] [Google Scholar]
- St Johnston D, Beuchle D, Nüsslein-Volhard C ( 1991) Staufen, a gene required to localize maternal RNAs in the Drosophila egg. Cell 66: 51-63. [DOI] [PubMed] [Google Scholar]
- Steward O, Schuman EM ( 2001) Protein synthesis at synaptic sites on dendrites. Annu Rev Neurosci 24: 299-325. [DOI] [PubMed] [Google Scholar]
- Tang SJ, Meulemans D, Vazquez L, Colaco N, Schuman E ( 2001) A role for a rat homolog of Staufen in the transport of RNA to neuronal dendrites. Neuron 32: 463-475. [DOI] [PubMed] [Google Scholar]
- Tetzlaff MT, Jaeckle H, Pankratz MJ ( 1996) Lack of Drosophila cytoskeletal tropomyosin affects head morphogenesis and the accumulation of oskar mRNA required for germ cell formation. EMBO J 15: 1247-1254. [PMC free article] [PubMed] [Google Scholar]
- Tomasetto C, Regnier C, Moog-Lutz C, Mattei MG, Chenard MP, Lidereau R, Basset P, Rio MC ( 1995) Identification of four novel human genes amplified and overexpressed in breast carcinoma and localized to the q11–q21.3 region of chromosome 17. Genomics 28: 367-376. [DOI] [PubMed] [Google Scholar]
- van Eeden FJ, Palacios IM, Petronczki M, Weston MJ, St Johnston D ( 2001) Barentsz is essential for the posterior localization of oskar mRNA and colocalizes with it to the posterior pole. J Cell Biol 154: 511-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varis A, Wolf M, Monni O, Vakkari ML, Kokkola A, Moskaluk C, Frierson Jr H, Powell SM, Knuutila S, Kallioniemi A, El-Rifai W ( 2002) Targets of gene amplification and overexpression at 17q in gastric cancer. Cancer Res 62: 2625-2629. [PubMed] [Google Scholar]
- Wilhelm JE, Mansfield J, Hom-Booher N, Wang S, Turck CW, Hazelrigg T, Vale RD ( 2000) Isolation of a ribonucleoprotein complex involved in mRNA localization in Drosophila oocytes. J Cell Biol 148: 427-440. [DOI] [PMC free article] [PubMed] [Google Scholar]