Abstract
The creation of technology that affords for the design of artificial enzymes is a new branch of biochemical engineering with the objective to solve the looming global catastrophe including food shortages, energy crisis, novel diseases, climate change and environmental degradation. However, the development of science and technology that will lead to the design of artificial enzymes depends on availability of scientists with a broad range of expertise including chemistry and physics of chemical bonding, structural biochemistry of macromolecular interactions, theoretical physics and mathematics with the focus on computer modeling of dynamic docking of macromolecules. Our previous experience in university STEM education led us to conclude that in order to train future scientists with a broad expertise in STEM, it is critical for high school students to learn interdisciplinary concepts of STEM courses at an earlier age. In this article, we describe the first phase of a STEM project that involved introducing students to STEM curriculum designed to steer high school students’ interest towards biochemical engineering and pharmacology. In addition, we present the outline of the STEM curriculum, along with user-friendly tutorials of AutoDock Vina, AutoDock Tools and PyMol programs that we designed to teach secondary STEM students computer modeling and docking of macromolecules. STEM high school students performed multiple exercises to understand how the potential pharmacological agents, cardiotoxins from cobra venom, interact with mitochondrial phospholipids in order to gain a deep understanding of elevated biophysical and biochemical concepts in protein drug interactions with biomembranes. We also present the results of evaluative assessments that tested students’ knowledge and skills that students gained following the completion of our pilot STEM course. In brief, the assessment results showed that the students successfully acquired a high level of understanding in structural biophysics and biochemistry. Importantly, this paper provides strong proof-of-concept that our pilot STEM curriculum can be successfully integrated in the traditional American and Chinese high school classroom. The curriculum and tutorials presented in this article could be used by college and high school teachers and students in STEM classes and to support undergraduate university courses in Pharmacology, Inorganic and Organic Chemistry, Biochemistry and Structural Biology for classroom instructions and homework assignments.
Keywords: STEM curriculum focused on Biochemistry and Pharmacology, Tutorials on docking of pharmacologically relevant cardiotoxins with mitochondrial phospholipids, Assessment of conceptual understanding of intermolecular interactions and mechanisms of pharmacologically potent natural toxins
4. Introduction
STEM education, which aims at steering young people towards careers in science, technology, engineering and mathematics, is becoming increasingly important because of rapid need and continuous development of new concepts in science to support technological breakthroughs needed to solve the looming global catastrophe including food shortages, energy crisis, novel diseases, climate change and environmental degradation. One of the fields of science which currently attracts a high level of attention is biochemical engineering. Experts in biochemical engineering are working on developing new technology capable of creating in vitro (outside the cell) analogues of important biological molecules. The technologies that afford for the creation of artificial enzymes seems to be one of the most important directions in this field [1–3].
Enzymes are macromolecular biological catalysts that accelerate chemical reactions in cells. Cells need metabolic pathways to increase the rates of reactions to a very high level in order to sustain life [4]. Most enzymes are proteins, although a few of them are ribozymes, which are catalytic RNA molecules. Enzymes catalyze more than 5,000 biochemical reaction types in cells [5] and they allow a reaction to occur in milliseconds that would otherwise take millions of years [6, 7]. Indeed, the incredible catalytic power of enzymes draws a high interest of the chemical industry and other industrial applications for developing artificial enzymes for biotechnological applications. However, the commercial use of enzymes faces challenges because enzymes catalyze the cellular reactions, which enzymes have been evolved to catalyze, and because the catalytic activity of enzymes work within a very narrow range of temperatures and pH, plus enzymes can denature in organic solvents. As a consequence, the engineering of artificial enzymes designed to catalyze non-cellular reactions of industrial importance is an active area of research that involves rational design of new enzymes capable of catalyzing specific reactions of commercial importance [1–3]. The research efforts in this field require a broad range of expertise including 1) structural biochemistry with a focus on basic physical and chemical principles of covalent and ionic bonds and wan der Waals forces including intra- and intermolecular interactions via hydrogen bonding, 2) theoretical physics and mathematics with a focus on computer modeling of macromolecular interactions, and 3) sophisticated computer programming skills. Contemporary computer programs allow to generate high-quality 3-D structures of proteins of up to about 150 amino acid residues [8] and can dock these proteins with smaller biological molecules termed “ligands” [9]. Although in silico docking of proteins with ligands does aid in visualizing the binding of a substrate to an enzyme, modern docking programs allow the docking of proteins containing rigid non-rotatable bonds [9]. This is a severe limitation of most docking programs given that enzymes are not rigid structures but have complex internal dynamic motions which are important in formation of the enzyme-substrate complexes [10].
In addition, the progress in the development of artificial enzymes engineering requires an effort in creating a set of algorithms that describe complex physico-chemical properties and parameters of intra- and intermolecular bonds and a complex set of factors that affect the motions of all individual bonds. Hence, a high level of understanding is needed for writing mathematical formulas describing changes in overall structural dynamics of the enzyme’s binding site(s) and of the active center in a process of binding to and complexing with a substrate. In addition, advanced computer programs have to be written for computing complex mathematical formulas that describe the complex dynamic interactions within the substrate-enzyme complex. This task requires a team of scientists each having an advanced training and expertise not only in structural biochemistry and physical chemistry of macromolecular bonding, but also in advanced computer programming.
Recently, we, Dr. E. S. Gasanoff and Dr. R. K. Dagda, were involved in an educational pilot STEM program in which university students in their third and fourth years of studies in the Applied Mathematics and Informatics Department (AMID) of the Lomonosov Moscow State University were taking two courses, 1) Physics of Chemical Bonding and 2) Structural Biochemistry, to supplement the standard courses in the AMID. The aim of the project was to assess whether senior students of the AMID can achieve high level of knowledge and skills in a field of science outside the expertise of the AMID, including physics of chemical bonding and structural biochemistry. After the AMID students have successfully completed the two aforementioned courses, the top ranked students were involved in performing research studies that analyzed interactions between the proteins with rigid, non-rotatable bonds and phospholipids with limited number of rotatable bonds using AutoDock Vina program [9] in collaboration between the University of Nevada Reno Medical School and the Lomonosov Moscow State University. In addition, the students were given the task to write a computer program in order to compute the docking of phospholipids and proteins with rotatable bonds. Although the students have made an ample contribution to the research, resulting in a series of research articles published in the top scientific journals in the field [11–14], students failed to formulate mathematical equations that describe physico-chemical parameters involved in dynamic interactions of macromolecules that contained rotatable covalent bonds. Overall, this project informed STEM curriculum experts that: although the selected senior students in the AMID have received excellent marks in the Physics of Chemical Bonding and Structural Biochemistry courses, the students were not able to achieve the high level of conceptual understanding related to the physical terms describing the dynamics of protein’s rotatable bonds and to the relevant macromolecular interactions. Overall, this project suggested that senior university students have already developed highly conceptualized analytical and critical thinking skills in an area of their main expertise – applied mathematics and informatics, which interfered with the development of high-level conceptual understanding in another area of specialized expertise – physics of chemical bonding. We further learned that: should the students develop the high-level conceptual understanding and advanced skills in several disciplines, the students should learn different areas of mathematics and science simultaneously at an earlier age – perhaps at the early years of undergraduate university or even in high/secondary school; that is, when students have not yet developed the high-level conceptualized understanding in one area of specialized expertise.
At the beginning of academic year 2018–2019, we launched the pilot STEM project in the secondary school of Chaoyang KaiWen Academy (Beijing, China) – a bilingual (Chinese and English) school that follows the Cambridge International Examinations curriculum. The aim of the project was to determine whether the academically advanced secondary students can achieve the high-level of understanding required in structural biochemistry/biophysics and computer programming by providing these courses consecutively in the secondary school with easy-to-understand, applied content. In the first phase of this pilot STEM project, we introduced secondary school STEM students (grades 7, 8 and 9) to a curriculum designed for the academically advanced secondary school students. This curriculum provides 72 hours of instruction and is based on a series of concepts, such as models of atomic structure, electronegativity of atoms, physical and chemical features of chemical bonding, 3-D structure of macromolecules, intermolecular interactions, computer modeling of proteins and in silico docking of pharmacologically relevant proteins with mitochondrial phospholipids. We then assessed the students’ knowledge and skills, which they gained after completing this curriculum designed for the first phase of pilot STEM project. The STEM students were given an assessment test that included docking of cardiotoxins with mitochondrial phospholipids, analysis of intermolecular attraction forces and discussion on potential physiological/pharmacological role(s) of cardiotoxins, which students successfully completed. The details of the test are given in the Results section of this article.
In this article we present an easy-to-follow curriculum that introduces STEM students to physics and chemistry of chemical bonding and structural biochemistry along with tutorial and protocols for performing molecular docking in order to gain skills in computational biology, chemistry and biophysics as part of our STEM pilot program. We also present here the user-friendly tutorials on AutoDock Vina, AutoDock Tools and PyMol programs. We have prepared these tutorials for teaching our STEM students to dock macromolecules and to analyze the docked sites on the surfaces of macromolecules. The curriculum and tutorials which we present in this STEM article could be used by other college and high school teachers and students in STEM classes and to support courses in Inorganic and Organic Chemistry, Biochemistry and Structural Biology for classroom instructions and homework assignments. We also present the results of assessment of students’ knowledge and skills that they have acquired after completing the first phase of our pilot STEM program.
5. Materials and Methods
5.1. Outline of the pilot STEM curriculum
We have developed a curriculum for the first phase of the pilot STEM project. The curriculum is designed for academically advanced secondary school students and is meant to supplement the curricula of the IGCSE Biology, IGCSE Chemistry and IGCSE Physics courses. To select academically advanced students, we screened 72 secondary school students from the grades 7, 8, 9 and 10 in the Chaoyang KaiWen Academy (Beijing, China). We selected 15 students who received high marks in science and mathematics. After interviewing these 15 students, we selected five students, two from grade 9, one from grade 8 and two from grade 7, who had strong motivation to take the STEM course with a curriculum we developed for the first phase of our pilot STEM project. From the selected five students, students from grade 7 have completed the Cambridge Lower Secondary Science, a student from grade 8 has completed the first year IGCSE Biology and was taking the second year IGCSE Biology and the first year IGCSE Physics, and students from grade 9 have completed IGCSE Biology and were taking the second year IGCSE Physics and the first year IGCSE Chemistry. After two weeks of studies one student from grade 9 has dropped the STEM course.
The content of the STEM curriculum we developed for the secondary school students in the first year of our pilot STEM project is given below:
THE STRUCTURE OF THE ATOM
Subatomic particles
Atomic nucleus and electron energy shells
The Periodic Table of the Elements and electron energy shells
The Periodic Table of the Elements and electron configurations
BACIS CONCEPTS OF CHEMICAL BONDS
Ionic bond
- Covalent bond
- Exceptions to the Octet Rule
- Co-ordinate (or dative) covalent bond
SHAPES OF MOLECULES WITH COVALENT BONDS
Electron-pair repulsion theory
The shape and bond angles of covalently bonded molecules
Lone pair, bond pair and the order of repulsion
ELECTRONEGATIVITY AND CHEMICAL BONDS
Pauling scale of electronegativity
The Periodic Table of electronegativity by Pauling scale
Covalent non-polar bond
Covalent polar bond
Ionic bond
The effect of electronegativity difference on type of bond
The continuum of bond types
Polarity in molecules
Polarity and chemical reactivity
INTERMOLECULAR FORCES AND BONDS
Van der Waals forces of intermolecular attraction
Permanent dipole-dipole forces
Hydrogen bonding
Intramolecular and intermolecular hydrogen bonding
INTRODUCTION TO ORGANIC CHEMISTRY
Alkanes
Molecular, empirical and structural formula
Functional groups of organic molecules
Alkenes and alkynes
Alcohols and ethers
Aldehydes and ketones
Carboxylic acids and esters
Amines and amides
Structural isomerism
ACIDS, BASES AND THE pH SCALE
Mineral and organic acids
Alkalis and bases
The pH scale and the pH indicators
Acid and alkali solutions and importance of water
BIOLOGICAL MOLECULES
- Functional groups of biological molecules
- Hydroxyl group
- Carbonyl group and carboxyl (ionized form) group
- Amino group
- Sulfhydryl group
- Phosphate (ionized form) group
- Methyl group
- Major classes of biological molecules
- Carbohydrates
- Lipids
- Nucleic acids
- Proteins
- Proteins are polymers made of amino acids
- Structure of amino acids
- Amino acids are zwitterions
- Amino acids are pH buffers in aqueous solutions
- Isoelectric points (pK values) of amino acids
- Hydrophobic (nonpolar), hydrophilic (polar) and charged amino acids
- Condensation reaction – formation of peptide bond
- Primary structure of polypeptide
- Secondary structures (alpha helix and betta sheet) of polypeptide
- Tertiary and quaternary structures of protein
- Molecular surface and cavities
- Lipids
- Glycerol, fatty acid and triglyceride
- Saturated and unsaturated triglycerides
- Phospholipids
- Cholesterol
BIOLOGICAL MEMBRANES
Bilayer lipid packing of biological membranes
Fluidity and passive diffusion across biological membranes
Proteins of biological membranes
Protein-facilitated diffusion across biological membranes
Active transport across biological membranes
Non-bilayer structures of biological membranes
COMPUTER MODELING OF BIOLOGICAL MACROMOLECULES
AutoDock Vina and AutoDock Tools programs to study interaction of macromolecules
PyMol program to analyze binding sites of docked macromolecules
Tabulating the types of intermolecular bonds in macromolecules.
Identifying molecular surface, cavities and subunits
Analyzing docking energies of ligand-receptor structures.
5.2. The user-friendly AutoDock Vina, AutoDoc Tools and PyMol tutorials
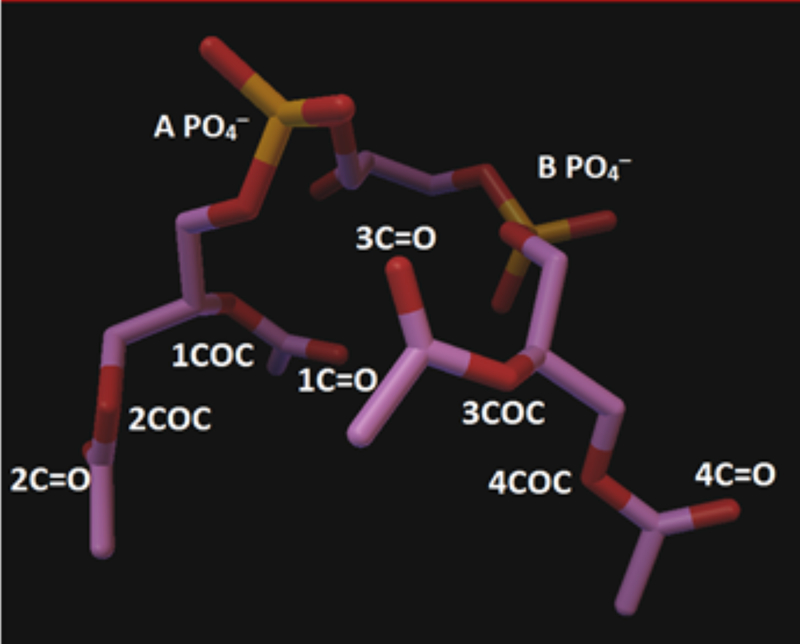
These tutorials describe docking of a macromolecular receptor – cobra venom protein cardiotoxin with the high pharmacological potential [14] (PDB ID# 1KBT), with a ligand – phosphatidylcholine (PC). For the PyMol analysis and visualization of docked conformations, we used another cardiotoxin (PDB ID# 1CDT) as a receptor and polar head of cardiolipin (CL) as a ligand (Fig. 1). The docking and PyMol analysis was performed by using Dell workstation (M4800, CPU i7–4800MQ, memory 8G, Windows 10). The PDB coordinates of CL were extracted from the bovine heart oxidoreductase crystal structure bound to CL (PDB ID# 1V54). The PDB coordinates of PC were extracted from the structure of PITP complexed to DOPC (PDB ID# 1T27). The CL was further edited to remove the alkyl chains using Avogadro as previously published [15], and the overall charges were checked and energy minimized using AutoDock. Alternatively, one can use Swiss Modeler to edit the structures of macromolecules (e.g. alkyl chains). The files of 1KBT, 1CDT, CL polar head and PC were initially used in PDB format. The ligands, PC or polar head of CL (Fig. 1), contained 32 rotatable bonds, which is a maximum number allowed by AutoDock Tools, whereas the receptor, a cardiotoxin, was kept as rigid molecule for each run. A small ligand with a maximum of 32 rotatable bonds and relatively small size of a receptor (no more than 150 amino acid residues) with rigid (non-rotatable bonds) are required by AutoDock Tools in order to ensure the computing reliability and simplify of the data interpretation. It is worth noting that the complete molecular surface of cardiotoxin should be considered for these docking studies in order to reduce bias on the location of the binding sites of the lipid on the cardiotoxins, a technique known as blind docking as further described in the Supplemental materials. A grid box was set up with dimensions indicated in the AutoDock tutorial in the Supplemental materials. The setting for exhaustiveness was set up as 16, which gave us consistent results in at least three sets of docking for each ligand and cardiotoxin pair in this study. It is important to note that the grid box was large enough to do a “blind” docking by encompassing the entire surface of the cardiotoxin and a phospholipid “ligand” for each molecular docking simulation. The user-friendly and easy to follow AutoDock Vina, AutoDoc Tools and PyMol tutorials that include computer creen shots for each operational step in using these programs are given in the Supplemental material 1. Following each Autodock run, the best nine docked conformations were analyzed for ionic, ion-polar, and hydrogen bond interactions between phospholipid polar head groups and charged and polar amino acids of cardiotoxin by using Python Molecular Viewer (MGL Tools, The Scripps Research Institute). A sample summary of all bond types in the docked complexes between cardiotoxin and cardiolipin’s polar head is given in Table 1. Designation of PO4– groups and carbon atoms in COδ−, COδ−C and C=Oδ− groups in cardiolipin’s polar head is shown in Figure 1.
Figure 1.
Designation of two PO4– groups and carbon atoms in COδ−, COδ−C and C=Oδ− groups in cardiolipin’s polar head.
Table 1.
A summary of the charged and polar groups of amino acid residues (a.a.r.) within each of the receptor’s (cardiotoxin – 1CDT 1/1)) nine binding states induced by the contact with polar head groups of a ligand (cardiolipin polar head – 1CDT_CL_HeadOnly) is shown in the Table 1, which was recently published [16]. Pb in NHpbδ+ denotes a peptide bond. Designation of PO4– groups and carbon atoms in COδ−, COδ−C and C=Oδ− in CL polar head is shown in Figure 1.
| Site number | Polar head groups of CL | 1CDT a.a.r. polar groups | Bond type |
|---|---|---|---|
| Binding site 1 Affinity: –4.2 kcal/mol |
A PO4− | K12(N+H3) | Ionic |
| A PO4− | C38 (NHpbδ+) | Ion-polar | |
| B PO4− | K12(N+H3) | Ionic | |
| 1 COδ−C | K35(N+H3) | Ion-polar | |
| 1 COδ−C | Y22(OHδ+) | Hydrogen | |
| 2 C=Oδ− | K35(N+H3) | Ion-polar | |
| Binding site 2 Affinity: –4.1 kcal/mol |
A PO4− | Y51(OHδ+) | Ion-polar |
| B PO4− | K35(N+H3) | Ionic | |
| B PO4− | Y22(OHδ+) | Ion-polar | |
| 1 C=Oδ− | K44(NHpbδ+) | Hydrogen | |
| COδ− | K35(N+H3) | Ion-polar | |
| 3 COδ−C | K18(N+H3) | Ion-polar | |
| 3 C=Oδ− | K18(N+H3) | Ion-polar | |
| 4 C=Oδ− | K12(N+H3) | Ion-polar | |
| 4 C=Oδ− | C38(NHpbδ+) | Hydrogen | |
| Binding site 3 Affinity: –4.1 kcal/mol |
A PO4− | K35(N+H3) | Ionic |
| B PO4− | C38(NHpbδ+) | Ion-polar | |
| B PO4− | K12(N+H3) | Ionic | |
| COδ− | K35(N+H3) | Ion-polar | |
| COδ− | Y22(OHδ+) | Hydrogen | |
| 2 C=Oδ− | N40(NH2δ+) | Hydrogen | |
| 3 C=Oδ− | K12(N+H3) | Ion-polar | |
| Binding site 4 Affinity: –3.7 kcal/mol |
B PO4− | K35(N+H3) | Ionic |
| 2 C=Oδ− | K5(N+H3) | Ion-polar | |
| 3 C=Oδ− | K12(N+H3) | Ion-polar | |
| 4 C=Oδ− | L6(NHpbδ+) | Hydrogen | |
| Binding site 5 Affinity: –3.7 kcal/mol |
A PO4− | K18(N+H3) | Ionic |
| B PO4− | K12(N+H3) | Ionic | |
| 2 COδ−C | K12(N+H3) | Ion-polar | |
| 3 COδ−C | L6(NHpbδ+) | Hydrogen | |
| Binding site 6 Affinity: –3.7 kcal/mol |
A PO4− | K35(N+H3) | Ionic |
| A PO4− | Y22(OHδ+) | Ion-polar | |
| B PO4− | Y51(OHδ+) | Ion-polar | |
| 1 C=Oδ− | N40(NH2δ+) | Hydrogen | |
| 1 C=Oδ− | K18(N+H3) | Ion-polar | |
| 2 C=Oδ− | K18(N+H3) | Ion-polar | |
| COδ− | Y51(OHδ+) | Hydrogen | |
| 3 C=Oδ− | S46(OHδ+) | Hydrogen | |
| 4 C=Oδ− | K44(N+H3) | Ionic-polar | |
| Binding site 7 Affinity: –3.6 kcal/mol |
A PO4− | K12(N+H3) | Ionic |
| B PO4− | L6(NHpbδ+) | Ion-polar | |
| 2 C=Oδ− | K12(N+H3) | Ion-polar | |
| 3 C=Oδ− | K12(N+H3) | Ion-polar | |
| 3 C=Oδ− | C38(NHpbδ+) | Hydrogen | |
| Binding site 8 Affinity: –3.5 kcal/mol |
B PO4− | Y51(OHδ+) | Ion-polar |
| 1 C=Oδ− | K35(N+H3) | Ion-polar | |
| 2 C=Oδ− | K35(N+H3) | Ion-polar | |
| 3 C=Oδ− | Y51(OHδ+) | Hydrogen | |
| Binding site 9 Affinity: –3.5 kcal/mol |
A PO4− | R36(=N+H2) | Ionic |
| A PO4− | R36(NH2δ+) | Ion-polar | |
| COδ− | R36(NH2δ+) | Hydrogen | |
| 4 COδ−C | R58(NH2δ+) | Hydrogen |
5.3. Assessment of the students’ knowledge and skills on docking. Rubric to evaluate students’ discussion on effects of docking
After the completion of the first year of the pilot STEM program, the docking skills and understanding on intermolecular interactions of STEM students were assessed. STEM students were asked to use AutoDock Vina and AutoDoc Tools to dock cardiotoxin 1KBT with the polar head of cardiolipin (CL) to generate nine best docked conformations. These nine best docked conformations were also given to the control group students from grade 10 (G10) who did not attend the pilot STEM program. Following the docking exercises, students from the STEM and control groups were assessed for 45 minutes to evaluate their knowledge in the areas of biophysics and biochemistry of macromolecular interactions and their skills in analysis of in silico data and discussion on potential pharmacological consequnses based on simulated cardiotoxin-CL polar head conformations. Students were asked to use PyMol to identify intermolecular contacts and classify the contacts as either ionic, ion-polar or hydrogen bonds. In order to get statistical data on students’ understanding in identification and classification of intermolecular contacts, students were asked to take another test a week later to identify and classify intermolecular contacts in the 1KBT-CL polar head docked conformations. The tabulation of results on students’ identification and classification of intermolecular bonds is shown in Table 2. In addition, the students from STEM and control groups were asked to discuss the effect(s) of the 1KBT-CL polar head docking modes on CL packing in mitochondrial membrane and protentional roles of 1KBT and CL in inducing structural changes in mitochondrial membranes of pharmacological importance. In order to grade on a scale 1 to 5 the quality of the discussions that each of the STEM or control (G10) student provided following the computer docking exercises, we employed the following rubric: 1 – student failed to offer a discussion; 2 – student showed some understanding of intermolecular bonding offering a discussion with a limited view on complexity of 1KBT and CL binding and without suggesting any functional roles for 1KBT and CL in binding states; 3 – student showed good understanding of intermolecular bonding and offered a discussion describing most of the events in binding of 1KBT and CL suggesting functional roles of 1KBT and CL; 4 – student showed comprehensive understanding of intermolecular bonding and offered a discussion describing complete sets of events in binding of 1KBT and CL leading to physiologically important effects of cardiotoxin induced changes in packing of mitochondrial phospholipids; 5 – student showed excellent understanding of intermolecular bonding and offered an in-depth discussion on the molecular mechanisms of how 1KBT interacts with CL leading to changes in packing of phospholipids in mitochondrial membranes which may have pharmacological importance. The text of discussions of the STEM and control group students is given in the Supplemental materials 2 and 3.
Table 2.
Identification and classification of intermolecular bonds by the students from STEM and G10 (non-STEM) groups in the binding states of 1KBT with the polar head of CL. The total number of intermolecular bonds in the nine binding states of 1KBT-CL polar head was 52.
| STEM students | G10 (non-STEM) students | ||||
|---|---|---|---|---|---|
|
Correctly identified bonds
(means from two separate tests) | |||||
| Student # | Bonds | Standard Deviation | Student # | Bonds | Standard Deviation |
| 1. | 52.0 | 0.0 | 1. | 52.0 | 0.0 |
| 2. | 52.0 | 0.0 | 2. | 52.0 | 0.0 |
| 3. | 52.0 | 0.0 | 3. | 52.0 | 0.0 |
| 4. | 52.0 | 0.0 | 4. | 40.5 | 0.5 |
|
Correctly classified bonds
(means from two separate tests) | |||||
| Student # | Bonds | Standard Deviation | Student # | Bonds | Standard Deviation |
| 1. | 51.5 | 0.5 | 1. | 45.0 | 3.0 |
| 2. | 52.0 | 0.0 | 2. | 40.5 | 3.5 |
| 3. | 52.0 | 0.0 | 3. | 35.5 | 6.8 |
| 4. | 52.0 | 0.0 | 4. | 21.0 | 0.0 |
6. Results and Discussion
6. Assessment of the STEM students’ knowledge and skills
The knowledge and skills the students acquired after the completion of the first phase of STEM project was assessed by giving the students a comprehensive test as further describe below. The test consisted a series of instructions (as described in this paper) that asked students to dock cardiotoxin 1KBT – a pharmacologically potent protein that at very low concentrations increases mitochondrial ATP-synthase activity – with a polar head of cardiolipin (CL, Fig. 1), which is a key phospholipid in mitochondrial inner membrane – a membrane where ATP-synthase is found [14]. Students were then asked to identify and classify all intermolecular ionic, ion-polar and hydrogen bonds (contacts) for each of the nine binding states between 1KBT and polar head of CL. A final task given to students in this test was to predict, based on analysis of binding states, the mode by which cardiotoxin 1KBT binds to the membrane surface and to discuss the possible effects of cardiotoxin on the packing of cardiolipin molecules in the inner mitochondrial membrane and what potential pharmacological effecst cardiotoxin may cause while binding to mitochondrial membrane. During the STEM course, we taught STEM students that the transition of phospholipids in biological membranes from a bilayer packing of phospholipids to a non-bilayer packing is affected by cationic proteins (e.g. cardiotoxins), and that this process largely depends on phospholipid composition of the lipid bilayer. Cardiotoxins, depending on their molecular surface, may bind to the surface of biomembranes with the long molecular axis of cardiotoxin perpendicular or parallel to the normal of the membrane surface. Depending on phospholipid composition of biomembrane and the mode by which the cardiotoxin interacts with phospholipid bilayers, the interaction of cardiotoxin with biomembrane may or may not induce the formation of non-bilayer structures. We have also taught the STEM students that small amount of non-bilayer structures in inner mitochondrial membranes (IMM) facilitate activity of ATP-synthase enzyme localized in IMM. Ability of certain cobra venom cardiotoxins to trigger the formation of small amount of non-bilayer structures in IMM may represent a noval pharmacological avenue in using cardiotoxins for rejuvenation of ATP production in aging mitochondria.
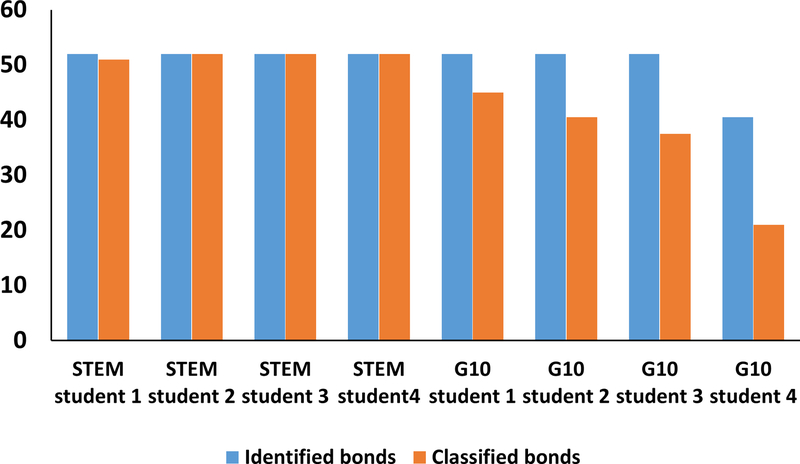
In brief, in the STEM test the students were asked to dock the cardiotoxin (1KBT) to polar head of CL a day before the test. On the next day, students were required to identify and classify intermolecular bonds for each of the nine binding docked structures and discuss the effect(s) of cardiotoxin on molecular packing of cardiolipin in inner mitochondrial membranes. For the purpose of statistical analysis, students have taken the same test on identification and classification of intermolecular bonds twice with a one week period between the tests. Overall, all four STEM students successfully completed the docking exercises and correctly identified 52 intermolecular bonds (contacts) in the nine binding states. Regarding the classification of inter-molecular contacts, all STEM students achieved the same results in two tests: three out of four students correctly classified all 52 bonds whereas one student correctly classified 51 bonds (Fig. 2).
Figure 2.
Assessment data on the correct number and type of intermolecular bonds identified for each of the 1KBT-CL polar head docked structures by STEM and G10 (non-STEM) students. Note that the total number of bonds (contacts) for all docked 1KBT-CL structures were 52.
Importantly, all four STEM students offered their discussions on the effect(s) of 1KBT on cardiolipin packing in the inner mitochondrial membrane (see Supplemental material 2).
For comparison, the same task was given to four “control” students from grade 10 who did not attend the first phase of STEM pilot course but completed the IGCSE Physics and Chemistry. In addition to IGCSE courses, these four students were also introduced to the concepts of electronegativity of atoms, covalent polar and non-polar bonds, computer 3-D macromolecule modeling and in silico docking of macromolecules. As part of an assessment to test their biophysics and biochemical knowledge, the “control” students were given the nine docked conformations of 1KBT with CL polar head and were asked to identify and classify all contacts between 1KBT and CL polar head for each of the nine binding states, and to discuss how the 1KBT binding with CL polar head may affect the packing of CL molecules in inner mitochondrial membranes. For the purpose of statistical analysis, students have taken the same test twice with a one week period between the tests. The test assessment results of the grade 10 students are shown in figures 2 and 3 above and the text of discussions is given in Supplemental material 3.
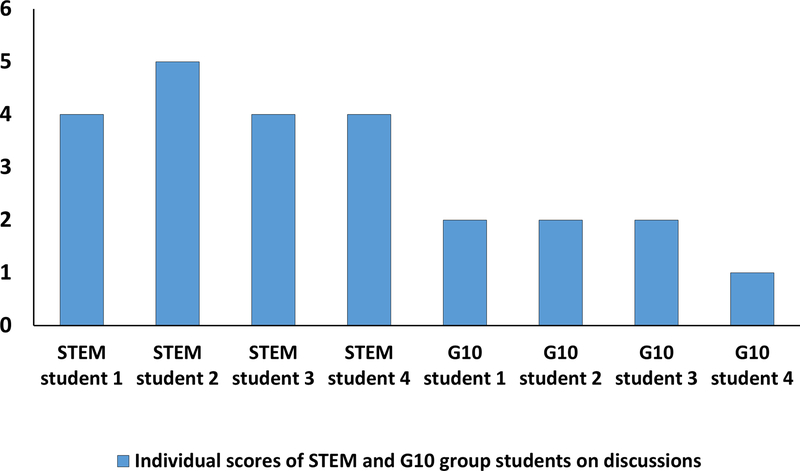
Figure 3.
Discussion scores on scale 1 to 5 of STEM and G10 (non-STEM) students. The graph shows individual scores for each of the groups of students. The mean score of STEM students (4.25) is statistically significant (p<0.0004) compared to that of G10 students (1.75) as assessed by an unpaired student t-test. The rubric used to score the quality of students’ discussion can be found in the Materials and Methods section.
Overall, the STEM group students performed significantly better on the assessment test compared to the G10 group students. In addition all STEM group students identified all intermolecular contacts for all nine binding states and correctly classified all contacts, with the exception of one student who made one mistake in classifying one contact. In addition, three of G10 students correctly identified all intermolecular contacts whereas only one G10 student identified only 40 contacts. However, none of the G10 students correctly classified all intermolecular contacts (Fig. 2). The quality of discussion based on analysis of the docked data was significantly higher for the STEM group students relative to the discussions offered by the G10 group students (Fig. 3, p<0.0004). To read the discussions of the G10 group students, please refer to the Supplemental material 3.
6.2. Discussion
STEM education becomes increasingly important because of the urgent need for the creation of new technologies to counteract the looming global catastrophe caused by food shortages, energy crisis, novel diseases, climate change and overall degradation of environment. Biochemical engineering, and particularly, the engineering of enzymes seems to be one of the most important directions in creation of new technologies. This is because technology used to engineer enzymes to catalyze reactions for the in vitro production of food molecules, pollution free fuel, novel pharmaceuticals, new catalysts for digesting plastic waste and to support environmentally friendly industrial processes may solve many, if not most, global issues. It should be noted however, that advances in artificial enzyme engineering, a new field of science that emerged about two decades ago [1–3], has not been showing any progress in terms of technological development. That is because this field of science and technology requires a broad range of expertise including structural biochemistry with extensive knowledge in physics and chemistry of intra- and intermolecular bonding, physical mathematics of macromolecular structure and interactions, and a high-level computer programming with extensive expertise in physics and chemistry of macromolecular structure and interactions. Nowadays, it is very critical to assemble large multi-collaborative STEM teams from across the globe in order to solve today’s biotechnological challenges. Hence, it is of paramount importance to bring experts as one collaborative team to work on a research problem. Another problem is that scientists in these different fields of science and technology, each with a top and highly developed expertise in one of these fields, may not understand each other. A highly specialized scientist develops highly conceptualized thinking skills that helps him/her to successfully use a complex methods and approaches in his/her area of expertise but lacks experience in another area of expertise. Thus, the successful development of artificial enzymes engineering relies on the availability of scientists each having a high-level of multidisciplinary training in physics and chemistry of macromolecular structure, computer programming with a high knowledge in physics, chemistry and mathematics of macromolecular structure and interactions. Hence, to foster scientists with such a broad area of expertise, students have to be taught STEM courses at an early age. Finally, to synergize collaborations, it is important that STEM team members develop inter-professional skills that can synergize collaborations while leveraging knowledge, expertise and resources among experts.
Dr. Edward S. Gasanoff, one of the authors of this article, has taught Physics of Chemical Bonding and Structural Biochemistry courses to the senior students of the Applied Mathematics and Informatics Department (AMID) of the Lomonosov Moscow State University. Dr. Gasanoff with the help of Dr. Ruben K. Dagda, another author of this article, successfully involved AMID students in research studies that entailed performing computer simulations of 3-D structure and docking of macromolecules with rigid non-rotatable bonds. However, the AMID students failed to create a computer program capable of simulating 3-D structure and docking of macromolecules with rotatable bonds because they were unable to formulate the physical equations in mathematical terms that describe physical and chemical factors that regulate the interactions of macromolecules with rotatable bonds. This is likely due to the fact that senior students have already developed a high-level conceptualized thinking in computer sciences, and they were unable to achieve a high-level conceptual understanding in physics and chemistry of chemical bonding and structure of macromolecules. Given this information, we then identified the earliest age by which students could be taught the physical and chemical concepts of chemical bonding and macromolecular structure followed by computer programming course.
In this article we described a pilot STEM course which introduces the physical and chemical concepts of chemical bonding, macromolecular structure and intra- and intermolecular forces of attraction. We introduced this course, which is designed for 72 hours of instruction, to a highly motivated group of secondary school students from grades 7, 8 and 9 (ages 13 to 16). The evaluative assessment of this group of students showed that these students acquired high level of understanding and skills as they were able to successfully simulate docking of macromolecules in silico, identify and classify intermolecular bonds, show how to identify the best docked structure based on the analysis of interactive bonds, discuss the underlying real-world implications of the data they analyzed, and how to synthesize a hypothesis to follow up on a biochemistry/ molecular biology lab to corroborate the newfound data. Also, a thorough analysis on the types and modes of intermolecular bonding in the docked conformations showed that STEM students were able predict the functional activities of macromolecules. By contrast, a “control” group that consisted of secondary school students from grade 10 (ages 16 to 17) - which completed the IGCSE Physics and IGCSE Chemistry and were only taught basic concepts in chemistry without completing the STEM pilot program - did not show a high-level of understanding regarding the types of intermolecular bonds in the docked conformations of macromolecules. Students from this group have also failed to successfully discuss the protentional functional activities of macromolecules involved in the docked conformations.
Although this pilot STEM course has been tested on a very small group of students, we believe that we can conclude that this pilot STEM course has served its purpose as it showed that motivated secondary school students (ages 13 to 16) successfully learned concepts including chemical bonding, the spatial structure of macromolecules and intra- and intermolecular bonding in macromolecules. In our view, delivering this STEM content builds critical knowledge and the necessary skills that can aid in the training of future scientists in the field of biochemical engineering. However, we do recognize that this pilot STEM course has to be taught to a larger group of students to further corroborate our findings in this study. Overall, the aforementioned curriculum gives high school students a very concrete and genuine experience that allows them to apply concepts of biochemistry, physics, organic chemistry and computer biology in one exercise. However, it is worth noting that these computational biology exercises can be complemented with benchwork. For instance, these in silico docking activities described in this paper can conceivably be part of a summer residential STEM program that teaches high school students basic concepts of biochemistry, pharmacology, chemistry, physics and engineering and affords students the ability to perform benchwork in a biochemistry or pharmacology lab in order to corroborate the data obtained with the molecular docking experiments.
Cardiotoxins 1KBT and 1CDT, which STEM students used in their docking studies, along with other homologous cardiotoxins from cobra venom possess unique physiological properties of a great pharmacological importance [17]. One of the well-established pharmacological activities of cardiotoxins is their ability to kill cancer cells in vitro [17] by targeting acidic phospholipids exposed on the outer leaflet of cell membranes [18–19]. In vivo, cardiotoxins interact with extracellular PLA2 which activates membrane lipid hydrolysis to harm not only cancer cells but also healthy cells [17–20]. Hence, it is conceivable that in silico docking of cardiotoxin with PLA2 may reveal crucial amino acid residues on cardiotoxin’s surface, which could be edited by site directed mutagenesis, to prevent cardiotoxin’s binding to PLA2, and thereby enhance its specificity toward killing cancer cells in vivo. In addition, certain cardiotoxins at very low concentrations increase production of ATP in mitochondrial membranes by generating non-bilayer structures [11, 13, 14]. It is conceivable that projects that involve high school students to perform computer-aided analysis on the molecular interactions between cardiotoxins and mitochondrial phospholipids may open a pharmacological avenue for engineering an efficient drug that can be used to treat brain-degenerative diseases (Parkinson’s disease) to reverse neurodegeneration caused by decline in production of ATP in neuronal mitochondria [16]. Rattlesnake venom is another source of protein toxins with a great pharmacological potential. Metalloproteinases – rattlesnake venom enzymes – can digest important proteins on surface of blood capillaries [17, 21, 22]. Analysis of metalloproteinase molecular surface in silico that relates to metalloproteinase enzymatic activities may help in designing novel drugs capable of dissolving blood clots [22]. Hence, we plan on involving our STEM students in using computer modeling along with biochemical bench lab studies, so the students can gain first-hand experience in understanding the complex structure-function relationships of biological molecules. Therefore, the involvement of students in this kind of studies at an early age will increase their expertise and skills in understanding the relationship between macromolecular structure and function of non-catalytic and catalytic proteins, a fundamental attribute of knowledge needed for development of science and technology in biochemical engineering and creation of artificial enzymes.
As a next phase of our pilot STEM project, we will teach the same group of secondary school students (aged 13 to 16) in the Chaoyang KaiWen Academy and at the University of Nevada, Reno STEM bilingual students, the basics of computer programming so that students can gain a deep understanding of the physico-chemical properties of macromolecules when docked to ligands. Following the in silico docking exercises, students can conceivably generate a testable hypothesis, and corroborate the newfound data on the lab by collaborating with international toxinologists (e.g. mutagenize residues in cardiotoxin to see if it fails to bind cardiolipin in vitro). Although we recognize that the acquisition of a high level of understanding in biophysics and biochemistry can be challenging to high school students, our small pilot educational program presented in this paper provides strong proof-of-concept that this program is feasible and can be successfully integrated in the traditional American and Chinese high school classroom.
7. Conclusion
This study has shown that interdisciplinary concepts of physics and chemistry of intra- and intermolecular bonding, structural biochemistry and computer modeling could be taught to the academically advanced and science motivated students of 13 to 16 years of age in a single STEM curriculum that included tutorials of computer modeling of intermolecular interactions. Although this curriculum was tried on a very small group of academically advanced students, all students showed after completing this curriculum a great deal of understanding of concepts in structural biochemistry and macromolecular interaction and demonstrated skills in relating structural modes of macromolecular complexes to the potential physiological mechanisms of interacting macromolecules. Use of natural toxins of pharmacological relevance in simulating interactions with membrane phospholipids supports building of knowledge and skills in engineering of novel pharmaceuticals. Although natural toxins used in this study were not enzymes, thorough analysis of the toxins’ molecular surface, cavities and subunits that complemented the molecular surface of “ligands” helps in development of understanding and skills about structural mechanisms of enzyme-substrate complexes. Overall, this pilot STEM curriculum seemed to work well in supplementing secondary/high school science courses for academically advanced students. We would also recommend to introduce this pilot STEM curriculum as an undergraduate university course for students who wish to major in Pharmacology, Pharmaceuticals Engineering, and Structural Biochemistry.
Supplementary Material
8. Acknowledgements
We thank all secondary school students of the Chaoyang KaiWen Academy (Beijing, China) who attended the course of the first phase of our pilot STEM program and who took the assessment tests related to our pilot STEM program. This study was partially funded by an NIH/SEPA grant R25 OD023795–02 (CBESS) and the University of Nevada at Reno International Activities Grant 2014.
3. Abbreviations
- STEM
Science, technology, engineering and mathematics
- AMID
Applied mathematics and informatics department
- IGCSE
International general certificate of secondary education
- PDP
protein data bank
- DOPC
dodecyl-oleyl-phosphatidylcholine
- PC
phosphatidylcholine
- CL
cardiolipin
- IMM
inner mitochondrial membrane
Footnotes
Conflick of interest
The authors declare no conflict of interest. The funders had no role in the design of the study, in the collection, analysis, or interpretation of data, in the writing of the manuscript or in the decision to publish the results.
10. Bibliography
- 1.Renugopalakrishnan V, Garduño-Juárez R, Narasimhan G, Verma CS, Wei X, Li P “Rational design of thermally stable proteins: relevance to bionanotechnology”. Journal of Nanoscience and Nanotechnology 511 (November 2005): 1759–1767. doi: 10.1166/jnn.2005.441. [DOI] [PubMed] [Google Scholar]
- 2.Hult K, Berglund P “Engineered enzymes for improved organic synthesis”. Current Opinion in Biotechnology 14 4 (August 2003): 395–400. doi: 10.1016/S0958-1669(03)00095-8. [DOI] [PubMed] [Google Scholar]
- 3.Jiang L, et al. “De novo computational design of retro-aldol enzymes”. Science 319 5868 (March 2008): 1387–91. doi: 10.1126/science.1152692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stryer L,, Berg JM, Tymoczko JL Biochemistry (5th ed.) (2002): San Francisco: W.H. Freeman; ISBN 0–7167-4955–6. [Google Scholar]
- 5.Schomburg I, et al. “BRENDA in 2013: integrated reactions, kinetic data, enzyme function data, improved disease classification: new options and contents in BRENDA”. Nucleic Acids Research 41 (Database issue) (January 2013): D764–72. doi: 10.1093/nar/gks1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Radzicka A, Wolfenden R “A proficient enzyme”. Science 2675194 (January 1995): 90–931. doi: 10.1126/science.7809611. [DOI] [PubMed] [Google Scholar]
- 7.Callahan BP, Miller BG “OMP decarboxylase—An enigma persists”. Bioorganic Chemistry 356 (December 2007): 465–9. doi: 10.1016/j.bioorg.2007.07.004. [DOI] [PubMed] [Google Scholar]
- 8.Nakamoto RK, Scanlon JAB, Al-Shawi MK “The rotary mechanism of the ATP synthase”. Arch Biochem Biophys 476 (2008): 43–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Trott O, Olson AJ “AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading”. Journal of Computational Chemistry 312 (2010): 455–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ramanathan A, Savol A, Burger V, Chennubhotla CS, Agarwal PK “Protein conformational populations and functionally relevant substates”. Acc Chem Res 47.1 (2014): 149–56. doi: 10.1021/ar400084s. [DOI] [PubMed] [Google Scholar]
- 11.Gasanov SE, Kim AA, Dagda RK “The possible role of nonbilayer structures in regulating ATP synthase activity in mitochondrial membranes”. Biophysics (Oxf) 61(4) (2016): 596–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gasanov SE, et al. “Naja naja oxiana cobra venom cytotoxins CTI and CTII disrupt mitochondrial membrane integrity: implications for basic three-fingered cytotoxins”. PLoS One 106 (2015): p. e0129248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gasanov SE, Kim AA, Dagda RK “Possible role of non-bilayer structures in regulating the activity of ATP synthase in mitochondria”. Biofizika 61.4 (2016): 705–710. [Google Scholar]
- 14.Gasanov SE, et al. “Non-bilayer structures in mitochondrial membranes regulate ATP synthase activity”. Biochim Biophys Acta Biomembr 18602 (2018): 586–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hanwell MD, et al. “Avogadro: an advanced semantic chemical editor, visualization, and analysis platform”. J Cheminform 4.1 (2012): 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang B et al. , “Naja mossambica mossambica cobra cardiotoxin targets mitochondria to disrupt mitochondrial membrane structure and function”. Toxins 11152 (2019): doi: 10.3390/toxins11030152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gasanov SE, Dagda RK, Rael ED “Snake venom cytotoxins, phospholipase A2s, and Zn2+-dependent metalloproteinases: Mechanisms of action and pharmacological relevance”. J Clin Toxicol (2014): 4 1000181. [DOI] [PMC free article] [PubMed]
- 18.Gasanov SE, Alsarraj MA, Gasanov NE, Rael ED “Cobra venom cytotoxin free of phospholipase A2 and its effect on model membranes and T leukemia cells”. J Membr Biol 155 (1997): 133–142. [DOI] [PubMed] [Google Scholar]
- 19.Gasanov SE, Rael ED, Gasanov NE, Vernon LP “In vitro evaluation of Pyrularia thionin-anti-CD5 immunotoxin”. Cancer Immunol Immunother 141 (1995): 122–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gasanov SE, Gasanov NE, Rael ED “Phospholipase A2 and cobra venom cytotoxin Vc5 interactions and membrane structure”. Gen Physiol Biophys 14 (1995): 107–123. [PubMed] [Google Scholar]
- 21.Rael ED, Lieb CS, Maddux N, Varela-Ramirez A. Perez J “Hemorrhagic and Mojave toxins in the venoms of the offspring of two Mojave rattlesnakes (Crotalus scutulatus scutulatus)”. Comp Biochem Physiol 1063 (1993): 595–600. [DOI] [PubMed] [Google Scholar]
- 22.Dagda RK, Gasanov SE, Zhang B, Welch W, Rael ED “Molecular models of the Mojave rattlesnake (Crotalus scutulatus scutulatus) venom metalloproteinases reveal a structural basis for differences in hemorrhagic activities”. J Biol Phys (2014): DOI 10.1007/s10867-013-9339-3. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.