Abstract
The budding yeast, Saccharomyces cerevisiae, has been widely used for genetic studies of fundamental cellular functions. The isolation and analysis of yeast mutants is a commonly used and powerful technique to identify the genes that are involved in a process of interest. Furthermore, natural genetic variation among wild yeast strains has been studied for analysis of polygenic traits by quantitative trait loci (QTL) mapping. Whole genome sequencing, often combined with bulk-segregant analysis, is a powerful technique that helps determine the identity of mutations causing a phenotype. Here, we describe a protocol for the construction of libraries for S. cerevisiae whole genome sequencing. We also present a bioinformatic pipeline to determine the genetic variants in a yeast strain using whole genome sequencing data. This pipeline can also be used for analyzing Schizosaccharomyces pombe mutants.
Keywords: Whole genome sequencing, bulk segregant analysis, Saccharomyces cerevisiae, Schizosaccharomyces pombe
Introduction
The budding yeast, Saccharomyces cerevisiae, is a well-established model organism for conducting genetic analyses. Genetic selections and screens in yeast are used to identify genes required for a particular function or process (Forsburg, 2001). Such screens and selections begin with the isolation of mutants that exhibit a desired phenotype. This is followed by the identification of candidates for causal mutations. Classically, this identification has been accomplished by cloning by complementation, using a recombinant library (Lundblad, 2001). However, this technique has a number of limitations. First, it is low throughput as it involves transforming each mutant with a yeast recombinant library, followed by screening thousands of transformants for complementation of the mutant phenotype. While the number of mutants to be analyzed can be condensed by complementation tests, such tests are time-consuming and can yield ambiguous results. Second, cloning will not work if more than one mutation is required for the mutant phenotype. Finally, cloning only works for recessive mutations unless one is willing to construct a new recombinant library from a dominant mutant. With the advent of next-generation sequencing, whole genome sequencing, sometimes paired with bulk-segregant analysis, has emerged as an alternative and powerful approach to identify mutations (Birkeland et al., 2010; Brauer et al., 2006; Wenger et al., 2010).
Whole genome sequencing is a technique used to determine the entire DNA sequence of an organism in a single experiment (Ng and Kirkness, 2010). To use the Illumina platform, whole genome sequencing involves the isolation of genomic DNA and shearing the DNA to obtain smaller fragments of approximately 100–500 basepairs. These DNA fragments are then end repaired and ligated to sequencing adapters, which contain the DNA sequence complementary to the sequencing primer as well as to the DNA sequences required for binding to the sequencing flow cell. The adapter-ligated DNA fragments are amplified by PCR and sequenced. The resulting DNA sequences are then aligned to a reference genome. Given the well assembled and annotated S. cerevisiae reference genome (Cherry et al., 1997; Cherry et al., 2012), single-nucleotide polymorphisms (SNPs) and small insertions/deletions (indels) (collectively referred to as variants) can be identified easily using computational tools. Given the small size of the S. cerevisiae genome, approximately 12.5 Mb, it is possible to sequence the genomes of several different strains together (a process known as multiplexing), with the number of strains depending upon the capacity of the sequencer. Multiplexing is carried out by including a barcode sequence specific to each sample in the sequencing adapters (Table 1) (Meyer et al., 2007; Current Protocols article: Wong et al., 2013).
Table 1:
Barcode sequences that can be used for multiplexing sequencing libraries
| ATCACG |
| CGATGT |
| TTAGGC |
| TGACCA |
| ACAGTG |
| GCCAAT |
| CAGATC |
| ACTTGA |
| GATCAG |
| TAGCTT |
| GGCTAC |
| CTTGTA |
| ATATAGGA |
| AACCGTGT |
| AGGTCAGT |
| CTCTGTCT |
| CCATACAC |
| CGCATTAA |
| GTCTACAT |
| GAGTTAAC |
| GCAGCCTC |
| TCGCGTAC |
| TATACCGT |
| TGCGGTTA |
| AACACCTAC |
| CCTTTACAG |
| GGTCCTTGA |
| TTGAGTGT |
| ACTAACTGC |
| CAGGAGGCG |
| GTTGTCCCA |
| TGACGCAT |
| ATCGCCAGC |
| CATTCCAAG |
| GCAAGTAGA |
| TGATCCGA |
| ACGTAGCTC |
| CGAACTGTG |
| TAGCTAGTA |
| GTGGGATA |
| ATCCTATTC |
| CGGACGTGG |
| GCGTTTCGA |
| TATCTCCG |
| ACAGTGCAC |
| CACAGTTGG |
| GTGACTACA |
| TGAGAGTG |
| AATGCTGAC |
| CCGTCTGAG |
| GGCAGACGA |
| TTCTGATG |
| AGTAGTGGC |
| CTAGTCATG |
| GACACTCTA |
| TCATTAGG |
| TCCAGCCTC |
| CTAGATTCG |
| GAACGCTGA |
| AGAACACC |
Preliminary genetic analyses can be done prior to sequencing to understand whether one or more than one mutation contributes to a phenotype. Regardless of this number, multiple variants are usually identified upon sequencing, with spontaneous mutants generally having far fewer variants than mutagenized strains, which can have over 100 variants. It is important to be able to identify the one or more causal mutations among this background of non-causal variants.
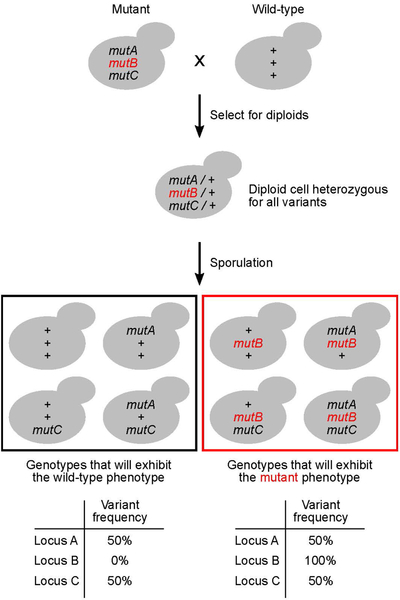
To enrich for the causal variants, bulk segregant analysis (also known as pooled linkage analysis) is often done prior to whole genome sequencing. This method involves crossing the mutant to a wild-type strain, sporulating the resulting diploid, and identifying haploid spores that exhibit the mutant phenotype (Figure 1). Approximately 50 of the mutant progeny are grown and pooled prior to genomic DNA extraction. A similar pool is prepared for progeny with the wild-type phenotype. The DNA extracted from each group is then sequenced. In each pool, non-causal variants will segregate randomly and be found at a similar frequency (~50%). However, causal variants will be present at a high frequency in the mutant pool and at a low frequency in the wild-type pool. This technique has been used successfully to identify causal variants that contribute to both Mendelian and polygenic traits (Brauer et al., 2006; Ehrenreich et al., 2010).
Figure 1.
A schematic showing the steps involved in bulk segregant analysis. mutA, mutB, and mutC represent DNA sequence variants at three hypothetical genomic loci A, B and C. ‘+’ indicates the wild-type allele. The causal variant, mutB, is highlighted in red.
In this protocol, we describe the steps for conducting bulk segregant analysis followed by whole genome sequencing on an Illumina platform. The protocol is divided into four steps: (1) construction of the yeast strains for bulk segregant analysis; (2) extracting genomic DNA from yeast cells; (3) shearing the DNA using sonication; and (4) preparing the sequencing libraries. We then present a bioinformatic pipeline for identification of variants starting from the raw sequencing data generated using this protocol. This pipeline can be used for variant identification from S. cerevisiae and S. pombe whole genome sequencing data.
Basic Protocol 1: Generation of haploid spores for bulk segregant analysis
The starting point for bulk segregant analysis is a yeast mutant of interest that is phenotypically different from a wild-type strain, with the goal of determining the causal mutation(s).
Materials:
Yeast strain with the mutant phenotype (referred to as mutant)
Yeast strain of the opposite mating type without the mutant phenotype (referred to as wild-type)
Test tubes
YPD
Inoculation sticks
30°C incubator
Test tube rotator
50 ml conical tubes
1.7 ml microfuge tubes
Materials required for yeast tetrad dissection (see Current Protocols article:Treco and Winston, 2008)
Isolating progeny for bulk segregant analysis
-
1
Cross the yeast mutant to a wild-type strain and identify diploids by selection or by micromanipulation of zygotes. Sporulate the diploids and dissect the resulting tetrads.
A protocol for yeast mating and tetrad dissection can be found here: (Current Protocols article:Treco and Winston, 2008). The number of tetrads to be dissected will depend on the number of genes that regulate the phenotype, as well as the frequency of obtaining complete tetrads from the cross (see next step for more details).
-
2
Analyze the progeny generated from tetrad dissection and score them for the phenotype of interest.
Generally, 30–50 progeny with the wild-type phenotype and an equal number of progeny with the mutant phenotype are sufficient to move forward. However, bulk segregant analysis has been conducted successfully with as few as 10 progeny per group (Thurtle-Schmidt et al., 2016). A polygenic trait will require dissection of a greater number of tetrads as compared to a Mendelian trait in order to obtain a sufficient number of progeny with the phenotype of interest.
An alternative approach to tetrad dissection is to use the marker system used for synthetic genetic arrays (Tong and Boone, 2006; Tong et al., 2001). Following sporulation of diploids, cells are plated on media that permits selection of haploid strains that have the phenotype of interest. A control group of haploid strains are also obtained in the absence of selection for the phenotype. The mutant group and control group are pooled separately, and processed using similar steps outlined hereafter. This technique has the advantage of generating a large number of progeny for bulk segregant analysis without tetrad dissection and it has been used for QTL mapping among wild yeast strains (Ehrenreich et al., 2010).
Growing cells for bulk segregant analysis
-
3
Inoculate each of 50 progeny that exhibit the mutant phenotype (mutant group) and 50 progeny that exhibit the wild-type phenotype (control group) into 5 ml of YPD (100 tubes total). Incubate the tubes in a test tube rotator for 24–36 hours at 30°C or until the cultures are saturated.
This protocol begins with using 50 progeny in each group. The exact number of progeny inoculated can vary based on the results from the tetrad dissection.
-
4
For each group of 50 cultures, pool 500 μl from each culture in a 50 ml conical tube. After pooling, you will have 25 ml of cells per tube. The downstream processing of samples for the mutant and control pools will be the same.
At this step, 5 μl of the pooled cultures can be spotted on a YPD plate and struck out to obtain single colonies. This can be used for replica plating to verify that the mutant and control groups display the expected phenotypes.
-
5
Pellet the cells in the conical tubes by centrifugation for 2.5 minutes at 3500 rpm, 4°C. Discard the supernatant.
-
6
Resuspend the cell pellets in 5 ml of water and divide them equally into 5 microfuge tubes.
-
7
Pellet the cells by centrifugation for 20 seconds at 10,000 rpm, 4°C. Discard the supernatant. Proceed with one tube for genomic DNA isolation. Store the other four cell pellets at −70°C.
If an insufficient amount of genomic DNA is obtained, you can return to this step and use the frozen cell pellets for a second round of DNA isolation.
Basic Protocol 2: Extraction of genomic DNA from yeast cells
The next step in the protocol involves the extraction of genomic DNA from yeast cells for preparing libraries for next-generation sequencing. Any other equivalent protocol that yields genomic DNA at a sufficient concentration for library preparation can also be used.
Materials:
1.7 ml microfuge tubes
Smash and Grab solution (see recipe)
Acid washed glass beads (Sigma G8772)
Phenol-chloroform-isoamyl alcohol (in a ratio of 25:24:1 by volume) solution
Tube mixer (Eppendorf 5432 or equivalent)
TE solution
3M sodium acetate
100% ethanol
70% ethanol (chilled at −20°C)
Sterile distilled water
RNase A/T1 (2 mg/ml)
Chloroform
Qubit dsDNA HS assay kit (Thermo Fisher Scientific)
Extracting genomic DNA from cells
-
1
Suspend the cell pellets in 200 μl of Smash and Grab solution. Add 300 μl of acid washed glass beads. Add 200 μl of phenol-chloroform-isoamyl alcohol solution. Place tubes in a tube mixer at 4°C and vortex the tubes for 30 minutes.
Phenol-chloroform-isoamyl alcohol should be handled only in the fume hood, wearing appropriate personal protective equipment.
The volume of glass beads can be measured approximately using the markings on a microfuge tube.
-
2
Add 200 μl of TE solution to the tubes. Mix by vortexing for 10 seconds. Centrifuge the tubes for 15 minutes at 12,500 rpm, 4°C. Carefully transfer the top aqueous layer to a new microfuge tube.
Following centrifugation, the organic phase collects at the bottom of the tube and the aqueous phase stays at the top. Take care not to transfer any liquid from the bottom organic phase to the new tube. It is okay to sacrifice a small amount of aqueous phase to avoid transferring any organic phase or any material that accumulates at the interface between the two phases.
-
3
Add 40 μl of 3M sodium acetate to the tubes. Mix briefly by inverting the tubes a few times. Add 1 ml of 100% ethanol. Precipitate DNA for at least 1 hour at −20°C or place in a dry ice-ethanol bath for a few minutes.
Immediate precipitation of nucleic acids should be visible upon the addition of ethanol. This is because of the large amount of starting material as well as the presence of RNA. The tubes may also be left at −20°C overnight at this step.
-
4
Centrifuge the tubes for 30 minutes at 12,500 rpm, 4°C. Discard the supernatant. Add 500 μl of chilled 70% ethanol to the tubes.
-
5
Centrifuge the tubes for 5 minutes at 12,500 rpm, 4°C. Discard the supernatant. Centrifuge the tubes for 10 seconds at 12,500 rpm, room temperature. Using a pipetman fitted with a fine tip, remove any residual ethanol in the tube. Leave the tubes open at room temperature for 5 minutes to allow the final trace amounts of ethanol in the tube to evaporate.
-
6
Suspend the DNA pellet in 200 μl of sterile distilled water.
The DNA pellet should not be dried for too long or else it becomes difficult to resolubilize. If you are having difficulty suspending the pellet, let the tubes sit for 5–10 minutes at room temperature after the addition of water. This will make the pellet softer and easier to get into solution.
RNase treatment of genomic DNA
-
7
Add 10 μl of RNase A/T1 to each tube. Incubate at 37°C for 1 hour.
-
8
Add 200 μl of TE solution followed by 400 μl of phenol-chloroform-isoamyl alcohol solution. Mix by vortexing for 10 seconds. Spin the tubes for 5 minutes at 12,500 rpm, 4°C. Transfer the top aqueous layer to a new microfuge tube.
-
9
Add 400 μl of chloroform to the tubes. Mix by vortexing for ~10 seconds. Spin the tubes for 5 minutes at 12,500 rpm, 4°C. Transfer the top aqueous layer to a new microfuge tube.
-
10
Repeat the ethanol precipitation of DNA (steps 3–5).
-
11
Suspend the DNA pellet in 100 μl of sterile distilled water. Measure the concentration of DNA using the Qubit dsDNA HS assay kit. The DNA can be stored at −20°C until the next step.
Basic Protocol 3: Shearing of genomic DNA for library preparation
Genomic DNA has to be sheared to obtain fragments that are small enough for library preparation and sequencing on an Illumina platform. We present a protocol for fragmentation of genomic DNA to a size range of 100 – 500 bp using sonication. This enables unbiased and inexpensive fragmentation of DNA. Alternate methods such as enzymatic fragmentation can also be used, although this would come with the caveat of biased cleavage at certain DNA sequences.
Materials:
0.3 ml thin-walled PCR tubes
QSonica 800R sonicator or equivalent
BioAnalyzer
Sonication of genomic DNA
-
Dilute the DNA to a final concentration of 15 ng/μl in a volume of 100 μl.
Concentrations of DNA as low as 1 ng/μl can also be used. However, this will result in the need for a higher number of PCR cycles for DNA amplification at a later step, which will increase the likelihood of obtaining biased coverage and introducing errors during amplification.
- Transfer the sample to 0.2 ml tubes and sonicate the DNA in a QSonica sonicator using the following settings:
- Sonicator amplitude setting: 40%
- Sonication pulse rate: 15 seconds on, 15 seconds off
- Total sonication time: 15 minutes
-
Sample process temperature: 4°CThe sonication conditions above are a guide; however, the precise conditions will need to be optimized to produce sheared DNA with a size range of 100–500 bp. Sonication can be done in any sonicator using equivalent settings.
-
Analyze 1 μl of a 1:10 diluted sonicated DNA sample on a BioAnalyzer to measure the size of the sonicated DNA fragments. The sonicated DNA will ideally have a size range of 100–500 bp (Figure 2).
The sonication of DNA can also be checked by running 10 μl of the sonicated DNA on a 0.8% agarose gel, using size markers.
The sonicated DNA can be stored at −20°C before proceeding to the next step.
Figure 2.
Analysis of sonicated genomic DNA. (A) Pseudogel and (B) electropherogram showing the size distribution of DNA fragments observed upon running sheared genomic DNA on an Agilent BioAnalyzer.
Basic Protocol 4: Construction and amplification of DNA libraries
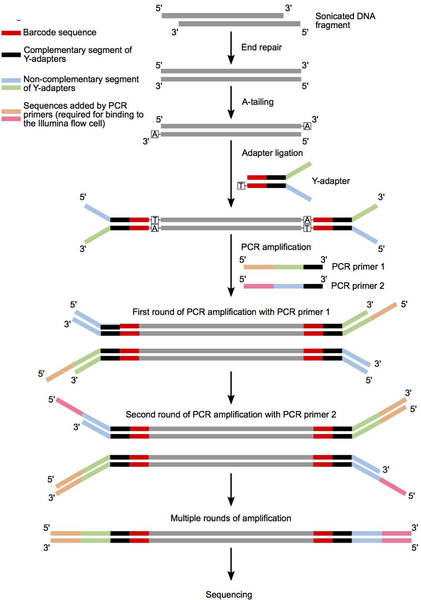
The basic steps involved in the construction and amplification of DNA libraries for whole genome sequencing are shown in Figure 3.
Figure 3.
A schematic showing the different steps involved in construction of DNA libraries for whole genome sequencing starting from sonicated genomic DNA. The sequences of the different color-coded regions are highlighted in Table 2.
Materials:
Qiagen GeneRead Library I Core Kit (Qiagen #180434)
Thermocycler
1.7 ml microfuge tubes
10 μl annealed adapters (see Support Protocol 1)
SPRI magnetic beads
Magnetic stand holding 1.7 ml microfuge tubes
Elution buffer (10 mM Tris-Cl, pH 8.5 – composition of elution buffer in most DNA column purification kits)
0.8% Agarose gel (with ethidium bromide diluted 25,000x)
Phusion polymerase
100 mM dNTPs
5 μM primer mix (5 μM each of PCR primer 1 and PCR primer 2, see Table 2)
Table 2:
Oligo sequences for PCR amplification and forming Y-adapters
| Oligo | Sequence |
|---|---|
| Barcoded Oligo 1 | 5’ACACTCTTTCCCTACACGACGCTCTTCCGATCT-barcode-T 3’ |
| Barcoded Oligo 2 | 5’p-barcode(reverse complement)-AGATCGGAAGAGCGGTTCAGCAGGAATGCCGAG 3’ |
| PCR primer 1 | CAAGCAGAAGACGGCATACGAGATCGGTCTCGGCATTCCTGCTGAACCGCTCTTCCGATC*T |
| PCR primer 2 | AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATC*T |
indicates phosphorothioate bond
5’ p indicates a 5’ phosphate bond
Table adapted from (Wong et al., 2013; see Current Protocols article). The colors correspond to the diagram in Figure 3
PCR tubes
Sterile distilled water
End repair, A-tailing and adapter ligation of DNA
The steps listed in the sections are based on the guidelines provided by Qiagen as part of the GeneRead Library I core kit (https://www.qiagen.com/us/products/next-generation-sequencing/library-preparation/generead-dna-l-core-kit/). Other kits that perform equivalent steps resulting in adapter ligated DNA fragments can also be used.
-
1End repair of DNA fragments: Set up each end repair reaction using solutions from the Qiagen GeneRead Library I core kit as follows:
Sonicated DNA 10.25 μl 10x End repair buffer 1.25 μl End repair enzyme mix 1 μl Total 12.5 μl When doing end repair for more than one sample, make a master mix of the reagents excluding the sonicated DNA. Distribute the mix to each tube, then add the sonicated DNA. Incubate the tubes in a thermocycler with the following conditions:25°C 30 minutes 75°C 20 minutes 4°C Infinite hold Set the lid of the thermocycler to 105°C.
-
2A-tailing of DNA: Set up each A-tailing reaction using solutions from the Qiagen GeneRead Library I core kit as follows:
End-repaired DNA 12.5 μl 10x A-addition buffer 1.5 μl Klenow fragment (3’->5’ exo) 1.5 μl Total 15.5 μl When doing A-tailing for more than one sample, make a master mix of the reagents excluding the end-repaired DNA. Distribute the mix to each tube containing the end-repaired DNA. Incubate the tubes in a thermocycler with the following conditions:37°C 30 minutes 75°C 10 minutes 4°C Infinite hold Set the lid of the thermocycler to 105°C.
-
3Adapter ligation: Set up each adapter ligation reaction using solutions from the Qiagen GeneRead Library I core kit as follows:
A-tailed DNA 15.5 μl Adapters (from 10 μM stock) 1 μl 2x Ligation buffer 22.5 μl T4 Ligase 2 μl RNase free water 4 μl Total 45 μl When doing adapter ligation for more than one sample, make a master mix of the reagents excluding the A-tailed DNA and the adapters. Distribute the mix to each tube containing A-tailed DNA. Add 1 μl of adapters containing the appropriate barcode sequence to each sample. Incubate the tubes in a thermocycler with the following conditions:25°C 10 minutes 4°C Infinite hold Do not use a heated lid for this step.
Samples may be stored at −20°C at this point.
Clean-up and size selection of adapter ligated DNA fragments
-
4
Bring up the volume of each sample from the previous step to 60 μl using water. Perform one round of SPRI bead purification (see support protocol 2). Elute the DNA by adding 100 μl of Elution buffer to the beads. Transfer 95 μl of the eluted DNA to a new tube.
The transfer of DNA after elution should be done in two steps – first transfer 90 μl using a 200μl pipette, next transfer 5 μl using a 10 μl pipette. This will help avoid contamination from SPRI beads.
-
5
Bring up the volume of sample to 100 μl using water. Perform a second round of SPRI bead purification (see Support Protocol 2). Elute the DNA by adding 16 μl of Elution buffer to the beads. Transfer 15 μl of the eluted DNA to a new tube.
The transfer of DNA after elution should be done in two steps of 7.5 μl each using a 10 μl pipette. This will help avoid the contamination from SPRI beads.
Determination of PCR cycle numbers for library amplification
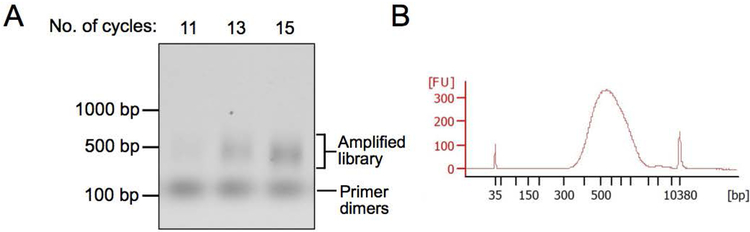
The number of PCR cycles required for sufficient amplification of the DNA libraries has to be determined empirically. At least 6–8 cycles are minimally required to incorporate the Illumina adapters. Beyond that, the goal is to choose the minimum number of cycles required to obtain sufficient material for sequencing. This is determined by setting up three 10 μl PCR reactions for each sample and carrying out each reaction for a different number of cycles. The DNA products are then run on a gel and the optimal cycle number for amplification is determined as the cycle number where amplification products are just visible on an agarose gel stained with ethidium bromide. For DNA libraries starting with 150 ng of DNA for end repair, the final number of PCR cycles chosen for amplification is in the range of 11–15. Hence, three different cycle numbers of 11, 13 and 15 are tested for PCR amplification. However, depending on the sample, a different range of PCR cycles might be required for obtaining optimal amplification.
-
6Set up three PCR reactions for each sample as follows:
Per reaction 3 reactions
(3.3x to account for pipetting error)Size-selected DNA 0.3 0.99 Water 6.4 21.12 5x HF buffer 2 6.6 5 μM primer mix 1 3.3 10 mM dNTPs 0.2 0.66 Phusion polymerase 0.1 0.33 Total 10 33 When doing PCR for more than one sample, make a master mix of the reagents excluding the template DNA. Distribute the mix to each tube and then add the template DNA. Divide the PCR reaction into three different tubes (10 μl each). Incubate the tubes in a thermocycler set to the following cycling conditions:98°C 30 seconds 98°C 10 seconds 65°C 30 seconds 11, 13 or 15 cycles 72°C 30 seconds 72°C 5 minutes 4°C Infinite hold For amplifying samples for a different number of cycles, place all samples in one thermocycler set to cycle for 15 cycles. Have another thermocycler set to hold temperature at 72°C. When the required number of cycles for each sample is completed, quickly transfer the sample to the 72°C thermocycler for 5 minutes, then place the sample on ice.
-
7
Run all of the sample on a 0.8% agarose gel. Examples of amplified libraries are shown in Figure 4A. Determine the optimal number of cycles for each sample.
-
8Scale up the DNA amplification for each library by setting up PCR reactions for each sample as follows:
Per sample Size-selected DNA 3 Water 64 5x HF buffer 20 5 μM primer mix 10 10 mM dNTPs 2 Phusion polymerase 1 Total 100 When doing PCR for more than one sample, make a master mix of the reagents excluding the template DNA. Distribute the mix to each tube and then add the template DNA. Incubate the tubes set to the same conditions as in step 6.
-
9
Run 10 μl of the amplified DNA on a 0.8% agarose gel to check for proper amplification.
Samples may be store at −20°C at this step.
Figure 4.
PCR amplification of sequencing libraries. (A) An agarose gel (0.8%) showing a single library PCR amplified for varying number of cycles. Thirteen cycles were chosen for the final amplification. (B) Electropherogram showing the size distribution of PCR-amplified DNA fragments after SPRI bead purification.
Final clean up and size selection of amplified libraries
-
10
Bring up the sample volume to 100 μl using water. Perform one round of SPRI bead purification (see support protocol 2). Elute the DNA by adding 50 μl of Elution buffer to the beads. Transfer 49 μl of the eluted DNA to a new tube.
The transfer of DNA after elution should be done in two steps – first transfer 42 μl using a 200μl pipette, next transfer 7 μl using a 10 μl pipette. This will help avoid the contamination from SPRI beads.
-
11
Bring up the sample volume to 50 μl using water. Perform a second round of SPRI bead purification (see support protocol 2). Elute the DNA by adding 16 μl of Elution buffer to the beads. Transfer 15 μl of the eluted DNA to a new tube.
The transfer of DNA after elution should be done in two steps of 7.5 μl each using a 10 μl pipette. This will help avoid the contamination from SPRI beads.
-
12
Mix 4 μl of the DNA with 16 μl of water to make a 1:5 dilution of the purified DNA. Analyze 1 μl of this sample on a BioAnalyzer to determine its concentration and ensure absence of primer dimers (see troubleshooting section). A sample BioAnalyzer trace is shown in Figure 4B.
Pooling libraries for next generation sequencing
-
Pool the different samples that have been barcoded with distinct adapter sequences such that the final concentration of each library in the pool is equal.
When pooling libraries for sequencing, use DNA from the 1:5 diluted sample since its concentration has been measured directly. Typically, 2–20 nM of pooled DNA is submitted for a Hi-Seq or a Next-Seq run.
Confirm the concentration of the pooled libraries by analyzing 1 μl of the pooled sample on a BioAnalyzer. Submit samples for sequencing.
Support Protocol 1: Annealing oligonucleotides for forming Y-adapters
This section describes a protocol for annealing partially complementary oligonucleotides that are then ligated to the fragmented genomic DNA and used as adaptors. The end product of this reaction is a double-stranded DNA molecule with a single ‘T’ overhang at one end, and non-complementary single-stranded DNA at the other end, resulting in a Y-shaped adapter.
Materials:
100 μM barcoded oligo 1 (see Tables 1 and 2)
100 μM barcoded oligo 2 (see Tables 1 and 2)
PCR tubes
Thermocycler
1.7 ml microfuge tubes
- Mix 10 μl of barcoded oligos 1 and 2 in a PCR tube. Incubate the tubes in a thermocycler set to the following cycling conditions:
95°C 5 minutes 94°C 1 minute 93°C 1 minute ⁞
Decrease 1°C every 1 minute
⁞26°C 1 minute 25°C 1 minute 4°C Infinite hold -
Transfer the annealed oligos to a microfuge tube. Make a 1:10 dilution of the annealed oligos (1 μl oligos + 9 μl water) to get the oligos at a concentration of 10 μM. Store the 100 μM and 10 μM primers at −20°C.
Aliquot the annealed adapters into smaller volumes depending on their usage. This will help to minimize the number of freeze thaw cycles.
Support Protocol 2: Size selection and cleanup using SPRI beads
The following protocol is described for low-throughput processing of samples. If a large number of samples are being processed, the same protocol can be adapted to a 96-well plate format. The only additional equipment required would be a magnetic stand suitable for a 96-well plate.
Materials:
SPRI beads
1.7 ml microfuge tubes
Magnetic stand that holds 1.7 ml microfuge tubes
70% ethanol
37°C incubator
Elution buffer (10 mM Tris-Cl, pH 8.5 – composition of elution buffer in most DNA column purification kits)
Incubate the tube containing SPRI beads for 30 minutes at room temperature. Mix the bead suspension by inverting the tube a few times.
Add 0.7x volume of SPRI beads to 1 volume of DNA. Mix by pipetting and incubate at room temperature for 5 minutes.
Place the tubes on the magnetic stand and incubate at room temperature for 10 minutes. Carefully pipette out and discard the supernatant.
-
Add 500 μl of 70% ethanol to the tubes while still on the stand. Incubate the tubes on the magnetic stand for 1 minute. Carefully pipette out and discard the supernatant. Repeat this step once.
If using a 96-well plate, use 200 μl of ethanol for this step.
-
Incubate the tubes for 20 minutes at 37°C while on the magnetic stand with tube caps open. Allow the beads to dry completely.
Dried beads will have a cracked pellet resembling sand.
-
Suspend the beads in the required volume of elution buffer. Remove the tubes from the magnetic stand and mix the beads by pipetting. Incubate the tubes at room temperature for 5 minutes.
Do not remove the tubes from the magnetic stand before addition of elution buffer.
-
Place the tubes on the magnetic stand and incubate at room temperature for 10 minutes. Carefully pipette out the supernatant into a new microfuge tube taking care not to disturb the beads.
It is recommended to sacrifice a small amount of sample while pipetting out the eluted DNA. This will help reduce bead contamination in downstream steps.
Basic Protocol 5: Identification of genomic variants from sequencing data
There are multiple tools and packages available for analysis of sequencing data and identification of genomic variants (Sandmann et al., 2017). We have developed a custom workflow using Snakemake (Koster and Rahmann, 2012) for analysis of whole genome sequencing data that uses GATK (DePristo et al., 2011; McKenna et al., 2010; Van der Auwera et al., 2013) for variant calling and filtering. This pipeline is available at https://github.com/winston-lab/wgs-pipeline, and can be used for processing S. cerevisiae and S. pombe samples. The basic steps involved in the pipeline are listed below.
Demultiplex the FASTQ files and perform quality trimming of reads. The reads from the sequenced fastq file are demultiplexed according to the inline barcode sequences using the fastq-multx command (Aronesty, 2011). Each demultiplexed fastq file is then subjected to quality trimming using cutadapt (Martin, 2010).
Align reads to the reference genome. The sequenced reads are aligned to the reference genome using Bowtie2 using default parameters (Langmead and Salzberg, 2012). Other alignment tools such as bwa (Li and Durbin, 2009) can also be used for this step.
Remove multiple aligners and add read groups. Reads that map to more than one location are removed using samtools (Li et al., 2009). Read groups are added to the BAM file using picard (http://broadinstitute.github.io/picard/). This is required to get the data in the right format for GATK to call variants.
Call and filter variants. Variant calling is done using the HaplotypeCaller command as part of the GATK suite of tools. This is followed separating indels and SNPs and filtering each set of variants according to standard recommended settings (see Current Protocols article: Van der Auwera et al., 2013).
Remove variants present in the parent sample. The variants identified so far are with respect to the reference genome. The variants that are present in the mutant samples as well as the parental strains are identified and filtered out. These are not likely to be causal variants.
Annotate the variants. Using an annotation file derived from SGD (https://www.yeastgenome.org/; for S. cerevisiae) or from Pombase (https://www.pombase.org/; for S. pombe), the variants are annotated as being intergenic or within the coding regions of genes.
The final output of the pipeline is a list of SNPs that are ordered according to the quality score of the variant call generated by GATK. This gives a short list of candidate variants that can be tested for causality by reconstructing the mutation in the parental strain. A recent study that conducted bulk segregant analysis reported the probability of finding a non-causal variant with an allele frequency greater than or equal to 0.67 in the mutant pool to be 5% (Coelho et al., 2019), which can be used as a threshold to further shortlist candidates. This number may vary based on the strain background, number of spores pooled and sample preparation methods.
Reagents and Solutions
YPD
1% Yeast extract
2% Peptone
2% Glucose
Smash and grab solution (Hoffman and Winston, 1987)
10mM Tris pH 8.0
100mM NaCl
1mM EDTA
1% SDS
2% Triton-X100
TE solution
10mM Tris pH 8.0
1mM EDTA
Commentary
Background Information
Bulk segregant analysis is useful if a large number of background mutations are expected to be found, or a polygenic trait is being studied. However, if the mutations have been obtained spontaneously and preliminary genetic analyses suggest that the phenotype of interest segregates in a Mendelian fashion, the mutant strain can be sequenced directly or after it has been put through a single back-cross. Alternatively, if multiple mutations are in the same complementation group or if they are tightly linked, the strains can be sequenced without being subjected to additional crosses (Gopalakrishnan et al., 2019). At the analysis step after whole genome sequencing, the causative variants can be identified as those present at the same genetic locus in all the mutants sequenced.
Critical Parameters
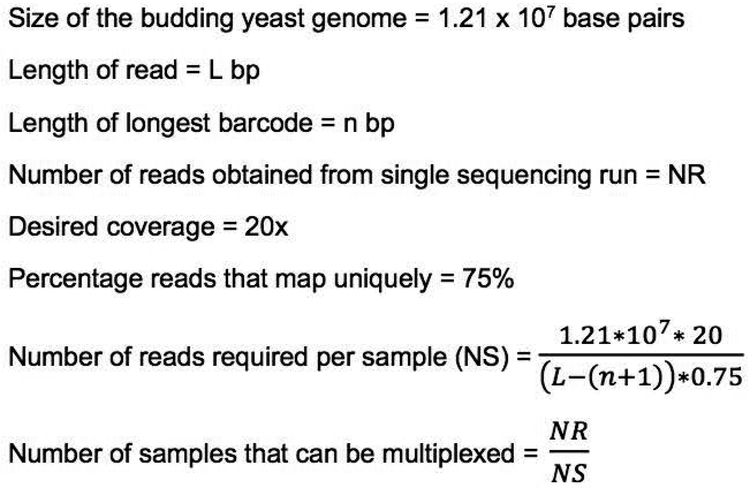
The number of sequencing libraries that can be multiplexed depends on the length of the read being sequenced and total number of reads obtainable from a single sequencing run. An estimate of the number of samples that can be multiplexed for obtaining an average coverage of 20x, which we have found to be sufficient for most experiments in our lab, can be obtained using the calculations outlined in Figure 5. The actual coverage required to identify causal variants can vary based on the experiment. This calculation includes an assumption of percentage of reads in a sample that map uniquely to the reference genome. The number presented here is based on the sequencing data analyzed in our lab and should be treated as a rough estimate. The actual percentage of uniquely mapping reads will differ between sequencing runs and yeast strain backgrounds.
Figure 5.
Calculation of the number of libraries that can be multiplexed in a single whole genome sequencing experiment.
It is critical to maintain the diversity of bases in the barcode sequences and to maintain a similar proportion of each nucleotide at every position in the barcode. This is especially important when multiplexing a small number of samples. If balancing of barcode sequences is difficult, then sequence diversity may be increased by spiking-in exogenous DNA such as a PhiX DNA library (https://www.illumina.com/products/by-type/sequencing-kits/cluster-gen-sequencing-reagents/phix-control-v3.html) (Mitra et al., 2015).
An alternate method to multiplex samples is to introduce the barcode sequences at the PCR amplification stage instead of including them in the adapters. The barcode sequences (also called indexes) are included as part of the primers used for PCR and are identified using a separate indexing primer during an Illumina sequencing run (Meyer and Kircher, 2010). Following demultiplexing, the downstream steps for identification of variants are the same as those listed here. An advantage of using this technique for multiplexing is that the sequence information from the entire read can be used for variant analysis and no portion of it is lost to barcoded sequences.
Troubleshooting
Low yields of genomic DNA
Genomic DNA yields can vary between yeast strains. If low yields of DNA are obtained, a larger volume of cells can be used as a starting material. Alternatively, DNA isolation kits that do not use glass beads or enzymatic steps can be used. Such protocols would avoid the use of phenol chloroform extractions and thereby prevent the loss of DNA associated with those steps.
Low yield of DNA following PCR amplification
The cause for this low yield could be either due to low levels of input DNA or due to inefficient ligation of adapters. The DNA adapters can get denatured over repeated freeze thaw cycles. Repeat the library preparation with a higher quantity of input DNA and freshly annealed adapters. This is recommended over choosing a higher number of PCR cycles as the latter can lead to biased coverage and increase the probability of introducing mismatches during amplification.
Contamination from primer dimers after PCR amplification and SPRI bead cleanup
Following PCR amplification and size selection using SPRI beads, if a sharp peak around 100 bp is observed upon analyzing the sample on an Agilent BioAnalyzer, it might be indicative of contamination from primer dimers. An additional round of SPRI bead purification or alternate size selection methods such as gel or column purification with the appropriate molecular weight cutoff can be used to resolve this problem.
Understanding Results
The coverage obtained for each sample may be different from the expected value. This might be due to pipetting error when pooling the barcoded samples as well as run-to-run variability in cluster formation during sequencing. Additionally, the coverage across the genome may not be uniform due to biases introduced at different steps during library preparation. However, we have been able to identify variants from sequencing data having an average coverage as low as 10x. On average, we find that 75% - 85% of reads map uniquely to the reference genome. The number of variants identified will depend on the sequencing depth as well as the mutagenesis rate prior to obtaining the yeast mutants. Spontaneous mutants typically tend to have a lower frequency of non-causal variants.
Time Considerations
A rough time frame for the steps listed in this protocol is provided in Table 3 below. The actual time for each step will depend on the number of samples being processed simultaneously.
Table 3:
Time Considerations
| Step | Approximate time | Notes |
|---|---|---|
| Bulk segregant analysis | 2 weeks | The time taken will depend on sporulation efficiency of the back-crossed diploid and growth rate of spores exhibiting the mutant phenotype |
| Growing colonies for genomic DNA isolation | 2 days | |
| Genomic DNA isolation | 7 hours | Samples can be stored at −20°C until further processing during DNA precipitation |
| Sonication and checking fragmentation | 2 hours | The time taken will depend on the sonicator being used and the ability to process multiple samples in parallel |
| End-repair, A-tailing and adapter ligation | 2 hours | Samples can be stored at −20°C after each enzymatic step |
| One round of SPRI bead purification | 2 hours | |
| Annealing oligos to form Y-adapters | 1.5 hours | |
| PCR amplification | 1.5 hours | The time for this step will depend on the number of PCR cycles used for amplification |
Acknowledgments
This work was supported by NIH grant R01GM120038 to F.W. We thank Catherine Weiner for critical reading of the manuscript, Francheska Lopez Rivera for contributing the gel images used in Figure 4, Dan Pagano for helping develop the bioinformatic pipeline, and Zarmik Moqtaderi and Kevin Struhl for the barcode sequences.
Literature Cited
- Aronesty E (2011). ea-utils : “Command-line tools for processing biological sequencing data”.
- Birkeland SR, Jin N, Ozdemir AC, Lyons RH Jr., Weisman LS, and Wilson TE (2010). Discovery of mutations in Saccharomyces cerevisiae by pooled linkage analysis and whole-genome sequencing. Genetics 186, 1127–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brauer MJ, Christianson CM, Pai DA, and Dunham MJ (2006). Mapping novel traits by array-assisted bulk segregant analysis in Saccharomyces cerevisiae. Genetics 173, 1813–1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry JM, Ball C, Weng S, Juvik G, Schmidt R, Adler C, Dunn B, Dwight S, Riles L, Mortimer RK, et al. (1997). Genetic and physical maps of Saccharomyces cerevisiae. Nature 387, 67–73. [PMC free article] [PubMed] [Google Scholar]
- Cherry JM, Hong EL, Amundsen C, Balakrishnan R, Binkley G, Chan ET, Christie KR, Costanzo MC, Dwight SS, Engel SR, et al. (2012). Saccharomyces Genome Database: the genomics resource of budding yeast. Nucleic Acids Res 40, D700–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coelho MC, Pinto RM, and Murray AW (2019). Heterozygous mutations cause genetic instability in a yeast model of cancer evolution. Nature 566, 275–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, et al. (2011). A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 43, 491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrenreich IM, Torabi N, Jia Y, Kent J, Martis S, Shapiro JA, Gresham D, Caudy AA, and Kruglyak L (2010). Dissection of genetically complex traits with extremely large pools of yeast segregants. Nature 464, 1039–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsburg SL (2001). The art and design of genetic screens: yeast. Nat Rev Genet 2, 659–668. [DOI] [PubMed] [Google Scholar]
- Gopalakrishnan R, Marr SK, Kingston RE, and Winston F (2019). A conserved genetic interaction between Spt6 and Set2 regulates H3K36 methylation. Nucleic Acids Res 47, 3888–3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman CS, and Winston F (1987). A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene 57, 267–272. [DOI] [PubMed] [Google Scholar]
- Koster J, and Rahmann S (2012). Snakemake--a scalable bioinformatics workflow engine. Bioinformatics 28, 2520–2522. [DOI] [PubMed] [Google Scholar]
- Langmead B, and Salzberg SL (2012). Fast gapped-read alignment with Bowtie 2. Nat Methods 9, 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, and Durbin R (2009). Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, and Genome Project Data Processing, S. (2009). The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundblad V (2001). Cloning yeast genes by complementation. Curr Protoc Mol Biol Chapter 13, Unit13 18. [DOI] [PubMed] [Google Scholar]
- Martin M (2010). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnetjournal. [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, et al. (2010). The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20, 1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer M, and Kircher M (2010). Illumina sequencing library preparation for highly multiplexed target capture and sequencing. Cold Spring Harb Protoc 2010, pdb prot5448. [DOI] [PubMed] [Google Scholar]
- Meyer M, Stenzel U, Myles S, Prufer K, and Hofreiter M (2007). Targeted high-throughput sequencing of tagged nucleic acid samples. Nucleic Acids Res 35, e97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra A, Skrzypczak M, Ginalski K, and Rowicka M (2015). Strategies for achieving high sequencing accuracy for low diversity samples and avoiding sample bleeding using illumina platform. PLoS One 10, e0120520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng PC, and Kirkness EF (2010). Whole genome sequencing. Methods Mol Biol 628, 215–226. [DOI] [PubMed] [Google Scholar]
- Sandmann S, de Graaf AO, Karimi M, van der Reijden BA, Hellstrom-Lindberg E, Jansen JH, and Dugas M (2017). Evaluating Variant Calling Tools for Non-Matched Next-Generation Sequencing Data. Sci Rep 7, 43169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurtle-Schmidt DM, Dodson AE, and Rine J (2016). Histone Deacetylases with Antagonistic Roles in Saccharomyces cerevisiae Heterochromatin Formation. Genetics 204, 177–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong AH, and Boone C (2006). Synthetic genetic array analysis in Saccharomyces cerevisiae. Methods Mol Biol 313, 171–192. [DOI] [PubMed] [Google Scholar]
- Tong AH, Evangelista M, Parsons AB, Xu H, Bader GD, Page N, Robinson M, Raghibizadeh S, Hogue CW, Bussey H, et al. (2001). Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science 294, 2364–2368. [DOI] [PubMed] [Google Scholar]
- Treco DA, and Winston F (2008). Growth and manipulation of yeast. Curr Protoc Mol Biol Chapter 13, Unit 13 12. [DOI] [PubMed] [Google Scholar]
- Van der Auwera GA, Carneiro MO, Hartl C, Poplin R, Del Angel G, Levy-Moonshine A, Jordan T, Shakir K, Roazen D, Thibault J, et al. (2013). From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics 43, 11 10 11–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenger JW, Schwartz K, and Sherlock G (2010). Bulk segregant analysis by high-throughput sequencing reveals a novel xylose utilization gene from Saccharomyces cerevisiae. PLoS Genet 6, e1000942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong KH, Jin Y, and Moqtaderi Z (2013). Multiplex Illumina sequencing using DNA barcoding. Curr Protoc Mol Biol Chapter 7, Unit 7 11. [DOI] [PubMed] [Google Scholar]