Summary:
Despite the worldwide success of vaccination, newborns remain vulnerable to infections. While neonatal vaccination has been hampered by maternal antibody-mediated dampening of immune responses, enhanced regulatory and toleragenic mechanisms, and immune system immaturity, maternal pre-natal immunization aims to boost neonatal immunity via antibody transfer to the fetus. However, emerging data suggest that antibodies are not transferred equally across the placenta. To understand this, we used systems serology to define Fc features associated with antibody transfer. The Fc-profile of neonatal and maternal antibodies differed, skewed towards NK cell-activating antibodies. This selective transfer was linked to di-galactosylated Fc-glycans that selectively bind FcRn and FCGR3A, resulting in transfer of antibodies able to efficiently leverage innate immune cells present at birth. Given emerging data that vaccination may direct antibody glycosylation, our study provides insights for the development of next-generation maternal vaccines designed to elicit antibodies that will most effectively aid neonates.
Keywords: Antibody functionality, antibody glycosylation, trans-placental transfer, maternal vaccination, ADCC, neonatal immunology
Graphical Abstract
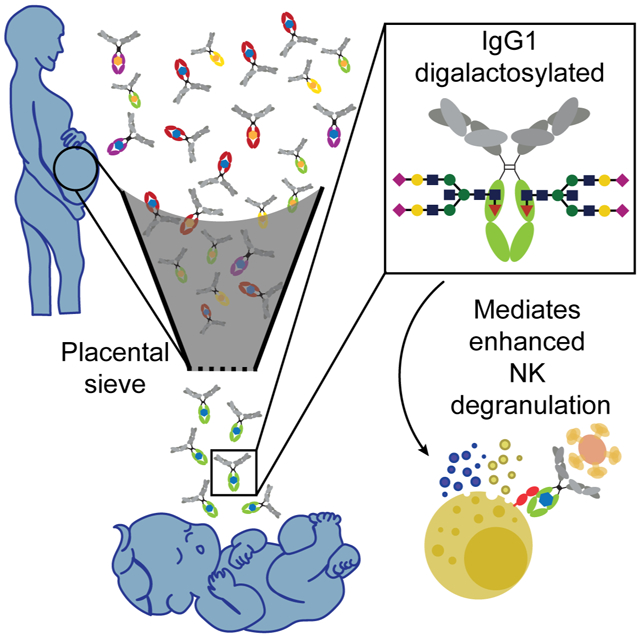
eTOC:
Antibodies with a specific glycan modification and with the ability to activate NK cells are selectively transferred across the placenta to the neonate.
Introduction
Vaccines, one of the most impactful public health interventions, have reduced global morbidity and mortality against infectious disease (Centers for Disease and Prevention, 1999; Pulendran and Ahmed, 2011). However, in newborns vaccines have been less effective at reducing infection-related deaths (Amenyogbe et al., 2015). Compromised vaccine-induced immunity in infants has been attributed to the potentially toleragenic nature of the neonatal immune system (Yu et al., 2018), the reduced functionality of newborn immune cells (Lee and Lin, 2013; Yu et al., 2018), as well as dampened immunity from pre-existing maternal antibodies (Saso and Kampmann, 2017). Strategies have been proposed to drive more effective immunity in newborns, including designing vaccines and adjuvants tailored to neonates (Saso and Kampmann, 2017; Whittaker et al., 2018). Maternal immunization, aimed at enhancing maternal-to-fetal transfer of antibodies, has shown significant promise in boosting newborn immunity (Forsyth et al., 2015), providing a non-invasive strategy to enhance immunity in this vulnerable population. However, epidemiologic studies focusing on matched mother:fetus pairs, have found that the extent of immunity transferred varies significantly by antigen (Fu et al., 2016; Palmeira et al., 2012). Specifically, while measles-specific antibodies are transferred efficiently (>100%), antibodies targeting other pathogens, including poliovirus and coxsackie viruses, are less efficiently transferred (Fu et al., 2016).
The neonatal Fc receptor, FcRn, is responsible for receptor-mediated trans-placental transport of IgG (Roopenian and Akilesh, 2007). FcRn binds IgG in a pH-dependent manner within acidified endosomes in syncytiotrophoblasts and transits IgG to the interstitial space between the maternal and fetal circulation (Jennewein et al., 2017; Roopenian and Akilesh, 2007). While FcRn binds to the CH3 domain of all IgG subclasses (Vidarsson et al., 2014), differences in IgG subclass transfer have been noted (Einarsdottir et al., 2014; Vidarsson et al., 2014), including enhanced binding to IgG1 and differential transfer efficiencies of allotypic variants of IgG3 known to bind FcRn with different affinities (Stapleton et al., 2011; Vidarsson et al., 2014). Given that FcRn binding occurs in the CH3 domain, IgG transport, and particularly antigen-specific IgG1 transport, should occur at the same rate. However, a transfer hierarchy exists across antigen-specific antibody subpopulations (Fu et al., 2016). Thus, other qualitative antibody features are likely to govern differential transfer of particular antibody populations.
With the re-emergence of pertussis infections among newborns (Winter et al., 2014), efforts have emerged to understand the transfer of pertussis immunity to neonates. Thus, we aimed to dissect the profile of transferred antibodies, focused on defining whether Fc features influence pertussis-specific antibody placental transfer, to help inform maternal vaccine campaigns and next-generation vaccine design. A global, unbiased, and antigen-specific systems serology antibody profiling approach was applied (Chung and Alter, 2017) to deeply and comprehensively define the specific qualitative Fc features of antibodies found in maternal and cord blood on the day of birth. Striking differences were observed in the functional profile of antibodies transferred to neonates, with preferential transfer of NK cell-activating antibodies. This preferential transfer, observed across many antigens, was linked to antigen-specific Fc-glycan profiles on the Fc-domain of antigen-specific antibodies, as well as to enhanced binding to FcRn and FCGR3A, two receptors found to be co-localized on syncytiotrophoblasts. The transfer of FCGR3A-binding, NK cell-activating antibodies coincided with the presence of fully competent NK cells in the cord blood, compared to less competent cord blood neutrophils. These data suggest an evolution of the placenta to selectively transfer antibodies with the most functional potential in the neonatal immune context to better provide protection to neonates.
Results
Unique signatures of placental transfer of Fc function
The placenta preferentially transfers IgG antibodies (Vidarsson et al., 2014). Though FcRn binds to all IgG subclasses, differences in transfer efficiencies have been noted across the subclasses (IgG1>4>3>2) (Palmeira et al., 2012; Vidarsson et al., 2014; Wilcox et al., 2017) and across IgG1 populations (Fu et al., 2016), raising the possibility for FcRn-mediated preferential selection of IgG1 transfer. Thus, to begin to define the characteristics of antibodies that are preferentially transferred across the placenta, we aimed to define whether the transferred antibodies possessed any qualitative functional differences from those in mothers, focusing on pertussis-specific immunity. Using samples drawn from a cohort of 14 mother:cord pairs on the day of birth (Table S1, Figure S1), we compared the functional activity of antibodies specific to the four pertussis antigens included in the Tdap vaccine–Pertactin (PTN), Filamentous Hemagglutinin (FHA), Fimbriae 2/3 (FIM) and Pertussis Toxin (PTX) (Edwards and Berbers, 2014).
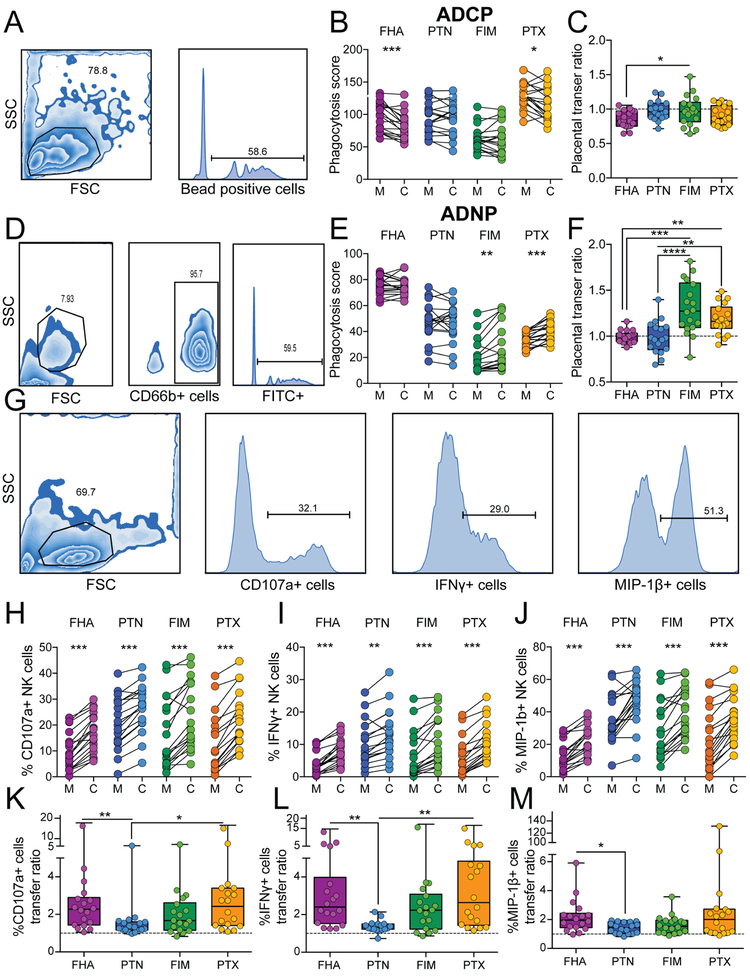
While significant differences existed in the magnitude of antibody-dependent monocyte phagocytosis (ADCP) across the pertussis antigens, including higher levels of PTX compared to FIM specific phagocytic antibodies in mothers (Figure 1A and B), overall transfer of phagocytic antibodies was relatively stable across all specificities (Figure 1B and C). Similarly, heterogeneous magnitudes of antibody-dependent neutrophil phagocytosis (ADNP) were observed across antigen specificities (Figure 1D and E). However significant differences were observed in antigen-specific ADNP transfer across specificities (Figure 1F), where FIM- and PTX-ADNP inducing antibodies were transferred with high efficiency compared to FHA and PTN specific antibodies. These data point to heterogeneity in overall levels and transfer efficiencies across pertussis specific monocyte and neutrophil recruiting antibodies, driven in an antigen-specific manner.
Figure 1: Placental transfer of antibody function.
Antibodies against pertussis derived filamentous haemagglutinin (FHA), pertactin (PTN), fimbriae (FIM), and pertussis toxin (PTX) antigens were compared in 14 mother:cord pairs. A. The flow cytometric plots depict the gating strategy for antibody dependent cellular phagocytosis (ADCP). B. The connected dot-plot shows the phagocytic activity across mother:cord pairs. C. The box and whisker plot shows the transfer ratio of ADCP. The dotted line indicates a 100% transfer efficiency (equivalent levels across both compartments). D. The flow plots highlight the gating strategy for antibody dependent neutrophil phagocytosis (ADNP). E. The dot-plot shows the relationship between ADNP activity across mother:cord pairs for each antigen-specificity. F. The whisker plots show the transfer ratio for ADNP. G. The flow plots highlighting the gating strategy for the NK cell activation assay. H-J. The dot-line plots show NK-dependent degranulation plotted as the percentage of NK cells positive for CD107a (H), IFNγ (I) and MIP-1β (J). K-M The whisker plots depict the transfer ratio across the NK cell activation assays. Significance for dot-line plots was calculated using a Wilcoxon test with a Bonferroni correction, where p<0.0125 was considered significant, *p<0.0125, **p<0.0025, ***p<0.00025. Significance across transfer rations was evaluated using a Friedman test with Dunn’s post-test correction **p<0.01, ***p<0.001, ****p<0.0001. See also Table S1 and Figure S1
To define whether the same variability would be observed across NK cell activating antibodies, we examined the ability of the maternal and cord antibodies to drive NK cell degranulation (CD107a upregulation), NK cell cytokine secretion (interferon-γ, IFNγ), and chemokine secretion (macrophage inflammatory protein-1β, MIP-1β). Strikingly, while lower levels of FHA specific NK cell-activating antibodies were observed across all mothers, all mothers transferred elevated levels of NK cell-activating antibodies (across all three degranulation readouts) (Figures 1H, I, and J), albeit at different transfer ratios (Figures 1K, L, and M). Across all antigen specificities and mother:cord pairs, enhanced antibody transfer was consistently observed for NK cell-activating antibodies. Thus, our data demonstrate the preferential transfer of antibodies involved in NK cell activation to neonates, providing the first evidence of a functional, potentially Fc-specific, sieving across the placenta.
Validation of the placental functional sieve
To confirm that the preferential transfer of NK cell recruiting antibodies could be extended beyond pertussis-specific immunity, the functional characteristics of viral-specific antibodies were interrogated. Similar to the results observed for pertussis, significant variability existed in the levels of functional antibodies to respiratory syncytial virus (RSV) and influenza (Flu) across mothers (Figure 2A and B), accompanied by variable ADCP and ADNP transfer across each antigen (Figure 2A-D). Conversely, enhanced transfer of NK cell IFNγ and MIP-1β inducing antibodies were transferred across both specificities, as were degranulation (CD107a) inducing antibodies to RSV (Figure 2E and F and Figure S2 C-F). Additionally, the NK cell transfer signature was further validated in a second geographically distinct cohort across pertussis, RSV, and measles, highlighting again, preferential transfer of NK-activating antibodies to neonates (Figure S2 A,B). Thus, despite heterogenous baseline pathogen-specific functional antibody levels, NK cell-activating antibodies are consistently preferentially transferred across the placenta.
Figure 2: Confirmatory antiviral-antibody placental transfer profiles.
To validate the pertussis specific placental transfer signature across additional specificities, the functional transfer profile was assessed across two viral targets, respiratory syncytial virus (RSV) and influenza (Flu). A. The dot-line plot show the ADCP levels across paired mother:cord samples. B. The dot-line plot highlights ADNP levels across paired mother:cord samples. C,D. The whisker plots show the transfer ratios across cord/mother for ADCP and ADNP, respectively. E. The dot-line plots show the ability of RSV and Flu specific antibodies across mother:cord pairs to recruit NK cell activation quantified as the level of CD107a, IFNγ and MIP-1β upregulation. F. The whisker plots show the transfer ratios for each NK cell function across each specificity. Significance for dot-line plots was evaluated using a Wilcoxon test with a Bonferroni correction, where p<0.025 was considered significant, *p<0.025, **p<0.005 F. Additionally, significant differences in transfer ratios were evaluated using a Friedman’s test with a Dunn’s post-test correction. See also Figure S2.
Antibody Fc glycans, rather than subclass, contribute to placental sieving
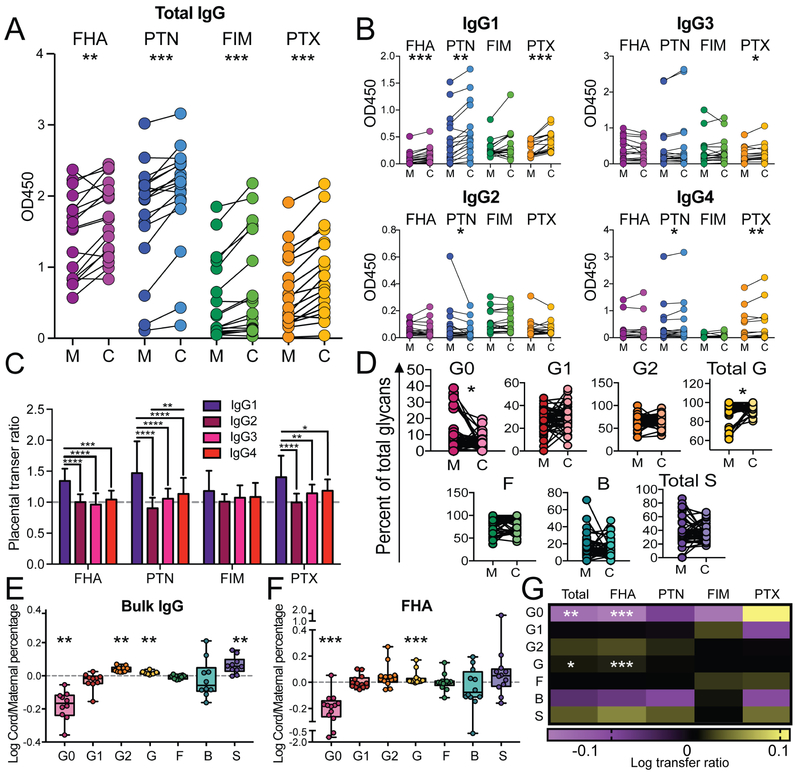
To begin to define the specific biophysical characteristics of antibodies that are selectively sieved via the placenta, we next examined the transfer of both antigen-specific IgG subclass and Fc glycosylation, which both influence Fc effector functions (Davies et al., 2001; Jennewein and Alter, 2017). As expected, total IgG was enhanced in the cords across the four pertussis antigens for all pairs (Figure 3A). IgG1 were transferred preferentially (Figure 3B). Whereas IgG2, IgG3, and IgG4 levels were largely equivalent across the mother and cord, with equivalent or lower transfer ratios across antigens (Figure 3B). As previously described (Fu et al., 2016), differences were detected in transfer ratios across antigen specificities, with reduced transfer of FIM specific IgG1 responses relative to other antigens, again highlighting qualitative differences in antigen specific antibody transfer, beyond subclass differences, that may account for differences in transfer ratios. Importantly, while these data confirm the preferential transfer of IgG1, they fail to explain why some antigen-specificities and functions transfer more efficiently than others.
Figure 3: Subclass and glycosylation transfer profiles.
To define the biophysical signatures that drive selective placental transfer, subclass and Fc-glycosylation profiles of pertussis-specific antibodies were interrogated across the mother:cord samples. A. The dot-line plot shows the level of total IgG antibodies for each of the pertussis vaccine antigens (FHA, PTN, FIM, and PTX) across mother:cord pairs. B. The dotline plots highlight the paired IgG1, IgG2, IgG3, and IgG4 levels for each of the 4 pertussis antigens. C. The bar graph depicts the overall transfer efficiency (cord/mother) for each subclass across all 4 antigens. D. The dot-line plot shows the major Fc-glycan profile levels (G0=agalactosyalted, G1=mono galactosylated, G2=digalactosylated, Total G=both G1 and G2, F=fucosylated, B=bisecting GLcNAc, Total S=total sialylated including both S1 and S2) for all antigen specificities across mother:cord pairs. E. The whisker box plots show the transfer efficiency (cord/mother) for each of the major Fc-glycan profiles for total circulating antibodies (bulk IgG) and FHA (right). G. The heat map shows the transfer ratios for antigen-specific glycosylation, across all specificities combined (total) or for individual antigen-specific antibody populations, where positive correlations are depicted in yellow, and inverse correlations are plotted in purple. Significance for dot/line analysis included a Wilcoxon test with a Bonferroni correction, where p<0.0125 was considered significant, *p<0.0125, **p<0.0025, ***p<0.00025. For the bar graph, significance was evaluated using a two-way ANOVA with a Tukey’s post-test correction. For transfer ratios, significance was evaluated using a Wilcoxon signed rank test, after variation of median was assessed from zero, and was corrected with a Hochberg’s step-up procedure, with **p<0.01, ***p<0.001. See also Figure S4
Beyond IgG subclass effects on Fc receptor (FcR) binding, Fc glycosylation is also linked to changes in regulating antibody effector function via altered FcR affinity (Jennewein and Alter, 2017). While univariate analyses pointed to significant changes in Fc-galactosylation across mother:cord pairs (Figure 3D, Figure S3), we explored Fc-glycan driven sieving using transfer efficiencies (Figure 3E-G and Figure S4). Both bulk (all IgG in plasma) and FHA-specific agalactosylated (G0) antibodies were transferred poorly (Figure 3D-F). Conversely, di-galactosylated (G2) and total galactosylated (G) bulk and total galactosylated (G) FHA-specific antibodies were transferred preferentially to the cord (Figure 3E and F). Further comparison across antibody populations highlighted consistent inhibition of the transfer of agalactosylated and bisected antibodies and enhanced transfer of galactosylated and sialylated antibodies across bulk and most antigen-specific antibody populations, except PTX-specific antibodies (Figure 3G and Figure S4). Combined, these data point to a potentially critical role for Fc glycosylation, in addition to subclass, in placental sieving of antibodies.
Defining the critical Fc-features of placental transfer
To gain a deeper appreciation for the key Fc signatures linked to antibody transfer, a multi-level partial least squares discriminant analysis (MLPLSDA) was used. This analysis was aimed at defining the antibody features, independent of overall mother:cord pair variation, that contributed to the sieving effect (Westerhuis et al., 2010). Clear separation was observed across the maternal and cord antibody profiles (Figure 4A). Features were then ranked based on their overall contribution to separating the mother:cord Fc-profiles (Figure 4B). In agreement with the univariate analysis and non-paired conventional orthogonalized PLSDA analysis (Figure S5 A,B), the ability of antibodies to drive NK cell activation were among the top 3 enriched features in the cord blood antibody profile. Additionally, the ability to drive neutrophil phagocytosis, IgG1 levels, and particular antigen-specific glycans were among the top features enriched in the cord antibody profiles. Conversely, mono-galactosylated glycans (G1F/GIFB) and the ability to drive monocyte phagocytosis (ADCP) were retained in the mother’s plasma. Furthermore, validation on RSV specific and Flu specific antibody profiles, separately, as well as simultaneously across all seven tested antigens, replicated the clear split between intra-pair mother:cord antibody profiles, always driven by NK cell function and glycan patterns (G2S1F predominantly) (Figure S2 C-F). These data provide a first glimpse of the individual functional and biophysical characteristics associated with placental transfer, across an array of antigen-specificities, and point to galactosylation as a key predictor of placental sieving.
Figure 4: Multivariate signatures of selective functional transfer.
Computational analysis was used to identify predictors of functional transfer. A. A multilevel PLSDA (MLPLSDA), which accounts for variance across mother:cord pairs, was used to define the specific features that most effectively provided resolution between mother:cord antibody profiles. Dots represent individual samples (mother=purple or cord=pink) across all four antigens. The orthogonalized approach ensured that latent variable 1 (LV1) captured the separation between mother and cord antibody profiles, while LV2 captured the antibody profile variances that do not contribute to this separation. 5-fold cross validation was performed, resulting in 94% Cross Validation accuracy. B. The bar graph shows the loadings on LV1, ordered by their enrichment in either cord (right/pink) or maternal (left/purple) profiles. Features were ordered based on their Variable Importance in Projection (VIP) scores, in such a way that features at the top of the plot provided the greatest resolution in antibody profiles, where the size of the bar reflected the impact on resolving differences between maternal:cord samples. C. The heat map shows the percentage of change in median and median absolute deviation (MAD), which are respectively robust estimates of the average and spread of variables in cord samples compared to their corresponding measurements in mothers. Significance of these changes was assessed using Wilcoxon signed-rank test. *p<0.05, , **p<0.005, and ***p<0.0005 D. A PLSR model was developed to define the maternal features associated with enhanced antibody-titer transfer. Dots represent single maternal:cord pairs across all four antigens and are colored according to their rank of low to high total IgG transfer (dark=high and light=low transfer). Features associated with highest transfer are captured on LV1, accounting for 51% of the variance of titer transfer. This model out-performed 97% of random models (Wilcoxon p=0.03) E. The bar graph depicts the loading plot for LV1, in which all maternal features were ranked based on their VIP score, importance in driving antibody transfer. F. A PLSR model was also generated to define the maternal antibody features associated with NK cell-activating antibody transfer. The dots were colored based on their rank of NK-activating antibody transfer (dark=high, light=low transfer). Transfer correlates are captured on LV1, explaining 53% of the variance of titer transfer order. This model out-performed 86% of random models (Wilcoxon p=0.14) G. The bar graph shows the loading plot associated with NK cell activating antibody functional transfer. See also Figure S5.
To hone in on the specific variables that were actively transferred, we next compared the median and median absolute deviation (MAD) of the antibody features in maternal samples to their corresponding cord blood samples. The median and MAD are robust estimators of the average and spread of individual variables across samples, respectively. Equivalent spread would indicate non-selective transfer, whereas constricted spread in the cord would point to a sieve effect. Thus two sets of variables were of highest interest; variables with an increased or decreased median in cord blood compared to their matched maternal plasma, indicating active transport or exclusion of the corresponding properties (Figure 4C, top row), and variables with decreased spread in cord blood, indicating selection for specific amounts of those variables in infants (Figure 4C, bottom row). Using this framework, CD107a, IFNγ, MIP-1β, ADNP, as well as G2S1F, IgG1, IgG4, C1q binding, and FcRn binding exhibited enhanced median levels toward the cord and are thus likely actively transferred across the placenta. Conversely, the median FCGR2A binding was decreased in cord blood indicating potential blockade of antibodies able to bind to this receptor (Figure 4C). By contrast, while there was no shift in the median G2F value, the spread of G2F was significantly decreased in the cord. These data point to a tight selection of antibodies, specifically linked to the directed transfer of G2F glycans. Moreover, most glycan species demonstrated a decrease in spread in cord compared to matched maternal samples, suggestive of Fc glycan-based selection through the placenta.
To specifically define the determinants of transfer beyond titer as well as that selectively predict NK cell functional transfer, two orthogonalized partial least squares regression (OPLSR) models were developed. First, antibody features that were enriched among maternal:cord pairs with the highest IgG titer transfer efficiencies were defined (Figure 4D, E). As expected, the top determinants of IgG transfer included elevated digalactosylated and sialylated antibodies and lower levels of agalactosylated glycans. Interestingly, while functional features did not track with total IgG transfer, binding to FcRs and complement were among the top predictors of transfer, pointing to clear qualitative processes involved in sieving antibodies across the placenta. Along the same lines, galactosylation and Fc-receptor binding profiles were among the most critical features that predicted transfer of NK cell activating antibodies in the second model (Figure 4F and G). Surprisingly, IgG1 titer and total IgG titer transfer were negatively associated with CD107a functional transfer, highlighting that not all antibodies are functional or selected for transfer, which points to the critical role of qualitative sieving activity of the placenta. Thus these two PLSR models clearly point to the critical nature of antigen-specific antibody Fc-glycosylation and Fc-receptor binding, beyond FcRn, as key determinants of placental antibody transfer.
Fc-galactosylation affects antibody binding to FcRn and other Fc Receptors
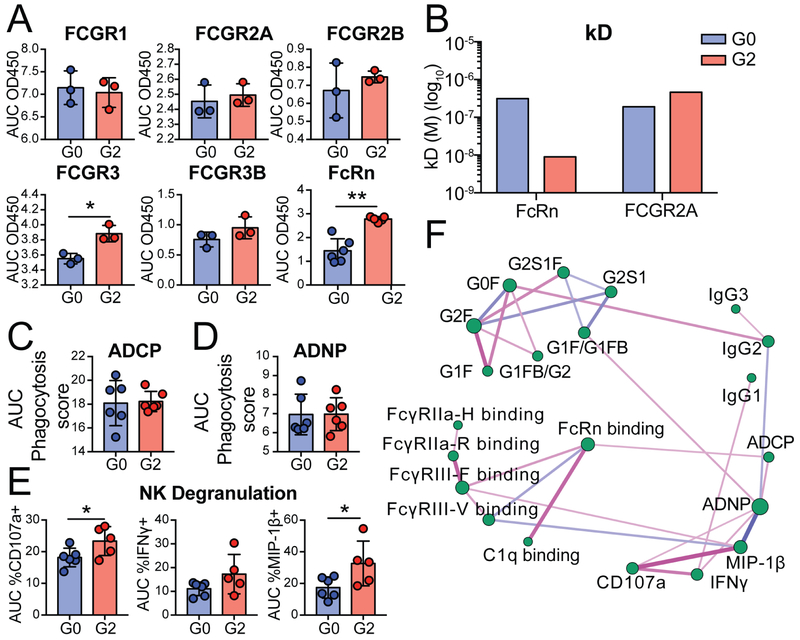
Contradictory roles for Fc glycosylation on FcRn binding have been reported (Bakchoul et al., 2013; Dashivets et al., 2015; Einarsdottir et al., 2013; Jansen et al., 2016b; Jefferis, 2012), including reports documenting negligible effects of Fc glycosylation on FcRn binding, with no change in antibody glycosylation across the placenta (Bakchoul et al., 2013; Einarsdottir et al., 2013), as well as studies pointing to significant antibody galactosylation-driven changes in FcRn affinity (Dashivets et al., 2015). Thus, to determine whether galactosylation impacts FcR and FcRn binding, a monoclonal antibody was synthesized that was either fully agalactosylated or digalactosylated (Figure S3). As previously described (Mimura et al., 2001; Thomann et al., 2015), galactosylation did not impact FCGR1, FCGR2A, FCGR2B or FCGR3B binding (Figure 5A). Conversely, the digalactosylated antibody exhibited improved binding to both FCGR3A and FcRn (Figure 5A). Moreover, while, no difference in binding was observed by bio-layer inferometry to FCGR2A by the galactosylated variants (Figure 5B), for FcRn binding the kD for the digalactosylated monoclonal was significantly lower than for the agalactosylated monoclonal variant, validating the observed influence of galactosylation on FcRn binding (Figure 4B).
Figure 5: Galactosylation selectively enhances FcRn, FCGR3 binding, and NK cell activation.
A monoclonal antibody was glycoengineered to either be fully agalactosylated (G0F) or fully di-galactosylated (G2F). See Figure S3. A. The bar graphs show the binding levels of the glycovariant monoclonal antibody to distinct human Fc-receptors by ELISA. Data are presented as area under the curve (AUC) of four five-fold dilutions or AUC of five two-fold dilutions (FcRn). B. The bar graph depicts the dissociation constant (kD) determined by bio-layer inferometry for binding to FcRn and FCGR2A. C. The bar graphs highlight the ability of the distinct monoclonal glycovariants to recruit ADCP (AUC divided by 103 of three five-fold dilutions), D. ADNP (AUC divided by 104 for three five-fold dilutions), and E. NK degranulation (AUC for three five-fold dilutions). F. The partial correlation network depicts pair-wise correlation between 22 antibody features across mother:cord pairs and pertussis antigens after removing the confounding effects (compare to Figure S5 C). Edges between nodes are weighted using significant partial correlation coefficients, after correcting for multiple comparisons (Benjamini-Hochberg q value < 0.05,). Line thicknesses and colors are proportional to the partial correlation strengths. Node sizes are proportional to their degree defined as their number of significant (p value < 0.05). Differences between glycan variants were assessed using an unpaired t-test *p<0.05, **p<0.01, ***p<0.001.
Beyond binding, glycan modified monoclonal antibodies also emulated the expected antibody functional profiles observed in maternal:cord samples. Specifically, while galactosylation did not alter ADCP or ADNP (Figure 5C and D), digalactosylated antibodies, which are more efficiently trapped on FcRn and FCGR3A, also drove enhanced NK cell degranulation and chemokine secretion (Figure 5E). Moreover, network analysis of all antibody features from the pertussis-specific response of the mother:cord pairs further highlighted the direct association of FcRn binding antibodies with FCGR3A binding antibodies that were in turn linked to NK cell activation, further pointing to the similar glycan binding preferences of these two Fc-receptors (Figure 5F, Figure S5). Thus, these data support the hypothesis that digalactosylated antibodies may selectively bind and become enriched on FcRn, resulting in enhanced transfer across the placenta aimed at deliberately arming the newborn with NK cell activating antibodies able to bind effectively to FCGR3A.
Fc receptor expression in the placenta
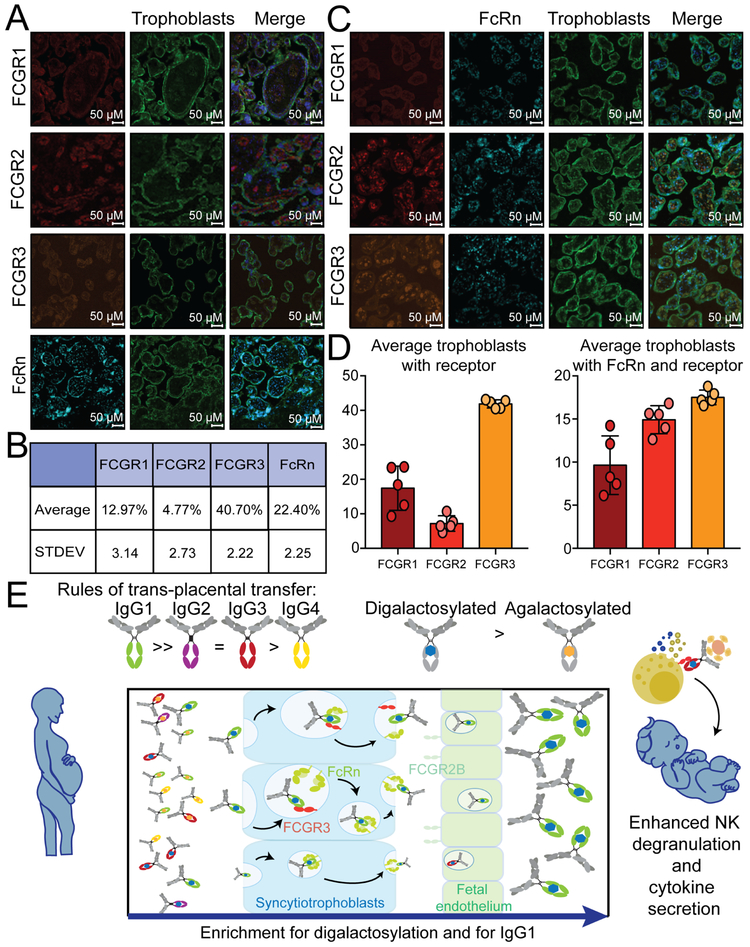
The presence of Fc-receptor binding predictors of placental transfer (Figure 4D-G) along with monoclonal-galactosylation mediated FCGR3A binding differences (Figure 5E) suggested that Fc-receptors, beyond FcRn, may contribute to placental sieving. Given our emerging appreciation for the expression of FcRs across tissues, including the placenta (Fouda et al., 2018; Martinez et al., 2018), we next aimed to determine whether FcRs could contribute to FcRn-mediated antibody sieving at the placental level. The localization of FcRs and FcRn was examined across placental tissue (Figure 6 A-D, Figure S6). As previously reported, low levels of FCGR1 and FCGR2 were observed on trophoblasts (12.97% and 4.7% positive, respectively) (Figure 6A). FCGR2, while present, was largely absent on trophoblasts, presumably enriched on fetal epithelial cells. Conversely, high levels of both FCGR3 and FcRn (40.70% and 22.40% positives, respectively) were observed on trophoblasts. Given the low expression levels, limited colocalization of FCGR1 with FcRn was observed (Figure 6C,D). However, FCGR3 was co-localized with FcRn on the trophoblast layer (Figure 6D). These data support a potential role for FcRs, and particularly FCGR3, not only in regulating immune cell function, but potentially also in synergizing with FcRn in the selective transfer of NK cell-activating antibodies across the placenta (Figure 6E).
Figure 6: FcR expression in the placenta.
To explore the potential role of additional Fc-receptors in shaping placental antibody transfer, healthy placentas were stained for Fc-receptor expression. A. Term placentas (n=5) were analyzed by immunofluorescence for Fc-receptor expression (red or orange), trophoblast staining (green), and nuclear localization (DAPI:blue). B. The table shows the average receptor expression on trophoblasts. C. Costaining of FcRs with FcRn (light blue), trophoblasts (green) and nuclei (dark blue) was also interrogated. D. The bar graph shows the averages of FcRs on trophoblasts or that colocalize with FcRn. E. Finally, the model depicts the potentially involvement of FcRn and FCGR3A collaboration with the trophoblasts, on selective antibody capture and subclass and Fc-glycan sieving, aimed at selectively transferring NK cell activating antibodies to newborns. See also Figure S6.
Implications for the transfer of NK-degranulation inducing antibodies in neonates
Finally, to gain insights into the potential evolutionary benefit of preferential transfer of NK cell-activating antibodies, we next aimed to determine whether the preferential transfer of NK cell activating antibodies could confer any immunological advantage to newborns. Accumulating data have documented attenuated macrophage, neutrophil, NK cell, and T and B cell function in early life (Yu et al., 2018). Neutrophils and NK cells are among the most abundant innate immune cells on the first day of life (Lee and Lin, 2013; Yu et al., 2018). Thus, the ability of cord antibodies to drive ADNP was assessed using both adult- and cord-derived neutrophils (Figure 7A and B). While the results were variable across the samples, cord-derived neutrophils exhibited largely decreased activity compared to adult neutrophils. Conversely, while FHA-specific NK cell degranulation trended towards lower in cord-derived NK cells, no difference was observed in antibody-dependent NK cell degranulation across adult and cord NK cells, suggesting that some antibody-directed innate immune functions may be conserved in early life (Figure 7C and D). However, striking differences were observed in the ability of cord antibodies to recruit NK cell cytokine production, with significantly elevated levels of IFNγ secretion in cord NK cells compared to adult NK cells (Figure 7C and D). Similar patterns were observed with adult-derived antibodies (Figure S7), highlighting the unique capacity of neonatal NK cells, but not neutrophils, to respond to antibody-opsonized targets. While emerging data suggest that cord blood cells may harbor distinct phenotypes/function compared to neonatal immune cells (Olin et al., 2018), these data are in agreement with previous reports suggesting that neutrophils, monocytes, dendritic cells, and macrophages require a period of time following birth to acquire full functionality, while NK cells are functionally primed to fight disease immediately post-partum, even though they are less cytotoxic (Lee and Lin, 2013). Thus, the placenta appears to transfer antibodies able to selectively leverage innate immune effector functions present on the very first day of life.
Figure 7: Differences in Fc-functional activity of adult and cord innate immune cells.
The ability of adult and cord innate immune cells to mediate ADNP and NK cytotoxicity in response to PTX and FHA was evaluated. A. The flow plots show the gating strategy for cord neutrophils. B. The line-dot plots depict the phagocytosis score for cord antibody samples in adult (dark pink) and cord (light pink) neutrophils. C. The flow plots highlight the gating strategy for neonatal NK cell function. D. The line-dot plots represent the percent maternal (dark pink) or cord (light pink) NK cell responses (CD107a and IFNγ) following stimulation with cord antibodies. Significance was evaluated using a two-way ANOVA with Sidak’s test.
Discussion
Emerging data point to significant differences in newborn and adult immunity, including the existence of attenuated immune inflammatory activity, reduced adaptive immune responsiveness, and enhanced regulatory functions in newborns (Saso and Kampmann, 2017; Whittaker et al., 2018; Yu et al., 2018), that collectively contribute to diminished vaccine responses in this population. To overcome the limitations in immune priming in newborns, vaccination of mothers in the third trimester of pregnancy has been suggested as a strategy to increase antibody titers in neonates. While successful, significant disparities exist in transfer rates in the setting of disease, including HIV (Martinez et al., 2019), or other co-morbidities (Palmeira et al., 2012), and across antibody specificities (Fu et al., 2016). Thus, these studies point to our limited understanding of the mechanism of placental antibody transfer (Fu et al., 2016; Palmeira et al., 2012; Wilcox et al., 2017). Defining the mechanics of placental transfer may offer novel insights for the rational design of maternal vaccines able to drive transfer of protective antibodies to fetuses and reduce their window of vulnerability. Here, we demonstrate that the placenta preferentially sieves pertussis-specific antibodies based on Fc qualities, transferring NK cell-activating, rather than neutrophil or monocyte activating, antibodies to the fetus. This selection is based on Fc glycosylation, where digalactosylated antibodies that interact effectively with FcRn and FCGR3A may be selectively captured and transferred, arming the fetus with antibodies able to access NK cell functions immediately upon birth (Figure 6E).
Pregnancy-specific changes in glycosylation have been observed across both the Fc and the Fab domain of antibodies (Bondt et al., 2014; Jansen et al., 2016a; Sonneveld et al., 2016). However, glycosylated Fabs do not have the capacity to interact with FcRn, which binds to antibodies in the CH3 domain (Jensen et al., 2017; Piche-Nicholas et al., 2018), suggesting that only changes in Fc-glycosylation are likely to regulate placental selection. The potentially critical role of galactosylation for FcRn binding has been previously noted, likewise demonstrating enhanced FcRn binding to digalactosylated antibodies (Dashivets et al., 2015). Moreover, large studies of worldwide antibody glycosylation data confirm enhanced transfer of digalactosylated antibodies across the placenta, as well as lower transfer rates of bisected glycans (Jansen et al., 2016a). Thus, FcRn-mediated preferences for specific Fc-glycovariants may influence the quality of antibodies that are transported across the trophoblast layer.
Changes in IgG galactosylation have been implicated in inflammatory diseases, with agalactosylated species accumulating in autoimmune, infectious and oncological diseases (Decker et al., 2016; Moore et al., 2005; Pasek et al., 2006). By contrast, less-inflammatory galactosylated IgG antibodies accumulate during pregnancy (Bondt et al., 2013; Jansen et al., 2016a). Whether this shift occurs stochastically, due to the toleragenic immune state in pregnancy, or in a deliberate manner to dampen inflammation during pregnancy and limit potential immunopathology during infection, is unclear. However, in HIV-exposed uninfected infants, antibody transfer is altered, suggesting that inflammation may interfere with antibody transfer, and that avoiding inflammation may be benefitical for transfer (Martinez et al., 2019; Wilcox et al., 2017). In healthy pregnancy, generation of highly galactosylated antibodies may correspond to a deliberate effort to generate antibodies able to transfer more effectively across the placenta. However, whether this change occurs across all antigen-specificities is unclear, but could provide insights into the type of IgG that may be transferred most effectively to fetuses. Moreover, whether maternal vaccination specifically results in the induction of digalactosylated antibodies is unclear. Yet, given that adjuvant-driven inflammatory cues at vaccination can shape antibody Fc function via altered glycosylation (Mahan et al., 2016; Vestrheim et al., 2014), the design of next-generation maternal vaccines able to selectively skew IgG glycosylation towards Fc profiles that are transferred more effectively to fetuses may result in enhanced immunity in neonates.
The selective transfer of NK cell-activating antibodies in the setting of functional NK cells in newborns points to the deliberate selection of antibodies able to leverage effective immunity at birth. Neonates are born not just deficient in IgG production, but deficient in other immune functions. Specifically, most innate and adaptive cell subsets are immature at birth, particularly in their ability to produce cytokines and extravasate (Yu et al., 2018). For example, neutrophils are unable to form neutrophil extracellular traps, lack many adhesion molecules, exhibit low expression of toll like receptors, mediate impaired phagocytosis, and exhibit attenuated neutrophil burst and microbial degradation. By contrast, while NK cells also exhibit functional attenuation of cytotoxic killing activity, which is acquired through NK cell education post-birth, NK cells are among the most abundant and functional innate immune cells post-partum (Lee and Lin, 2013) Though emerging data indicate that cord blood and infant innate immune cellular phenotype and function are distinct (Olin et al., 2018), neonatal NK cells have been shown to respond robustly to target cells and able to secrete copious inflammatory cytokines from birth (Lee and Lin, 2013). Along these lines, here we show that while monocyte and neutrophil phagocytic antibodies are not transferred actively, the placenta preferentially transfers NK cell activating antibodies from mother to cord. Moreover, these preferentially transferred antibodies are able to drive equivalent recruitment of NK cell degranulation across maternal and cord NK cells, and are responsible for significantly enhanced recruitment of NK cell IFNγ and MIP-1β secretion from cord NK cells (Figure 7). These data point to a potentially unique evolutionary placental bias aimed at selectively transferring antibodies to the newborn that are able to leverage the most competent immune cells.
FcR expression has been documented broadly in the placenta, particularly prominently on Hofbauer cells, located between the maternal trophoblast layer and the fetal endothelium, thought to contribute to immunity (Simister, 2003). Importantly, the tight fetal endothelial layer does not express FcRn (Lyden et al., 2001). However, the fetal endothelium expresses FCGR2B, which is thought to play a role in antibody transfer to the cord blood (Ishikawa et al., 2015; Takizawa et al., 2005). FCGR2B is the sole inhibitory, low-affinity, FcR in humans and, like FCGR3, interacts poorly with monomeric antibodies (Bruhns et al., 2009). Moreover, given the homology in the extracellular region of the activating FCGR2A and the inhibitory FCGR2B (Nimmerjahn and Ravetch, 2006), FCGR2B-driven sieving would result in enhanced transfer of antibodies with ADCP activity, which is largely mediated through FCGR2A (Nimmerjahn and Ravetch, 2008). However, limited sieving was observed for ADCP inducing antibodies, suggesting a limited role for FCGR2B mediated selection in the healthy placenta.
The colocalization of FCGR3 and FcRn on the trophoblasts raises the question of whether these two FcRs may collaborate to select antibodies for transfer. The expression of FCGR3 on trophoblasts may additionally provide some mechanistic understanding for the efficient transfer of IgG3 antibodies, which bind more effectively to FCGR3 but less efficiently to FcRn (Stapleton et al., 2011; Vidarsson et al., 2014). However, FCGR3 exhibits low-affinity binding to monomeric IgG (Boesch et al., 2014), and FCGR3 has a preference for afucosylated antibodies (Shields et al., 2002), a signature that is not preferentially transferred across the placenta. Thus, it is likely that FCGR3 participates post-FcRn capture, participating in the enrichment of FcRn trapped antibodies, potentially further skewing the shift of antibodies to those with the greatest functional potency for the newborn. Yet, whether additional FcRs contribute to selection following trophoblast selection, including sieving by Hofbauer cells or the fetal endothelium, remains unknown. FcR changes occur during inflammation and infection of the placenta (Wilcox et al., 2017) that could lead to differential antibody sieving in the diseased or inflamed placenta, as has been previously observed in children of HIV-infected mothers (Abu-Raya et al., 2016; Martinez et al., 2019). While the data here suggest that FcRn contributes most actively to the selection of NK cell-activating antibodies in the healthy placenta, it is plausible that alterations in the quality of antibodies in disease (Abu-Raya et al., 2016; Jennewein, 2017; Palmeira et al., 2012) could also be driven by changes in FcR expression on syncytiotrophoblasts or other immune cells of the placenta.
These data open up many options for improving the health of neonates worldwide. While the CDC has already recommended third trimester vaccination with Tdap and flu vaccines, neonates continue to suffer from many other infectious diseases. With our emerging appreciation for the unique immune states that arise during pregnancy and the unique selective function of the placenta, next-generation rational vaccine design for pregnant women may hold the key to delivering enhanced immunity to newborns. Moreover, for diseases for which vaccines have yet to be developed, similar design principles may be engineered onto monoclonal therapeutics to transfer immunity passively to prevent deadly or debilitating diseases. Collectively, these data point to a deeper understanding of mechanisms of placental transfer of immunity, offering insights for the development of novel strategies to enhance immunity in our vulnerable newborns.
STAR Methods
Contact for Reagent and Resource Sharing
Further information and requests for resources and reagents should be directed to, and will be fulfilled by, the Lead Contact, Galit Alter (galter@mgh.harvard.edu).
Experimental Model and Subject details:
Human serum samples:
Matched cord blood and peripheral maternal blood samples were collected at birth and during the first 3 days post-partum, respectively, from two mother:cord cohorts. Cohort 1 was recruited in the US and included eighteen mother:cord pairs. Cohort 2 was recruited in Belgium and included 28 mother:cord pairs. Plasma was separated from whole blood and cryopreserved until required. Immunization with Tdap was performed during pregnancy in fourteen mothers of cohort 1. Because equivalent titers and similar overall antibody profiles existed in cords from vaccinated and unvaccinated mothers (Supplemental Figure 1) all mother:cord pairs from cohort 1 were analyzed together. A full history of vaccination during the current pregnancy was obtained from each participant and, where possible, from before pregnancy. Clinical characteristics are included in Supplemental Table 1. All subjects were HIV, HBV, and HCV negative and provided written informed consent before sampling. The study was conducted according to the principles expressed in the declaration of Helsinki, reviewed by the Partners Internal Review Board, and approved by the Partners Human Research Committee (Approval Number 2011P001763). Cohort 2 is part of a study of maternal determinants of infant immunity (Goetghebuer et al., 2018) and was reviewed by the ethics Committee of the Hôpital Saint-Pierre, Belgium and Partners Human Research Committee (Approval Number 2011P001763).
Human placental samples:
Placental sections were obtained from term pregnancies of adult health women at Massachusetts General Hospital. The study was conducted according to the principles expressed in the declaration of Helsinki, reviewed by the Partners Internal Review Board, and approved by the Partners Human Research Committee (Approval Number 2010P000632).
Cell lines:
THP-1 cells (cell line isolated from a 1-year old male) were grown in R10 (RPMI plus 10% fetal bovine serum, L-glutamine and penicillin/streptomycin) supplemented with 0.01% β-mercaptoethanol.
Method details:
Heat inactivation of plasma
All plasma samples were heat inactivated at 56°C for one hour. Precipitate was spun down at 20,000×g for 10 minutes. The supernatant was stored at −80°C.
Phagocytosis
Antigen coupling to beads:
Antigens; the four pertussis antigens, RSV-post fusion protein (McLellan et al., 2013) or a mix of the H3N2 and HAB, were coupled to 1 μm yellow-green fluorescent, carboxylate-modified microspheres at a ratio of 10 μg of protein to 5 μl of beads. Microspheres were first activated with 100 mM monobasic sodium phosphate, pH 6.2 in the presence of 5 mg/mL EDC and 5 mg/mL sulfo-NHS. Beads were then washed in 0.05 M 2[N-Morpholino]ethanesulfonic acid (MES) pH 5.0 and incubated with antigen for two hours. The coupling reaction was quenched with 500 mM glycine for 30 minutes. Then beads were washed with 0.05% PBS-tween 20 and blocked in PBS-2% BSA for two hours. After washing with 0.05% PBS-tween 20, the 5 μl of beads were resuspended in a final volume of 1 mL PBS-0.1% BSA and stored at 4°C in the dark for up to one week.
Formation of immune complexes:
Ten microliters of protein-coated beads were incubated with an equal volume of either heat-inactivated plasma diluted in R10 or PBS in a 96-well U-bottom culture plate. Optimal plasma dilutions were predetermined by titration. Following a two-hour incubation at 37°C, 5% CO2, the immune complexes were spun down and washed in PBS to remove any lingering plasma components or cytokines and incubated with either THP-1 cells or neutrophils as described below.
Monocyte antibody-dependent cellular phagocytosis (ADCP) assay:
The monocyte ADCP assay was adapted from (Ackerman et al., 2011). Briefly, immune complexes were incubated with 25,000 THP-1 cells per well at a concentration of 1.25×105 cells/ml in R10 for 16 hours at 37°C, 5% CO2. After the incubation, the cells were fixed in 4% paraformaldehyde. Data were collected on a BD LSR II flow cytometer (BD Biosciences) equipped with FACS Diva software. All flow cytometry data were analyzed using Flowjo (TreeStar). Negative or unstimulated controls were used to set gates. Data is reported as the mean of 2-3 independent replicates. Phagocytosis score was calculated as the percentage of bead positive cells, multiplied by geometric mean fluorescence intensity of bead positive cells, divided by 10,000.
Antibody-dependent neutrophil phagocytosis (ADNP) assay:
For the ADNP assays comparing maternal and cord antibodies with adult neutrophils, granulocytes were isolated from freshly drawn peripheral blood. Erythrocytes were precipitated by adding a 3% Dextran solution, before separating the granulocytes and peripheral blood mononuclear cells (PBMC) by density centrifugation with Ficoll-paque PLUS (Sigma Aldrich). The isolated granulocytes, contained in the pelleted fraction, were washed briefly in water to remove remaining erythrocytes before tonicity was restored by the addition of an equal volume of 1.8% NaCl solution.
For neutrophils used to compare function in adult blood and cord blood, granulocytes were isolated from whole blood by lysing erythrocytes in ACK lysis buffer for 5 minutes before precipitation by centrifugation. Granulocytes were washed twice with PBS.
Granulocytes isolated via both methods were resuspended at 2.5×105 cells/ml in R10 and 50,000 cells per well were incubated with immune complexes for 1 hour at 37°C, 5% CO2. Neutrophils were stained with anti-CD66b-Alexafluor647 or anti-CD66b-Pacific blue and cells were fixed with 4% paraformaldehyde prior to flow cytometry. Phagocytosis scores were calculated as above in ADCP assay.
NK cell activation assay
For comparisons of NK degranulation activity between maternal and cord serum, PBMCs were isolated from freshly drawn buffy coats by density centrifugation and then incubated overnight at a concentration of 3×106 cells/ml in R10 supplemented with 1 ng/ml rhIL15. The next day, NK cells were isolated using EasySep NK cell Enrichment Kit.
For comparison of adult and cord blood NK cell degranulation, cord blood NK cells were isolated within 8 hours of birth from cord blood and adult NK cells were isolated from buffy coat’s drawn that day. NK cell isolation was performed with RosetteSep NK cell Enrichment Kit per the manufacturer’s instructions. Purified NK cells were separated by density centrifugation, washed twice with PBS and used immediately without rhIL-15 supplementation.
ELISA plates were prepared by coating plates with protein at a concentration of 1 μg/ml and incubated overnight at 4°C. Plates were then blocked with PBS-5% BSA overnight at 4°C. Plates were washed in PBS, and plasma was plated at dilutions pre-determined by titration. Human gamma globulin isotype was plated at 1 mg/ml on uncoated wells as a positive control and PBS was used as a negative control. Plates were incubated at 37°C 5% CO2 for two hours. The isolated NK cells were then added to each well at a concentration of 2.5×105 cells/ml, 50,000 cells per well, in R10 in the presence of anti-CD107a, Brefeldin A (2.5 μg/ml) and Golgistop and incubated at 37°C, 5% CO2 for five hours. After the incubation, cells were stained with anti-CD3, anti-CD56 and anti-CD16. Cells were fixed and permeabilized using the Fix&Perm cell permeabilization kit and intercellular staining was performed with anti-IFNγ and anti-MIP-1β. Data were collected on a BD LSR II flow cytometer equipped with FACS Diva software. All flow cytometry data were analyzed using Flowjo. Negative or unstimulated controls were used to set gates. Data are reported as the percentage of NK cells positive for a given marker minus the mean of the protein-matched PBS-only control.
Antigen-specific antibody isotype and subclass analysis
Antigen-specific IgG subclass levels were measured by ELISA. ELISA plates were coated with 1 μg/ml of FHA, Pertactin, Fimbriae 2/3 or Pertussis toxin overnight at 4°C. The next day, plates were blocked for two hours with PBS-5% BSA. Plasma was added at predetermined dilutions for each subclass and antigen, and incubated for two hours at 37°C. To account for background binding of antibodies, PBS-0.05% tween-20 control was included for each protein, and furthermore, control wells with no pertussis protein were blocked as above and included for each sample at each dilution (both in duplicate). Following a one-hour room temperature incubation with an anti-human IgG-horseradish peroxidase (HRP) secondary antibody), the ELISA was developed using ultra-TMB. The reaction was stopped with 2M H2SO4. Optical density values were read immediately at a wavelength of 450 nm (reference wavelength of 592 nm) using a Tecan Infinite M1000 Pro. Data are reported as reference value corrected, and BSA background corrected OD450 values.
Glycan analysis
Proteins were biotinylated and coupled to 1 μm neutravidin-coated magnetic beads; 2.5 μg of protein was coupled to 25 μl of beads for each sample. Heat-inactivated sample (200 μl) was incubated with 25 μl of un-coupled magnetic beads to clear non-specific bead binding for 30 minutes. Plasma was then removed from these beads and added to the 25 μl of protein-coupled beads and incubated for one hour at 37°C. The resulting immune complexes were washed, and the antibody Fc was cleaved off by incubating with 1 μl of IDEZ in a total volume of 20 μl of PBS at 37°C for 1 hour. The resulting Fc fragments were deglycosylated and fluorescently labeled using a GlycanAssure APTS Kit according to manufacturer’s instructions. Glycans were analyzed on 3500xL genetic analyzer (Applied Biosystems). Samples were run with N-glycan fucosyl, afucosyl, bisecting and mannose N-glycan libraries to enable identification of twenty-four discrete glycan species (Supplemental Figure 2). The relative frequencies of each of these glycans as percentages of total glycans were calculated using GlycanAssure software.
Fc Receptor binding
FcR binding was preformed using a multiplexed Luminex assay as described in Brown et al., 2012 (Brown et al., 2012). Pertussis antigens were coupled to magplex microspheres as described above in coupling for phagocytosis assays. Recombinant FcRs with an AviTag were biotinylated on their avidin tag using a Bir500 kit according to manufacturer’s instructions. FcRs were then incubated with streptavidin-PE (ProZyme, Hayward, CA) for ten minutes, and any residual streptavidin was quenched with an excess of 20 μM biotin from the Bir500 kit for a further ten minutes. Plasma samples were diluted to three dilutions (1:50, 1:100 and 1:500) in luminex wash buffer (PBS-0.05% BSA-0.001% tween-20) and incubated with microspheres for two hours. Samples were washed in Luminex wash buffer and then incubated with PE-labeled FcRs for one hour. Samples were washed again and resuspended in xMAP sheath fluid. Samples were analyzed on a Bioplex 3D system. Data is calculated as area under the curve of the median fluorescent intensity of PE.
Glyco-modified monoclonals
293T derived wild type PGT128 was predominantly agalactosylated and fucosylated (G0F) (Supplemental Figure 3). The di-galactosylated form was made by mixing 0.5 mg of PGT128 with 15 μg of β-1,4-Galactosyltransferase and 488 μg of UDP-Galactose in a total volume of 400 μl of 20 mM MnCl2, 100 mM MES and was incubated at 37°C for 24 hours. After galactosylation, antibodies were purified using the Nab Protein G spin column according to the manufacturer’s protocol.
FcR affinity ELISA
384-well ELISA plates were coated with FcRs at 10 μg/mL overnight at 4°C. Plates were blocked for two hours at 37°C with 2X assay buffer A. Samples were diluted in 1X Assay Buffer A and added in serial dilutions, then incubated for two hours. Secondary antibody, anti-Fab HRP, was diluted 1:10,000 in 1X Assay buffer A and incubated on plates for one hour. Binding was detected with TMB for 10 minutes, and then quenched with 2N H2SO4. Plates were read on a Tecan Infinite M1000 Pro at 450 nm (with reference at 570 nm). Values are reported as reference-background corrected, area under the curve.
FcR affinity by BLI
An Octet RED96 system (Fortebio) was used to analyze dissociation constants (kD) for glycosylation modified monoclonals. FcR receptors biotinylated as for FcRs binding were loaded onto streptavidin coated biosensors in assay buffer (PBS 0.05% Tween 20 and 1% BSA). Immobilization levels above 2.0 nm were reached, and biosensors were washed in assay buffer. For association phase, PGT128 G0F and G2F samples were diluted in assay buffer across five two-fold dilutions starting at 25 μg/mL and allowed to bind for 300 s. The dissociation phase was recorded using assay buffer for 800 s. The sensorgrams were plotted and evaluated using the Octet data analysis software.
Immunohistochemistry and Immunofluorescence
Placental sections were fixed for 24 hours in formalin and then embedded in paraffin. A microtome was used to cut the placenta sections into 0.5 μm sections which were mounted onto glass slides Immunohistochemistry was used to select dilutions for immunofluorescence analysis (Supplemental Figure 4). Slides were heated for 25 minutes at 60°C then deparaffinized in xylene for 10 minutes. Slides were rehydrated with graded ethanol washes. Antigen retrieval was done in Nuclear Decloaker. Sections were blocked with Background Sniper then washed in PBS. Sections were blocked with normal goat serum at a 1:10 dilution for 15 minutes then incubated with primary antibody for one hour. Primary antibodies used for immunohistochemistry included: Anti-Placental Alkaline Phosphatase to label trophoblasts, anti-FcRn to label FcRn receptors, Anti-CD64 to label FCGR1 receptors, Anti-CD32 to label FCGR2 receptors, and CD16 to label FCGR3 receptors. For immunohistochemistry, following primary incubation, slides were washed and stained with universal HRP secondary antibody for one hour. Samples were stained with DAB + chromogen substrate system, then washed and counterstained in hematoxylin. Samples were washed, dehydrated, then sealed with mounting medium and coverslips. After 24 hours of drying, samples were imaged on a TissueFAXS whole slide scanning system and analyzed with HistoQuest. For slides analyzed for immunofluorescence, following primary incubation slides were washed and incubated for one hour with a fluorescent secondary antibody. Secondary antibodies used included: Goat anti-Mouse IgG2b; anti-Mouse IgG1 Antibody; and Goat Anti-Rabbit IgG H&L. For co-stains between FcRn and FCGRI, FCGR2 or FCGR3, FcRn staining was done as a tertiary and quaternary stain using streptavidin/biotin binding, so streptavidin and biotin binding was blocked prior to secondary incubation using an Avidin/Biotin blocking kit. Biotinylated anti-FcRn was prepared using the same anti-FcRn as above and biotinylated using the SiteClick Biotin Antibody Labeling Kit. For these slides, anti-FcRn, was incubated on the slides for one hour following the secondary incubation. After washing, FcRn was detected with a quaternary labeling step of streptavidin-linked Brilliant Violet 421. For all slides, following antibody labeling samples were washed and incubated for five minutes with Hoechst. Samples were washed and mounted with Prolong Gold Antifade and coverslips. After drying for 24 hours, slides were imaged with fluorescence microscopy then analyzed with TissueQuest.
Quantification and statistical analysis:
Univariate Statistical Analysis:
A Wilcoxon matched-pairs signed rank test was used to examine differences in paired maternal:cord samples and a Bonferonni correction was used to correct for multiple corrections. A Wilcoxon rank test with Hochberg’s step-up method to correct for multiple comparisons was used to examine difference from baseline in the glycan transfer efficiency test. A Mann-Whitney test was used for comparisons between two groups. For more than two groups comparison, a Friedman’s test with a Dunn’s post-test or a two-way ANOVA with Sidak’s post hoc analysis was used. Spearman’s rank correlation was used to examine bivariate associations. P values are two-sided. Statistical analyses were conducted using GraphPad Prism.
Statistical analysis of change in median and median absolute deviation:
This analysis was used to identify the features that change in median and spread (MAD) from mothers to cords in a statistically significant manner (Figure 4C). A preprocessing step was performed, where we removed the measured glycan species with zero median, indicating low-abundance features, in both maternal and cord blood. For the remaining 22 features, the median and median absolute deviation (MAD) were calculated as robust measures of average and spread of data, respectively. The percentage change in median and MAD were calculated according to the following formula:
Here, xmother and xinfant are the values of the variable × in mothers and cords, respectively.
To assess whether the mothers and cords, which consist of matched samples, have different population mean ranks, the nonparametric Wilcoxon signed-rank test was performed, and p-values were calculated using the MATLAB function signrank. Significant p-values are judged at three levels of p<0.0005, p<0.005, and p<0.05 as denoted with ***, **, and * in Figure 4C, respectively. The statistical significance of the change in the spread was calculated by performing Wilcoxon signed-rank test on the absolute value of (x – median(x)) using the MATLAB function signrank.
Multivariate Analysis--Summary
Principal Component Analysis (PCA) to visualize if there is any subpopulation in the overall population of cord samples in the two-dimensional principal components (PC) space that is enriched in cord from vaccinated or non-vaccinated mothers.
To find correlates that contribute majorly to the separation of maternal and cord serology profiles, two sets of analyses were performed; (i) Orthogonalized Partial Least Squares Discriminant Analysis (OPLSDA) to find correlates that contribute majorly to the separation of mothers and cord serology profiles (Figure S5), (ii) multi-level partial least squares discriminant analysis (MLPLSDA) was used as the preferred alternative that utilizes the paired structure of the data leading to improved separation of maternal and cord serology profiles (Figure 4A, B). For these set of analyses, a preprocessing step was performed, where we removed the measured glycan species with zero median, indicating low-abundance features, in both maternal and cord blood, resulting in 22 features. MATLAB was used to perform all multivariate analysis.
To investigate the determinants of high transfer of antibody titer as well as NK-activating antibodies across the placenta, Orthogonalized Partial Least Squares Regression (OPLSR) modeling was performed to investigate (I) the relationship between the maternal features and the transfer of antibody titer (ordered from low to high titer transfer), and (II) the relationship between the transfer of NK-activating antibodies and the transfer ratios of biophysical features of antibodies including subclass titers, Fc-glycans, and binding to FcRs (Figure 4D-G).
Partial correlation network analysis was utilized to assess the pair-wise correlation of features along all the samples (mothers and cord). The analysis was performed in MATLAB. Cytoscape 3.6.1 was used for visualization.
Principal Component Analysis (PCA):
A PCA model was constructed using 22 variables with non-zero median in either maternal or cord blood. Variables were centered and scaled to a standard deviation of 1. In the two-dimensional space of PC1 vs. PC2, no apparent subpopulation with enrichment in cords from vaccinated or non-vaccinated mothers was observed.
Orthogonalized Partial Least Squares Discriminant Analysis (OPLSDA):
To mathematically identify the key features contributing to profile differences between maternal and cord blood, an OPLSDA framework was used (Arnold et al., 2016; Lau et al., 2011). An OPLSDA model was constructed using 22 variables with non-zero median in either maternal or cord blood. Variables were centered and scaled to a standard deviation of 1.5-fold cross validation was performed on the data (Venetian blinds), obtaining a Cross Validation (CV) accuracy of 64%. To assess model significance, a permutation test was performed by randomly shuffling the labels. The OPLSDA model performed significantly better than random (Wilcoxon p=0.017).
Multi-Level Partial Least Squares Discriminant Analysis (MLPLSDA):
To mathematically identify the key features contributing to the profile differences between maternal and cord blood, a MLPLSDA (Westerhuis et al., 2010). MLPLSDA uses the same principles as OPLSDA analysis for multivariate data (Arnold et al., 2007; Lau et al., 2011) but also takes advantage of the paired structure of the data (paired maternal and cord blood). Intuitively, this analysis subtracts the effect of heterogeneity between maternal:cord samples (inter-pair variability) and focuses on the effects within maternal:cord samples. The model was constructed using 22 variables with non-zero median in either maternal or cord blood. Variables were centered and scaled to a standard deviation of 1. 5-fold cross validation was performed on the data (Venetian blinds), obtaining a Cross Validation (CV) accuracy of 94%. To assess model significance, a permutation test was performed by randomly shuffling labels. The MLPLSDA model performed significantly better than random (Wilcoxon p=2×10−14).
Orthogonalized Partial Least Squares Regression (OPLSR):
PLSR is a multivariate regression technique (Lau et al., 2011) where linear combinations of features are used to predict the variance in the dependent variables. The model is then orthogonalized such that Latent Variable 1 (LV1) captures the variance in features that are in the direction of the dependent variable, while other latent variables describe the variation orthogonal to the predictive component (LV1). Here, OPLSR was applied to analyze (I) the relationship between the maternal features and the transfer of titer, and (II) the relationship between the maternal biophysical features and the transfer ratio of the NK cell-activating antibodies.
I. Here, Y = total IgG titer in cord blood relative to the maternal blood. These were ranked such that the lowest transfer ratio was ranked 1. X = All antibody features of maternal blood (Figure 4D,E).
II. Here, Y = total CD107a by NK cells in cord blood relative to the maternal blood. These were ranked such that the lowest transfer ratio was ranked 1. X = The maternal antibody biophysical features (Figure 4F,G).
Partial correlation network:
While correlation networks offer insights into the pairwise correlation of variables (Supplemental Figure 5C), they are sensitive to indirect effects. As an alternative, partial correlation networks, also known as Gaussian Graphical Models (GGM), are used to alleviate this effect. Previous studies of the metabolomics data and glycomics data in cohorts have shown that highly correlated pairs in GGMs appear predominantly between structures that are one enzymatic step apart (Benedetti et al., 2017; Krumsiek et al., 2011; Shin et al., 2014). Thus, we chose partial correlation network analysis, as it is a stricter measure of the degree of association between two variables. A partial correlation network was generated, where the nodes represent the 22 variables consisting of the biophysical features and functional responses--antibody subclasses, Fc-glycan structures, their binding to FcRs and the induced antibody-dependent functions. The edges represent their pairwise Pearson correlation coefficients corrected for the confounding effects of all other variables. The analysis was done in MATLAB using the partialcorr function. Edges between nodes are weighted using significant partial correlation coefficients, after correcting for multiple comparisons (Benjamini-Hochberg q value < 0.05, testing the hypothesis of zero correlation). Line thicknesses and colors are proportional to the partial correlation strengths with dark magenta and dark blue depicting a correlation coefficient of 1 and −1, respectively. Node sizes are proportional to their degree defined as their number of significant (false discovery rate < 0.05) connections. For example, ADNP with six connections has the highest degree, while G2F has the second highest number of connections, which is five.
Supplementary Material
Supplemental Figure 1. Related to Figure 1: Relationship of antibody profiles across vaccinated and non-vaccinated mothers. The principal component analysis includes all pertussis-specific antibody features across all mother:cord pairs. A. No differences were observed across antigen-specific antibody profiles (left) irrespective of vaccination. B. Similarly, no overall differences in antibody profiles were observed across vaccinated or un-vaccinated mothers (right).
Supplemental Figure 2. Related to Figure 2: NK degranulation selection across cohorts. A. The dot-plots show NK-dependent degranulation to RSV pre-fusion F antigen plotted as the percentage of NK cells positive for CD107a, IFNγ and MIP-1β. B. The dot-plots show 28 mother:cord pairs from cohort 2 ability to drive NK degranulation (CD107a expression) in response to Pertussis toxin (PTX), Pertactin (PTN) and measles virus antigens. Significance in A, B evaluated for NK degranulation with paired t-test. *p<0.05, **p<0.01, ***p<0.001. C. A MLPLSDA was used to analyze the features separating mother and cord blood samples for RSV pre-fusion, RSV post-fusion and flu-specific antibody profiles. Each dot represents an individual blood sample (mother or cord) tested for one of these three antigens. LV1 and LV2 account for 18% and 22% of the variability in functional and glycan profiles across the antigens. The separation of mothers and cord is largely captured on LV1, which explains 36% of the Y variance in the direction of the maternal:cord separation. 5-fold cross validation was performed on the data, obtaining a Cross Validation (CV) accuracy of 96%. D. The bar graph represents the loading plot for LV1 of the MLPLSDA, that captures variation across mother and cord. The predictors are ordered according to their VIP scores. E. MLPLSDA was used to define the features that separate mother and cord blood samples for all seven antigens tested. Each dot represents an individual blood sample (mother or cord) tested for one of these three antigens. LV1 and LV2 account for 14% and 17% of the variability. The separation between mothers and cords is largely captured on LV1, which explains 37% of the Y variance in the direction of the maternal:cord separation. 5-fold cross validation was performed on the data, resulting in a CV accuracy of 94%. F. The bar graph represents the loading plot for LV1 of the MLPLSDA, that captures variation across mother and cord. The predictors are ordered according to their VIP scores. Significance across functional comparisons was defined using a paired t-test, *p<0.05, **p<0.01, ***p<0.001.
Supplemental Figure 3. Related to Figure 3 and Figure 5: Capillary electrophoresis analysis of Fc-glycans. Fluorescently-labeled glycans were run on capillary electrophoresis. These example traces show a matched cord (A) and maternal (B) sample, the fucosylated (C) and afucosylated (C) glycan libraries used to assign glycan peaks and the agalactosylated PGT128 (primarily G0F), (E) and the glycosylation-modified digalactosylated PGT128 (primarily G2F) (F).
Supplemental Figure 4. Related to Figure 3: Transfer ratio of antigen-specific Fc-glycans. A. The whisker plots show the transfer efficiency (cord/mother) for each of the major Fc-glycan profiles for FIM, PTX and PTN specific antibodies. B,C. Representative raw CE plots highlight differences in mother:cord glycan profiles. A dotted line is centered at the top of the G0F peak to show relationships between the G0F, G2S1F and G2F peaks. D-G. The dot-plot shows mother to cord transfer of one of the most variable peaks, G2S1F., across all four pertussis antigens (D), FHA specific (E), PTN specific (F), and PTX specific (G). Statistics were evaluated using a Wilcoxon signed rank test, variation of median from zero with Bonferroni correction. P values below 0.007 were considered significant. *p<0.007, **p<0.0014. For G2S1F transfer statistics, a Mann-Whitney test was used, *p<0.05, **p<0.01, ***p<0.001.
Supplemental Figure 5. Related to Figure 5, Computational analysis of paired maternal:cord samples. An OPLSDA was used to analyze the features separating mother and cord blood samples. Each dot represents an individual blood sample (mother or cord) tested for one of the four antigens. Latent variable 1 (LV1) and LV2 account for 17.5% and 20.8% of the variability in the analysis, respectively. The separation of mothers and cords was mostly captured on LV1, capturing 21% of Y-variation. Conversely, LV2 largely captured the variability in the antibody profiles that do not contribute to the difference in maternal and cord blood. 5-fold CV was performed on the data, obtaining a CV accuracy of 64%. B. The bar graph represents the loading plot for LV1 of the OPLSDA, that captured variation across mother:cord. The predictors are ordered according to their VIP scores. Features are colored according to feature type; functions (pink), glycans (red), subclasses (purple) and FcR binding (blue). C. A network was constructed based on the pairwise correlation coefficients between the 22 biophysical features and functional responses. Edges are weighted using the significant correlation coefficients, ρij, after removing the one with p-value >0.05.
Supplemental Figure 6. Related to Figure 6: Immunohistochemical analysis of placental sections. Immunohistochemistry was used to select antibody dilutions for further immunofluorescence analysis. Representative images are shown for each primary antibody and the concentration used.
Supplemental Figure 7 Related to Figure 7: Comparison of functional potential of cord and adult innate immune cells. The ability of primary adult and cord blood innate immune cells to mediate ADNP and NK cytotoxicity was compared across three primary cord-donors. Antibodies from maternal samples were tested against pertussis toxin (PTX) and filamentous hemagglutinin (FHA) A. The dot-line plot shows the ability of maternal antibodies to drive ADNP in cord or adult neutrophils. For each maternal sample, the adult and cord ADNP levels are connected. B. The dot-line plot shows the ability of maternal antibodies to drive NK cell degranulation (CD107a) and cytokine (IFNγ) secretion. For each maternal sample, the adult and cord NK cell activation are connected. For both analyses, a two-way ANOVA with multiple comparisons was used with a post-hoc Sidak’s test for multiple comparisons. **p<0.01, ***p<0.001, ****p<0.0001.
Highlights:
NK cell–activating antibodies are selectively transferred across the placenta
Di-galactosylated Fc glycans are preferentially transferred across the placenta
Digalactosylated antibodies bind more effectively to FcRn and FCGR3A
Though immature, neonatal NK cells are highly responsive to immune complexes
Acknowledgements
This work was supported by the Ragon Institute, the Samana Cay MGH scholar program, and NIH grants AI080289 (5R37AI080289-09), AI102660-01 (5R01AI102660-04) and AI129797-01 (1R01A1129797-01). We would also like to acknowledge Harvard CFAR for ongoing support P30 AI060354-02. A thank you to Adam Barb for his insightful thoughts.
Footnotes
Declaration of Interests
The authors declare no competing interests
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References:
- Abu-Raya B, Smolen KK, Willems F et al. (2016). Transfer of Maternal Antimicrobial Immunity to HIV-Exposed Uninfected Newborns. Frontiers in immunology 7, 338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ackerman ME, Moldt B, Wyatt RT, et al. (2011). A robust, high-throughput assay to determine the phagocytic activity of clinical antibody samples. Journal of immunological methods 366, 8–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amenyogbe N, Levy O, and Kollmann TR (2015). Systems vaccinology: a promise for the young and the poor. Philos Trans R Soc Lond B Biol Sci 370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold JN, Wormald MR, Sim RB, et al. (2007). The impact of glycosylation on the biological function and structure of human immunoglobulins. Annu Rev Immunol 25, 21–50. [DOI] [PubMed] [Google Scholar]
- Arnold KB, Burgener A, Birse K, et al. (2016). Increased levels of inflammatory cytokines in the female reproductive tract are associated with altered expression of proteases, mucosal barrier proteins, and an influx of HIV-susceptible target cells. Mucosal Immunol 9, 194–205. [DOI] [PubMed] [Google Scholar]
- Bakchoul T, Greinacher A, Sachs UJ et al. (2013). Inhibition of HPA-1a alloantibody-mediated platelet destruction by a deglycosylated anti-HPA-1a monoclonal antibody in mice: toward targeted treatment of fetal-alloimmune thrombocytopenia. Blood 122, 321–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedetti E, Pucic-Bakovic M, Keser T, et al. (2017). Network inference from glycoproteomics data reveals new reactions in the IgG glycosylation pathway. Nat Commun 8, 1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boesch AW, Brown EP, Cheng HD, et al. (2014). Highly parallel characterization of IgG Fc binding interactions. MAbs 6, 915–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondt A, Rombouts Y, Selman MH, et al. (2014). Immunoglobulin G (IgG) Fab glycosylation analysis using a new mass spectrometric high-throughput profiling method reveals pregnancy-associated changes. Molecular & cellular proteomics : MCP 13, 3029–3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondt A, Selman MH, Deelder AM, et al. (2013). Association between galactosylation of immunoglobulin G and improvement of rheumatoid arthritis during pregnancy is independent of sialylation. J Proteome Res 12, 4522–4531. [DOI] [PubMed] [Google Scholar]
- Brown EP, Licht AF, Dugast AS, et al. (2012). High-throughput, multiplexed IgG subclassing of antigen-specific antibodies from clinical samples. Journal of immunological methods 386, 117–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruhns P, Iannascoli B, England P, et al. (2009). Specificity and affinity of human Fcgamma receptors and their polymorphic variants for human IgG subclasses. Blood 113, 3716–3725. [DOI] [PubMed] [Google Scholar]
- Centers for Disease, C., and Prevention (1999). Impact of vaccines universally recommended for children--United States, 1990–1998. MMWR Morb Mortal Wkly Rep 48, 243–248. [PubMed] [Google Scholar]
- Chung AW, and Alter G (2017). Systems serology: profiling vaccine induced humoral immunity against HIV. Retrovirology 14, 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dashivets T, Thomann M, Rueger P, et al. (2015). Multi-Angle Effector Function Analysis of Human Monoclonal IgG Glycovariants. PLoS One 10, e0143520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies J, Jiang L, Pan LZ, et al. (2001). Expression of GnTIII in a recombinant anti-CD20 CHO production cell line: Expression of antibodies with altered glycoforms leads to an increase in ADCC through higher affinity for FC gamma RIII. Biotechnology and bioengineering 74, 288–294. [PubMed] [Google Scholar]
- Decker Y, Schomburg R, Nemeth E, et al. (2016). Abnormal galactosylation of immunoglobulin G in cerebrospinal fluid of multiple sclerosis patients. Mult Scler 22, 1794–1803. [DOI] [PubMed] [Google Scholar]
- Edwards KM, and Berbers GA (2014). Immune responses to pertussis vaccines and disease. The Journal of infectious diseases 209 Suppl 1, S10–15. [DOI] [PubMed] [Google Scholar]
- Einarsdottir HK, Selman MH, Kapur R, et al. (2013). Comparison of the Fc glycosylation of fetal and maternal immunoglobulin G. Glycoconj J 30, 147–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Einarsdottir HK, Stapleton NM, Scherjon S et al. (2014). On the perplexingly low rate of transport of IgG2 across the human placenta. PLoS One 9, e108319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsyth K, Plotkin S, Tan T, et al. (2015). Strategies to decrease pertussis transmission to infants. Pediatrics 135, e1475–1482. [DOI] [PubMed] [Google Scholar]
- Fouda GG, Martinez DR, Swamy GK, et al. (2018). The Impact of IgG transplacental transfer on early life immunity. Immunohorizons 2, 14–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu C, Lu L, Wu H, et al. (2016). Placental antibody transfer efficiency and maternal levels: specific for measles, coxsackievirus A16, enterovirus 71, poliomyelitis I-III and HIV-1 antibodies. Sci Rep 6, 38874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetghebuer T, Smolen KK, Adler C, et al. (2018). Initiation of anti-retroviral therapy before pregnancy reduces the risk of infection-related hospitalization in HIV-exposed uninfected infants born in a high-income country. Clin Infect Dis. [DOI] [PubMed] [Google Scholar]
- Ishikawa T, Takizawa T, Iwaki J, et al. (2015). Fc gamma receptor IIb participates in maternal IgG trafficking of human placental endothelial cells. Int J Mol Med 35, 1273–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen BC, Bondt A, Reiding KR, et al. (2016a). Pregnancy-associated serum N-glycome changes studied by high-throughput MALDI-TOF-MS. Sci Rep 6, 23296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen BC, Bondt A, Reiding KR, et al. (2016b). MALDI-TOF-MS reveals differential N-linked plasma- and IgG-glycosylation profiles between mothers and their newborns. Sci Rep 6, 34001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferis R (2012). Isotype and glycoform selection for antibody therapeutics. Arch Biochem Biophys 526, 159–166. [DOI] [PubMed] [Google Scholar]
- Jennewein MF, Abu-Raya B, Jiang Y, et al. (2017). Transfer of maternal immunity and programming of the newborn immune system. Semin Immunopathol. [DOI] [PubMed] [Google Scholar]
- Jennewein MF, and Alter G (2017). The Immunoregulatory Roles of Antibody Glycosylation. Trends Immunol 38, 358–372. [DOI] [PubMed] [Google Scholar]
- Jensen PF, Schoch A, Larraillet V, et al. (2017). A two-pronged binding mechanism of IgG to the neonatal Fc receptor controls complex stability and IgG serum half-life. Molecular & cellular proteomics : MCP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krumsiek J, Suhre K, Illig T, et al. (2011). Gaussian graphical modeling reconstructs pathway reactions from high-throughput metabolomics data. BMC Syst Biol 5, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau KS, Juchheim AM, Cavaliere KR, et al. (2011). In vivo systems analysis identifies spatial and temporal aspects of the modulation of TNF-alpha-induced apoptosis and proliferation by MAPKs. Sci Signal 4, ra16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YC, and Lin SJ (2013). Neonatal natural killer cell function: relevance to antiviral immune defense. Clin Dev Immunol 2013, 427696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyden TW, Robinson JM, Tridandapani S, et al. (2001). The Fc receptor for IgG expressed in the villus endothelium of human placenta is Fc gamma RIIb2. Journal of immunology (Baltimore, Md : 1950) 166, 3882–3889. [DOI] [PubMed] [Google Scholar]
- Mahan AE, Jennewein MF, Suscovich T et al. (2016). Antigen-Specific Antibody Glycosylation Is Regulated via Vaccination. PLoS pathogens 12, e1005456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez DR, Fong Y, Li SH, et al. (2019). Fc characteristics mediate selective placental transfer of IgG in HIV-infected women. Cell. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez DR, Fouda GG, Peng X, et al. (2018). Noncanonical placental Fc receptors: What is their role in modulating transplacental transfer of maternal IgG? PLoS pathogens 14, e1007161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLellan JS, Chen M, Joyce MG, et al. (2013). Structure-based design of a fusion glycoprotein vaccine for respiratory syncytial virus. Science (New York, NY) 342, 592–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimura Y, Sondermann P, Ghirlando R, et al. (2001). Role of oligosaccharide residues of IgG1-Fc in Fc gamma RIIb binding. The Journal of biological chemistry 276, 45539–45547. [DOI] [PubMed] [Google Scholar]
- Moore JS, Wu X, Kulhavy R, et al. (2005). Increased levels of galactose-deficient IgG in sera of HIV-1-infected individuals. AIDS (London, England) 19, 381–389. [DOI] [PubMed] [Google Scholar]
- Nimmerjahn F, and Ravetch JV (2006). Fcgamma receptors: old friends and new family members. Immunity 24, 19–28. [DOI] [PubMed] [Google Scholar]
- Nimmerjahn F, and Ravetch JV (2008). Fcgamma receptors as regulators of immune responses. Nature reviews Immunology 8, 34–47. [DOI] [PubMed] [Google Scholar]
- Olin A, Henckel E, Chen Y et al. (2018). Stereotypic Immune System Development in Newborn Children. Cell 174, 1277–1292 e1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmeira P, Quinello C, Silveira-Lessa AL, et al. (2012). IgG placental transfer in healthy and pathological pregnancies. Clin Dev Immunol 2012, 985646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasek M, Duk M, Podbielska M, et al. (2006). Galactosylation of IgG from rheumatoid arthritis (RA) patients--changes during therapy. Glycoconj J 23, 463–471. [DOI] [PubMed] [Google Scholar]
- Piche-Nicholas NM, Avery LB, King AC, et al. (2018). Changes in complementarity-determining regions significantly alter IgG binding to the neonatal Fc receptor (FcRn) and pharmacokinetics. MAbs 10, 81–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulendran B, and Ahmed R (2011). Immunological mechanisms of vaccination. Nat Immunol 12, 509–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roopenian DC, and Akilesh S (2007). FcRn: the neonatal Fc receptor comes of age. Nature reviews Immunology 7, 715–725. [DOI] [PubMed] [Google Scholar]
- Saso A, and Kampmann B (2017). Vaccine responses in newborns. Semin Immunopathol 39, 627–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shields RL, Lai J, Keck R et al. (2002). Lack of fucose on human IgG1 N-linked oligosaccharide improves binding to human Fcgamma RIII and antibody-dependent cellular toxicity. The Journal of biological chemistry 277, 26733–26740. [DOI] [PubMed] [Google Scholar]
- Shin SY, Fauman EB, Petersen AK, et al. (2014). An atlas of genetic influences on human blood metabolites. Nat Genet 46, 543–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simister NE (2003). Placental transport of immunoglobulin G. Vaccine 21, 3365–3369. [DOI] [PubMed] [Google Scholar]
- Sonneveld ME, Natunen S, Sainio S, et al. (2016). Glycosylation pattern of anti-platelet IgG is stable during pregnancy and predicts clinical outcome in alloimmune thrombocytopenia. Br J Haematol 174, 310–320. [DOI] [PubMed] [Google Scholar]
- Stapleton NM, Andersen JT, Stemerding AM, et al. (2011). Competition for FcRn-mediated transport gives rise to short half-life of human IgG3 and offers therapeutic potential. Nat Commun 2, 599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takizawa T, Anderson CL, and Robinson JM (2005). A novel Fc gamma R-defined, IgG-containing organelle in placental endothelium. Journal of immunology (Baltimore, Md : 1950) 175, 2331–2339. [DOI] [PubMed] [Google Scholar]
- Thomann M, Schlothauer T, Dashivets T et al. (2015). In Vitro Glycoengineering of IgG1 and Its Effect on Fc Receptor Binding and ADCC Activity. PLoS One 10, e0134949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vestrheim AC, Moen A, Egge-Jacobsen W, et al. (2014). A pilot study showing differences in glycosylation patterns of IgG subclasses induced by pneumococcal, meningococcal, and two types of influenza vaccines. Immun Inflamm Dis 2, 76–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidarsson G, Dekkers G, and Rispens T (2014). IgG subclasses and allotypes: from structure to effector functions. Frontiers in immunology 5, 520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerhuis JA, van Velzen EJ, Hoefsloot HC, et al. (2010). Multivariate paired data analysis: multilevel PLSDA versus OPLSDA. Metabolomics 6, 119–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittaker E, Goldblatt D, McIntyre P, et al. (2018). Neonatal Immunization: Rationale, Current State, and Future Prospects. Frontiers in immunology 9, 532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilcox CR, Holder B, and Jones CE (2017). Factors Affecting the FcRn-Mediated Transplacental Transfer of Antibodies and Implications for Vaccination in Pregnancy. Frontiers in immunology 8, 1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter K, Glaser C, Watt J, et al. (2014). Pertussis epidemic--California, 2014. MMWR Morb Mortal Wkly Rep 63, 1129–1132. [PMC free article] [PubMed] [Google Scholar]
- Yu JC, Khodadadi H, Malik A, et al. (2018). Innate Immunity of Neonates and Infants. Frontiers in immunology 9, 1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. Related to Figure 1: Relationship of antibody profiles across vaccinated and non-vaccinated mothers. The principal component analysis includes all pertussis-specific antibody features across all mother:cord pairs. A. No differences were observed across antigen-specific antibody profiles (left) irrespective of vaccination. B. Similarly, no overall differences in antibody profiles were observed across vaccinated or un-vaccinated mothers (right).
Supplemental Figure 2. Related to Figure 2: NK degranulation selection across cohorts. A. The dot-plots show NK-dependent degranulation to RSV pre-fusion F antigen plotted as the percentage of NK cells positive for CD107a, IFNγ and MIP-1β. B. The dot-plots show 28 mother:cord pairs from cohort 2 ability to drive NK degranulation (CD107a expression) in response to Pertussis toxin (PTX), Pertactin (PTN) and measles virus antigens. Significance in A, B evaluated for NK degranulation with paired t-test. *p<0.05, **p<0.01, ***p<0.001. C. A MLPLSDA was used to analyze the features separating mother and cord blood samples for RSV pre-fusion, RSV post-fusion and flu-specific antibody profiles. Each dot represents an individual blood sample (mother or cord) tested for one of these three antigens. LV1 and LV2 account for 18% and 22% of the variability in functional and glycan profiles across the antigens. The separation of mothers and cord is largely captured on LV1, which explains 36% of the Y variance in the direction of the maternal:cord separation. 5-fold cross validation was performed on the data, obtaining a Cross Validation (CV) accuracy of 96%. D. The bar graph represents the loading plot for LV1 of the MLPLSDA, that captures variation across mother and cord. The predictors are ordered according to their VIP scores. E. MLPLSDA was used to define the features that separate mother and cord blood samples for all seven antigens tested. Each dot represents an individual blood sample (mother or cord) tested for one of these three antigens. LV1 and LV2 account for 14% and 17% of the variability. The separation between mothers and cords is largely captured on LV1, which explains 37% of the Y variance in the direction of the maternal:cord separation. 5-fold cross validation was performed on the data, resulting in a CV accuracy of 94%. F. The bar graph represents the loading plot for LV1 of the MLPLSDA, that captures variation across mother and cord. The predictors are ordered according to their VIP scores. Significance across functional comparisons was defined using a paired t-test, *p<0.05, **p<0.01, ***p<0.001.
Supplemental Figure 3. Related to Figure 3 and Figure 5: Capillary electrophoresis analysis of Fc-glycans. Fluorescently-labeled glycans were run on capillary electrophoresis. These example traces show a matched cord (A) and maternal (B) sample, the fucosylated (C) and afucosylated (C) glycan libraries used to assign glycan peaks and the agalactosylated PGT128 (primarily G0F), (E) and the glycosylation-modified digalactosylated PGT128 (primarily G2F) (F).
Supplemental Figure 4. Related to Figure 3: Transfer ratio of antigen-specific Fc-glycans. A. The whisker plots show the transfer efficiency (cord/mother) for each of the major Fc-glycan profiles for FIM, PTX and PTN specific antibodies. B,C. Representative raw CE plots highlight differences in mother:cord glycan profiles. A dotted line is centered at the top of the G0F peak to show relationships between the G0F, G2S1F and G2F peaks. D-G. The dot-plot shows mother to cord transfer of one of the most variable peaks, G2S1F., across all four pertussis antigens (D), FHA specific (E), PTN specific (F), and PTX specific (G). Statistics were evaluated using a Wilcoxon signed rank test, variation of median from zero with Bonferroni correction. P values below 0.007 were considered significant. *p<0.007, **p<0.0014. For G2S1F transfer statistics, a Mann-Whitney test was used, *p<0.05, **p<0.01, ***p<0.001.
Supplemental Figure 5. Related to Figure 5, Computational analysis of paired maternal:cord samples. An OPLSDA was used to analyze the features separating mother and cord blood samples. Each dot represents an individual blood sample (mother or cord) tested for one of the four antigens. Latent variable 1 (LV1) and LV2 account for 17.5% and 20.8% of the variability in the analysis, respectively. The separation of mothers and cords was mostly captured on LV1, capturing 21% of Y-variation. Conversely, LV2 largely captured the variability in the antibody profiles that do not contribute to the difference in maternal and cord blood. 5-fold CV was performed on the data, obtaining a CV accuracy of 64%. B. The bar graph represents the loading plot for LV1 of the OPLSDA, that captured variation across mother:cord. The predictors are ordered according to their VIP scores. Features are colored according to feature type; functions (pink), glycans (red), subclasses (purple) and FcR binding (blue). C. A network was constructed based on the pairwise correlation coefficients between the 22 biophysical features and functional responses. Edges are weighted using the significant correlation coefficients, ρij, after removing the one with p-value >0.05.
Supplemental Figure 6. Related to Figure 6: Immunohistochemical analysis of placental sections. Immunohistochemistry was used to select antibody dilutions for further immunofluorescence analysis. Representative images are shown for each primary antibody and the concentration used.
Supplemental Figure 7 Related to Figure 7: Comparison of functional potential of cord and adult innate immune cells. The ability of primary adult and cord blood innate immune cells to mediate ADNP and NK cytotoxicity was compared across three primary cord-donors. Antibodies from maternal samples were tested against pertussis toxin (PTX) and filamentous hemagglutinin (FHA) A. The dot-line plot shows the ability of maternal antibodies to drive ADNP in cord or adult neutrophils. For each maternal sample, the adult and cord ADNP levels are connected. B. The dot-line plot shows the ability of maternal antibodies to drive NK cell degranulation (CD107a) and cytokine (IFNγ) secretion. For each maternal sample, the adult and cord NK cell activation are connected. For both analyses, a two-way ANOVA with multiple comparisons was used with a post-hoc Sidak’s test for multiple comparisons. **p<0.01, ***p<0.001, ****p<0.0001.