Abstract
The diversity in antibody repertoire relies on different B cell populations working efficiently to fulfil distinct specific functions. We recently described an innate-like CD19+CD45R-/lo (19+45Rlo) cell population in postnatal unstimulated adult mice, a heterogeneous population containing cells expressing immunoglobulin M (IgM) and others behaving as differentiated mature B lymphocytes (intracytoplasmic IgG1, AID+, Blimp-1+RAG2-). In the present study, we characterized the Ig repertoire expressed by splenic 19+45Rlo cells, assuming that they would bear a restricted repertoire biased for germline rearrangements and low mutation rates similar to other innate-like cells. Sequences from 19+45Rlo cells displayed a variety of V, D and J regions, and the analysis of the CDR-H3 region revealed an intermediate overall CDR-H3 length and moderate hydrophobicity. Both IgM and switched sequences of PD15 19+45Rlo cells had shorter CDR-H3 region and fewer non-template N nucleotides than adult sequences, as expected for profiles that correspond to an immature phenotype. Regarding the mutation rate in the VH regions, IgG1 sequences already carried a high rate of replacement mutations at PD15, which increased further in the sequences obtained from adult mice. Moreover, statistical models suggest that a proportion of the switched sequences in adult 19+45Rlo cells had experienced antigen selection, unlike other innate-like B cell compartments.
Key Words: B lymphocytes, CDR-H3, Homeostasis, Innate-like lymphocytes, Immunoglobulin class switching
Introduction
The first committed B lymphocytes arise in the fetal liver early in embryonic development (11 days post coitum) as CD19+CD45R-/lo (19+45Rlo) pro-B cells [1]. Soon after birth, 19+45Rlo cells colonize the spleen, accounting for up to 11% of the total B lymphocyte population by postnatal day (PD) 7, and their numbers stabilize from PD30 onwards [2]. The first IgG-secreting plasma cells are found at PD15 among 19+45Rlo cells [3], suggesting the early differentiation of these cells. The differentiation of mature peripheral B lymphocytes to plasma cells requires the coordinated activity of different transcription factors, including B lymphocyte-induced maturation protein-1 (Blimp-1), X-box-binding protein 1 (Xbp-1) and activation-induced cytidine deaminase (AID). AID is required to introduce diversification through somatic hypermutation (SHM), somatic hyperconversion and class switch recombination (CSR) [4, 5, 6]. During the course of the reactions that take place in the germinal center, AID is activated after B lymphocyte encounter with antigen (Ag) with the help of T cells [7, 8]. However, AID activation and SHM can occur independently of T cells, outside the germinal center, in response to TLR ligation in an Ag-independent context [6, 9, 10, 11].
We previously demonstrated that adult splenic 19+45Rlo cells display features of innate-like B lymphocytes such as B1 cells and marginal zone (MZ) B lymphocytes. Indeed, not only are they preferentially derived from embryonic liver hematopoietic precursors but they also exhibit a pre-activated status (CD43+, CD44+), have a high turnover rate (Ki-67+; 5-BrdU uptake in vivo), and are maintained and expanded in immune-deficient mice [2, 3]. These cells form a heterogeneous population with around 60% of the cells expressing surface immunoglobulin M (IgM), as well as exhibiting some traits of pre-plasmablasts (CD138+, Xbp-1+), and a pronounced ability to release IgG1 and IgA [3]. Furthermore, we found that 19+45Rlo cells were located perifollicularly in the spleen close to the MZ metallophilic macrophages and not in the red pulp. This distribution suggests they have a potential role in the initial phases of Ag recognition, as described for MZ B lymphocytes. More recently, we demonstrated that the surface phenotype, functional features and molecular signature of 19+45Rlo cells separate them from B1 cells [2]. These cells can undergo CSR in vitro after TLR-dependent and B cell-activating factor and IL-4 activation [2], and they can secrete IL-10. By contrast, other innate-like B cell subsets, such as innate response activator B cells and age-associated B cells, display a distinct cytokine secretion pattern and do not secrete isotype-switched Igs after in vivo or in vitro stimulation [12, 13].
B lymphocytes from the innate-like branch of the immune system secrete low affinity Igs, mainly of the IgM isotype bearing germ-line IgH rearrangements. In the present study, we examined the IgH repertoire expressed by 19+45Rlo cells under homeostatic conditions and we compared it with that of naïve CD19+CD45R+ (19+45R+) cells from PD15 and adult (PD60) mice. Our results demonstrate that the repertoire of the V/D/J region in 19+45Rlo cells is not restricted and underwent SHM, and that at PD60, this region accommodated N insertions. AID expression was detected in 19+45Rlo cells, increasing at PD60. An analysis of isotype-switched V region sequences in 19+45Rlo cells revealed the accumulation of replacement mutations, mainly in CDRs, suggesting the selection of these mutations through Ag encounters. Finally, a detailed analysis of the features of CDR-H3 in adult splenic 19+45Rlo cells revealed an unrelated auto-antibody (auto-Ab)-prone profile, which distinguishes this population from those in other recently described innate-like B cell compartments.
Materials and Methods
Ethics Statement
All procedures were carried out in accordance with protocols and guidelines established by the Institutional Animal Care and Use Committee of the Instituto de Salud Carlos III.
Mice
BALB/c mice were bred and maintained in the specific pathogen-free animal facilities at the Instituto de Salud Carlos III. The mice were housed individually in ventilated cages and sacrificed by cervical dislocation to obtain cell suspensions from the spleen.
Flow Cytometry and Cell Sorting
Single cell suspensions were prepared in staining buffer (2.5% FCS in Dulbecco's phosphate-buffered saline; BioWhittaker, Lonza Group, Switzerland) and non-specific binding was blocked with Fc-Block™ (BD Biosciences, San Jose, Calif., USA). Staining was performed using standard protocols with the following Abs and secondary reagents: phycoerythrin-labelled anti-CD19 (clone 1D3, Affymetrix eBiosciences, San Diego, Calif., USA), FITC-labelled anti-CD45R (clone RA3-6B2) and biotinylated anti-IgG1 (clone A85-1), anti-IgG2a/2b (clone R2-40), anti-IgA (clone C10-1), anti-Ig light chain κ (clone 187.1), goat anti-mouse Ig light chain λ and isotype controls (clone R3-34, goat Igs), and allophycocyanin-labelled streptavidin (BioLegend, San Diego, Calif., USA). FITC-labelled and biotinylated Abs were obtained from BD Biosciences, and polyclonal antisera were purchased from SouthernBiotech (Birmingham, Ala., USA). The cells were analysed on a FACSCanto-I (BD Biosciences) using the FlowJo v6.3.4 (Tree Star, Ashland, Oreg., USA) and Infinicyt (Cytognos SL, Salamanca, Spain) software packages, and they were FACS purified using a FACSAria-I apparatus with DIVA v6.1 software (BD Biosciences). For intracytoplasmic Ig detection, fresh adult cells were stained for CD19 and CD45R surface expression. Stained cells were fixed/permeabilized using the Cytofix/Cytoperm kit (BD Biosciences) and they were assayed with the Live/Dead Exclusion Fixable Violet Dead Cell Stain kit (Invitrogen, Carlsbad, Calif., USA) prior to intracellular staining with anti-IgG1, anti-IgG2a/2b, anti-IgA and the isotype control.
RT-qPCR, Cloning and V Region Sequencing
Total RNA was extracted from sorted splenic 19+45Rlo, 19+45R+ and CD19loCD45Rlo (19lo45Rlo) cells, and oligo(dT)-primed cDNA samples were prepared with avian myeloblastosis virus reverse transcriptase as described previously [1]. Quantitative real-time PCR (RT-qPCR) was performed on a CFX96™ Real-Time System using the SsoFast™ Supermix EvaGreen (Bio-Rad, Hercules, Calif., USA), as indicated elsewhere [14]. Bio-Rad CFX Manager software was used to calculate the CT of each reaction and the relative amount of specific cDNA in each sample was determined by the ΔCT method. The results are presented as the expression of each transcript relative to that of the hypoxanthine phosphoribosyl transferase 1 gene (HPRT). The primers used for Blimp-1, AID, TdT, RAG2 and HPRT have been described previously [3, 4, 14, 15].
To detect VDJ-C rearrangement, preparations of 19+45Rlo and 19+45R+ cells were subjected to PCR amplification with 1 U of FastStart DNA polymerase (Roche, Mannheim, Germany) in a PTC-200 DNA Engine cycler (Bio-Rad). Rearranged alleles were amplified using a degenerate V primer (5′-AGGTSMARCTKCWSSAGTCWGG-3′: [16] and CH-specific primers: Cµ (5′-GGGAAATGGTGCTGGGCAGGAA-3′: [1], Cγ1R (5′-GGATCCAGAGTTCCAGGTCACT-3′) and CαR (5′-GAGCTGGTGGGAGTGTCAGTG-3′) [4]. The reaction products were resolved by electrophoresis on 2% agarose gels and visualized by Pronasafe Nucleic Acid Staining (Condalab, Madrid, Spain). The PCR products were cloned using the Dual Promoter TA cloning kit (PCR® II vector; Invitrogen) and transformed into JM109 Escherichia coli competent cells (Promega, Madison, Wisc., USA). After plating, the plasmids from IPTG selected colonies [17] were PCR amplified and the products were separated by electrophoresis to identify positive clones. The PCR products were cleaned with PCR clean-up kits (Mo Bio Laboratories, Carlsbad, Calif., USA) and the purified plasmids were sequenced in an ABI3500 automatic sequencer using the BigDye sequencing mixture (Applied Biosystems, Carlsbad, Calif., USA). The background error rate of the Taq polymerase was estimated as 1 × 10-3 by sequencing clones expressing HPRT.
Sequence Analysis
The sequences analysed in this study have been deposited in the EMBL database with the accession numbers HF569379-HF569839. The sequences obtained were aligned and studied using IgBLASTN® (http://www.ncbi.nlm.nih.gov/igblast/) and the Immunogenetics (IMGT) information system® (http://www.imgt.org/HighV-QUEST/) [18]. Comparative sequence analysis between populations and CDR-H3 amino acid alignments were performed using the Immunoglobulin Analysis Tool software (IgAT) [19]. Ambiguous sequences and repeated rearrangements with an identical V region and CDR-H3 segments were excluded, as we could not ascertain whether they corresponded to clonal expanded cells bearing such rearrangements or to PCR-amplified transcripts in rare cell populations. The features of the sequences are summarized in table 1. Sequences are considered clonally related when they use the same VH gene, and when they have a highly homologous CDR-H3 and an identical CDR-H3 length. The individual amino acid usage was compared among the different groups of sequences using the χ2 test, as described elsewhere [20]. The Shannon entropy value [21] was used to analyse the amino acid variability in the CDR-H3 region. Phylogenetic trees of the sequences were generated using the neighbour-joining method with BioEdit v7.2.0 (http://www.mbio.ncsu.edu/bioedit/bioedit.html) and MEGA5 (http://www.megasoftware.net) software.
Table 1.
Summary of the sequences
| Total | Functional | Sequence functionality (functional/total) | Unique | Sequence diversity (unique/functional) | Clonotypes | Clonotypic diversity (clonotypes/functional) | |
|---|---|---|---|---|---|---|---|
| PD15 | |||||||
| IgM 19+45Rlo | 36 | 32 | 0.89 | 32 | 1.0 | 30 | 0.94 |
| IgM 19+45R+ | 48 | 45 | 0.94 | 42 | 0.93 | 42 | 1.0 |
| Switched 19+45Rlo | 118 | 91 | 0.77a | 67 | 0.73 | 23 | 0.34a |
| Switched 19+45R+ | 16 | 9 | 0.56a | 8 | 0.88 | 6 | 0.75 |
| PD60 | |||||||
| IgM 19+45Rlo | 72 | 68 | 0.94 | 64 | 0.94 | 58 | 0.96 |
| IgM 19+45R+ | 66 | 62 | 0.94 | 61 | 0.98 | 61 | 1.0 |
| Switched 19+45Rlo | 81 | 55 | 0.68a | 51 | 0.92 | 42 | 0.82 |
| Switched 19lo45Rlo | 45 | 22 | 0.49a | 22 | 1 | 21 | 0.95 |
Sequence functionality represents the number of functional sequences divided by the total number of sequences. Sequence diversity is defined as the number of unique sequences divided by the number of functional sequences. Each clonotype includes sequences that use the same VH gene, and that have a highly homologous CDR-H3 and an identical CDR-H3 length. Clonotypic diversity is defined as the number of clonotypes divided by the number of functional sequences.
p < 0.05: data were compared in contingency tables with the χ2 test.
Statistical Analysis
The data are presented as the means ± SEM. Statistical analyses were performed with Prism 5.0 (GraphPad) software after testing the data distribution using Kolmogorov-Smirnov, Shapiro-Wilk and D'Agostino-Pearson normality tests. Comparisons were performed with the two-tailed Student t test and the χ2 test for contingency tables.
Results
Unstimulated Adult Splenic 19+45Rlo Cells Include Ig Class-Switched Cells
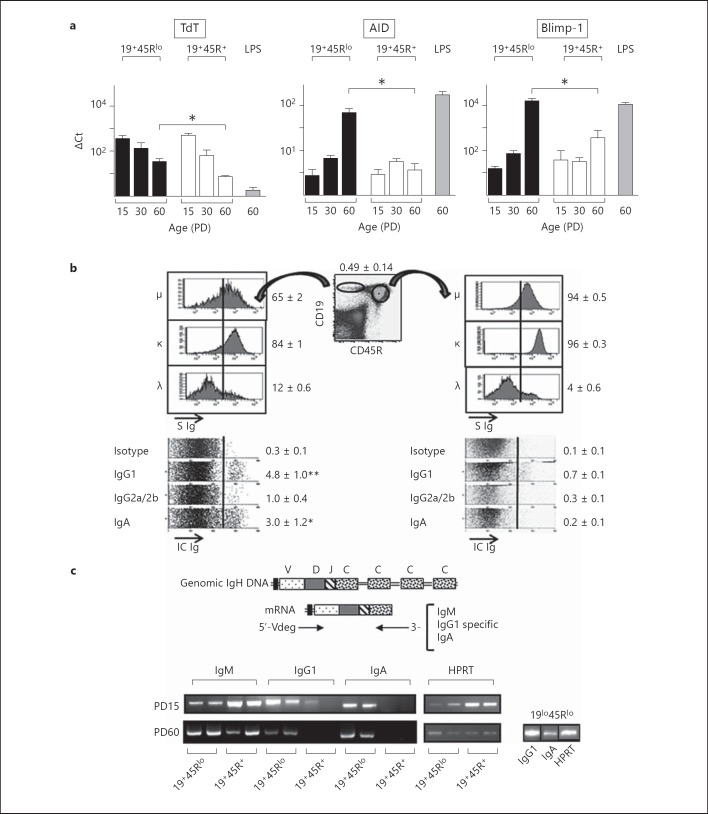
We previously demonstrated that 19+45Rlo cells colonize the spleen soon after birth and that they already release IgG by PD15. Moreover, adult splenic 19+45Rlo cells express transcription factors and phenotypic features characteristic of a highly differentiated B cell compartment [3]. RT-qPCR for Blimp-1, AID, RAG2 and TdT in FACS-sorted splenic 19+45Rlo cells, and in conventionally purified 19+45R+ cells, revealed similar transcript expression in both cell populations at PD15 (fig. 1a). However, there was a stronger increase in the expression of AID and Blimp-1 transcripts between PD15 and PD60 in 19+45Rlo than in 19+45R+ cells. As expected, TdT and RAG2 transcripts were detected in PD15 samples, and they disappeared progressively with age. Nevertheless, there were more TdT transcripts in adult 19+45Rlo cells than in adult 19+45R+ cells, while RAG2 was undetectable in both adult B cell subpopulations (online suppl. fig. 1a; for all online suppl. material, see www.karger.com/doi/10.1159/000358237). We next quantified the Ig class-switched 19+45Rlo cells present in the spleen under homeostatic conditions using intracytoplasmic and surface detection of Igs by flow cytometry (fig. 1b). We found that around 60% of the 19+45Rlo cells expressed a surface IgM, which is in agreement with our previous data [3], and 96% of these cells expressed either κ or λ light chains, indicating that most of them were mature B cells or pre-plasmablasts. Accordingly, the frequency of clonal progenitors that could be established in stromal cultures in the presence of IL-7 was extremely low (online suppl. fig. 1b), as described previously [22]. However, we cannot completely exclude that 19+45Rlo cells contain small fractions of pre-B cells, B1 cell progenitors (bearing a 19+45Rlo phenotype, as recently described in the bone marrow and spleen [23, 24]) or even rare CD19+ dendritic cells [25]. Finally, there was more intracytoplasmic IgG1, IgG2a/2b and IgA in 19+45Rlo than in 19+45R+ cells, with the latter expressing only minimal intracytoplasmic levels of these Igs. Together, these data show that 19+45Rlo cells undergo Ig class switching in homeostatic conditions and that these cells possess the molecular machinery responsible for generating Ig variability, including CSR, SHM and N nucleotide insertion.
Fig. 1.
19+45Rlo cells display traits of highly differentiated B lymphocytes. a Bar graphs show the RT-qPCR analyses for the genes indicated in sorted 19+45Rlo and 19+45R+ cells at PD15, PD30 and PD60. The results represent the expression of each transcript relative to that of HPRT as the mean ± SEM (n = 4 performed in duplicates). Bio-Rad CFX Manager software was used to calculate the CT of each reaction, and the relative amount of specific cDNA in each sample was determined using the ΔCT method. LPS, splenocytes stimulated with LPS (25 µg/ml) for 72 h. * p < 0.05: represent the statistical significance calculated by an unpaired two-tailed Student t test. b Flow cytometry analyses of surface and intracytoplasmic Igs from PD60 splenocytes stained with anti-CD19, anti-CD45R and the indicated isotype-specific Abs. The dot plot shows representative staining to indicate the gates for 19+45Rlo cells (ellipse) and 19+45R+ cells (circle). Histograms of IgM (μ), kappa (κ), and lambda (λ) surface expression are shown in the top panels. Fluorescence intensity of the indicated parameters is shown (S Ig). Isotype background fluorescence is indicated by the vertical lines. The panels below represent the intracytoplasmic staining of the gated populations displayed as the so-called parameter band dot plots (Infinicyt software, Cytognos SL), which represent events as dots distributed along an arbitrarily short y-axis (33% height of the x-axis length) for each parameter analysed, with the corresponding fluorescence intensity displayed in the x-axis (IC Ig). Positive events are those to the right of the vertical line and the numbers represent the frequencies as percentages (mean ± SEM, n = 6). * p < 0.05, ** p < 0.01. c Scheme of the PCR design used to analyse IgM, IgG1 and IgA rearrangements, with the primers used indicated below. A representative example of the results obtained for purified 19+45Rlo, 19+45R+ and 19lo45Rlo cells at PD15 and PD60 is shown (n = 4 different samples per population).
Clonotypic Diversity in the IgH Repertoire of Class-Switched 19+45Rlo Cells
To analyse the IgH repertoire of postnatal and adult 19+45Rlo cells in an unbiased manner, we amplified the VDJ-C rearrangements from 19+45Rlo cells by RT-PCR using a mixture of degenerate 5′-primers from the VH-leader regions and 3′-primers specific to each C region isotype analysed (IgM, IgG1 and IgA). IgM-specific products were detected in samples from both 19+45Rlo and 19+45R+ cells at PD15 and PD60 (fig. 1c). IgG1- and IgA-specific products were also detected in PD15 and PD60 19+45Rlo cells, yet they were detected only weakly or not at all in adult 19+45R+ cells. Based on this observation, we used cDNA samples from PD60 19lo45Rlo cells for comparisons, which yielded strong IgG1- and IgA-specific PCR signals (fig. 1c, right panel).
The isotype-specific VDJ-C rearrangements detected by RT-PCR from sorted 19+45Rlo, 19+45R+ and 19lo45Rlo cells at PD15 and PD60 were cloned and sequenced. A total of 482 sequences were analysed (table 1), of which 307 corresponded to 19+45Rlo cells and 175 to 19+45R+/19lo45Rlo cells. We obtained sequences for IgM (n = 222), IgG1 (n = 165) and IgA rearrangements (n = 95). Due to the limited number of IgA sequences obtained in some of the groups, they were analysed jointly with those of IgG1 (referred to henceforth as switched sequences) to obtain more robust comparisons, unless otherwise indicated. Nevertheless, since IgG1 was amplified from only 3 of the 13 sorted PD15 19+45R+ samples and no IgA was detected, the number of sequences analysed from this group was low when compared to switched sequences from 19+45Rlo cells, and therefore we cannot perform a meaningful statistically analysis in this case. Most IgM sequences were functional and very diverse (sequence diversity defined as the proportion of unique sequences among all the functional sequences). By contrast, a large number of switched sequences were non-functional and although they exhibited considerable sequence diversity, clonotypic diversity (the number of clonotypes relative to the number of functional sequences) was significantly lower at PD15 than for the IgM sequences.
Broad V and D Region Gene Family Usage in IgH Sequences from 19+45Rlo Cells
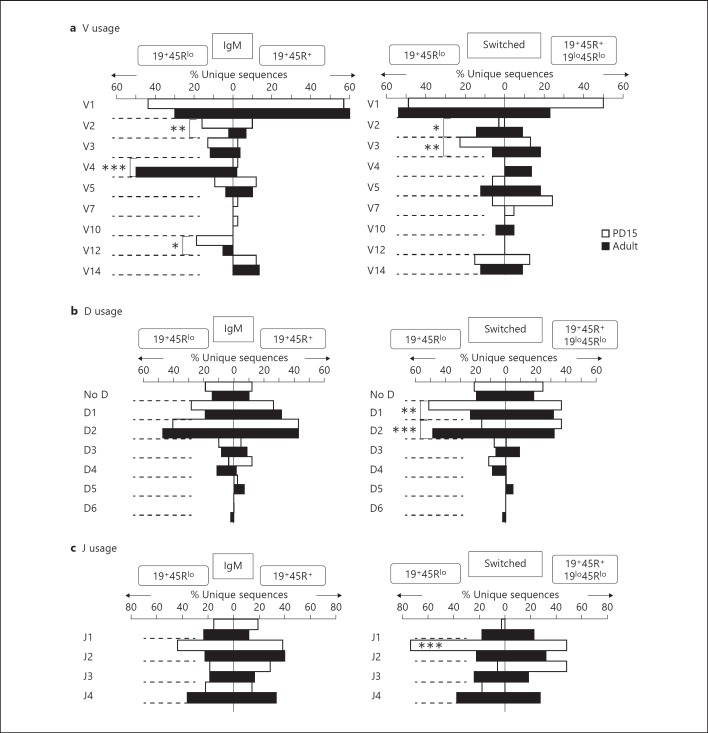
We compared the V region usage in 19+45Rlo cells from PD15 and PD60 spleens with that of 19+45R+ or 19lo45Rlo cells at the same ages (fig. 2a). Genes in the V1 family were those most often used in each group of sequences, except in the adult IgM sequences from 19+45Rlo cells. However, V2 (VHX24 and Q52), V3 and V12 were also used often at PD15 in the IgM sequences, and V3 and V14 accumulated in switched sequences at PD15. By contrast, the V4 and V1 families were preferentially used by IgM sequences from PD60 19+45Rlo cells, whereas the V2/V12 gene families were almost totally excluded from these IgM sequences. Interestingly, V2 usage was preserved in switched sequences from PD60 19+45Rlo cells. We also observed little usage of other V region families, including the most J-proximal V5 segment (VH7183) family and the V14 family, which are frequently expressed in newborn IgM sequences and in B1 cells [26], as well as V11 (the latter detected by specific V11-Cµ RT-PCR amplification; data not shown).
Fig. 2.
Analysis of the IgH repertoires expressed by 19+45Rlo, 19+45R+ and 19lo45Rlo cells. Sorted PD15 and PD60 samples were examined by RT-PCR isotype-specific amplification as in fig. 1c. The procedure for cloning and sequencing has been described in Materials and Methods. The results for IgM and switched (IgG1 and IgA) sequences are represented as percentages of the unique sequences (white bars, PD15 sequences; black bars, PD60 sequences). The data shown on the left correspond to the sequences from 19+45Rlo cells, while those on the right correspond to 19+45R+ cells (PD15 and PD60 IgM and PD15 switched sequences) and 19lo45Rlo cells (PD60 switched sequences). The total numbers analysed are indicated in table 1. a V region family usage. b D region usage. c J region usage. Statistical analysis was performed using the χ2 test comparing the groups indicated by the brackets. J region use comparison was performed in a contingency table of the 4 J regions among the different groups. * p < 0.05, ** p < 0.01, *** p < 0.001: represents statistical significance.
Analysis of B cell repertoires based on PCR amplification of Ig cDNAs may introduce a bias in favour of cells expressing large amounts of Igs, such as the plasmablasts that are prevalent among 19+45Rlo cells, thereby hampering the interpretation of the frequency data. Yet, our results reveal a diverse profile of V region usage in sequences from 19+45Rlo cells, which varies with age and isotype switching and is distinct from that of 19+45R+ and 19lo45Rlo cells. This reflects a different scenario from the initial hypothesis of a restricted repertoire.
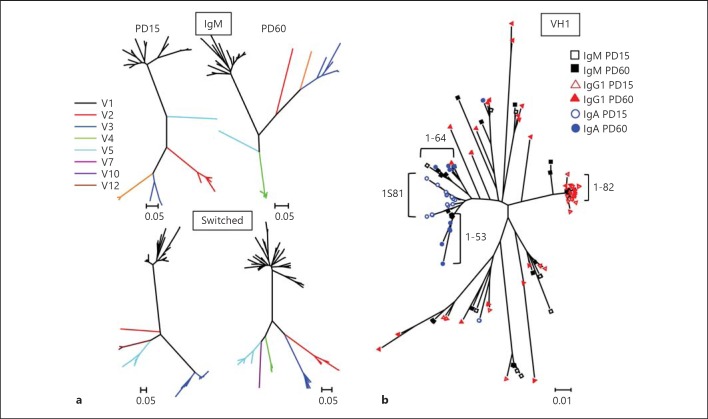
We generated phylogenetic trees to better define the genetic relationships between the postnatal and adult 19+45Rlo cells (fig. 3a). As expected, the samples clustered into different V region clonotypes, with more branches and duplications in the adult samples, and there was a more restricted clonotype diversity for switched postnatal samples, as described above. Phylogenetic trees generated for postnatal and adult samples of the V1 group (fig. 3b) revealed that the IgM repertoires expressed by them were closely related, indicating a progression in the repertoires and the generation of new branches in adult samples. Switched repertoires exhibited some clusters that were expressed in either postnatal (groups IGHV1-82 and IGHV1S81) or adult (IGHV1-53 and IGHV1-64) samples, suggesting that the primary low-affinity switched repertoire [27] is progressively replaced by other more operative Abs. Interestingly, individualization of IgG1 and IgA sequences revealed that some of them belonged to the same clusters as IgM-bearing sequences and, therefore, they could have originated through a sequential switch process (online suppl. fig. 2).
Fig. 3.
Phylogenetic trees of the sequences from 19+45Rlo cells. Sequences were analysed using the BioEdit and MEGA5 software. a The phylogenetic relationship is shown for the IgM (upper trees) and switched (bottom trees) sequences from PD15 and PD60 19+45Rlo cells. b Overlaid analysis of the V1 family results for the IgM and switched sequences from PD15 and PD60 19+45Rlo cells. Black squares indicate IgM, red triangles indicate IgG1 and blue circles show IgA sequences (PD15 and PD60 samples are shown as empty and filled symbols, respectively). Bars represent the nucleotide substitutions per site.
Between 10 and 20% of the sequences in all the groups contained no identifiable D region gene segments, indicating that exonucleolytic nibbling occurred to a similar extent in all groups [28, 29]. The most frequently used D regions were those of the D1 and D2 groups (fig. 2b). As expected, comparing switched sequences from PD15 and PD60 19+45Rlo cells revealed that D1 (mostly D1-1, DFL16.1) predominated at PD15, whereas a broadening of the other D region groups was evident in PD60 samples, as observed in the other sequence groups analysed (data not shown). Interestingly, 25% of PD15 switched sequences from 19+45Rlo cells contained a DFL16.1-DST4 rearrangement, in which the second D region is inverted (online suppl. fig. 3). These sequences used J2 and the IGHV1-82·01 V region, and they correspond to one of the postnatal switched 19+45Rlo cell clonotypes. In fact, the preferential use of J2 in PD15 switched sequences from 19+45Rlo cells (fig. 2c) mostly corresponds to these sequences. Apart from that, no differences in J region usage were observed in any of the other groups analysed. Taken together, our data show that 19+45Rlo cells display a broad profile of V, D and J region genes and that, interestingly, postnatal switched sequences displayed lower clonotypic diversity and a predominant use of J2.
CDR-H3 Sequences in Postnatal 19+45Rlo Cells Are Shorter and Contain Fewer N Insertions
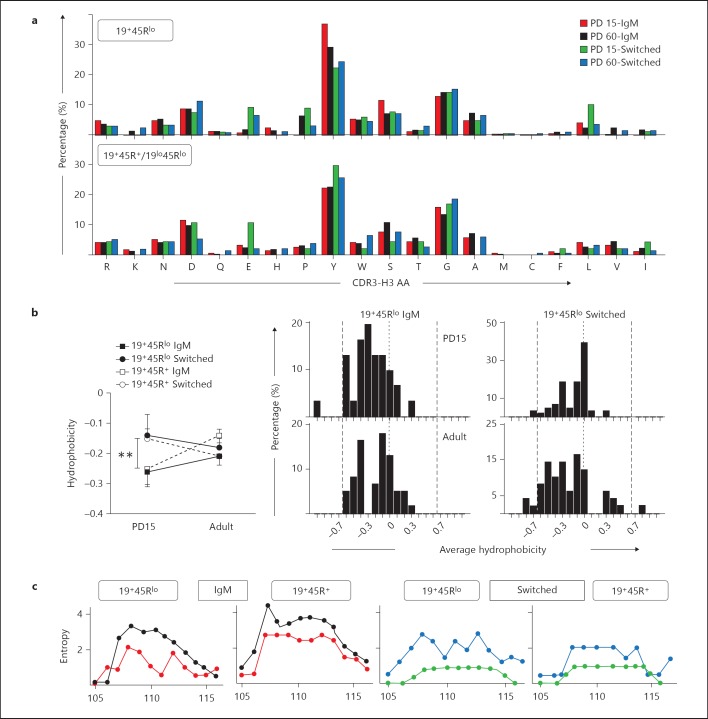
In adult mice, the average length of the CDR-H3 segment increases during development, from that found in immature pro-B progenitors to that of the mature Hardy fraction F in the bone marrow [20]. The distribution of CDR-H3 length has been used as a measure of the diversity of the Ig repertoire. Indeed, the presence of a long CDR-H3 segment (above 45 nucleotides), more aromatic residues and an elevated isoelectric point have been reported in auto-Abs and in pathogenic autoimmune disorders [30, 31]. Innate-like B cell subsets (B1 and MZ cells) have been associated with the generation of natural polyreactive Abs in response to internal auto-Ag recognition and early encounters with common bacterial Ags [32, 33, 34]. To determine whether the sequences obtained from the innate-like 19+45Rlo cells might also be related to the circulating pool of auto-Abs, we analysed the length and amino acid composition of the CDR-H3 domain.
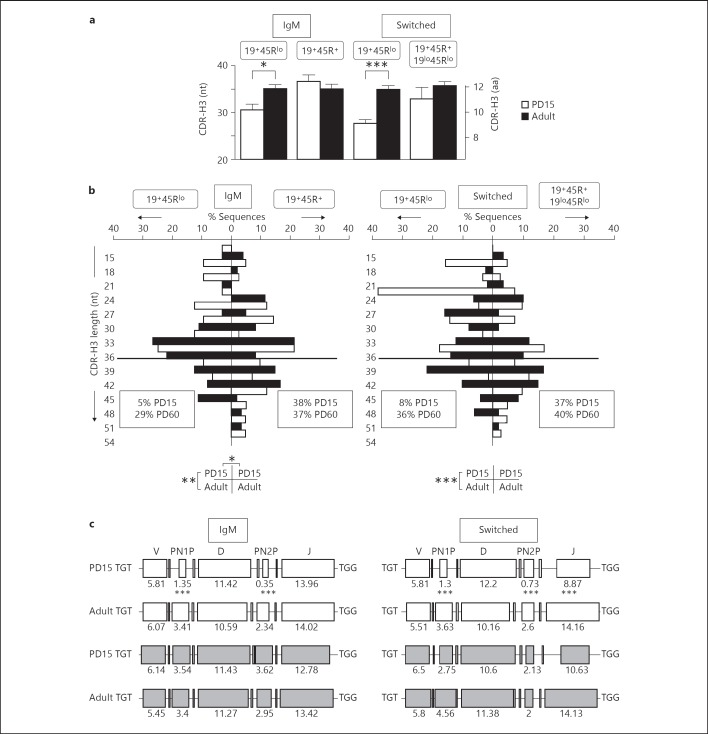
The mean CDR-H3 length in 19+45Rlo cells was below 35 nucleotides in all the groups analysed (fig. 4a). Moreover, the CDR-H3 length in both the IgM and switched sequences from PD15 19+45Rlo cells was 1 and 2 amino acids shorter, respectively, than in all the other sequences analysed. A normal distribution of CDR-H3 lengths was observed in IgM and switched sequences from 19+45R+/19lo45Rlo cells, and in those from adult 19+45Rlo cells (fig. 4b). By contrast, the sequence distributions from PD15 19+45Rlo cells were skewed towards shorter lengths than in all the other groups analysed (only 5% of the IgM and 8% of the switched sequences were longer than 36 nucleotides; fig. 4b). This was mainly due to the shorter N nucleotide insertions at both the V-D and D-J junctions, as depicted in the deconstructed CDR-H3 schemes shown for D region-containing sequences (fig. 4c; online suppl. fig. 4). However, the shorter J regions observed in PD15 switched sequences also influence the short CDR-H3 length in these switched 19+45Rlo cells. The D region could also contribute to the differences in length between sequences, given that the DFL16 D1-1 gene is larger than other genes and it is used differentially among the groups analysed. Surprisingly, no significant differences in the CDR-H3 length were found in sequences containing either DFL16.1 or other D region families (online suppl. table I), as the variation in D region length was compensated for by N nucleotide addition or V or J region deletions (online suppl. table II). Finally, and as expected, the few sequences that did not contain an identifiable D region were shorter (online suppl. table I).
Fig. 4.
Analysis of the CDR-H3 region of IgM and switched sequences. nt = Nucleotides; aa = amino acids. a Average length (±SEM) of the CDR-H3 segment. Left scale is in nucleotides and right scale in amino acids from PD15 (white bars) and PD60 (black bars) sequences. Significance was calculated using an unpaired two-tailed Student t test. * p < 0.05, *** p < 0.001. b Length distribution (in nucleotides) of the unique and functional sequences analysed. The results are displayed as in figure 2 and the statistical analyses were performed using the χ2 test for contingency tables as indicated below the histograms. * p < 0.05, ** p < 0.01, *** p < 0.001. The frequencies of sequences larger than 36-nucleotide sequences are boxed (this limit is indicated by the thick horizontal line). c Deconstruction of the components of the CDR-H3 segment in sequences containing identifiable D regions. Sequences from 19+45Rlo cells are shown in white and those from 19+45R+/19lo45Rlo cells in grey. The numbers indicate the average length of each segment in nucleotides. Comparisons among the values were performed using an unpaired two-tailed Student t test. *** p < 0.001.
Functional Sequences from Postnatal Switched 19+45Rlo Cells Exhibit an Unusual Use of Reading Frame 3
In agreement with the literature [20], there was a preponderance of reading frame 1 (RF1; table 2) in the sequences. Nevertheless, although we performed these analyses using only functional and unique sequences, there was a striking frequency of RF3 use in switched PD15 samples (≥40%), largely due to the aforementioned D-D region rearrangement in the sequences of 19+45Rlo cells (online suppl. fig. 3). Switched sequences from 19+45R+ cells also use RF3, although the low number of sequences obtained at PD15 precludes performing statistical analysis, as mentioned above. RF3 is used less frequently in productive IgH transcripts as it is associated with stop codons [28]. Accordingly, a large number of the non-functional sequences initially included in the study (from both 19+45Rlo and 19lo45Rlo cells) used RF3 (table 2).
Table 2.
RF usage of the sequences bearing an identifiable D region
| Population | Productive sequences |
Non-productive sequences |
||||||
|---|---|---|---|---|---|---|---|---|
| total numbera | RF1 | RF2 | RF3 | total number | RF1 | RF2 | RF3 | |
| PD15 | ||||||||
| IgM 19+45Rlo | 26 | 22 (84.6) | 1 (3.8) | 3 (11.5) | 3 | 0 | 0 | 3 (100) |
| IgM 19+45R+ | 37 | 28 (75.7) | 5 (10.8) | 4 (16.7) | 1 | 0 | 0 | 1 (100) |
| Switched 19+45Rlo | 54 | 27 (50) | 4 (7.4) | 23 (42.6)b | 15 | 3 (20) | 3 (20) | 9 (60) |
| Switched 19+45R+ | 8 | 4 (50) | 1 (12.5) | 3 (37.5) | 1 | 1 (100) | 0 | 0 |
| PD60 | ||||||||
| IgM 19+45Rlo | 55 | 42 (76.4) | 9 (16.4) | 4 (7.3) | 3 | 0 | 0 | 3 (100) |
| IgM 19+45R+ | 55 | 33 (60) | 18 (33) | 4 (7.3) | 1 | 0 | 0 | 1 (100) |
| Switched 19+45Rlo | 42 | 32 (76.2) | 5 (11.9) | 5 (11.9) | 15 | 1 (7) | 0 | 14 (93) |
| Switched 19lo45Rlo | 18 | 13 (72.2) | 2 (11.1) | 3 (16.7) | 13 | 2 (15) | 1 (8) | 10 (77) |
Amino Acid Variability and Mild Hydrophobicity in the CDR-H3 Loop Region of 19+45Rlo Cells
We analysed the amino acid composition and hydrophobicity of the CDR-H3 loop. Tyrosine (ranging from 22 to 37%) and glycine (from 12 to 18%) were highly frequent amino acids (fig. 5a), which is in agreement with published results from different B cell populations [20, 35, 36]. In our study (fig. 5a), the overall amino acid comparison of IgM sequences differed significantly between PD15 19+45Rlo cells and PD15 19+45R+ cells and between PD15 19+45Rlo cells and PD60 19+45Rlo cells (p = 0.0108 and p = 0.0496, respectively, χ2 test, 19 d.f.). This was mainly due to the significantly higher proportion of tyrosine residues used by PD15 19+45Rlo sequences (61 tyrosine residues of 165 amino acid residues = 36.97%) than by sequences from PD15 19+45R+ cells (68/304, 22.36%, p < 0.001, χ2 test). However, this change did not reach significance for PD60 19+45Rlo cells (124/426, 29.11%). On the other hand, switched sequences from PD15 19+45Rlo cells and PD60 19+45Rlo cells also had a different overall amino acid composition (p = 0.0264, χ2 test, 19 d.f.), in this case primarily due to a higher use of leucine in PD15 19+45Rlo cells (24/269 = 8.92%) than PD60 19+45Rlo cells (12/335 = 3.58%, p < 0.01, χ2 test). Other changes in individual amino acid usage did not achieve significance.
Fig. 5.
Amino acid frequencies, hydrophobicity and variability profiles of the CDR-H3 region. The CDR-H3 loop was defined by amino acids 107-114, according to the IMGT unique numbering system. a Overall amino acid frequency distributions in the CDR-H3 loop of the indicated samples. Amino acids are arranged by polarity from arginine (left) to isoleucine (right). PD15 IgM sequences (red), PD60 (black), PD15 switched sequences (green) and PD60 switched (blue). b Left: average hydrophobicity of the CDR-H3 loop from the groups shown in the figure was calculated using the normalized Kyte-Doolittle index [37]. Right: histograms show the hydrophobicity distribution profile of the CDR-H3 loop of the indicated 19+45Rlo sequences. The dashed vertical lines mark the preferred range in average hydrophobicity described for Hardy's fraction F [20]. The dotted lines indicate the zero value. p values were calculated using the unpaired two-tailed Student t test. ** p < 0.01. c CDR-H3 region variability represented by the Shannon entropy [21] was calculated for each position using the 12 amino acid long CDR-H3 sequences (positions 105-117). Overlaid representations of the PD15 and PD60 results are shown for the groups indicated. The colour code is as in a.
We used a normalized Kyte-Doolittle scale [37] to calculate the average hydrophobicity of the CDR-H3 loops (fig. 5b). In all groups, the average hydrophobicity of the sequences was close to neutral, in contrast to the more hydrophilic pattern described for self-reactive repertoires [38]. However, IgM sequences in PD15 19+45Rlo cells were more hydrophilic than the switched sequences. To measure the amino acid variability we calculated the Shannon entropy of the CDR-H3 loop sequences from positions 105 to 117 (according to the IMGT unique numbering; fig. 5c). There was less CDR-H3 variability in the Shannon entropy pattern in the IgM sequences of PD15 19+45Rlo than in those of 19+45R+ cells, while comparable values were obtained for both these groups at PD60. Interestingly, the variability in the Shannon entropy pattern of the CDR-H3 in switched sequences from PD15 19+45Rlo and 19lo45Rlo cells was very low compared with the corresponding values obtained at PD60.
In summary, our data show that IgM and switched sequences from postnatal 19+45Rlo cells frequently use tyrosine and leucine, respectively. Likewise, although all the sequences have low overall hydrophobicity, those from IgM are more hydrophilic. Moreover, these cells exhibit weak variability in CDR-H3 loops at PD15. Together with the short CDR-H3 length, these observations define a pattern that differs from that associated with autoimmune and other known pathological disorders. In addition, the amino acid variability of adult switched sequences from 19+45Rlo cells suggests an increasing diversity of the Ig repertoire of this innate-like cell population.
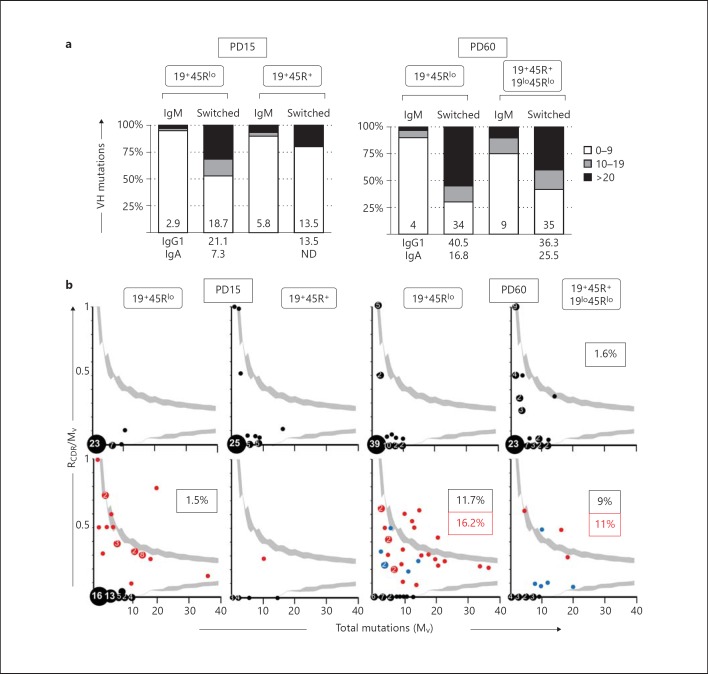
Increased Somatic Mutations in the V region of Sequences from Adult 19+45Rlo Cells
The SHM process is driven by both AID and CSR. As we found that 19+45Rlo cells actively produce IgG and IgA, and that AID is expressed by these cells, we analysed the mutations in their V regions. For the ease of interpretation, we classified the sequences as low-mutated (0-9 mutations in the V region), intermediate-mutated (10-19 mutations) and highly mutated (≥20 mutations; fig. 6a). As expected, the mutation profile of the IgM sequences was skewed towards germline/low-mutated, and these sequences had an overall mutation rate ranging from 2.9 × 10-3 to 9 × 10-3 nucleotides. By contrast, 47% of the switched sequences from PD15 19+45Rlo cells were intermediate/highly mutated, with a mutation rate of 18.7 × 10-3 nucleotides and a maximum mutation rate reached at PD60 (34 × 10-3 nucleotides). Interestingly, the highest mutation rate corresponded to PD15 and PD60 IgG1 sequences.
Fig. 6.
Quantitative analysis of the mutations found in the V and CDR-H3 regions of PD15 and PD60 from 19+45Rlo and 19+45R+/19lo45Rlo sorted cells, as indicated in table 1. a Vertical bars represent the frequency of the mutated sequences in each group, classified as low-mutated (0-9 mutations in the V region), intermediate-mutated (10-19 mutations) and highly mutated (≥20 mutations). The numbers inside the bars indicate the corresponding somatic mutation rates, determined as the number of mutations × 10-3 nucleotides. The numbers below show the mutation rates of IgG1 and IgA samples. ND = Not determined. b Inference of Ag selection in sequences from 19+45Rlo, 19+45R+ and 19lo45Rlo at PD15 and PD60. The ratio of replacement mutations on CDR-H1 and CDR-H2 (RCDR), relative to the total number of mutations in the V region (Mv), is shown plotted against Mv. The 90–95% confidence interval is shown in light grey. Data points are accompanied by their observed frequencies. Upper plots (black dots) represent results for IgM sequences and lower plots represent results for switched sequences (red dots, IgG1 sequences; blue dots, IgA sequences). Bottom far right plot shows the results obtained from adult switched 19lo45Rlo cells. The dots above the upper confidence limits represent the sequences with a high proportion of R mutations in the CDR, and the probability that this is a random process is <0.05. Boxed numbers represent the frequency of Ag selected sequences (black, IgM or switched sequences, respectively; red, IgG1 sequences).
The number of replacement mutations in CDRs (RCDR) relative to the total number of mutations in the V region (Mv) can be used to identify sequences undergoing antigenic selection [19]. We plotted the RCDR/Mv ratio against the Mv and calculated the 95% confidence interval of the distributions, as described previously [39, 40]. Sequences lying outside these limits (see light grey shading in fig. 6b) were considered to be undergoing antigenic selection, while those in the origin of the axes are non-mutated germline sequences. For 19+45Rlo cells at PD15 and PD60, this analysis revealed that 71.9 and 60.9% of the IgM sequences were germline and non-mutated, whereas 23.8 and 11.7% of the switched sequences were non-mutated (p < 0.05 and p < 0.001 for PD15 and PD60 sequences, respectively), confirming the results obtained for the complete V region. Moreover, most of PD15 sequences were located within the confidence limits of the plots, as would be expected if antigenic selection did not occur and as observed for adult IgM sequences. By contrast, 11.7 and 9.1% of the switched sequences from PD60 19+45Rlo and 19lo45Rlo cells, respectively, exceeded the 95% limit, indicating that sequences from adult switched cells were Ag selected. In fact, this was preferentially due to IgG1 mutated sequences, 16.2 and 11% of which were situated above the 95% limit. Based on these findings, we conclude that the adult IgG1 repertoire of 19+45Rlo cells from normal unmanipulated mice is established by positive selection.
Discussion
In this study, we sought to characterize the immunoglobulin repertoire expressed by novel innate-like 19+45Rlo cells at postnatal and adult stages under homeostatic conditions. Innate-like lymphoid cell compartments differ from conventional lymphoid cell populations in terms of their generation, maintenance, activating pathways and immunoregulatory functions [12, 13, 41]. We previously described the 19+45Rlo cell population as a pre-plasmablast compartment that develops in perifollicular areas within the primary lymphoid follicles from the first week of age, and that has a pronounced capacity to secrete IgG1 and IgA [3]. Moreover, we recently demonstrated the ability of these cells to respond to TLR ligands and to secrete IL-10 but not IL-6 or IL-12 properties that may confer specific functions to this population [2]. Because other innate-like B cell compartments share some of the features exhibited by 19+45Rlo cells and as they are present only minimally in CBA/CaHN (Btk) mice [3], we speculated that genetic restrictions may potentially restrict V region usage and germline rearrangements in 19+45Rlo cells, as described for the B1 compartment [42]. Alternatively, these cells may receive different antigenic signals that induce a more open Ig repertoire in unmanipulated mice.
In PD15 mice, cellular components are still immature and the responses of germinal centres have not yet fully developed [43]. Nevertheless, ELISPOT assays of sorted purified 19+45Rlo and 19+45R+ cells have demonstrated that it is mainly 19+45Rlo cells that can release IgG1 at PD15, implying that the first IgGs are secreted by 19+45Rlo cells [3]. In the present study, we found that the clonotypic diversity of switched postnatal sequences was lower than that of adult sequences, suggesting an immature and limited Ig repertoire in switched postnatal sequences. The Ig repertoire of 19+45Rlo cells at PD15 is characterized by a short CDR-H3 segment and few N nucleotides, suggestive of an immature repertoire as described previously in neonates [27, 44]. Moreover, the differential usage of V/D/J regions in switched sequences from 19+45Rlo cells (V3, D1, tandem D-D and J2) may reflect the initial low affinity IgG repertoire that is enriched in the adult through the generation of a variety of new specificities.
The transition to adult life leads to major changes in the Ig repertoire of 19+45Rlo cells. For example, we found that levels of AID and Blimp-1 were significantly higher in adult as opposed to postnatal 19+45R+ cells, favouring CSR events. Likewise, we observed differences in V, D and J region usage in adult and postnatal sequences (V4 in IgM and V1 in switched, few D1 and random J region ratios in both IgM and switched sequences). As indicated above, the population of splenic 19+45Rlo cells is heterogeneous, and it may include pre-lambda 5 progenitors and B1 cell progenitors that have been described in bone marrow and spleen as 19+45Rlo [23, 24, 45]. However, 96% of the 19+45Rlo cells are mature B cells since they express surface light chains. Splenic 19+45Rlo cells are close to the B1 lineage since they share some features: their embryonic origin, their absence from the spleens of adult CBA/CaHN mice, constitutive IL-10 expression and spontaneous ERK phosphorylation. Nevertheless, 19+45Rlo cells differ from B1 cells not only in the spontaneous release of isotype-switched Igs but also, because they exhibit no CD5 or CD11b expression in the adult spleen, they are found in Peyer's patches, in the circulation, and also in the spleen of young CBA/CaHN mice, and they present a distinctive transcriptional fingerprint [2]. Furthermore, unlike 19+45Rlo cells, the CDR-H3 features described for early B cell progenitors and B1 cells involve a prevalence for VH7183 and DFL16.1 together with a shorter CDR-H3 (on B cell progenitors) and high DQ52, RF2 usage and a hydrophobicity profile (on B1 cells) [20, 35]. Therefore, although we cannot exclude that the samples analysed in this paper contain different cell subsets, sequences derived from B1 cells and B cell progenitors do not seem to contribute fundamentally to our results. Finally, RF1 was the preferred RF in all the groups analysed, followed by RF2, which is frequently used in the MZ and the peritoneal cavity [46], and lastly RF3. RF3 contains stop codons [28], although it is used heavily in switched PD15 cells but not in adult switched sequences. Interestingly, many of these switched PD15 sequences from 19+45Rlo cells bear a specific V/D/D/J2 rearrangement that may contribute to the function of these sequences. It has also been proposed that in fetal/neonatal samples, an Ag-independent mechanism can induce a random pattern of RF usage in the D region [29], which may explain the RF3 frequencies observed in switched functional PD15 samples.
The presence of circulating polyreactive auto-Abs has been attributed to encounters with common bacterial Ags or to internal auto-Ag recognition [34, 47]. The B1 lineage and the MZ compartments have been associated with the generation of these natural auto-Abs [32, 33], which have a large hydrophilic CDR-H3 loop [48]. By contrast, a detailed analysis of the CDR-H3 loop from 19+45Rlo cells defined a non-autoimmune pattern in IgM and switched Igs in terms of CDR-H3 length, amino acid content and hydrophobicity (close to neutral). Restrictions on Ig rearrangements and low levels of SHM have classically been described in the B1 compartment [42] but not in MZ [49] or 19+45Rlo cells (data herein). Nonetheless, the CSR ability of B1 and MZ cells was limited compared to that of 19+45Rlo cells [48]. The CDR-H3 analysis of another recently described innate-like B cell population associated with autoimmune events [13] is still unknown, although they differ from 19+45Rlo cells in terms of their cytokine and Ig isotype secretion.
PD15 switched sequences exhibited more V region mutations than the corresponding IgM sequences, suggesting postnatal selection. The same was found for RCDR mutations in 19+45Rlo cells at PD15, which probably constitutes the initial baseline mutation rate. IgG1 sequences from PD60 19+45Rlo cells contained a remarkable number of sequences with high mutation rates in CDRs, which may indicate Ag-driven selection. Although this ratio was lower than that obtained for IgE in allergic children (29%) using the same analytical IgAT software [50], it still indicates that a significant degree of Ag selection takes place in 19+45Rlo cells under homeostatic conditions. The increased mutational rate of 19+45Rlo cells and their ability to secrete IgG1 and IgA are also characteristics of the memory cell compartment. However, their pre-activated status and high turnover rate under homeostatic conditions [3] distinguish these cells from classical memory cells [51].
Several conclusions can be drawn from the data presented here. Firstly, it appears that 19+45Rlo cells exhibit no genetic restrictions at the level of IgH rearrangements between PD15 and adulthood, in contrast to what occurs in other innate-like B cell compartments. In addition, SHM appears to be driven by AID expression in 19+45Rlo cells, as witnessed by the mutational rate of switched sequences, indicating that Ag selection occurs actively in 19+45Rlo cells. We hypothesize that the driving force behind mutations in innate-like 19+45Rlo cells is the early and continuous contact with common microbial Ags, TLR ligands, or a combination of both. Indeed, 19+45Rlo cells can respond to TLRs by inducing IgG1, IgA and IL-10 secretion [2], suggesting that this population is involved in conventional bacterial and viral recognition through the induction of a rapid and efficient isotype-switched Ab response. In summary, here we describe unique features displayed by innate-like 19+45Rlo cells, and we propose that these cells represent a new ‘first line’ working B cell population that specializes in the secretion of IgG and IgA under the control of immunoregulatory cytokines like IL-10. Moreover, we propose that the maintenance of homeostasis involves the active participation of different innate-like cell populations (e.g. 19+45Rlo cells) that can respond to tonic antigenic encounters by activating the genetic machinery controlling SHM and CSR expression and by generating a broad spectrum of Abs.
Supplementary Material
Supplementary data
Acknowledgements
We thank B. Palacios for assisting with the cloning and sequencing, A.G. de la Campa, A. Trento, I. Cuesta and L. Garcia-Albert for their help with the bioinformatics software analyses, and M.A.R. Marcos for his contribution to the original planning of the experiments. This work was supported by grants from the Fondo de Investigaciones Sanitarias (FIS, MPY 1450/11) and from the Ministerio de Ciencia e Innovación (MICINN, SAF2009-12596) and Ministerio de Economia y competitividad (MINECO, SAF2012-33916). I. Cortegano received a postdoctoral fellowship from the MICINN.
References
- 1.de Andres B, Gonzalo P, Minguet S, Martinez-Marin JA, Soro PG, Marcos MA, Gaspar ML. The first 3 days of B-cell development in the mouse embryo. Blood. 2002;100:4074–4081. doi: 10.1182/blood-2002-03-0809. [DOI] [PubMed] [Google Scholar]
- 2.de Andres B, Prado C, Palacios B, Alia M, Jagtap S, Serrano N, Cortegano I, Marcos MA, Gaspar ML. Dynamics of the splenic innate-like CD19+CD45Rlo cell population from adult mice in homeostatic and activated conditions. J Immunol. 2012;189:2300–2308. doi: 10.4049/jimmunol.1200224. [DOI] [PubMed] [Google Scholar]
- 3.de Andres B, Cortegano I, Serrano N, del Rio B, Martin P, Gonzalo P, Marcos MA, Gaspar ML. A population of CD19highCD45R-/lowCD21low B lymphocytes poised for spontaneous secretion of IgG and IgA antibodies. J Immunol. 2007;179:5326–5334. doi: 10.4049/jimmunol.179.8.5326. [DOI] [PubMed] [Google Scholar]
- 4.Honjo T, Nakai S, Nishida Y, Kataoka T, Yamawaki-Kataoka Y, Takahashi N, Obata M, Shimizu A, Yaoita Y, Nikaido T, Ishida N. Rearrangements of immunoglobulin genes during differentiation and evolution. Immunol Rev. 1981;59:33–67. doi: 10.1111/j.1600-065x.1981.tb00455.x. [DOI] [PubMed] [Google Scholar]
- 5.Neuberger MS, Milstein C. Somatic hypermutation. Curr Opin Immunol. 1995;7:248–254. doi: 10.1016/0952-7915(95)80010-7. [DOI] [PubMed] [Google Scholar]
- 6.Reynaud CA, Garcia C, Hein WR, Weill JC. Hypermutation generating the sheep immunoglobulin repertoire is an antigen-independent process. Cell. 1995;80:115–125. doi: 10.1016/0092-8674(95)90456-5. [DOI] [PubMed] [Google Scholar]
- 7.Kelsoe G. The germinal center reaction. Immunol Today. 1995;16:324–326. doi: 10.1016/0167-5699(95)80146-4. [DOI] [PubMed] [Google Scholar]
- 8.Muramatsu M, Sankaranand VS, Anant S, Sugai M, Kinoshita K, Davidson NO, Honjo T. Specific expression of activation-induced cytidine deaminase (AID), a novel member of the RNA-editing deaminase family in germinal center B cells. J Biol Chem. 1999;274:18470–18476. doi: 10.1074/jbc.274.26.18470. [DOI] [PubMed] [Google Scholar]
- 9.Toellner KM, Jenkinson WE, Taylor DR, Khan M, Sze DM, Sansom DM, Vinuesa CG, MacLennan IC. Low-level hypermutation in T cell-independent germinal centers compared with high mutation rates associated with T cell-dependent germinal centers. J Exp Med. 2002;195:383–389. doi: 10.1084/jem.20011112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.William J, Euler C, Christensen S, Shlomchik MJ. Evolution of autoantibody responses via somatic hypermutation outside of germinal centers. Science. 2002;297:2066–2070. doi: 10.1126/science.1073924. [DOI] [PubMed] [Google Scholar]
- 11.Aranburu A, Ceccarelli S, Giorda E, Lasorella R, Ballatore G, Carsetti R. TLR ligation triggers somatic hypermutation in transitional B cells inducing the generation of IGM memory B cells. J Immunol. 2010;185:7293–7301. doi: 10.4049/jimmunol.1002722. [DOI] [PubMed] [Google Scholar]
- 12.Hao Y, O'Neill P, Naradikian MS, Scholz JL, Cancro MP. A B-cell subset uniquely responsive to innate stimuli accumulates in aged mice. Blood. 2011;118:1294–1304. doi: 10.1182/blood-2011-01-330530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rauch PJ, Chudnovskiy A, Robbins CS, Weber GF, Etzrodt M, Hilgendorf I, Tiglao E, Figueiredo JL, Iwamoto Y, Theurl I, Gorbatov R, Waring MT, Chicoine AT, Mouded M, Pittet MJ, Nahrendorf M, Weissleder R, Swirski FK. Innate response activator B cells protect against microbial sepsis. Science. 2012;335:597–601. doi: 10.1126/science.1215173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gozalbo-Lopez B, Andrade P, Terrados G, de Andres B, Serrano N, Cortegano I, Palacios B, Bernad A, Blanco L, Marcos MA, Gaspar ML. A role for DNA polymerase μ in the emerging DJH rearrangements of the postgastrulation mouse embryo. Mol Cell Biol. 2009;29:1266–1275. doi: 10.1128/MCB.01518-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li YS, Hayakawa K, Hardy RR. The regulated expression of B lineage associated genes during B cell differentiation in bone marrow and fetal liver. J Exp Med. 1993;178:951–960. doi: 10.1084/jem.178.3.951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Orlandi R, Gussow DH, Jones PT, Winter G. Cloning immunoglobulin variable domains for expression by the polymerase chain reaction. Proc Natl Acad Sci USA. 1989;86:3833–3837. doi: 10.1073/pnas.86.10.3833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martinez MJ, Minguet S, Gonzalo P, Soro PG, de Andres B, Izcue A, Marcos MA, Gaspar ML. Long-lived polyclonal B-cell lines derived from midgestation mouse embryo lymphohematopoietic progenitors reconstitute adult immunodeficient mice. Blood. 2001;98:1862–1871. doi: 10.1182/blood.v98.6.1862. [DOI] [PubMed] [Google Scholar]
- 18.Alamyar E, Giudicelli V, Li S, Duroux P, Lefranc MP. Imgt/highv-quest: The IMGT(R) web portal for Immunoglobulin (Ig) or antibody and T cell receptor (TR) analysis from NGS high throughput and deep sequencing. Immunome Res. 2012;8:26. [Google Scholar]
- 19.Rogosch T, Kerzel S, Hoi KH, Zhang Z, Maier RF, Ippolito GC, Zemlin M. Immunoglobulin analysis tool: a novel tool for the analysis of human and mouse heavy and light chain transcripts. Front Immunol. 2012;3:176. doi: 10.3389/fimmu.2012.00176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ivanov II, Schelonka RL, Zhuang Y, Gartland GL, Zemlin M, Schroeder HW., Jr Development of the expressed Ig CDR-H3 repertoire is marked by focusing of constraints in length, amino acid use, and charge that are first established in early B cell progenitors. J Immunol. 2005;174:7773–7780. doi: 10.4049/jimmunol.174.12.7773. [DOI] [PubMed] [Google Scholar]
- 21.Shannon CE. The mathematical theory of communication. 1963. MD Comput. 1997;14:306–317. [PubMed] [Google Scholar]
- 22.Rolink A, Haasner D, Nishikawa S, Melchers F. Changes in frequencies of clonable pre-B cells during life in different lymphoid organs of mice. Blood. 1993;81:2290–2300. [PubMed] [Google Scholar]
- 23.Montecino-Rodriguez E, Leathers H, Dorshkind K. Identification of a B-1 B cell-specified progenitor. Nat Immunol. 2006;7:293–301. doi: 10.1038/ni1301. [DOI] [PubMed] [Google Scholar]
- 24.Ghosn EE, Sadate-Ngatchou P, Yang Y, Herzenberg LA. Distinct progenitors for B-1 and B-2 cells are present in adult mouse spleen. Proc Natl Acad Sci USA. 2011;108:2879–2884. doi: 10.1073/pnas.1019764108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bjorck P, Kincade PW. CD19+ pro-B cells can give rise to dendritic cells in vitro. J Immunol. 1998;161:5795–5799. [PubMed] [Google Scholar]
- 26.Yancopoulos GD, Desiderio SV, Paskind M, Kearney JF, Baltimore D, Alt FW. Preferential utilization of the most JH-proximal VH gene segments in pre-B-cell lines. Nature. 1984;311:727–733. doi: 10.1038/311727a0. [DOI] [PubMed] [Google Scholar]
- 27.Schallert N, Pihlgren M, Kovarik J, Roduit C, Tougne C, Bozzotti P, Del Giudice G, Siegrist CA, Lambert PH. Generation of adult-like antibody avidity profiles after early-life immunization with protein vaccines. Eur J Immunol. 2002;32:752–760. doi: 10.1002/1521-4141(200203)32:3<752::AID-IMMU752>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 28.Gu H, Kitamura D, Rajewsky K. B cell development regulated by gene rearrangement: arrest of maturation by membrane-bound D mu protein and selection of DH element reading frames. Cell. 1991;65:47–54. doi: 10.1016/0092-8674(91)90406-o. [DOI] [PubMed] [Google Scholar]
- 29.Marshall AJ, Doyen N, Bentolila LA, Paige CJ, Wu GE. Terminal deoxynucleotidyl transferase expression during neonatal life alters DH reading frame usage and Ig-receptor-dependent selection of V regions. J Immunol. 1998;161:6657–6663. [PubMed] [Google Scholar]
- 30.Shiokawa S, Mortari F, Lima JO, Nunez C, Bertrand FE, 3rd, Kirkham PM, Zhu S, Dasanayake AP, Schroeder HW., Jr IgM heavy chain complementarity-determining region 3 diversity is constrained by genetic and somatic mechanisms until two months after birth. J Immunol. 1999;162:6060–6070. [PubMed] [Google Scholar]
- 31.Wakui M, Kim J, Butfiloski EJ, Morel L, Sobel ES. Genetic dissection of lupus pathogenesis: Sle3/5 impacts IgH CDR3 sequences, somatic mutations, and receptor editing. J Immunol. 2004;173:7368–7376. doi: 10.4049/jimmunol.173.12.7368. [DOI] [PubMed] [Google Scholar]
- 32.Hayakawa K, Hardy RR, Parks DR, Herzenberg LA. The ‘Ly-1 B’ cell subpopulation in normal immunodefective, and autoimmune mice. J Exp Med. 1983;157:202–218. doi: 10.1084/jem.157.1.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martin F, Kearney JF. Positive selection from newly formed to marginal zone B cells depends on the rate of clonal production, CD19, and Btk. Immunity. 2000;12:39–49. doi: 10.1016/s1074-7613(00)80157-0. [DOI] [PubMed] [Google Scholar]
- 34.Dighiero G, Lymberi P, Guilbert B, Ternynck T, Avrameas S. Natural autoantibodies constitute a substantial part of normal circulating immunoglobulins. Ann NY Acad Sci. 1986;475:135–145. doi: 10.1111/j.1749-6632.1986.tb20863.x. [DOI] [PubMed] [Google Scholar]
- 35.Vale AM, Tanner JM, Schelonka RL, Zhuang Y, Zemlin M, Gartland GL, Schroeder HW., Jr The peritoneal cavity B-2 antibody repertoire appears to reflect many of the same selective pressures that shape the B-1a and B-1b repertoires. J Immunol. 2010;185:6085–6095. doi: 10.4049/jimmunol.1001423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zemlin M, Klinger M, Link J, Zemlin C, Bauer K, Engler JA, Schroeder HW, Jr, Kirkham PM. Expressed murine and human CDR-H3 intervals of equal length exhibit distinct repertoires that differ in their amino acid composition and predicted range of structures. J Mol Biol. 2003;334:733–749. doi: 10.1016/j.jmb.2003.10.007. [DOI] [PubMed] [Google Scholar]
- 37.Eisenberg D. Three-dimensional structure of membrane and surface proteins. Annu Rev Biochem. 1984;53:595–623. doi: 10.1146/annurev.bi.53.070184.003115. [DOI] [PubMed] [Google Scholar]
- 38.Wardemann H, Yurasov S, Schaefer A, Young JW, Meffre E, Nussenzweig MC. Predominant autoantibody production by early human B cell precursors. Science. 2003;301:1374–1377. doi: 10.1126/science.1086907. [DOI] [PubMed] [Google Scholar]
- 39.Chang B, Casali P. The CDR1 sequences of a major proportion of human germline Ig VH genes are inherently susceptible to amino acid replacement. Immunol Today. 1994;15:367–373. doi: 10.1016/0167-5699(94)90175-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dahlke I, Nott DJ, Ruhno J, Sewell WA, Collins AM. Antigen selection in the IgE response of allergic and nonallergic individuals. J Allergy Clin Immunol. 2006;117:1477–1483. doi: 10.1016/j.jaci.2005.12.1359. [DOI] [PubMed] [Google Scholar]
- 41.Hayday AC, Pennington DJ. Key factors in the organized chaos of early T cell development. Nat Immunol. 2007;8:137–144. doi: 10.1038/ni1436. [DOI] [PubMed] [Google Scholar]
- 42.Carmack CE, Shinton SA, Hayakawa K, Hardy RR. Rearrangement and selection of VH11 in the Ly-1 B cell lineage. J Exp Med. 1990;172:371–374. doi: 10.1084/jem.172.1.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pihlgren M, Tougne C, Bozzotti P, Fulurija A, Duchosal MA, Lambert PH, Siegrist CA. Unresponsiveness to lymphoid-mediated signals at the neonatal follicular dendritic cell precursor level contributes to delayed germinal center induction and limitations of neonatal antibody responses to T-dependent antigens. J Immunol. 2003;170:2824–2832. doi: 10.4049/jimmunol.170.6.2824. [DOI] [PubMed] [Google Scholar]
- 44.Astori M, Finke D, Karapetian O, Acha-Orbea H. Development of T-B cell collaboration in neonatal mice. Int Immunol. 1999;11:445–451. doi: 10.1093/intimm/11.3.445. [DOI] [PubMed] [Google Scholar]
- 45.Esplin BL, Welner RS, Zhang Q, Borghesi LA, Kincade PW. A differentiation pathway for B1 cells in adult bone marrow. Proc Natl Acad Sci USA. 2009;106:5773–5778. doi: 10.1073/pnas.0811632106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schelonka RL, Zemlin M, Kobayashi R, Ippolito GC, Zhuang Y, Gartland GL, Szalai A, Fujihashi K, Rajewsky K, Schroeder HW., Jr Preferential use of DH reading frame 2 alters b cell development and antigen-specific antibody production. J Immunol. 2008;181:8409–8415. doi: 10.4049/jimmunol.181.12.8409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen C, Stenzel-Poore MP, Rittenberg MB. Natural auto- and polyreactive antibodies differing from antigen-induced antibodies in the H chain CDR3. J Immunol. 1991;147:2359–2367. [PubMed] [Google Scholar]
- 48.Schelonka RL, Tanner J, Zhuang Y, Gartland GL, Zemlin M, Schroeder HW., Jr Categorical selection of the antibody repertoire in splenic B cells. Eur J Immunol. 2007;37:1010–1021. doi: 10.1002/eji.200636569. [DOI] [PubMed] [Google Scholar]
- 49.Kretschmer K, Jungebloud A, Stopkowicz J, Kleinke T, Hoffmann R, Weiss S. The selection of marginal zone B cells differs from that of B-1a cells. J Immunol. 2003;171:6495–6501. doi: 10.4049/jimmunol.171.12.6495. [DOI] [PubMed] [Google Scholar]
- 50.Kerzel S, Rogosch T, Struecker B, Maier RF, Zemlin M. IgE transcripts in the circulation of allergic children reflect a classical antigen-driven B cell response and not a superantigen-like activation. J Immunol. 2010;185:2253–2260. doi: 10.4049/jimmunol.0902942. [DOI] [PubMed] [Google Scholar]
- 51.Dogan I, Bertocci B, Vilmont V, Delbos F, Megret J, Storck S, Reynaud CA, Weill JC. Multiple layers of B cell memory with different effector functions. Nat Immunol. 2009;10:1292–1299. doi: 10.1038/ni.1814. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data