Abstract
Innate immunity induces rapid responses to fight invading pathogens. To eliminate intracellular bacteria or viruses, innate cellular responses lead to the production of nuclear factor-κB-dependent inflammatory cytokines, inflammasome activation, type I interferon synthesis, and/or eventually death of the infected cells. Autophagy emerged as another component of innate immunity, as it offers an immediate autonomous cell defense mechanism by degrading intracellular pathogens. In addition, autophagy participates in the regulation of immune and inflammatory cell responses. Instead of providing a comprehensive status of the art that has already been addressed elsewhere, we chose to highlight some recent issues brought up in the field.
Key Words: Virus, Bacteria, Cell death, Inflammation, Autophagy, Innate immunity
Autophagy and Pathogen Degradation
Xenophagy (from Greek xenos ‘of a stranger’ and phagein ‘to eat’) defines the autophagy pathway induced upon membrane wrapping of extracellular pathogenic bacteria. Macroautophagy defines a process related to degrading altered organelles such as mitochondria. Both xenophagy and macroautophagy share the properties to target large materials for degradation upon fusion with lysosomes. Autophagy relies on autophagy-related genes (ATG) at the molecular level. The ‘ATG protein’ core of the autophagic molecular machineries is conserved between xenophagy and macroautophagy. Pathogens have evolved mechanisms to target the regulation of the autophagy pathway. These regulations operate early at the cell surface, which is the initial donor membrane compartment, and also at further membrane trafficking steps down the internalization pathway. Several pathogenic bacteria, such as Mycobacteriaceae, Streptococcus pyogenes and Salmonella Thyphimurium, are conveyed by the autophagy machinery to lysosomal degradation in order to restrict growth (xenophagy), whereas other bacteria can benefit from autophagy to elaborate replication niches [1]. Bacteria can exploit autophagy in many ways. For instance, at the Shigella flexneri entry site, ATG16L1 is recruited by the nucleotide-binding oligomerization domain (NOD) pattern recognition receptors (PRRs) [2]. The ATG16L1-containing membranes that bud off the cell surface undergo homotypic fusion leading to the formation of LC3-negative and ATG12-ATG5-ATG16L1-positive phagophore precursor vesicles [3]. Such involvement of ATG16L1 is also of particular interest in the context of inflammatory diseases linked to luminal bacteria [see review in this issue by Nguyen et al. [4]]. At the cell surface, activation of autophagy depends at least on the role of the ADP ribosylation factor 6 (ARF6) to produce phosphatidylinositol 4,5-bisphosphate [5]. ARF6 also regulates the CpG oligodeoxynucleotide endosomal uptake through its upstream molecule class III phosphatidylinositol-3 kinases. CpG oligodeoxynucleotide uptake is the rate-limiting step of Toll-like receptor 9 (TLR9) immune signaling [6]. After Shigella entry, the vacuolar compartment is disrupted, which represents a huge membrane stress [7], and the membrane remnants undergo autophagy-mediated degradation (fig. 1) [8].
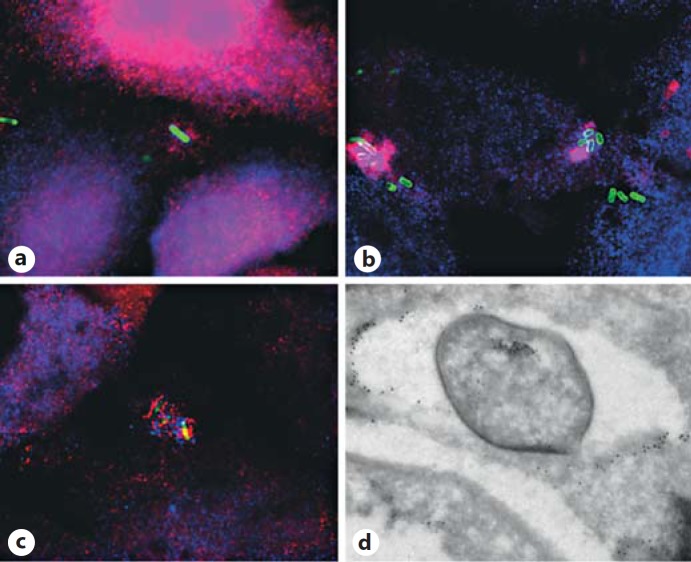
Fig. 1.
S. flexneri membrane remnants recruit inflammasome components and ubiquitin. HeLa cells were infected with S. flexneri M90T-green fluorescent protein (green) for 30 min (a-c) at 37°C and then processed for labeling. Specific primary antibodies were used, by immunostaining, to visualize endogenous NOD1 (red) and galectin-3 (blue; a), transfected Ipaf-Flag (red) and galectin-3 (blue; b), and galectin-3 (blue; b, c) and the FLICA-1 reagent was used to detect caspase-1 (red). d HeLa cells were infected by S. flexneri for 45 min at 37°C before processing for immunoelectron microscopy for detection of polyubiquitin (FK1 antibody) and galectin-3 using 6- and 10-nm gold-conjugated secondary antibodies, respectively [see also ref. [6]].
Pathogen-Induced Autophagy Membrane Trafficking
Deciphering the signaling associated with membrane trafficking of infectious microorganisms in relation to autophagy during infection will help to better understand the tuning of the immune response. In recent years, autophagy membrane trafficking itself has been analyzed in terms of targeting, docking and fusion membrane events. The formation of phagophores (ATG12-ATG5-ATG16L1 positive and LC3 positive) depends on vesicle-associated membrane proteins 7 (VAMP7)-mediated fusion events [9]. VAMP7 is not the only soluble N-ethylmaleimide-sensitive fusion attachment protein receptor (SNARE) involved in the fusion of autophagosomes with lysosomes. The synatxin7:VAMP8 pair and VTI1b (vesicle transport through interaction with t-SNAREs homolog 1B) were also involved in this fusion, whereas VAMP3 and Rab11 operate during the fusion of multivesicular bodies with autophagosomes to generate amphisomes [10, 11, 12]. Interestingly, VAMP7 has been involved in the autophagy-dependent secretion of ATP [13], a process that participates in interleukin (IL)-1β secretion [see below and review in this issue by Jiang et al. [14]], in response to inflammation. This highlights the increased interest in deciphering the fusion machinery involved in the autophagy pathway, as few studies related to SNAREs are available [15]. The small GTPase Rab proteins also play a crucial role in membrane trafficking. Rab5 has been involved in the endosome invasion step, Rab23 in the formation of isolation membranes upon group A streptococcus (GAS) targeting, and Rab9A has been shown to be recruited on GAS-containing autophagosome-like vacuoles for enlargements and eventual lysosome fusion [16, 17, 18]. For Rab7, multifunctions have been proposed, stemming from invasion to autophagosomal formation [16, 17, 18]. Interestingly, some regulators of Rab proteins playing a role in autophagy have started to be unveiled. Using RNA silencing and yeast two-hybrid screens, a comprehensive set of TBC (Tre2, Bub2, Cdc16) containing the domain Rab GTPase-activating proteins which interact with ATG8 modifiers were identified to contribute to autophagy dynamics [19]. TBC1D5 was proposed to bridge endosomes to LC3-positive autophagosomes upon starvation, though GTPase-activating protein activity did not seem to always be mandatory, at least for TBC1D14 that perturbed ULK1 recycling in the endosomal traffic [19, 20]. Due to their role, it is not surprising that Rab proteins constitute targets for bacterial effectors. Thus, the VirA effector of S. flexneri and the EspG effector of enteropathogenic Escherichia coli were demonstrated to harbor TBC Rab GTPase-activating protein activities [21]. Indeed, both effectors inactivate Rab1. Upon S. flexneri infection, VirA disrupts the cis-Golgi structure though Golgi disruption is also observed independently of VirA, suggesting that it is not this activity that is targeted [21]. Rather, Rab1 and functional endoplasmic reticulum exit are involved in autophagosome formation [22], and Rab1 is required for the antibacterial autophagy observed in Salmonella-infected cells [23]. EspG also leads to Golgi fragmentation interfering with the biosynthetic secretory route, in particular upon tumor necrosis factor (TNF)-α stimulation leading to inhibition of IL-8 production (a hallmark of infection by enteric bacteria such as Shigella and enteropathogenic E. coli) [21]. EspG has been shown to interact with the GTP-bound form of (ADP ribosylation) ARF1/ARF6 [24]. This interaction (possibly upon GM130 binding as well) [25] targets EspG for Rab1 inactivation [21]. EspG contains a nonoverlapping surface to bind to Rab1 and ARF6 [21]. In response, the host develops innate immune defense mechanisms which will be discussed below. Always in an evolutionary-driven escalation, pathogens select hijacking (conveying through the same membrane flow) and subversion infection strategies (rerouting of membranes).
An important issue is to identify bacterial effectors that can directly target autophagy components. In this context, it has been shown that Legionella pneumophila Philadelphia 1 can inhibit autophagy through the RavZ effector [26]. RavZ cleaves the amide bond between the tyrosine and the phycoerythrin-conjugated glycine residue of ATG8, rendering it resistant to conjugation and preventing its association in the membrane of the phagophore [26].
Virophagy
Although autophagy may be induced immediately upon viral infections [27], to date, no viral particles were observed within autophagosomes immediately after virus entry. This is certainly due to the rapid disassembly of infectious viral particles following entry in order to deliver viral genomes to the host and to start replication. Moreover, although autophagy often persists within cells infected by a virus, few reports have described the entrapment of complete viral particles within the autophagy machinery [28, 29, 30]. Nevertheless, autophagy could control viral infection through the selective destruction of viral components essential for viral replication or assembly, a process referred to as virophagy [30, 31] (fig. 2). For instance, p62/SQSTM1 (hereafter referred to as p62) was found to bind to the capsid of the Sindbis virus [30]. Other autophagy-related proteins might play similar roles, as these proteins have a high tendency to interact with viral proteins [32]. Furthermore, a genetic screen revealed more than a hundred cellular genes with the potency to target the capsid of the Sindbis virus to autophagy. These genes do not influence ongoing autophagy, supporting their role in selective virophagy [30]. Among them, SMURF1 contributes to the targeting of recombinant herpes simplex virus type 1 (HSV-1), suggesting that conserved pathways might trigger virophagy towards components of viruses belonging to different families. However, a direct control of viral infections by virophagy/xenophagy might rather be adapted to nonmitotic cells, such as neurons, than to mitotic cells for which virophagy might mainly serve to trigger innate antiviral immune responses [33].
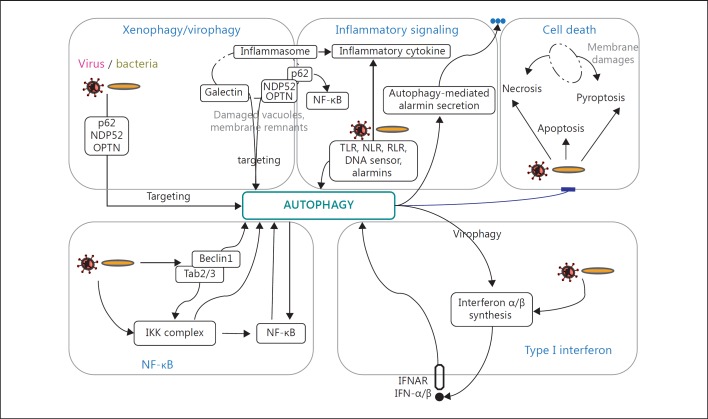
Fig. 2.
General scheme of potential activating events involved in pathogen-induced autophagy signaling in innate immunity. The crosstalk between xenophagy/virophagy, NF-κB activation, IFN-I synthesis or inflammatory signaling with autophagy are schematically represented. Pathogen-induced cell death mechanisms can be prevented/delayed by autophagy. Note that other inhibitory pathways are not represented. See text for details.
Autophagy in Nuclear Factor-κB Signaling
The nuclear factor-κB (NF-κB) family of transcription factors is essential for the expression of numerous genes involved in diverse biological processes, including cell survival, differentiation, inflammation and immunity. Although yet poorly studied in the context of innate immunity, crosstalk between NF-κB and autophagy pathways was unveiled recently in different models.
The formation of the IκB kinase (IKK) complex turns out to be central in canonical activation of NF-κB. The IKK complex is composed of two catalytic subunits, IKKα and IKKβ, associated with the regulatory subunit IKKγ (also called ‘NEMO’). Many stimuli activate the IKK complex, including TNF, PRRs of the TLR/NOD-like receptor (NLR) families or DNA damages. Upon activation, IKKγ binds to the inhibitory IκBα, which is then phosphorylated by IKKβ, and subsequently ubiquitinated and fragmented by the proteasome. IκBα degradation releases cytosolic NF-κB, which translocates to the nucleus and binds to the promoter of target genes, including those encoding for proinflammatory cytokines such as TNF-α and IL-1β. These cytokine signals further potentiate NF-κB activation, in an autocrine and paracrine manner, contributing to the amplification of an inflammatory response. A pathway noncanonical for NF-κB activation exists, which is independent of IκBα degradation, but relies on the activation of IKKα kinase by the NF-κB-inducing kinase.
Regulation of Autophagy by NF-κB Activation
Pioneer work reporting a crosstalk between the autophagy and NF-κB pathways showed that TNF-α-induced NF-κB activation suppresses autophagy in tumor cells. More specifically, in cells deficient in NF-κB, TNF-α signaling led to an accumulation of reactive oxygen species (ROS) responsible for autophagy induction [34]. Cells of patients suffering from a myelodysplastic syndrome display a constitutive activation of the NF-κB pathway, whereas autophagy is compromised. However, inhibition of NF-κB reactivates the autophagic process, arguing for an inhibitory activity of NF-κB on autophagy in these cells [35]. Interestingly, beyond cancer cells, autophagy blockage by NF-κB activation was also observed in infected cells. Upon Escherichia coli infection, a sustained NF-κB activation suppresses autophagy, as demonstrated by the fact that the inhibition of long-term NF-κB activation upon E. coli challenge leads to extensive autophagy, which improves cell survival and cytokine production [36].
Conversely, in certain conditions, NF-κB activation potentiates autophagy (fig. 2). For example, during heat shock response, activation of NF-κB leads to the induction of autophagy, which is required to eliminate accumulated aggregates. This process involves transcription of NF-κB target genes which regulate autophagy, such as BAG3 and heat shock protein B8 (HSPB8) [37]. Similarly, DNA damage signals to activate NF-κB, which induces expression of SKP2, that in turn induces autophagy via a pathway involving p27KIP1 [38]. Interestingly, a direct regulation of autophagy induction by NF-κB was also shown to rely on the presence of an NF-κB binding site on the promoter of the beclin1 (Atg6 gene). As a consequence, NF-κB upregulates both the expression of Beclin 1 and the level of autophagy [39]. This last study displayed a direct link between both pathways, which was shown to be effective in T-cell activation, but remains to be tested in other contexts.
The NF-κB pathway also contributes to the activation of autophagy upon infection. Human CD4 T-cell infection with human T-cell leukemia virus type 1 may lead to adult T-cell leukemia/lymphoma. Infection with the closely related human T-cell leukemia virus type 2 retrovirus is also suspected to induce leukemia. Both viruses encode for a highly homologous transforming protein, Tax. Both Tax1 and Tax2 induce the constitutive activation of NF-κB by targeting the IKK complex, which leads to the immortalization of human primary CD4 T cells. Recently, it was found that Tax proteins constitutively activate autophagy through a pathway involving IKKα and IKKβ. Furthermore, Tax2 interacts with Beclin 1 and the phosphatidylinositol-3 kinase class III Vps34 and recruits them together with the IKK complex, in order to form the autophagosome. As a consequence, Tax mediates cellular transformation and immortalization of infected T cells [40, 41]. However, it is unknown whether the NF-κB activation is involved in this process or whether the IKK complex acts on autophagy independently of its role in NF-κB activation.
Recent evidences indeed highlighted a role for the IKK complex in the regulation of autophagy, independently of NF-κB. Both catalytic subunits IKKα and IKKβ can activate autophagy upon several stimuli (fig. 2). The depletion of any of the IKK complex subunits restricts the induction of autophagy. In contrast, the expression of an active IKK complex is sufficient to trigger activation of autophagy [42]. In order to achieve this activation, the IKK complex upregulates the expression of the Atg5 and Lc3 (Atg8) genes [43]. Interestingly, IKKβ activity can enhance autophagy when associate with an IKKγ form unable to bind IκBα, suggesting that the IKK complex could be selectively oriented on one or the other of the two pathways [44]. However, the precise substrates of IKKβ for autophagy induction remain to be determined.
Interestingly, autophagy and NF-κB activation could be regulated simultaneously in certain situations. In the signal transduction pathway of NF-κB inducers (some PRRs, TNF-α), transforming growth factor β-activated protein kinase 1 (TAK1)-binding protein 2 (TAB2) and TAB3 are two molecular intermediates that recruit the kinase TAK1, which then activates the IKK complex. TAB2 and TAB3 were recently found to be associated with the autophagy protein Beclin 1 [45] (fig. 2). These interactions inhibit autophagy, whereas their dissociation regulates both NF-κB activation (by releasing TAB2/3 for TAK1 recruitment) and autophagy (by releasing Beclin 1). Thus, TAB2 and TAB3 might be at the cornerstone of the coregulation of these two essential cellular pathways, in response to PRR engagement.
Regulation of NF-κB Activation by Autophagy
Autophagy deficiency can impair NF-κB activation in several cellular models. In mouse embryonic fibroblasts (MEFs), autophagy induction leads to NF-κB activation, whereas in MEFs deficient for the Atg5 gene, its activation is prevented. Similarly, in two cancer epithelial cell lines, autophagy is required for NF-κB activation upon TNF-α stimulation [46]. It was also reported that in the context of infections, NOD2 signaling links autophagy to the NF-κB pathway. Upon binding of muramyl dipeptide, a bacterial wall product, NOD2 transduces signals leading to NF-κB activation and autophagy. Whereas NOD2-induced autophagy does not rely on NF-κB [2], autophagy regulates NOD2-dependent NF-κB activation. Indeed, other reports show that the reduced expression of Beclin 1 decreased the NOD2-dependent NF-κB activation [47]. The crosstalk between NF-κB and autophagy could be extended to other PRRs. Indeed, several TLRs can induce autophagy upon specific pathogen-associated molecular pattern (PAMP) recognition [48]. However, the role of NF-κB in this pathway has not yet been extensively studied. Of note, in Drosophila melanogaster, peptidoglycan-recognition protein-LE signaling induces autophagy independently of activation of the NF-κB pathway [49]. The understanding of the interplay between autophagy and NF-κB activation in the context of PRR engagement requires further specific investigations.
Autophagy was found to attenuate lipopolysaccharide (LPS)-induced inflammatory response in intestinal epithelium. Indeed, in Atg7−/− intestinal epithelium, the activation of NF-κB is enhanced upon bacterial endotoxin stimulation, leading to an increased synthesis of proinflammatory cytokines [50]. Strikingly, in the same cell types, TNF-α signaling induces two waves of NF-κB activation through two independent mechanisms, which both govern the degradation of IκBα. Early after stimulation, IκB degradation is promoted by the proteasome, whereas later on, IκB is degraded by autophagy, thus dampening the inflammatory response [51]. Furthermore, the signal transduction emanating from TNF-α receptor 1 (TNFR1) can be terminated by clearance of the receptor within the autophagic machinery, subsequently to its endocytosis. Strikingly, inherited mutations in TNFR1, responsible for TNFR-associated periodic syndrome, lead to accumulation of the receptor due to its defective targeting towards the autophagic pathway. As a consequence, the NF-κB pathway is overstimulated, and proinflammatory cytokine production exacerbated in TNFR-associated periodic syndrome patients [52].
Similarly, M2 macrophage (macrophage with anti-inflammatory properties, which often infiltrates tumors) differentiation in the presence of a tumor-conditioned media requires a TLR2-dependent signal, which in turn activates NF-κB and promotes the synthesis of proinflammatory cytokines. However, to avoid extensive inflammatory response, NF-κB is subsequently aggregated within the cytosol and selectively targeted to autophagy for degradation [53]. Interestingly, similar strategies might be used by pathogens to dampen NF-κB-mediated inflammatory innate response. Indeed, the murine cytomegalovirus encodes for a protein, M45, which prevents NF-κB activation by interacting with and mediating the autophagic degradation of IKKγ [54].
Another mechanism by which autophagy dampens NF-κB activation is through the recurrent degradation of p62. p62 is an autophagic receptor which targets cellular cargo to autophagy for degradation and which can induce innate immunity mechanisms via proinflammatory signaling [55]. Indeed, p62 regulates several signaling pathways by interacting with a large number of proteins. Among them, p62 interacts with ERK1 and represses its signaling. Moreover, p62 promotes ubiquitination of TNFR-associated factor 6 (TRAF6) in response to several inflammatory signals including IL-1 and receptor activator of NF-κB ligand. Thus, a receptor activator of NF-κB-p62/TRAF6/NF-κB axis is thought to be of relevance in human physiology: mutations in p62 are associated with activation of NF-κB in Paget's disease of the bone, a genetic disorder characterized by enhanced osteoclastogenic activity. IL-1R signaling stimulates p62 to recruit a TRAF6-IL-1 receptor-associated kinase 1 complex, which triggers NF-κB activation. In addition, p62 interacts with the Nrf2-binding site of Keap1, which leads to the expression of genes encoding antioxidant proteins and detoxification enzymes. p62 is also involved in the activation of the IKK complex [56]. Interestingly, the stimulation of TLR on keratinocytes induces p62 expression, which activates NF-κB and increases proinflammatory cytokine production, and this process is amplified when autophagy is inhibited due to the accumulation of cytosolic p62 [57]. Thus, autophagy negatively regulates NF-κB activation upon TLR engagement, via the degradation of p62. In autophagy-deficient cells, p62 upregulation may lead to NF-κB pathway deregulation and promotes tumorigenesis [58].
Finally, autophagy could dampen NF-κB activation through the degradation of subunits of the IKK complex. It was recently described that Keap1 binds to and promotes the autophagic degradation of IKKβ, downregulating TNF-α-stimulated NF-κB activation [59]. Conversely, the chaperone HSP90 also interacts with the IKK complex, but this interaction stabilizes the NF-κB activation complex. Furthermore, HSP90 protects the IKK complex from an autophagy-mediated degradation [60]. Interestingly, Keap1 and Hsp90 compete for IKKβ binding, suggesting that upstream signals at this level might control the autophagy/NF-κB crosstalk [59]. Furthermore, Hsp90 also protects NF-κB-inducing kinase from an autophagic degradation, suggesting similar regulation of the noncanonical pathway of NF-κB activation by autophagy [61].
The crosstalk between NF-κB and autophagy is far from revealing all its secrets. Furthermore, this crosstalk is not exclusive to innate immunity, as specific T-cell receptor-dependent NF-κB activation in primary effector T cells is also regulated by autophagy [62].
Autophagy in Inflammatory Signaling
In the context of its innate immunological functions, autophagy can regulate PRR signaling and vice versa. Autophagy also regulates secretion of damage-associated molecular patterns (DAMPs) or alarmins [see review in this issue by Jiang et al. [14]] and affects the onset of inflammatory-linked disease through immunity-related GTPase M and guanylate-binding protein (GBP) [see review in this issue by Nguyen et al. [4]]. Initial interaction of microorganisms with innate immune cells, macrophages and dendritic cells (DCs) is crucial in eliciting immune responses to invading pathogens. Thus, autophagy can protect against bacterial burden and inflammation. For instance, in vivo mycobacteria infection of autophagy-deficient mice leads to exacerbation of bacillary burden and pulmonary inflammation characterized by neutrophil infiltration and IL-17 response with increased IL-1α levels [63]. Interestingly, isoniazid and pyrazinamide, the current standard treatment against tuberculosis, activate cellular and mitochondrial ROS and autophagy [64]. Moreover, this activation of autophagy dampens the proinflammatory response consecutive to Mycobacterium tuberculosis infection in macrophages [64]. Detection of the intruders involves PRRs that gather TLRs, NLR inflammasomes that include sequestosome 1/p62-like receptors (SLRs) [65], retinoic acid inducible gene I (RIG-I)-like receptors (RLRs), C-type lectin receptors, and absence in melanoma-2-like receptors [66, 67] (fig. 2). Autophagy acts as a topological inverter to induce lysosomal degradation of cytoplasmic exposed materials and SLRs studied either in the context of autophagy or as part of inflammatory signaling platforms [66, 67].
Toll-Like Receptors
The membrane TLRs were the first to be shown to be involved in autophagy, and this link can be exemplified by TLR4. TLR4 stimulation activates the VPS34-dependent enhancement of mycobacteria-selective degradation via autophagy [68]. The TLR4 engagement leads to TRAF6-mediated K63 ubiquitination of Beclin 1 followed by Bcl-2 dissociation from the BH3 domain of Beclin 1 [69]. This pathway is negatively regulated by the deubiquitinating enzyme A20 limiting the autophagy response which turns off activation of NF-κB signaling [69, 70]. Once internalized, TLR4 requires the adaptor protein TRAM (translocating chain-associating membrane protein) to activate TRIF [TIR-domain-containing adapter-inducing interferon (IFN)-β] through self-oligomerization and directly binds to (1) TRAF6 to activate TAK1-dependent NF-κB signaling and (2) TRAF3 that forms a complex with TANK-binding kinase 1 (TBK1) and IKKε [71]. Optineurin (OPTN), an essential mediator for TLR4-mediated autophagic elimination of microbes downstream of TRIF, is phosphorylated by TBK1 upon Salmonella infection [72]. OPTN then binds to LC3, thereby bridging bacteria to autophagosomes where OPTN colocalizes with TBK1 and NDP52 (nuclear dot protein 52 kDa), but not with p62 [72]. NDP52 binds to TBK1 through interaction with SINTBAD (similar to nucleosome assembly protein 1 TBK1 adaptor) and nucleosome assembly protein 1, as well as to LC3C [73, 74]. In macrophages infected with mycobacteria, OPTN binds to Rab8b and, in turn, to huntingtin [75]. TBK knockdown causes a deficit in M. bovis bacillus Calmette-Guérin killing, whereas huntingtin is dispensable [75]. TBK1 is necessary for autophagosome maturation, phosphorylation of p62 on Ser-403 (a critical residue for autophagic function of p62) and is required for IL-1β-induced killing of mycobacteria in macrophages [75].
Nucleotide-Binding Oligomerization Domains
The role of ATG16L1 in inflammation is reinforced by its involvement in inflammatory bowel disease [see review in this issue by Nguyen et al. [4]]. NLRC4 (NLR family, CARD domain containing 4), NLRP3 (NOD-like receptor family, pyrin domain containing 3), NLRP4 and NLRP10 interact with Beclin 1 [76]. RNA silencing of NLRP4 results in enhancement of the autophagic bactericidal process of GAS [76]. NLRP4 recruits GAS-containing phagosomes and dissociates transiently from Beclin 1 [76]. In this case, NLRP4 would act as a sensor and would permit autophagy initiation. Moreover, NLRP4 interacts with class C vacuolar protein-sorting complex, thereby negatively regulating maturation of autophagosomes and endosomes [76]. NLRC4 has been involved in discriminating between commensal and pathogenic bacteria, such as Salmonella, due to the constitutive expression of pro-IL-1β that facilitates the production of IL-1β without TLR activation [77]. Whether autophagy is involved in this production remains to be investigated. On the other hand, NLRP3 and NLRC4 inflammasomes cooperate in vivo during S. Typhimurium infection, but deficiency in either NLRP3 or NLRC4 does not change the bacterial infection in mice [78].
Sequestosome 1/p62-Like Receptors
SLRs exert immune functional duality in bacterial clearance through autophagy and as a proinflammatory response along the TRAF6-NF-κB pathway. The Ospl effector of Shigella has been shown to deamidate the glutamine residue at position 100 in ubiquitin-conjugating enzyme UBC13. This leads to the generation of a glutamic acid residue leading to inhibition of the E2 ubiquitin-conjugating activity required for TRAF6 activation [79] and dampens the diacylglycerol-CBM (CARD-BCL10-MALT1) complex- TRAF6-NF-κB signaling pathway [79]. However, SLRs can act completely differently by initiating the formation of bactericidal peptides from cytoplasmic precursors that are transferred into autophagolysosomes. This microbicidal property has been illustrated with p62-mediated killing of the H37rv stain of mycobacteria in macrophages by antimicrobial peptides derived from ubiquitin and ribosomal innocuous precursors [80]. Similarly, IFN-γ-inducible 65-kDa GTPase GBP1 binds p62 and GBP7-captured ATG4b to promote bacillus Calmette-Guérin mycobacteria oxidative killing and deliver antimicrobial peptides to autophagolysosomes [81]. Also related to antimicrobial peptides is the production of the human cathelicidin antimicrobial peptide, which can be induced by 1α,25-dihydroxycholecalciferol, the active form of vitamin D [82]. Cathelicidin is required for 1α,25-dihydroxycholecalciferol to activate the autophagic flux in the human immunodeficiency virus 1 (HIV-1) and M. tuberculosis coinfected human macrophages, which inhibits mycobacterial growth and the replication of HIV [82]. Single or coinfection inhibition is not only due to autophagosomal impairment but also to phagosomal maturation leaving the exact relationship between the two pathways to be clarified [82]. It has also been proposed that SLR participates in autophagy targeting of membrane remnants after intracytosolic replicating bacteria have gained access to their replication niche, hereby damping danger signals that trigger inflammatory responses [83]. Often, danger signals are released in response to bacterial invasion that activates host cell receptors. However, host cell components associated with bacterial invasion have also been proposed to act as danger signals. The polarity of glycosylated proteins along the biosynthetic pathway moves to the cell surface glycan chains to sit in the extracellular environment. During Shigella and Listeria internalization, the vacuolar membrane is disrupted, and these chains are exposed to the cytosolic environment. After bacterial escape, these exposed extracellular glycosylated moieties recruit galectin-3, −8 and −9, among which galectin-8 binds to NDP52 which mediates further recruitment of LC3 [7, 74]. Figure 1 shows the distribution of galectin-3 on remnant membranes after S. flexneri escape. Galectin-3 codistributes with several molecular components linked to several host cell response pathways, i.e. inflammasomes NOD1, NLRC4, caspase-1 () and autophagy via ubiquitination (fig. 1d) and LC3 [7].
DAMPs - Alarmins
The response to exogenous pathogens leads to activation of innate immunity through the release of alarmins and their binding to PRRs. A classic example is septic shock where TLR4 recognizes PAMPs. More generally, DAMPs can activate autophagy as demonstrated for the high mobility group box 1 (HMGB1) protein that translocates from the nucleus into the cytoplasm where it displaces Bcl-2 from Beclin 1 [84, 85]. Upon further injury of the cell, HMGB1 is released from dying cells to act in a paracrine manner to further induce autophagy [84, 85]. miR-34c and miR-214 are significantly expressed in fresh human peripheral blood mononuclear cells exposed to DAMP-containing freeze-thaw lysates or to conditioned media from serum-starved and glucose-deprived cells [86]. miR-34c and miR-214 expression in human peripheral blood mononuclear cells is dependent on the presence of HMGB1 [86]. Both HMGB1 and these miRs may play a common role in driving autophagy and the response to inflammation. It also suggests that cytokine stimulation of immune cells by potent agents such as IL-2 enhances autophagy as a global response to cell stress [87, 88]. ATP is another alarmin that can be released, as demonstrated for lymphoma cells dying by apoptosis that are associated with autophagy in response to IL-3 withdrawal. In these cells, NALP3 (NACHT, LRR and PYD domain-containing protein 3) and caspase-1 are activated while the dying cell is taken up, leading to IL-1β release [13]. When injected into the peritoneal cavity of mice, autophagic dying cells are more potent in recruiting neutrophils than necrotic dying cells [13]. Autophagic dying cell phagocytosis also induces IL-1β secretion through a mechanism that depends on the ATP receptor P2X7 and NALP3 [89]. In the context of autophagy, the secretion of IL-1β and IL-18 was observed in LPS-primed Salmonella-infected macrophages from ATG16L1 knock-out mice [90]. This is associated with pro-IL-1β targeting for degradation [91]. Similarly, LPS stimulation leads to IL-1β secretion in Map1lc3b−/− or Becn1+/- bone marrow-derived macrophages [92]. In macrophages depleted for autophagic proteins, LPS stimulation induces an enhanced level of abnormal mitochondria, which produce more ROS [92, 93]. ROS can activate the inflammasome and the excess of NALP3-mediated mitochondrial DNA in the cytosol then activates capsase-1 and induces IL-1β secretion [92, 93]. Secretion of IL-1β can be observed in the response of bone marrow-derived cells silenced for Beclin 1 expression to irradiated M. tuberculosis strain H37Rv [94]. Whereas inflammasome activity is controlled by basal autophagy in these examples involving bacteria and bacteria products, induction of autophagy promotes unconventional secretion of IL-1β involving Rab8a and the Golgi protein GRASP [95]. Altogether, these results argue for autophagy to negatively regulate inflammasome activation by triggering degradation of inflammatory agents, and thus, lowering the basal level of inflammation. Moreover, these results suggest a fine tuning between the anti-inflammatory effects of autophagy and a proeffect on acute short-term secretion of alarmins with rapid downregulation. The mechanisms by which pathogens control this balance remains to be elucidated with regard to the microorganism and the host cell.
M. marinum can be targeted by autophagy after escape into the cytoplasm [96]. For M. tuberculosis, phagosomal membrane damage is mediated by the ESX-1 secretion system through the inflammasome NALP3/ASC (apoptosis-associated speck-like protein containing a CARD) activator ESAT-6 (early secreted antigen 6 kDa) protein [97, 98]. This allows bacterial DNA to be recognized by the stimulator of IFN gene (STING)-dependent cytosolic pathway of the host required for p62 and NDP52 recruitment leading to autophagy activation [99]. This mechanism allows further ubiquitination of the bacterium and targeting to fusion with lysosomes. Consistently, monocytes from autophagy-deficient mice are more prone to infection [99].
Inflammation Downregulation
An intriguing question concerns the mechanism through which the proinflammatory response induced by pathogens is downregulated. At an early stage of nonmyeloid cell infection by S. flexneri, there is a balance between necrosis and the NOD1 receptor-interacting protein-2 (RIP2)-NF-κB-dependent cell survival, whereas at a later stage, pyroptosis occurs releasing bacteria for other cycles of infection [7, 100]. Shigella membrane remnants associated with inflammasome components are cleared by autophagy to dampen the inflammatory response [7]. Remarkably, THP-1 cells transfected with double-stranded DNA (poly dA-dT) induce absence in melanoma and NLRP3 to trigger the activation of the small G protein RALB G (RalA-binding G) and autophagosome formation [101]. RALB triggers the assembly of EXO84-Beclin and EXO-VPS34 complexes leading to autophagy induction [102]. Once recruited on autophagosomes, the inflammasome adaptor protein ASC undergoes K63-mediated polyubiquitination and p62 is in turn recruited leading to autophagy-mediated clearance [101]. These results suggest that inflammatory response tempering depends on autophagy elimination of the activated inflammasome. How the balance between inflammasome activation and autophagosome assembly for further autophagy-mediated degradation is regulated remains to be challenged. Nonetheless, both Shigella and transfected double-stranded DNA autophagy potentiate inflammation that is associated with autophagy limiting the inflammation.
Autophagy in Type I IFN Signaling
Type I IFNs (IFN-I, IFN-α and IFN-β) are potent innate cytokines whose synthesis mostly relies on detection of viral patterns or tumor-linked factors. Following the recognition of a ligand, several PRRs, including TLRs, NLR, RLRs and DNA sensors, can induce IFN-I synthesis: TLR4 and TLR3 signals via the adaptor TRIF, and TLR7/8/9 via the adaptor MyD88, which leads to the activation of IFN regulatory factor (IRF) transcription factors for IFN-I synthesis. RIG-I, MDA-5 (melanoma differentiation-associated protein 5) and NOD2 interact with the mitochondrial intermediate IFN promoter-stimulating factor 1 (IPS-1; also known as MAVS), which signals for IFN-I synthesis. Due to the activation of the constitutively expressed IRF3 and IRF7, plasmacytoid DCs (pDCs) produce the highest quantities per cell of both IFN-α and IFN-β, early after stimulation. In contrast, in most other cell types, only IRF3 is constitutively expressed, which regulates IFN-β induction. Following its secretion, IFN-β binds to the IFN-I receptor (IFNAR) in an autocrine/paracrine-positive feedback pathway, which amplifies the response by inducing a large amount of IFN-α. Thus, potent expression of IFN-α occurs after the primary induction of IFN-β. Both IFN-α and IFN-β share the same IFNAR, which transduces signals to activate the signal transducer and activator of transcription 1 (STAT1) and STAT2 transcription factors, leading to the upregulation of hundreds of IFN-stimulating genes that have a direct antiviral/antitumoral effect.
Regulation of IFN-I Synthesis by Autophagy
The pioneer observation of a crosstalk between autophagy and IFN-I has shown that autophagy potentiates IFN-I synthesis. Murine pDCs display a high level of ongoing autophagy that is not significantly increased upon vesicular stomatitis virus (VSV) infection. However, autophagy delivers viral RNA genome of replicating VSV to TLR7-containing endosomes, which signals for antiviral IFN-I synthesis [103] (fig. 2). A similar observation was shown in human pDCs, infected by HIV-1, although here, viral replication was not required to induce autophagy-dependent IFN-I synthesis [104]. The use of wortmannin also led to IFN-I synthesis modulation during infection of human pDC by the Epstein-Barr virus, a DNA virus [105]. The contribution of autophagy in IFN-I induction seems a specific functional characteristic of pDC, as demonstrated by the ability of these cells, but not of conventional DCs, to secrete IFN-α in response to simian virus 5 infection via an autophagy-dependent process [106]. Thus, in pDC, the high level of constitutive autophagy might contribute to keeping cells continually alert to optimize innate responses against infections by different families of viruses. Nevertheless, autophagy might contribute to the low production of IFN-β upon viral infection in other cell types as shown in DCs infected by the respiratory syncytial virus [107].
In contrast to pDCs, in most other cell types, autophagy negatively regulates IFN-I production. This regulation might contribute to homeostasis of the innate response by cells whose feature it is not to produce IFN-I. Thus, in epithelial and fibroblastic cells, autophagy proteins can prevent IFN-I production induced upon RLR signaling. IPS-1 was shown to interact with the ATG5-ATG12 complex under physiological conditions. This interaction is enhanced upon VSV infection and contributes to downregulate VSV-induced IFN-I production, probably by interfering with the RIG-I/IPS-1 signaling [108]. Furthermore, the increased production of IFN-I in autophagy-defective cells was shown to be due to the accumulation of IPS-1, because of the defective autophagy-mediated clearance of mitochondrion. This signaling is further increased by ROS accumulation, which potentiates RIG-I signaling [109]. Hepatitis C virus (HCV) infection also induces autophagy, via a pathway that may rely on the activation of the unfolded protein response. HCV-induced autophagy was shown to prevent IFN-β/α synthesis in infected human hepatocytes and to promote cell survival: these two autophagy-mediated effects could contribute to HCV-persistent infection [110]. Thus, it could also be possible that the HCV infection may suppress IFN-I synthesis by directly inhibiting the RIG-I/IPS-1 interactions via induction of autophagy. Interestingly, upon VSV infection, the mitochondrial protein TUFM was recently shown to prevent IFN-β synthesis by interacting with NLRX1, an IPS-1-interacting/inhibitory cellular protein. Interestingly, TUFM also interacts with the ATG5-ATG12 complex and, by this way, contributes to induce autophagy [111]. Although this dual function (IFN-I inhibition versus autophagy activation) facilitates VSV replication, it remains to be determined whether IFN-I induction is ‘physically’ prevented by TUFM-dependent autophagy induction [111]. Interestingly, another mitochondrial protein might present similar characteristics. Indeed, mitofusin 2 contributes to the supply of mitochondria membranes for the biogenesis of autophagosomes [112], as well as interacting with IPS-1, and blocks IFN-β synthesis and facilitates VSV replication [113].
Similarly, the DNA genome of viruses such as HSV-1 was reported to induce autophagy via a molecular pathway which involves the transmembrane cellular protein STING [114]. Upon double-stranded DNA stimulation, STING can also trigger IFN-I induction, which is however dampened by the autophagy protein ATG9a. This inhibition does not involve another autophagy protein, ATG7, suggesting that it may rely on an ATG9a-specific, autophagy-independent mechanism [115]. However, STING-mediated autophagy upon HSV-1 infection was shown to facilitate IFN-β synthesis [114]; further studies are required to decipher this discrepancy. Thus, viruses might exploit cellular proteins to induce autophagy in order to prevent IFN-I production. Strikingly, tumor cells might also exploit the negative regulation of IFN-I synthesis by autophagy. Thus, in a model of breast cancer, the deletion of FIP200 (a component of the complex essential for autophagosome formation), prevents autophagy in tumor cells, by releasing the inhibition of IFN-β production upon RLR stimulation [116]. Since IFN-I contribute to the recruitment of effector T cells required for the elimination of tumors, induction of autophagy might prevent antitumoral T-cell functions.
Regulation of Autophagy by IFN-I Signaling
Recently, the ability of IFN-I to activate autophagy was described (fig. 2). IFN-α is widely used for treatment in chronic myeloid leukemia (CML), a cancer linked to the aberrant expression of the leukemic oncogene BCR-ABL1. However, resistances of CML to IFN-α treatment frequently emerge, and autophagy was suspected to play a role in this resistance. IFN-α indeed activates the autophagic flux in CML cells through a pathway involving a STAT1 and NF-κB-dependent pathway, both of which may contribute to upregulate the expression of Beclin 1. Furthermore, inhibition of autophagy enhanced the anticancer potency of IFN-α, possibly by preventing autophagy-mediated caspase-8-dependent CML apoptosis [117]. Similarly, IFN-β can induce the autophagy flux in breast cancer cells which contributes to cell survival; this regulation relies on the inhibition of mammalian target of rapamycin (mTOR) and activation of STAT1 signaling. Importantly, IFN-β also induce LC3-II conversion in MEFs, suggesting that IFN-β might induce autophagy in different cell contexts [118].
Interestingly, distinct autophagic pathways were found to result in IFN-β or IFN-α signaling, in a model of viral infection [119]. IFN-β, which is first produced by hepatocytes upon viral detection (VSV or a synthetic HCV genome), was found to signal in order to orientate viral proteins towards an autophagic degradation, within autolysosomes. However, IFN-α, produced later, leads to a different autophagic pathway, since autophagosomes do not fuse with lysosomes but possibly only with endosomes. This IFN-α-mediated autophagic pathway might further induce antiviral IFN-I production by delivering viral components to TLR-containing endosomes. Since IFN-α and IFN-β signal via the same IFNAR, it would be important to understand the molecular discrepancy of both cytokines in terms of autophagic trafficking.
It is not clear whether IFN-I could signal to dampen autophagy. However, a recent work has shown that whereas cytotoxic lymphocytes (cytotoxic T lymphocytes, natural killer cells) induce lysis of most target cancer cells, some cells escape from lysis through the induction of autophagy. Interestingly, IFN-α treatment can inhibit autophagy in target cells and can limit their resistance to cytotoxicity [120].
The autophagy/IFN-I molecular crosstalk is far from being fully understood. As such, several genes regulating autophagy for selective virophagy were found to also contribute to IFN-I pathways, such as STAT2, although further investigations are required to depict the molecular significance of these interplays [30].
Autophagy in Cell Death
Cell death has two sides in the context of immunity towards pathogens [121]. On the one hand, it offers a way to dampen danger signals emitted by infected cells and allows degradation in macrophages and DCs of apoptotic bodies with infected material for presentation to major histocompatibility class I molecules. On the other hand, cell death provides benefit to bacteria spreading after replication within the host cell. Autophagy has been linked to cell death and the autophagy proteins Beclin 1, ATG5 and ATG3, and GABARAP interact with cell death-related proteins including caspases, FADD, c-FLIP and NIX, respectively [for a review see, ref. [122]]. Antiapoptotic proteins Bcl-2 and BCL-xl also interact with Beclin 1 and inhibit autophagy [123]. We will discuss few examples shown below.
Shigella
PAMPs such as LPS, flagelin and peptidoglycans activate PRR to induce inflammatory responses [121]. Moreover, infection leads to the production of danger signals activating proinflammatory signals after vacuolar escape of Shigella[7]. The danger signals, such as galectins, associated with remnant membranes, target the whole set of membrane and associated signaling to autophagy-mediated degradation dampening the inflammatory response [7, 74]. Moreover, autophagy can regulate the host cell fate in part through shutting down signaling cascades associated with these membranes [7]. In nonmyeloid cells, at early stages, infection outcome relies on a balance between RIP2-independent necrosis and cell survival signals while, at later stages, pyroptosis cell death allows further propagation of the microbes [7, 100] (fig. 2). IpaB is a bacterial effector part of the type 3 secretion system that can induce pyroptosis and the release of IL-1β and IL-18, the typical inflammatory response hallmark of shigellosis symptoms [124]. In Hep2b cells, IpaB induces membrane stress of the Golgi compartment [125]. The link between Golgi fragmentation and inflammatory response remains to be elucidated. The NLRC4 NLR protein and the NLR adaptor ASC participate in the induction of pyroptosis in macrophages [126]. NLRC4 is associated with the remnant membranes, constituting a drastic membrane stress upon Shigella infection [7]. On the other hand, Shigella and Salmonella induce membrane stress that was shown to induce amino acid starvation [127]. Shigella downregulates mTORC1 activity upon displacement of mTOR from LAMP2-positive late endosomes/lysosomes [under RAPTOR (regulatory associated proteinof mTOR) targeting to a protein complex including Rag GTPase and regulator] to the cytosol [127]. The amino acid starvation triggers the GCN2/elF2α/ATF4/ATF3-dependent integrated stress response pathway and mRNA stress granule starvation [127]. The sustained membrane damage could be due to the release of IpaB. In contrary to the observation obtained with Shigella, amino acid starvation in Salmonella-infected cells is rapidly normalized with mTOR reactivation on Salmonella-containing vesicles dependent on the RAPTOR/RAG GTPase/regulator axis [127].
Salmonella
Unlike the situation in epithelial cells infected by Shigella[2], fibroblasts infected with Salmonella activate a NOD2- and RIP2-depdendent signaling pathway [128]. However, in the latter case, RIP2 tyrosine kinase activity plays an essential dual-faceted role as it provides an activating signal through stimulation of MAPK p38 and relieves PP2Ac-PPP2R1A-mediated repression of autophagy [128]. The PP2Ac-PPP2R1A complex interacts with NOD2-RIP2 and becomes tyrosine-phosphorylated on a negative regulatory site (Tyr-307) upon muramyl dipeptide stimulation [128]. PP2A is a ubiquitous serine/threonine phosphatase involved in many pathways including cell death. Autophagy ameliorates the cellular stress induced by accumulation of misfolded or aggregated proteins that can lead to cell death [129, 130]. Salmonella infection in macrophages is associated with ubiquitinated aggregates [131]. These aggregates and aggresome-like induced structures are counteracted by the Salmonella pathogenicity island 2 type 3 secretion system deubiquitinase SseL [131]. In the absence of SseL activity, ubiquitinated structures are recognized by p62, which recruits LC3 and targets them for autophagic degradation [131]. Moreover, SseL activity lowers autophagic flux and favors intracellular Salmonella replication [131]. This suggests that the reduction in autophagic flux mediated by SseL accounts for its ability to induce delayed cytotoxicity in macrophages [132].
Wolbachia
The role of autophagy in cell death associated with infection may yet be underestimated due to the lack of research. For instance, xenophagy has been associated with nerve cell and adipocytes of the central nervous system cell death induced by high virulence of the endosymbiont Wolbachia upon horizontal transfer from its natural host Armadillidium vulgare to another species Porcellio dilatatus[133]. Wolbachia and host cell factors involved remain to be identified.
Conclusion
Recent works highlight that autophagy does not only regulate degradation but also participates in immune and inflammatory cell responses. With the advent of data on the molecular regulation of the autophagy process, it appeared easier to analyze the interplay between autophagy and innate immunity signaling. Thus, the picture emerging is becoming more complex. The positive and/or negative loops contributing to these crosstalks, as well as the involvement in these processes of autophagy-related proteins independently of their role in canonical autophagy contribute to this complexity, but still challenge our curiosity.
Acknowledgments
We thank N. Dupont and S. Lacas-Gervais for sharing data. We thank the reviewers for their very constructive comments on the review, and Dr. Uzma Hasan for checking the English language of this manuscript. We apologize to colleagues whose works were not cited herein owing to space limitation. This work was supported by grants from the ANR (ANR-09-MIEN-020 and ANR-08-JCJC-0064-01), Institut Universitaire de France (IUF, MF) and FINOVI.
References
- 1.Cemma M, Brumell JH. Interactions of pathogenic bacteria with autophagy systems. Curr Biol. 2012;22:R540–R545. doi: 10.1016/j.cub.2012.06.001. [DOI] [PubMed] [Google Scholar]
- 2.Travassos LH, Carneiro LA, Ramjeet M, Hussey S, Kim YG, et al. Nod1 and Nod2 direct autophagy by recruiting ATG16L1 to the plasma membrane at the site of bacterial entry. Nat Immunol. 2010;11:55–62. doi: 10.1038/ni.1823. [DOI] [PubMed] [Google Scholar]
- 3.Ravikumar B, Moreau K, Jahreiss L, Puri C, Rubinsztein D. Plasma membrane contributes to the formation of pre-autophagosomal structures. Nat Cell Biol. 2010;12:747–757. doi: 10.1038/ncb2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nguyen HTT, Lapaquette P, Bringer M-A, Darfeuille-Michaud A. Autophagy and Crohn's disease. J Innate Immun. 2013 doi: 10.1159/000345129. DOI: 10.1159/000345129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moreau K, Ravikumar B, Puri C, Rubinsztein D. Arf6 promotes autophagosome formation via effects on phosphatidylinositol 4,5-bisphosphate and phospholipase D. J Cell Biol. 2012;196:483–496. doi: 10.1083/jcb.201110114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu J, Kuo C. Pivotal role of ADP-ribosylation factor 6 in Toll-like receptor 9-mediated immune signaling. J Biol Chem. 2012;287:4323–4334. doi: 10.1074/jbc.M111.295113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Randow F, Münz C. Autophagy in the regulation of pathogen replication and adaptive immunity. Trends Immunol. 2012;33:475–487. doi: 10.1016/j.it.2012.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dupont N, Lacas-Gervais S, Bertout J, Paz I, Freche B, et al. Shigella phagocytic vacuolar membrane remnants participate in the cellular response to pathogen invasion and are regulated by autophagy. Cell Host Microbe. 2009;6:137–149. doi: 10.1016/j.chom.2009.07.005. [DOI] [PubMed] [Google Scholar]
- 9.Moreau K, Ravikumar B, Renna M, Puri C, Rubinsztein D. Autophagosome precursor maturation requires homotypic fusion. Cell. 2011;146:303–317. doi: 10.1016/j.cell.2011.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fader C, Sánchez D, Mestre M, Colombo M. TI-VAMP/VAMP7 and VAMP3/cellubrevin: two v-SNARE proteins involved in specific steps of the autophagy/multivesicular body pathways. Biochim Biophys Acta. 2009;1793:1901–1916. doi: 10.1016/j.bbamcr.2009.09.011. [DOI] [PubMed] [Google Scholar]
- 11.Furuta N, Fujita N, Noda T, Yoshimori T, Amano A. Combinational soluble N-ethylmaleimide-sensitive factor attachment protein receptor proteins VAMP8 and Vti1b mediate fusion of antimicrobial and canonical autophagosomes with lysosomes. Mol Biol Cell. 2010;21:1001–1010. doi: 10.1091/mbc.E09-08-0693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Itakura E, Kishi-Itakura C, Mizushima M. The hairpin-type tail-anchored SNARE syntaxin 17 targets to autophagosomes for fusion with endosomes/lysosomes. Cell. 2012;151:1256–1269. doi: 10.1016/j.cell.2012.11.001. [DOI] [PubMed] [Google Scholar]
- 13.Ayna G, Krysko DV, Kaczmarek A, Petrovski G, Vandenabeele P, et al. ATP release from dying autophagic cells and their phagocytosis are crucial for inflammasome activation in macrophages. PLoS One. 2012;7:e40069. doi: 10.1371/journal.pone.0040069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiang S, Dupont N, Castillo EF, Deretic V. Secretory versus degradative autophagy: unconventional secretion of inflammatory mediators. J Innate Immun. 2013 doi: 10.1159/000346707. DOI: 10.1159/000346707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nair U, Jotwani A, Geng J, Gammoh N, Richerson D, et al. SNARE proteins are required for macroautophagy. Cell. 2011;146:290–302. doi: 10.1016/j.cell.2011.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sakurai A, Maruyama F, Funao J, Nozawa T, Aikawa C, et al. Specific behavior of intracellular Streptococcuspyogenes that has undergone autophagic degradation is associated with bacterial streptolysin O and host small G proteins Rab5 and Rab7. J Biol Chem. 2010;285:22666–22675. doi: 10.1074/jbc.M109.100131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yamaguchi H, Nakagawa I, Yamamoto A, Amano A, Noda T, et al. An initial step of GAS-containing autophagosome-like vacuoles formation requires Rab7. PLoS Pathog. 2009;5:e1000670. doi: 10.1371/journal.ppat.1000670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nozawa T, Aikawa C, Goda A, Maruyama F, Hamada S, et al. The small GTPases Rab9A and Rab23 function at distinct steps in autophagy during Group A Streptococcus infection. Cell Microbiol. 2012;14:1149–1165. doi: 10.1111/j.1462-5822.2012.01792.x. [DOI] [PubMed] [Google Scholar]
- 19.Popovic D, Akutsu M, Novak I, Harper J, Behrends C, et al. Rab GTPase-activating proteins in autophagy: regulation of endocytic and autophagy pathways by direct binding to human ATG8 modifiers. Mol Cell Biol. 2012;32:1733–1744. doi: 10.1128/MCB.06717-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Longatti A, Lamb C, Razi M, Yoshimura S, Barr F, et al. TBC1D14 regulates autophagosome formation via Rab11- and ULK1-positive recycling endosomes. J Cell Biol. 2012;197:659–675. doi: 10.1083/jcb.201111079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dong N, Zhu Y, Lu Q, Hu L, Zheng Y, et al. Structurally distinct bacterial TBC-like GAPs link Arf GTPase to Rab1 inactivation to counteract host defenses. Cell. 2012;150:1029–1041. doi: 10.1016/j.cell.2012.06.050. [DOI] [PubMed] [Google Scholar]
- 22.Zoppino F, Militello R, Slavin I, Alvarez C, Colombo M. Autophagosome formation depends on the small GTPase Rab1 and functional ER exit sites. Traffic. 2010;1:1246–1261. doi: 10.1111/j.1600-0854.2010.01086.x. [DOI] [PubMed] [Google Scholar]
- 23.Huang J, Birmingham C, Shahnazari S, Shiu J, Zheng Y, et al. Antibacterial autophagy occurs at PI(3)P-enriched domains of the endoplasmic reticulum and requires Rab1 GTPase. Autophagy. 2011;7:17–26. doi: 10.4161/auto.7.1.13840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Selyunin A, Sutton S, Weigele B, Reddick L, Orchard R, et al. The assembly of a GTPase-kinase signalling complex by a bacterial catalytic scaffold. Nature. 2011;469:107–111. doi: 10.1038/nature09593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clements A, Smollett K, Lee S, Hartland E, Lowe M, et al. EspG of enteropathogenic and enterohemorrhagic E. coli binds the Golgi matrix protein GM130 and disrupts the Golgi structure and function. Cell Microbiol. 2011;13:1429–1439. doi: 10.1111/j.1462-5822.2011.01631.x. [DOI] [PubMed] [Google Scholar]
- 26.Choy A, Dancourt J, Mugo B, O'Connor T, Isberg R, et al. The Legionella effector RavZ inhibits host autophagy through irreversible Atg8 deconjugation. Science. 2012;338:1072–1076. doi: 10.1126/science.1227026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Joubert PE, Meiffren G, Gregoire IP, Pontini G, Richetta C, et al. Autophagy induction by the pathogen receptor CD46. Cell Host Microbe. 2009;6:354–366. doi: 10.1016/j.chom.2009.09.006. [DOI] [PubMed] [Google Scholar]
- 28.Talloczy Z, Virgin HWT, Levine B. PKR-dependent autophagic degradation of herpes simplex virus type 1. Autophagy. 2006;2:24–29. doi: 10.4161/auto.2176. [DOI] [PubMed] [Google Scholar]
- 29.Blanchet FP, Moris A, Nikolic DS, Lehmann M, Cardinaud S, et al. Human immunodeficiency virus-1 inhibition of immunoamphisomes in dendritic cells impairs early innate and adaptive immune responses. Immunity. 2010;32:654–669. doi: 10.1016/j.immuni.2010.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Orvedahl A, Sumpter R, Jr, Xiao G, Ng A, Zou Z, et al. Image-based genome-wide siRNA screen identifies selective autophagy factors. Nature. 2011;480:113–117. doi: 10.1038/nature10546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Richetta C, Faure M. Autophagy in antiviral innate immunity. Cell Microbiol. 2013;15:368–376. doi: 10.1111/cmi.12043. [DOI] [PubMed] [Google Scholar]
- 32.Gregoire IP, Richetta C, Meyniel-Schicklin L, Borel S, Pradezynski F, et al. IRGM is a common target of RNA viruses that subvert the autophagy network. PLoS Pathog. 2011;7:e1002422. doi: 10.1371/journal.ppat.1002422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yordy B, Iijima N, Huttner A, Leib D, Iwasaki A. A neuron-specific role for autophagy in antiviral defense against herpes simplex virus. Cell Host Microbe. 2012;12:334–345. doi: 10.1016/j.chom.2012.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Djavaheri-Mergny M, Amelotti M, Mathieu J, Besancon F, Bauvy C, et al. NF-kappaB activation represses tumor necrosis factor-alpha-induced autophagy. J Biol Chem. 2006;281:30373–30382. doi: 10.1074/jbc.M602097200. [DOI] [PubMed] [Google Scholar]
- 35.Fabre C, Carvalho G, Tasdemir E, Braun T, Ades L, et al. NF-kappaB inhibition sensitizes to starvation-induced cell death in high-risk myelodysplastic syndrome and acute myeloid leukemia. Oncogene. 2007;26:4071–4083. doi: 10.1038/sj.onc.1210187. [DOI] [PubMed] [Google Scholar]
- 36.Schlottmann S, Buback F, Stahl B, Meierhenrich R, Walter P, et al. Prolonged classical NF-kappaB activation prevents autophagy upon E. coli stimulation in vitro: a potential resolving mechanism of inflammation. Mediators Inflamm. 2008;2008:725854. doi: 10.1155/2008/725854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nivon M, Abou-Samra M, Richet E, Guyot B, Arrigo AP, et al. NF-kappaB regulates protein quality control after heat stress through modulation of the BAG3-HspB8 complex. J Cell Sci. 2012;125:1141–1151. doi: 10.1242/jcs.091041. [DOI] [PubMed] [Google Scholar]
- 38.Barre B, Perkins ND. The Skp2 promoter integrates signaling through the NF-kappaB, p53, and Akt/GSK3beta pathways to regulate autophagy and apoptosis. Mol Cell. 2010;8:524–538. doi: 10.1016/j.molcel.2010.03.018. [DOI] [PubMed] [Google Scholar]
- 39.Copetti T, Bertoli C, Dalla E, Demarchi F, Schneider C. p65/RelA modulates BECN1 transcription and autophagy. Mol Cell Biol. 2009;29:2594–2608. doi: 10.1128/MCB.01396-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cheng H, Ren T, Sun SC. New insight into the oncogenic mechanism of the retroviral oncoprotein Tax. Protein Cell. 2012;3:581–589. doi: 10.1007/s13238-012-2047-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ren T, Dong W, Takahashi Y, Xiang D, Yuan Y, et al. HTLV-2 Tax immortalizes human CD4+ memory T lymphocytes by oncogenic activation and dysregulation of autophagy. J Biol Chem. 2012;287:34683–34693. doi: 10.1074/jbc.M112.377143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Criollo A, Senovilla L, Authier H, Maiuri MC, Morselli E, et al. The IKK complex contributes to the induction of autophagy. EMBO J. 2010;29:619–631. doi: 10.1038/emboj.2009.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Comb WC, Cogswell P, Sitcheran R, Baldwin AS. IKK-dependent, NF-kappaB-independent control of autophagic gene expression. Oncogene. 2011;30:1727–1732. doi: 10.1038/onc.2010.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schrofelbauer B, Polley S, Behar M, Ghosh G, Hoffmann A. NEMO ensures signaling specificity of the pleiotropic IKKbeta by directing its kinase activity toward IkappaBalpha. Mol Cell. 2012;47:111–121. doi: 10.1016/j.molcel.2012.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Criollo A, Niso-Santano M, Malik SA, Michaud M, Morselli E, et al. Inhibition of autophagy by TAB2 and TAB3. EMBO J. 2011;30:4908–4920. doi: 10.1038/emboj.2011.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Criollo A, Chereau F, Malik SA, Niso-Santano M, Marino G, et al. Autophagy is required for the activation of NFkappaB. Cell Cycle. 2012;11:194–199. doi: 10.4161/cc.11.1.18669. [DOI] [PubMed] [Google Scholar]
- 47.Homer CR, Richmond AL, Rebert NA, Achkar JP, McDonald C. ATG16L1 and NOD2 interact in an autophagy-dependent antibacterial pathway implicated in Crohn's disease pathogenesis. Gastroenterology. 2010;139:1630–1641. doi: 10.1053/j.gastro.2010.07.006. 1641e1-e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Delgado MA, Elmaoued RA, Davis AS, Kyei G, Deretic V. Toll-like receptors control autophagy. EMBO J. 2008;27:1110–1121. doi: 10.1038/emboj.2008.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yano T, Mita S, Ohmori H, Oshima Y, Fujimoto Y, et al. Autophagic control of Listeria through intracellular innate immune recognition in Drosophila. Nat Immunol. 2008;9:908–916. doi: 10.1038/ni.1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fujishima Y, Nishiumi S, Masuda A, Inoue J, Nguyen NM, et al. Autophagy in the intestinal epithelium reduces endotoxin-induced inflammatory responses by inhibiting NF-kappaB activation. Arch Biochem Biophys. 2011;506:223–235. doi: 10.1016/j.abb.2010.12.009. [DOI] [PubMed] [Google Scholar]
- 51.Colleran A, Ryan A, O'Gorman A, Mureau C, Liptrot C, et al. Autophagosomal IkappaB alpha degradation plays a role in the long term control of tumor necrosis factor-alpha-induced nuclear factor-kappaB (NF-kappaB) activity. J Biol Chem. 2011;286:22886–22893. doi: 10.1074/jbc.M110.199950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bachetti T, Chiesa S, Castagnola P, Bani D, Di Zanni E, et al. Autophagy contributes to inflammation in patients with TNFR-associated periodic syndrome (TRAPS) Ann Rheum Dis. 2012 doi: 10.1136/annrheumdis-2012-201952. E-pub ahead of print. [DOI] [PubMed] [Google Scholar]
- 53.Chang CP, Su YC, Hu CW, Lei HY. TLR2-dependent selective autophagy regulates NF-kappaB lysosomal degradation in hepatoma-derived M2 macrophage differentiation. Cell Death Differ. 2013;20:515–523. doi: 10.1038/cdd.2012.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fliss PM, Jowers TP, Brinkmann MM, Holstermann B, Mack C, et al. Viral mediated redirection of NEMO/IKKgamma to autophagosomes curtails the inflammatory cascade. PLoS Pathog. 2012;8:e1002517. doi: 10.1371/journal.ppat.1002517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Moscat J, Diaz-Meco M. p62 at the crossroads of autophagy, apoptosis, and cancer. Cell. 2009;137:1001–1004. doi: 10.1016/j.cell.2009.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Duran A, Linares JF, Galvez AS, Wikenheiser K, Flores JM, et al. The signaling adaptor p62 is an important NF-kappaB mediator in tumorigenesis. Cancer Cell. 2008;13:343–354. doi: 10.1016/j.ccr.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 57.Lee HM, Shin DM, Yuk JM, Shi G, Choi DK, et al. Autophagy negatively regulates keratinocyte inflammatory responses via scaffolding protein p62/SQSTM1. J Immunol. 2011;186:1248–1258. doi: 10.4049/jimmunol.1001954. [DOI] [PubMed] [Google Scholar]
- 58.Mathew R, Karp CM, Beaudoin B, Vuong N, Chen G, et al. Autophagy suppresses tumorigenesis through elimination of p62. Cell. 2009;137:1062–1075. doi: 10.1016/j.cell.2009.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kim JE, You DJ, Lee C, Ahn C, Seong JY, et al. Suppression of NF-kappaB signaling by KEAP1 regulation of IKKbeta activity through autophagic degradation and inhibition of phosphorylation. Cell Signal. 2010;22:1645–1654. doi: 10.1016/j.cellsig.2010.06.004. [DOI] [PubMed] [Google Scholar]
- 60.Qing G, Yan P, Xiao G. Hsp90 inhibition results in autophagy-mediated proteasome-independent degradation of IkappaB kinase (IKK) Cell Res. 2006;16:895–901. doi: 10.1038/sj.cr.7310109. [DOI] [PubMed] [Google Scholar]
- 61.Qing G, Yan P, Qu Z, Liu H, Xiao G. Hsp90 regulates processing of NF-kappa B2 p100 involving protection of NF-kappa B-inducing kinase (NIK) from autophagy-mediated degradation. Cell Res. 2007;17:520–530. doi: 10.1038/cr.2007.47. [DOI] [PubMed] [Google Scholar]
- 62.Paul S, Kashyap AK, Jia W, He YW, Schaefer BC. Selective autophagy of the adaptor protein Bcl10 modulates T cell receptor activation of NF-kappaB. Immunity. 2012;36:947–958. doi: 10.1016/j.immuni.2012.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Castillo E, Dekonenko A, Arko-Mensah J, Mandell M, Dupont N, et al. Autophagy protects against active tuberculosis by suppressing bacterial burden and inflammation. Proc Natl Acad Sci USA. 2012;109:E3168–E3176. doi: 10.1073/pnas.1210500109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kim J, Lee H, Shin D, Kim W, Yuk J, et al. Host cell autophagy activated by antibiotics is required for their effective antimycobacterial drug action. Cell Host Microbe. 2012;11:457–468. doi: 10.1016/j.chom.2012.03.008. [DOI] [PubMed] [Google Scholar]
- 65.Deretic V. Autophagy in immunity and cell-autonomous defense against intracellular microbes. Immunol Rev. 2011;240:92–104. doi: 10.1111/j.1600-065X.2010.00995.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Deretic V. Autophagy: an emerging immunological paradigm. J Immunol. 2012;189:15–20. doi: 10.4049/jimmunol.1102108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Deretic V. Autophagy as an innate immunity paradigm: expanding the scope and repertoire of pattern recognition receptors. Curr Opin Immunol. 2012;24:21–31. doi: 10.1016/j.coi.2011.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xu Y, Jagannath C, Liu X, Sharafkhaneh A, Kolodziejska K, et al. Toll-like receptor 4 is a sensor for autophagy associated with innate immunity. Immunity. 2007;27:135–144. doi: 10.1016/j.immuni.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shi C, Kehrl J. TRAF6 and A20 regulate lysine 63-linked ubiquitination of Beclin-1 to control TLR4-induced autophagy. Sci Signal. 2010;3:ra42. doi: 10.1126/scisignal.2000751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Boone D, Turer E, Lee E, Ahmad R, Wheeler M, et al. The ubiquitin-modifying enzyme A20 is required for termination of Toll-like receptor responses. Nat Immunol. 2004;5:1052–1060. doi: 10.1038/ni1110. [DOI] [PubMed] [Google Scholar]
- 71.Into T, Inomata M, Takayama E, Takigawa T. Autophagy in regulation of Toll-like receptor signaling. Cell Signal. 2012;24:1150–1162. doi: 10.1016/j.cellsig.2012.01.020. [DOI] [PubMed] [Google Scholar]
- 72.Wild P, Farhan H, McEwan D, Wagner S, Rogov V, et al. Phosphorylation of the autophagy receptor optineurin restricts Salmonella growth. Science. 2011;333:228–233. doi: 10.1126/science.1205405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.von Muhlinen N, Akutsu M, Ravenhill B, Foeglein Á, Bloor S, et al. LC3C, bound selectively by a noncanonical LIR motif in NDP52, is required for antibacterial autophagy. Mol Cell. 2012;48:329–342. doi: 10.1016/j.molcel.2012.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Thurston T, Wandel M, von Muhlinen N, Foeglein A, Randow F. Galectin 8 targets damaged vesicles for autophagy to defend cells against bacterial invasion. Nature. 2012;482:414–418. doi: 10.1038/nature10744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pilli M, Arko-Mensah J, Ponpuak M, Roberts E, Master S, et al. TBK-1 promotes autophagy-mediated antimicrobial defense by controlling autophagosome maturation. Immunity. 2012;37:223–234. doi: 10.1016/j.immuni.2012.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jounai N, Kobiyama K, Shiina M, Ogata K, Ishii K, et al. NLRP4 negatively regulates autophagic processes through an association with beclin1. J Immunol. 2011;186:1646–1655. doi: 10.4049/jimmunol.1001654. [DOI] [PubMed] [Google Scholar]
- 77.Franchi L, Kamada N, Nakamura Y, Burberry A, Kuffa P, et al. NLRC4-driven production of IL-1β discriminates between pathogenic and commensal bacteria and promotes host intestinal defense. Nat Immunol. 2012;13:449–456. doi: 10.1038/ni.2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Broz PNK, Lamkanfi M, Mariathasan S, Dixit VM, et al. Redundant roles for inflammasome receptors NLRP3 and NLRC4 in host defense against Salmonella. J Exp Med. 2010;207:1745–1755. doi: 10.1084/jem.20100257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sanada T, Kim M, Mimuro H, Suzuki M, Ogawa M, et al. The Shigella flexneri effector OspI deamidates UBC13 to dampen the inflammatory response. Nature. 2012;483:623–626. doi: 10.1038/nature10894. [DOI] [PubMed] [Google Scholar]
- 80.Ponpuak M, Davis AS, Roberts EA, Delgado MA, Dinkins C, et al. Delivery of cytosolic components by autophagic adaptor protein p62 endows autophagosomes with unique antimicrobial properties. Immunity. 2010;32:329–341. doi: 10.1016/j.immuni.2010.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kim B, Shenoy A, Kumar P, Das R, Tiwari S, et al. A family of IFN-γ-inducible 65-kD GTPases protects against bacterial infection. Science. 2011;332:717–721. doi: 10.1126/science.1201711. [DOI] [PubMed] [Google Scholar]
- 82.Campbell G, Spector SA. Vitamin D inhibits human immunodeficiency virus type 1 and Mycobacterium tuberculosis infection in macrophages through the induction of autophagy. PLoS Pathog. 2012;8:e1002689. doi: 10.1371/journal.ppat.1002689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Dupont N, Temime-Smaali N, Lafont F. How ubiquitination and autophagy participate in the regulation of the cell response to bacterial infection. Biol Cell. 2010;102:621–634. doi: 10.1042/BC20100101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tang D, Kang R, Cheh C, Livesey K, Liang X, et al. HMGB1 release and redox regulates autophagy and apoptosis in cancer cells. Oncogene. 2010;29:5299–5310. doi: 10.1038/onc.2010.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tang D, Kang R, Livesey K, Cheh C, Farkas A, et al. Endogenous HMGB1 regulates autophagy. J Cell Biol. 2010;190:881–892. doi: 10.1083/jcb.200911078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Unlu S, Tang S, Wang E, Martinez I, Tang D, et al. Damage associated molecular pattern molecule-induced microRNAs (DAMPmiRs) in human peripheral blood mononuclear cells. PLoS One. 2012;7:e38899. doi: 10.1371/journal.pone.0038899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Buchser W, Laskow T, Pavlik P, Lin H, Lotze M. Cell-mediated autophagy promotes cancer cell survival. Cancer Res. 2012;72:2970–2979. doi: 10.1158/0008-5472.CAN-11-3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Liang X, de Vera ME, Buchser WJ, Romo de Vivar Chavez A, Loughran P, et al. Inhibiting autophagy during interleukin 2 immunotherapy promotes long term tumor regression. Cancer Res. 2012;72:2791–2801. doi: 10.1158/0008-5472.CAN-12-0320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Petrovski G, Ayna G, Majai G, Hodrea J, Benko S, et al. Phagocytosis of cells dying through autophagy induces inflammasome activation and IL-1beta release in human macrophages. Autophagy. 2011;7:321–330. doi: 10.4161/auto.7.3.14583. [DOI] [PubMed] [Google Scholar]
- 90.Saitoh T, Fujita N, Jang MH, Uematsu S, Yang BG, et al. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1beta production. Nature. 2008;456:264–268. doi: 10.1038/nature07383. [DOI] [PubMed] [Google Scholar]
- 91.Harris J, Hartman M, Roche C, Zeng SG, O'Shea A, et al. Autophagy controls IL-1beta secretion by targeting pro-IL-1beta for degradation. J Biol Chem. 2011;286:9587–9597. doi: 10.1074/jbc.M110.202911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Nakahira K, Haspel JA, Rathinam VA, Lee SJ, Dolinay T, et al. Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. Nat Immunol. 2011;12:222–230. doi: 10.1038/ni.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhou R, Yazdi AS, Menu P, Tschopp J. A role for mitochondria in NLRP3 inflammasome activation. Nature. 2011;469:221–225. doi: 10.1038/nature09663. [DOI] [PubMed] [Google Scholar]
- 94.Peral de Castro C, Jones SA, Ni Cheallaigh C, Hearnden CA, Williams L, et al. Autophagy regulates IL-23 secretion and innate T cell responses through effects on IL-1 secretion. J Immunol. 2012;189:4144–4153. doi: 10.4049/jimmunol.1201946. [DOI] [PubMed] [Google Scholar]
- 95.Dupont N, Jiang S, Pilli M, Ornatowski W, Bhattacharya D, et al. Autophagy-based unconventional secretory pathway for extracellular delivery of IL-1beta. EMBO J. 2011;30:4701–4711. doi: 10.1038/emboj.2011.398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Collins C, De Mazière A, van Dijk S, Carlsson F, Klumperman J, et al. Atg5-independent sequestration of ubiquitinated mycobacteria. PLoS Pathog. 2009;5:e1000430. doi: 10.1371/journal.ppat.1000430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Mishra BB, Moura-Alves P, Sonawane A, Hacohen N, Griffiths G, et al. Mycobacterium tuberculosis protein ESAT-6 is a potent activator of the NLRP3/ASC inflammasome. Cell Microbiol. 2010;12:1046–1063. doi: 10.1111/j.1462-5822.2010.01450.x. [DOI] [PubMed] [Google Scholar]
- 98.Simeone R, Bobard A, Lippmann J, Bitter W, Majlessi L, et al. Phagosomal rupture by Mycobacterium tuberculosis results in toxicity and host cell death. PLoS Pathog. 2012;8:e1002507. doi: 10.1371/journal.ppat.1002507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Watson R, Manzanillo P, Cox J. Extracellular M. tuberculosis DNA targets bacteria for autophagy by activating the host DNA-sensing pathway. Cell. 2012;150:803–815. doi: 10.1016/j.cell.2012.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Carneiro LA, Travassos LH, Soares F, Tattoli I, Magalhaes JG, et al. Shigella induces mitochondrial dysfunction and cell death in nonmyleoid cells. Cell Host Microbe. 2009;5:123–136. doi: 10.1016/j.chom.2008.12.011. [DOI] [PubMed] [Google Scholar]
- 101.Shi C, Shenderov K, Huang N, Kabat J, Abu-Asab M, et al. Activation of autophagy by inflammatory signals limits IL-1β production by targeting ubiquitinated inflammasomes for destruction. Nat Immunol. 2012;13:255–263. doi: 10.1038/ni.2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Bodemann B, Orvedahl A, Cheng T, Ram R, Ou Y, et al. RalB and the exocyst mediate the cellular starvation response by direct activation of autophagosome assembly. Cell. 2011;144:253–267. doi: 10.1016/j.cell.2010.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lee HK, Lund JM, Ramanathan B, Mizushima N, Iwasaki A. Autophagy-dependent viral recognition by plasmacytoid dendritic cells. Science. 2007;315:1398–1401. doi: 10.1126/science.1136880. [DOI] [PubMed] [Google Scholar]
- 104.Zhou D, Kang KH, Spector SA. Production of interferon alpha by human immunodeficiency virus type 1 in human plasmacytoid dendritic cells is dependent on induction of autophagy. J Infect Dis. 2012;205:1258–1267. doi: 10.1093/infdis/jis187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Severa M, Giacomini E, Gafa V, Anastasiadou E, Rizzo F, et al. EBV stimulates TLR- and autophagy-dependent pathways and impairs maturation in plasmacytoid dendritic cells: implications for viral immune escape. Eur J Immunol. 2013;43:147–158. doi: 10.1002/eji.201242552. [DOI] [PubMed] [Google Scholar]
- 106.Manuse MJ, Briggs CM, Parks GD. Replication-independent activation of human plasmacytoid dendritic cells by the paramyxovirus SV5 requires TLR7 and autophagy pathways. Virology. 2010;405:383–389. doi: 10.1016/j.virol.2010.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Morris S, Swanson MS, Lieberman A, Reed M, Yue Z, et al. Autophagy-mediated dendritic cell activation is essential for innate cytokine production and APC function with respiratory syncytial virus responses. J Immunol. 2011;187:3953–3961. doi: 10.4049/jimmunol.1100524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Jounai N, Takeshita F, Kobiyama K, Sawano A, Miyawaki A, et al. The Atg5 Atg12 conjugate associates with innate antiviral immune responses. Proc Natl Acad Sci USA. 2007;104:14050–14055. doi: 10.1073/pnas.0704014104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Tal MC, Sasai M, Lee HK, Yordy B, Shadel GS, et al. Absence of autophagy results in reactive oxygen species-dependent amplification of RLR signaling. Proc Natl Acad Sci USA. 2009;106:2770–2775. doi: 10.1073/pnas.0807694106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ke PY, Chen SS. Activation of the unfolded protein response and autophagy after hepatitis C virus infection suppresses innate antiviral immunity in vitro. J Clin Invest. 2011;121:37–56. doi: 10.1172/JCI41474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lei Y, Wen H, Yu Y, Taxman DJ, Zhang L, et al. The mitochondrial proteins NLRX1 and TUFM form a complex that regulates type I interferon and autophagy. Immunity. 2012;36:933–946. doi: 10.1016/j.immuni.2012.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Hailey DW, Rambold AS, Satpute-Krishnan P, Mitra K, Sougrat R, et al. Mitochondria supply membranes for autophagosome biogenesis during starvation. Cell. 2010;141:656–667. doi: 10.1016/j.cell.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Yasukawa K, Oshiumi H, Takeda M, Ishihara N, Yanagi Y, et al. Mitofusin 2 inhibits mitochondrial antiviral signaling. Sci Signal. 2009;2:ra47. doi: 10.1126/scisignal.2000287. [DOI] [PubMed] [Google Scholar]
- 114.Rasmussen SB, Horan KA, Holm CK, Stranks AJ, Mettenleiter TC, et al. Activation of autophagy by alpha-herpesviruses in myeloid cells is mediated by cytoplasmic viral DNA through a mechanism dependent on stimulator of IFN genes. J Immunol. 2011;187:5268–5276. doi: 10.4049/jimmunol.1100949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Saitoh T, Fujita N, Hayashi T, Takahara K, Satoh T, et al. Atg9a controls dsDNA-driven dynamic translocation of STING and the innate immune response. Proc Natl Acad Sci USA. 2009;106:20842–20846. doi: 10.1073/pnas.0911267106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Wei H, Wei S, Gan B, Peng X, Zou W, et al. Suppression of autophagy by FIP200 deletion inhibits mammary tumorigenesis. Genes Dev. 2011;25:1510–1527. doi: 10.1101/gad.2051011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Zhu S, Cao L, Yu Y, Yang L, Yang M, et al. Inhibiting autophagy potentiates the anticancer activity of IFN1@/ IFNalpha in chronic myeloid leukemia cells. Autophagy. 2013;9:317–327. doi: 10.4161/auto.22923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ambjorn M, Ejlerskov P, Liu Y, Lees M, Jaattela M, et al. IFNB1/interferon-beta-induced autophagy in MCF-7 breast cancer cells counteracts its proapoptotic function. Autophagy. 2013;9:287–302. doi: 10.4161/auto.22831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Desai MM, Gong B, Chan T, Davey RA, Soong L, et al. Differential, type I interferon-mediated autophagic trafficking of hepatitis C virus proteins in mouse liver. Gastroenterology. 2011;141:674–685. doi: 10.1053/j.gastro.2011.04.060. 685.e671-e676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Buchser WJ, Laskow TC, Pavlik PJ, Lin HM, Lotze MT. Cell-mediated autophagy promotes cancer cell survival. Cancer Res. 2012;72:2970–2979. doi: 10.1158/0008-5472.CAN-11-3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ashida H, Mimuro H, Ogawa M, Kobayashi T, Sanada T, et al. Cell death and infection: a double-edged sword for host and pathogen survival. J Cell Biol. 2011;195:931–942. doi: 10.1083/jcb.201108081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Kuballa P, Nolte W, Castoreno A, Xavier R. Autophagy and the immune system. Annu Rev Immunol. 2012;30:611–646. doi: 10.1146/annurev-immunol-020711-074948. [DOI] [PubMed] [Google Scholar]
- 123.Pattingre S, Tassa A, Qu XP, Garuti R, Liang XH, et al. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell. 2005;122:927–939. doi: 10.1016/j.cell.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 124.Sansonetti PJ, Phalipon A, Arondel J, Thirumalai K, Banerjee S, et al. Caspase-1 activation of IL-1beta and IL-18 are essential for Shigella flexneri-induced inflammation. Immunity. 2000;12:581–590. doi: 10.1016/s1074-7613(00)80209-5. [DOI] [PubMed] [Google Scholar]
- 125.Mounier J, Boncompain G, Senerovic L, Lagache T, Chretien F, et al. Shigella effector IpaB-induced cholesterol relocation disrupts the Golgi complex and recycling network to inhibit host cell secretion. Cell Host Microbe. 2012;12:381–389. doi: 10.1016/j.chom.2012.07.010. [DOI] [PubMed] [Google Scholar]
- 126.Suzuki T, Franchi L, Toma C, Ashida H, Ogawa M, et al. Differential regulation of caspase-1 activation, pyroptosis, and autophagy via Ipaf and ASC in Shigella-infected macrophages. PLoS Pathog. 2007;3:e111. doi: 10.1371/journal.ppat.0030111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Tattoli I, Sorbara M, Vuckovic D, Ling A, Soares F, et al. Amino acid starvation induced by invasive bacterial pathogens triggers an innate host defense program. Cell Host Microbe. 2012;11:563–575. doi: 10.1016/j.chom.2012.04.012. [DOI] [PubMed] [Google Scholar]
- 128.Homer C, Kabi A, Marina-García N, Sreekumar A, Nesvizhskii A, et al. A dual role for receptor-interacting protein kinase 2 (RIP2;) kinase activity in nucleotide-binding oligomerization domain 2 (NOD2)-dependent autophagy. J Biol Chem. 2012;287:25565–25576. doi: 10.1074/jbc.M111.326835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Jia K, Hart A, Levine B. Autophagy genes protect against disease caused by polyglutamine expansion proteins in Caenorhabditis elegans. Autophagy. 2007;3:21–25. doi: 10.4161/auto.3528. [DOI] [PubMed] [Google Scholar]
- 130.Sarkar S, Krishna G, Imarisio S, Saiki S, O'Kane C, et al. A rational mechanism for combination treatment of Huntington's disease using lithium and rapamycin. Hum Mol Genet. 2008;17:170–178. doi: 10.1093/hmg/ddm294. [DOI] [PubMed] [Google Scholar]
- 131.Mesquita F, Thomas M, Sachse M, Santos A, Figueira R, et al. The Salmonella deubiquitinase SseL inhibits selective autophagy of cytosolic aggregates. PLoS Pathog. 2012;8:e1002743. doi: 10.1371/journal.ppat.1002743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Rytkonen A, Poh J, Garmendia J, Boyle C, Thompson A, et al. SseL, a Salmonella deubiquitinase required for macrophage killing and virulence. Proc Natl Acad Sci USA. 2007;104:3502–3507. doi: 10.1073/pnas.0610095104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Le Clec'h W, Braquart-Varnier C, Raimond M, Ferdy J, Bouchon D, et al. High virulence of Wolbachia after host switching: when autophagy hurts. PLoS Pathog. 2012;8:e1002844. doi: 10.1371/journal.ppat.1002844. [DOI] [PMC free article] [PubMed] [Google Scholar]