Abstract
Tumor necrosis factor receptor-associated factor (TRAF) signaling plays a central role in many biological activities, such as the regulation of immune and inflammatory responses and control of apoptosis, which are key events in the pathogenesis of the human immunodeficiency virus (HIV)-1 and the hepatitis C virus (HCV) infections. Here we show that TRAF2, TRAF5 and TRAF6 interact with the HIV-1 Nef protein, an immunomodulatory viral protein expressed and released by cells infected by the virus. We also found that TRAF2 and TRAF5 interact with the HCV Core protein. Interestingly, we observed that HIV-1 Nef interacts with HCV Core. The activation of TRAF (2, 5, 6) - mediated by HIV-1 Nef and HCV Core - enhanced the activation of the nuclear factor-kappa B (NF-κB) and increased HIV-1 replication in monocyte- derived macrophages (MDMs). The knockdown of TRAF2, TRAF5 and TRAF6 resulted in decreased NF-κB activation and reduced HIV-1 replication in MDMs. Our results reveal a mechanism by which the activation of the TRAF pathway by HIV-1 Nef and HCV Core favors the replication of HIV-1 in macrophages and could be a critical factor for optimal replication of HIV-1 in macrophages of HIV-HCV-coinfected patients.
Key Words: Human immunodeficiency virus, Hepatitis C virus, Macrophage, Nef protein, Core protein
Introduction
Tumor necrosis factor (TNF)-α exerts a variety of biological effects, including the production of inflammatory cytokines, the upregulation of adhesion molecules, proliferation, differentiation and apoptosis by binding to its cognate receptors, TNF receptor (TNFR) 1 and TNFR2 [1, 2]. One of the protein families that bind to both TNFRs is the TNF receptor-associated factors (TRAFs) [3, 4]. Up to now, seven members of the TRAF family have been described [4, 5]. All the TRAFs contain a C-terminal TRAF domain that mediates the interaction with TNF receptors and hetero- or homodimerization among the TRAF family members [3]. In addition to the TRAF domain, the N-terminus of the TRAF proteins, with the exception of TRAF1, contains a RING domain and multiple Zinc finger structures, which are essential for their effector functions [3]. The TRAF family not only mediates TNF receptor family-mediated signaling but may also link the downstream signaling via other receptors like the interleukin-1 receptor [6]. TRAFs interact with the nuclear factor-kappa B (NF-κB)-inducing kinase, the MAP kinase (MAPK)/ERK kinase kinase 1, the transforming growth factor β-activated kinase, or the atypical protein kinase C, and these kinases phosphorylate IKKs resulting in the activation of the NF-κB [7, 8].
TRAF proteins are thought to be important regulators of cell death and cellular responses to stress, and TRAF2, TRAF5 and TRAF6 have been demonstrated to mediate activation of NF-κB and JNK [3]. Among the TRAF members, TRAF2 is a prototypical member of the TRAF family and transduces signals from the TNFR superfamily members [9]. Double knockouts of TRAF2 and TRAF5 suggest some functional redundancy between these two molecules in the context of TNFα-induced NF-κB activation and HIV-1 gene expression [9, 10, 11, 12]. Whereas TRAF5-null mouse embryonic fibroblasts respond normally to TNFα-induced JNK and NF-κB activation, TRAF2 and TRAF5 double knockout mouse embryonic fibroblasts exhibit an almost complete loss of TNFα-induced NF-κB activation [9]. TRAF6 not only mediates the signaling from the members of the TNF receptor superfamily, but also from the members of the Toll/interleukin-1 family, and interacts with various protein kinases including IRAK1/IRAK, SRC and PKCzeta, which provides a link between distinct signaling pathways [13]. TRAF6 also functions as a signal transducer in the NF-κB pathway that activates IKK in response to proinflammatory cytokines [3]. TRAF3 is a highly versatile regulator that positively controls the production of type I interferon, but negatively regulates the activation of MAPK and alternative NF-κB signaling [4]. The molecular mechanisms of action of TRAF4 remain evasive as it was found to interact with diverse types of proteins, leading either to proapoptotic or antiapoptotic functions [14]. TRAF1 can associate with multiple TNFR family members and can also bind several protein kinases and adaptor proteins, suggesting that it likely possesses multiple functions in cytokine signaling networks [15]. TRAF7 is the last member of the TRAF family that has been identified and recent data indicate that TRAF7 regulates the activation of cellular stress pathways, as well as unconventional ubiquitination events and differentiation of muscle tissue [5]. TRAF2, TRAF3 and TRAF6 are E3 ubiquitine ligases that play a pivotal role in the control of NF-κB activation by innate and adaptive immunity stimuli [16].
The Nef protein of human immunodeficiency virus (HIV)-1 is an important factor in AIDS pathogenesis. In addition to downregulating CD4 and the major histocompatibility complex class I molecules from the cell surface, as well as increasing virion infectivity, Nef triggers the activation of the T cell receptor cascade to facilitate the spread of the virus and modulates cellular signal transduction pathways [17]. Besides endogenous Nef protein expressed within infected cells, the exogenous Nef protein is detected in the serum of HIV-infected subjects [18]. The exogenous Nef protein has been shown to enter the cell by absorptive endocytosis after binding to the surface of CD4+ T cells, primary macrophages and U937 promonocytic cells [19], and to activate the signal transducer and activator of transcription 1 and NF-κB in human monocytes/macrophages [20, 21]. HIV-1 replication is tightly regulated at the transcription level through the specific interaction of viral regulatory proteins and cellular transcription factors, such as NF-κB, binding to the HIV long terminal repeat (LTR) [12].
Hepatitis C virus (HCV) infection is common in HIV-1-infected patients and each of these infections may affect the other [22]. Thus, several reports have found that HIV infection accelerates the development of severe liver diseases [23]. An association between HCV coinfection and progression of the HIV disease has also been reported [24, 25]. Among HCV structural proteins, the HCV Core protein interacts with several cellular components, thereby influencing lipid metabolism, signal transmission, and the regulation of gene expression and transcription from several viral and cellular promoters [26, 27]. HCV Core affects cell proliferation, apoptosis and host defense mechanisms by interfering with both innate and adaptive immunity [27]. Besides endogenous HCV Core expressed within infected hepatocytes, exogenous HCV Core is detected in the serum of HCV-infected subjects [28, 29]. HCV Core has been shown to enter the cell and to activate both MAPK and NF-κB signaling in several cell types [30, 31].
Since HIV Nef, HCV Core and several of the TRAF proteins activate NF-κB, we investigated their respective role in HIV-1 replication in the macrophage, which is a cell type infected by HIV-1 and a potential target for exogenous HCV Core in HIV/HCV-coinfected patients. We observed that HIV-1 Nef binds to HCV Core and that the two proteins recruit TRAF2, TRAF5 and TRAF6, then activating NF-κB and enhancing the replication of HIV-1 in monocyte-derived macrophages (MDMs).
Materials and Methods
Reagents
The recombinant myristoylated Nef protein derived from the SF-2 HIV-1 strain (HIV-1 rNef) was purchased from Jena Bioscience (Jena, Germany)and the recombinant HCV Core protein (HCV rCore) was purchased from US Biological (Swampscott, Mass., USA). Anti-p50, anti-p65, anti-RelB, anti-c-Rel, anti-p52, and the single-stranded NF-κB oligonucleotide and mutated oligonucleotide were purchased from Santa Cruz Biotechnology (Santa Cruz, Calif., USA). Anti-IκBα, anti-IKKα, anti-IKKβ, and anti-pIKKβ/pIKKα antibodies were purchased from Cell Signaling Technologies (Beverly, Mass., USA), and peroxidase-conjugated secondary anti-rabbit and anti-mouse immunoglobulin were obtained from Jackson ImmunoResearch (West Grove, Pa., USA). Abs against HIV-1 Nef and HCV Core proteins were provided by Chemicon (Temecula, Calif., USA) and US Biological, respectively. TNFα was purchased from R&D Systems (Minneapolis, Minn., USA). Abs against TRAF1, TRAF2, TRAF3 and TRAF6 were provided by Cell Signaling Technologies. Abs against TRAF4 and TRAF5 were purchased from Santa Cruz Biotechnology.
Cell Culture
The studies were performed with the promonocytic cells U937 obtained from the American Tissue Cell Culture Collection (Manassas, Va., USA). The promonocytic cell line U1, derived from cells surviving acute infection of the U937 cell line, contains two integrated HIV copies per cells. U1 cells were a gift from Dr. C. Van Lint (Université Libre de Bruxelles, Belgium). U937 and U1 cells were cultivated in RPMI-1640 (Sigma-Aldrich, St. Louis, Mo., USA) supplemented with 10% fetal bovine serum (Sigma-Aldrich), 1% L-glutamine and 1% penicillin/streptomycin. Primary MDMs (>94% CD14+ by flow cytometric analysis) were prepared from peripheral blood of healthy donors and were cultured in RPMI-1640 medium supplemented with 10% (v/v) pooled AB human serum (Sigma-Aldrich), as previously described [32]. Peripheral blood mononuclear cells (PBMCs), monocytes and peripheral blood lymphocytes (PBLs) were isolated from fresh whole blood (10 ml) of viremic HIV-1-infected patients (n = 13), HCV-infected patients (n = 6) and HIV-1-HCV-coinfected individuals (n = 6) followed up at the Besancon University Hospital, as reported previously [33, 34]. Ethical approval was granted by the human subjects ethics committee (Comité de Protection des Personnes EST-II).
GST Pull-Down Assay
HIV-1 Nef wild type, N-terminus (1-60), or C-terminus (55-206) was cloned into pGEX 4T-1 expression vector (kindly provided by Dr. Fackler, University of Heidelberg, Germany) and GST Core constructs were kindly provided by Dr. N. Pavio, INSERM U785, Villejuif, France. For the GST pull-down assay, each construct was transformed into bacteria (BL21DE from Novagen, Bad Soden, Germany). A single colony was inoculated in 5 ml LB broth containing 50 μg/ml ampicillin at 37°C, 250 rpm in a shaker overnight. A 1-ml aliquot of the overnight bacterial culture was used to inoculate 300 ml LB broth containing 50 μg/ml ampicillin and grown for 2.5 h at 37°C in a shaker (until OD600 = 0.5). IPTG was added at a final concentration of 0.1 mM and the culture grown for an additional 3 h at 37°C in the shaker. The bacteria were pelleted, washed with ice-cold TBS and lysed in bacterial protein extraction reagents (Pierce, Rockford, Ill., USA). The bacterial-expressed protein was immobilized on glutathione beads and washed five times with 1 × PBS. Twenty micrograms of purified GST fusion proteins were incubated with 1,500 µg protein lysates overnight at 4°C with gentle shaking. Following incubation the reaction mixture was centrifuged and washed five times in 1 × PBS. After a final wash, the bound protein was eluted by boiling in 2 × SDS-PAGE sample buffer and then subjected to SDS-PAGE and immunoblotted.
Western Blot
Cellular extracts of cells treated with HIV-1 rNef and/or rHCV Core were used to examine the different protein expression by Western blot according to previously described procedures [21]. Cellular extracts were resolved by 10% SDS-PAGE using a Mini-PROTEAN 3 Cell (Bio-Rad, Hercules, Calif., USA). The proteins were electrotransferred onto a PVDF membrane (Amersham Biosciences, Little Chalfont, UK) using Mini Trans-Blot Electrophoretic Transfer Cell (Bio-Rad). The membranes were probed with primary antibodies (Abs) followed by HRP-conjugated secondary immunoglobulin raised against the appropriate species; bands were detected using the ECL Plus kit (Amersham Biosciences).
Immunoprecipitation
MDMs were left untreated or were treated with HIV-1 rNef (100 ng/ml) and HCV rCore (100 ng/ml) for 30 min. Alternatively, MDMs were transfected with a Nef-expressing plasmid and/or aHCV-Core-expressing plasmid for 24 h. For in vitro coinfections, MDMs were infected with HIV-89.6 (10 ng p24/4 × 105 cells) overnight, cells were washed and infected with HCV (19.106 IU/ml) and in total 5 × 106 cell lysates were prepared after 48 h. Cell lysates were precleared by adding 50 μl of Protein G Plus/Protein A-agarose (Calbiochem-Novabiochem, Bad Soden, Germany) for 1 h at 4°C. The cleared supernatants were removed, combined with 10 μg/ml anti-HIV-1 Nef Ab (Chemicon) or isotype control Ab and incubated overnight at 4°C. Immune complexes were washed in the presence of protease inhibitors and the bound proteins were eluted with sample buffer and run on 10% SDS-PAGE gels. SDS-PAGE and Western blot analysis using an anti-HCV Core Ab were performed according to standard procedure [21]. Western blots were developed with the ECL detection kit (Amersham Biosciences, Piscataway, N.J., USA).
Isolation of Nuclear and Cytoplasmic Extracts
Isolation of nuclear and cytoplasmic extracts was performed as previously described [21]. Cells were scraped from the plastic surface of the culture dishes and washed with wash buffer (10 mM HEPES, pH 7.6, 10 mM KCl, 2 mM MgCl2, 1 mM EDTA). Cell pellets were then incubated on ice using a cytoplasmic isolation buffer (10 mM HEPES, pH 7.6, 10 mM KCl, 2 mM MgCl2, 1 mM EDTA, 0.02% NP-40). Cytoplasmic extracts were collected by centrifugation and the nuclear pellets were washed twice in wash buffer, spun and incubated for 15 min on ice with nuclear isolation buffer (20 mM HEPES, pH 7.6, 420 mM NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 25% glycerol). Supernatants containing nuclear extracts were collected by centrifugation and stored at −80°C. Protease inhibitors (1 mM DTT, 1 mM PMSF, 1 µg/ml aprotinin, 1 µg/ml leupeptin, 1 µg/ml pepstatin) were added to all solutions. Protein concentration in nuclear and cytoplasmic extracts was determined by the Bradford method using a BioPhotometer (Eppendorf, Hamburg, Germany).
Electrophoretic Mobility Shift Assay
To measure NF-κB activation, electrophoretic mobility shift assay (EMSA) was carried out as previously described [21]. Briefly, the cells were treated with HIV-1 rNef and/or HCV rCore during different periods of time. Alternatively, PBMCs, monocytes and PBLs were isolated from fresh whole blood (10 ml) of HIV-1-infected patients, HCV-infected patients, HIV-1-HCV-coinfected individuals and normal healthy donors. Nuclear extracts prepared from cells were incubated with 20 fmol of biotin-end-labeled 45-mer double-stranded NF-κB oligonucleotide.
5′-TTGTTACAAGGGACTTTCCGCTGGGGACTTTCCAGGGAGGCGTGG-3′ (bold letters indicate NF-κB binding sites) in the presence of binding buffer [10 mM Tris, 50 mM KCl, 1 mM DTT at pH 7.5 and 50 ng/μl Poly (dI∙dC)]. A double-stranded mutated oligonucleotide 5′-TTGTTACAACTCACTTTCCGCTGCTCACTTTCCAGGGAG GCGTGG-3′ was used to examine the specificity of NF-κB binding to the DNA. The specificity of binding was also examined by competition with the unlabeled oligonucleotide and a heterologous unlabeled oligonucleotide. NF-κB oligonucleotide was labeled with biotin using the Biotin 3′ End DNA Labeling kit (Pierce) and complementary pairs were annealed by heating in boiling water for 5 min and then cooling slowly to room temperature. DNA protein complexes were resolved from free oligonucleotide on a 6% native polyacrylamide gel in 1 × Tris-borate-EDTA buffer using a Mini-PROTEAN 3 Cell (Bio-Rad) and were transferred to a Biodyne precut nylon membrane (Pierce) using the Mini Trans-Blot Electrophoretic Transfer Cell (Bio-Rad). Biotin-end-labeled DNA was detected using the LightShift Chemiluminescent EMSA kit (Pierce). Control Epstein-Barr Nuclear Antigen (EBNA) System (Pierce) containing biotin-EBNA control DNA and EBNA extract were assayed in parallel with the sample to ensure that the components of the kit and the overall procedure was working properly.
Reporter Gene Expression Assays
To examine the NF-κB LTR-driven gene expression following treatment with HIV-1 rNef and/or HCV rCore, U937 cells were transiently transfected with 20 µg of pLTR-Luc or 20 µg of pLTRmut-NF-κB-Luc using an electroporation system, according to the manufacturer's instructions (Bio-Rad) [21]. Twenty-four hours later, the cells were stimulated with HIV-1 rNef (100 ng/ml), HCV rCore (100 ng/ml) or HIV-1 rNef (100 ng/ml) + HCV rCore (100 ng/ml). At 48 h posttransfection, luciferase activity was measured in cell lysates using a luminometer (TD-20/20; Promega, Madison, Wisc., USA) as previously described [21]. Values normalized to protein concentrations were expressed in fold increase over unstimulated control values.
RNA Interference
MDM cultures (0.5 × 106 cells) were transfected with a scrambled control, TRAF1 siRNA, TRAF2 siRNA, TRAF3 siRNA, TRAF4 siRNA, TRAF5 siRNA and TRAF6 siRNA duplexes (Dharmacon RNAi Technologies) using lipofectamine RNAiMAX (Invitrogen, Carlsbad, Calif., USA). MDMs were treated with HIV-1 rNef and/or HCV rCore 48 h posttransfection, and EMSA and HIV-1 replication follow-up were performed as reported previously. The efficiency of transfection was monitored using a fluorescein-conjugated scrambled control duplex and exceeded 50% in MDMs.
Flow Cytometry Analysis
Following treatment of MDMs (>94% CD14+ by flow cytometric analysis) with rNef (100 ng/ml) and rCore (100 ng/ml) for 30 min, cells were fixed with 4% PFA for 15 min and permeabilized or not with cytofix/cytoperm solution (BD Bioscience, San Jose, Calif., USA). Cells were incubated for 1 h at room temperature with a goat anti-Nef Ab (Santa Cruz Biotechnology) or a mouse anti-HCV Core Ab (Euromedex, Souffelweyersheim, France). Cells were washed three times with a BD wash buffer. Nef and Core detections were performed after incubating with secondary Abs, PE-labeled anti-goat IgG and FITC-labeled anti-mouse IgG, respectively (Santa Cruz Biotechnology). FITC-conjugated mouse IgG (BD Bioscience) and PE-conjugated goat IgG (Santa Cruz Biotechnology) were used as isotype controls. Labeled cells were analyzed by flow cytometry with a FACSCalibur flow cytometer (Becton Dickinson, Franklin Lakes, N.J., USA). Data from 5 × 103 cells were collected, stored and analyzed with CellQuest software (Becton Dickinson).
Assessments of HIV-1 Replication
MDMs were infected overnight with HIV-1 89.6 (10 ng p24/4 × 105 cells). Cells were then washed three times with PBS to remove the unabsorbed inoculum and reincubated in fresh culture medium at 37°C. The infected MDMs were treated with HIV-1 rNef (100 ng/ml), HCV rCore (100 ng/ml) or HIV-1 rNef (100 ng/ml) + HCV rCore (100 ng/ml) added every 3 days. U1 cells were treated with native or boiled HIV-1 rNef (100-1,000 ng/ml) and/or HCV rCore (10-500 ng/ml). Where specified, a neutralizing anti-Nef mAb (10 μg/ml) and an anti-HCV Core mAb (10 μg/ml) were used. Culture supernatants were collected and assessed for p24 antigen using a microELISA assay (Organon Teknika, Boxtel, The Netherlands).
Statistical Analysis
The figures show the means of independent experiments and standard deviations. Statistical analyses were performed using the analysis of variance test and the Mann-Whitney U test. Differences were considered significant at a value of * p < 0.05, ** p < 0.01, *** p < 0.001. The program used for plotting was Microsoft Excel and SPSS (SPSS Inc., Chicago, Ill., USA) for statistical analysis.
Results
HIV-1 Nef Interacts with HCV Core
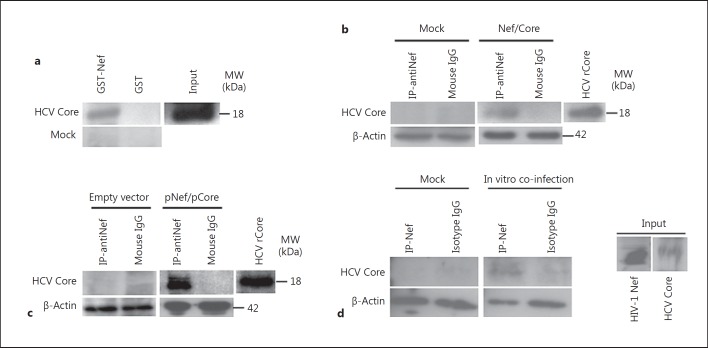
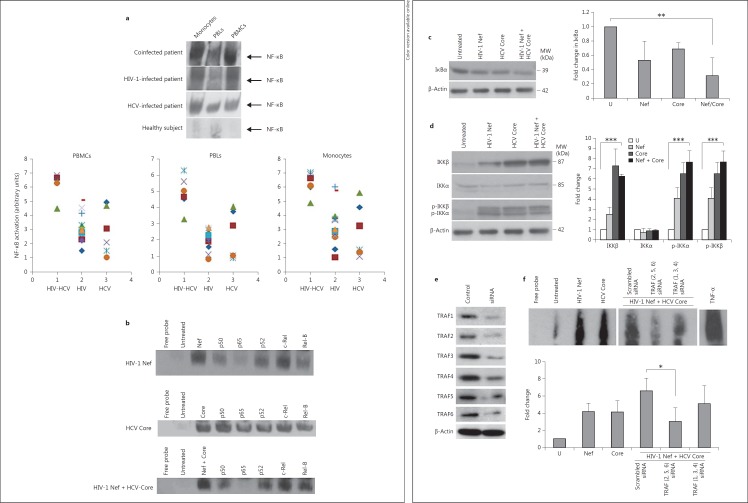
Since both HIV-1 Nef and HCV Core proteins have been reported to activate NF-κB in several cell types [21, 31], we assessed the potential HIV-1 Nef/HCV Core interaction. We expressed Nef as a GST fusion protein in Escherichia coli and tested its ability to interact with HCV rCore using U937 cells treated with HCV rCore as a source of lysates. HIV-1 Nef bound to HCV Core (fig. 1a). HIV-1 Nef coimmunoprecipitated with HCV Core in promonocytic U937 cells treated with the two recombinant viral proteins whereas the isotype control immunoprecipitation showed no associated HCV Core (fig. 1b). The interaction of HIV-1 Nef and HCV Core was also demonstrated by transient cotransfection of primary MDMs with pHIV-1 Nef and pHCV Core plasmids (fig. 1c). Following in vitro coinfection of MDMs with HIV-1 89.6 and with HCV, we observed the interaction of HIV-1 Nef and HCV Core by coimmunoprecipitation (fig. 1d). Our results indicate that the Nef-Core interaction does not only occur in the U937 cells treated with recombinant viral proteins, but can also be observed in MDMs infected with the two viruses, making it more relevant in regard to the HIV-HCV pathogenesis.
Fig. 1.
HIV-1 Nef and HCV Core interact. a Binding of HIV-1 Nef to HCV Core was measured in GST pull-down assays using U937 cells treated with HCV rCore (100 ng/ml) as a source of lysates. Input corresponds to 10% of the material used for pull-down. Untreated U937 cells (Mock) were used as a negative control. Results are representative of three independent experiments. b Total cellular extracts from U937 cells treated with HIV-1 rNef (100 ng/ml) and HCV rCore (100 ng/ml)for 30 min were immunoprecipitated with an anti-Nef Ab or with an isotype control Ab. Immunoprecipitated material was analyzed by Western blotting with an anti-HCV Core mAb. Untreated U937 cells (Mock) were used as a negative control. β-actin was used as a loading control. Results are representative of three independent experiments. c Cellular extracts of MDMs cotransfected for 48 h with a Nef-expressing plasmid (pNef) and a core-expressing plasmid (pCore) were immunoprecipitated with an anti-Nef Ab or isotype control Ab. Immunoprecipitated material was analyzed by Western blotting with an anti-HCV Core mAb. MDMs transfected with empty vectors were used as a negative control. β-actin was used as a loading control. Results are representative of two independent experiments. d Cellular extracts of MDMs coinfected in vitro with HIV-1 89.6 and with HCV, as described in Materials and Methods, were immunoprecipitated with an anti-Nef Ab or isotype control Ab. Immunoprecipitated material was analyzed by Western blotting with an anti-HCV Core mAb. Uninfected MDMs isolated from the peripheral blood of healthy subjects (Mock) was used as a negative control. β-actin was used as a loading control. Results are representative of two independent experiments.
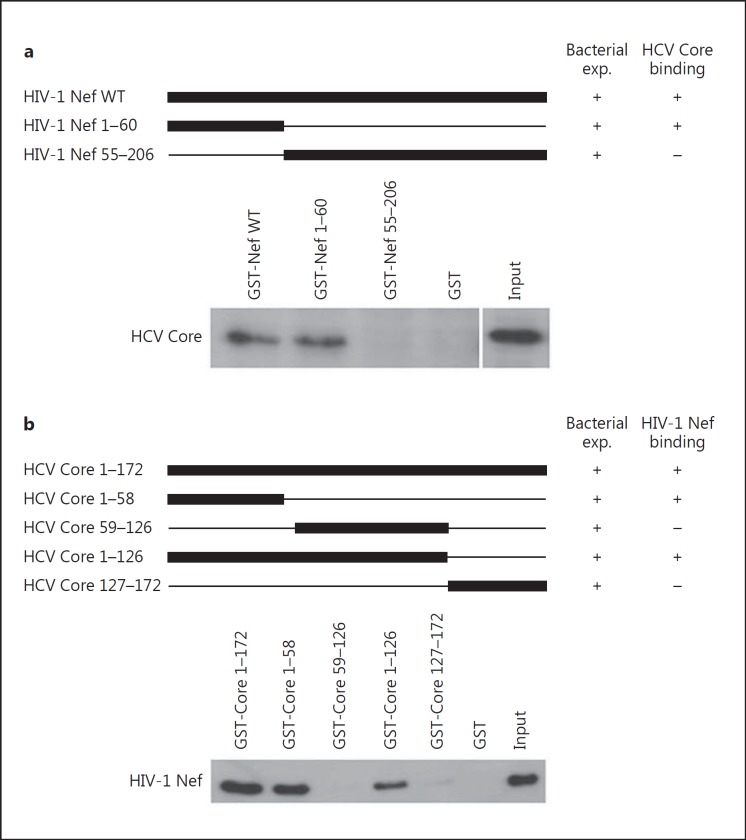
To determine the extremity of HIV-1 Nef that binds HCV Core, we tested GST-Nef plasmids truncated at their C-terminal or N-terminal extremity for their binding to HCV Core using a pull-down assay (fig. 2a). HCV binding capacity was totally annihilated in GST pull-down assay using a GST-tagged HIV-1 Nef construct truncated at its N-terminus (fig. 2a), indicating that HIV-1 Nef interacts with HCV Core via its N-terminal region.
Fig. 2.
The N-terminal region of HIV-1 Nef binds to the N-terminal region of HCV Core. a The N-terminal region of HIV-1 Nef (aa 1-60) is sufficient for binding to HCV Core. Upper panel: schematic diagram of GST-Nef constructs. Lower panel: HIV-1 rNef interacts with HCV Core via its N-terminus extremity (aa 1-60). Using WT and truncated GST-Nef constructs, the binding of HCV Core present in lysates of MDMs treated with HCV rCore (100 ng/ml) was measured in GST pull-down assays. Input corresponds to 10% of the material used for pull-down. Results are representative of two independent experiments. b The N-terminal region (aa 1-58) of HCV Core is sufficient for binding to HIV-1 Nef. Upper panel: schematic diagram of GST-HCV-core constructs. The names of the mutants are shown to the left; numbers refer to the amino acid residues retained by the deletion mutants. Lower panel: HCV Core interacts with HIV-1 Nef via its N-terminal region (aa 1-58). Using WT and mutated GST-HCV Core constructs, binding of HIV-1 Nef present in lysates of MDMs treated with rNef (100 ng/ml) was measured in GST pull-down assays. Input corresponds to 10% of the material used for pull-down. Results are representative of two independent experiments.
To determine the extremity of HCV Core that binds HIV-1 Nef, we tested several GST-HCV Core plasmids for their binding to HIV-1 Nef using a pull-down assay (fig. 2b). HIV-1 Nef binding capacity was totally annihilated in GST pull-down assay using GST-tagged HCV Core constructs truncated upstream of residue 59 (fig. 2b), indicating that HCV Core interacts with HIV-1 Nef via its N-terminal region (residues 1-58).
HIV-1 Nef and HCV Core Recruit TRAF2, TRAF5 and TRAF6
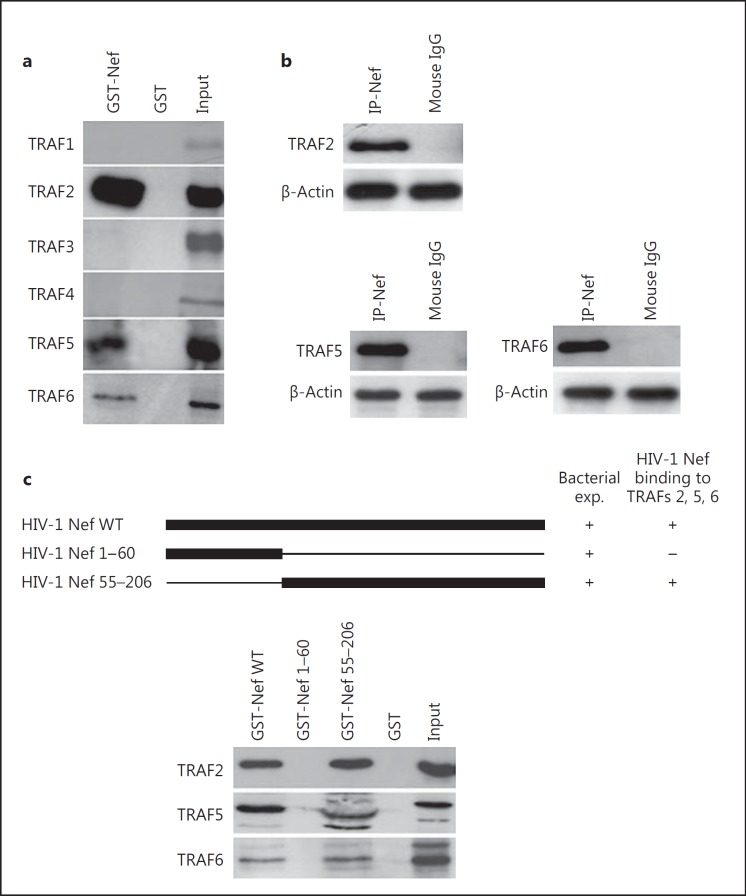
Since TRAF proteins play a critical role in the signal transduction delivered through the CD40 receptor and TNFRs, we investigated the potential interaction of HIV-1 Nef and HCV Core with TRAF proteins. Therefore, we tested the ability of GST-Nef fusion proteins to interact with TRAF proteins present in MDM lysates. HIV-1 Nef bound to TRAF2, TRAF5 and TRAF6 proteins (fig. 3a). HIV-1 Nef did not interact with other TRAF members (TRAF1, TRAF3, TRAF4; fig. 3a), indicating that the HIV-1 Nef/TRAF (2, 5, 6) interaction was specific. Endogenous TRAF2, TRAF5 and TRAF6 were coimmunoprecipitated with HIV-1 Nef in U937 cells treated with rNef (fig. 3b). Other TRAF members (TRAF1, TRAF3, TRAF4) did not interact with HIV-1 Nef in U937 cells treated with rNef (data not shown), indicating that HIV-1 Nef/TRAF (2, 5, 6) interaction was specific.
Fig. 3.
The C-terminal region of HIV-1 Nef binds to TRAF2, TRAF5 and TRAF6. a HIV-1 Nef binds to TRAF2, TRAF5 and TRAF6. Using a WT GST-Nef construct, binding of TRAF1 to TRAF6 present in lysates of MDMs was measured in GST pull-down assays. Input corresponds to 10% of the material used for pull-down. Results are representative of two independent experiments. b Total cellular extracts from U937 cells treated with HIV-1 rNef (100 ng/ml)for 30 min were immunoprecipitated with an anti-HIV-1 Nef mAb or with an isotype control Ab. Immunoprecipitated material was analyzed by Western blotting with anti-TRAF2, anti-TRAF5 and anti-TRAF6 mAbs. β-actin was used as a loading control. Results are representative of three independent experiments. c The C-terminal region of HIV-1 Nef (aa 55-206) is sufficient for binding to TRAF2, TRAF5 and TRAF6. Using WT and truncated GST-Nef constructs, binding of TRAF2, TRAF5 and TRAF6 present in lysates of MDMs was measured in GST pull-down assays. Input corresponds to 10% of the material used for pull-down. Results are representative of two independent experiments.
To determine the extremity of HIV-1 Nef that binds TRAF proteins, we tested GST-Nef plasmids truncated at their C-terminal or N-terminal extremity for their binding to TRAF proteins using a pull-down assay (fig. 3c). HIV-1 Nef binding capacity was totally annihilated in GST pull-down assay using a GST-tagged HIV-1 Nef construct truncated at its C-terminus (fig. 3c), indicating that HIV-1 Nef interacts with TRAF2, TRAF5 and TRAF6 via its C-terminal region (residues 55-206).
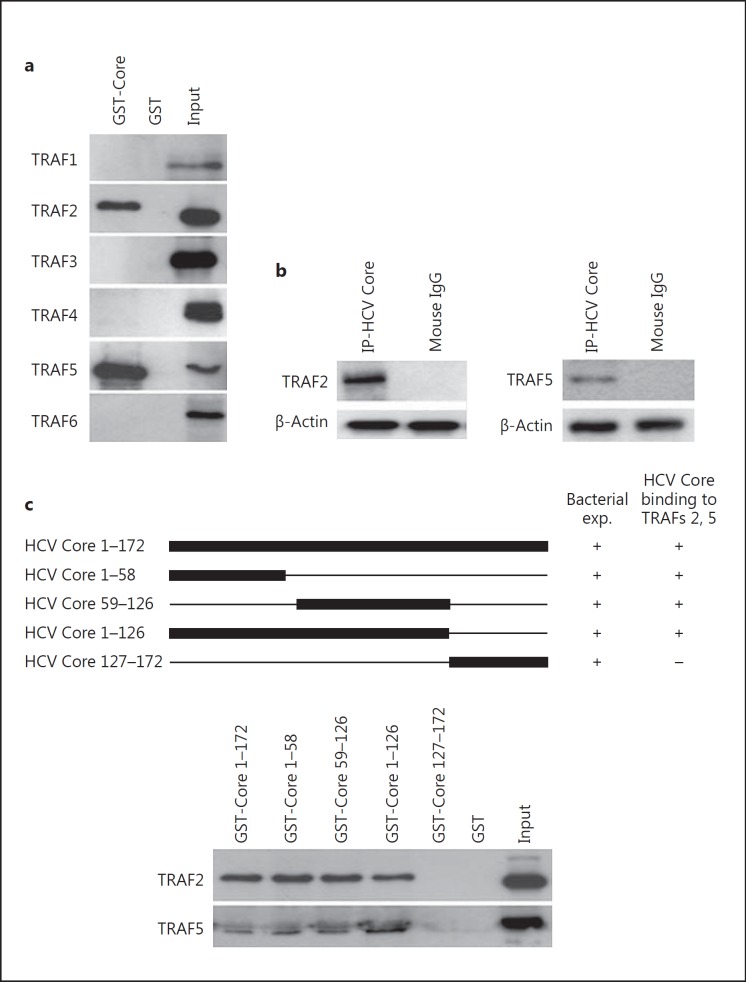
We also tested the ability of GST-HCV Core fusion proteins to interact with TRAF proteins present in MDM lysates. HCV Core bound to TRAF2 and TRAF5 proteins (fig. 4a). HCV Core did not interact with other TRAF members (TRAF1, TRAF3, TRAF4, TRAF6; fig. 4a), indicating that HCV Core/TRAF (2, 5) interaction was specific. Endogenous TRAF2 and TRAF5 were coimmunoprecipitated with HCV Core in U937 cells treated with HCV rCore (fig. 4b). Other TRAF members (TRAF1, TRAF3, TRAF4, TRAF6) did not interact with HCV Core in U937 cells treated with HCV rCore (data not shown), which indicates that the HCV Core/TRAF (2, 5) interaction was specific. To determine the extremity of HCV Core that binds TRAF2 and TRAF5 proteins, we tested GST-HCV Core plasmids truncated at their C-terminal or N-terminal extremity for their binding to TRAF2 and TRAF5 proteins using a pull-down assay (fig. 4c). HCV Core binding capacity was totally annihilated during the GST pull-down assay using a GST-tagged HCV Core construct truncated at its N-terminus (GST Core 127-172; fig. 4c), indicating that HCV Core interacts with TRAF2 and TRAF5 via its N-terminal region (residues 1-126). We also observed an upregulated expression of TRAF2 and TRAF6 in MDMs treated with HIV-1 rNef and HCV rCore (p < 0.01 and p < 0.001, respectively; fig. 5).
Fig. 4.
The N-terminal region of HCV Core (residues 1-126) binds to TRAF2 and TRAF5. a HCV Core binds to TRAF2 and TRAF5. Using a WT GST-HCV Core construct, binding of TRAF1 to TRAF6 present in lysates of MDMs was measured in GST pull-down assays. Input corresponds to 10% of the material used for pull-down. Results are representative of two independent experiments. b Total cellular extracts from U937 cells treated with HCV rCore (100 ng/ml)for 30 min were immunoprecipitated with an anti-HCV Core mAb or with an isotype control Ab. Immunoprecipitated material was analyzed by Western blotting with anti-TRAF2 and anti-TRAF5 mAbs. β-actin was used as a loading control. Results are representative of three independent experiments. c The N-terminal region of HCV Core (aa 1-126) is sufficient for binding to TRAF2 and TRAF5. Using WT and truncated GST-HCV Core constructs, binding of TRAF2 and TRAF5 present in lysates of MDMs was measured in GST pull-down assays. Input corresponds to 10% of the material used for pull-down. Results are representative of two independent experiments.
Fig. 5.
Upregulation of TRAF2 and TRAF6 in MDMs treated with HIV-1 rNef and HCV rCore. Total cellular extracts from MDMs untreated or treated with HIV-1 rNef (100 ng/ml), HCV rCore (100 ng/ml) or HIV-1 rNef (100 ng/ml) and HCV rCore (100 ng/ml)for 30 min were analyzed by Western blotting with anti-TRAF1-TRAF6 mAbs. β-actin was used as a loading control. Protein levels of TRAFs were quantified by densitometry using ImageJ 1.40 software (the level of TRAFs in mock cells was arbitrarily established at 1). Results represent means ± SD of threeindependent experiments. ** p < 0.01, *** p < 0.001.
HIV-1 Nef and HCV Core Activate the NF-κB Canonical Pathway in Primary Macrophages through a TRAF (2, 5, 6) Pathway
We compared the level of NF-κB activation in autologous PBMCs, monocytes and PBLs isolated from the peripheral blood of HIV-infected patients (n = 13), HCV-infected patients (n = 6) and HIV-HCV-coinfected patients (n = 6) (table 1). The highest levels of NF-κB activation were observed in peripheral blood cells of HIV-HCV-coinfected subjects, mostly in PBMCs, monocytes and to a lesser extent in PBLs (fig. 6a; table 1). When compared to coinfected subjects, 2-fold lower levels of NF-κB activation were measured in peripheral blood cells of HIV-infected subjects and HCV-infected subjects, in particular in monocytes (p < 0.05; fig. 6a; table 1). Since HIV-1 Nef and HCV Core are independent activators of NF-κB in several cell types [21, 31], we investigated their respective role on NF-κB activation in MDMs. Treatment of MDMs with HIV-1 Nef and/or HCV Core resulted in NF-κB activation (fig. 6b). The gel shift bands were specific; the formation of the complex was diminished by inclusion of an unlabeled NF-κB oligonucleotide but not by inclusion of a mutated NF-κB oligonucleotide (data not shown). To identify the subunits in the NF-κB complexes, nuclear extracts from MDMs treated with rNef, rCore or the two recombinant proteins together were incubated with Abs against different NF-κB subunits and DNA-binding activity was assayed by EMSA. Interference of NF-κB activation by anti-p65 and anti-p50 Ab suggested that p65 and p50 were the major components of the activated NF-κB complex in MDMs treated with rHIV-1 Nef and rHCV Core (fig. 6b). In agreement with the activation of the canonical p65/p50 NF-κB complex in MDMs treated with rHIV-1 Nef and rHCV Core, we observed the degradation of IkBα following treatment with the two viral proteins alone or together, as typically occurs during the activation of the canonical NF-κB pathway (fig. 6c). We observed an increased expression of IKKβ in MDMs treated with rHIV-1 Nef and rHCV Core alone and together (fig. 6d). In addition both IKKα and IKKβ were highly phosphorylated in MDMs treated with rHIV-1 Nef and rHCV Core alone or together as compared with untreated MDMs (fig. 6d). Phosphorylation of IKKγ was not observed in either treatment (data not shown).
Table 1.
NF-κB activation (arbitrary units) measured in PBMCs, monocytes and PBLs isolated from the peripheral blood of coinfected patients, HIV-infected patients and HCV-infected patients
| PBMCs | Monocytes | PBLs | ||
|---|---|---|---|---|
| Coinfected patients (n = 6) | 1 | 6.33 | 6.04 | 4.56 |
| 2 | 6.68 | 6.65 | 4.66 | |
| 3 | 4.53 | 4.92 | 3.29 | |
| 4 | 6.88 | 6.94 | 5.61 | |
| 5 | 6.45 | 7.06 | 6.30 | |
| 6 | 6.32 | 6.12 | 5.04 | |
| Mean | 6.20 | 6.29 | 4.91 | |
| HIV-infected patients (n = 13) | 7 | 1.55 | 1.65 | 1.59 |
| 8 | 2.37 | 1.05 | 1.92 | |
| 9 | 3.32 | 3.98 | 2.47 | |
| 10 | 2.50 | 3.68 | 1.09 | |
| 11 | 3.36 | 3.78 | 2.61 | |
| 12 | 2.95 | 2.49 | 0.82 | |
| 13 | 4.18 | 6.06 | 1.92 | |
| 14 | 5.13 | 5.81 | 1.78 | |
| 15 | 3.18 | 2.70 | 2.40 | |
| 16 | 2.25 | 2.75 | 2.12 | |
| 17 | 2.83 | 2.92 | 2.30 | |
| 18 | 2.98 | 3.04 | 2.76 | |
| 19 | 4.57 | 2.59 | 2.75 | |
| Mean | 3.16 | 3.26 | 2.04 | |
| HCV-infected patients (n = 6) | 20 | 4.98 | 4.62 | 3.79 |
| 21 | 3.13 | 3.28 | 2.90 | |
| 22 | 4.72 | 5.63 | 4.06 | |
| 23 | 2.13 | 1.13 | 1.04 | |
| 24 | 1.55 | 1.59 | 0.90 | |
| 25 | 1.09 | 1.44 | 1.03 | |
| Mean | 2.93 | 2.94 | 2.28 |
Fig. 6.
Enhanced NF-κB activation in MDMs treated with HIV-1 Nef and HCV Core is mediated through TRAF2, TRAF5 and TRAF6. a Highest NF-κB activation in monocytes and PBMCs isolated from HIV-HCV-coinfected subjects. Upper panel: data are representative of NF-κB activation determined by EMSA in PBMCs, monocytes and PBLs isolated from the peripheral blood of coinfected patients (n = 6), HIV-infected patients (n = 13), HCV-infected patients (n = 6) and normal healthy donors (n = 3). Lower panel: NF-κB activation measured by EMSA and quantified by densitometry using ImageJ 1.40 software in PBMCs, monocytes and PBLs isolated from the peripheral blood of HIV-HCV-coinfected patients (n = 6), HIV-infected patients (n = 13), and HCV-infected patients (n = 6). The level of NF-κB in cells of healthy donors was arbitrarily established at 1. b Enhanced NF-κB activation in MDMs treated with HIV-1 rNef, HCV rCore and HIV-1 rNef + HCV rCore. MDM cultures were treated with HIV-1 rNef (100 ng/ml), HCV rCore protein (100 ng/ml) or HIV-1 rNef (100 ng/ml) + HCV rCore protein (100 ng/ml) and nuclear extracts were prepared at 30 min posttreatment. NF-κB activation was determined by EMSA. Interference of NF-κB activation in response to HIV-1 rNef and HCV rCore by Abs against NF-κB subunits: nuclear extracts from treated MDMs were incubated for 20 min with anti-p50, anti-p65, anti-p52, anti-c-Rel and anti-RelB Abs, and then assayed for NF-κB binding activity by EMSA. Results are representative of two independent experiments. c IkBα degradation in MDMs treated with HIV-1 rNef and HCV rCore. Cytoplasmic extracts were prepared from MDMs treated with HIV-1 rNef (100 ng/ml), HCV rCore (100 ng/ml) or HIV-1 rNef (100 ng/ml) + HCV rCore protein (100 ng/ml) for 30 min and then assayed for the expression of IkBα by Western blot. β-actin was used as a loading control. Protein levels were quantified by densitometry using ImageJ 1.40 software (the level of protein in mock cells was arbitrarily established at 1). Results represent means ± SD of threeindependent experiments. ** p < 0.01. d IKKα and IKKβ expression and phosphorylation in MDMs treated with HIV-1 rNef and HCV rCore. Cytoplasmic extracts were prepared from MDMs treated with HIV-1 rNef (100 ng/ml), HCV rCore (100 ng/ml) or HIV-1 rNef (100 ng/ml) + HCV rCore protein (100 ng/ml) for 30 min and then assayed for the corresponding protein by Western blot. β-actin was used as a loading control. Protein levels were quantified by densitometry using ImageJ 1.40 software (the level of protein in mock cells was arbitrarily established at 1). Results represent means ± SD of threeindependent experiments. *** p < 0.001. e Knockdown of TRAF1-TRAF6 proteins by siRNA in MDMs. MDM cultures were transfected with TRAF1-TRAF6 siRNAs or scrambled control. Total cellular extracts were prepared 48 h posttransfection. Protein expression was analyzed by Western blot. β-actin was used as a loading control. f NF-κB activation in MDMs treated with HIV-1 rNef and HCV rCore depends on TRAF2, TRAF5 and TRAF6. 48 h before treatment with HIV-1 rNef (100 ng/ml) and HCV rCore (100 ng/ml), MDMs were transfected with a mixture of TRAF2/TRAF5/TRAF6 siRNAs, a mixture of TRAF1/TRAF3/TRAF4 siRNAs, or a scrambled control. NF-κB activation was determined by EMSA. Nuclear extracts of MDMs treated with TNFα (10 ng/ml) were used as a positive control. The histogram represents the quantification of NF-κB activity by densitometry using ImageJ 1.40 software (the level of NF-κB activity in mock cells was arbitrarily established at 1). Results represent means ± SD of three independent experiments. * p < 0.05.
To further confirm the role of TRAF proteins in NF-κB activation, MDMs were transfected with TRAFs siRNA 48 h prior to treatment with rNef and rCore. The knockdown of the TRAF1 to TRAF6 proteins in MDMs was monitored by Western blot (fig. 6e). The knockdown of TRAF2, TRAF5 and TRAF6 decreased NF-κB activation, whereas the knockdown of TRAF1, TRAF3 and TRAF4 did not modify NF-κB activation (fig. 6f). Our results indicate that in MDMs treated with rNef and rCore, the canonical pathway of NF-κB is activated through an upstream signal mediated through TRAF2, TRAF5 and TRAF6.
HIV-1 Nef and HCV Core Enhance HIV-1 Replication in MDMs through a TRAF (2, 5, 6) Pathway
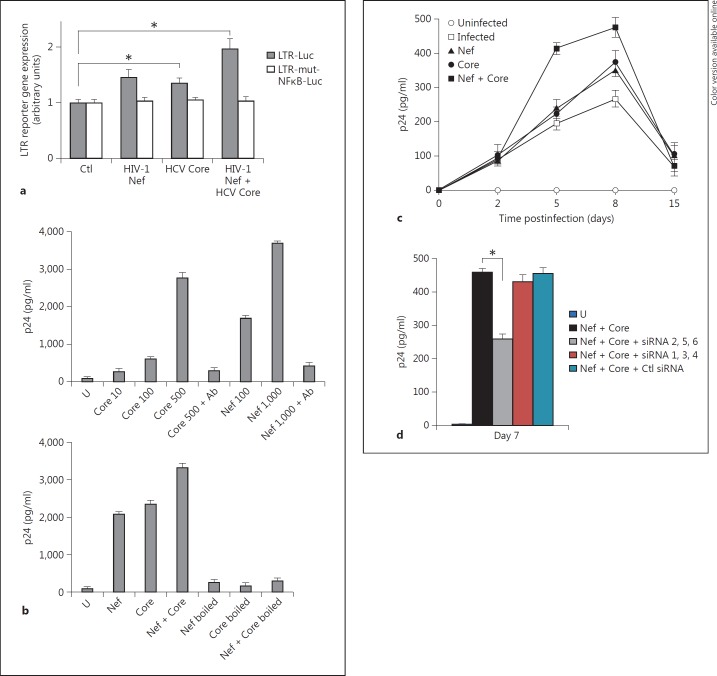
We assessed the effect of HIV-1 Nef, HCV Core or both proteins together on NF-κB-dependent LTR stimulation. U937 cells were transiently transfected with a target plasmid that contains the luciferase reporter gene under the control of the HIV-1 LTR promoter, pLTR-Luc. Twenty-four hours later, transfected cells were treated for 24 h with HIV-1 rNef, HCV rCore, or both proteins together, harvested, and luciferase activity was measured in cell lysates. The HIV-1 rNef treatment resulted in a 1.5-fold LTR stimulation over untreated control cells (fig. 7a). HCV rCore treatment of transfected U937 cells increased HIV-1 LTR stimulation by 1.4-fold (fig. 7a). The cotreatment of transfected U937 cells with HIV-1 rNef and HCV rCore resulted in the highest LTR stimulation with 2-fold luciferase activity over untreated control cells (fig. 7a). LTR activation induced by HIV-1 rNef, HCV rCore, or both proteins together, was not observed when a plasmid containing mutated NF-κB sites, pLTR-mut-NF-κB-Luc, was used instead of pLTR-Luc (fig. 7a). These data indicate that HIV-1 rNef and HCV rCore additionally activate the LTR via NF-κB stimulation in promonocytic cells U937.
Fig. 7.
Enhanced HIV-1 replication in MDMs treated with HIV-1 Nef and HCV Core depends on TRAF2, TRAF5 and TRAF6. a Enhanced LTR stimulation in transfected U937 cells treated by HIV-1 rNef and HCV rCore. U937 cells (2 × 106) were transiently transfected with 20 µg of pLTR-Luc, or 20 µg of pLTRmut-NF-κB-Luc. 24 h later, transfected cells were treated with HIV-1 rNef (100 ng/ml), HCV rCore (100 ng/ml) or HIV-1 rNef (100 ng/ml) + HCV rCore (100 ng/ml). 48 h after transfection, luciferase activity was measured in cell lysates as described in ‘Materials and Methods’. Values normalized to protein concentrations were expressed as fold increase over unstimulated control values. Results represent mean values ± SD of three independent experiments. * p < 0.05. b Enhanced HIV-1 replication in chronically infected U1 promonocytic cells treated with HIV-1 rNef and HCV rCore. Upper panel: U1 cells were treated with HIV-1 rNef (at the concentration specified) or HCV rCore (at the concentration specified) in the presence and absence of neutralizing anti-Nef and anti-HCV Core mAbs. Lower panel: U1 cells were treated with HIV-1 rNef (500 ng/ml), HCV rCore (500 ng/ml) or HIV-1 rNef (500 ng/ml) + HCV rCore (500 ng/ml) boiled at 100°C for 10 min or not. p24 was measured in culture supernatants 72 h after treatment. Representative results of three independent experiments are shown. c HIV-1 replication in MDMs is enhanced by treatment with HIV-1 rNef and HCV rCore. MDMs were infected overnight with HIV-89.6 (10 ng p24/4 × 105 cells). After extensive washes with PBS to remove the unadsorbed inoculum, MDMs were treated with HIV-1 rNef (100 ng/ml), HCV rCore (100 ng/ml) or HIV-1 rNef (100 ng/ml) + HCV rCore (100 ng/ml) every 3 days and p24 was measured in culture supernatants up to 15 days postinfection. Results represent mean values ± SD of two independent experiments. d Enhanced HIV-1 replication in MDMs treated with HIV-1 rNef and HCV rCore involves TRAF2, TRAF5 and TRAF6. 48 h before infection with HIV-1 89.6 in the absence and presence of treatment with HIV-1 rNef and HCV rCore, MDMs were transfected with a mixture of TRAF2/TRAF5/TRAF6 siRNAs, a mixture of TRAF1/TRAF3/TRAF4 siRNAs, or a scrambled control. Transfected MDMs were infected overnight with HIV-89.6 (10 ng p24/4 × 105 cells). After extensive washes with PBS to remove the unadsorbed inoculum, MDMs were treated with HIV-1 rNef (100 ng/ml) + HCV rCore (100 ng/ml) every 3 days and p24 was measured in culture supernatants at day 7 postinfection. Results represent means ± SD of two independent experiments. * p < 0.05.
Since NF-κB DNA-binding sites are present in the HIV-1 LTR, we determined the effects of HIV-1 Nef and HCV Core, alone and together, on provirus transcription in the promonocytic cell line U1, U937 cells that contain two integrated HIV copies per cell. We measured HIV-1 replication in U1 cells following treatment with HIV-1 rNef and HCV rCore, alone or together. Both HIV-1 rNef and HCV rCore proteins stimulated viral replication in U1 cells in a dose-dependent manner as measured by p24 assay (fig. 7b). The highest viral replication was detected in culture supernatants of U1 cells cotreated with HIV-1 rNef and HCV rCore (fig. 7b), indicating an additional effect of the two proteins on HIV-1 transcription. To rule out the possibility that a TNFα inducer, such as lipopolysaccharide, enhanced viral replication in U1 cells, we boiled HIV-1 rNef and HCV rCore at 100°C. Boiling abolished viral replication in U1 cells stimulated with the two proteins either alone or together (fig. 7b), indicating that both HIV-1 Nef and HCV Core, but not lipopolysaccharide contamination, were responsible for enhanced replication.
Since HIV-1 Nef and HCV Core additionally stimulated viral growth in chronically infected U1 cells, we assessed their effect on viral growth following acute infection of primary human MDMs with HIV-1. First, we measured the presence of intracellular Nef and Core proteins by flow cytometric analysis following in vitro treatment of primary human MDMs with rNef and rCore. We observed that following treatment of MDMs with recombinant Nef and Core proteins, only very low amounts of viral proteins were detected on the cell surface, whereas most of the viral proteins were detected intracellularly (online suppl. fig. 1; for all online supplementary material, see www.karger.com/doi/10.1159/000350517). Then we measured viral growth in primary MDMs infected with HIV-89.6 (10 ng p24/4 × 105 cells) and treated with HIV-1 Nef and HCV Core. HIV-1 rNef and HCV rCore enhanced HIV-1 replication in MDMs by 1.5-fold at day 8after infection versus untreated infected cells (fig. 7c). When acutely infected MDMs were treated with the two proteins together, the viral growth was enhanced by approximately 2-fold at day 8 after infection versus untreated infected cells (fig. 7c), indicating an additional action of HIV-1 Nef and HCV Core on HIV-1 replication in MDMs. The knockdown of TRAF2, TRAF5 and TRAF6 repressed HIV-1 replication in acutely infected MDMs treated with HCV rCore and HIV-1 rNef, whereas the knockdown of TRAF1, TRAF3 and TRAF4 had no effect (fig. 7d). Our results indicate that in MDMs acutely infected with HIV-1 and treated with HCV rCore and HIV-1 rNef, the HIV-1 replication depends on an upstream signal mediated through TRAF2, TRAF5 and TRAF6.
Discussion
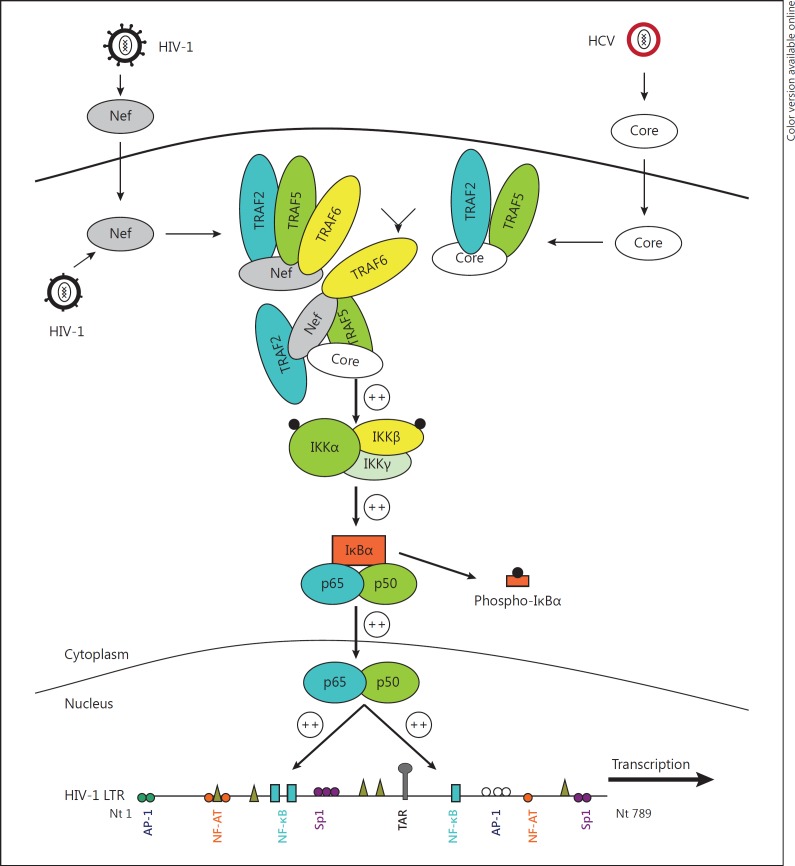
We observed that TRAF2, TRAF5 and TRAF6 interact with HIV-1 Nef and that HCV Core interacts with TRAF2 and TRAF5. We also found that HIV-1 Nef interacts with HCV Core. The TRAF (2, 5, 6) recruitment mediated by HIV-1 Nef and HCV Core enhanced the activation of NF-κB and increased HIV-1 replication in MDMs. Our results reveal a mechanism by which the activation of the TRAF pathway by HIV-1 Nef and HCV Core favors the replication of HIV-1 in MDMs (fig. 8) and could be a critical factor for optimal replication of HIV-1 in MDMs of HIV-HCV-coinfected patients.
Fig. 8.
Potential enhancing effect of HIV-1 Nef, HCV Core and TRAF (2, 5, 6) on HIV-1 replication in MDMs. TRAF2, TRAF5 and TRAF6 interact with HIV-1 Nef, HCV Core interacts with TRAF2 and TRAF5, and HIV-1 Nef interacts with HCV Core. The TRAF (2, 5, 6) activation mediated by HIV-1 Nef and HCV Core enhances NF-κB activation and increases HIV-1 replication in MDMs, and could be a critical factor for optimal replication of HIV-1 in MDMs of HIV-HCV-coinfected patients.
We previously reported that HIV-1 rNef activates NF-κB and induces IκBα phosphorylation and degradation in promonocytic cells U937. In agreement with our results, since phosphorylation of IκBα at serine is induced, myristoylated rNef probably activates IKK-β, the only kinase known to phosphorylate IκBα directly. We previously observed that the rNef-triggered activation of NF-κB was mediated via the canonical NF-κB complex p50/p65 in monocytic cells [21]. Our data now indicate that both HIV-1 rNef and HCV rCore activate NF-κB in MDMs, and that the canonical NF-κB pathway is involved in this phenomenon. The amount of HIV-1 Nef and HCV Core proteins detected in the serum of infected subjects is around 1-10 ng/ml and 0.1-10 ng/ml, respectively [29, 35]. Nevertheless, we cannot rule out that higher amounts of exogenous Nef and HCV Core proteins could be present in tissue compartments such as lymph nodes where macrophages and infected lymphocytes tightly interact [36, 37]. Additionally, the amounts of Nef detected in the serum of patients may be even higher than 10 ng/ml when taking into account the presence of Nef/anti-Nef immune complexes [35, 38, 39]. Circulating immune complexes are detected in HCV chronic infection and the HCV Core protein is a constitutive component of these immune complexes. Additionally, the HCV Core protein can self-assemble to multimeric complexes [40, 41].
We observed the highest levels of NF-κB activation in PBMCs and monocytes isolated from the peripheral blood of coinfected subjects. High donor-linked variability between monocytes/macrophages from different donors, in particular for their in vitro HIV-1 susceptibility, has been reported previously [42, 43]. Although we observed variability between peripheral blood cells (PBMCs, monocytes, PBLs) from different donors for NF-κB activation, NF-κB activation was 2-fold higher in cells isolated from coinfected patients than in cells isolated from monoinfected patients. Additionally, our results indicate that, although both HIV-1 Nef and HCV Core activate NF-κB independently, the two viral proteins act additionally on the NF-κB activation in MDMs. The optimal activation of NF-κB observed following the treatment of promonocytic cells and MDMs with HIV-1 rNef and HCV rCore could be initiated at the cell's surface, or most probably just below the plasma membrane. In agreement with this hypothesis, we observed previously that HIV-1 rNef activates NF-κB in MDMs via its myristoylation domain and via lipid rafts [32]. Additionally, HCV Core, TRAF2, TRAF5 and TRAF6 can be recruited to lipid rafts and have been reported to activate NF-κB [31, 44]. Recently, HIV-1 Nef has been reported to induce a proinflammatory state in macrophages through its acidic cluster domain and the involvement of TRAF2 [45]. Our results expand these previous observations by indicating that both HIV-1 Nef and HCV Core bind to several members of the TRAF family, namely TRAF2, TRAF5 and TRAF6. In addition to the acidic cluster domain of HIV-1 Nef involved in binding to TRAF2 (AxEE) already reported by others [45], we observed a minor TRAF-binding consensus sequence (PxxQxxT) which could bind TRAF5 and a TRAF6-binding consensus sequence (PxExxD/E/F/W/Y) in the C-terminal region of HIV-1 Nef (aa 55-206; online suppl. fig. 2). We also detected putative TRAF-binding motifs in the N-terminal region of HCV Core (aa 1-126; online suppl. fig. 3).
We observed a direct binding between HIV-1 Nef and HCV Core, indicating a potential crosstalk between the two viruses resulting in an optimal NF-κB activation, especially in macrophages. The plasma membrane via lipid rafts and/or detergent-resistant membranes could be a critical platform where HIV-1 Nef and HCV Core proteins colocalize within the cell leading to an optimal replication of HIV-1 in macrophages [46, 47]. HIV Nef anchors to the membrane through its myristoylation signal [48] and HCV Core contains a C-terminal anchor sequence [49]. Although HIV-1 Nef has been reported to bind to the CXCR4 receptor on the cell surface [50], in MDMs treated in vitro with rNef and rCore, we detected the two viral proteins within the cells rather than bound on the cell's surface using flow cytometric analysis (online suppl. fig. 1). Therefore, we believe that increased NF-κB activation and enhanced HIV replication in MDMs results primarily from intracellular Nef-Core interaction rather than from viral proteins binding to the cell's surface.
HIV-1 Nef has been reported to expand the cellular reservoir of HIV-1 by permitting the infection of resting CD4+ T lymphocytes and by enhancing HIV-1 replication within CD4+ memory T cells [51]. Our results suggest that soluble HCV Core released from HCV-infected cells could favor HIV-1 replication in macrophages of HIV-HCV-coinfected subjects, thereby favoring the formation of macrophage reservoirs of HIV-1 [52].
In conclusion, our results indicate that HIV-1 Nef and HCV Core enhance NF-κB activation in HIV-1-infected macrophages through the involvement of TRAF2, TRAF5 and TRAF6. Our results underline the critical role of HIV-1 Nef, HCV Core and several members of the TRAF family in the development of AIDS pathogenesis in HIV/HCV-coinfected patients, but also indicate that new therapeutic approaches to curtail the formation of viral reservoirs in mononuclear phagocytes of HIV/HCV-coinfected subjects have to be developed.
Disclosure Statement
The authors have no financial conflicts of interest.
Supplementary Material
Supplementary data
Acknowledgments
This work was supported by grants to G.H. from the University of Franche-Comté (UFC) and the Région Franche-Comté (RECH-FON12-000013). W.A. and K.A.K. are recipients of doctoral scholarships from the Higher Education Commission, Pakistan. A.K. is a recipient of a postdoctoral fellowship of the Region Franche-Comté. We thank Diasorin SA and Roche for their financial support.
References
- 1.Baud V, Karin M. Signal transduction by tumor necrosis factor and its relatives. Trends Cell Biol. 2001;11:372–377. doi: 10.1016/s0962-8924(01)02064-5. [DOI] [PubMed] [Google Scholar]
- 2.Wajant H, Scheurich P. TNFR1-induced activation of the classical NF-κB pathway. FEBS J. 2011;278:862–876. doi: 10.1111/j.1742-4658.2011.08015.x. [DOI] [PubMed] [Google Scholar]
- 3.Bradley JR, Pober JS. Tumor necrosis factor receptor-associated factors (TRAFs) Oncogene. 2001;20:6482–6491. doi: 10.1038/sj.onc.1204788. [DOI] [PubMed] [Google Scholar]
- 4.Hacker H, Tseng PH, Karin M. Expanding TRAF function: TRAF3 as a tri-faced immune regulator. Nat Rev Immunol. 2011;11:457–468. doi: 10.1038/nri2998. [DOI] [PubMed] [Google Scholar]
- 5.Scudiero I, Zotti T, Ferravante A, Vessichelli M, Reale C, Masone MC, Leonardi A, Vito P, Stilo R. Tumor necrosis factor (TNF) receptor-associated factor 7 is required for TNFα-induced Jun NH2-terminal kinase activation and promotes cell death by regulating polyubiquitination and lysosomal degradation of c-FLIP protein. J Biol Chem. 2012;287:6053–6061. doi: 10.1074/jbc.M111.300137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cao Z, Xiong J, Takeuchi M, Kurama T, Goeddel DV. TRAF6 is a signal transducer for interleukin-1. Nature. 1996;383:443–446. doi: 10.1038/383443a0. [DOI] [PubMed] [Google Scholar]
- 7.Korchnak AC, Zhan Y, Aguilar MT, Chadee DN. Cytokine-induced activation of mixed lineage kinase 3 requires TRAF2 and TRAF6. Cell Signal. 2009;21:1620–1625. doi: 10.1016/j.cellsig.2009.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Song HY, Regnier CH, Kirschning CJ, Goeddel DV, Rothe M. Tumor necrosis factor (TNF)-mediated kinase cascades: bifurcation of nuclear factor-κB and c-Jun N-terminal kinase (JNK/SAPK) pathways at TNF receptor-associated factor 2. Proc Natl Acad Sci USA. 1997;94:9792–9796. doi: 10.1073/pnas.94.18.9792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Au PY, Yeh WC. Physiological roles and mechanisms of signaling by TRAF2 and TRAF5. Adv Exp Med Biol. 2007;597:32–47. doi: 10.1007/978-0-387-70630-6_3. [DOI] [PubMed] [Google Scholar]
- 10.Nakano H, Sakon S, Koseki H, Takemori T, Tada K, Matsumoto M, Munechika E, Sakai T, Shirasawa T, Akiba H, Kobata T, Santee SM, Ware CF, Rennert PD, Taniguchi M, Yagita H, Okumura K. Targeted disruption of TRAF5 gene causes defects in CD40- and CD27-mediated lymphocyte activation. Proc Natl Acad Sci USA. 1999;96:9803–9808. doi: 10.1073/pnas.96.17.9803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yeh WC, Shahinian A, Speiser D, Kraunus J, Billia F, Wakeham A, de la Pompa JL, Ferrick D, Hum B, Iscove N, Ohashi P, Rothe M, Goeddel DV, Mak TW. Early lethality, functional NF-κB activation, and increased sensitivity to TNF-induced cell death in TRAF2-deficient mice. Immunity. 1997;7:715–725. doi: 10.1016/s1074-7613(00)80391-x. [DOI] [PubMed] [Google Scholar]
- 12.Cullen BR. Regulation of HIV-1 gene expression. FASEB J. 1991;5:2361–2368. doi: 10.1096/fasebj.5.10.1712325. [DOI] [PubMed] [Google Scholar]
- 13.Xing L, Venegas AM, Chen A, Garrett-Beal L, Boyce BF, Varmus HE, Schwartzberg PL. Genetic evidence for a role for Src family kinases in TNF family receptor signaling and cell survival. Genes Dev. 2001;15:241–253. doi: 10.1101/gad.840301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kedinger V, Rio MC. TRAF4, the unique family member. Adv Exp Med Biol. 2007;597:60–71. doi: 10.1007/978-0-387-70630-6_5. [DOI] [PubMed] [Google Scholar]
- 15.Lee SY, Choi Y. TRAF1 and its biological functions. Adv Exp Med Biol. 2007;597:25–31. doi: 10.1007/978-0-387-70630-6_2. [DOI] [PubMed] [Google Scholar]
- 16.Megas C, Hatzivassiliou EG, Yin Q, Marinopoulou E, Hadweh P, Vignali DA, Mosialos G. Mutational analysis of TRAF6 reveals a conserved functional role of the ring dimerization interface and a potentially necessary but insufficient role of ring-dependent TRAF6 polyubiquitination towards NF-κB activation. Cell Signal. 2011;23:772–777. doi: 10.1016/j.cellsig.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Foster JL, Garcia JV. HIV-1 Nef: at the crossroads. Retrovirology. 2008;5:84. doi: 10.1186/1742-4690-5-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qiao X, He B, Chiu A, Knowles DM, Chadburn A, Cerutti A. Human immunodeficiency virus 1 Nef suppresses CD40-dependent immunoglobulin class switching in bystander B cells. Nat Immunol. 2006;7:302–310. doi: 10.1038/ni1302. [DOI] [PubMed] [Google Scholar]
- 19.Alessandrini L, Santarcangelo AC, Olivetta E, Ferrantelli F, d'Aloja P, Pugliese K, Pelosi E, Chelucci C, Mattia G, Peschle C, Verani P, Federico M. T-tropic human immunodeficiency virus (HIV) type 1 Nef protein enters human monocyte-macrophages and induces resistance to HIV replication: a possible mechanism of HIV T-tropic emergence in AIDS. J Gen Virol. 2000;81:2905–2917. doi: 10.1099/0022-1317-81-12-2905. [DOI] [PubMed] [Google Scholar]
- 20.Federico M, Percario Z, Olivetta E, Fiorucci G, Muratori C, Micheli A, Romeo G, Affabris E. HIV-1 Nef activates STAT1 in human monocytes/macrophages through the release of soluble factors. Blood. 2001;98:2752–2761. doi: 10.1182/blood.v98.9.2752. [DOI] [PubMed] [Google Scholar]
- 21.Varin A, Manna SK, Quivy V, Decrion AZ, Van Lint C, Herbein G, Aggarwal BB. Exogenous Nef protein activates NF-κB, AP-1, and c-Jun N-terminal kinase and stimulates HIV transcription in promonocytic cells: role in AIDS pathogenesis. J Biol Chem. 2003;278:2219–2227. doi: 10.1074/jbc.M209622200. [DOI] [PubMed] [Google Scholar]
- 22.Ridzon R, Gallagher K, Ciesielski C, Ginsberg MB, Robertson BJ, Luo CC, DeMaria A., Jr Simultaneous transmission of human immunodeficiency virus and hepatitis C virus from a needle-stick injury. N Engl J Med. 1997;336:919–922. doi: 10.1056/NEJM199703273361304. [DOI] [PubMed] [Google Scholar]
- 23.Kim AY, Chung RT. Coinfection with HIV-1 and HCV - a one-two punch. Gastroenterology. 2009;137:795–814. doi: 10.1053/j.gastro.2009.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lesens O, Deschenes M, Steben M, Belanger G, Tsoukas CM. Hepatitis C virus is related to progressive liver disease in human immunodeficiency virus-positive hemophiliacs and should be treated as an opportunistic infection. J Infect Dis. 1999;179:1254–1258. doi: 10.1086/314720. [DOI] [PubMed] [Google Scholar]
- 25.Piroth L, Duong M, Quantin C, Abrahamowicz M, Michardiere R, Aho LS, Grappin M, Buisson M, Waldner A, Portier H, Chavanet P. Does hepatitis C virus coinfection accelerate clinical and immunological evolution of HIV-infected patients? Aids. 1998;12:381–388. doi: 10.1097/00002030-199804000-00006. [DOI] [PubMed] [Google Scholar]
- 26.Cerutti A, Maillard P, Minisini R, Vidalain PO, Roohvand F, Pecheur EI, Pirisi M, Budkowska A. Identification of a functional, CRM-1-dependent nuclear export signal in hepatitis C virus core protein. PLoS One. 2011;6:e25854. doi: 10.1371/journal.pone.0025854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lai MM, Ware CF. Hepatitis C virus core protein: possible roles in viral pathogenesis. Curr Top Microbiol Immunol. 2000;242:117–134. doi: 10.1007/978-3-642-59605-6_6. [DOI] [PubMed] [Google Scholar]
- 28.Dolganiuc A, Chang S, Kodys K, Mandrekar P, Bakis G, Cormier M, Szabo G. Hepatitis C virus (HCV) Core protein-induced, monocyte-mediated mechanisms of reduced IFN-α and plasmacytoid dendritic cell loss in chronic HCV infection. J Immunol. 2006;177:6758–6768. doi: 10.4049/jimmunol.177.10.6758. [DOI] [PubMed] [Google Scholar]
- 29.Pivert A, Payan C, Morand P, Fafi-Kremer S, Deshayes J, Carrat F, Pol S, Cacoub P, Perronne C, Lunel F. Comparison of serum hepatitis C virus (HCV) RNA and core antigen levels in patients coinfected with human immunodeficiency virus and HCV and treated with interferon plus ribavirin. J Clin Microbiol. 2006;44:417–422. doi: 10.1128/JCM.44.2.417-422.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Park KJ, Choi SH, Koh MS, Kim DJ, Yie SW, Lee SY, Hwang SB. Hepatitis C virus core protein potentiates c-Jun N-terminal kinase activation through a signaling complex involving TRADD and TRAF2. Virus Res. 2001;74:89–98. doi: 10.1016/s0168-1702(00)00251-3. [DOI] [PubMed] [Google Scholar]
- 31.Yoshida H, Kato N, Shiratori Y, Otsuka M, Maeda S, Kato J, Omata M. Hepatitis C virus core protein activates nuclear factor κB-dependent signaling through tumor necrosis factor receptor-associated factor. J Biol Chem. 2001;276:16399–16405. doi: 10.1074/jbc.M006671200. [DOI] [PubMed] [Google Scholar]
- 32.Herbein G, Varin A, Larbi A, Fortin C, Mahlknecht U, Fulop T, Aggarwal BB. Nef and TNFα are coplayers that favor HIV-1 replication in monocytic cells and primary macrophages. Curr HIV Res. 2008;6:117–129. doi: 10.2174/157016208783884985. [DOI] [PubMed] [Google Scholar]
- 33.Reuse S, Calao M, Kabeya K, Guigen A, Gatot JS, Quivy V, Vanhulle C, Lamine A, Vaira D, Demonte D, Martinelli V, Veithen E, Cherrier T, Avettand V, Poutrel S, Piette J, de Launoit Y, Moutschen M, Burny A, Rouzioux C, De Wit S, Herbein G, Rohr O, Collette Y, Lambotte O, Clumeck N, van Lint C. Synergistic activation of HIV-1 expression by deacetylase inhibitors and prostratin: implications for treatment of latent infection. PLoS One. 2009;4:e6093. doi: 10.1371/journal.pone.0006093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Decrion AZ, Varin A, Estavoyer JM, Herbein G. CXCR4-mediated T cell apoptosis in human immunodeficiency virus infection. J Gen Virol. 2004;85:1471–1478. doi: 10.1099/vir.0.79933-0. [DOI] [PubMed] [Google Scholar]
- 35.Fujii Y, Otake K, Tashiro M, Adachi A. Soluble Nef antigen of HIV-1 is cytotoxic for human CD4+ T cells. FEBS Lett. 1996;393:93–96. doi: 10.1016/0014-5793(96)00859-9. [DOI] [PubMed] [Google Scholar]
- 36.Herbein G, Khan KA. Is HIV infection a TNF receptor signalling-driven disease? Trends Immunol. 2008;29:61–67. doi: 10.1016/j.it.2007.10.008. [DOI] [PubMed] [Google Scholar]
- 37.Pal S, Sullivan DG, Kim S, Lai KK, Kae J, Cotler SJ, Carithers RL, Jr, Wood BL, Perkins JD, Gretch DR. Productive replication of hepatitis C virus in perihepatic lymph nodes in vivo: implications of HCV lymphotropism. Gastroenterology. 2006;130:1107–1116. doi: 10.1053/j.gastro.2005.12.039. [DOI] [PubMed] [Google Scholar]
- 38.Prost S, Le Dantec M, Augé S, Le Grand R, Derdouch S, Auregan G, Déglon N, Relouzat F, Aubertin AM, Maillere B, Dusanter-Fourt I, Kirszenbaum M. Human and simian immunodeficiency viruses deregulate early hematopoiesis through a Nef/PPARγ/STAT5 signaling pathway in macaques. J Clin Invest. 2008;118:1765–1775. doi: 10.1172/JCI33037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Raymond AD, Campbell-Sims TC, Khan M, Lang M, Huang MB, Bond VC, Powell MD. HIV type 1 Nef is released from infected cells in CD45+ microvesicles and is present in the plasma of HIV-infected individuals. AIDS Res Hum Retroviruses. 2011;27:167–178. doi: 10.1089/aid.2009.0170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sansonno D, Lauletta G, Nisi L, Gatti P, Pesola F, Pansini N, Dammacco F. Non-enveloped HCV core protein as constitutive antigen of cold-precipitable immune complexes in type II mixed cryoglobulinaemia. Clin Exp Immunol. 2003;133:275–282. doi: 10.1046/j.1365-2249.2003.02204.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Santolini E, Migliaccio G, La Monica., N Biosynthesis and biochemical properties of the hepatitis C virus core protein. J Virol. 1994;68:3631–3641. doi: 10.1128/jvi.68.6.3631-3641.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Martinez FO, Gordon S, Locati M, Mantovani A. Transcriptional profiling of the human monocyte-to-macrophage differentiation and polarization: new molecules and patterns of gene expression. J Immunol. 2006;177:7303–7311. doi: 10.4049/jimmunol.177.10.7303. [DOI] [PubMed] [Google Scholar]
- 43.Bol SM, van Remmerden Y, Sietzema JG, Kootstra NA, Schuitemaker H, van't Wout., AB Donor variation in in vitro HIV-1 susceptibility of monocyte-derived macrophages. Virology. 2009;390:205–211. doi: 10.1016/j.virol.2009.05.027. [DOI] [PubMed] [Google Scholar]
- 44.Chung YM, Park KJ, Choi SY, Hwang SB, Lee SY. Hepatitis C virus core protein potentiates TNF-α-induced NF-κB activation through TRAF2-IKKβ-dependent pathway. Biochem Biophys Res Commun. 2001;284:15–19. doi: 10.1006/bbrc.2001.4936. [DOI] [PubMed] [Google Scholar]
- 45.Mangino G, Percario ZA, Fiorucci G, Vaccari G, Acconcia F, Chiarabelli C, Leone S, Noto A, Horenkamp FA, Manrique S, Romeo G, Polticelli F, Geyer M, Affabris E. HIV-1 Nef induces proinflammatory state in macrophages through its acidic cluster domain: involvement of TNF alpha receptor associated factor 2. PLoS One. 2011;6:e22982. doi: 10.1371/journal.pone.0022982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Alexander M, Bor YC, Ravichandran KS, Hammarskjold ML, Rekosh D. Human immunodeficiency virus type 1 Nef associates with lipid rafts to downmodulate cell surface CD4 and class I major histocompatibility complex expression and to increase viral infectivity. J Virol. 2004;78:1685–1696. doi: 10.1128/JVI.78.4.1685-1696.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Boulant S, Vanbelle C, Ebel C, Penin F, Lavergne JP. Hepatitis C virus core protein is a dimeric alpha-helical protein exhibiting membrane protein features. J Virol. 2005;79:11353–11365. doi: 10.1128/JVI.79.17.11353-11365.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bentham M, Mazaleyrat S, Harris M. Role of myristoylation and N-terminal basic residues in membrane association of the human immunodeficiency virus type 1 Nef protein. J Gen Virol. 2006;87:563–571. doi: 10.1099/vir.0.81200-0. [DOI] [PubMed] [Google Scholar]
- 49.McLauchlan J, Lemberg MK, Hope G, Martoglio B. Intramembrane proteolysis promotes trafficking of hepatitis C virus core protein to lipid droplets. EMBO J. 2002;21:3980–3988. doi: 10.1093/emboj/cdf414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.James CO, Huang MB, Khan M, Garcia-Barrio M, Powell MD, Bond VC. Extracellular Nef protein targets CD4+ T cells for apoptosis by interacting with CXCR4 surface receptors. J Virol. 2004;78:3099–3109. doi: 10.1128/JVI.78.6.3099-3109.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Choi J, Walker J, Talbert-Slagle K, Wright P, Pober JS, Alexander L. Endothelial cells promote human immunodeficiency virus replication in nondividing memory T cells via Nef-, Vpr-, and T-cell receptor-dependent activation of NFAT. J Virol. 2005;79:11194–11204. doi: 10.1128/JVI.79.17.11194-11204.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Trono D, Van Lint C, Rouzioux C, Verdin E, Barre-Sinoussi F, Chun TW, Chomont N. HIV persistence and the prospect of long-term drug-free remissions for HIV-infected individuals. Science. 2010;329:174–180. doi: 10.1126/science.1191047. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data