Abstract
Many bacterial pathogens have developed methods to overcome the defences of the host innate immune system. One such defence is the release of antimicrobial peptides (AMPs). Histones have been found to function as AMPs, in addition to their main biological function of packaging and organising DNA into nucleosomes. In this study, the Gram-positive anaerobic coccus Finegoldia magna was found to bind histones by Western blot and immunoprecipitation analysis. F. magna, which is normally a commensal of the skin and mucous membranes, is also known to act as an opportunistic pathogen and has been isolated from various clinical infection sites. It was found to bind to histones extracted from human skin epidermis through its surface and extracellular adhesion protein FAF. Through FAF binding, F. magna was protected from histone bactericidal activity. Furthermore, the histones were found to be degraded by SufA, a subtilisin-like extracellular serine protease of F. magna. Hence, the results of the present study will give more insight into how F. magna persists both as a commensal organism at the basement membrane of the skin and as an opportunistic pathogen during infection.
Key Words: Finegoldia magna, Gram-positive anaerobic cocci, Histones, Innate immunity, SufA
Introduction
Finegoldia magna (formerly Peptostreptococcus magnus) is a member of the normal microbiota that colonizes the skin, mucous membranes, oral cavity and the gastrointestinal and urogenital tracts [1, 2]. It is an obligately anaerobic bacterium that is a member of the Gram-positive anaerobic cocci (GPAC). As well as being a commensal, F. magna is also an opportunistic pathogen and is the most common GPAC isolated from anaerobic specimens [1, 2]. Infections from which it is isolated include soft tissue abscesses, wound infections, bone and joint infections and vaginoses [1, 2, 3, 4, 5, 6]. Most likely, the incidence of F. magna in clinical infection is highly underestimated due to problems obtaining good-quality anaerobic specimens and difficulties associated with handling and growing of these bacteria. Currently, F. magna is susceptible to most antibiotics used for the treatment of anaerobic infections. However, there have been reports of an increase in resistance rates to erythromycin and tetracycline [7, 8, 9].
Investigations on F. magna mechanisms of infection have identified possible virulence factors, such as the production of collagenase and gelatinase enzymes [10] and capsule formation [11]. In addition, most strains have been found to express SufA, a subtilisin-like enzyme that has already been shown to have multiple functions in the bacteria [12]. It protects the bacteria from antimicrobial peptides (AMPs), such as LL-37 and MIG/CXCL9, and cleaves fibrinogen, resulting in delayed wound healing and clotting [12, 13, 14]. Protein L is a surface protein expressed by approximately 10% of F. magna isolates that has an affinity for immunoglobulin κ light chains and induces the release of pro-inflammatory mediators such as IL-4 and IL-13 from human FcεRI+ cells [15, 16]. Protein PAB is another surface protein of F. magna and has an affinity for albumin through the GA module [17]. PAB-expressing F. magna strains have an up to 50% increase in growth rate, giving them a selective advantage during infection [18].
Protein FAF is a recently described surface protein that is expressed by more than 90% of F. magna isolates [19]. As well as being present on the surface, it is also released in the form of a 53-kDa fragment by SufA and forms large protein aggregates in solution. FAF has already been found to have multiple functions, including binding to BM-40 at the basement membrane of the skin and neutralising the bactericidal activity of AMPs such as LL-37, MIG/CXCL9, midkine, BRAK/CXCL14 and hBD-3 [19, 20].
Histones are largely known to be involved in transcription regulation and DNA repair, replication and condensation. They undergo extensive post-translational modifications that effect transcription and other DNA-related mechanisms [21, 22]. In addition, studies have shown that histones also have antimicrobial properties and histone-derived peptides with antimicrobial activity have been identified [23, 24]. Histone H2A was found to kill Shigella flexneri, Salmonella typhimurium and Staphylococcus aureus at a concentration of only 140 nM within 30 min, proving itself to be an effective antibacterial protein [25]. Furthermore, histones have been found to be released in the skin by sebum-secreting sebocytes as part of the innate immune response to pathogens, resulting in exposure of commensals, such as F. magna, to their antimicrobial action [26].
In this study, protein FAF was found to bind to histones H2A, H2B and H4 that were extracted from epidermal skin extracts. Furthermore, SufA was shown to degrade these histones, resulting in a dual barrier protective function for F. magna. Through the binding and degradation of histones in the skin epidermis by FAF and SufA, F. magna is protected from their antimicrobial activity.
Methods
Bacteria and Growth Conditions
The F. magna strain 505 used in this study was isolated from a clinical infection site in the urethra of a patient at the Department of Clinical Microbiology, Skåne University Hospital, Sweden. F. magna strain ALB8 isolate was from Lund University Hospital, Lund, Sweden, and has been described earlier [17]. The bacteria were grown under strict anaerobic conditions in Todd-Hewitt broth (Difco) supplemented with 0.5% Tween-80 at 37°C.
Proteins, Plasma, Antibodies and Reagents
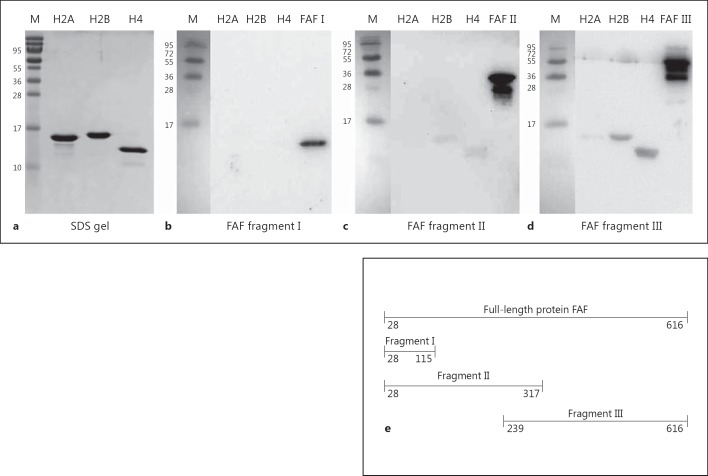
Histones H1, H2A, H2B, H3 and H4 were purchased from New England Biolabs. Anti-FAF antibodies were raised in rabbits as described [19]. Fresh-frozen citrated plasma was obtained from healthy individuals from the blood bank at the Lund University Hospital and stored at −80°C until use. Rabbit anti-human H2B was purchased from Abcam. Horseradish peroxidase (HRP) conjugated goat anti-rabbit IgG was purchased from Pierce. SufA was purified natively from F. magna ALB8 treated with papain as described previously [12]. Recombinantly expressed protein FAF [amino acid (AA) 28-616], and FAF fragments I (AA 28-115), II (AA 28-317) and III (AA 239-616), schematically depicted in figure 3e, were obtained as fusion proteins with glutathione S-transferase (GST) as described [19]. The GST-tag was removed with PreScission protease according to the manufacturer's instructions (Amersham Biosciences).
Fig. 3.
Mapping of binding of histones to the COOH terminus of FAF. H2A, H2B and H4 were subjected to SDS-PAGE and Western blot analysis. Membranes containing histones were incubated with a 15-μg/ml solution of FAF fragment I/II/III (corresponding to concentrations of 1.6/0.5/0.36 μM, respectively) and probed with a rabbit anti-FAF antibody to determine binding. M = BioRad broad-range molecular weight marker. a SDS-PAGE gel containing 2 μg of H2A, H2B and H4, to act as a size comparison with the histones detected in the Western blots. b Western blot analysis of FAF fragment I (AA 28-115 of FAF) binding to H2A, H2B and H4. c Western blot analysis of FAF fragment II (AA 28-317 of FAF) binding to H2A, H2B and H4. d Western blot analysis of FAF fragment III (AA 239-616 of FAF) binding to H2A, H2B and H4. H2A, H2B, H4: 3 μg; FAF I/II/III: 2 μg. e Schematic representation of length and location of recombinant FAF fragments.
Epidermal Lysis from Human Skin
Skin specimens were obtained as excess healthy tissue from skin surgery, under protocols approved by the Ethics Committee at Lund University (permit No. LU 762-02). Subcutaneous fat tissue was removed using scissors and the skin was cut into small pieces measuring approximately 1 cm2. The skin was incubated overnight under rotation at 4°C in epidermolysis buffer (50 mM Tris, 20 mM EDTA, pH 4.0) in the presence of Roche cOmplete Mini protease inhibitor cocktail (Roche; one inhibitor cocktail tablet per 10 ml solution). Solid NaCl was added to a final concentration of 1 M and incubation was continued for a further 3 h at 4°C. The epidermis could then be manually removed from the dermis using a tweezer.
Extraction of Proteins from Epidermal Skin Extracts
The epidermis from a 1-cm2 skin segment was transferred to 500 μl of 20% SDS in an Eppendorf tube and boiled for 20 min. Insoluble fragments were removed by spinning at 13,000 g for 20 min at room temperature. The supernatant was transferred to a fresh tube and spun again to remove all insoluble fragments.
Slot-Binding, SDS-PAGE and Western Blot Analysis
SDS-PAGE was performed as described by Neville [27]. Samples were prepared for boiling in sample buffer containing 2% SDS and 5% β-mercapto-ethanol for 5 min and 15% SDS-PAGE or 4–20% Tris-glycine gradient gels (Thermo Scientific) were used to study FAF-histone interactions. Separated proteins were visualised by Coomassie blue staining. For Western blot analysis, proteins were electrophoretically transferred to a polyvinylidene difluoride (PVDF) membrane (Amersham Biosciences). Proteins were also directly applied to PVDF membranes using a Milliblot-D system (Millipore). Membranes were blocked in a phosphate-buffered saline-Tween [PBS containing 0.1% Tween-20 (PBS-T)] solution containing 5% (wt/vol) skim milk powder at 37°C for 30 min. The membranes were then incubated with FAF (0.23 μM) for 1 h at 37°C in blocking buffer. Membranes were washed three times with PBS-T for 5 min followed by incubation with primary antibodies (rabbit anti-FAF 1:1,000 dilution) in blocking buffer at 37°C for 30 min. Membranes were washed three times with PBS-T for 5 min followed by incubation with HRP-conjugated goat anti-rabbit secondary antibody (1:3,000 dilution) in blocking buffer at 37°C for 30 min. Following a repeat of the wash steps, bound antibodies were detected by chemiluminescence as described by Nesbitt and Horton [28].
Immunoprecipitation of FAF in Complex with Histones
Epidermal skin extract (150 μl) was incubated with recombinant FAF (1.55 μM) for 3 h at room temperature. Rabbit anti-H2B antibody was added at a concentration of 1 μg/ml and incubated for a further hour at room temperature with end-over-end rotation. Fifty microlitres of protein A Sepharose (2 mg/ml; GE Healthcare Bio-Sciences), pre-washed 4 times with PBS, were added and incubated for a further 4 h at room temperature with end-over-end rotation. The Sepharose beads were collected by spinning at 3,000 g for 5 min, the supernatant was discarded and the Sepharose washed 4 times with PBS. Following the final wash step, the Sepharose was boiled in 20 μl SDS sample buffer for 5 min to elute any bound protein FAF. The sample was spun again and the supernatant analysed by SDS-PAGE and Western blot.
Bactericidal Assay
F. magna 505 and ALB8 were grown to mid-log phase (A620 ≈ 0.4-0.6) in Todd-Hewitt broth supplemented with Tween-80. Bacteria were washed and diluted in 10 mM Tris-HCl, pH 7.5. Fifty microlitres of a 2 × 106-cfu/ml solution of bacteria was added to various concentrations of histones H2A, H2B and H4 for 1 h at 37°C under strict anaerobic conditions. Bactericidal activity was quantified by plating serial dilutions of the incubation mixtures on blood agar in duplicate, which were incubated anaerobically for 3 days, and the number of cfu was determined. In later experiments, 505 bacteria were incubated with histones together with different amounts of FAF (0.009-0.9 μM) for 1 h at 37°C and the bactericidal activity was determined.
Cleavage of Histones by SufA
SufA (0.11 μM) was incubated with H1, H2A, H2B, H3 and H4 (3.5 μg of each histone) in 50 mM Tris-HCl (pH 7.5). As a control, SufA was substituted with buffer. Following incubation for 3 h at 37°C, the samples were subjected to SDS-PAGE analysis using a polyacrylamide concentration of 15%. Further investigation into SufA cleavage of histones was carried out by adding citrated human plasma or human skin epidermal extract to the reaction to imitate physiological conditions. To investigate SufA cleavage in human plasma, 5 or 10% plasma were incubated with 0.11 μM SufA in 50 mM Tris-HCl (pH 7.5). As no H2B could be detected in human plasma, 3.3 μM of H2B were added and the reaction was incubated for 3 h at 37°C and subsequently analysed by SDS-PAGE. To examine H2B cleavage in epidermal skin extracts, 5.3 μg of epidermal skin extract were incubated with 0.11 μM SufA in 50 mM Tris-HCl (pH 7.5). The reaction was supplemented with 3.3 μM of H2B and the incubation was carried out under the same conditions as above.
Results
F. magna Binds to Histones H2B and H4 Extracted from Human Epidermis
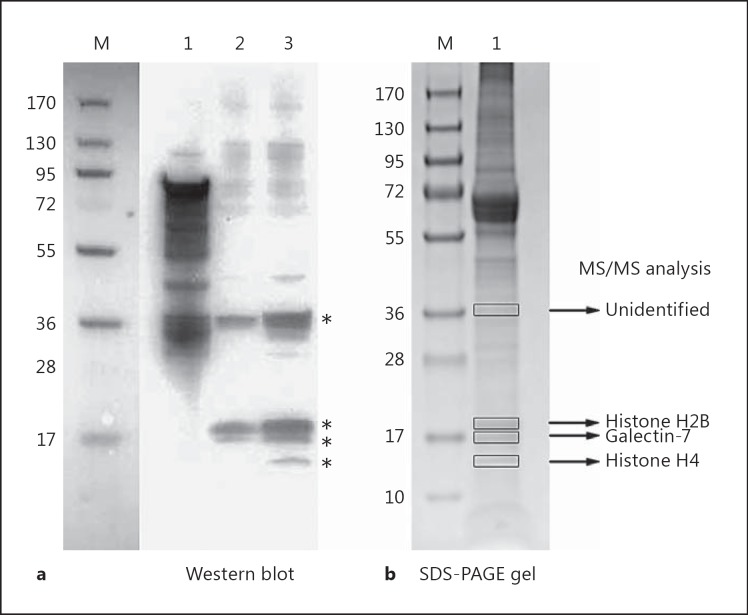
To investigate if the surface adhesion protein FAF can mediate F. magna binding to epidermal skin proteins, protein extracts from human skin epidermis were subjected to SDS-PAGE. Separated proteins were transferred to a PVDF membrane, which was incubated with a solution containing FAF. Binding of FAF to epidermal components was detected by using antibodies against FAF. As shown in figure 1a, FAF bound four proteins from the human skin epidermis. These protein bands were excised from an SDS-PAGE gel and identified by tandem mass spectrometry (MS/MS). The 14-kDa protein band was identified as histone H4, the 17-kDa band as galectin-7 and the 19-kDa band as histone H2B. The 36-kDa band could not be identified due to a too low concentration. Galectin-7 is a member of the family of animal lectins and is involved in epithelial cell migration [29]. One of the ligands to galectin-7 is FAF [Murphy et al., in preparation], which is further emphasized by the MS/MS analysis of the FAF-interacting bands shown in figure 1. The aim of this study will be to investigate the biological role of F. magna binding to histones.
Fig. 1.
Investigation of FAF binding to human epidermal skin proteins. M = BioRad broad-range molecular weight marker. a Human epidermal skin proteins were subjected to SDS-PAGE using a 4–20% Tris-glycine gradient gel followed by Western blot analysis. The membrane was incubated with a 0.23-μM solution of FAF in blocking buffer followed by probing with a rabbit anti-FAF antibody to determine binding. Lane 1 = Protein FAF 1 μg; lane 2 = human epidermal skin extract proteins 5.3 μg; lane 3 = human epidermal skin extract proteins 10.6 μg. The heterogeneity of the FAF-positive control is due to FAF degradation, which cannot be removed by gel filtration chromatography due to aggregation of the fragments to each other. b Epidermal skin proteins which bound to FAF in a Western blot were excised from an SDS-PAGE gel and identified by MS/MS analysis. Lane 1 = Epidermal skin protein extract.
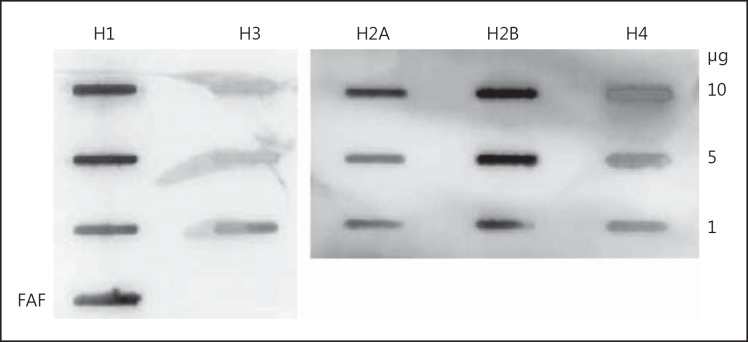
Binding of FAF to Purified Histones H1, H2A, H2B, H3 and H4
To investigate the binding of FAF to histones H2B and H4, purified forms of the histones were used and a slot-binding analysis was carried out. Histone H2A was added to the study at this point, considering its high structural similarity with H2B [30]. Binding of FAF to H1 and H3 was also investigated to determine if FAF could interact with all histones (fig. 2). This blot clearly shows strong binding of FAF to H2B and H1, with a slightly less intense binding to H2A and H4 and no binding to H3. This confirms the findings from the epidermal protein extract study and MS/MS analysis.
Fig. 2.
Binding of whole FAF to purified forms of H1, H2A, H2B, H3 and H4 by slot blot analysis. Various amounts of histones were applied to a PVDF membrane. 1 μg FAF was loaded as a positive control. The membrane was incubated with a 0.23-μM solution of FAF in blocking buffer followed by probing with a rabbit anti-FAF antibody to determine binding.
In order to map the binding of H2A, H2B and H4 to a particular part of the FAF molecule, binding studies by Western blotting were carried out using recombinant FAF fragments that were constructed in a previous study [19]. Fragments I and II correspond to the NH2-terminal part of the protein and consist of AA 28-115 (10 kDa) and 28-317 of FAF (32 kDa), respectively. Fragment III corresponds to the COOH-terminal part of FAF from AA 239-616 (42 kDa). A schematic depiction of FAF and the various fragments is shown in figure 3e. As shown in figure 3d, FAF fragment III demonstrates binding to H2B and H4, with a weaker binding to H2A. There is no binding by FAF fragment I to any of the histones and a very weak binding by FAF fragment II to H2B and H4. The secondary structure of FAF is largely α-helical and is predicted to have a high probability of forming a coiled-coil structure in the most extracellular region of the molecule (AA 28-417) [19]. Secondary structure analysis using Jpred (not shown) predicts that the most COOH-terminal part of the molecule (AA 417-616) does not adopt a coiled-coil structure and is a mixture of α-helix and β-sheet [31]. The COOH-terminal half of FAF (AA 239-616) seems to be an important adhesion site on the FAF molecule and has previously been shown to be responsible for binding to the skin basement membrane protein BM-40 [19]. However, efficient binding to host proteins probably requires a combination of fragments II and III of FAF where there will be a complete coiled-coil structure in the intact protein.
In summary, these results demonstrate an interaction between FAF and histones, where the major part of the binding occurs in the COOH-terminal half of protein FAF.
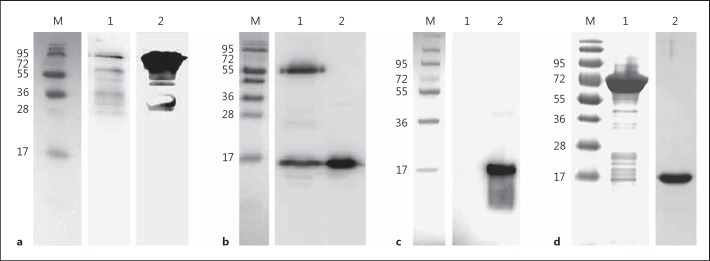
FAF Associates with Histone H2B in Solution
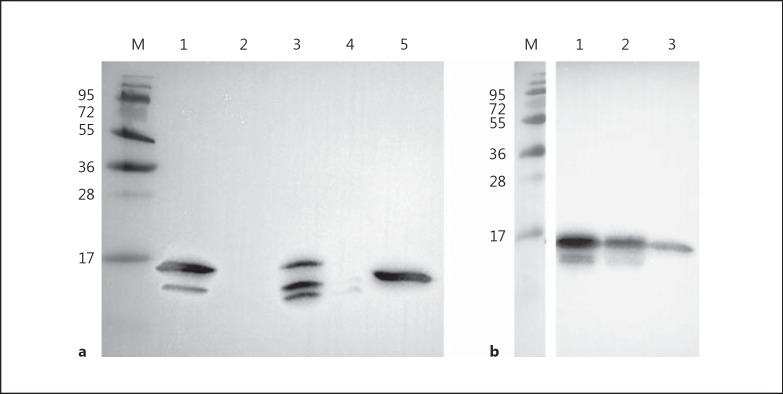
To demonstrate that FAF also associates with histones in solution, immunoprecipitation experiments were performed. Western blot analysis of immunoprecipitated proteins using an anti-FAF antibody clearly showed that FAF has associated with histone H2B in solution (fig. 4a). The presence of smaller immunoreactive bands in the FAF sample is due to breakdown of FAF into smaller fragments. Western blot analysis was also carried out using an anti-H2B antibody to confirm that the bands seen in figure 4a were not background binding (fig. 4b). Lane 1 shows two protein bands that have been bound by the anti-H2B antibody. The highest band is approximately 55 kDa and could represent H2B in complex with a FAF fragment. The lower band represents histone H2B that has not formed a complex with FAF. In order to ensure that the higher band seen in lane 1 is not just the result of cross-reactivity between the anti-H2B antibody and FAF, a further blot was carried out where pure FAF was probed directly with the anti-H2B antibody (fig. 4c, lane 1). This blot shows no cross-reactivity of the antibody with FAF, strongly suggesting that the high-molecular-weight band seen in figure 4b is actually an FAF fragment in complex with H2B. Figure 4d exhibits an SDS-PAGE gel with pure FAF and H2B for size comparisons. This experiment demonstrates that in solution the native form of FAF is able to associate with the native form of H2B, thereby protein-protein complexes are formed. The biological function of this interaction is further investigated below.
Fig. 4.
FAF forms complex with H2B in solution. Human skin epidermal extract was incubated with FAF (1.55 μM) and then immunoprecipitated using an anti-histone H2B antibody. M = BioRad broad-range molecular weight marker. a Western blot analysis on immunoprecipitated material was carried out using rabbit anti-FAF antibody to determine binding of FAF by H2B in solution. Lane 1 = Immunoprecipitation of FAF in complex with H2B; lane 2 = FAF 1.5 μg. b Western blot analysis was carried out using rabbit anti-H2B antibody, to confirm binding of H2B by FAF in solution. Lane 1 = Immunoprecipitation of H2B in complex with FAF; lane 2 = H2B 1 μg. c Western blot analysis using rabbit anti-H2B antibody to check cross-reactivity with protein FAF. Lane 1 = FAF 1.5 μg; lane 2 = H2B 1 μg. d SDS-PAGE gel displaying FAF and H2B. Lane 1 = FAF 1.5 μg; lane 2 = H2B 1 μg.
Bactericidal Activity of H2A, H2B and H4 against F. magna and the Protective Effect of FAF
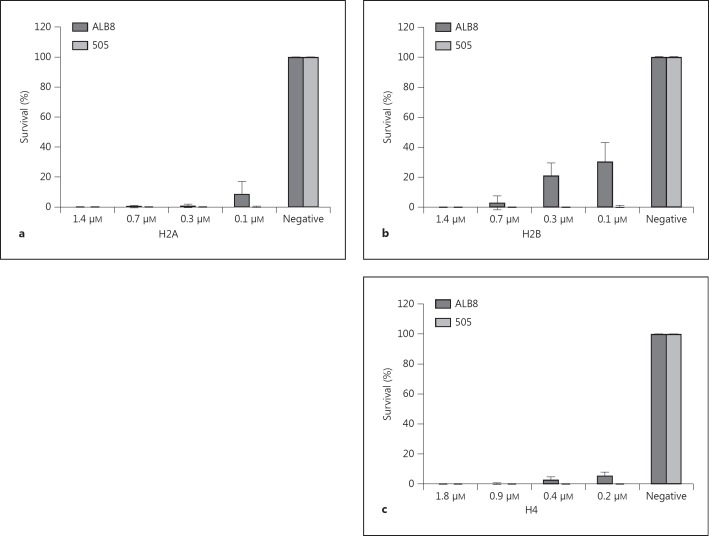
Previous studies have demonstrated that histones possess antimicrobial activity [23, 25]. Due to the strong association between protein FAF and histones seen in several binding assays, it was investigated whether histones exert a bactericidal activity against F. magna and if this effect can be neutralised by FAF. Initially, a concentration gradient of H2A, H2B and H4 [from 0.1 to 1.0 μg (H2A, H2B: 0.1-1.4 μM; H4: 0.2-1.8 μM)] was used to determine the bactericidal activity against F. magna 505 (a non-FAF-expressing strain of F. magna) and ALB8 (an FAF-expressing strain). A 100% killing of the 505 strain occurred at all histone concentrations (fig. 5). In contrast, survival of the ALB8 strain increases as the concentration of histones decreases (fig. 5). Survival of ALB8 is highest against H2B, with the least percent survival for H4. This could be due to FAF being able to bind H2B and H2A stronger than it does to H4 (fig. 2). Therefore, it has a diminished capacity to neutralise the bactericidal activity of H4.
Fig. 5.
The bactericidal effect of histones against F. magna. Percent survival of F. magna strains ALB8 and 505 in the presence of different concentrations of H2A (a), H2B (b) and H4 (c). The negative control contains no histones. Bars represent means ± SE of at least 3 experiments.
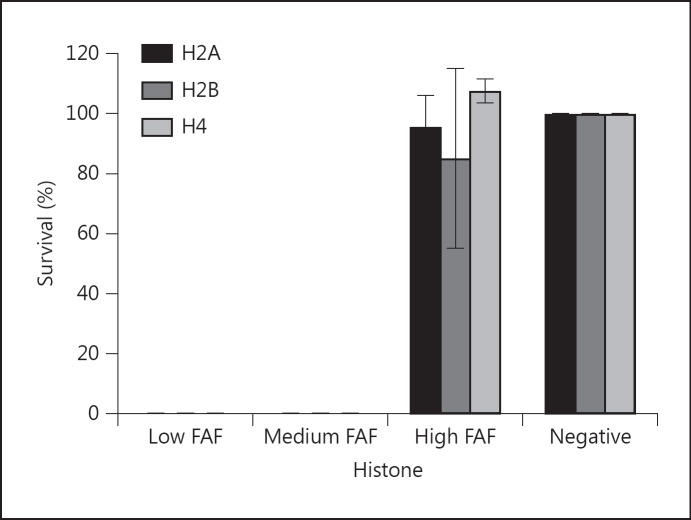
To further investigate the effect of FAF binding on the antimicrobial effect of histones, different concentrations of FAF were tested for its inhibitory effect at bactericidal concentrations of the histones (0.5 μg of H2A/H2B/H4 corresponding to 0.7 μM of H2A/H2B and 0.9 μM of H4). In the presence of H2A, H2B or H4 with 0.009 and 0.09 μM FAF, there is no survival of F. magna 505 (fig. 6). However, the addition of 0.9 μM FAF almost completely protects the bacterium from the antimicrobial effect of the histones and there is almost 100% survival when compared to a negative control (no histones added).
Fig. 6.
FAF protects F. magna against the antibacterial activity of histones. Bactericidal assay showing percent survival of F. magna strain 505 by addition of 0.5 μg H2A, H2B or H4 (corresponding to 0.7 μM of H2A/H2B and 0.9 μM of H4) in the presence of low (0.009 μM), medium (0.09 μM) and high amounts (0. 9 μM) of FAF. The negative control contains no histones. Bars represent means ± SE of at least 3 experiments.
These results show that the FAF-histone interaction has important biological implications for F. magna. FAF binding appears to protect the bacteria from the antimicrobial activity of H2A, H2B and H4, which is of great advantage to the bacteria during infection, when they are exposed to these peptides.
Histones Are Cleaved and Degraded by SufA
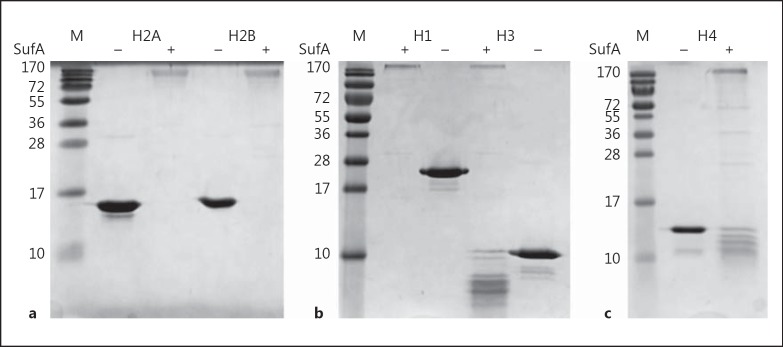
SufA was the first proteinase to be isolated and characterised from F. magna. Studies have found it to have several important functions, including the cleavage and inactivation of MIG/CXCL9, a chemokine with antibacterial activity, and the antibacterial peptide LL-37 [12, 32]. Therefore, its interaction with histones was also investigated as part of this study (fig. 7). SufA was found to completely degrade H1, H2A and H2B and cleavage of H3 and H4 can also be clearly seen. In order to examine cleavage of histones by SufA under more physiological-like conditions, human citrated plasma and human skin epidermal extracts were used to examine SufA cleavage of H2B. H2B cleavage was carried out in reactions supplemented with 5 or 10% human citrated plasma (fig. 8a). Higher concentrations of plasma could not be separated on an SDS-PAGE gel and so could not be tested. H2B was found to be completely degraded by SufA under these conditions. As can be seen from the gel, H2B is partly degraded in human plasma in the absence of SufA due to the presence of proteases in the plasma. In human skin epidermis, H2B was already present; however, SufA was unable to cleave it, which could possibly be due to altering of the histone structure during the epidermal skin extract preparation, or by being bound by another protein, and blocking the cleavage site. Hence, 3.3 μM of recombinant H2B was added to the sample. As seen in figure 8b, SufA appears to have cleaved the recombinant H2B, while the pre-existing H2B is unaffected. Degradation of histones by extracellular SufA could act as an important defence mechanism during infection, when F. magna are exposed to activated neutrophils. Inactivation of the bactericidal activity of histones would help F. magna to persist at an infection site. Taking into account FAF inactivation of histone bactericidal activity through binding, SufA acts as a double barrier to histone bactericidal activity against F. magna.
Fig. 7.
SufA cleaves histones. SufA (0.11 μM) was incubated with H1, H2A, H2B, H3 and H4 (3.5 μg of each histone) in 50 mM Tris-HCl (pH 7.5). As a control, SufA was substituted with buffer. Following incubation for 3 h at 37°C, the samples were subjected to analysis by SDS-PAGE using a polyacrylamide concentration of 15%. M = BioRad broad-range molecular weight marker. a SufA cleavage of H2A and H2B. b SufA cleavage of H1 and H3. c SufA cleavage of H4.
Fig. 8.
Cleavage of H2B by SufA in human plasma and epidermal skin extracts. SufA (0.11 μM) was incubated with H2B (3.3 μM) in the presence of human citrated plasma or skin epidermal extract in 50 mM Tris-HCl (pH 7.5). As a control, SufA was substituted with buffer. Following incubation for 3 h at 37°C, the samples were subjected to analysis by SDS-PAGE, followed by Western blotting using a rabbit anti-H2B antibody. M = BioRad broad-range molecular weight marker. a H2B cleavage in 5 or 10% citrated plasma. Lane 1 = 5% plasma positive control - SufA; lane 2 = 5% plasma + SufA; lane 3 = 10% plasma positive control - SufA; lane 4 = 10% plasma + SufA; lane 5 = H2B 0.7 μg. b H2B cleavage in human epidermal skin extract. Lane 1 = Skin extract positive control - SufA; lane 2 = skin extract + SufA; lane 3 = H2B 0.7 μg.
Discussion
F. magna is an opportunistic pathogen and its clinical incidence in infection is rising due to the increased use of foreign materials (such as catheters and joint replacement), a growing number of immunocompromised individuals and an increase in the elderly population [1, 2]. Among the GPAC, F. magna is probably the most pathogenic species and it is the organism most isolated in pure culture from different clinical infection sites [33]. Once it passes through the host's protective barriers, it must contend with the whole gambit of the host's immune response. One such response is the release of AMPs - effector molecules of innate immunity that rapidly respond to invading microorganisms. In the present investigation, F. magna was found to interact with and modulate the bactericidal effects of one particular type of AMPs, namely histones.
In the present study, the histones H2A, H2B and H4 were found to efficiently kill F. magna bacteria lacking the surface adhesion protein FAF. This protein is expressed on the surface of the majority of F. magna isolates, but it can also be released in the form of a 53-kDa fragment by the subtilisin-like protease SufA [14, 19]. Previous studies have shown that soluble FAF is able to neutralise the action of a variety of AMPs, including LL-37, midkine, BRAK/CXCL14 and hBD-3 [19, 20]. Here, we find that soluble FAF also interferes with the antibacterial function of the histones. Moreover, F. magna expressing FAF on its surface was more protected against the bactericidal activity of the histones. This protection was most obvious against histone H2B, which might be explained by a stronger binding to FAF or, possibly, a more effective antimicrobial activity of H2A and H4 against F. magna. Histones have a broad bactericidal activity, but some are more effective at killing certain types of bacteria than others. For example, the histone H2B isolated from Schegel's green tree frog Rhacophorus schlegelii was found to inhibit the growth of Escherichia coli but to have no effect on the growth of the Gram-positive bacterium S. aureus [34]. Additionally, SufA was found to cleave histones, which is in line with previous reports on SufA degradation of other AMPs, such as LL-37, MIG/CXCL3, midkine and BRAK/CXCL14, thereby reducing their antimicrobial activity [12, 14, 20].
FAF mediates bacterial adherence during colonization through binding to BM-40 at the basement membrane of the skin via its highly conserved COOH-terminal end [19]. Its ability to bind to BM-40 may also result in delayed wound healing and skin proliferation as BM-40 is found in soluble form in wound fluid and plays an important role in wound closure [35]. This may help explain why F. magna is such a prominent pathogen in chronic wounds. The interaction between FAF and histones was also localised to the conserved COOH-terminal part of the protein. Binding and blocking bactericidal activity of AMPs may help F. magna survive as a commensal in the human skin epidermis and could also prevent killing during infection.
The antimicrobial function of histones can be seen quite dramatically when neutrophils exude neutrophil extracellular traps, consisting of DNA and histones. Such traps function as both physical and antimicrobial barriers to pathogens during infection and are effective against both Gram-negative and Gram-positive bacteria [25]. A recent study showed that histone H4 is a major antimicrobial component released by sebocytes of the sebaceous gland in the skin [26]. These cells release a lipid-rich sebum onto the skin surface, but they are also thought to have an innate immune defence function through the AMP release [26]. Histone H4 was found to have antimicrobial activity against S. aureus and Propionibacterium acnes, both of which are potential pathogens of the human pilosebaceous unit [26]. This study suggests that commensals living in the skin, such as F. magna, may be periodically exposed to AMPs such as histones, so the ability to neutralise their effects is necessary to be an effective coloniser. Another study reported that histones H2A, H2B, H3 and H4 are detectable in the cell lysate of Jurkat and HeLa cells as a result of apoptosis induced by a death receptor [36]. They discovered that core nucleosomal histones are separated from chromatin during apoptosis. However, in some cases, DNA fragmentation occurred, but no histone release could be detected. Their results suggested that not every feature of apoptosis could be detected in every apoptotic setting [36]. Nevertheless, this study showed that histones are released under some apoptotic settings, suggesting that commensals, such as F. magna, could be exposed to histones during cell turnover. In a study by Frick et al. [20], protein FAF was found to be more competent at binding and neutralising the antibacterial proteins midkine and BRAK/CXCL14 than protein SIC from Streptococcus pyogenes. This is an important survival mechanism for commensal bacteria such as F. magna, as these AMPs are constitutively expressed in the basal parts of the epidermal layer as a protective mechanism during normal non-inflamed conditions [37]. Unlike pathogenic bacteria, such as S. pyogenes, it does not interfere with host homeostasis, cause inflammation or induce the release of AMPs such as β-defensins [20]. Most likely, F. magna deals in a similar way to histones as to constitutively expressed AMPs. Exposure to histones through their release from the sebaceous gland and/or cell turnover is dealt with by neutralisation with SufA and FAF, allowing F. magna to maintain its protected ecological niche at the basement membrane.
The ability of F. magna to combat histone bactericidal action may mean that it has an increased chance of survival compared to other bacteria. Furthermore, the protective mechanism of FAF and SufA as a double barrier against histone antimicrobial activity could help F. magna survive as a commensal in the human skin epidermis, but it is also a distinct advantage during opportunistic infections.
Acknowledgements
This work was supported by the Swedish Research Council (project 7480), the Foundations of Crafoord, Bergvall and Österlund, Kungliga Fysiografiska Sällskapet of Lund, the Royal Physiographic Society and Hansa Medical AB. We acknowledge the help with the protein analysis from the Proteomics Resource Centre of the Swegene Centre for Integrative Biology at Lund University.
References
- 1.Murdoch DA. Gram-positive anaerobic cocci. Clin Microbiol Rev. 1998;11:81–120. doi: 10.1128/cmr.11.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Murphy EC, Frick IM. Gram-positive anaerobic cocci - commensals and opportunistic pathogens. FEMS Microbiol Rev. 2013;37:520–553. doi: 10.1111/1574-6976.12005. [DOI] [PubMed] [Google Scholar]
- 3.Hansson C, Hoborn J, Moller A, Swanbeck G. The microbial flora in venous leg ulcers without clinical signs of infection. Repeated culture using a validated standardised microbiological technique. Acta Derm Venereol. 1995;75:24–30. doi: 10.2340/00015555752430. [DOI] [PubMed] [Google Scholar]
- 4.Stephens P, Wall IB, Wilson MJ, Hill KE, Davies CE, Hill CM, Harding KG, Thomas DW. Anaerobic cocci populating the deep tissues of chronic wounds impair cellular wound healing responses in vitro. Br J Dermatol. 2003;148:456–466. doi: 10.1046/j.1365-2133.2003.05232.x. [DOI] [PubMed] [Google Scholar]
- 5.Bowler PG, Davies BJ. The microbiology of infected and noninfected leg ulcers. Int J Dermatol. 1999;38:573–578. doi: 10.1046/j.1365-4362.1999.00738.x. [DOI] [PubMed] [Google Scholar]
- 6.Levy PY, Fenollar F, Stein A, Borrione F, Raoult D. Finegoldia magna: a forgotten pathogen in prosthetic joint infection rediscovered by molecular biology. Clin Infect Dis. 2009;49:1244–1247. doi: 10.1086/605672. [DOI] [PubMed] [Google Scholar]
- 7.Aldridge KE, Ashcraft D, Cambre K, Pierson CL, Jenkins SG, Rosenblatt JE. Multicenter survey of the changing in vitro antimicrobial susceptibilities of clinical isolates of Bacteroides fragilis group, PrevotellaFusobacteriumPorphyromonas, and Peptostreptococcus species. Antimicrob Agents Chemother. 2001;45:1238–1243. doi: 10.1128/AAC.45.4.1238-1243.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brazier J, Chmelar D, Dubreuil L, Feierl G, Hedberg M, Kalenic S, Könönen E, Lundgren B, Malamou-Ladas H, Nagy E, Sullivan A, Nord CE. European surveillance study on antimicrobial susceptibility of gram-positive anaerobic cocci. Int J Antimicrob Agents. 2008;31:316–320. doi: 10.1016/j.ijantimicag.2007.11.006. [DOI] [PubMed] [Google Scholar]
- 9.Brazier JS, Hall V, Morris TE, Gal M, Duerden BI. Antibiotic susceptibilities of Gram-positive anaerobic cocci: results of a sentinel study in England and Wales. J Antimicrob Chemother. 2003;52:224–228. doi: 10.1093/jac/dkg316. [DOI] [PubMed] [Google Scholar]
- 10.Krepel CJ, Gohr CM, Walker AP, Farmer SG, Edmiston CE. Enzymatically active Peptostreptococcus magnus: association with site of infection. J Clin Microbiol. 1992;30:2330–2334. doi: 10.1128/jcm.30.9.2330-2334.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brook I. Encapsulated anaerobic bacteria in synergistic infections. Microbiol Rev. 1986;50:452–457. doi: 10.1128/mr.50.4.452-457.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Karlsson C, Andersson ML, Collin M, Schmidtchen A, Björck L, Frick IM. SufA - a novel subtilisin-like serine proteinase of Finegoldia magna. Microbiology. 2007;153:4208–4218. doi: 10.1099/mic.0.2007/010322-0. [DOI] [PubMed] [Google Scholar]
- 13.Karlsson C, Mörgelin M, Collin M, Lood R, Andersson ML, Schmidtchen A, Björck L, Frick IM. SufA - a bacterial enzyme that cleaves fibrinogen and blocks fibrin network formation. Microbiology. 2009;155:238–248. doi: 10.1099/mic.0.021311-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karlsson C, Eliasson M, Olin AI, Mörgelin M, Karlsson A, Malmsten M, Egesten A, Frick IM. SufA of the opportunistic pathogen Finegoldia magna modulates actions of the antibacterial chemokine MIG/CXCL9, promoting bacterial survival during epithelial inflammation. J Biol Chem. 2009;284:29499–29508. doi: 10.1074/jbc.M109.025957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Björck L. Protein l. A novel bacterial cell wall protein with affinity for Ig L chains. J Immunol. 1988;140:1194–1197. [PubMed] [Google Scholar]
- 16.Genovese A, Borgia G, Björck L, Petraroli A, de Paulis A, Piazza M, Marone G. Immunoglobulin superantigen protein L induces IL-4 and IL-13 secretion from human Fc epsilon RI+ cells through interaction with the kappa light chains of IgE. J Immunol. 2003;170:1854–1861. doi: 10.4049/jimmunol.170.4.1854. [DOI] [PubMed] [Google Scholar]
- 17.de Château M, Björck L. Protein PAB, a mosaic albumin-binding bacterial protein representing the first contemporary example of module shuffling. J Biol Chem. 1994;269:12147–12151. [PubMed] [Google Scholar]
- 18.de Château M, Holst E, Björck L. Protein PAB, an albumin-binding bacterial surface protein promoting growth and virulence. J Biol Chem. 1996;271:26609–26615. doi: 10.1074/jbc.271.43.26609. [DOI] [PubMed] [Google Scholar]
- 19.Frick IM, Karlsson C, Mörgelin M, Olin AI, Janjusevic R, Hammarstrom C, Holst E, de Château M, Björck L. Identification of a novel protein promoting the colonization and survival of Finegoldia magna, a bacterial commensal and opportunistic pathogen. Mol Microbiol. 2008;70:695–708. doi: 10.1111/j.1365-2958.2008.06439.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frick IM, Nordin SL, Baumgarten M, Mörgelin M, Sørensen OE, Olin AI, Egesten A. Constitutive and inflammation-dependent antimicrobial peptides produced by epithelium are differentially processed and inactivated by the commensal Finegoldia magna and the pathogen Streptococcus pyogenes. J Immunol. 2011;187:4300–4309. doi: 10.4049/jimmunol.1004179. [DOI] [PubMed] [Google Scholar]
- 21.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 22.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 23.Kim HS, Park CB, Kim MS, Kim SC. cDNA cloning and characterization of buforin I, an antimicrobial peptide: a cleavage product of histone H2A. Biochem Biophys Res Commun. 1996;229:381–387. doi: 10.1006/bbrc.1996.1814. [DOI] [PubMed] [Google Scholar]
- 24.Kawasaki H, Iwamuro S. Potential roles of histones in host defense as antimicrobial agents. Infect Disord Drug Targets. 2008;8:195–205. doi: 10.2174/1871526510808030195. [DOI] [PubMed] [Google Scholar]
- 25.Brinkmann V, Reichard U, Goosmann C, Fauler B, Uhlemann Y, Weiss DS, Weinrauch Y, Zychlinsky A. Neutrophil extracellular traps kill bacteria. Science. 2004;303:1532–1535. doi: 10.1126/science.1092385. [DOI] [PubMed] [Google Scholar]
- 26.Lee DY, Huang CM, Nakatsuji T, Thiboutot D, Kang SA, Monestier M, Gallo RL. Histone H4 is a major component of the antimicrobial action of human sebocytes. J Invest Dermatol. 2009;129:2489–2496. doi: 10.1038/jid.2009.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Neville DM., Jr Molecular weight determination of protein-dodecyl sulfate complexes by gel electrophoresis in a discontinuous buffer system. J Biol Chem. 1971;246:6328–6334. [PubMed] [Google Scholar]
- 28.Nesbitt SA, Horton MA. A nonradioactive biochemical characterization of membrane proteins using enhanced chemiluminescence. Anal Biochem. 1992;206:267–272. doi: 10.1016/0003-2697(92)90365-e. [DOI] [PubMed] [Google Scholar]
- 29.Saussez S, Kiss R. Galectin-7. Cell Mol Life Sci. 2006;63:686–697. doi: 10.1007/s00018-005-5458-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 31.Cole C, Barber JD, Barton GJ. The Jpred 3 secondary structure prediction server. Nucleic Acids Res. 2008;36:W197–W201. doi: 10.1093/nar/gkn238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Egesten A, Eliasson M, Johansson HM, Olin AI, Mörgelin M, Mueller A, Pease JE, Frick IM, Björck L. The CXC chemokine MIG/CXCL9 is important in innate immunity against Streptococcus pyogenes. J Infect Dis. 2007;195:684–693. doi: 10.1086/510857. [DOI] [PubMed] [Google Scholar]
- 33.Bourgault AM, Rosenblatt JE, Fitzgerald RH. Peptococcus magnus: a significant human pathogen. Ann Intern Med. 1980;93:244–248. doi: 10.7326/0003-4819-93-2-244. [DOI] [PubMed] [Google Scholar]
- 34.Kawasaki H, Isaacson T, Iwamuro S, Conlon JM. A protein with antimicrobial activity in the skin of Schlegel's green tree frog Rhacophorus schlegelii (Rhacophoridae) identified as histone H2B. Biochem Biophys Res Commun. 2003;312:1082–1086. doi: 10.1016/j.bbrc.2003.11.052. [DOI] [PubMed] [Google Scholar]
- 35.Brekken RA, Sage EH. SPARC, a matricellular protein: at the crossroads of cell-matrix. Matrix Biol. 2000;19:569–580. doi: 10.1016/s0945-053x(00)00105-0. [DOI] [PubMed] [Google Scholar]
- 36.Wu D, Ingram A, Lahti JH, Mazza B, Grenet J, Kapoor A, Liu L, Kidd VJ, Tang D. Apoptotic release of histones from nucleosomes. J Biol Chem. 2002;277:12001–12008. doi: 10.1074/jbc.M109219200. [DOI] [PubMed] [Google Scholar]
- 37.Inazumi T, Tajima S, Nishikawa T, Kadomatsu K, Muramatsu H, Muramatsu T. Expression of the retinoid-inducible polypeptide, midkine, in human epidermal keratinocytes. Arch Dermatol Res. 1997;289:471–475. doi: 10.1007/s004030050223. [DOI] [PubMed] [Google Scholar]