Abstract
Interferon (IFN)-α is able to stimulate many cellular genes and inhibit the replication of various viruses. However, it is unknown whether some IFN-stimulated genes (ISGs) specifically inhibit hepatitis B virus (HBV) replication. Therefore, we attempted to identify ISGs with antiviral activities against HBV. Knockdown of IFN-induced proteins with tetratricopeptide repeats 1 and 2 (IFIT1 and IFIT2) in HepG2.2.15 led to markedly increased HBV replication. Consistently, this effect was verified by transient transfection with a replication-competent HBV clone in HepG2 and Huh7. However, IFN-α stimulation could override the knockdown by siRNAs and enhance the expression of IFIT1 and IFIT2, leading to reduced HBV replication. Silencing of IFIT1 or IFIT2 decreased the expression of the corresponding genes while other ISGs like MxA were not affected. Northern blot analysis showed that IFIT1 and IFIT2 knockdown slightly increased the levels of HBV 3.5, 2.4 and 2.1 kb transcripts, while IFIT1 and IFIT2 overexpression did not change their levels. Consistently, the reporter assays with HBV promoters demonstrated that IFIT1 and IFIT2 differentially but only modestly regulated HBV promoter activity. Thus, IFIT1 and IFIT2 contribute significantly to the regulation of HBV replication, likely at both transcriptional and posttranscriptional steps.
Key Words: IFIT1, IFIT2, HBV
Introduction
The hepatitis B virus (HBV) is a double-stranded DNA virus belonging to the family of Hepadnaviridae [1]. Chronic HBV infection is a global public health problem affecting more than 350 million people in the world, of whom approximately 600,000 die annually due to HBV-related liver diseases [2]. Patients with chronic hepatitis B have a high risk of developing liver cirrhosis and hepatocellular carcinoma [1]. Acute HBV infection is mainly self-limiting in adults. It is evident that HBV-specific T cell responses play a pivotal role in the control of HBV infection via noncytopathic mechanisms involving antiviral cytokines such as interferon (IFN)-γ and tumor necrosis factor (TNF)-α, which contribute significantly to HBV clearance [3, 4, 5].
Several studies suggest that HBV does not trigger an innate response in infected cells [6, 7, 8]. However, IFN-α and -β have been shown to be able to suppress HBV replication [9, 10] and recombinant IFN-α has been approved by the FDA as a standard treatment for chronic hepatitis B. Significant efforts have been expended in elucidating the mechanisms of the IFN-mediated anti-HBV effect [11, 12, 13]. Accumulated evidence shows that IFN-α and -β can inhibit HBV replication by preventing the formation of the pregenomic RNA-containing capsid [11] and promoting the decay of replication-competent nucleocapsids [12]. In hepatoma cells and in a mouse model, IFN-α was shown to be able to stimulate a number of cellular genes and inhibit HBV replication. However, it is not known whether some IFN-stimulated genes (ISGs) are able to directly inhibit HBV replication. In an HBV transgenic mouse model, twenty-nine genes were identified to be associated with the IFN-induced inhibition of HBV replication [14] and four ISGs, APOBEC3C [15, 16, 17], MxA [18, 19, 20], TRIM22 [21] and IDO [22], are reported to mediate the anti-HBV effect of type I IFN.
IFN-induced proteins with tetratricopeptide repeats 1 and 2 (IFIT1 and IFIT2) are related genes which can be strongly induced by type I IFN, dsRNA and many viruses [23, 24]. Both genes contain multiple tetratricopeptide repeat (TPR) motifs and are located on chromosome 10. The TPR domains mediate the protein-protein interaction. IFIT1 was found to interact with eIF3e and eEF1A and block the initiation of translation [25, 26, 27]. IFIT2 could bind to both eIF3c and eIF3e subunits and inhibit the initiation of translation [24]. Recently, IFIT1 was identified as a sensor for 5′pppRNA. It also forms complexes with other IFIT family members and inhibits virus replication [28]. IFIT1 was reported to mediate IFN blockage of hepatitis C virus replication through translational control programs together with protein kinase R [29]. It can also inhibit human papilloma virus replication by interaction with the viral E1 protein [30]. However, some viruses have developed strategies to escape the inhibition by IFIT family genes. For example, the 2′-O methylation of the 5′ cap of viral RNA functions to subvert innate host antiviral responses through escape of IFIT-mediated suppression [31]. In this report we identify IFIT1 and IFIT2 as genes which are involved in the control of HBV replication in human hepatoma cells.
Material and Methods
Cell Lines and Constructs
Huh7 cells were maintained in Dulbecco's modified Eagle medium supplemented with 10% fetal bovine serum, 100 U/ml penicillin and 100 μg/ml streptomycin. HepG2 and HepG2.2.15 cells were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum, 100 U/ml penicillin and 100 μg/ml streptomycin. HepG2.2.15 cells were cultured in the presence of 500 μg/ml of G418 to maintain the stably transfected dimeric HBV genome.
The pSM2 plasmid, harboring a head-to-tail tandem dimer of the HBV genome, (GenBank accession number: V01460) was provided by Dr. Hans Will (Heinrich-Pette-Institute, Hamburg, Germany). Full-length cDNAs for IFIT1 (NM_001548) and IFIT2 (NM_001547) were amplified by RT-PCR using the primer pairs: IFIT1-sense: 5′-gcg gat ccc atg agt aca aat ggt gat gatc-3′, IFIT1-antisense: 5′-cgc tcg agc taa gga cct tgt ctc aca g-3′, and IFIT2-sense: 5′-gcg gat ccc atg agt gag aac aat aag-3′, IFIT2-antisense: 5′- cgc tcg agt cat tcc cca ttc cag ctt g −3′. The RT-PCR fragments containing the coding regions of the IFIT1 and IFIT2 genes were inserted into expression plasmid pcDNA3.1 (Invitrogen, Karlsruhe, Germany) by the restriction sites BamHI and XhoI tagged to the primers. The expression plasmids for IFIT1 and IFIT2 were entirely sequenced to verify the correctness of the sequences.
Reporter Plasmids and Reporter Assay
The luciferase reporter vectors with HBV promoters, pSP1, pSP2, pCP, pXP and pEN2/CP were described previously [32]. The renilla luciferase reporter plasmid pRL-TK was purchased from Promega (Madison, Wisc., USA) and used as an internal control. The firefly luciferase reporter plasmid pISRE-luc was purchased from Clontech (Mountain View, Calif., USA). For the reporter assay, Huh7 cells (8 × 104 cells/well in 24-well plates) were first transfected with siRNAs targeting IFIT1 or IFIT2 at a concentration of 20 μM and then transfected with 0.5 μg of HBV promoter reporter plasmid together with 0.01 μg of pRL-TK by using lipofectamine 2000 (Invitrogen) 48 h later. The expression plasmids of IFIT1 and IFIT2 were cotransfected with HBV promoter reporter plasmid and pRL-TK. The firefly luciferase and renilla luciferase activities were measured by the dual luciferase reporter assay system (Promega). All reporter assays were repeated at least three times.
Western Blot Analysis
Total cell lysates were subjected to SDS-PAGE and transferred electrophoretically to a polyvinylidene difluoride membrane. The membrane was blocked with 5% nonfat milk in PBS with 0.1% Tween 20 (PBST) and then incubated with primary antibodies recognizing IFIT1, IFIT2 (Abnova) or β-actin (Sigma-Aldrich) in appropriate dilutions. After 3 washes with PBST, peroxidase-conjugated secondary antibodies matched to the primary antibodies were added. After further incubation and washing with PBST, the proteins to be detected were visualized using ECL Western blot detection reagents (GE Healthcare, Little Chalfont, UK).
HBsAg and HBeAg Chemiluminescent Microparticle Immunoassay
HBsAg and HBeAg levels in cell culture supernatants were determined using the Architect system and HBsAg and HBeAg chemiluminescent microparticle immunoassay (CMIA) kits (Abbott Laboratories, Wiesbaden-Delkenheim, Germany) according to the manufacturer's instructions.
Purification and Analysis of HBV DNA from Intracellular Core Particles and Southern Blot Analysis
Encapsidated HBV replicative intermediates were purified and subjected to Southern blot analysis as described previously [33, 34]. Briefly, cells were washed in ice-cold PBS and lysed in 0.4 ml of lysis buffer containing 50 mM Tris-HCl, pH 7.4, 1 mM EDTA, 1% NP-40 at 4°C for 15 min. Nuclei were pelleted by centrifugation and intracellular core particles were purified from the supernatants, and the HBV replicative intermediate DNA was extracted from the core particles as described before [34]. The isolated HBV DNA was electrophoresed onto a 1% agarose gel followed by denaturation and blotted onto a positive nylon membrane. After fixing at 150 J/cm2, the membrane was prehybridized in QuikHyb solution at 68°C for 10-20 min and then hybridized with a 32P-labeled full-length HBV DNA probe in QuikHyb solution at 68°C overnight. Hybridization signals were visualized and analyzed by a Phospho-Imager (Cyclon, Parkard Instrument).
Quantitative HBV-PCR and Real-Time RT-PCR
HBV-DNA was quantified by real-time PCR on a Light Cycler instrument using QuantiTect SYBR Green PCR Kits (Qiagen, Hilden, Germany). Plasmid DNA was series diluted and used as standard. Primers RC-FW5′-gttgcccgtttgtcctctaattc-3′ and RC-REV5′-ggagggatacatagaggttcctt-3′ were used for the detection of HBV RC (relaxed circular) genomes.
To detect the mRNA level in cells described in figures 3 and , 6, total RNA was extracted from cells by Trizol reagent (Invitrogen) according to the manufacturer's instruction, and digested with DNaseI (Qiagen). The quantification of IFIT1, IFIT2, MxA and β-actin mRNA was performed by one-step real-time RT-PCR using a QuantiFast SYBR Green RT-PCR kit (Qiagen). The copy numbers of β-actin mRNA were used for normalization.
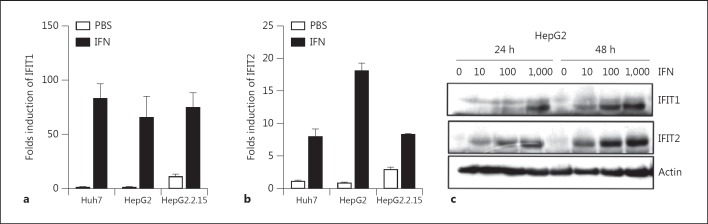
Fig. 3.
The baseline expression and induction of IFIT1 and IFIT2 by IFN in hepatoma cell lines. Huh7, HepG2 and HepG2.2.15 cells were treated with IFN-α or left untreated for 24 h. The total RNA and protein were extracted. The IFIT1 and IFIT2 expressions were analyzed by real-time RT-PCR (a, b) and Western blot (HepG2 cells; c).
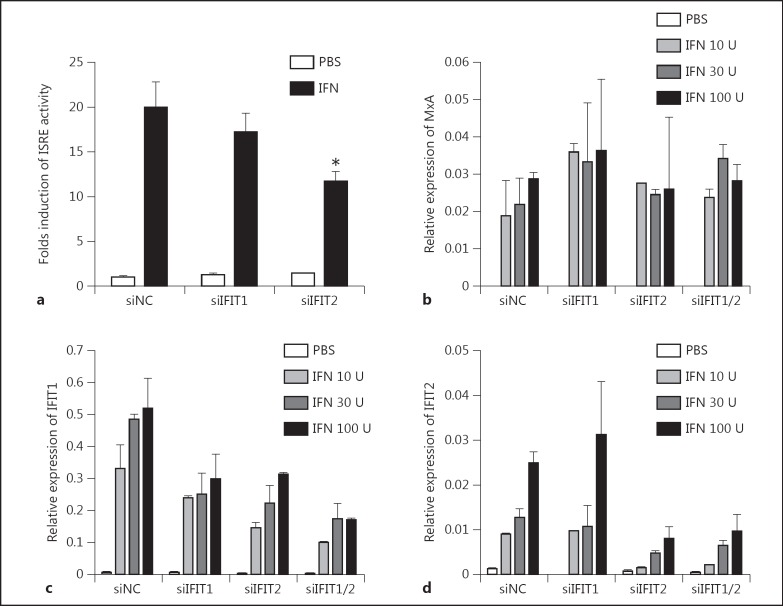
Fig. 6.
Silencing IFIT1 and IFIT2 did not affect IFN-α-induced ISG expression. a Huh7 cells were transfected with siControl, siIFIT1 or siIFIT2, incubated for 24 h, then reseeded into 24-well plates. Second transfections with pISRE-luc and pRL-TK were performed the next day. IFN-α was added to the medium 24 h later and luciferase activity was measured 6 h later. b-d Huh7 cells were transfected with siControl, siIFIT1 or siIFIT2 and incubated for 72 h. IFN-α was added to the medium and incubated for 6 h. Total RNA was extracted and IFIT1, IFIT2 and MxA mRNA levels were detected by real-time RT-PCR.
Formaldehyde-Based Northern Blot Analysis
Formaldehyde-based northern blot analysis was performed using NorthernMax kit (Ambion) according to the manufacture's instruction. Briefly, 10 μg of total RNAs were electrophoresed on a 1% agarose gel and transferred to a positively charged nylon membrane. After fixing at 150 J/cm2, the membrane was prehybridized in ULTRAhyb solution at 42°C for 1 h and then hybridized with a 32P-labeled full-length HBV DNA probe in ULTRAhyb solution at 42°C overnight. After hybridization, the signals on the nylon membrane were visualized and analyzed by a Phospho-Imager (Cyclon, Parkard Instrument).
RNA Interference
Control siRNA and validated siRNAs targeting IFIT1 (SI03224284, target sequence TACATGGGAGTTATCCATTGA) and IFIT2 (SI04295851, target sequence CACGGTATGCTTGGAACGATT) were purchased from Qiagen (Hilden, Germany). The siRNA were transfected into hepatoma cells using Lipofectamine 2000 according to the manufacturer's instructions.
Statistical Analysis
The statistical significance of the obtained data in this study was analyzed using a two-tail unpaired t test implemented in the GraphPadPrism software package (GraphPad Software Inc., San Diego, Calif., USA). A p value <0.05 was considered to be statistically significant. Data are presented as means ± standard deviation.
Results
Silencing of IFIT1 and IFIT2 Expression in Hepatoma Cells Significantly Increased HBV Replication
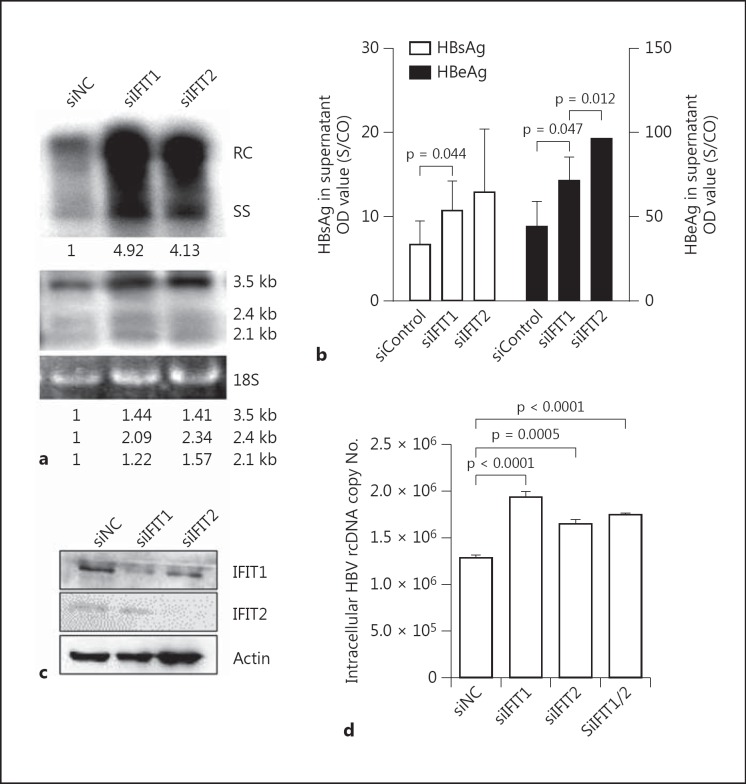
Gene array analysis using Affimetrix gene chips were performed to compare the expression level of ISGs in human hepatoma cell lines HepG2 and Huh7 upon IFN stimulation. According to the gene array results and published data from other groups, 26 genes strongly upregulated by IFN-α stimulation were screened by siRNA transfection for their role in the control of HBV replication (online suppl. table 1; for all online suppl. material, see www.karger.com/doi/10.1159/000353220). Silencing of IFIT1 and IFIT2 expression by validated siRNAs significantly increased HBV replication in Huh7 cells transfected with pSM2, while the others showed no or only a slight effect on HBV replication (data not shown). As shown in figure 1a (upper part), the amounts of core associated HBV DNA in Huh7 cells were significantly increased after transfection with siRNAs against IFIT1 and IFIT2 (siIFIT1 and siIFIT2). To a lesser extent, HBV RNA levels were elevated by IFIT1 and IFIT2 silencing as shown by Northern blot hybridization (fig. 1a, lower part). Consistently, higher HBsAg and HBeAg levels in culture supernatants of Huh7 cells transfected with siIFIT1 and siIFIT2 were measured (fig. 1b). Finally, the silencing efficiency of siIFIT1 and siIFIT2 in Huh7 cells could be verified by Western blot (fig. 1c). The effect of siIFIT1 and siIFIT2 on HBV replication was verified in HepG2.2.15 cells stably transfected with a dimeric HBV genome. The amounts of core-associated HBV DNA was isolated and quantified by real time PCR. Consistently, silencing of IFIT1 and IFIT2 in HepG2.2.15 cells increased the levels of HBV replication intermediates (fig. 1d).
Fig. 1.
Silencing of IFIT1 and IFIT2 increased HBV replication. To achieve a strong silencing effect by siRNAs, Huh7 cells were transfected with siNC, siIFIT1 or siIFIT2, incubated for 24 h and then reseeded into 6-well plates. Second transfections with pSM2 and siRNAs were performed the next day. Three days later, core-associated HBV DNA in cells was extracted and analyzed by Southern blot (a, upper part), and HBV RNAs were analyzed by Northern blot (a, lower part). The numbers below the blot images show the relative intensities of specific HBV DNA and RNA species in the blots. The 18S rRNA was used for the normalization of samples used for the Northern blot. b The HBsAg and HBeAg secreted in the supernatant were detected by CMIA. c Cell lysates were prepared and subjected to Western blot analysis for IFIT1 and IFIT2. d HepG2.2.15 cells were transfected with siControl, siIFIT1 or siIFIT2. The core-associated HBV DNA in cells was extracted and analyzed by real-time PCR 3 days later. The p value is indicated when the difference was statistically significant.
Overexpression of IFIT1 and IFIT2 Reduces HBV Replication in Hepatoma Cells and HBsAg and HBeAg Secretion
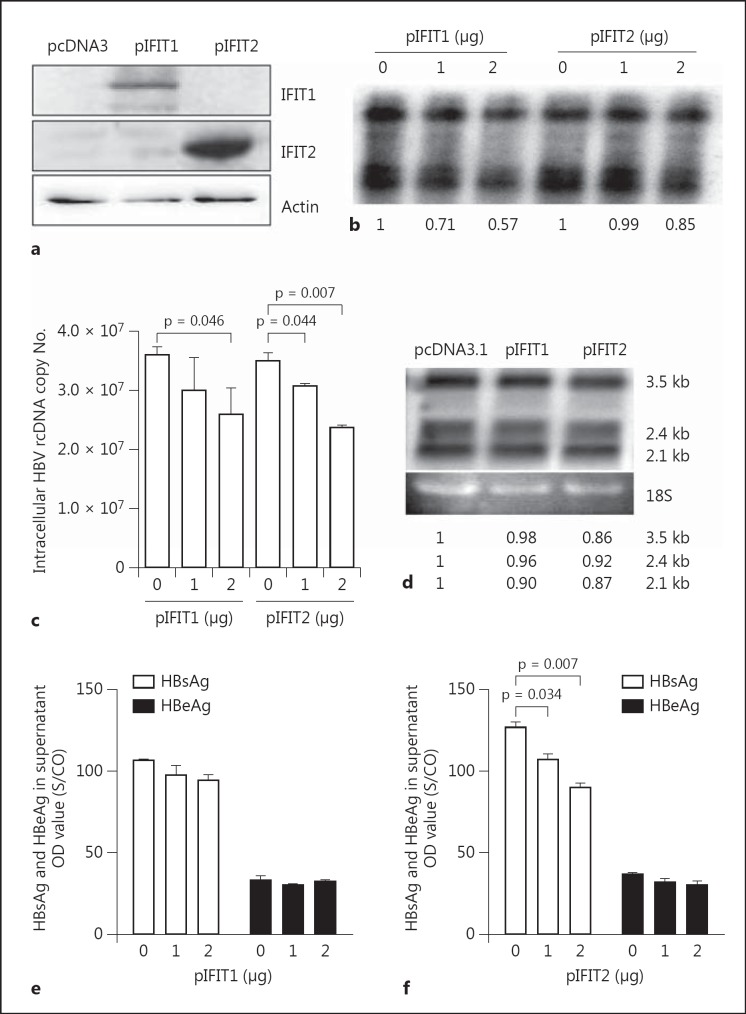
To further validate the antiviral functions of IFIT1 and IFIT2 in the context of HBV replication, two expression plasmids of IFIT1 and IFIT2, designated pIFIT1 and pIFIT2, were cotransfected into HepG2 cells with plasmid pSM2. The expression of IFIT1 and IFIT2 after pIFIT1 and pIFIT2 transfection in HepG2 could be detected by Western blot analysis, respectively (fig. 2a). The overexpression of both IFIT1 and IFIT2 reduced the accumulation of intracellular HBV replicative intermediates in a dose-dependent manner, respectively (fig. 2b). Using a real-time PCR assay designed for HBV rcDNA quantification, we showed that IFIT1 and IFIT2 overexpression reduced HBV rcDNA fractions, consistent with the results obtained by Southern blot hybridization (fig. 2c). The effect of the IFIT1 and IFIT2 overexpression on HBV RNA level was rather marginal, if at all, as shown by Northern blot analysis (fig. 2d). IFIT2 overexpression also resulted in a slight decrease of HBsAg levels in culture supernatants of transfected cells (fig. 2e, f).
Fig. 2.
IFIT1 and IFIT2 overexpression decreased HBV replication. HepG2 cells were cotransfected with pSM2 together with pcDNA3, pIFIT1 or pIFIT2. Cell lysates were prepared and analyzed by Western blot using IFIT1 or IFIT2 antibody (a). Core-associated HBV DNA in cells were extracted and analyzed by Southern blot (b) or real-time PCR (c). HBV RNAs were analyzed by Northern blot (d). The numbers below the blot images show the relative intensities of specific HBV DNA and RNA species in the blots. The 18S rRNA was used for the normalization of samples used for the Northern blot. The HBsAg and HBeAg secreted in the supernatant were detected by CMIA (e, f). The p value is indicated when the difference was statistically significant.
Expression and Induction of IFIT1 and IFIT2 by IFN in Hepatoma Cell Lines
IFIT1 and IFIT2 are two of the most potently induced genes by type I IFN. The baseline expression of IFIT1 and IFIT2 and the induction of these two genes by IFN-α in different hepatoma cell lines were analyzed next. Huh7, HepG2 and HepG2.2.15 cells were treated with IFN-α at a concentration of 100 units/ml or left untreated. The total RNA and protein were prepared from the cells 24 h later. IFIT1 and IFIT2 mRNA and protein levels were detected by real-time RT-PCR and Western blot, respectively (fig. 3a-c). The IFIT1 and IFIT2 RNAs were present at low levels in untreated hepatoma cells. However, the IFIT1 and IFIT2 RNA levels were found at higher levels in HepG2.2.15 with HBV replication, compared with those in HepG2 and Huh7. IFIT1 and IFIT2 expression were strongly induced to higher levels upon IFN-α stimulation in all cell lines. The IFIT1 and IFIT2 proteins were detected in all hepatoma cells, at least after IFN-α stimulation (fig. 3c; online suppl. fig. 1). Two more human hepatoma cell lines, Hep3B and pLC, were also tested for IFIT1 and IFIT2 expression with and without IFN-α stimulation. Similar results were obtained for these cell lines (online suppl. fig. 1).
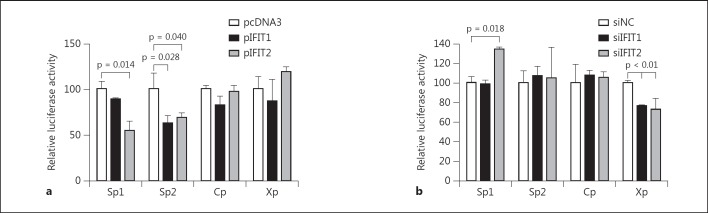
IFIT1 and IFIT2 Affect HBV S Promoter Activity
To examine the antiviral functions of IFIT1 and IFIT2 against HBV, we tested whether IFIT1 and IFIT2 expression regulates the transcriptional activity of HBV promoters. Four reporter plasmids containing the HBV promoter regions were constructed and used in this study [34]. Cotransfection of the expression vectors for IFIT1 and IFIT2 with reporter plasmids in Huh7 cells resulted in a decrease of HBV SP1 and SP2 promoter activity, while the activity of HBV C and X promoters was not affected by IFIT1 or IFIT2 overexpression (fig. 4a). On the other hand, when siIFIT2 was applied to Huh7 cells, the activity of the HBV SP1 promoter was elevated 1.5-fold compared to the control siRNA transfection, while siIFIT1 did not enhance HBV SP1 promoter activity (fig. 4b). Also, both siIFIT1 and siIFIT2 did not affect HBV SP2 promoter activity. Surprisingly, knockdown of IFIT1 and IFIT2 caused a slight reduction of the X promoter activity even though the X promoter was not affected by IFIT1 or IFIT2 overexpression, indicating that both IFIT1 and IFIT2 might play the role at very low expression levels. These results indicate that IFIT1 and IFIT2 are able to influence HBV promoter activity in different manners.
Fig. 4.
IFIT1 and IFIT2 affect HBV S promoter activity. a Huh7 cells were cotransfected with pRT-TK and HBV promoter report plasmid, pSp1, pSp2, pCp or pXp, together with pcDNA3, pIFIT1 or pIFIT2. pRL-TK was used as an internal control. Relative luciferase activity was measured by dual-luciferase assay 48 h after transfection. The promoter activity of cells transfected with pcDNA3 was set as 100. b Huh7 cells were first transfected with siRNAs as indicated, then transfected with HBV promoter report plasmid together with pRL-TK as the internal control the next day. Luciferase activity was measured 48 h after plasmid transfection. The relative luciferase activity in cells transfected with siControl was set as 100.
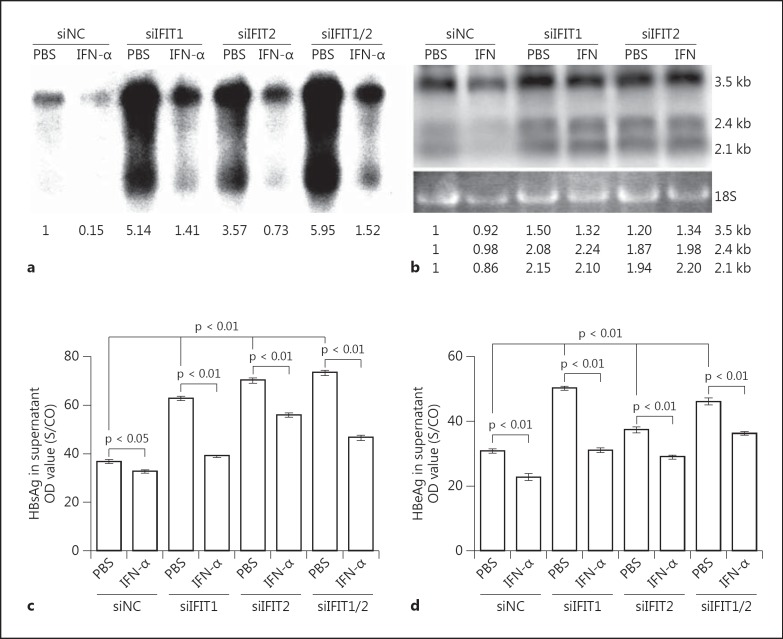
Role of IFIT1 and IFIT2 Expression in the Antiviral Effect of IFN-α
IFIT1 and IFIT2 are two of the most highly activated genes induced by IFN-α. Thus, we next considered whether IFIT1 and IFIT2 are essential for the antiviral effect of IFN-α against HBV. Huh7 cells were first transfected with siIFIT1, siIFIT2 or control siRNA. Cells were reseeded 24 h later and then transfected with the plasmid pSM2 together with corresponding siRNAs. IFN-α were added to the medium following pSM2 transfection at a concentration of 100 units/ml. The cells were maintained for 4 days and subjected to analysis of HBV replication. The cell culture supernatants were collected for HBsAg and HBeAg detection. HBV core-associated DNA and HBV RNAs were extracted from cells and analyzed by Southern blot and Northern blot hybridization. Silencing of IFIT1 and IFIT2 significantly increased the amounts of HBV replication intermediates and HBV transcripts, even in the presence of IFN-α (fig. 5a, b, lanes 2, 4, 6 and 8), and also enhanced the production of HBsAg and HBeAg (fig. 5c, d). However, siIFIT1 and siIFIT2 together did not have synergistic effects. siIFIT1 and/or siIFIT2 did not fully rescue HBV replication inhibited by IFN-α, indicating that other ISGs might also have anti-HBV activity.
Fig. 5.
Role of IFIT1 and IFIT2 in the antiviral effect of IFN-α. Huh7 cells were transfected with siControl, siIFIT1 or siIFIT2, incubated for 24 h, then reseeded into 6-well plates. Second transfections with pSM2 and siRNAs were performed the next day. IFN-α was added to the medium following pSM2 transfection and maintained for 4 days. Core-associated HBV DNA in cells were extracted and analyzed by Southern blot (a), and HBV RNAs were analyzed by Northern blot (b). The numbers below the blot images show the relative intensities of specific HBV DNA and RNA species in the blots. The 18S rRNA was used for the normalization of samples used for the Northern blot. The HBsAg and HBeAg secreted in the supernatant were detected by CMIA (c, d). The p value is indicated when the difference was statistically significant.
To accurately interpret the above results, we asked whether silencing of IFIT1 and IFIT2 influenced ISG expression. The ISRE promoter activity and the expression of MxA, IFIT1 and IFIT2 induced by IFN-α after siIFIT1 and siIFIT2 transfection were determined by luciferase assays and real-time RT-PCR, respectively (fig. 6). Transfection of Huh7 cells with siIFIT1 did not have a significant effect on the ISRE promoter activity, while a slight decrease of the IRES promoter activity was measured after siIFIT2 transfection (fig. 6a). The expression level of MxA stimulated by IFN-α was not changed by siIFIT1 or siIFIT2 (fig. 6b). IFN-α treatment could overcome the silencing effect of siIFIT1 and siIFIT2, and induced IFIT1 and IFIT2 expression (fig. 6c, d, online suppl. fig. 2). Nevertheless, the induction of IFIT1 and IFIT2 by IFN-α was reduced significantly by siIFIT1 and siIFIT2. Interestingly, the silencing of IFIT2 could somehow reduce the induction of IFIT1 expression by IFN stimulation, which might be explained by the fact that silencing of IFIT2 significantly reduced the ISRE-mediated gene expression for as yet unknown reasons (fig. 6a). The exact mechanism needs to be studied in further study. These results explain why siIFIT1 and siIFIT2 transfection still led to increased HBV replication in IFN-α treated hepatoma cells (fig. 5).
Discussion
In the present study, we showed that the ISGs IFIT1 and IFIT2 were involved in the control of HBV replication. Silencing of the IFIT1 and IFIT2 genes resulted in enhanced HBV replication in untreated and IFN-α-treated hepatoma cells. Conversely, IFIT1 and IFIT2 overexpression reduced HBV replication to some extent. Thus, IFIT1 and IFIT2 contribute to the control of HBV by limiting HBV replication and might play an important role during HBV infection by slowing down the replication and spread of HBV.
The potency of siIFIT1 and siIFIT2 on HBV replication differed in Huh7 and HepG2.2.15 cells. This apparent difference could be primarily due to the experimental systems: HBV replication in Huh7 cells started de novo after transient transfection, while a relatively high steady-state level of HBV replication was established in HepG2.2.15. Thus, increased HBV replication in Huh7 cells, which was more pronounced at the baseline, was low compared with in HepG2.2.15, which showed high baseline HBV replication.
It is clear that HBV replication still occurred in the presence of IFIT1 and IFIT2. The overexpression of IFIT1 and IFIT2 had only a limited inhibitory effect on HBV replication. As a reduction of the baseline expression of IFIT1 and IFIT2 by siRNAs led to more pronounced enhancement of HBV replication, we may assume that IFIT1 and IFIT2 fully exerted antiviral functions at the baseline expression level. The control of HBV replication by the cellular innate immune responses may not be adequately appreciated as they are not able to completely clear HBV from infected hepatocytes. Recently, we showed that silencing of the cellular adaptor proteins involved in TLR2 signaling led to significant enhancement of HBV replication [32]. Similarly, the activation of the TLR2 mediated signaling pathway by externally added ligands only reduced HBV replication to about 40% of the control. It should be assumed that a great number of cellular antiviral genes are readily functional at baseline and contribute decisively to the suppression of HBV replication. Once such an antiviral function is defective, HBV may replicate at a markedly higher level. In this light, host factors involved in the control of HBV replication may be identified by the gene silencing strategy and not be recognized by studies based on the overexpression of host genes [35].
In Huh7 cells transfected with siIFIT1 and siIFIT2, IFN-α strongly suppressed HBV replication in a pronounced way (fig. 5). This was certainly due to the high level of HBV replication after IFIT1 and IFIT2 silencing. We could show that IFN-α significantly upregulated the IFIT1 and IFIT2 mRNA levels despite the presence of siRNAs, although the IFIT1 and IFIT2 mRNAs did not reach the same levels as the controls. Thus, HBV replication in these hepatoma cells remained at higher levels compared with that in the Huh7 cells that received control siRNA. Silencing of IFIT1 did not have a detectable effect on the ISRE-dependent gene expression, which was also the case with MxA and IFIT2 expression. However, silencing of IFIT2 reduced slightly but statistically significantly the ISRE-dependent gene expression and IFIT1 expression (fig. 6). This finding requires further investigation to clarify its relevance. Nevertheless, it would be useful to examine HBV replication and the actions of ISGs in a background with IFIT1 and IFIT2 silenced. The antiviral effect of other ISGs may be better recognized in such an experimental approach.
The exact molecular mechanisms of how IFIT1 and IFIT2 contribute to the control of HBV replication remain to be defined. Although silencing of IFIT1 and IFIT2 specifically enhanced the activity of HBV S gene promoter but not the C and X gene promoters, the increased production of HBsAg may not directly facilitate the intracellular HBV replication process. As shown by the Northern blot analysis, the levels of HBV transcripts (3.5, 2.4 and 2.1 kb) were elevated maximally around 2-fold after silencing of IFIT1 and IFIT2. The overexpression of IFIT1 and IFIT2 had only very little effect on the levels of HBV transcripts, consistent with the limited effect on HBV replication intermediates. According to these results, IFITs affect HBV transcription to an extent which does not fully explain their effect on HBV replication. Many previous studies have suggested that IFN-α mainly regulates the posttranscriptional steps of the HBV life cycle [11, 36]. Based on these findings, it is reasonable and logical to propose the involvement of IFIT1 and IFIT2 in regulation of the posttranscriptional process of HBV replication. Wieland et al. [14] identified IFIT1 and IFIT2 in HBV transgenic mice as two members of the genes tightly associated with inhibition of HBV replication. The IFIT family contain TPR domains which can promote protein-protein interactions. IFIT1 and IFIT2 are able to bind to the eIF3 component and inhibit the initiation of protein translation. Though silencing of eIF3S6 reduced HBV replication (online suppl. fig. 3), indicating that HBV replication may require the eIF3 activity, we cannot exclude the possibility that eIF3 is essential for cell viability in general. The relevance of eIF3 for HBV replication and the relation to the antiviral effect of IFIT1 and IFIT2 needs to be examined in the future. It also remains to be examined whether IFIT proteins could interact with HBV RNAs.
HBV was believed to be a stealth virus, as it induces no or only relatively weak IFN responses during the acute phase of infection in chimpanzees, as well as in acutely infected humans [6, 8, 37]. It is reported that primary human hepatic cells exposed to HBV particles do not produce type I interferon [37]. Nonparenchymal liver cells, mainly Kupffer cells, respond to the exposure to HBV with transient release of interleukin 6 and other proinflammatory cytokines. In contrast, a strong initial HBV replication in hepatic cells may elicit IFN responses, as shown by transduction of HepRG cells with a recombinant baculovirus vector [38]. It remains to be clarified whether the baculovirus vector-mediated HBV replication is suitable to study the interaction of HBV and the type I IFN system. Although HBV infection usually does not trigger strong type I IFN responses, our results suggest that the ISGs at their baseline expression may still play a role in limiting initial HBV replication and spread, winning time for the adaptive immune responses that will ultimately clear HBV.
Supplementary Material
Supplementary data
Acknowledgements
We would like to thank Prof. Simon Rayner for critically reviewing the manuscript. This work was supported in part by the National Nature Science Foundation of China (Grant 31200315) and grants from the Deutsche Forschungsgemeinschaft (GRK1045/2 and DFG Transregio TRR60). Rongjuan Pei was supported by a joint scholarship of the Deutscher Akademischer Austauschdienst and the Chinese Academy of Science.
References
- 1.Seeger C, Mason WS. Hepatitis B virus biology. Microbiol Mol Biol Rev. 2000;64:51–68. doi: 10.1128/mmbr.64.1.51-68.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McMahon BJ. Epidemiology and natural history of hepatitis B. Semin Liver Dis. 2005;25((suppl 1)):3–8. doi: 10.1055/s-2005-915644. [DOI] [PubMed] [Google Scholar]
- 3.Guidotti LG, Ando K, Hobbs MV, Ishikawa T, Runkel L, Schreiber RD, Chisari FV. Cytotoxic t lymphocytes inhibit hepatitis B virus gene expression by a noncytolytic mechanism in transgenic mice. Proc Natl Acad Sci USA. 1994;91:3764–3768. doi: 10.1073/pnas.91.9.3764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guidotti LG, Ishikawa T, Hobbs MV, Matzke B, Schreiber R, Chisari FV. Intracellular inactivation of the hepatitis B virus by cytotoxic T lymphocytes. Immunity. 1996;4:25–36. doi: 10.1016/s1074-7613(00)80295-2. [DOI] [PubMed] [Google Scholar]
- 5.Guidotti LG, Rochford R, Chung J, Shapiro M, Purcell R, Chisari FV. Viral clearance without destruction of infected cells during acute HBV infection. Science. 1999;284:825–829. doi: 10.1126/science.284.5415.825. [DOI] [PubMed] [Google Scholar]
- 6.Wieland S, Thimme R, Purcell RH, Chisari FV. Genomic analysis of the host response to hepatitis B virus infection. Proc Natl Acad Sci USA. 2004;101:6669–6674. doi: 10.1073/pnas.0401771101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wieland SF, Chisari FV. Stealth and cunning: hepatitis B and hepatitis C viruses. J Virol. 2005;79:9369–9380. doi: 10.1128/JVI.79.15.9369-9380.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dunn C, Peppa D, Khanna P, Nebbia G, Jones M, Brendish N, Lascar RM, Brown D, Gilson RJ, Tedder RJ, Dusheiko GM, Jacobs M, Klenerman P, Maini MK. Temporal analysis of early immune responses in patients with acute hepatitis B virus infection. Gastroenterology. 2009;137:1289–1300. doi: 10.1053/j.gastro.2009.06.054. [DOI] [PubMed] [Google Scholar]
- 9.Guidotti LG, Borrow P, Hobbs MV, Matzke B, Gresser I, Oldstone MB, Chisari FV. Viral cross talk: intracellular inactivation of the hepatitis B virus during an unrelated viral infection of the liver. Proc Natl Acad Sci USA. 1996;93:4589–4594. doi: 10.1073/pnas.93.10.4589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wieland SF, Guidotti LG, Chisari FV. Intrahepatic induction of alpha/beta interferon eliminates viral RNA-containing capsids in hepatitis B virus transgenic mice. J Virol. 2000;74:4165–4173. doi: 10.1128/jvi.74.9.4165-4173.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wieland SF, Eustaquio A, Whitten-Bauer C, Boyd B, Chisari FV. Interferon prevents formation of replication-competent hepatitis B virus RNA-containing nucleocapsids. Proc Natl Acad Sci USA. 2005;102:9913–9917. doi: 10.1073/pnas.0504273102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu C, Guo H, Pan XB, Mao R, Yu W, Xu X, Wei L, Chang J, Block TM, Guo JT. Interferons accelerate decay of replication-competent nucleocapsids of hepatitis B virus. J Virol. 2010;84:9332–9340. doi: 10.1128/JVI.00918-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anderson AL, Banks KE, Pontoglio M, Yaniv M, McLachlan A. Alpha/beta interferon differentially modulates the clearance of cytoplasmic encapsidated replication intermediates and nuclear covalently closed circular hepatitis B virus (HBV) DNA from the livers of hepatocyte nuclear factor 1α-null HBV transgenic mice. J Virol. 2005;79:11045–11052. doi: 10.1128/JVI.79.17.11045-11052.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wieland SF, Vega RG, Muller R, Evans CF, Hilbush B, Guidotti LG, Sutcliffe JG, Schultz PG, Chisari FV. Searching for interferon-induced genes that inhibit hepatitis B virus replication in transgenic mouse hepatocytes. J Virol. 2003;77:1227–1236. doi: 10.1128/JVI.77.2.1227-1236.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nguyen DH, Hu J. Reverse transcriptase- and RNA packaging signal-dependent incorporation of APOBEC3G into hepatitis B virus nucleocapsids. J Virol. 2008;82:6852–6861. doi: 10.1128/JVI.00465-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baumert TF, Rosler C, Malim MH, von Weizsacker., F Hepatitis B virus DNA is subject to extensive editing by the human deaminase APOBEC3C. Hepatology. 2007;46:682–689. doi: 10.1002/hep.21733. [DOI] [PubMed] [Google Scholar]
- 17.Suspene R, Guetard D, Henry M, Sommer P, Wain-Hobson S, Vartanian JP. Extensive editing of both hepatitis B virus DNA strands by APOBEC3 cytidine deaminases in vitro and in vivo. Proc Natl Acad Sci USA. 2005;102:8321–8326. doi: 10.1073/pnas.0408223102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gordien E, Rosmorduc O, Peltekian C, Garreau F, Brechot C, Kremsdorf D. Inhibition of hepatitis B virus replication by the interferon-inducible MxA protein. J Virol. 2001;75:2684–2691. doi: 10.1128/JVI.75.6.2684-2691.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peltekian C, Gordien E, Garreau F, Meas-Yedid V, Soussan P, Willams V, Chaix ML, Olivo-Marin JC, Brechot C, Kremsdorf D. Human MxA protein participates to the interferon-related inhibition of hepatitis B virus replication in female transgenic mice. J Hepatol. 2005;43:965–972. doi: 10.1016/j.jhep.2005.06.019. [DOI] [PubMed] [Google Scholar]
- 20.Li N, Zhang L, Chen L, Feng W, Xu Y, Chen F, Liu X, Chen Z, Liu W. MxA inhibits hepatitis B virus replication by interaction with hepatitis B core antigen. Hepatology. 2012;56:803–811. doi: 10.1002/hep.25608. [DOI] [PubMed] [Google Scholar]
- 21.Gao B, Duan Z, Xu W, Xiong S. Tripartite motif-containing 22 inhibits the activity of hepatitis B virus core promoter, which is dependent on nuclear-located ring domain. Hepatology. 2009;50:424–433. doi: 10.1002/hep.23011. [DOI] [PubMed] [Google Scholar]
- 22.Mao R, Zhang J, Jiang D, Cai D, Levy JM, Cuconati A, Block TM, Guo JT, Guo H. Indoleamine 2,3-dioxygenase mediates the antiviral effect of gamma interferon against hepatitis B virus in human hepatocyte-derived cells. J Virol. 2011;85:1048–1057. doi: 10.1128/JVI.01998-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wacher C, Muller M, Hofer MJ, Getts DR, Zabaras R, Ousman SS, Terenzi F, Sen GC, King NJ, Campbell IL. Coordinated regulation and widespread cellular expression of interferon-stimulated genes (ISG) ISG-49, ISG-54, and ISG-56 in the central nervous system after infection with distinct viruses. J Virol. 2007;81:860–871. doi: 10.1128/JVI.01167-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Terenzi F, Hui DJ, Merrick WC, Sen GC. Distinct induction patterns and functions of two closely related interferon-inducible human genes, ISG54 and ISG56. J Biol Chem. 2006;281:34064–34071. doi: 10.1074/jbc.M605771200. [DOI] [PubMed] [Google Scholar]
- 25.Guo J, Hui DJ, Merrick WC, Sen GC. A new pathway of translational regulation mediated by eukaryotic initiation factor 3. EMBO J. 2000;19:6891–6899. doi: 10.1093/emboj/19.24.6891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hui DJ, Bhasker CR, Merrick WC, Sen GC. Viral stress-inducible protein p56 inhibits translation by blocking the interaction of eIF3 with the ternary complex eIF2.GTP.MET- tRNAi. J Biol Chem. 2003;278:39477–39482. doi: 10.1074/jbc.M305038200. [DOI] [PubMed] [Google Scholar]
- 27.Li HT, Su YP, Cheng TM, Xu JM, Liao J, Chen JC, Ji CY, Ai GP, Wang JP. The interaction between interferon-induced protein with tetratricopeptide repeats-1 and eukaryotic elongation factor-1A. Mol Cell Biochem. 2010;337:101–110. doi: 10.1007/s11010-009-0289-9. [DOI] [PubMed] [Google Scholar]
- 28.Pichlmair A, Lassnig C, Eberle CA, Gorna MW, Baumann CL, Burkard TR, Burckstummer T, Stefanovic A, Krieger S, Bennett KL, Rulicke T, Weber F, Colinge J, Muller M, Superti-Furga G. IFIT1 is an antiviral protein that recognizes 5′-triphosphate RNA. Nat Immunol. 2011;12:624–630. doi: 10.1038/ni.2048. [DOI] [PubMed] [Google Scholar]
- 29.Wang C, Pflugheber J, Sumpter R, Jr, Sodora DL, Hui D, Sen GC, Gale M., Jr Alpha interferon induces distinct translational control programs to suppress hepatitis C virus RNA replication. J Virol. 2003;77:3898–3912. doi: 10.1128/JVI.77.7.3898-3912.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Terenzi F, Saikia P, Sen GC. Interferon-inducible protein, p56, inhibits HPV DNA replication by binding to the viral protein E1. EMBO J. 2008;27:3311–3321. doi: 10.1038/emboj.2008.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Daffis S, Szretter KJ, Schriewer J, Li J, Youn S, Errett J, Lin TY, Schneller S, Zust R, Dong H, Thiel V, Sen GC, Fensterl V, Klimstra WB, Pierson TC, Buller RM, Gale M, Jr, Shi PY, Diamond MS. 2′-O methylation of the viral mRNA cap evades host restriction by IFIT family members. Nature. 2010;468:452–456. doi: 10.1038/nature09489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang X, Ma Z, Liu H, Liu J, Meng Z, Broering R, Yang D, Schlaak JF, Roggendorf M, Lu M. Role of Toll-like receptor 2 in the immune response against hepadnaviral infection. J Hepatol. 2012;57:522–528. doi: 10.1016/j.jhep.2012.05.004. [DOI] [PubMed] [Google Scholar]
- 33.Qiu J, Qin B, Rayner S, Wu CC, Pei RJ, Xu S, Wang Y, Chen XW. Novel evidence suggests hepatitis B virus surface proteins participate in regulation of HBV genome replication. Virol Sin. 2011;26:131–138. doi: 10.1007/s12250-011-3190-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang X, Zhang E, Ma Z, Pei R, Jiang M, Schlaak JF, Roggendorf M, Lu M. Modulation of hepatitis B virus replication and hepatocyte differentiation by microRNA-1. Hepatology. 2011;53:1476–1485. doi: 10.1002/hep.24195. [DOI] [PubMed] [Google Scholar]
- 35.Mao R, Zhang J, Jiang D, Cai D, Levy JM, Cuconati A, Block TM, Guo JT, Guo H. Indoleamine 2,3-dioxygenase mediates the antiviral effect of gamma interferon against hepatitis B virus in human hepatocyte-derived cells. J Virol. 2012;85:1048–1057. doi: 10.1128/JVI.01998-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guidotti LG, Morris A, Mendez H, Koch R, Silverman RH, Williams BR, Chisari FV. Interferon-regulated pathways that control hepatitis B virus replication in transgenic mice. J Virol. 2002;76:2617–2621. doi: 10.1128/JVI.76.6.2617-2621.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hosel M, Quasdorff M, Wiegmann K, Webb D, Zedler U, Broxtermann M, Tedjokusumo R, Esser K, Arzberger S, Kirschning CJ, Langenkamp A, Falk C, Buning H, Rose-John S, Protzer U. Not interferon, but interleukin-6 controls early gene expression in hepatitis B virus infection. Hepatology. 2009;50:1773–1782. doi: 10.1002/hep.23226. [DOI] [PubMed] [Google Scholar]
- 38.Lucifora J, Durantel D, Testoni B, Hantz O, Levrero M, Zoulim F. Control of hepatitis B virus replication by innate response of HepaRG cells. Hepatology. 2010;51:63–72. doi: 10.1002/hep.23230. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data