Abstract
Lactoferrin (LF) is a multifunctional glycoprotein that plays an important role in native immune defense against infections, including human herpetic viruses, such as cytomegalovirus and herpes simplex virus types 1 and 2. However, its anti-Epstein-Barr virus (EBV, a γ-herpesvirus) function has not been reported in the literature. EBV is widespread in all human populations and is believed to be linked to tumorigenesis, such as lymphomas and nasopharyngeal carcinoma (NPC). We previously reported that LF expressed a significantly lower level in NPC tissues and was a likely tumor suppressor. Since EBV infection is a major carcinogen of NPC development, we investigated the effect of LF on EBV infection and found that LF could protect human primary B lymphocytes and nasopharyngeal epithelial cells from EBV infection, but had no effect on EBV genome DNA replication. LF prevented EBV infection of primary B cells mediated by its direct binding to the EBV receptor (CD21) on the B-cell surface. Tissue array immunohistochemistry revealed that LF expression was significantly downregulated in NPC specimens, in which high EBV viral capsid antigen-IgA levels were observed. These data suggest that LF may inhibit EBV infection and that its downregulation could contribute to NPC development.
Key Words: Lactoferrin, Epstein-Barr virus, Nasopharyngeal carcionma, CD21
Introduction
Lactoferrin (LF) is an 80-kDa multifunctional extracellular iron-binding glycoprotein, which is expressed by polymorphonuclear leukocytes, secretory and epithelial cells and is secreted into most biological fluids including human milk, tears, nasal secretions, saliva, intestinal mucus and genital secretions. Multiple biological functions have now been described for LF, including iron homeostasis, anti-inflammatory activity, cancer protection and antimicrobials [1,2,3,4].
LF is strategically situated at the mucosa and plays a vital role in the first line of defense against microbial infections, since many pathogens tend to enter the body via the mucosa [1]. In addition to the antibacterial and antifungal effects, LF is active against a wide variety of viruses. From in vitro studies to date, it has been recognized as a potent inhibitor of different enveloped viruses such as human cytomegalovirus [5], herpes simplex virus types 1 and 2 (HSV-1 and HSV-2) [6,7], human immunodeficiency virus (HIV) [8,9], human hepatitis C virus [10] and hepatitis B virus [11]. The major mechanisms of LF in reducing virus infection are reported to be the result of LF binding to the virus particle directly or to the membrane-bound viral receptor candidate(s) to prevent virus adsorption to host cells [12]. LF is also known to inhibit cell-to-cell spread of HSV [6].
Epstein-Barr virus (EBV) is mainly transmitted via oral secretions but evidence exists that sometimes transmission occurs also, for example, sexually. EBV is widespread in human populations of all ages, causes infectious mononucleosis and has a strong association with various malignancies such as Burkitt's lymphoma, Hodgkin's disease and nasopharyngeal carcinoma (NPC) [13]. EBV exhibits a marked infection tropism for B lymphocytes, and its selective binding to B cells is initiated by interaction of the major viral envelope glycoprotein gp350 with the complement receptor CD21 on the B-cell surface [14]. After binding, passive virus endocytosis, de-envelopment, passage of the viral nucleocapsid into the cytoplasm and vectorial movement of the nucleocapsid to the nucleus follow within minutes, which lead to immortalize B cells. On infection of host cells, EBV can undergo lytic infection during which virus progeny is released or initiate active latency infection. EBV is harbored in resting memory B cells and induces B-cell proliferation [15]. The increased magnitude of EBV-induced reactivation in B cells raises the risk of development of lymphoma. EBV can be detected in two different tissues, B lymphocytes and epithelial cells, and is potentially oncogenic for both cell types [13]. In contrast to B cells, epithelial cells express little or no CD21, which implies that EBV assesses some epithelial cells via a CD21-independent process. Exposing epithelial cells to cell-free virus preparations in vitro gave very low rates of infection. It is only when some epithelial cells were cocultured with EBV-positive B cells, the infection rates reached quantifiable levels and epithelial cells were induced into lytic cycle [16].
NPC is rare in most parts of the world but shows a high incidence in South China and Southeast Asia. One of the unique features of this epithelial cancer is its consistent association with EBV infection [17]. Studies have shown that EBV plays a key role in the genesis and maintenance of B lymphocytes and NPC. Therefore, interference with the early infection of B cells and some epithelial cells by EBV is a good strategy to prevent malignant tumor. LF has been found to have antitumor activity to regulate tumorigenesis in solid tumors [18,19]. We and others previously reported that a deficient expression of LF was a common event in NPC and predicted poorer prognosis in NPC [20,21].
In this study, we demonstrate that LF can prevent EBV infection of B lymphocytes and interfere with EBV transfering from B cells to epithelial cells, rather than inhibiting the replication of EBV DNA. The present study also indicated that LF inhibited EBV infection mediated by binding to the B-lymphocytic EBV receptor CD21. By means of NPC tissue arrays, it was revealed that LF expression levels correlated inversely with the patients’ EBV viral capsid antigen (VCA)-IgA titers. We then discuss the role of LF in NPC.
Materials and Methods
Cell Culture and Reagents
Human peripheral blood mononuclear cells (PBMCs) were isolated from heparinized blood by density gradient centrifugation from healthy donors. CD19+ cells were obtained from fresh PBMCs by positive immunoselection with magnetic beads, according to the manufacturer's instructions (Miltenyi Biotec, Bergisch Gladbach, Germany). The resulting preparations were consistently >99% CD19+, as determined by FACS (MoFlox XDP flow cytometry, Beckman Coulter, Fullerton, Calif., USA). Established normal nasopharyngeal epithelial cell lines NP69 were grown in serum-free keratinocyte growth medium (Invitrogen, Carlsbad, Calif., USA). HEK293T cells were grown in DMEM supplemented with 10% fetal calf serum. B95-8, P3HR-1 and EBV-transformed lymphoid cells were cultured in RPMI-1640 with 10% fetal calf serum. Human LF (hLF) proteins were obtained from Sigma-Aldrich (St. Louis, Mo., USA), which is iron free.
Virus Preparations and Virus Binding
Purified EBV was obtained from the productive EBV B-cell lineage B95.8, as described in the literature [22]. Viral titers were evaluated as described and expressed as transforming units per milliliter [22]. Virus preparations were assayed for EBV genome content by quantitative (Q)-PCR amplification for the BamHI-W fragment [23]. For the virus-binding assay, primary B cells were preincubated with hLF (50 µg/ml) or BSA (50 µg/ml) as control at 4°C for 1 h; hLF or BSA was washed away with cold PBS, and then the cells were exposed to EBV (multiplicity of infection, MOI 50) at 4°C for 3 h with gentle rocking. To remove unbound viruses, cells were washed extensively in PBS. Then cells were labeled with gp350 antibody and counted by flow cytometry, or were detected by immunofluorescence with anti-gp350 antibody; gp350-positive cells were considered as EBV-bound cells.
Flow Cytometry
Flow cytometry was used to evaluate the EBV-binding ability to B cells by counting the percentage of stained gp350-positive cells. B-cell-bound EBV was labeled by mouse monoclonal anti-gp350 (Santa Cruz Biotechnology, Santa Cruz, Calif., USA) at 4°C for 1 h. Cells were then washed with PBS three times. Alex Flour 568-conjugated goat anti-mouse IgG antibody (Invitrogen) was added to the cells, incubated at 4°C for 1 h and washed with PBS. The cells were fixed with 0.5% formalin and counted using FACS. gp350-positive cells were considered as EBV-bound cells. BSA was used as control for hLF treatment. All experiments were done in triplicate.
Immunofluorescence
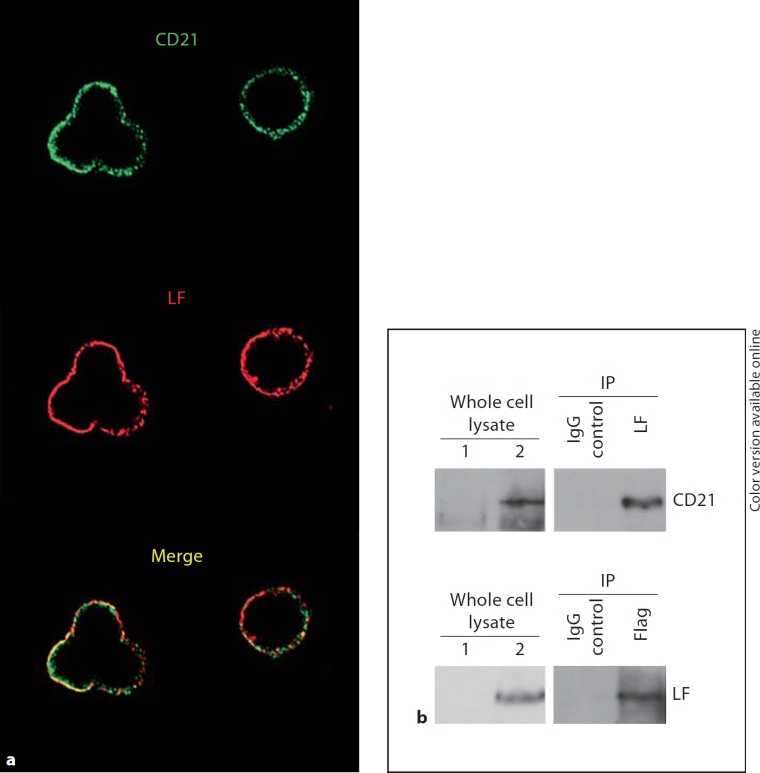
Infected B cells were washed in PBS, fixed in methanol and acetone (1:1) at −20°C for 15 min, and incubated with anti-gp350 antibody for 1 h at room temperature. After washing with PBS, the viral envelope glycoprotein was stained with Alex Flour 568-conjugated anti-mouse IgG antibody (Invitrogen) and observed by a laser scanning confocal microscope (FluoView FV1000 Confocal Microscope, Olympus, Tokyo, Japan). To assay the cellular localization of LF and CD21 proteins, lymphoblastoid cells were incubated with hLF (10 µg/ml) at 4°C for 1 h. The hLF-treated cells were fixed and then incubated with anti-LF (Sigma- Aldrich) and CD21 antibodies (Epitomics, Burlingame, Calif., USA). After washing with PBS, cells were incubated with Alex Flour 568-conjugated anti-rabbit IgG (target LF antibody) and Alex Flour 488-conjugated anti-mouse IgG antibody (target CD21 antibody) and then observed by confocal microscope. The merged image represents the extent of the colocalization of LF with CD21 in the cell surface (fig. 3a).
Fig. 3.
LF interacts with CD21. a Immunofluorescence colocalization of LF with CD21. Lymphoblastoid cells were incubated with hLF (10 µg/ml) at 4°C for 1 h. After washing, bound LF was detected by anti-LF antibody (red), and CD21 was detected by anti-CD21 antibody (green). The cells were observed using confocal fluorescent microscopy. The yellow color in the merged image represents the extent of the colocalization of LF with CD21 on the cell surface. The colors refer to the online version of the figure. b Coimmunoprecipitation of LF with CD21. 293T cells were cotransfected with pcDNA3.1-Flag-CD21 and pcDNA3.1-LF plasmids. The cell lysates were subjected to coimmunoprecipitation with either anti-LF or anti-Flag antibodies [an unrelated IgG antibody was used as negative immunoprecipitation (IP) control]. Immunoprecipitates were analyzed by Western blot with anti-CD21 or anti-LF antibodies. The left panel shows whole cell lysate from cotransfected cells (lane 2) or untransfected cells (lane 1, as negative control). The right panel shows immunoprecipitates with anti-CD21 or anti-LF or with unrelated IgG antibody as immunoprecipitation control.
Q-PCR and Virus Entry
To assay the effect of LF on EBV entry, primary B cells were incubated with hLF (50 µg/ml) or BSA as control at 4°C for 1 h, and hLF or BSA was then washed away. After that, the cells were infected with EBV (MOI 50) at 4°C for 3 h and then washed with PBS to remove unbound virus. Thereafter, cells were cultured for 3 more days. EBV DNA entering into infected B cells was isolated with virus DNA extraction kit (Omega Bio-Tek, Norcross, Ga., USA), and the EBV copy number per cell (i.e. EBV titers) was determined using Q-PCR for the BamHI-W fragment. The EBV gene EBER mRNA expression levels in the infected B cells were also assayed to monitor the virus entry efficiency. Total cellular RNA of the B cells was extracted with Trizol (Invitrogen), and cDNA was synthesized from 1 µg of total RNA by means of the reverse reaction kit, according to the manufacturer's instructions (Promega, Madison, Wisc., USA); then, mRNA expression levels of EBV gene EBER in infected cells were determined by Q-PCR. Primer sequence for Q-PCR: BamHI-W forward 5′-AGTCTCTGCCTCCAGGCA-3′, reverse 5′-ACAGAGGGCCTGTCCACCG-3′. EBER forward 5′-AGGACCTACGCTGCCCTA-3′, reverse 5′-AAAACATGCGGACCACCA-3′. β-Actin forward 5′-AGCGAGCATCCCCCAAAGTT-3′, reverse 5′-GGGCACGAAGGCTCATCATT-3′. The Q-PCR experiments were repeated 3 times.
Fluorscence in situ Hybridization
The standard fluorescence in situ hybridization (FISH) protocol was used to visualize EBV episomes in PBMCs prepared by conventional methanol-acetic acid fixation. For simultaneous FISH and immunofluorescence, hypotonically swollen cells were fixed as described above, permeabilized with blocking buffer containing RNase (100 µg/ml) for 1 h and then incubated in 50% formamide 2× SSC for 30 min at room temperature for equilibration. Each spot of cells was overlaid with 5 µl of hybridization mixture and covered by a cover slip. Sealed slides were directly immersed in water (85°C) for 5 min for denaturation, followed by hybridization. The EBV BamHI-W fragment (EBV genome 13232–16189, 2,957 bp) was obtained by PCR amplification, and the amplified products were labeled with biotin-labeled dUTP by random-primed labeling method (Roche, Mannheim, Germany). This probe has been used in our previous study [24]. The labeled probes were diluted in hybridization buffer (2× SSC, 50% formamide, 10% dextran sulfate) and denatured at 75°C for 5 min. Then, the probes were placed on a slide, covered with a coverslip and sealed with rubber cement. Slides were placed at 37°C overnight in a moist chamber. After hybridization, the slides were washed four times for 5 min in 4× SSC-50% formamide at 42°C. After a preincubation step in 4× SSC-3% bovine serum albumin at 37°C for 30 min, the hybridized probe was detected by incubation with avidin-conjugated fluorescein isothiocyanate (5 mg/ml; Vector Laboratories, Burlingame, Calif., USA) in 4× SSC-3% BSA-0.1% Tween 20 at 37°C for 30 min. Slides were washed three times for 5 min in 4× SSC-0.1% Tween 20 at 42°C. An amplification of the obtained signals was desirable, and the slides were incubated with anti-avidin antibody coupled with biotin in the same buffer as described above, followed by reincubation with avidin-fluorescein isothiocyanate. The mounted slides were observed with the fluorescence microscope.
Coimmunoprecipitation and Immunoblotting
293T cells were cotransfected with pcDNA3.1-LF and pcDNA3.1-Flag-CD21 vector. After 36 h, the cells were lysed in modified radioimmune precipitation buffer, and insoluble material was removed by centrifugation. Anti-LF antibody (Sigma- Aldrich) or anti-Flag beads (Sigma-Aldrich) were then added to lysates, incubated overnight at 4°C. An unrelated IgG antibody was used as immunoprecipitation control. Immune complexes added with anti-LF were bound to protein A/G-agarose beads (Merck Calbiochem, Darmstadt, Germany) for 1 h at 4°C. At the same time, respective negative control has been used in preliminary experiments to verify the specificity of immunoprecipitation assays. Immunobeads were washed four times with lysis buffer, and the precipitates were resuspended in SDS sample buffer. The immunoprecipitates were resolved on SDS-PAGE followed by Western blot analysis with anti-CD21 and LF antibodies. The bound primary antibody was detected with a horseradish peroxidase-conjugated secondary antibody and visualized.
Assay of EBV Replication in Cells
EBV-positive cells (lymphoblastoid and P3HR1 cells) were treated with hLF (10 or 100 µg/ml) or BSA (as negative control) or SPLUNC1 (10 µg/ml, as positive control) [25] for 3 to 21 days. hLF was presented throughout the reproduction process. Cells were harvested and viral DNA was extracted. The EBV copy number per cell (i.e. EBV titers) was determined using Q-PCR for the BamHI-W fragment and used as an index of EBV replication.
Assay of EBV Transfer from B Cells to Nasopharyngeal Epithelial Cells
Nasopharyngeal epithelial cells NP69 were pretreated with hLF (10 µg/ml) or BSA as control at 4°C for 1 h, then cocultured for 24 h with primary B cells (which had been pre-exposed to EBV at MOI 50), followed by washing extensively with cold PBS to remove the B cells. hLF was presented both before and throughout the transfer infection. Then, DNA of NP69 cells was isolated and viral DNA copy numbers (i.e. EBV titers) were determined by Q-PCR for the BamHI-W fragment and used as an index of EBV transfer. Total cellular RNA of the NP69 cells was also extracted and cDNA was synthesized; expression levels of the EBV gene EBER were measured by Q-PCR assay. Detection of EBV genomes by FISH was also employed to directly assay the EBV entry into NP69 cells.
NPC Tissue Microarray and LF Immunohistochemistry
Tissue microarray (TMA) for NPC and non-tumor pharyngeal tissue sections were constructed in our laboratory, as previously described [20]. A total of 703 tissue cores, containing 158 tissue cores from 75 nasopharyngeal epithelia with chronic inflammation, 440 cores from 316 NPC and 95 cores from 88 non-tumor epithelia adjacent to NPC, were placed in 2 TMAs [20]. The TMA sections were probed with LF antibody (1:1,000; Upstate, Chicago, Ill., USA) overnight at 4°C and stained with a secondary antibody at room temperature for 30 min. Tissue section staining was independently scored by 2 pathologists, and any discrepancy in scores was re-examined until a consensus score was reached for each core. The criteria for semiquantitative scoring were described previously [26].
Human Serum Samples and Enzyme-Linked Immunosorbent Assay
Serum samples from patients of the above TMA were collected after obtaining informed consent. Commercial enzyme-linked immunosorbent assay (ELISA) kits were used to determine EBV VCA-IgA levels in serum (Bio-Quant, San Diego, Calif., USA). The use of human material was approved by the Institutional Review Board of the Institute.
Statistical Analysis
Totally, EBV VCA-IgA and LF levels of 327 patients were obtained at the same time in this study. Spearman's correlation test was used to evaluate the pairwise association of patients’ EBV VCA-IgA levels and LF levels. Student's t test is used to compare LF treatment to controls. Two-way ANOVA is used to compare the difference among three or more experiment groups. Calculations were performed using the SPSS 13.0 statistical software. p < 0.05 is considered statistically significant.
Results
Inhibition of EBV Binding to Primary B Cells by hLF
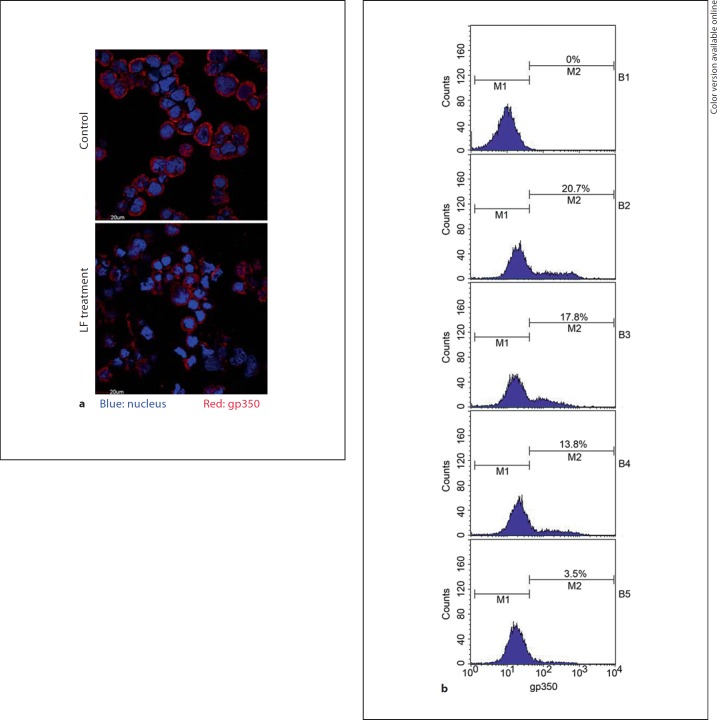
Since LF can inhibit HSV-1 binding to the host cell surface, we first investigated whether hLF has effects on anti-EBV binding to host cells (i.e. B cells). An EBV binding assay was performed with primary B cells in the presence of hLF. Primary B cells were preincubated with various concentrations of hLF, and then EBV was allowed to bind to the cells. Unbound virus was removed by washing, and the cells that bound with EBV were counted by flow cytometry and immunofluorescence. As shown in figure 1a, immunofluorescence with anti-gp350 antibody showed that surface levels of the EBV envelope glycoprotein gp350 were reduced markedly by hLF treatment (50 µg/ml) compared to control BSA treatment. Flow cytometry was used to evaluate the inhibitory effect of different concentrations of hLF (0–50 µg/ml) on the EBV binding ability to B cells. The EBV binding ability (as judged by gp350 positive percentage) was decreased by hLF from 20 to 3% in a dose-dependent manner (fig. 1b), while as a control, BSA (0–50 µg/ml) had no effect on EBV binding ability (data no shown). These results indicated that LF could inhibit the binding of EBV to primary B cells.
Fig. 1.
LF inhibits EBV binding to host cells. a Primary B cells were preincubated with hLF (50 µg/ml) or BSA as control at 4°C for 1 h; hLF was washed away with cold PBS, and then the cells were exposed to EBV (MOI 50) at 4°C for 3 h with gentle rocking. To remove unbound viruses, cells were washed extensively in PBS. Bound EBV particles on the cell membrane were detected by immunofluorescence with anti-gp350 antibody (red). b Primary B cells were preincubated with different concentrations of hLF (0–50 µg/ml) or with BSA as control (data not shown). The percentage of stained gp350-positive cells was counted by FACS as an index of EBV binding ability. B1 = Cells were not exposed to EBV; B2–B5 = cells were exposed to EBV; B2 = no hLF added; B3 = 12.5 µg/ml hLF; B4 = 25 µg/ml hLF; B5 = 50 µg/ml hLF. The colors refer to the online version of the figure.
Effects of LF on EBV Entry
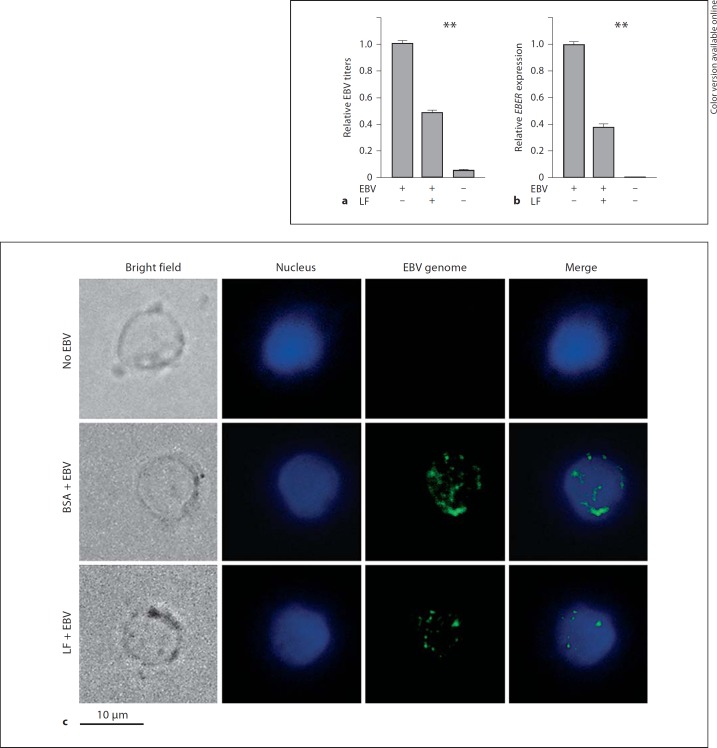
Beside its inhibition of binding EBV to primary B cells, the effect of LF on EBV entry into B cells was also examined. hLF-preincubated (or BSA as control) primary B cells were infected with EBV. Viral DNA copies including virus binding both to cell surface and inside the cells were determined by Q-PCR assay. As shown in figure 2a, the virus copies of the hLF-pretreated B cells decreased by 51% compared to cells without LF treatment, while the BSA pretreatment had no effect on EBV entry (data no shown). FISH with a sensitive EBV DNA probe also detected that viral copy numbers decreased in primary B cells preincubated with hLF, whereas the B cells preincubated with BSA had higher EBV entry efficiency after EBV infection (fig. 2c). In further experiments, we determined the mRNA expression levels of the EBV gene EBER. Figure 2b shows that the expression levels of EBER were reduced by 63% in hLF-treated cells, while control BSA had no effect on EBER expression (data not shown). These results suggested that LF treatment could reduce the efficiency of EBV entry into primary B cells.
Fig. 2.
LF inhibits EBV entry. Primary B cells were incubated with hLF (50 µg/ml) or BSA (50 µg/ml) as control at 4°C for 1 h and hLF was washed away. After that, the cells were infected with EBV (MOI 50) at 4°C for 3 h and were then washed with PBS to remove unbound virus. a Total cellular and bound viral DNA was extracted, and the EBV copy number per cell (i.e. EBV titers) was determined using the Q-PCR assay of the EBV BamHI-W fragment. The EBV titer of the control treatment group (first lane) was adjusted as 1. b Total cellular RNA of the treated primary B cells was extracted, and mRNA expression levels of EBV gene EBER were measured by Q-PCR assay. The expression level of the control group was adjusted as 1. Two-way ANOVA was used to evaluate differences between the three groups. ** p < 0.01. c Detection of EBV genomes by FISH. Primary B cells were treated as above and then fixed; intracellular viral genomes were detected by green fluorescence, and nuclei were identified by DAPI staining.
Interaction of EBV Receptor CD21 and LF
To investigate the possible reason that LF protects primary B cells from EBV infection, we presupposed that LF could interact with EBV receptor CD21 on B cells, and thus, disturb EBV binding to CD21. In an initial experiment, we used immunofluorescence assay to visualize the location of LF and CD21. For visualization of LF, hLF was added to the B cells for 1 h and then washed away. LF and CD21 were observed to colocalize on the cell surface at a certain extent (fig. 3a). To confirm the interaction between LF and CD21, we performed coimmunoprecipitation experiments. LF and Flag-CD21 vectors were cotransfected into 293T cells. Cell lysates were immunoprecipitated with anti-Flag or anti-LF antibodies, and then Western blot analysis was performed using anti-LF or anti-CD21 primary antibodies. We determined that LF can associate with CD21 (fig. 3b). These data implied that the inhibitory effects of LF on EBV binding to primary B cells may be due to the interaction between LF and CD21 on the B-cell surface.
Failure of LF to Inhibit Replication of EBV in EBV-Positive B Cells
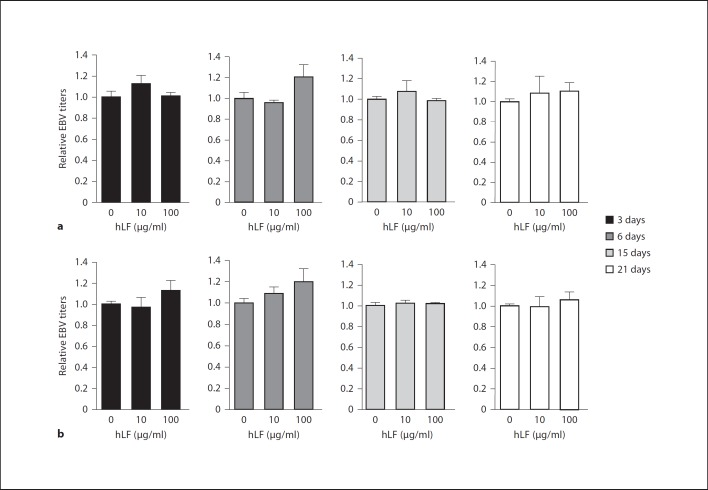
It has been reported that LF had a notable inhibitory effect on the replication of a few types of viruses [27]. To test whether LF could inhibit the replication of EBV genome, LF or BSA (as negative control) was added into EBV-transformed B cells and P3HR1 cells for 3, 6, 15 or 21 days, and then viral genome DNA was extracted for EBV copy number detection. Figure 4 showed that EBV copy numbers (i.e. EBV titers) were not decreased upon LF treatment in both cell types compared to control groups. This result demonstrated that LF did not have inhibitory effects on EBV replication in EBV-positive B cells. We also used 10 µg/ml SPLUNC1 protein (as a positive control) [25] instead of using LF to treat the above cells and found that SPLUNC1 can significantly inhibit the EBV replication, which confirms that the EBV replication experiment was done properly.
Fig. 4.
LF does not inhibit EBV replication. Lymphoblastoid cells (a) and P3HR1 cells (b) were treated with either hLF (10 or 100 µg/ml) or BSA (as control) for 3, 6, 15 or 21 days. hLF was presented throughout the reproduction process. The cells were harvested, and viral DNA copies (EBV titers) were determined by Q-PCR assay of the EBV BamHI-W fragment. The EBV titer of the control treatment group (first lane) was adjusted as 1 in both cell types.
Prevention of EBV Transfer from B Cells to Nasopharyngeal Epithelial Cells by LF
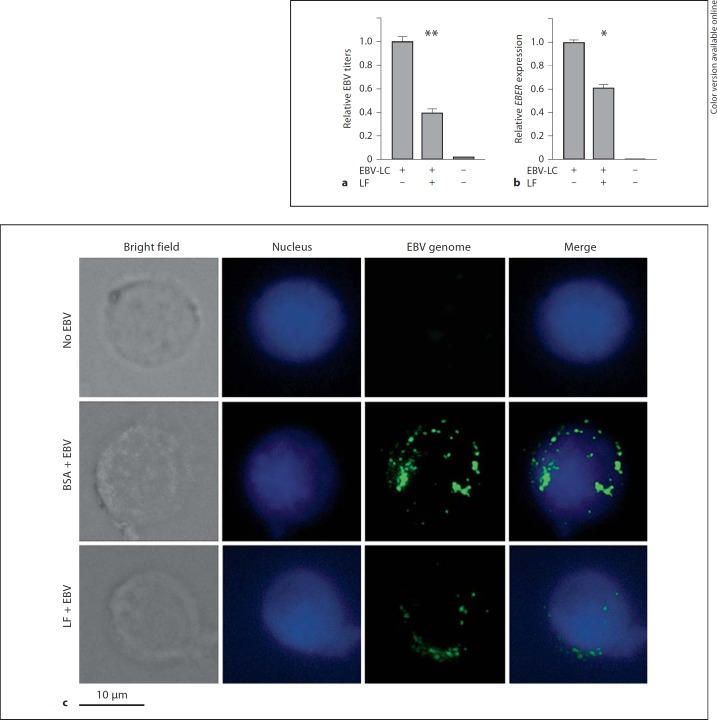
Since LF protects primary B cells from EBV infection, we then asked the question: what about human epithelial cells? Exposure of human epithelial cells to cell-free recombinant EBV gave a lower infection rate, whereas coculture with virus-loaded B cells increased the efficiency of infection [16]. Therefore, we set up a transfer infection assay coculturing NPC cells with virus-loaded B cells and assayed the EBV copies by Q-PCR and FISH. Figure 5a shows that the binding and entry of EBV to NP69 cells were decreased by 60% upon hLF treatment by means of the virus titer assay. Furthermore, the EBV gene EBER expression levels were inhibited by 39% upon hLF treatment (fig. 5b). FISH with an EBV DNA probe also confirmed that viral copy numbers in NP69 cells were clearly decreased by hLF (fig. 5c). These results indicated that hLF prevented the transmission of EBV from primary B cells to nasopharyngeal epithelial cells NP69.
Fig. 5.
LF inhibits EBV transfer infection from resting B cells to nasopharyngeal epithelial cells. NP69 cells were pretreated with hLF (10 µg/ml) or BSA as control at 4°C for 1 h, then cocultured for 24 h with primary B cells (which had been pre-exposed to EBV), followed by washing extensively with PBS to remove the B cells. hLF was presented both before and throughout the transfer infection. a DNA of NP69 cells was isolated and viral DNA copy numbers (i.e. EBV titers) were determined by Q-PCR assay of the EBV BamHI-W fragment. The EBV titer of the control treatment group (i.e. EBV-LC group) was adjusted as 1. b Total cellular RNA of the NP69 cells was extracted, and mRNA expression levels of EBV gene EBER were measured by Q-PCR assay. The expression level of the control group was adjusted as 1. EBV-LC = NP69 cells cocultured with EBV-positive B lymphocytes. Two-way ANOVA was used to evaluate differences between the three groups. * p < 0.05; ** p < 0.01. c Detection of EBV genomes by FISH. NP69 cells were treated as above and then fixed; intracellular viral genomes were detected by green fluorescence, and nuclei were identified by DAPI staining.
LF Expression Levels Correlated Inversely with EBV VCA-IgA Titers in NPC
We next took advantage of high-throughput NPC TMA [20] to assay the LF protein expression levels by immunohistochemistry with anti-LF antibody. LF protein expression was detected in nasopharyngeal mucosal glands (24/24, 100%), in epithelial tissues adjacent to NPC (68/83, 82%) and in non-tumor nasopharyngeal epithelial tissues with chronic inflammation (55/71, 77%); but only 46% of NPC tissues had LF protein expression (142/310). The LF-positive ratio was much lower in NPC specimens than in non-NPC specimens (p < 0.001). It is known that EBV VCA-IgA titers correlate with virus titer or tumor load in NPC patients. Thus, we carried out an ELISA assay to detect the EBV VCA-IgA titers in the serum of those patients. As shown in table 1, 66% (41/62) of VCA-IgA titers of normal controls were undetectable; 61% (20/33) in inflammation or hyperplasia patients and 13% (30/232) in NPC patients. On the other hand, 10% (6/62) of VCA-IgA titers of normal controls were 0:80; while this titer was suitable for 30% (10/33) of inflammation or hyperplasia patients as well as for 60% (139/232) of NPC patients. The VCA-IgA titer of NPC patients is obviously the highest among the three groups (p < 0.01). We have obtained EBV VCA-IgA and LF protein levels of 327 patients at the same time. Spearman's correlation test was used to evaluate the pairwise association of the patients’ VCA-IgA and LF levels. The pairwise association between LF levels and VCA-IgA titers is revealed in table 2 (Spearman correlation coefficient r = −0.179, p = 0.001). LF expression deficiency had significant associations with high VCA-IgA titers. However, its pathological significance still needs to be studied in the future.
Table 1.
EBV VCA-IgA titers for the patients of TMA by means of ELISA (n = 327)
| EBV VCA-IgA titers |
|||||
|---|---|---|---|---|---|
| Undetected | 0:10 or 0:20 | 0:40 | 0:80 | total specimens |
|
| Normal | 41 | 10 | 5 | 6 | 62 |
| Inflammation or hyperplasia | 20 | 2 | 1 | 10 | 33 |
| NPC | 30 | 26 | 37 | 139 | 232 |
| Total specimens | 101 | 46 | 54 | 198 | 327 |
There were significant differences between the EBV VCA-IgA titers of the three groups, evaluated by χ2 test (p < 0.01).
Table 2.
Pairwise association between LF protein expression levels and EBV VCA-IgA titers for the patients of TMA (n = 327)
| LF protein levels | |
|---|---|
| EBV VCA-IgA titers | |
| Spearman correlation coefficient | –0.179 |
| Significance (2-tailed) | 0.0011 |
Correlation is significant at the 0.01 level (2-tailed).
Discussion
We previously observed that frequent loss of heterozygosity on chromosome 3p21 occurs in patients with NPC [28]. Linkage analysis of 18 pedigrees from the Hunan province of China revealed that potential susceptibility loci linked to NPC are located on chromosome 3p21 [29]. Isolation and identification of new tumor suppressor genes for NPC from this region have been pursued and the LF gene (NCBI Entrez Gene ID 4057) is one result of this pursuit. LF is an abundantly expressed protein in human milk and its protection and nutrition roles for newborns are well known. LF has been wildly used in milk and many other preventative medicines or nutrition foods based on its multiple biological functions in innate immunity, anti-inflammation and antimicrobials.
Although the role of LF in NPC development is still unclear, its vital role in antivirus function has been widely appreciated for a long time. EBV infection has been linked to several tumors such as Burkitt's lymphoma and NPC. An important consequence of epithelial infection with EBV is malignant transformation, resulting in the development of NPC [13]. EBV is widespread in the human population as a life-long and largely asymptomatic infection. Acquired orally, EBV replicates in permissive cells in the oropharynx but persists as a latent infection of the B-cell pool, from which reactivations into the lytic cycle continually seed subclinical foci of oral shedding [30]. The dual character of EBV, as classical herpes virus and tumor virus, suggested that the innate immune system of our body against EBV must be involved in NPC development [31]. A link between EBV infection and the antivirus function of LF should somehow exist and has been ignored in the field of NPC research.
LF is presented in plasma and external secretions, like milk, tears and nasal secretions [4]. It is an important innate immune protein by suppressing a variety of infections including bacteria and viruses [1]. LF interferes with virus entry through binding directly to the virus particle or to its cell surface receptor. LF has a high net-positive charge and can interact with negatively charged big molecules located on target cells which can result in antivirus function [32]. The focus of this study was to investigate whether LF can inhibit EBV infection and its implication in NPC carcinogenesis.
In a virus-binding experiment, LF pretreatment can inhibit EBV binding to the human primary B-lymphocyte cell surface at a certain extent. These inhibitory effects were observed in both flow cytometry and immunofluorescence assay, as shown in figure 1.
Besides the inhibition of EBV binding to host cells, further studies revealed that LF also has the ability to inhibit EBV entering into host cells. The Q-PCR assay indicated that EBV titers in infected cells were significantly reduced by LF. The FISH assay showed that EBV genome staining was much weaker upon LF treatment. Furthermore, the EBV gene EBER expression was also downregulated by LF. The above observations provide evidence that LF can protect host cells from EBV entry at least partly.
LF can prevent infection of other human herpes virus family members such as HSV-1 and HSV-2 and human cytomegalovirus [33]. LF prevents the virus from infecting the host cells, either by interacting with cellular receptors to block the viral binding or by binding to the virus particles directly [34]. Since LF is known to bind cell surface glycosaminoglycans and low-density lipoprotein receptors [35], which in turn act as binding sites for HSV-1 and HIV [36,37], its inhibiting activity on these viruses has been ascribed to a competition for cell surface glycosaminoglycans and low-density lipoprotein receptors leading to the prevention of virus adsorption to host cells. LF is a glycoprotein with a large amount of positive charge, which enables it to bind to many receptors. Here, we demonstrate that LF can interact with EBV receptor CD21 on primary B-cell surface. Confocal microscopy experiments indicated that exogenous LF could bind to primary B-cell surface and in part colocalize with the EBV receptor CD21. The coimmunoprecipitation assay also showed that LF can interact with CD21. This finding suggests that LF is likely to interfere with EBV binding to its receptor CD21 and to prevent the virus from entering into primary B cells.
It has been reported that LF had a notable inhibitory effect on the replication of cytomegalovirus and HIV in vitro replication [6]. Thus, we next investigated whether LF could inhibit the replication of EBV in EBV-positive cells. We employed different concentrations of LF to treat transformed B cells and human lymphoma cells P3HR1 for 3, 6, 15 or 21 days and found that LF could not inhibit the replication of EBV. EBV has two alternative life styles: latent or lytic replication. In lymphoblastoid cells, EBV is generally in the latent state and the EBV genomic DNA exists as a closed circular plasmid, which duplicates with cell replication. Therefore, LF might not be able to intervene in the EBV replication process in lymphoblastoid cells.
The above studies showed that LF could prevent EBV infection of primary B cells. However, EBV targets not only B cells but also some epithelial cells, such as nasopharyngeal epithelial cells. Previous studies indicated that the pathway through which EBV entered into nasopharyngeal epithelial cells mainly depended on EBV-positive B cells as a transfer vehicle, especially virus-loaded resting B cells [16]. Using a cell and cell coculture system, we found that the amount of EBV entering into epithelial cells was decreased upon LF treatment, which implies that LF could inhibit the transmission of EBV from B lymphocytes to epithelial cells in some degree. The mechanism of inhibition still needs to be clarified in the future, but the biological meanings of this inhibition are quite obvious, i.e. LF can also protect nasopharyngeal epithelial cells from EBV infection and prevent the EBV-induced transformation process.
Finally, we explored the expression levels of LF and EBV VCA-IgA titers in NPC patients by means of NPC TMA and ELISA techniques. LF proteins are absent or downregulated in most NPC tissues and their expression levels are reversely correlated with EBV VCA-IgA titers. The tumor suppressor role of LF in NPC and its relationship to EBV infection is worth of ongoing study.
In summary, our findings provide evidence that LF protects both primary B cells and epithelial cells from EBV infection. LF prevents EBV infection of primary B cells through its interaction with EBV receptors. Since LF shows inhibitory potential in EBV infection, LF may be applied in preventative medicine or nutrition supplies for NPC or NPC-susceptive people.
Disclosure Statement
The authors declare that they have no conflicts of interest.
Acknowledgements
This work was supported by the China 111 Project (No. 111-2-12), National Nature Scientific Foundation of China (30871282, 81071756, 81171988). We thank Ke Tang, Li Cao, Jianhong Lu and Haibo Yu for their technical assistance.
References
- 1.Legrand D, Mazurier J. A critical review of the roles of host lactoferrin in immunity. Biometals. 2010;23:365–376. doi: 10.1007/s10534-010-9297-1. [DOI] [PubMed] [Google Scholar]
- 2.Teng CT. Lactoferrin: the path from protein to gene. Biometals. 2010;23:359–364. doi: 10.1007/s10534-010-9310-8. [DOI] [PubMed] [Google Scholar]
- 3.Rodrigues L, Teixeira J, Schmitt F, Paulsson M, Mansson HL. Lactoferrin and cancer disease prevention. Crit Rev Food Sci Nutr. 2009;49:203–217. doi: 10.1080/10408390701856157. [DOI] [PubMed] [Google Scholar]
- 4.Gonzalez-Chavez SA, Arevalo-Gallegos S, Rascon-Cruz Q. Lactoferrin: structure, function and applications. Int J Antimicrob Agents. 2009;33(301):e301–e308. doi: 10.1016/j.ijantimicag.2008.07.020. [DOI] [PubMed] [Google Scholar]
- 5.Beljaars L, van der Strate BW, Bakker HI, Reker-Smit C, van Loenen-Weemaes AM, Wiegmans FC, Harmsen MC, Molema G, Meijer DK. Inhibition of cytomegalovirus infection by lactoferrin in vitro and in vivo. Antiviral Res. 2004;63:197–208. doi: 10.1016/j.antiviral.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 6.Valimaa H, Tenovuo J, Waris M, Hukkanen V. Human lactoferrin but not lysozyme neutralizes HSV-1 and inhibits HSV-1 replication and cell-to-cell spread. Virol J. 2009;6:53. doi: 10.1186/1743-422X-6-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jenssen H, Sandvik K, Andersen JH, Hancock RE, Gutteberg TJ. Inhibition of HSV cell-to-cell spread by lactoferrin and lactoferricin. Antiviral Res. 2008;79:192–198. doi: 10.1016/j.antiviral.2008.03.004. [DOI] [PubMed] [Google Scholar]
- 8.Groot F, Geijtenbeek TB, Sanders RW, Baldwin CE, Sanchez-Hernandez M, Floris R, van Kooyk Y, de Jong EC, Berkhout B. Lactoferrin prevents dendritic cell-mediated human immunodeficiency virus type 1 transmission by blocking the DC-SIGN-gp120 interaction. J Virol. 2005;79:3009–3015. doi: 10.1128/JVI.79.5.3009-3015.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berkhout B, Floris R, Recio I, Visser S. The antiviral activity of the milk protein lactoferrin against the human immunodeficiency virus type 1. Biometals. 2004;17:291–294. doi: 10.1023/b:biom.0000027707.82911.be. [DOI] [PubMed] [Google Scholar]
- 10.Kaito M, Iwasa M, Fujita N, Kobayashi Y, Kojima Y, Ikoma J, Imoto I, Adachi Y, Hamano H, Yamauchi K. Effect of lactoferrin in patients with chronic hepatitis C: combination therapy with interferon and ribavirin. J Gastroenterol Hepatol. 2007;22:1894–1897. doi: 10.1111/j.1440-1746.2007.04858.x. [DOI] [PubMed] [Google Scholar]
- 11.Hara K, Ikeda M, Saito S, Matsumoto S, Numata K, Kato N, Tanaka K, Sekihara H. Lactoferrin inhibits hepatitis B virus infection in cultured human hepatocytes. Hepatol Res. 2002;24:228. doi: 10.1016/s1386-6346(02)00088-8. [DOI] [PubMed] [Google Scholar]
- 12.Drobni P, Naslund J, Evander M. Lactoferrin inhibits human papillomavirus binding and uptake in vitro. Antiviral Res. 2004;64:63–68. doi: 10.1016/j.antiviral.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 13.Young LS, Rickinson AB. Epstein-Barr virus: 40 years on. Nat Rev Cancer. 2004;4:757–768. doi: 10.1038/nrc1452. [DOI] [PubMed] [Google Scholar]
- 14.Nemerow GR, Wolfert R, McNaughton ME, Cooper NR. Identification and characterization of the Epstein-Barr virus receptor on human B lymphocytes and its relationship to the C3d complement receptor (CR2) J Virol. 1985;55:347–351. doi: 10.1128/jvi.55.2.347-351.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Babcock GJ, Decker LL, Volk M, Thorley-Lawson DA. EBV persistence in memory B cells in vivo. Immunity. 1998;9:395–404. doi: 10.1016/s1074-7613(00)80622-6. [DOI] [PubMed] [Google Scholar]
- 16.Shannon-Lowe CD, Neuhierl B, Baldwin G, Rickinson AB, Delecluse HJ. Resting B cells as a transfer vehicle for Epstein-Barr virus infection of epithelial cells. Proc Natl Acad Sci USA. 2006;103:7065–7070. doi: 10.1073/pnas.0510512103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tao Q, Chan AT. Nasopharyngeal carcinoma: molecular pathogenesis and therapeutic developments. Expert Rev Mol Med. 2007;9:1–24. doi: 10.1017/S1462399407000312. [DOI] [PubMed] [Google Scholar]
- 18.Kuhara T, Iigo M, Itoh T, Ushida Y, Sekine K, Terada N, Okamura H, Tsuda H. Orally administered lactoferrin exerts an antimetastatic effect and enhances production of IL-18 in the intestinal epithelium. Nutr Cancer. 2000;38:192–199. doi: 10.1207/S15327914NC382_8. [DOI] [PubMed] [Google Scholar]
- 19.Tsuda H, Sekine K, Fujita K, Ligo M. Cancer prevention by bovine lactoferrin and underlying mechanisms – a review of experimental and clinical studies. Biochem Cell Biol. 2002;80:131–136. doi: 10.1139/o01-239. [DOI] [PubMed] [Google Scholar]
- 20.Zhou Y, Zeng Z, Zhang W, Xiong W, Wu M, Tan Y, Yi W, Xiao L, Li X, Huang C, Cao L, Tang K, Shen S, Li G. Lactotransferrin: a candidate tumor suppressor-deficient expression in human nasopharyngeal carcinoma and inhibition of NPC cell proliferation by modulating the mitogen-activated protein kinase pathway. Int J Cancer. 2008;123:2065–2072. doi: 10.1002/ijc.23727. [DOI] [PubMed] [Google Scholar]
- 21.Zhang H, Feng X, Liu W, Jiang X, Shan W, Huang C, Yi H, Zhu B, Zhou W, Wang L, Liu C, Zhang L, Jia W, Huang W, Li G, Shi J, Wanggou S, Yao K, Ren C. Underlying mechanisms for LTF inactivation and its functional analysis in nasopharyngeal carcinoma cell lines. J Cell Biochem. 2011;112:1832–1843. doi: 10.1002/jcb.23101. [DOI] [PubMed] [Google Scholar]
- 22.Gaudreault E, Fiola S, Olivier M, Gosselin J. Epstein-Barr virus induces MCP-1 secretion by human monocytes via TLR2. J Virol. 2007;81:8016–8024. doi: 10.1128/JVI.00403-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Junying J, Herrmann K, Davies G, Lissauer D, Bell A, Timms J, Reynolds GM, Hubscher SG, Young LS, Niedobitek G, Murray PG. Absence of Epstein-Barr virus DNA in the tumor cells of European hepatocellular carcinoma. Virology. 2003;306:236–243. doi: 10.1016/s0042-6822(02)00027-2. [DOI] [PubMed] [Google Scholar]
- 24.Gao J, Li X, Li G. Identification of EBV chromosomal integration sites in Raji cells by fluorescence in situ hybridization. J Cent South Univ Med Sci. 2009;34:13–19. [PubMed] [Google Scholar]
- 25.Zhou HD, Li XL, Li GY, Zhou M, Liu HY, Yang YX, Deng T, Ma J, Sheng SR. Effect of SPLUNC1 protein on the Pseudomonas aeruginosa and Epstein-Barr virus. Mol Cell Biochem. 2008;309:191–197. doi: 10.1007/s11010-007-9659-3. [DOI] [PubMed] [Google Scholar]
- 26.Zeng ZY, Zhou YH, Zhang WL, Xiong W, Fan SQ, Li XL, Luo XM, Wu MH, Yang YX, Huang C, Cao L, Tang K, Qian J, Shen SR, Li GY. Gene expression profiling of nasopharyngeal carcinoma reveals the abnormally regulated Wnt signaling pathway. Hum Pathol. 2007;38:120–133. doi: 10.1016/j.humpath.2006.06.023. [DOI] [PubMed] [Google Scholar]
- 27.Ikeda M, Nozaki A, Sugiyama K, Tanaka T, Naganuma A, Tanaka K, Sekihara H, Shimotohno K, Saito M, Kato N. Characterization of antiviral activity of lactoferrin against hepatitis C virus infection in human cultured cells. Virus Res. 2000;66:51–63. doi: 10.1016/s0168-1702(99)00121-5. [DOI] [PubMed] [Google Scholar]
- 28.Deng L, Jing N, Tan G, Zhou M, Zhan F, Xie Y, Cao L, Li G. A common region of allelic loss on chromosome region 3p25.3–26.3 in nasopharyngeal carcinoma. Genes Chromosomes Cancer. 1998;23:21–25. doi: 10.1002/(sici)1098-2264(199809)23:1<21::aid-gcc4>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 29.Xiong W, Zeng ZY, Xia JH, Xia K, Shen SR, Li XL, Hu DX, Tan C, Xiang JJ, Zhou J, Deng H, Fan SQ, Li WF, Wang R, Zhou M, Zhu SG, Lu HB, Qian J, Zhang BC, Wang JR, Ma J, Xiao BY, Huang H, Zhang QH, Zhou YH, Luo XM, Zhou HD, Yang YX, Dai HP, Feng GY, Pan Q, Wu LQ, He L, Li GY. A susceptibility locus at chromosome 3p21 linked to familial nasopharyngeal carcinoma. Cancer Res. 2004;64:1972–1974. doi: 10.1158/0008-5472.can-03-3253. [DOI] [PubMed] [Google Scholar]
- 30.Shah KM, Young LS. Epstein-Barr virus and carcinogenesis: beyond Burkitt's lymphoma. Clin Microbiol Infect. 2009;15:982–988. doi: 10.1111/j.1469-0691.2009.03033.x. [DOI] [PubMed] [Google Scholar]
- 31.Merlo A, Turrini R, Dolcetti R, Martorelli D, Muraro E, Comoli P, Rosato A. The interplay between Epstein-Barr virus and the immune system: a rationale for adoptive cell therapy of EBV-related disorders. Haematologica. 2010;95:1769–1777. doi: 10.3324/haematol.2010.023689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jenssen H, Hancock RE. Antimicrobial properties of lactoferrin. Biochimie. 2009;91:19–29. doi: 10.1016/j.biochi.2008.05.015. [DOI] [PubMed] [Google Scholar]
- 33.Jenssen H. Anti herpes simplex virus activity of lactoferrin/lactoferricin – an example of antiviral activity of antimicrobial protein/peptide. Cell Mol Life Sci. 2005;62:3002–3013. doi: 10.1007/s00018-005-5228-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van der Strate BW, Beljaars L, Molema G, Harmsen MC, Meijer DK. Antiviral activities of lactoferrin. Antiviral Res. 2001;52:225–239. doi: 10.1016/s0166-3542(01)00195-4. [DOI] [PubMed] [Google Scholar]
- 35.Ji ZS, Mahley RW. Lactoferrin binding to heparan sulfate proteoglycans and the LDL receptor-related protein. Further evidence supporting the importance of direct binding of remnant lipoproteins to HSPG. Arterioscler Thromb. 1994;14:2025–2031. doi: 10.1161/01.atv.14.12.2025. [DOI] [PubMed] [Google Scholar]
- 36.WuDunn D, Spear PG. Initial interaction of herpes simplex virus with cells is binding to heparan sulfate. J Virol. 1989;63:52–58. doi: 10.1128/jvi.63.1.52-58.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Giroglou T, Florin L, Schafer F, Streeck RE, Sapp M. Human papillomavirus infection requires cell surface heparan sulfate. J Virol. 2001;75:1565–1570. doi: 10.1128/JVI.75.3.1565-1570.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]