Abstract
Advances in genetics have shed light on the molecular basis of Crohn's disease (CD) predisposition and pathogenesis, via linkage disequilibrium analysis to genome-wide association studies. The discovery of genetic variants of NOD2, an intracellular pathogen molecular sensor, as risk factors for CD has paved the way for further research on innate immunity in this disease. Remarkably, polymorphisms in autophagy genes, such as ATG16L1 and IRGM, have been identified, allowing the pivotal role of autophagy in innate immunity to be uncovered. In this review, we summarize recent studies on the CD-associated NOD2, ATG16L1 and IRGM risk variants and their contribution to the autophagy functions that have most influenced our understanding of CD pathophysiology.
Key Words: Autophagy, Crohn's disease, Adherent-invasive Escherichia coli, Nucleotide Oligomerization Domain 2, ATG16L1, IRGM
Introduction
Inflammatory bowel diseases (IBD), encompassing Crohn's disease (CD) and ulcerative colitis, are chronic inflammatory disorders of the gastrointestinal tract [1, 2]. IBD is a growing problem in developed countries, affecting 1.4 million people in the USA and 2.2 million in Europe. Extensive studies in the last decades have suggested that the etiology of IBD involves environmental and genetic factors that lead to dysfunction of the epithelial barrier with consequent deregulation of the mucosal immune response to gut microbiota. There is currently no curative therapy [3], indicating a need to fully elucidate the underlying causes of the disease.
Since the early 2000s, many genetic variants have been identified as CD susceptibility factors [4]. The first locus identified as a risk factor for CD was nucleotide oligomerization domain 2 (NOD2) [5, 6], a member of the NLR (NOD-like receptors) family of intracellular sensors of pathogen/microbe-associated molecular patterns that play crucial roles in innate immunity [7]. Subsequently, significant progress has been made in understanding the etiology of CD with the help of large-scale genomic analyses (linkage disequilibrium analysis, genome-wide association studies and deep sequencing analysis) [3, 8]. Polymorphisms in NOD2 remain the largest and most prominent genetic risk factor among over more than one hundred IBD-associated risk loci identified so far [9, 10]. Genome-wide association studies have revealed that polymorphisms in autophagy genes ATG16L1 (autophagy-related 16-like 1) [11, 12] and IRGM (immunity-related GTPase family M) [13, 14, 15] were genetic risk factors for CD and raised autophagy as a very attractive molecular pathway in current CD study [16]. Studies have linked the known CD-associated NOD2 mutations to autophagy induction [17, 18] via the physical interaction of NOD2 with ATG16L1. These studies link NOD2 and autophagy-related genes to innate immune responses to bacterial infection and subsequent induction of adaptive immunity, for the first time placing these important CD-associated genes directly within the same pathway. In addition to NOD2, ATG16L1 and IRGM genes, other autophagy-related genes have been identified as CD susceptibility genes. They encode: (i) upstream regulators of autophagy pathways such as PTPN2 (protein tyrosine phosphatase nonreceptor type 2) [19, 20] and LRRK2 (leucine-rich repeat kinase 2) [10], (ii) proteins essential for autophagy initiation such as the serine/threonine kinase ULK1 (Unc-51-like kinase 1) [21] or proteins involved in bacterial sensing and their subsequent uptake by specific selective autophagy (referred to as xenophagy) such as Toll-like receptor 4 (TLR4) [22, 23]. Defects in autophagy could be particularly detrimental for CD outcome since a number of stress signals, such as persistence of intracellular bacteria, nuclear factor (NF)-κB activation, proinflammatory cytokines, hypoxia, ER stress, ROS production and the associated genotoxic stress, are integrated and resolved by autophagy-dependent mechanisms.
ATG16L1, Autophagy and CD
A single-nucleotide polymorphism (SNP) in the ATG16L1 gene that results in the amino acid substitution of a polar threonine by a nonpolar alanine (Thr300Ala) has been identified as a CD risk allele [11, 12]. ATG16L1 forms a multimeric complex with the conjugate ATG12-ATG5, which associates with the outer membrane of the autophagosome, facilitates the phosphatidylethanolamine conjugation of LC3 and supports the elongation and completion of autophagosome [24, 25, 26]. This polymorphism resides in the carboxy-terminal WD-repeat domain of ATG16L1. The homolog of ATG16L1 in yeast lacks the entire WD-repeat domain, suggesting that it could be dispensable for conventional autophagy in mammals. Generally, WD-repeat domains are involved in protein-protein interaction, but there is no documented evidence of the association of this domain with other partner proteins. Efforts have been made to gain insights into the functional role of this allele, which is located at an evolutionary conserved region in mammals, in the pathogenesis of CD.
As autophagy is an innate defense mechanism that acts as a cell-autonomous system to eliminate intracellular pathogens, a number of studies have linked the CD-associated mutation Thr300Ala in ATG16L1 to autophagy activity in handling and clearing intracellular microorganisms, a process called xenophagy. In human epithelial cells, the CD-associated ATG16L1/Ala300 variant exhibits impaired efficiency of autophagy-mediated clearance of the intracellular enteric pathogen Salmonella Typhimurium [27]. The human immortalized lymphoblasts from donors homozygous for the risk-associated ATG16L1 variant showed impaired induction of autophagy when stimulated with the bacterial muramyl dipeptide (MDP) or Gram-positive peptidoglycan [18]. In addition, the CD-associated ATG16L1 variant was also shown to suppress MDP-induced autophagy and subsequent killing of Salmonella in epithelial cell lines, but not in macrophages or dendritic cells, suggesting a possible cell- or tissue-specific effect in MDP-induced autophagy [28]. Our group has shown the decreased efficiency of autophagy-mediated clearance of pathogenic adherent-invasive Escherichia coli (AIEC), which colonize ileal lesions of CD patients, in human cells expressing the ATG16L1 variant [29]. Of note, AIEC are distinguishable from conventional commensal E. coli in that they are able to adhere to and invade intestinal epithelial cells [29, 30, 31], to survive and replicate within macrophages [10, 11, 12, 13] and to induce production of proinflammatory cytokines and chemokines [32, 33]. Silencing expression of ATG16L1 by siRNA in human epithelial cells or macrophages leads to increased intracellular replication of AIEC [29, 33]. Thus, defects in autophagy could favor the persistence of AIEC within host cells and thereby have a profound effect on the outcome of proinflammatory responses. Dendritic cells from CD individuals expressing the ATG16L1 risk variant were shown to be defective in autophagy induction, bacterial trafficking and antigen presentation [17]. These studies together suggest that impaired capacity of autophagy to handle and clear bacteria could be a mechanism underlying the increased risk of CD patients harboring gut dysbiosis with increased numbers of proinflammatory bacteria.
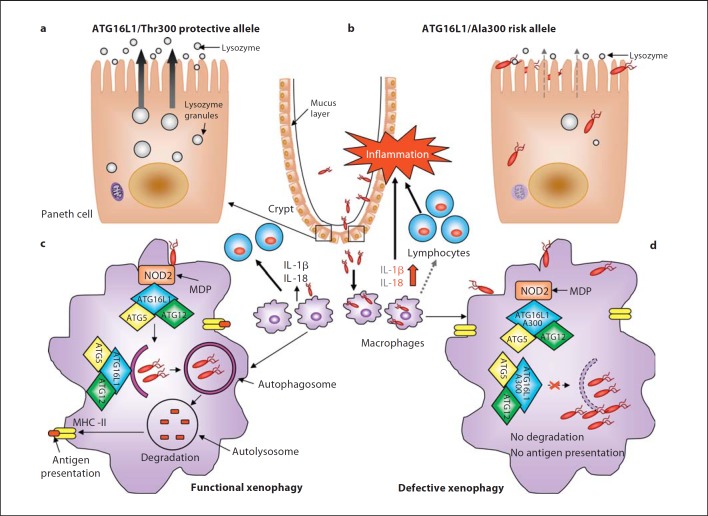
By generating mice with hypomorphic ATG16L1 expression (ATG16L1HM), Cadwell et al. [34] showed another mechanism by which ATG16L1 may be involved in the pathogenesis of CD. ATG16L1HM mice developed normally with no sign of intestinal inflammation but exhibited abnormalities in Paneth cell structure. Paneth cells are specialized epithelial cells located at the base of small intestinal crypts that normally store and secrete antimicrobial peptides, called defensins, within large intracellular granules, thereby contributing to host defense against microbes in the small intestine. In ATG16L1HM mice, Paneth cells exhibited disorganized granule compartments with a disrupted granule exocytosis pathway, which had decreased the number of granules and lacked lysosomes in the ileal mucus layer [34]. In addition, transcriptional profiling of the ATG16L1HM Paneth cells showed a large number of dysregulated genes, including the inflammatory mediators such as the chemokine (C-X-C motif) ligand 1, the acute-phase serum amyloid A1, which is highly expressed in response to inflammation, or the adipocytokines leptin and adiponectin, previously reported to be increased in CD [34]. CD patients homozygotes for the ATG16L1 risk allele exhibited similar structural aberrances in Paneth cells to those observed in ATG16L1HM mice [34], indicating that defects in intestinal barrier function in CD could involve dysfunction of Paneth cells related to ATG16L1 mutation (fig. 1).
Fig. 1.
Impact of the CD-associated ATG16L1/Ala300 variant on xenophagy-mediated intracellular bacteria clearance during CD onset. a, b Paneth cells with the ATG16L1/Ala300 risk allele exhibit abnormalities in structure and function compared to Paneth cells with the ATG16L1/Thr300 protective allele such as decreased and disorganized granule compartments with disrupted granule exocytosis pathway (grey dotted arrow) and degraded mitochondria, leading to reduced lysozymes in the mucus layer. This could probably confer the intestinal epithelial layer susceptibility to microbial infection, leading to bacterial overgrowth and the invasion of commensal bacteria, and predominantly of pathogenic bacteria with invasive properties. c Invading bacteria are detected by macrophages and dendritic cells. NOD2 in macrophages senses MDP and recruits ATG16L1 to the plasma membrane at the bacterial entry site, initiating functional xenophagy with the recruitment of the ATG16-ATG5-ATG12 complex to the membrane of autophagosomes. Bacteria are degraded inside autophagolysosomes and the produced peptides are loaded to the MHC class II for antigen presentation, which will be recognized by CD4+ T cells. Wild-type macrophages and dendritic cells detecting invasive bacteria also secrete inflammatory cytokines IL-1β and IL-18. d In macrophages and dendritic cells with the ATG16L1/Ala300 risk alleles, the mutated ATG16L1 protein is still able to bind to NOD2 upon MDP stimulation and able to recruit the ATG12-ATG5 complex, but fails to induce xenophagy. Autophagy-deficient macrophages fail to kill intracellular bacteria and to present antigen by MHC class II, leading to inappropriate activation of the adaptive immune system (grey dotted arrow). Autophagy-deficient macrophages produce high levels of IL-1β and IL-18 (red arrow). These result in severe inflammation and consequently chronic inflammatory status, which occur during CD development.
In another study, Saitoh et al. [35] observed an abnormal inflammatory response to lipopolysaccharide (LPS) stimulation in ATG16L1-deficient macrophages, characterized by an elevated production of IL-1β and IL-18 (fig. 1)[17]. Of note, this experiment was performed in ATG16L1-deficient fetal liver macrophages, since ATG16L1 knockout mice died within a day of birth [35]. In addition, ATG16L1-deficient macrophages produced a higher level of IL-1β in response to commensal noninvasive Gram-negative bacteria, such as Enterobacter aerogenes, E. coli and Klebsiella pneumoniae[35]. This was mediated through TRIF (a Toll/IL-1 receptor domain-containing adaptor inducing IFN-β), an essential adaptor molecule for TLR signaling pathways, as ATG16L1-TRIF double-deficient macrophages failed to produce IL-1β in response to LPS stimulation owing to a lack of caspase-1 activation [35]. Interestingly, through a bone marrow transplantation study, the authors also demonstrated that mice receiving the ATG16L1-deficient monocytes were more susceptible to dextran sulfate sodium (DSS)-induced acute colitis, shown by more severe colonic inflammation as evidenced by histological examination and increased levels of IL-1β and IL-18 in the sera [35]. It is noteworthy that the ATG16L1-deficient chimeric mice developed neither spontaneous colitis nor an increase in bacterial burden or bacterial translocation. Intraperitoneal administration of 3-methyladenine, an autophagy inhibitor, increased the level of IL-1β in serum and reduced the survival rate of DSS-treated mice. Injection of anti-IL-1β and anti-IL-18 antibodies reduced the mortality and body weight loss of DSS-treated ATG16L1-deficient chimeric mice, indicating the involvement of excessive cytokine production in the development of severe colitis [35]. This elegant study showed a protective role of ATG16L1 in innate immune responses against intestinal inflammation.
In addition, the findings of Cadwell et al. [36] have lent weight to the argument that genetic and environmental factors interplay in the etiology of CD since susceptibility gene and virus infection are both responsible and required for abnormalities in the functional structure and transcriptional profile of Paneth cells, observed in ATG16L1HM mice. Indeed, the structural and transcriptional aberrances in Paneth cells were shown to be dependent on persistent infection with a specific strain of murine norovirus MNV CR6, since mice raised in a germ-free facility or mice infected with a nonpersistent norovirus strain exhibited normal Paneth cell morphology [36]. In addition, MNV CR6 infection did not induce intestinal inflammation but on ATG16L1HM mice conferred increased susceptibility to DSS-induced colitis, which was abrogated by microbial flora depletion with broad-spectrum antibiotics. Interaction between the CD-associated ATG16L1 variant and MNV CR6 infection resulted in intestinal pathologies in mice. It was therefore concluded that susceptibility genes and virus infection are both responsible and required for abnormalities in the functional structure and transcriptional profile of Paneth cells. Although clinical studies have shown that norovirus infection in humans can aggravate IBD symptoms [37, 38], there is no evidence yet that supports the hypothesis that norovirus infection could be a causative factor of CD. The study by Cadwell et al. [36] importantly supports the notion that the host's microbiome (virus and bacteria) needs to be carefully taken into account in patients harboring CD-associated risk alleles.
NOD2, Autophagy and CD
NOD2 is a member of the NLR family of intracellular sensors of pathogen/microbe-associated molecular patterns that play important roles in innate immunity [7]. NOD2 recognizes MDP, a component of the peptidoglycan present in the bacterial cell wall. MDP-mediated stimulation of NOD2 activates NF-κB and triggers the production of various proinflammatory cytokines and chemokines. A cross-talk between NOD2 and TLRs, which plays a crucial role in host defense against microbial infection, has been explored and extensively studied [7]. Of particular interest, three mutations in NOD2, including a frameshift mutation (L1007fsinsC) that results in a truncated NOD2 protein and two amino acid substitutions (R702W and G908R), have been documented to be strongly associated with CD onset [5, 6]. These NOD2 risk mutations are located within the leucine-rich repeat domain, which is the microbe-associated molecular pattern recognition region and has a molecular structure similar to the leucine-rich repeat domains of TLRs [7]. The mechanism by which CD-associated NOD2 polymorphisms contribute to intestinal inflammation has been extensively studied but with conflicting results [7].
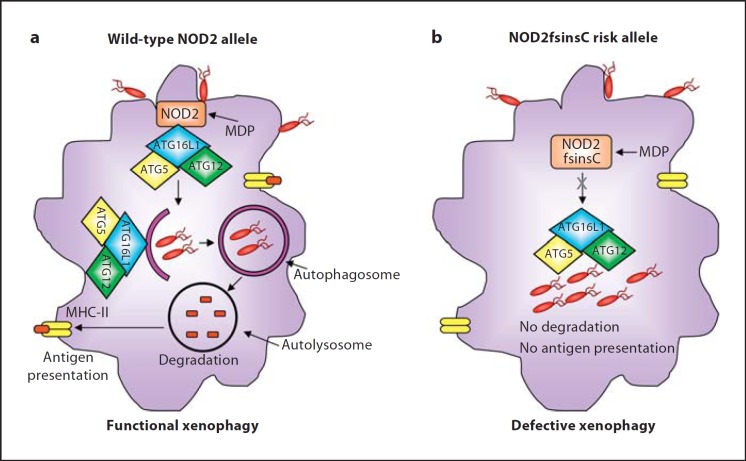
Current studies have revealed unexpected functions of NOD2 in autophagy, which will be discussed in this section. Pioneering studies showing the regulatory role of NOD2 in autophagy reported that MDP-mediated NOD2 activation induces autophagy [17, 18]. Of note, as discussed in the previous section, the CD-associated ATG16L1 risk variant exhibited impaired autophagy induction upon MDP stimulation [17]. The CD-associated NOD2 frameshift mutation resulted in impaired intracellular pathogenic bacterial killing owing to a defect in autophagic response (fig. 2) [17, 18]. Remarkably, recruitment of ATG16L1 to the plasma membrane and consequent wrapping of invading bacteria by autophagosomes failed in human cells homozygous for the NOD2 frameshift mutation (fig. 2)[18]. NOD2 recruits ATG16L1 to plasma membrane at the bacterial entry site, thereby initiating autophagosome formation to wrap invading pathogens (fig. 2). This autophagic function was shown to be dependent on NOD proteins but independent of NF-κB signaling and RIP2 [18]. Hence, these studies show a link between bacterial sensing by NOD proteins and autophagy induction, providing a functional interaction between NOD2 and ATG16L1 to mediate an efficient autophagy response against invasive bacteria that is altered by CD-associated risk variants.
Fig. 2.
Impact of the CD-associated NOD2 (L1007fsinsC) risk variant on xenophagy-mediated intracellular bacteria clearance during CD onset. a Induction of functional xenophagy in macrophages with wild-type NOD2 allele in response to bacterial invasion. MDP-activated NOD2 recruits and interacts with ATG16L1 at bacterial entry sites in the plasma membrane, inducing functional xenophagy with the consequent killing of intracellular bacteria and antigen presentation by MHC class II. b Impaired xenophagy in macrophages with the NOD2 (L1007fsinsC) risk variant in response to invasive bacteria. NOD2 (L1007fsinsC) fails to recruit ATG16L1 to the bacterial entry sites or to induce xenophagy. Consequently, intracellular bacterial degradation and MHC class II-mediated antigen presentation are defective.
The study by Cooney et al. [17] not only revealed the role of autophagy and ATG16L1 in innate immunity via NOD proteins, but also highlighted a role for adaptive immune responses to pathogens. The authors showed that MDP-mediated activation of NOD2 in dendritic cells induces autophagy and promotes major histocompatibility complex (MHC) class II antigen presentation dependently of RIP2, which was defective in dendritic cells from donors with either CD-associated NOD2 (L1007fsinsC) or ATG16L1 (Thr300Ala) risk variants (fig. 2)[17]. Autophagosomes were previously shown to fuse with multivesicular MHC class II-loading compartments to present their contents for recognition by CD4+ T cells [39]. Thus, defects in the autophagy pathway might lead to inappropriate activation of the adaptive immune system, which can impair bacterial handling and consequently result in chronic inflammatory status.
A recent study shed further light on the link between NOD2 and autophagy by showing that MDP stimulation of epithelial cells, macrophages and dendritic cells activated autophagy and NF-κB and mitogen-activated protein kinase signaling and increased pathogenic bacterial killing [28]. These responses appeared to require ATG16L1 and NOD2 expression and were impaired by CD-associated NOD2 variants. NOD2-dependent antibacterial signaling was impaired by the ATG16L1 (Thr300Ala) risk variant in epithelial cells, but not in macrophages or dendritic cells. Finally, NOD2 is required for efficient autophagy to kill CD-associated AIEC in macrophages, since NOD2-deficient macrophages displayed a reduced autophagy-mediated clearance of AIEC and a higher proinflammatory response compared to wild-type macrophages [33]. However, forced induction of autophagy by the use of pharmacological rapamycin inducers or by starving cells, concomitantly with AIEC infection, made it possible to amplify autophagic response in NOD2-deficient macrophages, overcoming defects in intracellular AIEC clearance [33]. The ATG16L1 and NOD2 risk variants could be implicated in autophagy-mediated antibacterial pathways and, further, in the pathogenesis of CD in a cell- and function-specific manner [28]. Since autophagy has been recognized as a bactericidal mechanism, it would be of interest to investigate the role of other pattern recognition receptors such as TLR, in addition to NLR, in the control of autophagy response to bacterial infection. A report published in 2007 demonstrated, for the first time, that bacterial LPS binding on TLR4 induces autophagy [40]. Since TLR4 has been identified as a CD susceptibility gene [22, 23], the impact of CD-associated TLR4 risk alleles on bacterial sensing and the subsequent autophagy activity warrants further investigation.
IRGM, Autophagy and CD
IRGM (Immunity-related GTPase family M) belongs to the p47 immunity-related guanosine triphosphatase family. The murine homolog LRG-47 is induced by IFN-γ and activates autophagy as a mechanism for the elimination of invasive pathogens such as Mycobacterium tuberculosis, Salmonella Typhimurium and Listeria monocytogenes [41, 42, 43, 44, 45].
Unlike the 23 murine Irg gene family, the unique human IRGM sequence lacks IFN-γ-responsive elements and is thought to encode five isoforms (IRGMa to IRGMe), although it is unknown whether all of these result in protein products [46]. Two polymorphisms of IRGM have been highly associated with CD risk, including a ‘silent’ SNP within the coding region (c.313C>T) and a 20-kb deletion upstream of the IRGM gene [13, 14, 15]. The deletion in the IRGM promoter region could have an impact on the level of IRGM protein expression by potentially altering the recruitment of transcription factors. Thus, McCarroll et al. [13] investigated the functional effect of IRGM expression level on autophagic processing of internalized Salmonella Typhimurium in HeLa cells with manipulated IRGM expression. SiRNA-mediated IRGM knockdown significantly attenuated the proportion of internalized bacteria captured within autophagosomes, whereas IRGM overexpression resulted in a plasmid dose-dependent increase in autophagic response with over 50% of internalized bacteria within autophagosomes [13]. Our group has shown that depletion of IRGM in human intestinal epithelial cells and macrophages resulted in defective autophagy and consequently increased intracellular replication of AIEC [29, 33]. Similar results have been obtained in Mycobacterium paratuberculosis-infected murine and human macrophages [42]. These studies suggest that the CD association at IRGM arises from an alteration in IRGM regulation that affects the efficacy of autophagy defense and identify a common deletion polymorphism as a likely causal variant. However, in cells heterozygous for the 20-kb deletion upstream of the IRGM gene, both alleles are expressed in variable proportions depending on cell lines [13], which suggests that this deletion is not able to reliably affect IRGM protein levels and that there may be a role for the synonymous exonic SNP (c.313C>T).
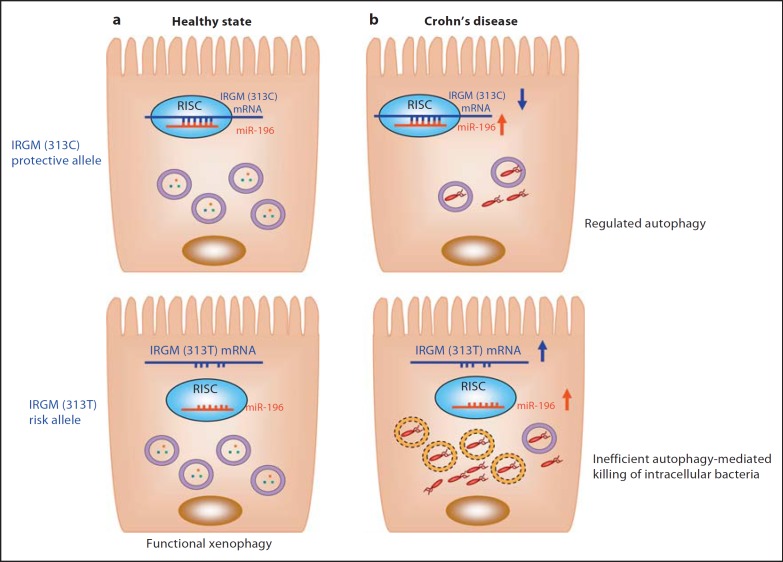
The common synonymous SNP (c.313C>T) in the IRGM gene has been found in strong linkage disequilibrium with the deletion polymorphism and classified as noncausative because it does not change the mature protein sequence. In a study investigating a potential noisy role of this silent polymorphism, in silico analysis using microRNA/mRNA interaction prediction algorithms revealed that the c.313C>T polymorphism of IRGM is located within the IRGM mRNA region that interacts with the microRNA-196 family. Importantly, the silent polymorphism is located within the ‘seed region’, an eight-base pair sequence, in which 100% matches between the microRNA and target mRNA sequence are usually required to allow target mRNA decay or degradation catalyses by the RISC (RNA-induced silencing complex) [47]. Of particular interest, the CD-associated IRGM (c.313C>T) risk allele resulted in loss of binding of microRNA-196 (fig. 3). In addition, microRNA-196 is overexpressed in the inflamed epithelium of CD patients and downregulates the IRGM protective variant (c.313C) but not the risk-associated allele (c.313T) [47]. Loss of tight regulation of IRGM expression by microRNA-196 resulted in defects in autophagy-mediated control of intracellular replication of AIEC, leading to an abnormal persistence of AIEC in host cells (fig. 3)[47]. This suggests that the association of IRGM with CD arises from abnormal microRNA-mediated IRGM regulation, which affects the efficacy of autophagy. In addition, overexpression of IRGM, and in particular that of the IRGMd isoform, led to a cell death presenting apoptotic features and a massive release of the danger-associated protein HMGB1 [33, 48].
Fig. 3.
Hypothetical model for the involvement of the CD-associated IRGM (c.313C>T) in autophagy, bacterial clearance and CD. a MicroRNA 196 (miR-196) normally targets IRGM mRNA within RISC for a negative regulation. The IRGM (c.313C>T)risk allele mRNA, however, lacks the binding site for miR-196 and therefore is not regulated by this microRNA. The IRGM risk variant does not cause alterations in autophagy during healthy states. b During CD, expression of miR-196 is increased, which leads to a downregulation of IRGM expression. This results in a decrease in autophagic flux accompanied by a decrease in autophagosome-associated intracellular bacteria (upper panel). In subjects with the IRGM risk variant (bottom panel), IRGM expression is not inhibited by miR-196 and is upregulated, leading to defective xenophagy with a decrease in the percentage of intracellular bacteria captured in functional acidic vacuoles (violet circle) in comparison with that in IRGM (c.313C) protective variant-bearing subjects. Most intracellular bacteria replication occurs in nonmature vacuoles (yellow dotted circle). This consequently results in intracellular bacteria overload, which could further worsen disease status.
In conclusion, perturbations in autophagy-protein-dependent functions in immunity could contribute to increased susceptibility to infection, chronic inflammation and CD. Owing to the presence of risk alleles in autophagy genes ATG16L1, IRGM and NOD2 in CD-unaffected individuals, and given that autophagy is a vital process for cells, we could hypothesize that CD-associated risk polymorphisms have no effect on the basal autophagic process, but in the event of infection with AIEC or other CD-associated bacteria, or environmental stress, it could lead to a less efficient autophagic response, allowing bacteria to persist in intracellular replicative niches. In addition, autophagic activity can deeply affect the outcome of proinflammatory responses to CD-associated bacterial infection. Induction of autophagy, by the use of rapalogs for example [49], makes it possible to target AIEC bacteria entering macrophages within autophagosomes. As a consequence, there is a significant decrease in the number of intramacrophagic AIEC bacteria and a drastic decrease in proinflammatory cytokine secretion. Restoring bacteria-induced autophagy in patients genotyped for risk alleles in ATG16L1, IRGM or NOD2 (or composite) could be a powerful therapeutic strategy to concomitantly restrain the number of intracellular AIEC bacteria and to slow down inflammatory response.
Conclusions and Perspectives
Genetic studies have highlighted the potential role of the autophagy pathway in CD onset with the identification of risk polymorphisms in various autophagy-related genes and those which regulate autophagy. Exciting recent progress has been made in understanding mechanistic links between the cause of these risk variants and CD pathogenesis, particularly in innate immune activation, defects in autophagy-mediated bacterial handling and antigen presentation and homeostasis of the epithelial barrier. However, further investigations are needed to better understand the functional relevance of the CD susceptibility genes, known or newly identified, in autophagy response to environmental triggers such as gut dysbiosis or infection with a particular pathogen. In addition, it is of particular interest to focus on the factors that regulate autophagy processes in a CD disease context. Translation of these findings into clinical benefits will help to establish novel autophagy-targeting therapeutic strategies for CD treatment. Of note, two reports have shown marked and sustained improvement in CD symptoms in patients with refractory CD treated with rapamycin analogues [50, 51], remissions that owe less to the immunosuppressive properties of these drugs than to their ability to induce autophagy. Studies have identified other classes of molecules, already FDA approved, that modulate the autophagic process [52, 53]. In CD, depending on the patient genotype for autophagy-related genes, the use of well-tolerated pharmacological autophagic inducers could achieve a double benefit by restraining the load of intracellular CD-associated bacteria and slowing down inflammatory response.
Acknowledgements
This work was supported by the Ministère de la Recherche et de la Technologie, INSERM and Université d'Auvergne (UMR1071), INRA (USC-2018) and by grants from the Association F. Aupetit (AFA). Hang Nguyen received a European Union FP7 People Marie Curie international incoming fellowship.
References
- 1.Blumberg R, Cho J, Lewis J, Wu G. Inflammatory bowel disease: an update on the fundamental biology and clinical management. Gastroenterology. 2011;140:1701–1703. doi: 10.1053/j.gastro.2011.03.013. [DOI] [PubMed] [Google Scholar]
- 2.Chassaing B, Darfeuille-Michaud A. The commensal microbiota and enteropathogens in the pathogenesis of inflammatory bowel diseases. Gastroenterology. 2011;140:1720–1728. doi: 10.1053/j.gastro.2011.01.054. [DOI] [PubMed] [Google Scholar]
- 3.Rosenstiel P, Sina C, Franke A, Schreiber S. Towards a molecular risk map - recent advances on the etiology of inflammatory bowel disease. Semin Immunol. 2009;21:334–345. doi: 10.1016/j.smim.2009.10.001. [DOI] [PubMed] [Google Scholar]
- 4.Cho JH, Brant SR. Recent insights into the genetics of inflammatory bowel disease. Gastroenterology. 2011;140:1704–1712. doi: 10.1053/j.gastro.2011.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hugot JP, Chamaillard M, Zouali H, Lesage S, Cezard JP, Belaiche J, Almer S, Tysk C, O'Morain CA, Gassull M, Binder V, Finkel Y, Cortot A, Modigliani R, Laurent-Puig P, Gower-Rousseau C, Macry J, Colombel JF, Sahbatou M, Thomas G. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn's disease. Nature. 2001;411:599–603. doi: 10.1038/35079107. [DOI] [PubMed] [Google Scholar]
- 6.Ogura Y, Bonen DK, Inohara N, Nicolae DL, Chen FF, Ramos R, Britton H, Moran T, Karaliuskas R, Duerr RH, Achkar JP, Brant SR, Bayless TM, Kirschner BS, Hanauer SB, Nunez G, Cho JH. A frameshift mutation in NOD2 associated with susceptibility to Crohn's disease. Nature. 2001;411:603–606. doi: 10.1038/35079114. [DOI] [PubMed] [Google Scholar]
- 7.Strober W, Watanabe T. NOD2, an intracellular innate immune sensor involved in host defense and Crohn's disease. Mucosal Immunol. 2011;4:484–495. doi: 10.1038/mi.2011.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.The Wellcome., Trust Case Control Consortium Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Franke A, McGovern DP, Barrett JC, Wang K, Radford-Smith GL, Ahmad T, Lees CW, Balschun T, Lee J, Roberts R, Anderson CA, Bis JC, Bumpstead S, Ellinghaus D, Festen EM, Georges M, Green T, Haritunians T, Jostins L, Latiano A, Mathew CG, Montgomery GW, Prescott NJ, Raychaudhuri S, Rotter JI, Schumm P, Sharma Y, Simms LA, Taylor KD, Whiteman D, Wijmenga C, Baldassano RN, Barclay M, Bayless TM, Brand S, Buning C, Cohen A, Colombel JF, Cottone M, Stronati L, Denson T, De Vos M, D'Inca R, Dubinsky M, Edwards C, Florin T, Franchimont D, Gearry R, Glas J, Van Gossum A, Guthery SL, Halfvarson J, Verspaget HW, Hugot JP, Karban A, Laukens D, Lawrance I, Lemann M, Levine A, Libioulle C, Louis E, Mowat C, Newman W, Panes J, Phillips A, Proctor DD, Regueiro M, Russell R, Rutgeerts P, Sanderson J, Sans M, Seibold F, Steinhart AH, Stokkers PC, Torkvist L, Kullak-Ublick G, Wilson D, Walters T, Targan SR, Brant SR, Rioux JD, D'Amato M, Weersma RK, Kugathasan S, Griffiths AM, Mansfield JC, Vermeire S, Duerr RH, Silverberg MS, Satsangi J, Schreiber S, Cho JH, Annese V, Hakonarson H, Daly MJ, Parkes M. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn's disease susceptibility loci. Nat Genet. 2010;42:1118–1125. doi: 10.1038/ng.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barrett JC, Hansoul S, Nicolae DL, Cho JH, Duerr RH, Rioux JD, Brant SR, Silverberg MS, Taylor KD, Barmada MM, Bitton A, Dassopoulos T, Datta LW, Green T, Griffiths AM, Kistner EO, Murtha MT, Regueiro MD, Rotter JI, Schumm LP, Steinhart AH, Targan SR, Xavier RJ, Libioulle C, Sandor C, Lathrop M, Belaiche J, Dewit O, Gut I, Heath S, Laukens D, Mni M, Rutgeerts P, Van Gossum A, Zelenika D, Franchimont D, Hugot JP, de Vos M, Vermeire S, Louis E, Cardon LR, Anderson CA, Drummond H, Nimmo E, Ahmad T, Prescott NJ, Onnie CM, Fisher SA, Marchini J, Ghori J, Bumpstead S, Gwilliam R, Tremelling M, Deloukas P, Mansfield J, Jewell D, Satsangi J, Mathew CG, Parkes M, Georges M, Daly MJ. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn's disease. Nat Genet. 2008;40:955–962. doi: 10.1038/NG.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hampe J, Franke A, Rosenstiel P, Till A, Teuber M, Huse K, Albrecht M, Mayr G, De La Vega FM, Briggs J, Gunther S, Prescott NJ, Onnie CM, Hasler R, Sipos B, Folsch UR, Lengauer T, Platzer M, Mathew CG, Krawczak M, Schreiber S. A genome-wide association scan of nonsynonymous SNPs identifies a susceptibility variant for Crohn disease in ATG16L1. Nat Genet. 2007;39:207–211. doi: 10.1038/ng1954. [DOI] [PubMed] [Google Scholar]
- 12.Rioux JD, Xavier RJ, Taylor KD, Silverberg MS, Goyette P, Huett A, Green T, Kuballa P, Barmada MM, Datta LW, Shugart YY, Griffiths AM, Targan SR, Ippoliti AF, Bernard EJ, Mei L, Nicolae DL, Regueiro M, Schumm LP, Steinhart AH, Rotter JI, Duerr RH, Cho JH, Daly MJ, Brant SR. Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis. Nat Genet. 2007;39:596–604. doi: 10.1038/ng2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McCarroll SA, Huett A, Kuballa P, Chilewski SD, Landry A, Goyette P, Zody MC, Hall JL, Brant SR, Cho JH, Duerr RH, Silverberg MS, Taylor KD, Rioux JD, Altshuler D, Daly MJ, Xavier RJ. Deletion polymorphism upstream of IRGM associated with altered IRGM expression and Crohn's disease. Nat Genet. 2008;40:1107–1112. doi: 10.1038/ng.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parkes M, Barrett JC, Prescott NJ, Tremelling M, Anderson CA, Fisher SA, Roberts RG, Nimmo ER, Cummings FR, Soars D, Drummond H, Lees CW, Khawaja SA, Bagnall R, Burke DA, Todhunter CE, Ahmad T, Onnie CM, McArdle W, Strachan D, Bethel G, Bryan C, Lewis CM, Deloukas P, Forbes A, Sanderson J, Jewell DP, Satsangi J, Mansfield JC, Cardon L, Mathew CG. Sequence variants in the autophagy gene IRGM and multiple other replicating loci contribute to Crohn's disease susceptibility. Nat Genet. 2007;39:830–832. doi: 10.1038/ng2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Prescott NJ, Dominy KM, Kubo M, Lewis CM, Fisher SA, Redon R, Huang N, Stranger BE, Blaszczyk K, Hudspith B, Parkes G, Hosono N, Yamazaki K, Onnie CM, Forbes A, Dermitzakis ET, Nakamura Y, Mansfield JC, Sanderson J, Hurles ME, Roberts RG, Mathew CG. Independent and population-specific association of risk variants at the IRGM locus with Crohn's disease. Hum Mol Genet. 2010;19:1828–1839. doi: 10.1093/hmg/ddq041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lapaquette P, Brest P, Hofman P, Darfeuille-Michaud A. Etiology of Crohn's disease: many roads lead to autophagy. J Mol Med (Berl) 2012;90:987–996. doi: 10.1007/s00109-012-0934-8. [DOI] [PubMed] [Google Scholar]
- 17.Cooney R, Baker J, Brain O, Danis B, Pichulik T, Allan P, Ferguson DJ, Campbell BJ, Jewell D, Simmons A. NOD2 stimulation induces autophagy in dendritic cells influencing bacterial handling and antigen presentation. Nat Med. 2010;16:90–97. doi: 10.1038/nm.2069. [DOI] [PubMed] [Google Scholar]
- 18.Travassos LH, Carneiro LA, Ramjeet M, Hussey S, Kim YG, Magalhaes JG, Yuan L, Soares F, Chea E, Le Bourhis L, Boneca IG, Allaoui A, Jones NL, Nunez G, Girardin SE, Philpott DJ. Nod1 and Nod2 direct autophagy by recruiting ATG16L1 to the plasma membrane at the site of bacterial entry. Nat Immunol. 2010;11:55–62. doi: 10.1038/ni.1823. [DOI] [PubMed] [Google Scholar]
- 19.Glas J, Wagner J, Seiderer J, Olszak T, Wetzke M, Beigel F, Tillack C, Stallhofer J, Friedrich M, Steib C, Goke B, Ochsenkuhn T, Karbalai N, Diegelmann J, Czamara D, Brand S. PTPN2 gene variants are associated with susceptibility to both crohn's disease and ulcerative colitis supporting a common genetic disease background. PLoS One. 2012;7:e33682. doi: 10.1371/journal.pone.0033682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scharl M, Mwinyi J, Fischbeck A, Leucht K, Eloranta JJ, Arikkat J, Pesch T, Kellermeier S, Mair A, Kullak-Ublick GA, Truninger K, Noreen F, Regula J, Gaj P, Pittet V, Mueller C, Hofmann C, Fried M, McCole DF, Rogler G. Crohn's disease-associated polymorphism within the PTPN2 gene affects muramyl-dipeptide-induced cytokine secretion and autophagy. Inflamm Bowel Dis. 2012;18:900–912. doi: 10.1002/ibd.21913. [DOI] [PubMed] [Google Scholar]
- 21.Henckaerts L, Cleynen I, Brinar M, John JM, Van Steen K, Rutgeerts P, Vermeire S. Genetic variation in the autophagy gene ULK1 and risk of Crohn's disease. Inflamm Bowel Dis. 2011;17:1392–1397. doi: 10.1002/ibd.21486. [DOI] [PubMed] [Google Scholar]
- 22.Franchimont D, Vermeire S, El Housni H, Pierik M, Van Steen K, Gustot T, Quertinmont E, Abramowicz M, Van Gossum A, Deviere J, Rutgeerts P. Deficient host-bacteria interactions in inflammatory bowel disease? The toll-like receptor (TLR)-4 Asp299gly polymorphism is associated with Crohn's disease and ulcerative colitis. Gut. 2004;53:987–992. doi: 10.1136/gut.2003.030205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oostenbrug LE, Drenth JP, de Jong DJ, Nolte IM, Oosterom E, van Dullemen HM, van der Linde K, te Meerman GJ, van der Steege G, Kleibeuker JH, Jansen PL. Association between Toll-like receptor 4 and inflammatory bowel disease. Inflamm Bowel Dis. 2005;11:567–575. doi: 10.1097/01.mib.0000161305.81198.0f. [DOI] [PubMed] [Google Scholar]
- 24.Mizushima N, Yamamoto A, Hatano M, Kobayashi Y, Kabeya Y, Suzuki K, Tokuhisa T, Ohsumi Y, Yoshimori T. Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. J Cell Biol. 2001;152:657–668. doi: 10.1083/jcb.152.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mizushima N, Kuma A, Kobayashi Y, Yamamoto A, Matsubae M, Takao T, Natsume T, Ohsumi Y, Yoshimori T. Mouse Apg16l, a novel WD-repeat protein, targets to the autophagic isolation membrane with the Apg12-Apg5 conjugate. J Cell Sci. 2003;116:1679–1688. doi: 10.1242/jcs.00381. [DOI] [PubMed] [Google Scholar]
- 26.Fujita N, Itoh T, Omori H, Fukuda M, Noda T, Yoshimori T. The Atg16l complex specifies the site of LC3 lipidation for membrane biogenesis in autophagy. Mol Biol Cell. 2008;19:2092–2100. doi: 10.1091/mbc.E07-12-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuballa P, Huett A, Rioux JD, Daly MJ, Xavier RJ. Impaired autophagy of an intracellular pathogen induced by a Crohn's disease associated Atg16l1 variant. PLoS One. 2008;3:e3391. doi: 10.1371/journal.pone.0003391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Homer CR, Richmond AL, Rebert NA, Achkar JP, McDonald C. ATG16L1 and NOD2 interact in an autophagy-dependent antibacterial pathway implicated in Crohn's disease pathogenesis. Gastroenterology. 2010;139:1630–1641. doi: 10.1053/j.gastro.2010.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lapaquette P, Glasser AL, Huett A, Xavier RJ, Darfeuille-Michaud A. Crohn's disease-associated adherent-invasive E. Coli are selectively favoured by impaired autophagy to replicate intracellularly. Cell Microbiol. 2010;12:99–113. doi: 10.1111/j.1462-5822.2009.01381.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Darfeuille-Michaud A, Boudeau J, Bulois P, Neut C, Glasser AL, Barnich N, Bringer MA, Swidsinski A, Beaugerie L, Colombel JF. High prevalence of adherent-invasive Escherichia coli associated with ileal mucosa in Crohn's disease. Gastroenterology. 2004;127:412–421. doi: 10.1053/j.gastro.2004.04.061. [DOI] [PubMed] [Google Scholar]
- 31.Barnich N, Bringer MA, Claret L, Darfeuille-Michaud A. Involvement of lipoprotein Nlpl in the virulence of adherent invasive Escherichia coli strain LF82 isolated from a patient with Crohn's disease. Infect Immun. 2004;72:2484–2493. doi: 10.1128/IAI.72.5.2484-2493.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carvalho FA, Barnich N, Sauvanet P, Darcha C, Gelot A, Darfeuille-Michaud A. Crohn's disease-associated Escherichia coli LF82 aggravates colitis in injured mouse colon via signaling by flagellin. Inflamm Bowel Dis. 2008;14:1051–1060. doi: 10.1002/ibd.20423. [DOI] [PubMed] [Google Scholar]
- 33.Lapaquette P, Bringer MA, Darfeuille-Michaud A. Defects in autophagy favour adherent-invasive Escherichia coli persistence within macrophages leading to increased pro-inflammatory response. Cell Microbiol. 2012;14:791–807. doi: 10.1111/j.1462-5822.2012.01768.x. [DOI] [PubMed] [Google Scholar]
- 34.Cadwell K, Liu JY, Brown SL, Miyoshi H, Loh J, Lennerz JK, Kishi C, Kc W, Carrero JA, Hunt S, Stone CD, Brunt EM, Xavier RJ, Sleckman BP, Li E, Mizushima N, Stappenbeck TS, Virgin HW. A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature. 2008;456:259–263. doi: 10.1038/nature07416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saitoh T, Fujita N, Jang MH, Uematsu S, Yang BG, Satoh T, Omori H, Noda T, Yamamoto N, Komatsu M, Tanaka K, Kawai T, Tsujimura T, Takeuchi O, Yoshimori T, Akira S. Loss of the autophagy protein Atg16l1 enhances endotoxin-induced IL-1beta production. Nature. 2008;456:264–268. doi: 10.1038/nature07383. [DOI] [PubMed] [Google Scholar]
- 36.Cadwell K, Patel KK, Maloney NS, Liu TC, Ng AC, Storer CE, Head RD, Xavier R, Stappenbeck TS, Virgin HW. Virus-plus-susceptibility gene interaction determines Crohn's disease gene Atg16l1 phenotypes in intestine. Cell. 2010;141:1135–1145. doi: 10.1016/j.cell.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gebhard RL, Greenberg HB, Singh N, Henry P, Sharp HL, Kaplan L, Kapikian AZ. Acute viral enteritis and exacerbations of inflammatory bowel disease. Gastroenterology. 1982;83:1207–1209. [PubMed] [Google Scholar]
- 38.Khan RR, Lawson AD, Minnich LL, Martin K, Nasir A, Emmett MK, Welch CA, Udall JN., Jr Gastrointestinal norovirus infection associated with exacerbation of inflammatory bowel disease. J Pediatr Gastroenterol Nutr. 2009;48:328–333. doi: 10.1097/mpg.0b013e31818255cc. [DOI] [PubMed] [Google Scholar]
- 39.Schmid D, Pypaert M, Munz C. Antigen-loading compartments for major histocompatibility complex class II molecules continuously receive input from autophagosomes. Immunity. 2007;26:79–92. doi: 10.1016/j.immuni.2006.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu Y, Jagannath C, Liu XD, Sharafkhaneh A, Kolodziejska KE, Eissa NT. Toll-like receptor 4 is a sensor for autophagy associated with innate immunity. Immunity. 2007;27:135–144. doi: 10.1016/j.immuni.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.MacMicking JD, Taylor GA, McKinney JD. Immune control of tuberculosis by IFN-gamma-inducible LRG-47. Science. 2003;302:654–659. doi: 10.1126/science.1088063. [DOI] [PubMed] [Google Scholar]
- 42.Singh SB, Davis AS, Taylor GA, Deretic V. Human IRGM induces autophagy to eliminate intracellular mycobacteria. Science. 2006;313:1438–1441. doi: 10.1126/science.1129577. [DOI] [PubMed] [Google Scholar]
- 43.Henry SC, Daniell X, Indaram M, Whitesides JF, Sempowski GD, Howell D, Oliver T, Taylor GA. Impaired macrophage function underscores susceptibility to salmonella in mice lacking IRGM1 (LRG-47) J Immunol. 2007;179:6963–6972. doi: 10.4049/jimmunol.179.10.6963. [DOI] [PubMed] [Google Scholar]
- 44.Collazo CM, Yap GS, Sempowski GD, Lusby KC, Tessarollo L, Woude GF, Sher A, Taylor GA. Inactivation of LRG-47 and IRG-47 reveals a family of interferon gamma-inducible genes with essential, pathogen-specific roles in resistance to infection. J Exp Med. 2001;194:181–188. doi: 10.1084/jem.194.2.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Feng CG, Collazo-Custodio CM, Eckhaus M, Hieny S, Belkaid Y, Elkins K, Jankovic D, Taylor GA, Sher A. Mice deficient in LRG-47 display increased susceptibility to mycobacterial infection associated with the induction of lymphopenia. J Immunol. 2004;172:1163–1168. doi: 10.4049/jimmunol.172.2.1163. [DOI] [PubMed] [Google Scholar]
- 46.Bekpen C, Hunn JP, Rohde C, Parvanova I, Guethlein L, Dunn DM, Glowalla E, Leptin M, Howard JC. The interferon-inducible p47 (IRG) GTPases in vertebrates: loss of the cell autonomous resistance mechanism in the human lineage. Genome Biol. 2005;6:R92. doi: 10.1186/gb-2005-6-11-r92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brest P, Lapaquette P, Souidi M, Lebrigand K, Cesaro A, Vouret-Craviari V, Mari B, Barbry P, Mosnier JF, Hebuterne X, Harel-Bellan A, Mograbi B, Darfeuille-Michaud A, Hofman P. A synonymous variant in IRGM alters a binding site for miR-196 and causes deregulation of IRGM-dependent xenophagy in Crohn's disease. Nat Genet. 2011;43:242–245. doi: 10.1038/ng.762. [DOI] [PubMed] [Google Scholar]
- 48.Singh SB, Ornatowski W, Vergne I, Naylor J, Delgado M, Roberts E, Ponpuak M, Master S, Pilli M, White E, Komatsu M, Deretic V. Human IRGM regulates autophagy and cell-autonomous immunity functions through mitochondria. Nat Cell Biol. 2010;12:1154–1165. doi: 10.1038/ncb2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dancey J. mTOR signaling and drug development in cancer. Nat Rev Clin Oncol. 2010;7:209–219. doi: 10.1038/nrclinonc.2010.21. [DOI] [PubMed] [Google Scholar]
- 50.Dumortier J, Lapalus MG, Guillaud O, Poncet G, Gagnieu MC, Partensky C, Scoazec JY. Everolimus for refractory Crohn's disease: a case report. Inflamm Bowel Dis. 2008;14:874–877. doi: 10.1002/ibd.20395. [DOI] [PubMed] [Google Scholar]
- 51.Massey DC, Bredin F, Parkes M. Use of sirolimus (rapamycin) to treat refractory Crohn's disease. Gut. 2008;57:1294–1296. doi: 10.1136/gut.2008.157297. [DOI] [PubMed] [Google Scholar]
- 52.Balgi AD, Fonseca BD, Donohue E, Tsang TC, Lajoie P, Proud CG, Nabi IR, Roberge M. Screen for chemical modulators of autophagy reveals novel therapeutic inhibitors of mTORC1 signaling. PLoS One. 2009;4:e7124. doi: 10.1371/journal.pone.0007124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang L, Yu J, Pan H, Hu P, Hao Y, Cai W, Zhu H, Yu AD, Xie X, Ma D, Yuan J. Small molecule regulators of autophagy identified by an image-based high-throughput screen. Proc Natl Acad Sci USA. 2007;104:19023–19028. doi: 10.1073/pnas.0709695104. [DOI] [PMC free article] [PubMed] [Google Scholar]