ABSTRACT
Constitutive photomorphogenesis 9 (COP9) signalosome 6 (CSN6) plays an essential role in tumor development. The present study aims to demonstrate that CSN6 is an important biomarker and has prognostic value for patients with pancreatic ductal adenocarcinoma (PDAC). We analyzed CSN6 expression levels in PDAC and adjacent non-cancerous tissues using immunohistochemistry (IHC) and quantitative real-time PCR (qPCR) analysis. We found that CSN6 was highly expressed in PDAC tissues, contrasting to adjacent non-cancerous tissues. Interestingly, CSN6 expression was positively associated with proliferating cell nuclear antigen (PCNA) expression. Further investigation indicated that CSN6 knockdown significantly suppressed the proliferation of PDAC cells and decreased the expression levels of PCNA, while CSN6 overexpression increased the proliferation, as well as the expression levels of PCNA in PDAC cells. Furthermore, a χ2 test indicated that the expression of CSN6 in PDAC tissues was markedly associated with tumor infiltration and serum carbohydrate antigen 19–9 levels. In addition, univariate and multivariate analyses showed that CSN6 levels were significantly correlated with poor clinical outcomes of patients with PDAC. Kaplan–Meier analysis showed that patients with high expression of CSN6 had shorter overall survival. These results suggest that the expression of CSN6 correlates with the progression of PDAC, resulting in poor prognosis. Thus, CSN6 may play a significant role in the development of PDAC and is a potential target to prevent and treat PDAC.
KEYWORDS: CSN6, pancreatic ductal adenocarcinoma, proliferation, PCNA, survival, prognosis
Introduction
Pancreatic ductal adenocarcinoma (PDAC) is one of the most aggressive and fatal human tumors, having a high propensity for local invasion and distant metastasis.1 It is the third leading cause of cancer death in the world and is predicted to be ranked second within a decade.2–4 PDAC has a median survival measured in months and the overall 5-year survival rate remains less than 6%.5 Although surgery and chemotherapy for PDAC have improved greatly, the prognosis for patients with PDAC is still unsatisfactory.6 This is because most patients with PDAC are asymptomatic until the disease progresses to the advanced stage4 and distant metastasis has already occurred at diagnosis.6 Autopsy reports show that 90% cases of PDAC have complex distant metastasis.6 Therefore, there is an urgent need for specific biomarkers related to early diagnosis and prognosis.
The fundamental cause of cancer is disordered gene expression systems, including oncogenes, tumor suppressor genes, and DNA damage repair related genes. When a gene is activated or enters a silent state, the growth of the mutant cell will become uncontrolled, and the cell will begin to endlessly divide and increase, thereby rapidly forming a tumor, which is also a major reason for the rapid development of malignant tumors.7 Pancreatic cancer is a tumor with a complex mutational landscape, which is associated with the unusual expression of multiple proteins.5 The ubiquitin-proteasome pathway is critical to control the turnover of various proteins and plays a vital role in maintaining normal cellular function. Disorders of ubiquitin-mediated proteolysis lead to the development of various human cancers.8
Constitutive photomorphogenesis 9 (COP9) signalosome (CSN) is a protein complex involved in protein degradation, transcriptional activation,9 signal transduction,10–14 DNA damage response,15–17 and tumorigenesis.13,15,18,19 Mammalian CSN contains eight subunits (CSN1–CSN8). CSN6 and CSN5 are the only two subunits containing an MPN (Mpr1, Pad1 N-terminal) domain, which has impact on Cullin-RING Ligases (CRL).20 CSN5 can deneddylate Cullin to regulate CRL activity; however, the function of CSN6 remains obscure.11
Recently, studies implicated CSN6 in cancer progression, because it is overexpressed in a variety of cancers,21 such as breast cancer, lung cancer, cervical cancer, thyroid cancer, colorectal cancer, glioblastoma, and hepatocellular carcinoma.19,21–25 CSN6 is involved in ubiquitin-mediated degradation of important proteins implicated in cell cycle progression and signal transduction.22,26 CSN6 can induce the degradation of cyclin-dependent kinase inhibitors p27 and p57.10,16,23 In addition, CSN6 assists the degradation of p53 via stabilization of MDM2 (the human homolog of mouse double minute 2).27–29 CSN6 can stabilize several important oncogenic proteins such as β-catenin,21,25 Myc,14,15 and the epidermal growth factor receptor (EGFR)24 to promote cancer development. Nevertheless, the impact of CSN6 in human PDAC remains unclear. Therefore, it remains to be determined whether CSN6 is a prognostic marker for PDAC. Previous research showed that CSN6 overexpression led to accelerated cell proliferation and promoted tumorigenicity.11 Therefore, in the present study, we aimed to determine the effect of CSN6 expression on the proliferation and prognosis of PDAC. The results showed that the expression of CSN6 and proliferating cell nuclear antigen (PCNA), a classic and widely using marker of cancer cell proliferation,30,31 were positively correlated in PDAC, suggesting that CSN6 might promote the malignant proliferation of PDAC cells by influencing PCNA expression. Thus, CSN6 might be a significant prognostic marker and play a potential role in antiproliferative therapy for PDAC.
Results
CSN6 protein expression is significantly higher in PDAC tissues than in adjacent non-cancerous
CSN6 protein levels were examined in 10 PDAC tissues and 10 paired adjacent non-tumor tissues. The cytoplasmic and nuclear expression levels of CSN6 were significantly higher in PDAC tissues compared with those in adjacent non-tumor tissues. The IHC staining of CSN6 in PDAC and adjacent non-tumor tissues was shown in Figure 1a. Of the 10 paired cancer and non-cancerous tissues, 7 (70%) PDAC tissue samples were CSN6 positive, whereas all adjacent non-cancerous tissue samples were CSN6 negative (P < .001; Figure 1b). The mRNA expression of CSN6 (in 15 PDAC tissues and 15 paired adjacent non-tumor tissues) agreed with the protein expression data, showing high expression in PDAC tissues compared with that in adjacent non-cancerous (P < .001) (Figure 1c). The Oncomine database further confirmed that the CSN6 mRNA expression levels in PDAC tissue samples were significantly higher than those in normal pancreas controls (P < .001)32 (Fig. S1). Subsequently, we performed IHC analysis for 94 patients with PDAC to test CSN6 expression levels and divided them into stages 1, 2, 3, and 4 according to the degree of tumor infiltration observed from the pathological examination. The expression of CSN6 in the patients in each stage was shown in Figure 1d, which showed that the expression level of CSN6 gradually increased with increasing T stage; however, the expression level of CSN6 was not related to the pTNM stage (Figure 1e). In summary, the expression level of CSN6 in PDAC tumor tissues was higher than that in normal control tissues, and the expression level of CSN6 was related to the degree of tumor infiltration (tumor size). We speculated that CSN6 might affect tumor proliferation.
Figure 1.
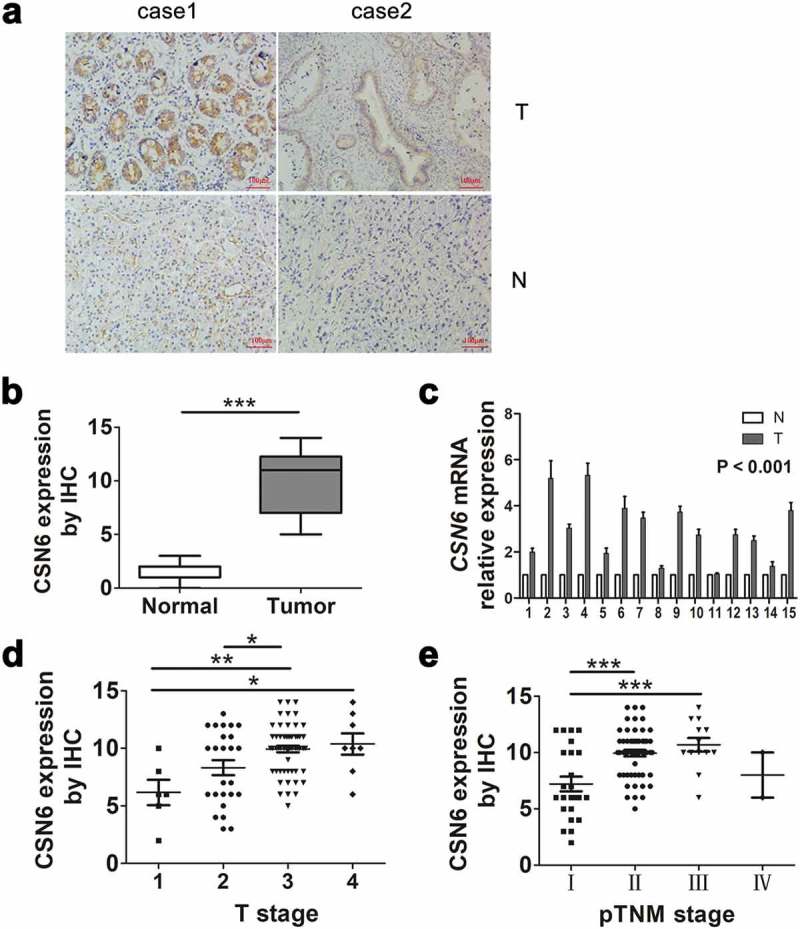
CSN6 protein expression is significantly higher in PDAC tissues than in adjacent non-cancerous tissue. a CSN6 expression in cancer tissues and adjacent non-cancerous tissue (magnification, 200×); T CSN6 expression in PDAC tissues, N CSN6 expression in adjacent non-cancerous tissues. b CSN6 IHC score in cancer tissues and adjacent non-cancerous tissues. c Fifteen representative paired samples of PDAC tissue (T) and adjacent normal tissues (N) were used for real-time qPCR analysis, and CSN6 expression levels were higher in 15 representative paired PDAC samples (p < .001). d The expression of CSN6 in each T stage (tumor infiltration) of PDAC patients (n = 94; T1, n = 6; T2, n = 25; T3, n = 55; T4, n = 8). e The expression of CSN6 in each pTNM stage of PDAC patients (n = 94; Ι, n = 24; ΙΙ, n = 55; ΙΙΙ, n = 12; ΙV, n = 3). Four groups of data were analyzed using ANOVA with post-test. PDAC, pancreatic ductal adenocarcinoma; CSN6, constitutive photomorphogenesis 9 (COP9) signalosome 6; qPCR, quantitative real-time PCR; IHC, immunohistochemistry. *P < .05; **P < .01; ***P < .001.
CSN6 expression has a positive interdependency with PCNA expression
An IHC examination showed that CSN6 was expressed in the cytoplasm and nucleus of PDAC. Examples of negative, weak, moderate and strong expression of CSN6 were shown in Figure 2a. Among the 94 PDAC specimens, we found that 58 (61.7%) cases exhibited positive CSN6 expression and 36 (38.3%) cases exhibited negative CSN6 expression, which demonstrated that CSN6 had a high expression in PDAC tissues. Interestingly, CSN6 and PCNA protein expression were positively correlated. Pearson correlation coefficient analysis was conducted to analyze the correlation between CSN6 expression and PCNA expression, which showed that the correlation was significant (r = 0.502, P < .001) (Figure 2b). The CSN6 and PCNA expression levels in serial sections were presented in Figure 2c. These results further strengthened that CSN6 might promote tumor proliferation by influencing the expression of PCNA. We also analyzed the correlation between CSN6 expression and another proliferative biomarker Ki67 expression were showed in Figure S2 (r = 0.317, P = .0018). Overall, these results demonstrated that CSN6 expression correlated positively with that of proliferative biomarker, especially PCNA.
Figure 2.
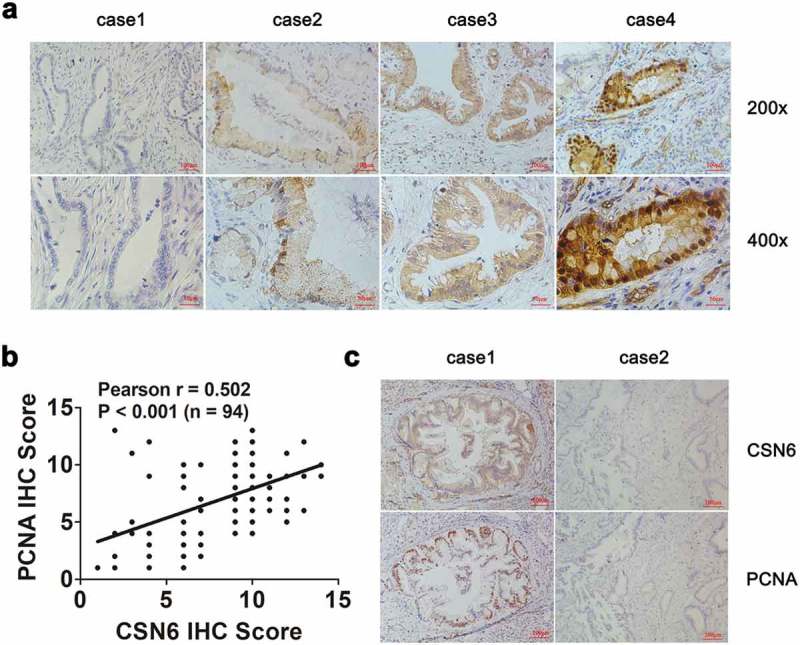
CSN6 expression is positively associated with PCNA expression. a Representative images of CSN6 protein expression in paraffin-embedded tissues from 94 patients with PDAC; Negative expression, low expression, moderate expression and high expression were shown (magnification, 200× and 400×). b The positive correlation between CSN6 expression and PCNA expression as assessed using Pearson correlation coefficient analysis (r = 0.502, P < .001, n = 94). c Staining of the same cohorts of pancreatic tumor sections for analysis of the related expression of CSN6 and PCNA, positive and negative expression of CSN6 and PCNA in cancer tissues were displayed (magnification, 100×). PCNA, proliferating cell nuclear antigen; PDAC, pancreatic ductal adenocarcinoma; CSN6, constitutive photomorphogenesis 9 (COP9) signalosome 6.
CSN6 expression is associated with the proliferative ability of PDAC cells
To investigate the biological significance of CSN6, we established CSN6 knockdown and overexpression in PDAC cells and the expression level of the CSN6 protein was assessed using western blotting analysis (Figure 3a,b). We then examined the effects of CSN6 knockdown and overexpression on cellular proliferation and colony formation in Capan-1 and PANC-1 cells. A CCK-8 assay revealed lower cell viability in Capan-1 cells transfected with the short interfering RNA (siRNA) targeting CSN6 (siCSN6) than in those transfected with the control siRNA (siNC) (Figure 3c), and clonal formation experiments showed similar results in Capan-1 siCSN6 cells. CSN6 knockdown suppressed the colony-forming potential of Capan-1 cells (Figure 3d). In contrast, when CSN6 was overexpressed in PANC-1 cells, the cells’ proliferative ability and clonality were enhanced compared with those of control cells (Figure 3e,f). To explore the underlying mechanisms of this effect, we examined the expression levels of PCNA. The results demonstrated that the expression levels of CSN6 and PCNA were positive correlated (Figure 3g,h). Taken together, these results suggested that CSN6 could evidently promote PDAC cells proliferation.
Figure 3.
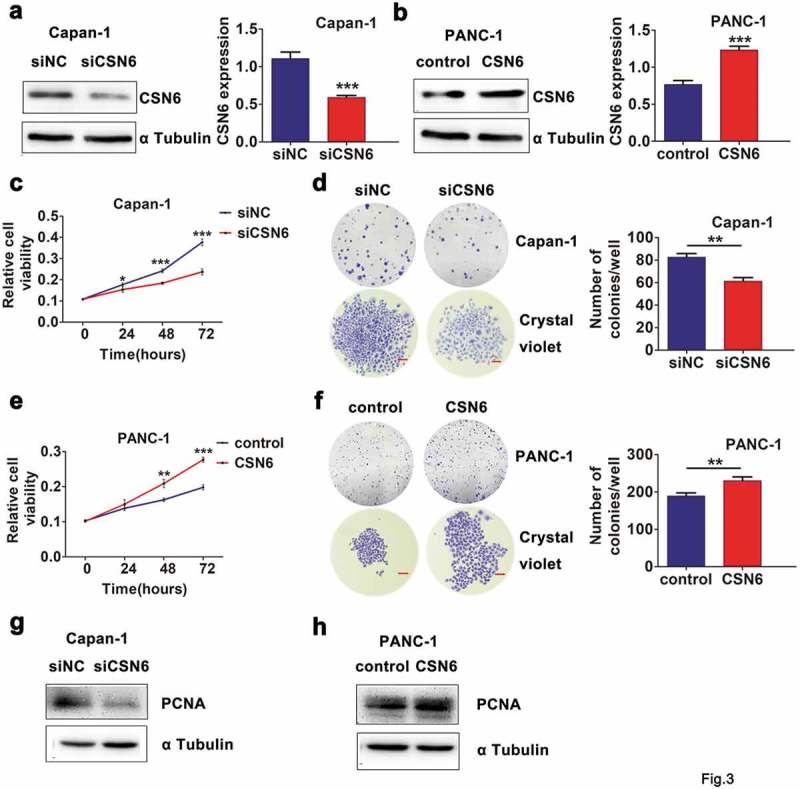
CSN6 expression is associated with proliferation ability of PDAC cells. a-b CSN6 knockdown in Capan-1 cells and CSN6 overexpression in PANC-1 cells were identified by western blot analysis (CSN6, 34kD; α Tubulin, 55kD). Western blotting data were quantified by densitometric analysis with ImageJ software. c Knockdown of CSN6 suppressed the viability of Capan-1 cells. d Knockdown of CSN6 inhibited colony formation of the Capan-1 cells. e Overexpression of CSN6 improved the viability of PANC-1 cells. f Overexpression of CSN6 enhanced colony formation of the PANC-1 cells. Images recorded under micro and macro view, indicating the size and the number of colonies in each group of cells, and statistical analysis of the colony numbers (>50 cells/colony) in the Capan-1 and PANC-1 cells. Scale bar = 200μm. g-h The expression levels of PCNA in CSN6 knockdown and overexpression cells were detected by western blot analysis (PCNA, 29kD). PDAC, pancreatic ductal adenocarcinoma; CSN6, constitutive photomorphogenesis 9 (COP9) signalosome 6; PCNA, proliferating cell nuclear antigen; si, small interfering; NC, negative control. *P < .05; **P < .01; ***P < .001.
Association between CSN6 and the clinicopathological features of PDAC
We next analyzed the association of CSN6 expression levels with the clinicopathological characteristics of patients with PDAC. We found that high CSN6 levels were related to tumor infiltration and serum carbohydrate antigen 19–9 (CA19-9) levels in PDAC (P = .021 and P = .028, respectively). However, CSN6 levels were not associated with sex, age, tumor sites, tumor sizes, or serum carcino-embryonic antigen (CEA) levels (P = .404, 0.471, 0.14, 0.689 and 0.717, respectively). Table 1 showed the detailed clinicopathological characteristics of these 94 patients. The pathological grade was determined according to the eighth edition of the American Joint Committee on Cancer (AJCC) staging system.
Table 1.
Correlation between CSN6 expression and clinicopathological features of patients with pancreatic cancer.
| CSN6 expression |
||||
|---|---|---|---|---|
| Variables | Clinicopathological feature | Low | High | P value |
| Age(years) | 0.404 | |||
| ≤50 | 23 | 32 | ||
| >50 | 13 | 26 | ||
| Sex | 0.471 | |||
| Male | 19 | 35 | ||
| Female | 17 | 23 | ||
| Tumor site | 0.14 | |||
| Pancreatic head | 23 | 28 | ||
| Pancreatic body and tail | 13 | 30 | ||
| Tumor size | 0.689 | |||
| ≤3cm | 21 | 35 | ||
| >3cm | 15 | 21 | ||
| Differentiation | 0.696 | |||
| High differentiation | 19 | 33 | ||
| Middle and low differentiation | 17 | 25 | ||
| T stage | 0.021* | |||
| T1-2 | 17 | 14 | ||
| T3-4 | 19 | 44 | ||
| N stage | 1.000 | |||
| N0 | 32 | 52 | ||
| N1 | 4 | 6 | ||
| M stage | 0.556 | |||
| M0 | 34 | 57 | ||
| M1 | 2 | 1 | ||
| AJCC (pTNM) | 0.151 | |||
| I-II | 33 | 46 | ||
| III-IV | 3 | 12 | ||
| CEA | 0.717 | |||
| Normal | 20 | 30 | ||
| Abnormal | 16 | 28 | ||
| CA19-9 | 0.028* | |||
| Normal | 18 | 16 | ||
| Abnormal | 18 | 42 | ||
High CSN6 expression is an unfavorable prognostic factor for PDAC
As shown in Table 2, univariate analyses identified CSN6 expression (P < .001), differentiation (P = .001), pTNM (P = .039), and serum CA19-9 levels (P = .013) as significant prognostic predictors. The expression of CSN6 was the most significant factor. Multivariate analysis identified that CSN6 expression (P < .001) and differentiation (P = .002) were independent prognostic factors (Table 2). Thus, the results indicated that positive expression of CSN6 was an adverse prognostic factor for patients with PDAC.
Table 2.
Univariate and multivariate analysis of clinicopathological factors in patients with pancreatic cancer Variable.
| Univariate analysis |
Multivariate analysis |
|||
|---|---|---|---|---|
| HR (95% CI) | P value | HR (95% CI) | P value | |
| Sex | 1.380 (0.874–2.178) | 0.167 | ||
| Age(years) | 1.154 (0.730–1.825) | 0.540 | ||
| CSN6 | 2.676 (1.634–4.383) | <0.001* | 2.561 (1.528–4.291) | <0.001* |
| Tumor site | 1.207 (0.773–1.886) | 0.408 | ||
| Differentiation | 0.472 (0.229–0.744) | 0.001* | 0.473 (0.296–0.756) | 0.002* |
| T stage | 1.481 (0.915–2.399) | 0.110 | ||
| N stage | 1.453 (0.739–2.859) | 0.279 | ||
| M stage | 1.255 (0.394–3.988) | 0.701 | ||
| pTNM | 1.881 (1.032–3.427) | 0.039* | 1.445 (0.771–2.708) | 0.250 |
| CEA | 0.829 (0.530–1.296) | 0.410 | ||
| CA19-9 | 1.818 (1.135–2.912) | 0.013* | 1.417 (0.858–2.339) | 0.173 |
Positive CSN6 expression is associated with a shorter OS
We observed these 94 patients with PDAC for 5.5 years of follow-up to investigate the correlation between CSN6 expression and their overall survival (OS). Kaplan–Meier survival analysis was performed to explore the association between CSN6 and the survival time in patients with PDAC. The median OS of patients with positive CSN6 expression (12.10 months) was markedly shorter than that for patients with negative CSN6 expression (16.67 months) (P < .001, n = 94; Figure 4). The Kaplan–Meier OS curves indicated that patients with negative CSN6 expression would have a longer survival time than those with positive CSN6 expression.
Figure 4.

The expression of CSN6 and survival time in patients with PDAC. The overall survival curves for the high-CSN6 expression group and the low-CSN6 expression group; the difference is statistically significant (P < .001). PDAC, pancreatic ductal adenocarcinoma; CSN6, constitutive photomorphogenesis 9 (COP9) signalosome 6.
Discussion
With a 5-year survival rate of less than 6%, PDAC is one of the most aggressive cancers and is the third leading cause of cancer related death.4,5 In spite of great efforts to understand the etiology and pathogenesis of PDAC, the underlying molecular mechanisms of PDAC development remain unclear.33 Hence, the primary mission in the field of PDAC therapy is to identify specific prognostic molecular markers.
The ubiquitin-proteasome pathway is vital for gene expression and maintaining normal cellular function.8 Ubiquitination plays a significant role in the localization, metabolism, regulation, function, and degradation of proteins.34 It is also involved in the regulation of proliferation, differentiation, apoptosis, cell cycle, metastasis, gene expression, transcription, signal transmission, and almost all life activities.35,36 Ubiquitination is a common endogenous protein degradation pathway. Dysregulation of ubiquitin-mediated proteolysis leads to the development of various cancers.37
CSN6 has an MPN domain, which may influence the Cullin-RING Ligases (CRL); however the precise function of CSN6 remains unclear. Recent studies showed that CSN6 is involved in ubiquitin-mediated degradation of the main proteins implicated in cell cycle progression and signal transduction.22,26 In addition, CSN6 was overexpressed in some certain cancers;38 however, the expression and function of CSN6 in PDAC are poorly understood.
The results of the present study revealed that CSN6 levels in PDAC tissues were notably higher than those in adjacent non-cancerous tissues, and the high expression of CSN6 predicted poor prognosis in PDAC. In addition, CSN6 expression was related to tumor infiltration (T-stage). However, the specific mechanisms by which CSN6 contributes to tumorigenesis remain obscure. Recently, research into CSN6 has concentrated on the signaling pathways involved in tumorigenesis,13,15,18,19 which identifies the MDM2-p53 signaling pathway as an important one, in which CSN6 plays a vital role in modulating MDM2 stability, p53-dependent apoptosis, and tumorigenesis.19 Mechanistic studies indicated that CSN6 prevented MDM2 auto-ubiquitination, leading to stability of MDM2 and the degradation of p53. Conversely, a lack of CSN6 resulted in decreased MDM2 levels and an increase in p53 levels, which enhanced p53-induced apoptosis and tumor suppression.19,29 These results indicated that CSN6 is an oncogene that positively regulates MDM2 and plays an important role in DNA damage-related apoptosis and carcinogenesis via the MDM2-p53 signaling pathway.19 Moreover, CSN6 could decrease the levels of carboxy terminus of Hsp70-interacting protein (CHIP) and increase those of EGFR in tumors, identifying the CSN6-CHIP-EGFR axis as a potential therapeutic target.24 The CSN–Cullin – F-Box and WD repeat domain containing 7 (FBXW7) axis regulates the stability of Myc and provides a clue for the relevance of CSN6 overexpression and Myc stabilization and activation during tumorigenesis.15 CSN6, via EGFR signaling, blocks the ubiquitin-proteasome pathway to stabilize β-catenin, and in the disordered β-catenin caused by ERK2-activation, CSN6 is essential for the development of colorectal cancer.21
Not all ubiquitination modifications lead to degradation, some cause changes in the activity of proteins, leading to other biological effects, such as DNA damage repair.39 Previously, human CSN6 was observed to take part in cell proliferation and the G2/M phase transition of the cell cycle.24 Our results showed that CSN6 and PCNA had similarly high expression levels in PDAC, and the expression of CSN6 was positively correlated with that of PCNA. We also found that high expression level of CSN6 promoted the proliferative ability of PDAC cells. However, to date, there has been no report of a relationship between CSN6 and PCNA. Determining the mechanism of this relationship is beyond the scope of the present study; therefore, identifying the regulation between CSN6 and PCNA in tumors will require further study.
PCNA is required for eukaryotic DNA synthesis, and its expression may be a marker for the expression of DNA polyploids and for the dysregulation of tumor cells.40 The detection of PCNA can objectively evaluate the proliferation state of tumor cells. Studies have shown that PCNA can better reflect the proliferative activity of cells,41 and some studies have shown that PCNA has a certain relationship with the diagnosis and prognosis of some tumors. In a prognostic study of breast cancer, proliferation markers, such as Ki67 and PCNA, are listed as the second most important prognostic indicators of breast cancer after hormone receptors and histological grade, together with c-erbB-2 and p53.42 PCNA has a wide range of applications, including differentiating benign and malignant tumors, determining the degree of malignancy, and evaluating prognosis. PCNA plays a significant role in DNA replication and several DNA damage-responsive pathways.43 Studies have shown that ubiquitin modification of PCNA plays an important role in DNA damage tolerance.43 We hypothesized that CSN6 might mediate a post-translational ubiquitination modification of PCNA, thereby promoting the proliferation of PDAC cells and exerting malignant biological functions. Thus, further studies are required to investigate the specific mechanism. PCNA is an important proliferative marker in tumor. CSN6 may also affect the expression of PCNA and endow malignant biological behavior to tumor by affecting the growth and proliferation related pathway of tumor.
Pancreatic ductal adenocarcinoma has a high degree of malignancy and poor prognosis. Currently, there is no effective prognostic marker or therapeutic target for PDAC. The results of the present study showed that CSN6 is an independent risk factor for PDAC. Patients with high CSN6 expression have a shorter OS and poorer prognosis. The present study also confirms that differentiation is an independent risk factor for PDAC, which agrees with the results of previous studies,44–46 confirming the credibility of our results. In addition, high expression of CSN6 may be associated with tumor infiltration; however, unexpectedly, the N-stage and M-stage did not correlate with the expression of CSN6. This contrasts with the results of previous research.47 One explanation is that because patients with advanced pancreatic cancer are ineligible for surgery, fewer cases of advanced N-stage and M-stage are detected and reported. Univariate and multivariate analyses demonstrated that high CSN6 expression is a risk factor for PDAC. However, the specific mechanism resulting in the high expression of CSN6 requires further study.
This study proved that patients with PDAC with positive CSN6 had a higher risk of poor prognosis. For these patients, we should evaluate whether more effective drugs should be prescribed, such as a combination of three drugs: fluorouracil, irinotecan and oxaliplatin, FOLFIRINOX, and combination of anti-proliferative targeted drugs, to improve the prognosis of these patients.
In summary, the present study identifies that CSN6 is highly expressed in PDAC and may have clinical significance for patients with PDAC. CSN6 activation might play a vital role in PDAC progression and prognosis, and may be a potential biomarker for PDAC. The CSN6 expression level was positively correlated with that of the proliferation related marker PCNA, indicating that CSN6 might have a key role in cell proliferation and cancer development by preventing PCNA degradation. However, the regulatory mechanism involving CSN6 in PDAC remains to be determined in future investigations.
Materials and methods
This study was approved by the Ethics Committee of Harbin Medical University Cancer Hospital, and patients provided signed informed consent. We used a retrospective design to analyze PDAC samples and all patients with PDAC underwent surgery between December 2003 and June 2010, and were confirmed pathologically to have the disease. A total of 94 PDAC specimens were paraffin-embedded. The clinicopathological characteristics of the patients were derived from their medical records (Table 1).
IHC and scoring
The expression of CSN6 in PDAC tissues and adjacent non-cancerous tissues was detected using immunohistochemistry (IHC). The pancreatic cancer tissues were cut into 4-μm-thick sections using a microtome, fixed on a glass slide, and deparaffinized. The slides were then heated in citrate buffer, incubated with 0.3% H2O2 for 10 minutes, and gradually hydrated with a gradient alcohol series. The slides were then incubated overnight at 4°C with anti-CSN6 antibodies (1:100 dilution, Origene, Rockville, MD, USA), anti-PCNA antibodies (1:50 dilution, Arigo, Taiwan, China) and Anti-Ki67 antibody (1:50 dilution, Arigo, Taiwan, China). After washing several times with phosphate-buffered saline (PBS), the sections were incubated in biotinylated secondary antibodies. After extensive washing, they were exposed to 3,3-diaminobenzidine and stained with Gill’s hematoxylin. After continuous dehydration, the slides were mounted under a coverslip and examined under a microscope. The blank control group and a negative control group comprised slides incubated with PBS and omitting the primary antibody, respectively. The positive control group was a positive immune-stained slide. Two pathologists independently evaluated and recorded the IHC results in all cases, without prior knowledge of the clinical data. The intensity of CSN6 staining was scored as 0 (no signal), 1 (weak), 2 (moderate), and 3 (marked). Percentage score was assigned as 1 (0–25%); 2 (26–50%); 3 (51–75%); and 4 (76–100%). The score of each sample was multiplied to give a final score of 0–14. The final CSN6 expression score was calculated as the sum of both grades (negative: total grade ≤ 9; positive: total grade > 9).
Quantitative real-time PCR (qpcr) assay
We used the Trizol reagent (Thermo Fisher Scientific, USA) to extract total RNA from 15 paired cancer and adjacent non-cancerous tissues, and the M-MLV Reverse Transcriptase kit (Invitrogen, USA) was used to perform the reverse-transcription reactions. The SYBR Green PCR kit (Toyobo, Osaka, Japan) was used to carry out the real-time PCR assay. Real-time PCR was performed for CSN6 and ACTB (encoding β-actin). The comparative cycle threshold (Ct) method was used, with β-actin expression as a reference, to determine the fold changes of the target gene. Primer sequence (5′ to 3′) were: CSN6-F: CTCATCTTGGAGTACGTCAAGGC, CSN6-R: CATGGTGTTGCACGTTTTGGT; β-actin-F: CATGTACGTTGCTATCCAGGC, β-actin-R: CTCCTTAATGTCACGCACGAT.
Cell culture, CSN6 knockdown and overexpression
Human PDAC cell lines PANC-1 and Capan-1 were purchased from the American Type Culture Collection in 2017. PANC-1 and Capan-1 were authenticated in March, 2018 using short tandem repeat (STR) profiling in an ABI Prism® 3500 Genetic Analyzer (Applied Biosystems, USA). Both of them were cultured in Dulbecco’s modified Eagle’s medium (DMEM, Gibco, USA) with 10% fetal bovine serum (FBS, Gibco) at 37°C and 5% CO2.
PDAC cells were seeded in 6-well plates at a concentration of 1 × 105 cells/well. We selected CSN6 low-expressing PANC-1 cells for CSN6 gene overexpression and Capan-1 cells with high CSN6 expression for CSN6 gene knockdown. Diluted siRNAs and plasmids were added to Lipofectamine® 2000 (Thermo Fisher Scientific, USA) for 20 min, and then the cells were transfected using the mixture and incubated at 4°C for 6 h. Subsequently, the transfection efficiency was evaluated using western blotting analysis.
Small interfering RNA (siRNA) targeting CSN6 were transfected at concentration of 80 nM. siRNA transfection efficiency was 50–70%, and siRNA duplex could persist for at least 5 days. All RNA oligoribonucleotides were purchased from Genepharma (Shanghai, China), and the selected siCSN6 (from 3 kinds of RNA oligoribonucleotides) had the following sequence: 5ʹ-GCCGAAAUAUCGAGGUGAUTTAUCACCUCGAUAUUUCGGCTT-3ʹ. A random sequence of RNA duplex was used as negative control for siRNA. The overexpression plasmid was the ORF of CSN6 in pEnter, purchased from Vigene Biosciences (Shandong, China), and an empty pEnter plasmid was used as control for CSN6 overexpression plasmid.
Cell viability assay
Cell viability was determined using the Cell Counting Kit-8 (Dojindo, Japan) according to the manufacturer’s instructions. Capan-1 and PANC-1 Cells (5 × 103 and 8 × 103 cells per well, respectively) were seeded separately into 96-well plates (at 30 to 40% confluence) and starved for 4 h. Then they were treated with siRNAs and overexpression plasmids. After treatment for 0, 24, 48, or 72 hours, 10 μL of CCK-8 solution was added to the cells, which were then incubated for another 2–3 hours in the dark. The absorbance of these wells was measured at 450 nm. All determinations were performed in triplicate.
Western blotting
Total proteins were harvested from cultured cells using an ice-cold lysis buffer. Proteins were separated by 10% SDS-polyacrylamide gel electrophoresis and then transferred to PVDF membranes. The membranes were blocked with 5% nonfat milk and then incubated with primary antibodies against CSN6 (Origene, Rockville, MD, USA), PCNA (Arigo, Taiwan, China), and α Tubulin (Santa Cruz Biotechnology Inc., Dallas, TX, USA), followed by horseradish peroxidase (HRP)-conjugated secondary antibodies (Proteintech Wuhan Sanying, Wuhan, China). Immunoreactive proteins were detected using a chemiluminescence solution (Thermo fisher Scientific, USA).
Colony formation assay
Following siRNA and plasmid transduction, Capan-1 (300 cells/well) and PANC-1 (500 cells/well) cells were seeded into 6-well plates for 7 days, respectively. When the colonies were formed, the plate was stained with crystal violet solution for 3 min and then washed with ddH2O. These cells were then photographed and counted under an optical microscope (the number of colonies containing >50 cells/colony were counted).
Database
The data of CSN6 levels of pancreatic ductal adenocarcinoma and pancreas tissues were obtained from Oncomine (https://www.oncomine.org/resource/login.html).
Statistical analysis
All stained slides were scanned using an Aperio Scan Scope scanner, and Image Scope software was used to obtain images of representative areas. All data were analyzed using GraphPad prism 7.0 (GraphPad Software, USA) and SPSS statistical software 24.0 (IBM Corp., Armonk, NY, USA). The relationships between CSN6 expression and other clinicopathological features, including gender, age, tumor location, tumor size, pathological grade, tumor infiltration, lymph node metastasis, distant metastasis, and preoperative serum CEA and CA19-9 levels, were studied using the χ2 test in 94 patients with PDAC. The Cox proportional hazard model was generated for univariate and multivariate regression analysis to identify prognostic factors. Kaplan–Meier survival curves were constructed using a log-rank test. Pearson correlation coefficient analysis was conducted to test for correlation between two quantitative measurements. The significant difference between multiple comparisons was analyzed by one-way analysis of variance (ANOVA) followed by Tukey’s multiple comparison. All data shown as mean ± SD were based on three independent experiments. All tests were bilateral and a P value < .05 was considered statistically significant. * means P < .05; ** means P < .01, *** means P < .001.
Abbreviations
| AJCC | American Joint Committee on Cancer |
| CA19-9 | carbohydrate antigen 19-9 |
| CCK-8 | Cell Counting Kit-8 |
| CEA | carcino-embryonic antigen |
| CI | confidence interval |
| CRL | Cullin-Ring Ligases |
| CSN6 | constitutive photomorphogenesis 9 (COP9) signalosome 6 |
| EGFR | epidermal growth factor receptor |
| HR | hazard ratio |
| IHC | immunohistochemistry |
| MDM2 | murine double minute 2 |
| MPN | Mpr1p and Pad1p N-terminal |
| OS | overall survival |
| PDAC | pancreatic ductal adenocarcinoma |
| PCNA | proliferating cell nuclear antigen |
Funding Statement
This work was supported by the National Natural Science Foundation of China under Grant numbers 81672428, 81773210, 81703000, and 81872427), the Beijing Xisike Clinical Oncology Research under Grant number Y-MX2016-027) and the Science Technological Research Funds of the Department of Education of Heilongjiang Province (No. 12541540) .
Acknowledgments
We thank Professor Tongsen Zheng in the Harbin Medical University Cancer Hospital for insightful comments and patient assistance during the writing process.
Disclosure statement
No potential conflict of interest was reported by the authors.
Ethics approval and consent to participate
For the use of these clinical materials for research purposes, prior patient consent and approval from the Institutional Research Ethics Committee were obtained.
Authors’ contributions
Jiaqi Shi conceived, designed, and performed the experiments; analyzed the data and wrote the manuscript. Xin Guan and Fei Zhan performed the experiments and analyzed the data. Chao Liu, Bojun Wang and Changjie Lou collected and analyzed the clinical samples. Yanqiao Zhang, Yuanfei Yao, and Zhiwei Li conceived and designed the study and participated in its design and coordination and wrote the manuscript. All authors read and approved the final version of the manuscript.
Supplemental material
Supplemental data for this article can be accessed on the publisher’s website.
References
- 1.Stathis A, Moore MJ.. Advanced pancreatic carcinoma: current treatment and future challenges. Nat Rev Clin Oncol. 2010;7:163–172. PMID:20101258. doi: 10.1038/nrclinonc.2009.236. [DOI] [PubMed] [Google Scholar]
- 2.Lomberk G, Blum Y, Nicolle R, Nair A, Gaonkar KS, Marisa L, Mathison A, Sun Z, Yan H, Elarouci N, et al. Distinct epigenetic landscapes underlie the pathobiology of pancreatic cancer subtypes. Nat Commun. 2018;9:1978 PMID:29773832. doi: 10.1038/s41467-018-04383-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rahib L, Smith BD, Aizenberg R, Rosenzweig AB, Fleshman JM, Matrisian LM.. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 2014;74:2913–2921. PMID:24840647. doi: 10.1158/0008-5472.CAN-14-0155. [DOI] [PubMed] [Google Scholar]
- 4.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30. PMID:29313949. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 5.Bailey P, Chang DK, Nones K, Johns AL, Patch AM, Gingras MC, Miller DK, Christ AN, Bruxner TJ, Quinn MC, et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature. 2016;531:47–52. PMID:26909576. doi: 10.1038/nature16965. [DOI] [PubMed] [Google Scholar]
- 6.Kamisawa T, Wood LD, Itoi T, Takaori K. Pancreatic cancer. Lancet. 2016;388:73–85. PMID:26830752. doi: 10.1016/S0140-6736(16)00141-0. [DOI] [PubMed] [Google Scholar]
- 7.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. PMID:21376230. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 8.Hoeller D, Hecker CM, Dikic I. Ubiquitin and ubiquitin-like proteins in cancer pathogenesis. Nat Rev Cancer. 2006;6:776–788. PMID:16990855. doi: 10.1038/nrc1994. [DOI] [PubMed] [Google Scholar]
- 9.Azzolin L, Zanconato F, Bresolin S, Forcato M, Basso G, Bicciato S, Cordenonsi M, Piccolo S. Role of TAZ as mediator of Wnt signaling. Cell. 2012;151:1443–1456. PMID:23245942. doi: 10.1016/j.cell.2012.11.027. [DOI] [PubMed] [Google Scholar]
- 10.Chen B, Zhao R, Su CH, Linan M, Tseng C, Phan L, Fang L, Yang HY, Yang H, Wang W, et al. CDK inhibitor p57 (Kip2) is negatively regulated by COP9 signalosome subunit 6. Cell Cycle. 2012;11:4633–4641. PMID:23187808. doi: 10.4161/cc.22887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Choi HH, Gully C, Su CH, Velazquez-Torres G, Chou PC, Tseng C, Zhao R, Phan L, Shaiken T, Chen J, et al. COP9 signalosome subunit 6 stabilizes COP1, which functions as an E3 ubiquitin ligase for 14-3-3σ. Oncogene. 2011;30:4791–4801. PMID:21625211. doi: 10.1038/onc.2011.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shin J, Phan L, Chen J, Lu Z, Lee MH. CSN6 positively regulates c-Jun in a MEKK1-dependent manner. Cell Cycle. 2015;14:3079–3087. PMID:26237449. doi: 10.1080/15384101.2015.1078030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xue Y, Chen J, Choi HH, Phan L, Chou PC, Zhao R, Yang H, Santiago J, Liu M, Yeung GE, et al. HER2-Akt signaling in regulating COP9 signalsome subunit 6 and p53. Cell Cycle. 2012;11:4181–4190. PMID:23095642. doi: 10.4161/cc.22413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang L, Liu Y, Wang M, Qian Y, Dong X, Gu H, Wang H, Guo S, Hisamitsu T. Quercetin-induced apoptosis of HT-29 colon cancer cells via inhibition of the Akt-CSN6-Myc signaling axis. Mol Med Rep. 2016;14:4559–4566. PMID:27748879. doi: 10.3892/mmr.2016.5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen J, Shin JH, Zhao R, Phan L, Wang H, Xue Y, Post SM, Ho CH, Chen JS, Wang E, et al. CSN6 drives carcinogenesis by positively regulating Myc stability. Nat Commun. 2014;5:5384 PMID:25395170. doi: 10.1038/ncomms6384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Choi HH, Guma S, Fang L, Phan L, Ivan C, Baggerly K, Sood A, Lee MH. Regulating the stability and localization of CDK inhibitor p27(Kip1) via CSN6-COP1 axis. Cell Cycle. 2015;14:2265–2273. PMID:25945542. doi: 10.1080/15384101.2015.1046655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Choi HH, CH S, Fang L, Zhang J, Yeung SC, Lee MH. CSN6 deregulation impairs genome integrity in a COP1-dependent pathway. Oncotarget. 2015;6:11779–11793. PMID:25957415. doi: 10.18632/oncotarget.3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Azzolin L, Panciera T, Soligo S, Enzo E, Bicciato S, Dupont S, Bresolin S, Frasson C, Basso G, Guzzardo V, et al. YAP/TAZ incorporation in the β-catenin destruction complex orchestrates the Wnt response. Cell. 2014;158:157–170. PMID:24976009. doi: 10.1016/j.cell.2014.06.013. [DOI] [PubMed] [Google Scholar]
- 19.Wang W, Tang M, Zhang L, Xu X, Qi X, Yang Y, Jin F, Chen B. Clinical implications of CSN6 protein expression and correlation with mutant-type P53 protein in breast cancer. Jpn J Clin Oncol. 2013;43:1170–1176. PMID:24106298. doi: 10.1093/jjco/hyt148. [DOI] [PubMed] [Google Scholar]
- 20.Echalier A, Pan Y, Birol M, Tavernier N, Pintard L, Hoh F, Ebel C, Galophe N, Claret FX, Dumas C. Insights into the regulation of the human COP9 signalosome catalytic subunit, CSN5/Jab1. Proc Natl Acad Sci U S A. 2013;110:1273–1278. PMID:23288897. doi: 10.1073/pnas.1209345110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fang L, Lu W, Choi HH, Yeung SC, Tung JY, Hsiao CD, Fuentes-Mattei E, Menter D, Chen C, Wang L, et al. ERK2-Dependent Phosphorylation of CSN6 Is Critical in Colorectal Cancer Development. Cancer Cell. 2015;28:183–197. PMID:26267535. doi: 10.1016/j.ccell.2015.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gao S, Fang L, Phan LM, Qdaisat A, Yeung SC, Lee MH. COP9 signalosome subunit 6 (CSN6) regulates E6AP/UBE3A in cervical cancer. Oncotarget. 2015;6:28026–28041. PMID:26318036. doi: 10.18632/oncotarget.4731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guo H, Jing L, Cheng Y, Atsaves V, Lv Y, Wu T, Su R, Zhang Y, Zhang R, Liu W, et al. Down-regulation of the cyclin-dependent kinase inhibitor p57 is mediated by Jab1/Csn5 in hepatocarcinogenesis. Hepatology. 2016;63:898–913. PMID:26606000. doi: 10.1002/hep.28372. [DOI] [PubMed] [Google Scholar]
- 24.Hou J, Deng Q, Zhou J, Zou J, Zhang Y, Tan P, Zhang W, Cui H. CSN6 controls the proliferation and metastasis of glioblastoma by CHIP-mediated degradation of EGFR. Oncogene. 2017;36:1134–1144. PMID:27546621. doi: 10.1038/onc.2016.280. [DOI] [PubMed] [Google Scholar]
- 25.Wen D, Liao T, Ma B, Qu N, Shi RL, Lu ZW, Wang YL, Wei WJ, Ji QH. Downregulation of CSN6 attenuates papillary thyroid carcinoma progression by reducing Wnt/β-catenin signaling and sensitizes cancer cells to FH535 therapy. Cancer Med. 2018;7:285–296. PMID:29341469. doi: 10.1002/cam4.1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang SN, Pei DS, Zheng JN. The COP9 signalosome subunit 6 (CSN6): a potential oncogene. Cell Div. 2013;8:14 PMID:24286178. doi: 10.1186/1747-1028-8-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Choi HH, Lee MH. CSN6-COP1 axis in cancer. Aging (Albany NY). 2015;7(7):461–462. PMID:26186957. doi: 10.18632/aging.100778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Iyer SV, Iwakuma T. A novel link between the HER2-Akt and MDM2-p53 pathways via CSN6. Cell Cycle. 2012;11:4112 PMID:23099920. doi: 10.4161/cc.22606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao R, Yeung SC, Chen J, Iwakuma T, Su CH, Chen B, Qu C, Zhang F, Chen YT, Lin YL, et al. Subunit 6 of the COP9 signalosome promotes tumorigenesis in mice through stabilization of MDM2 and is upregulated in human cancers. J Clin Invest. 2011;121:851–865. PMID:21317535. doi: 10.1172/JCI44111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Villafañez F, García IA, Carbajosa S, Pansa MF, Mansilla S, Llorens MC, Angiolini V, Guantay L, Jacobs H, Madauss KP, et al. AKT inhibition impairs PCNA ubiquitylation and triggers synthetic lethality in homologous recombination-deficient cells submitted to replication stress. Oncogene. 2019;38:4310–4324. PMID:30705406. doi: 10.1038/s41388-019-0724-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang SC. PCNA: a silent housekeeper or a potential therapeutic target. Trends Pharmacol Sci. 2014;35:178–186. PMID:24655521. doi: 10.1016/j.tips.2014.02.004. [DOI] [PubMed] [Google Scholar]
- 32.Badea L, Herlea V, Dima SO, Dumitrascu T, Popescu I. Combined gene expression analysis of whole-tissue and microdissected pancreatic ductal adenocarcinoma identifies genes specifically overexpressed in tumor epithelia. Hepatogastroenterology. 2008;55:2016–2027. PMID:19260470. [PubMed] [Google Scholar]
- 33.Whitcomb DC, Shelton CA, Brand RE. Genetics and Genetic Testing in Pancreatic Cancer. Gastroenterology. 2015;149:1252–64.e4. PMID:26255042. doi: 10.1053/j.gastro.2015.07.057. [DOI] [PubMed] [Google Scholar]
- 34.Ge Z, Leighton JS, Wang Y, Peng X, Chen Z, Chen H, Sun Y, Yao F, Li J, Zhang H, et al. Integrated Genomic Analysis of the Ubiquitin Pathway across Cancer Types. Cell Rep. 2018;23:213–26.e3. PMID:29617661. doi: 10.1016/j.celrep.2018.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schmid JA, Berti M, Walser F, Raso MC, Schmid F, Krietsch J, Stoy H, Zwicky K, Ursich S, Freire R, et al. Histone Ubiquitination by the DNA Damage Response Is Required for Efficient DNA Replication in Unperturbed S Phase. Mol Cell. 2018;71:897–910.e8. PMID:30122534. doi: 10.1016/j.molcel.2018.07.011. [DOI] [PubMed] [Google Scholar]
- 36.Seo JH, Agarwal E, Bryant KG, Caino MC, Kim ET, Kossenkov AV, Tang HY, Languino LR, Gabrilovich DI, Cohen AR, et al. Syntaphilin Ubiquitination Regulates Mitochondrial Dynamics and Tumor Cell Movements. Cancer Res. 2018;78:4215–4228. PMID:29898993. doi: 10.1158/0008-5472.CAN-18-0595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang X, Liu Z, Zhang L, Yang Z, Chen X, Luo J, Zhou Z, Mei X, Yu X, Shao Z, et al. Targeting deubiquitinase USP28 for cancer therapy. Cell Death Dis. 2018;9:186 PMID:29415985. doi: 10.1038/s41419-017-0208-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee MH, Zhao R, Phan L, Yeung SC. Roles of COP9 signalosome in cancer. Cell Cycle. 2011;10:3057–3066. PMID:21876386. doi: 10.4161/cc.10.18.17320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fraile JM, Quesada V, Rodríguez D, Freije JM, López-Otín C. Deubiquitinases in cancer: new functions and therapeutic options. Oncogene. 2012;31:2373–2388. PMID:21996736. doi: 10.1038/onc.2011.443. [DOI] [PubMed] [Google Scholar]
- 40.Shemesh A, Kundu K, Peleg R, Yossef R, Kaplanov I, Ghosh S, Khrapunsky Y, Gershoni-Yahalom O, Rabinski T, Cerwenka A, et al. NKp44-Derived Peptide Binds Proliferating Cell Nuclear Antigen and Mediates Tumor Cell Death. Front Immunol. 2018;9:1114 PMID:29875773. doi: 10.3389/fimmu.2018.01114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen LY, Cheng CS, Qu C, Wang P, Chen H, Meng ZQ, Chen Z. Overexpression of CBX3 in Pancreatic Adenocarcinoma Promotes Cell Cycle Transition-Associated Tumor Progression. Int J Mol Sci. 2018;19 PMID:29903985. doi: 10.3390/ijms19061768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Borgquist S, Anagnostaki L, Jirström K, Landberg G, Manjer J. Breast tumours following combined hormone replacement therapy express favourable prognostic factors. Int J Cancer. 2007;120:2202–2207. PMID:17278089. doi: 10.1002/ijc.22542. [DOI] [PubMed] [Google Scholar]
- 43.Qin Z, Lu M, Xu X, Hanna M, Shiomi N, Xiao W. DNA-damage tolerance mediated by PCNA*Ub fusions in human cells is dependent on Rev1 but not Polη. Nucleic Acids Res. 2013;41:7356–7369. PMID:23761444. doi: 10.1093/nar/gkt542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kardosh A, Lichtensztajn DY, Gubens MA, Kunz PL, Fisher GA, Clarke CA. Long-Term Survivors of Pancreatic Cancer: A California Population-Based Study. Pancreas. 2018;47:958–966. PMID:30074526. doi: 10.1097/MPA.0000000000001133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ma W, Li T, Wu S, Li J, Wang X, Li H. LOX and ACSL5 as potential relapse markers for pancreatic cancer patients. Cancer Biol Ther. 2019;1–12. PMID:30712446. doi: 10.1080/15384047.2018.1564565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hu D, Ansari D, Zhou Q, Sasor A, Said HK, Andersson R. Galectin 4 is a biomarker for early recurrence and death after surgical resection for pancreatic ductal adenocarcinoma. Scand J Gastroenterol. 2019;1–6. PMID:30663442. doi: 10.1080/00365521.2018.1561937. [DOI] [PubMed] [Google Scholar]
- 47.Aoyama T, Yamamoto N, Kamiya M, Murakawa M, Tamagawa H, Sawazaki S, Numata M, Shiozawa M, Kobayashi S, Ueno M, et al. The Lymph Node Ratio Is an Independent Prognostic Factor in Pancreatic Cancer Patients Who Receive Curative Resection Followed by Adjuvant Chemotherapy. Anticancer Res. 2018;38:4877–4882. PMID:30061263. doi: 10.21873/anticanres.12801. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


