Abstract
Heterorhabditis bacteriophora is an entomopathogenic nematode (EPN) which infects its host by accessing the hemolymph where it releases endosymbiotic bacteria of the species Photorhabdus luminescens. We performed a genome-wide transcriptional analysis of the Drosophila response to EPN infection at the time point at which the nematodes reached the hemolymph either via the cuticle or the gut and the bacteria had started to multiply. Many of the most strongly induced genes have been implicated in immune responses in other infection models. Mapping of the complete set of differentially regulated genes showed the hallmarks of a wound response, but also identified a large fraction of EPN-specific transcripts. Several genes identified by transcriptome profiling or their homologues play protective roles during nematode infections. Genes that positively contribute to controlling nematobacterial infections encode: a homolog of thioester-containing complement protein 3, a basement membrane component (glutactin), a recognition protein (GNBP-like 3) and possibly several small peptides. Of note is that several of these genes have not previously been implicated in immune responses.
Key Words: Thioester-containing proteins, Basement membrane, Coagulation, Hemocyte, Insect immunity, Nematodes, Recognition molecule
Introduction
Entomopathogenic nematodes (EPNs) are natural pathogens used to control insect pests. In most cases, they associate with symbiotic bacteria, which they release once inside their insect host [1]. Both nematode-derived [2] and bacterial factors [3] interfere to varying extent with host immune reactions often resulting in the ultimate death of the insect [1]. During recent years, EPNs have increasingly been used to probe the insect immune system and identify genes and factors that have the potential to protect against EPN infections [4]. Some of this work was performed on target pest species, but also on well-established models for insect immunity [4, 5, 6, 7, 8]. Surprisingly, when Drosophila melanogaster larvae were infected with the combination of the EPN Heterorhabditisbacteriophora and the bacterium Photorhabdus luminescens, the canonical Toll and imd pathways, which are protective against bacterial and fungal infections, had little effect [5]. Instead, the Drosophila coagulation system reduced nematode infectivity and further work established that this involves a close collaboration between humoral- and cell-derived factors, which together act against EPN infections [6, 9]. In accordance with previous work on other hosts [10], eicosanoids also appear to contribute to this response in fly larvae [6].
In addition to the interest in EPNs as pest control agents and as tools to study insect innate immunity, the study of infectious nematodes has also implications for medical research. Filarial nematodes use insects as vectors to infect vertebrates, such as cattle and humans, sometimes causing debilitating and fatal diseases such as river blindness, and heart and lymphatic filariasis [4]. This has stimulated several systematic studies on the transcriptome changes that occur in vector insects after infection with filarial nematodes [11, 12, 13]. For example, the innate immune responses against the causative agents of lymphatic filariasis (caused by Brugia pahangi) were characterized in their insect host (Armigeres subalbatus), and several putative immune-related molecules were found to be induced [12]. A closely related species (B. malayi) to which Armigeres is resistant showed a distinctly different pattern of induction [12]. Ultimately, the hope behind these studies is to increase our understanding of these responses and identify protective factors [4]. This may permit blocking or at least delaying transmission of the nematodes.
One obstacle for the functional studies in vector hosts is the more restricted repertoire of molecular techniques available for their genetic manipulation. The goal of the work presented here is that the extended knowledge of Drosophila immunity will advance our general understanding of the responses against both EPNs and filarial nematodes. To this end, we performed a genome-wide analysis of the Drosophila transcriptome response after EPN infection. In addition, we tested a panel of mutant Drosophila lines in representative genes detected in our screen together with some other candidates and identified several promising gene products which slow down EPN infections in Drosophila larvae.
Materials and Methods
Nematode Culture
EPNs H. bacteriophora (strain H222 isolated from Pouzdrany, Czech Republic) were cultured in vivo on larvae of the greater wax moth Galleria mellonella at room temperature. Released infective juveniles (IJs) were collected and stored at room temperature in tap water with 0.075% formaldehyde. Small pieces of sponge were placed to the storage dishes to provide a solid medium for nematodes.
Nematode Infections
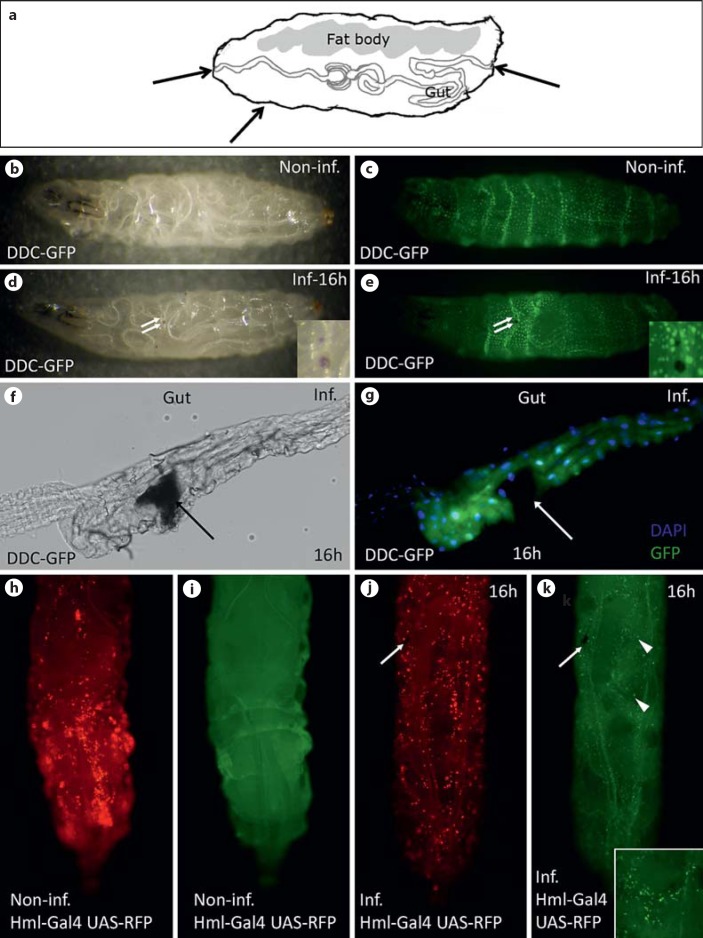
Nematode infections were performed according to the protocol described previously [14]. Here we further modified the microtiter plate infection assay to provide more consistent data and enable easy comparison of larger groups of genes and their knockdown in different tissues. For the infection assays, Drosophila eggs were collected 6 h after transferring flies to new food with addition of yeast; 60 h later, larvae were collected, rinsed briefly in 25°C tap water and placed individually in the wells of a microtiter plate. EPNs were used for infection 1 or 2 months after their release from G. mellonella cadavers to ensure optimal pathogenicity and were diluted to 25 IJs/larva. Ten microliters of the EPN suspension were added to each well; the plate was covered with Parafilm® and kept at 25°C at a 12-hour light/dark cycle. In all infection experiments, larval mortality caused by EPNs was scored 48 h after infection [14]. The mortality results of the screens were normalized to the respective control and the fold increase in mortality is shown. For microarray analysis and quantitative PCR verification, third instar larvae (88 h after egg laying) were infected with H. bacteriophora harboring green fluorescent protein (GFP)-expressing P. luminescens in microtiter plates (100 IJs/larva). After 2 h of infection, Drosophila larvae were rinsed briefly in water and transferred to vials with fly food. They were scored for a GFP signal after additional 6 h (confirmation of Photorhabdus septicemia) and used for RNA isolation. To improve visualization of EPN invasion into their hosts and the hemocyte response to the infection (fig. 1), we performed nematode infections in plastic bags [14]. Larvae of D. melanogaster expressing GFP under the control of DDC (dopa decarboxylase) promoter were infected with EPNs harboring the wild-type strain of P. luminescens. To simultaneously monitor the infection process and hemocyte recruitment, larvae expressing red fluorescent protein (RFP) in hemocytes were infected with H. bacteriophora harboring GFP-expressing P. luminescens. In both cases, the infection was performed using a high dose of EPNs (300 IJs/larva). The larvae were washed 2 h after infection and transferred to the new plastic bag without EPNs. Eight hours after infection, larvae were checked for the signs of infection (melanized wounds: fig. 1).
Fig. 1.
Infection routes of nematodes and hemocyte recruitment. a Infections can occur via the mouth, hindgut or by penetrating the cuticle of Drosophila larvae. Arrows indicate entry sites. b, c Noninfected DDC-GFP-expressing larva [visualized by stereomicroscopy (b) and fluorescence channel (c)]. d, e DDC-GFP larva infected by nematodes. Note that melanin spots (two of which are indicated by arrows and also shown at larger magnification) are visible accompanied by local DDC activation [stereomicroscopy (d) and fluorescence channel (e)]. f, g Upon entry via the mouth or hindgut, nematodes create wounds (arrow) in the gut; phase-contrast picture (f), fixed dissected gut stained with DAPI (g, the punctate signal) and the DDC-GFP signal (more diffuse signal in the gut epithelium, surrounding the wound site). h, i Sessile hemocytes (red) in hml-GAL4/UAS-RFP larva; noninfected larva shows hemocyte clusters (h) and after infection the clusters disperse (j). The punctate signal corresponds to hemocytes. i Noninfected larva seen in the fluorescence channel. To trace the infection the larva was infected with nematodes harboring GFP-expressing bacteria (green). Arrows indicate the wound (j, k) and arrowheads show bacteria (k). Inset shows bacteria at a higher magnification (all infected samples were analyzed 16 h after infection with nematodes). Inf./Non-inf. = Infected/noninfected.
Fly Strains
Fly strains were kept under standard conditions. All RNAi lines came from the Vienna Drosophila RNAi Center [15] and NIG-Fly Stock Center (see online suppl. table 1B for further details; see www.karger.com/doi/10.1159/000353734 for all online suppl. material). Two GD and one KK RNAi lines from the Vienna Drosophila RNAi Center were used for a single candidate gene primarily upon availability. Glutactin mutant GltEY22126, which contains insertion of P(EPgy2) transposable element within the second coding exon, was obtained from the Bloomington Stock Center. Bruno Lemaitre kindly provided the mutants GNBP-3hades[16], PGRP-SAseml[17], PGRP-LCE12[18], PGRP-LE112[19] and PGRP-LF200[20] (see online suppl. table 1B for further details). Infection of Drosophila larvae was compared with either wild-type w1118 or crosses between Gal4 drivers and w1118 (compare figure legends 4-6 for details). Gal4-expressing driver lines with specificity for either the fat body (ppl-Gal4) or hemocytes (he-Gal4 and hml-Gal4) were used. The DDC expression pattern or hemocyte localization was assessed in DDC-GFP and hml-GAL4 UAS-RFP flies, respectively (see online suppl. table 1B for details). The vkg-GFP (G00454) strain was obtained from the Flytrap collection. W; Idgf3/+; UAS-Idgf3/+ served as our wild-type stock for the microarray analysis. The data presented here are part of a larger study, which includes addressing the influence of Idgf3 on nematode infection. We have performed the controls for the heterozygous larvae and found no significant difference (see online suppl. fig. 4). In line with our previously published results [21], we observe that mortality rates are consistent among the wild-type strains used in this study, only Oregon R (not used otherwise in this study) shows slightly but not significantly (p = 0.102) increased mortality.
RNA Isolation
Total RNA from nematode-infected and naive third instar Drosophila larvae was extracted using RiboZol RNA extraction reagent (Amresco) according to the manufacturer's protocol and subsequently cleaned with NucleoSpin RNA II kit (Macherey-Nagel). Quality and concentration of the RNA were measured with a NanoDrop 2000 spectrophotometer (Thermo Scientific). RNA integrity was analyzed in an Agilent 2100 Bioanalyzer. We included only samples with an intact RNA profile.
Expression Profiling
The Affymetrix GeneChip® Drosophila Genome 2.0 Array System was used for microarray analysis following the standard protocol [100 ng RNA was amplified with GeneChip 3′ IVT Express Kit (Affymetrix) and 10 μg of labeled cRNA was hybridized to the chip according to the manufacturer's instructions].
Statistical Analysis of Array Data
Analysis was performed in three replicates. Data were preprocessed in Partek Genomic Suit (Partek). In short, the transcription profiles were background corrected using the GCRMA method, quantile normalized and variance stabilized using base-2 logarithmic transformation. Analysis of variance yielded transcripts differentially expressed between analyzed samples (within LIMMA [22]); Storey's q values [23] were used to select significantly differentially transcribed genes, q < 0.05. The transcription data are MIAME compliant and deposited in the ArrayExpress database (accession E-MTAB-1542).
All statistical analyses were performed in R (http://www.R-project.org) and within Bioconductor [24]. Differentially expressed genes were selected for gene set enrichment analysis (GSEA). We performed GSEA on genes that mapped to KEGG pathways [25] and have defined gene ontology (GO) terms [26] using the Fisher test and approach of Tian et al. [27]. For GSEA, pathways with q < 0.05 and |log FC| ≥0.4 (where FC = fold change) after hypergeometric testing were considered differentially expressed (online suppl. table 4) but pathways at q < 0.05 and |log FC| ≥0.4 are also shown (online suppl. table 4, right-most column part).
Quantitative PCR
Total RNA from infected and noninfected Drosophila larvae was isolated using the RiboZol RNA extraction reagent (Amresco). The RNA was further purified by NucleoSpin RNA II kit (Macherey-Nagel) including an on-column digestion step with rDNase I. Total RNA (1,000 ng) was applied for reverse transcription using PrimeScript Reverse Transcriptase (Takara) and oligo(dT)(17-mer). Quantitative PCR was performed using the HOT FIREPol EvaGreen qPCR Mix Plus (Solis BioDyne, Tartu, Estonia). The PCR reaction volume was 20 µl, containing 5 µl of diluted cDNA and 250 nM primers. The amplification was carried out in an Eco real-time PCR system (Illumina) for 45 cycles (95°C for 15 s; annealing temperature dependent on primer pair for 30 s; 72°C for 20 s) following an initial denaturation/Pol activation step (95°C for 15 min). Each sample was analyzed in triplicate. Primers (online suppl. table 1) were designed with Lasergene PrimerSelect Software (DNASTAR) to assure that each amplicon was specific. Melting analysis confirmed a single product for each primer pair reaction. The product size was verified by gel electrophoresis. Data were analyzed and quantified with the Illumina EcoStudy software. Relative mRNA levels were normalized to Rack1 and rp49 expression and standardized to the noninfected sample. The results are presented as the mean log2-transformed fold changes (infected/noninfected larvae) in transcript levels ±SEM of 2 independent biological replicates.
Microscopy
Leica MZ FLIII fluorescence stereomicroscope coupled to a Panasonic DMC-G2 camera was used to visualize wounds, DDC-GFP localization and hemocyte recruitment. Images of dissected guts and tissues stained with DAPI were taken with a Hamamatsu ORCA-ER camera (C4742-95) attached to a Zeiss Axioplan 2 microscope. Confocal images were taken in a Zeiss LSM 510 Meta microscope.
Statistical Evaluation of Infection Experiments
All experiments were run at least in triplicate using 48 Drosophila larvae per replicate. Each experiment was repeated independently at least twice. The results are expressed as the mean ± SD. Mortality rates were compared to the respective control cross between driver lines and w1118, which was always included to account for differences between different sets of experiments. The level of significance for the individual comparisons was determined using Student's t test (unpaired, two-sided).
Results
EPN Infections in Drosophila Occur via Two Entry Sites
To follow the infection of Drosophila larvae after incubation with EPNs, we used a combination of H. bacteriophora and its associated bacterium P. luminescens expressing GFP [5]. To assess the infection status of the larvae, we relied on a recently developed method that allows continuous monitoring based on the detection of GFP-expressing Photorhabdus [5, 14]. The entry of the nematodes into the hemocoel of Drosophila larvae causes wounds, which melanize (fig. 1d-e) and the release of bacterial symbionts subsequently leads to bacteremia (fig. 1k). Similar to other EPNs, Heterorhabditis was found to enter larvae both via the cuticle and the underlying epidermis and via the gut epithelium (fig. 1; online suppl. fig. 1A, B). When using a GFP-expressing Drosophila strain under the control of the DDC promoter, induction of the reporter in cells in the wider area surrounding the wound was observed in both locations (fig. 1e, g; online suppl. fig. 1C for comparison). Similar to other infection models that involve cellular immunity [28], hemocytes are recruited from their sessile compartments into the circulation (fig. 1h-k).
Transcriptome Changes upon EPN Infection
To obtain a complete picture of the larval immune response against EPNs, we performed a genome-wide transcriptional analysis comparing infected and noninfected animals at the same stage, feeding third instar larvae (approx. 96 h after egg laying). Since the duration of the infection and the multiple attempts of nematodes to infect larvae (fig. 1d, e) precluded an exact determination of the time point of infection, the infected samples were chosen such that the GFP signal due to the release of bacteria from the nematode intestine had just passed the detection threshold 6 h after exposure to EPNs. In this way, we ensured that the samples had reached the same stage of the infection and the sampled transcriptome represents the early stage of a developing bacteremia.
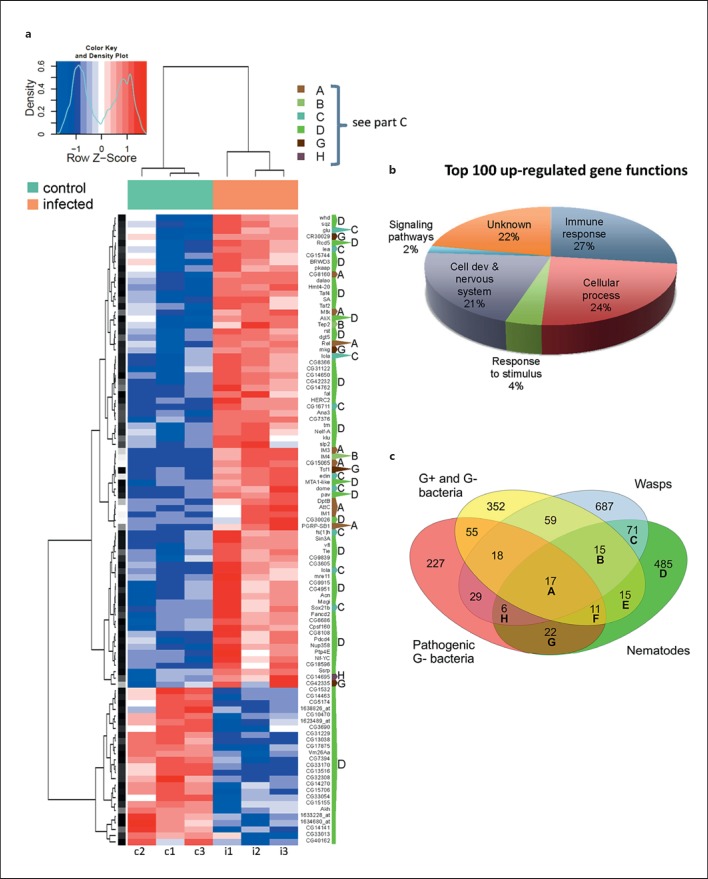
Samples were collected in triplicate and RNA was hybridized to Affymetrix GeneChip Drosophila Genome 2.0 arrays. After statistical analysis and setting the statistical threshold to a value of q < 0.05 and the threshold of induction at 2, a total of 642 transcripts were found differentially regulated upon nematode infection. A complete heatmap of the 100 most strongly regulated transcripts shows that the replicates from the infected and noninfected samples are well separated and that the majority of regulated genes are induced (fig. 2a). Exploratory analysis using principal component analysis confirms the similarity between the triplicates of induced and noninduced samples (fig. 2a and data not shown). The list of all transcripts induced upon infection with Heterorhabditis/Photorhabdus was compared to earlier studies, which included bacterial (both nonpathogenic and pathogenic)- and wasp-infected larvae and revealed a core set of 17 genes induced under all four conditions (fig. 2c, category A; online suppl. table 3). Additional transcripts were shared between EPN-infected larvae and the three other conditions (fig. 2a, c, additional categories B, C, E-H). The transcripts that are shared with the array results from earlier studies include the majority of the transcripts in the highly regulated fraction (fig. 2; online suppl. table 3). The majority of EPN-regulated transcripts (485) are not differentially regulated under the other conditions [fig. 2c, category D; online suppl. table 3 (complete survey of induced genes)].
Fig. 2.
The significantly regulated transcripts after nematobacterial infection are enriched for immune genes. a A heatmap representing the 100 most strongly regulated transcripts from the microarray (columns c1-c3 = Control: noninfected; i1-i3 = infected larvae). Each column represents an independent sample. Color key and density plot represent the level of regulation. Dark intensities indicate the most up- and down-regulated genes, respectively. The one-letter code to the right of the heatmap indicates whether the gene was previously detected in other genome-wide analysis of Drosophila larval immune response (category A-C, G, H) or is specifically regulated upon nematode infection (D); for a description of the categories compare the Venn diagram in c. b GO classification of the 100 most strongly up-regulated genes. Immune response molecules occupy a fourth of the top 100 genes (see also online suppl. table 2). c Venn diagram showing differentially regulated transcripts after infection with common Gram-negative (G-) and -positive (G+) bacteria [32, 34], pathogenic G- bacterial wasps [33, 35, 49] and nematodes (this work) in Drosophila larvae. 381 and 104 transcripts are specifically up- or down-regulated after nematobacterial infection (comprising altogether 485 differentially regulated transcripts in category D).
GO analysis using AmiGO (http://amigo.geneontology.org) of the top 100 list, which included the most strongly positively and negatively regulated genes, shows a highly significant enrichment for immune-related genes: antimicrobial peptides, immune-induced genes (IMs and Edin), a thioester-containing protein (TEP) 2 and Relish, the key transcriptional activator of the imd pathway (fig. 2b; online suppl. table 2). This is particularly true when GO analysis was performed on those genes that had previously been found to be induced in bacterial infection models (table 1, categories A, B, F). Nematode- and wasp-specific genes mapped to additional pathways, most of which are related to developmental processes. When all differentially regulated transcripts were mapped to the KEGG database, several additional developmental pathways were identified. Among those, the fly equivalent of vertebrate oocyte maturation, Wnt signaling, and ubiquitin-mediated proteolysis were the most significantly enriched (online suppl. table 4). Other pathways were found enriched with lower significance, although some of these are connected to the Wnt pathway (online suppl. fig. 2).
Table 1.
Gene ontology analysis of genes regulated by nematobacterial infection
| Region name | Gene No. | Category | Biological process |
|---|---|---|---|
| A | 17 | all | GO:0019731 antibacterial humoral response GO:0006952 defense response GO:0042742 defense response to bacterium |
| B | 15 | nematode + wasps + G+ and G– bacteria | GO:0006952 defense response GO:0050829 defense response to Gram– bacterium GO:0042742 defense response to bacterium |
| C | 71 | nematodes + wasps | GO:0048731 system development GO:0007275 multicellular organismal development GO:0009653 anatomical structure morphogenesis |
| D | 485 | nematode | GO:0030154 cell differentiation GO:0048699 generation of neurons GO:0048869 cellular developmental process |
| E | 15 | nematodes + G+ and G– bacteria | no significant hit |
| F | 11 | nematodes + G+ and G– bacteria + pathogenic G– bacteria | GO:0019731 antibacterial humoral response GO:0019731 antibacterial humoral response |
| G | 22 | nematodes + pathogenic G– bacteria | no significant hit |
| H | 6 | nematodes + wasps + pathogenic G– bacteria | no significant hit |
Significantly enriched pathways after GO analysis using AmiGO. The top list of enriched GO terms is shown for all the nematode-regulated categories shown in the Venn diagram in figure 2c. G+/G– = Gram-positive/Gram-negative.
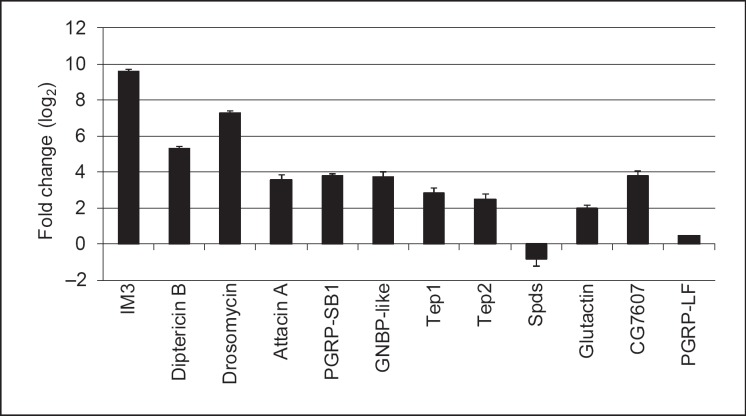
To confirm the data obtained by microarrays, we used real-time RT-PCR on genes selected from representatives of all categories in figure 2c covering different induction levels including repression. This analysis largely confirms the results from the array study, however one of the genes (PGRP-LF) displayed a lower fold change |log FC <1| (fig. 3, online suppl. table 6).
Fig. 3.
Validation of microarray data using quantitative RT-PCR. Selected genes from the different categories of genes regulated by nematobacterial infection were analyzed by quantitative PCR using RNA samples from naive and infected larvae. The relative expression level (ratio infected/noninfected larvae) is shown as log2 mean from 2 independent experiments ± SEM.
In summary, the genome-wide transcriptional analysis identifies both immune-related genes as well as members of other pathways. In general, immune genes appear most strongly regulated. The identification of the additional pathways requires further analysis, but a likely explanation for their induction is a requirement during closure and healing of the wounds afflicted by the nematodes.
Functional Tests of Candidate Genes in Response to Nematodes
To test their contribution to the response to nematodes, we used RNAi lines and mutants for candidate genes identified in the arrays and examined the viability of such larvae upon EPN infection. As a criterion used for selecting candidate defense genes from the list of induced transcripts, we focused on those for which a function in immunity had been established in other infection models [29] as well as on their paralogs, which had not yet been implicated in immune reactions.
Functional Tests Using RNAi Lines
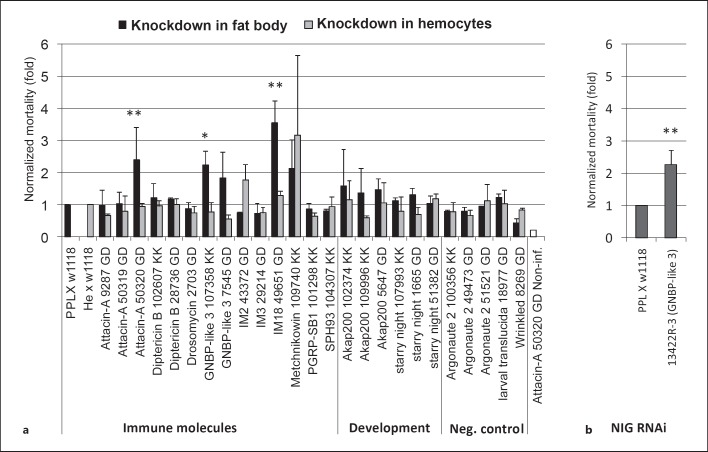
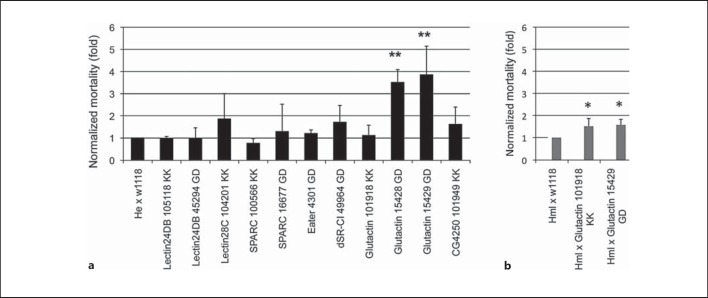
Since the transcriptional response against nematodes and their bacteria shows a strong immune signature, we decided to score mortality rates after knocking down candidate genes in the fat body, the key Drosophila immune organ. Having observed that upon EPN infection, hemocytes are recruited into the circulation (fig. 1j, k), we supplemented this approach using a second, hemocyte-specific driver (fig. 4a). Using a selection of significantly induced immune and developmental genes identified in our array, we failed to find any effects when the genes were knocked down in hemocytes. In contrast, when using gene silencing in the fat body, significant effects were observed for Gram-negative binding protein-like protein (GNBP-like 3), attacin A (one line), and a third one for an immune-induced peptide (IM18). For the GNPB-like 3 gene, a similar trend was found for a second line while two additional attacin A lines failed to reproduce the phenotype. Increased mortality was also seen for an independent GNBP-like 3 line from a different collection, providing additional confirmation of our results (fig. 4b). For IM18, only one line was available. These results indicate that the candidate genes obtained by transcriptional profiling, and in particular GNBP-like 3, likely play a role in the immunity against EPNs, although the data from RNAi studies require further verification.
Fig. 4.
Increased Drosophila mortality upon nematobacterial infection after knocking down the expression of genes selected from the array data. a Selected RNAi lines (see online suppl. table 1B for further details) were crossed with a fat body (ppl-GAL4) or hemocyte-specific driver (he-GAL4) and infected with H. bacteriophora at a dose of 25 IJs/larva at 25°C. Mortality was scored after 48 h and is presented after normalization to the respective control crosses, which were set to 1. Attacin A-noninfected (Non-inf.) is a parallel handling and developmental control. b The increase in mortality observed for GNBP-like 3 was confirmed using an independent RNAi line from the NIG-Fly collection. Data presented are means ± SD; t test: * p < 0.05; ** p < 0.01, only for lines with significantly increased mortality.
Functional Tests of Mutants
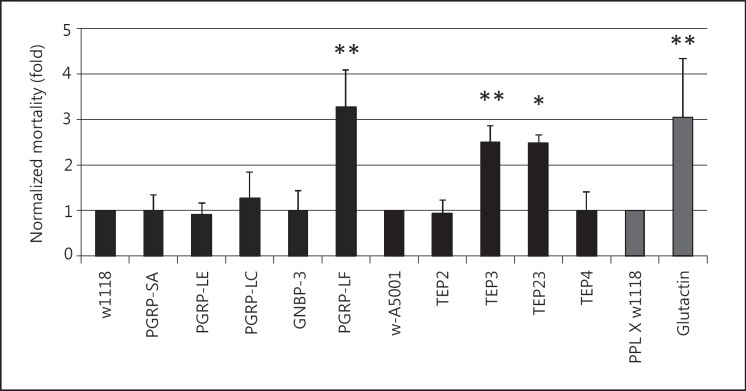
Based on the microarray results, we extended our assay to an additional set of key immune molecules, for which mutant fly strains were available (fig. 5), including members of well-characterized classes of pattern recognition receptors, such as GNBPs and peptidoglycan recognition proteins (PGRPs), as well as some members of Drosophila TEPs, three of which had also been found changed after EPN infection (see online suppl. table 3) and the basement membrane protein glutactin. Amongst the recognition proteins, a significant influence on EPN infections was observed for PGRP-LF and for TEP3. The most significant difference was obtained with TEP3 mutants either alone or in combination with TEP2, both of which had previously been tested in in vivo infections of Drosophila without any detectable effects [30]. Finally, we observed increased mortality of glutactin mutants, supporting a protective function of the basement membrane. This result is consistent with the idea that upon entry of EPNs into Drosophila larvae, the basement membrane has a protective function since it is encountered by nematodes crossing either gut or cuticular epidermis.
Fig. 5.
Candidate screen for Drosophila genes involved in defense against nematobacterial infection using available mutants. Mutants for recognition molecules (see online suppl. table 1B for further details) and the basement membrane component glutactin were infected with H. bacteriophora at a dose of 25 IJs/larva at 29°C. Mortality was scored as in figure 4. * p < 0.05; ** p < 0.01.
Glutactin Is Required in Hemocytes
Since hemocytes are known to contribute to the formation of the basement membrane [31], we speculated that interfering with glutactin production specifically in these cells might also have an impact on EPN infection. This idea is further supported by the observation that glutactin shows enriched expression in hemocytes compared to whole larvae [32]. As controls, we also knocked down expression of several other genes with enriched expression in hemocytes, including two C-type lectins, SPARC and two phagocytic receptors (see online suppl. table 5 for a list of the genes and the ratio of hemocyte enrichment). Indeed, while none of the control knockdowns led to increased susceptibility, two out of three glutactin RNAi lines crossed with the first hemocyte-specific driver (Hemese) showed an increase in mortality after EPN infection, while the third available line failed to do so. When this line was crossed with a different hemocyte GAL4 line (Hemolectin), it turned out that it presented higher susceptibility, too (fig. 6b). This means that the expression of glutactin in hemocytes contributes to protection against EPNs. To assess the damage afflicted to the basement membrane during nematode infection, we infected larvae that express a GFP-tagged version of collagen IV (in Drosophila also called Viking), one of the major components of the basement membrane. While Viking normally forms a fibrous network surrounding the gut, transmigration of nematodes creates wounds that melanize as visible in figure 1 and lack Viking in an area that extends beyond melanization (online suppl. fig. 3). This underlines the need for creating a replacement for the basement membrane.
Fig. 6.
Screen for genes involved in the defense against nematobacterial infection using candidate genes with hemocyte-enriched transcription. a RNAi lines for selected hemocyte-enriched genes (see online suppl. table 1B and text for further details) were crossed with a hemocyte-specific driver (he-Gal4) and infected with H. bacteriophora at a dose of 25 IJs/ larva at 29°C. The effects for glutactin knockdown were confirmed with an early hemocyte driver (hml-Gal4, b). Data presented are means ± SD; t test: * p < 0.05; ** p < 0.01. Mortality was scored as in figure 4.
Discussion
Here we provide a genome-wide characterization of the transcriptional changes that occur upon infection of Drosophila larvae with the EPN H. bacteriophora and its associated bacteria P. luminescens. Our initial characterization of the infection process shows that the gut epithelium, the cuticle and the underlying epidermis are targeted, leading to activation of a wound DDC-GFP reporter construct (fig. 1). Our microarray analysis therefore covers all major events that occur as a consequence of the entry of the nematode and the influence of both the nematode and its bacteria on the host immune system.
A highly significant enrichment for immune genes obtained in the set of most strongly induced genes (fig. 2) supports the notion that an immune response against Photorhabdus has been initiated at the time when the GFP signal indicating the presence of bacteria appeared in the hemolymph. In addition to immune genes shared with earlier reports on septic injury due to bacteria or wasp infestation [32, 33, 34, 35], some genes with proposed immune functions are specifically induced in the presence of EPNs (fig. 2c). When genes with weaker levels of induction are included and mapped to known pathways, three pathways turn up as most significantly enriched: an oocyte maturation pathway, Wnt signaling and ubiquitin-mediated proteolysis. Additional pathways are identified at lower significance levels. It is worth noting that some of these pathways are connected, such as the ubiquitin and mitogen-activated protein kinase pathways, both of which are linked to Wnt signaling (online suppl. fig. 2). In addition, upstream of mitogen-activated protein kinases, the receptor tyrosine kinase stitcher has been shown to be involved in wound healing in the embryo [36] and may play a similar role in larvae.
One possible explanation for the activation of Drosophila equivalent to the mammalian oocyte maturation pathway might be that this pathway includes mainly a number of general cell cycle regulators and such proteins are also induced in a proliferative response of various somatic tissues upon nematode wounding (online suppl. fig. 2). These include Polo-like kinases, which are key regulators of cell division, the mitotic cyclin B and protein kinase A. An effect of wounding is also supported by the fact that genes in Wnt and hedgehog signaling, as well as components of the extracellular matrix, are enriched in our gene set. All three pathways have been shown to be important during wound healing acting at different stages in vertebrates and may play comparable roles in flies [37, 38]. For example, the Wnt pathway may support the establishment of planar cell polarity during epidermal wound healing similar to its role during development and the extracellular matrix may play a role similar to the granulation tissue that forms after wounding in vertebrates [39]. Hemocytes may be crucial during this process by contributing collagen IV [40] and glutactin (this work) in a localized fashion. Ultimately, wound healing converges on the activation of small ρ GTPases, which have a well-defined role during wound healing in Drosophila [41, 42]. However, this occurs mostly at the posttranslational level and is less amenable to transcriptome studies.
For a preliminary functional analysis of the genes involved in the nematobacterial response, we focused on several of the induced genes from the array including the basement membrane component glutactin and a set of known recognition molecules (fig. 5). The data confirm earlier observations from others and our group that the imd and Toll pathways that are required for induction of immune genes are dispensable for the response against EPNs, since mutants in the receptors required for their activation do not show increased mortality [5, 21]. We attribute the effects we observed with PGRP-LF, which is a negative regulator of the imd pathway, to the previously observed developmental defects [20]. Alternatively, PGRP-LF, which acts as a negative regulator of imd signaling [20, 43], may aid to combat nematodes indirectly by positively regulating other effector pathways. Most interestingly, mutants in one of the TEP3 survive EPN infections significantly less than controls and other TEP mutants. Of note, the same mutants had previously failed to show any developmental or immune phenotype [30]. A trend to increased susceptibility was also found for the recognition molecule GNBP-like 3.
We also tried to interfere with the induction of some of the highly regulated immune genes identified in the array by their specific knockdown in both hemocytes and the fat body. Since robust knockdowns are difficult to achieve in this setting, it is not too surprising that these data are more variable than our previous findings. Nevertheless, two RNAi lines with specificity for small immune peptides (attacin A and IM18) did also show an effect. Although these data require verification, for example due to possible off-target effects (online suppl. table 1B), they seem to contradict the concept that the imd pathway is not required during the anti-EPN response. However, they are consistent with the possible attacin function implicated from its differential induction in a different worm infection model involving a tapeworm and the flour beetle as a host [44]. Possible explanations for this discrepancy may be a JNK-dependent induction and/or a developmentally regulated constitutive expression of this gene. Recent data regarding infections of adult flies with EPNs suggest that AMPs can be induced by bacteria-free (axenic) nematodes, most likely also as a part of wounding defense [8]. Notably, the mammalian AMP LL-37 contributes to wound healing [45] as part of its pleiotropic effects [46].
Finally, consistent with our notion that one of the pathways we identified in the GSEA was the synthesis of extracellular matrix, we found that the ECM protein glutactin protects against EPN infections. This extends our previous findings that clot components reduce EPN mortality. We propose that the basement membrane components cooperate with the clot matrix to seal the wounds afflicted by nematodes. This potentially occurs by linking the clot to the wound edges and/or sealing it off on the hemolymph side, such a process is akin to the formation of the granulation tissue during wound healing in vertebrates [39]. Similarly, the recent observation that the complement system interacts with the clotting cascade is consistent with our observation that both the clotting and the C3 homolog TEP3 act against nematobacterial infections [47]. Taken together, both the list of genes we found to be regulated by EPN infection and the genes we have tested functionally show specific signatures, which are not shared with other infection models. Both sets of genes provide a rich source for further testing in different settings such as septic and aseptic wounding and infection with bacteria that are pathogenic to Drosophila such as Pseudomonas entomophila and Serratia marcescens [34, 48].
Supplementary Material
Supplementary data
Supplementary data
Supplementary data
Acknowledgments
The authors' work is supported by the Swedish Research Council (U.T.), STINT (Swedish Foundation for International Cooperation in Research and Higher Education), the Knut and Alice Wallenberg Foundation (U.T.), the Swedish Cancer Foundation (U.T.), the European Social Fund and the state budget of the Czech Republic (P.H. and P.D.) and the European Union Seventh Framework Program (FP7/2007-2013) under grant agreement No. 316304 (M.Z.). We thank Dominique Ferrandon for providing the TEP mutant lines, Zdenek Mracek for the wild-type strain of H. bacteriophora and Todd Ciche for GFP-expressing P. luminescens. Authors are grateful to the service laboratory at the Institute of Molecular Genetics and especially to Martina Chmelikova for technical assistance.
References
- 1.Eleftherianos I, ffrench-Constant RH, Clarke DJ, Dowling AJ, Reynolds SE. Dissecting the immune response to the entomopathogen Photorhabdus. Trends Microbiol. 2010;18:552–560. doi: 10.1016/j.tim.2010.09.006. [DOI] [PubMed] [Google Scholar]
- 2.Balasubramanian N, Nascimento G, Ferreira R, Martinez M, Simoes N. Pepsin-like aspartic protease (Sc-ASP155) cloning, molecular characterization and gene expression analysis in developmental stages of nematode Steinernema carpocapsae. Gene. 2012;500:164–171. doi: 10.1016/j.gene.2012.03.062. [DOI] [PubMed] [Google Scholar]
- 3.Lang AE, Schmidt G, Schlosser A, Hey TD, Larrinua IM, Sheets JJ, Mannherz HG, Aktories K. Photorhabdus luminescens toxins ADP-ribosylate actin and RhoA to force actin clustering. Science. 2010;327:1139–1142. doi: 10.1126/science.1184557. [DOI] [PubMed] [Google Scholar]
- 4.Castillo JC, Reynolds SE, Eleftherianos I. Insect immune responses to nematode parasites. Trends Parasitol. 2011;27:537–547. doi: 10.1016/j.pt.2011.09.001. [DOI] [PubMed] [Google Scholar]
- 5.Hallem EA, Rengarajan M, Ciche TA, Sternberg PW. Nematodes, bacteria, and flies: a tripartite model for nematode parasitism. Curr Biol. 2007;17:898–904. doi: 10.1016/j.cub.2007.04.027. [DOI] [PubMed] [Google Scholar]
- 6.Hyrsl P, Dobes P, Wang Z, Hauling T, Wilhelmsson C, Theopold U. Clotting factors and eicosanoids protect against nematode infections. J Innate Immun. 2011;3:65–70. doi: 10.1159/000320634. [DOI] [PubMed] [Google Scholar]
- 7.Castillo JC, Shokal U, Eleftherianos I. A novel method for infecting Drosophila adult flies with insect pathogenic nematodes. Virulence. 2012;3:339–347. doi: 10.4161/viru.20244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Castillo JC, Shokal U, Eleftherianos I. Immune gene transcription in Drosophila adult flies infected by entomopathogenic nematodes and their mutualistic bacteria. J Insect Physiol. 2013;59:179–185. doi: 10.1016/j.jinsphys.2012.08.003. [DOI] [PubMed] [Google Scholar]
- 9.Wang R, Liang Z, Hal M, Söderhäll K. A transglutaminase involved in the coagulation system of the freshwater crayfish, Pacifastacus leniusculus. Tissue localisation and cDNA cloning. Fish Shellfish Immunol. 2001;11:623–637. doi: 10.1006/fsim.2001.0341. [DOI] [PubMed] [Google Scholar]
- 10.Kim Y, Ji D, Cho S, Park Y. Two groups of entomopathogenic bacteria, Photorhabdus and Xenorhabdus, share an inhibitory action against phospholipase A2 to induce host immunodepression. J Invertebr Pathol. 2005;89:258–264. doi: 10.1016/j.jip.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 11.Aliota MT, Fuchs JF, Mayhew GF, Chen CC, Christensen BM. Mosquito transcriptome changes and filarial worm resistance in Armigeres subalbatus. BMC Genomics. 2007;8:463. doi: 10.1186/1471-2164-8-463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aliota MT, Fuchs JF, Rocheleau TA, Clark AK, Hillyer JF, Chen CC, Christensen BM. Mosquito transcriptome profiles and filarial worm susceptibility in Armigeres subalbatus. PLoS Negl Trop Dis. 2010;4:e666. doi: 10.1371/journal.pntd.0000666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Erickson SM, Xi Z, Mayhew GF, Ramirez JL, Aliota MT, Christensen BM, Dimopoulos G. Mosquito infection responses to developing filarial worms. PLoS Negl Trop Dis. 2009;3:e529. doi: 10.1371/journal.pntd.0000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dobes P, Wang Z, Markus R, Theopold U, Hyrsl P. An improved method for nematode infection assays in Drosophila larvae. Fly (Austin) 2012;6:75–79. doi: 10.4161/fly.19553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dietzl G, Chen D, Schnorrer F, Su KC, Barinova Y, Fellner M, Gasser B, Kinsey K, Oppel S, Scheiblauer S, Couto A, Marra V, Keleman K, Dickson BJ. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 2007;448:151–156. doi: 10.1038/nature05954. [DOI] [PubMed] [Google Scholar]
- 16.Gottar M, Gobert V, Matskevich AA, Reichhart JM, Wang C, Butt TM, Belvin M, Hoffmann JA, Ferrandon D. Dual detection of fungal infections in Drosophila via recognition of glucans and sensing of virulence factors. Cell. 2006;127:1425–1437. doi: 10.1016/j.cell.2006.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Michel T, Reichhart JM, Hoffmann JA, Royet J. Drosophila Toll is activated by Gram-positive bacteria through a circulating peptidoglycan recognition protein. Nature. 2001;414:756–759. doi: 10.1038/414756a. [DOI] [PubMed] [Google Scholar]
- 18.Gottar M, Gobert V, Michel T, Belvin M, Duyk G, Hoffmann JA, Ferrandon D, Royet J. The Drosophila immune response against Gram-negative bacteria is mediated by a peptidoglycan recognition protein. Nature. 2002;416:640–644. doi: 10.1038/nature734. [DOI] [PubMed] [Google Scholar]
- 19.Takehana A, Yano T, Mita S, Kotani A, Oshima Y, Kurata S. Peptidoglycan recognition protein (PRGP)-LE and PGRP-LC act synergistically in Drosophila immunity. EMBO J. 2004;23:4690–4700. doi: 10.1038/sj.emboj.7600466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maillet F, Bischoff V, Vignal C, Hoffmann J, Royet J. The Drosophila peptidoglycan recognition protein PGRP-LF blocks PFRP-LC and IMD/JNK pathway activation. Cell Host Microbe. 2008;3:293–303. doi: 10.1016/j.chom.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 21.Wang Z, Wilhelmsson C, Hyrsl P, Loof TG, Dobes P, Klupp M, Loseva O, Mörgelin M, Ikle J, Cripps RM, Herwald H, Theopold U. Pathogen entrapment by transglutaminase - a conserved early innate immune mechanism. PLoS Pathog. 2010;6:e1000763. doi: 10.1371/journal.ppat.1000763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3:article3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 23.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci USA. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini AJ, Sawitzki G, Smith C, Smyth G, Tierney L, Yang JY, Zhang J. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kanehisa M, Goto S. Kegg: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology. The gene ontology consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tian L, Greenberg SA, Kong SW, Altschuler J, Kohane IS, Park PJ. Discovering statistically significant pathways in expression profiling studies. Proc Natl Acad Sci USA. 2005;102:13544–13549. doi: 10.1073/pnas.0506577102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zettervall CJ, Anderl I, Williams MJ, Palmer R, Kurucz E, Ando I, Hultmark D. A directed screen for genes involved in Drosophila blood cell activation. Proc Natl Acad Sci USA. 2004;101:14192–14197. doi: 10.1073/pnas.0403789101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lemaitre B, Hoffmann J. The host defense of Drosophila melanogaster. Annu Rev Immunol. 2007;25:697–743. doi: 10.1146/annurev.immunol.25.022106.141615. [DOI] [PubMed] [Google Scholar]
- 30.Bou Aoun R, Hetru C, Troxler L, Doucet D, Ferrandon D, Matt N. Analysis of thioester-containing proteins during the innate immune response of Drosophila melanogaster. J Innate Immun. 2011;3:52–64. doi: 10.1159/000321554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lavine MD, Strand MR. Insect hemocytes and their role in immunity. Insect Biochem Mol Biol. 2002;32:1295–1309. doi: 10.1016/s0965-1748(02)00092-9. [DOI] [PubMed] [Google Scholar]
- 32.Irving P, Ubeda JM, Doucet D, Troxler L, Lagueux M, Zachary D, Hoffmann JA, Hetru C, Meister M. New insights into Drosophila larval haemocyte functions through genome-wide analysis. Cell Microbiol. 2005;7:335–350. doi: 10.1111/j.1462-5822.2004.00462.x. [DOI] [PubMed] [Google Scholar]
- 33.Wertheim B, Kraaijeveld AR, Schuster E, Blanc E, Hopkins M, Pletcher SD, Strand MR, Partridge L, Godfray HC. Genome-wide gene expression in response to parasitoid attack in Drosophila. Genome Biol. 2005;6:R94. doi: 10.1186/gb-2005-6-11-r94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vodovar N, Vinals M, Liehl P, Basset A, Degrouard J, Spellman P, Boccard F, Lemaitre B. Drosophila host defense after oral infection by an entomopathogenic Pseudomonas species. Proc Natl Acad Sci USA. 2005;102:11414–11419. doi: 10.1073/pnas.0502240102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schlenke TA, Morales J, Govind S, Clark AG. Contrasting infection strategies in generalist and specialist wasp parasitoids of Drosophila melanogaster. PLoS Pathog. 2007;3:1486–1501. doi: 10.1371/journal.ppat.0030158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang S, Tsarouhas V, Xylourgidis N, Sabri N, Tiklova K, Nautiyal N, Gallio M, Samakovlis C. The tyrosine kinase stitcher activates grainy head and epidermal wound healing in Drosophila. Nat Cell Biol. 2009;11:890–895. doi: 10.1038/ncb1898. [DOI] [PubMed] [Google Scholar]
- 37.Razzell W, Wood W, Martin P. Swatting flies: modelling wound healing and inflammation in Drosophila. Dis Model Mech. 2011;4:569–574. doi: 10.1242/dmm.006825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shaw TJ, Martin P. Wound repair at a glance. J Cell Sci. 2009;122:3209–3213. doi: 10.1242/jcs.031187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sonnemann KJ, Bement WM. Wound repair: toward understanding and integration of single-cell and multicellular wound responses. Annu Rev Cell Dev Biol. 2011;27:237–263. doi: 10.1146/annurev-cellbio-092910-154251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Adachi T, Tomita M, Yoshizato K. Synthesis of prolyl 4-hydroxylase alpha subunit and type IV collagen in hemocytic granular cells of silkworm, Bombyx mori: Involvement of type IV collagen in self-defense reaction and metamorphosis. Matrix Biol. 2005;24:136–154. doi: 10.1016/j.matbio.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 41.Stramer B, Wood W, Galko MJ, Redd MJ, Jacinto A, Parkhurst SM, Martin P. Live imaging of wound inflammation in Drosophila embryos reveals key roles for small GTPases during in vivo cell migration. J Cell Biol. 2005;168:567–573. doi: 10.1083/jcb.200405120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lesch C, Jo J, Wu Y, Fish GS, Galko MJ. A targeted UAS-RNAi screen in Drosophila larvae identifies wound closure genes regulating distinct cellular processes. Genetics. 2010;186:943–957. doi: 10.1534/genetics.110.121822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Basbous N, Coste F, Leone P, Vincentelli R, Royet J, Kellenberger C, Roussel A. The Drosophila peptidoglycan-recognition protein LF interacts with peptidoglycan-recognition protein LC to downregulate the Imd pathway. EMBO Rep. 2011;12:327–333. doi: 10.1038/embor.2011.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhong D, Wang MH, Pai A, Yan G. Transcription profiling of immune genes during parasite infection in susceptible and resistant strains of the flour beetles (Tribolium castaneum) Exp Parasitol. 2013;134:61–67. doi: 10.1016/j.exppara.2013.01.014. [DOI] [PubMed] [Google Scholar]
- 45.Ramos R, Silva JP, Rodrigues AC, Costa R, Guardao L, Schmitt F, Soares R, Vilanova M, Domingues L, Gama M. Wound healing activity of the human antimicrobial peptide LL37. Peptides. 2011;32:1469–1476. doi: 10.1016/j.peptides.2011.06.005. [DOI] [PubMed] [Google Scholar]
- 46.Vandamme D, Landuyt B, Luyten W, Schoofs L. A comprehensive summary of LL-37, the factotum human cathelicidin peptide. Cell Immunol. 2012;280:22–35. doi: 10.1016/j.cellimm.2012.11.009. [DOI] [PubMed] [Google Scholar]
- 47.Nikolajsen CL, Scavenius C, Enghild JJ. Human complement C3 is a substrate for transglutaminases. A functional link between non-protease-based members of the coagulation and complement cascades. Biochemistry. 2012;51:4735–4742. doi: 10.1021/bi3004022. [DOI] [PubMed] [Google Scholar]
- 48.Cronin SJ, Nehme NT, Limmer S, Liegeois S, Pospisilik JA, Schramek D, Leibbrandt A, Simoes Rde M, Gruber S, Puc U, Ebersberger I, Zoranovic T, Neely GG, von Haeseler A, Ferrandon D, Penninger JM. Genome-wide RNAi screen identifies genes involved in intestinal pathogenic bacterial infection. Science. 2009;325:340–343. doi: 10.1126/science.1173164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee MJ, Mondal A, Small C, Paddibhatla I, Kawaguchi A, Govind S. A database for the analysis of immunity genes in Drosophila: Padma database. Fly (Austin) 2011;5:155–161. doi: 10.4161/fly.5.2.14674. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data
Supplementary data
Supplementary data